Introduction
Gastric cancer (GC) is a major unresolved digestive
problem that affects >1 million patients worldwide (1,2). Due
to the increasing incidence rate (5.6%) and mortality rate (7.7%),
GC is now considered the fifth most common and fourth most lethal
tumor worldwide (3). While the
5-year survival rate for early GC is >90%, most patients present
with advanced-stage GC due to the low early diagnosis rate
(4). Treatment strategies for GC
are rapidly evolving, and systemic chemotherapy, immunotherapy,
radiotherapy and targeted therapy have all been effective in curing
the tumor. Among them, surgery is the most efficient, but the
recurrence rate for patients with GC is high (5). A previous study reported that patients
with GC who undergo surgery have a poor 5-year survival rate of
60–80% (1). These findings
highlight the importance of developing efficacious targeted
therapies (1).
Cisplatin is widely used to treat various types of
solid cancers, including ovarian, non-small-cell lung, breast and
muscle-invasive bladder cancer (6–9).
Studies have revealed that cisplatin plays an anticancer role
through a number of mechanisms. The most widely accepted mechanism
is that cisplatin inhibits the synthesis of DNA, mRNA and proteins,
thereby leading to cell death (10–12).
However, the major challenges associated with cisplatin are
resistance and toxicity (13).
Therefore, deciphering the effect of cisplatin response is helpful
in identifying novel potential targets for combinatory therapies in
cancer.
Anti-silencing function 1 (ASF1), an important
chaperone protein of histones H3/H4, performs an important role in
DNA replication, damage response and transcription (14). ASF1 has two paralogs, ASF1A and
ASF1B. Previous studies have suggested that ASF1A is primarily
involved in DNA repair and cell senescence, whereas ASF1B is
associated with cell proliferation (15,16).
Other studies have shown that dysregulation of ASF1B expression is
linked to the progression and metastasis of multiple cancers,
including cervical cancer, thyroid cancer, hepatocellular carcinoma
and clear cell renal cell carcinoma (15,17–19).
However, the functional role of ASF1B in GC requires further
investigation.
In the present study, the aim was to evaluate the
function and possible mechanisms of ASF1B in GC. Here, we
constructed ASF1B knockdown cells and detected the biological
behavior changes using colony formation, wound healing and
Transwell assays, and flow cytometry. The present study may provide
a therapeutic strategy for patients with GC experiencing cisplatin
resistance.
Materials and methods
Bioinformatics analysis
The RNA-sequencing expression profiles and clinical
data were obtained from The Cancer Genome Atlas-Stomach
Adenocarcinoma (TCGA-STAD) database (https://portal.gdc.cancer.gov) to examine the
expression of ASF1B. The Kaplan-Meier Plotter online database
(https://kmplot.com/analysis/index.php?p=service&cancer=gastric)
was used for the Kaplan-Meier analysis (20). The Kaplan-Meier method followed by
the log-rank test was employed to plot and analyze the survival
curves of STAD patients with high/low expression of ASF1B.
According to the autoselected best cutoff value of 285, patients
were divided into two groups (high and low) for the survival
analysis. The underlying mechanism of ASF1B in GC and pathway
enriched genes were explored using Gene Set Enrichment Analysis
(GSEA) based on the clinical data of TCGA database (21). GSEA parameter settings were as
follows: Number of permutations=1000, permutation type=gene_set,
enrichment statistic=weighted, metric for ranking
genes=Signal2Noise. Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/) was used
to highlight the intersecting genes. Pearson's correlation analysis
was used to explore the correlation between the expression of the
ASF1B gene and that of other key genes, such as Myc, minichromosome
maintenance (MCM)4 and MCM5.
Tissue samples from patients with
GC
A total of 25 pairs of GC and adjacent healthy
tissues (>5 cm from the tumor tissues) from patients with GC
(age, 26–73 years; female, 9; male, 16) were acquired between July
2021 and June 2022, and tissues were immediately frozen in liquid
nitrogen and stored at −80°C. The inclusion criteria were as
follows: i) Patients were diagnosed with GC by pathology; ii)
before surgery, patients didn't receive any treatment, such as
chemotherapy and/or radiotherapy. The exclusion criteria were as
follows: i) Patients had other malignant tumors or critical
illness, such as coronary heart disease or diabetes; ii) patients
didn't sign informed consent for the use of their tissues. All
samples were obtained from Cangzhou Central Hospital (Cangzhou,
China) in accordance with the ethical and legal standards of the
Ethics Committee of Cangzhou Central Hospital (approval no.
2021-018-01).
Cell culture
The human gastric epithelial cell line GES-1 and the
GC cell lines HGC-27 and AGS were purchased from The Cell Bank of
Type Culture Collection of The Chinese Academy of Sciences. All
cells were maintained in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin solution (Gibco; Thermo Fisher
Scientific, Inc.) in a humidified atmosphere at 37°C and 5%
CO2.
Cell transfection and treatment
Small interfering (si)RNAs against ASF1B and
scrambled siRNA negative control (si-NC) were obtained from
Shanghai GenePharma Co. Ltd. as follows: si1-ASF1B sense,
5′-GUUGUGAUUGCUGUUUGUAUA-3′ and antisense,
5′-UACAAACAGCAAUCACAACAG-3′; si2-ASF1B sense,
5′-UGUGGAUGCUGUUGUGAUUGC-3′ and antisense,
5′-AAUCACAACAGCAUCCACAUG-3′; and si-NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. The Myc gene was cloned into the
pcDNA3.1 vector (Guangzhou RiboBio Co., Ltd.) to overexpress Myc.
The pcDNA3.1 empty vector (Guangzhou RiboBio Co., Ltd.) was
employed as NC. HGC-27 and AGS cells were divided into the
following groups: i) si-NC (cells were transfected with 50 nM si-NC
for 48 h at 37°C); ii) si1-ASF1B (cells were transfected with 50 nM
si1-ASF1B for 48 h at 37°C); iii) si2-ASF1B (cells were transfected
with 50 nM si2-ASF1B for 48 h at 37°C); iv) si-NC + cisplatin
(HGC-27 cells were transfected with 50 nM si-NC for 48 h at 37°C
and then treated with 6.8 µM cisplatin for 48 h at 37°C; AGS cells
were transfected with 50 nM si-NC for 48 h at 37°C and then treated
with 13.3 µM cisplatin for 48 h at 37°C); and v) si2-ASF1B +
cisplatin (HGC-27 cells were transfected with 50 nM si2-ASF1B for
48 h at 37°C and then treated with 6.8 µM cisplatin for 48 h at
37°C; AGS cells were transfected with 50 nM si2-ASF1B for 48 h at
37°C and then treated with 13.3 µM cisplatin for 48 h at 37°C).
Additionally, AGS cells were also divided into the following
groups: i) Vector (cells were transfected with 2 µg pcDNA3.1 empty
vector for 48 h at 37°C); ii) Myc (cells were transfected with 2 µg
pcDNA3.1-Myc for 48 h at 37°C); iii) si2-ASF1B + Vector (cells were
transfected with 50 nM si2-ASF1B and 2 µg pcDNA3.1 empty vector for
48 h at 37°C); iv) si2-ASF1B + Myc (cells were transfected with 50
nM si2-ASF1B and 2 µg pcDNA3.1-Myc for 48 h at 37°C); v) si2-ASF1B
+ Vector + cisplatin (cells were transfected with 50 nM si2-ASF1B
and 2 µg pcDNA3.1 empty vector for 48 h at 37°C and then treated
with 13.3 µM cisplatin for 48 h at 37°C); and vi) si2-ASF1B + Myc +
cisplatin (cells were transfected with 50 nM si2-ASF1B and 2 µg
pcDNA3.1-Myc for 48 h at 37°C and then treated with 13.3 µM
cisplatin for 48 h at 37°C). Cisplatin was purchased from
MedChemExpress. Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for transfection. After
transfection for 48 h, cells were harvested for subsequent
experiments.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) reagent was used to isolate total RNA from GC
cells and tissues. Complementary DNA was prepared using
PrimeScript™ RT Reagent Kit (Takara Bio, Inc.) according to the
manufacturer's protocol. Subsequently, RT-qPCR was performed using
SYBR Premix Ex Taq II (Takara Bio, Inc.) and the 7900HT PCR system.
The following thermocycling conditions were used: Initial
denaturation at 95°C for 1 min, followed by 39 cycles of 95°C for
30 sec, 60°C for 30 sec and 72°C for 30 sec. The mRNA expression of
Myc and ASF1B was normalized to that of the reference gene, GAPDH,
and quantified using the 2−ΔΔCq method (22). All primers used were purchased from
Shanghai GenePharma Co. Ltd. The primer sequences were as follows:
ASF1B forward, 5′-ATGTTTGTCTTTCAGGCCGAC-3′ and reverse,
5′-GCTCAGGGTTGAGGTACTCG-3′; Myc forward, 5′-TGGAAAACCAGCCTCCCGC-3′
and reverse, 5′-CGAAGGGAGAAGGGTGTGAC-3′); and GAPDH forward,
5′-GTTGCAACCGGGAAGGAAAT-3′ and reverse,
5′-GCCCAATACGACCAAATCAGA-3′.
Western blotting
HGC-27 and AGS cells were lysed using RIPA buffer
(MilliporeSigma) containing 1% protease inhibitors. Total protein
was quantified using a BCA assay and 30 µg protein/lane was
separated by SDS-PAGE on 12% gels. The separated proteins were
subsequently transferred onto PVDF membranes. After blocking with
5% non-fat dry milk for 1 h at 37°C, the membranes were incubated
with primary antibodies against ASF1B (1:1,000, cat. no. 2902, Cell
Signaling Technology, Inc.), MMP-7 (1:1,000, cat. no. 3801, Cell
Signaling Technology, Inc.), MMP-9 (1:1,000, cat. no. ab76003,
Abcam), anti-Myc (1:1,000, cat. no. SAB4501941, Sigma-Aldrich),
MCM4 (1:1,000, cat. no. 12973, Cell Signaling Technology, Inc.),
MCM5 (1:1,000, cat. no. ab75975, Abcam) and GAPDH (1:1,000, cat.
no. ab9485, Abcam) at 4°C overnight. Membranes were washed three
times with PBS with 0.5% Tween 20. Following the primary
incubation, membranes were incubated with HRP-conjugated goat
anti-rabbit IgG secondary antibodies (1:5,000, cat. no. ab97051,
Abcam) for 2 h at room temperature. Protein bands were visualized
using enhanced chemiluminescence detection kit (MilliporeSigma),
and semi-quantified using ImageJ v1.8.0 software (National
Institutes of Health) with GAPDH as the loading control.
Cell viability assay
HGC-27 and AGS cells in the logarithmic growth phase
were maintained in 96-well plates with a density of 1,000
cells/well. Following incubation for 0, 24, 48 and 72 h at 37°C, 10
µl Cell Counting Kit-8 (CCK-8) reagent (Beyotime Institute of
Biotechnology) was placed into each well and cells were incubated
for another 1.5 h. The absorbance at 450 nm was examined using a
microplate reader (Tecan Group, Ltd.).
Colony formation assay
During the logarithmic growth phase, HGC-27 and AGS
cells were harvested to examine their clonogenic capability. The
cells (1×103 cells per well) were inoculated into 30-mm
dishes with DMEM +10% FBS and cultured at 37°C for 14 days. The
culture medium was refreshed every 2 days. The cells were
subsequently fixed in 4% paraformaldehyde for 30 min at room
temperature, and stained using 0.1% crystal violet for 20 min at
room temperature. Finally, the number of colonies with >50 cells
was calculated with blinded manual counting.
Wound healing assay
HGC-27 and AGS cells (2×106 cells/well)
were plated into 6-well plates and cultured in serum-free medium. A
200 µl sterile pipette tip was used to scratch the cell monolayer
when cell confluence was >90%. All detached cells were removed
with PBS and the remaining cells were imaged at 0 and 48 h under a
light microscope (Olympus Corporation). The wound area was
determined using ImageJ v1.8.0 software (National Institutes of
Health) according to the following formula: Wound healing rate
(%)=(A0-At)/A0, where
A0 indicates the initial wound area and At
indicates the wound area after 48 h.
Transwell assay
Cell invasive ability was detected using 24-well
Transwell chambers (0.8 µm; Corning, Inc.). Transwell chambers were
pre-coated with Matrigel (BD Biosciences) for 45 min at 37°C HGC-27
and AGS cells at a density of 2×104 in 100 µl DMEM were
plated into the upper chambers. A total of 500 µl DMEM including
10% FBS were used to fill the lower chamber. After incubation for
48 h at 37°C, the non-invaded cells were removed using swabs, while
the remaining cells were fixed in 100% methanol for 10 min at room
temperature and stained using 0.1% crystal violet for 20 min at
room temperature. All cells were washed with PBS twice and counted
from five randomly selected fields of view. Images were captured
using a light microscope.
Apoptosis analysis
Annexin V-FITC Apoptosis Detection Kit (Beyotime
Institute of Biotechnology) was applied to detect HGC-27 and AGS
cell apoptosis. Briefly, cells were collected in 5 ml tubes and
centrifuged at 1,000 × g for 5 min at 4°C. Pre-chilled PBS was used
to resuspend cells prior to being centrifuged as aforementioned.
After the supernatant had been removed, 1X binding buffer was used
to resuspend cells at a density of 1×106 cells/ml. Cells
were stained with 5 µl Annexin V-FITC in the dark for 5 min at room
temperature, followed by incubation with 10 µl PI for 15 min at
room temperature. Apoptotic cells were subsequently analyzed using
a BD Accuri C6 flow cytometer (BD Biosciences). Cell apoptosis rate
(upper right + lower right) were analyzed by FlowJo V7.6 software
(FlowJo LLC).
Statistical analysis
For each assay, 3 independent repeats were
performed. Data were analyzed using GraphPad Prism (version 8.0;
Dotmatics). All data are presented as the mean ± standard
deviation. The comparison between two groups was analyzed using
Student's t-test (paired or unpaired), while differences between
multiple groups were analyzed using one-way ANOVA followed by
Tukey's post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
ASF1B expression is increased in GC
tissues and cells and is associated with an unfavorable outcome in
patients with GC
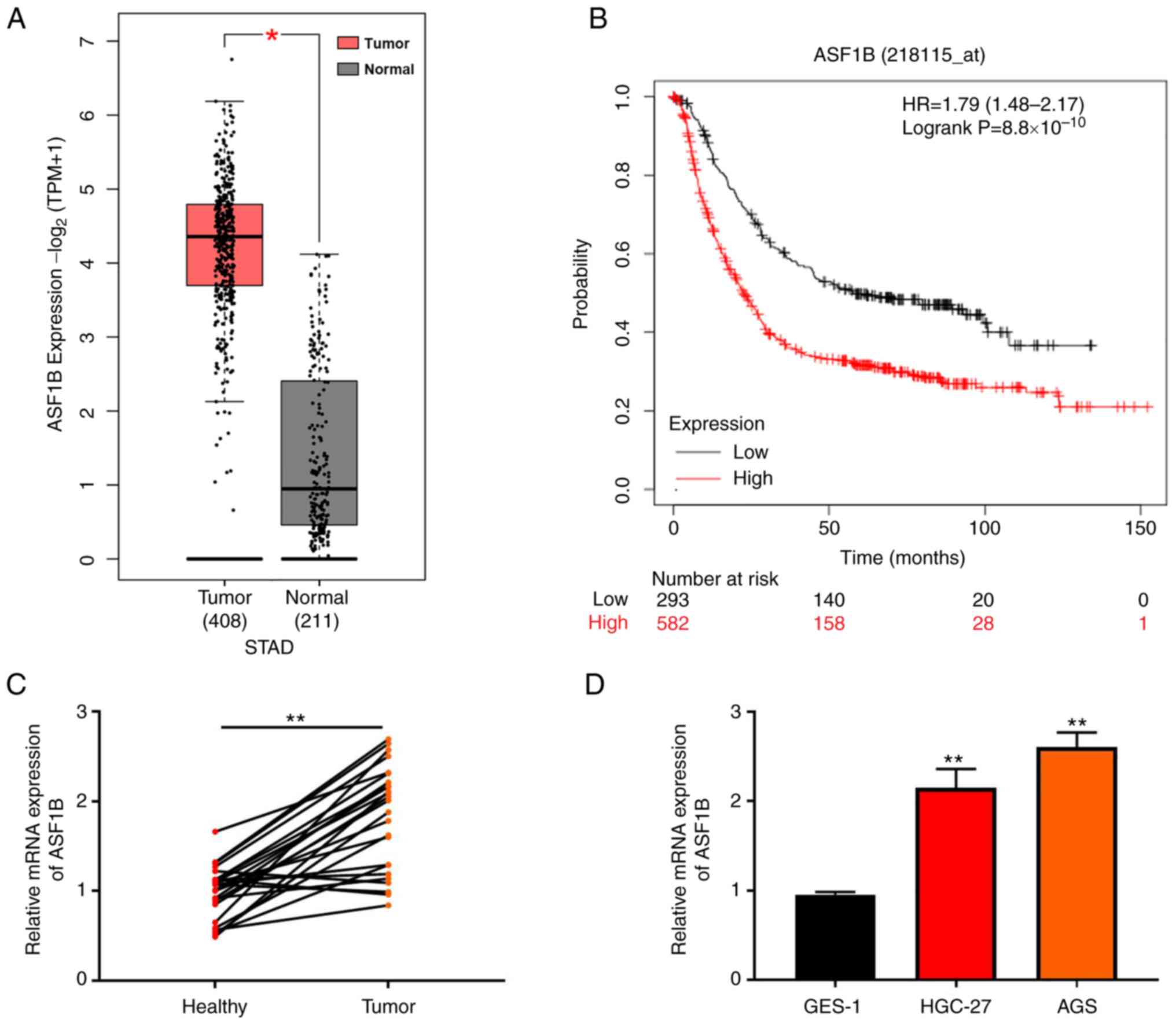
Analysis of data from TCGA database showed that
ASF1B exhibited higher expression in GC tissues compared with
normal tissues (Fig. 1A). Survival
curve analysis showed that high ASF1B expression was indicative of
a poor prognosis in patients with GC (Fig. 1B). RT-qPCR was performed to examine
the expression of ASF1B in tumor and healthy tissues, indicating
that ASF1B expression was increased in GC compared with adjacent
healthy tissues (Fig. 1C).
Moreover, compared with GES-1 cells, ASF1B expression was elevated
both in HGC-27 and AGS GC cell lines (Fig. 1D).
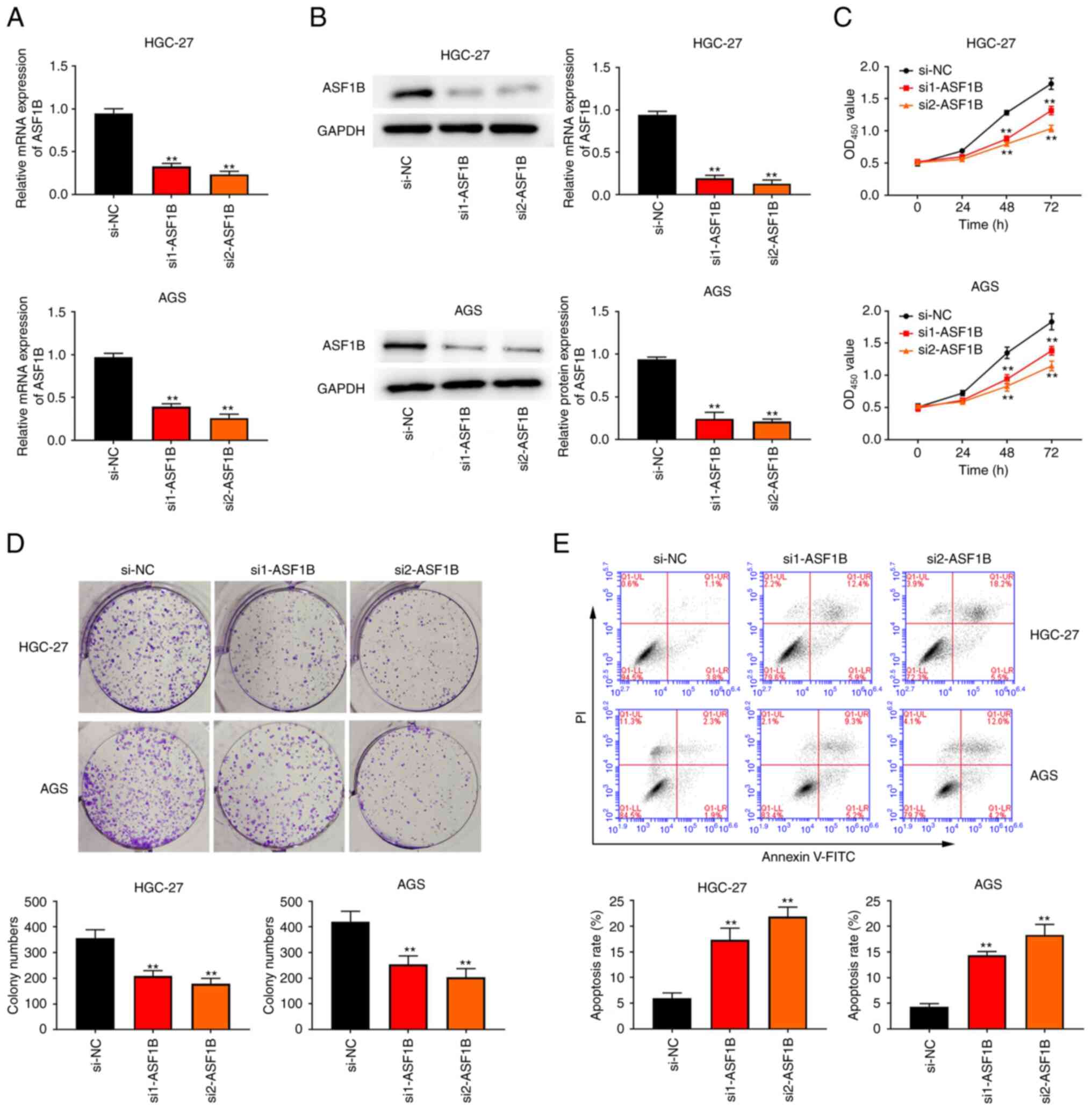
Silencing of ASF1B suppresses
proliferation and increases apoptosis in GC cells
To explore the role of ASF1B, HGC-27 and AGS GC
cells were transfected with siRNAs against ASF1B and scrambled
si-NC. First, it was confirmed that HGC-27 and AGS cell models with
silenced ASF1B expression were successfully established (Fig. 2A and B). Furthermore, in both HGC-27
and AGS cells, ASF1B expression in the si2-ASF1B group was lower
than that in the si1-ASF1B group (Fig.
2A and B). The CCK-8 and colony formation assays indicated that
cell proliferation was significantly reduced after ASF1B knockdown
(Fig. 2C and D). Additionally,
attenuation of ASF1B expression remarkably improved the apoptotic
rate of HGC-27 and AGS cells compared with the si-NC group
(Fig. 2E).
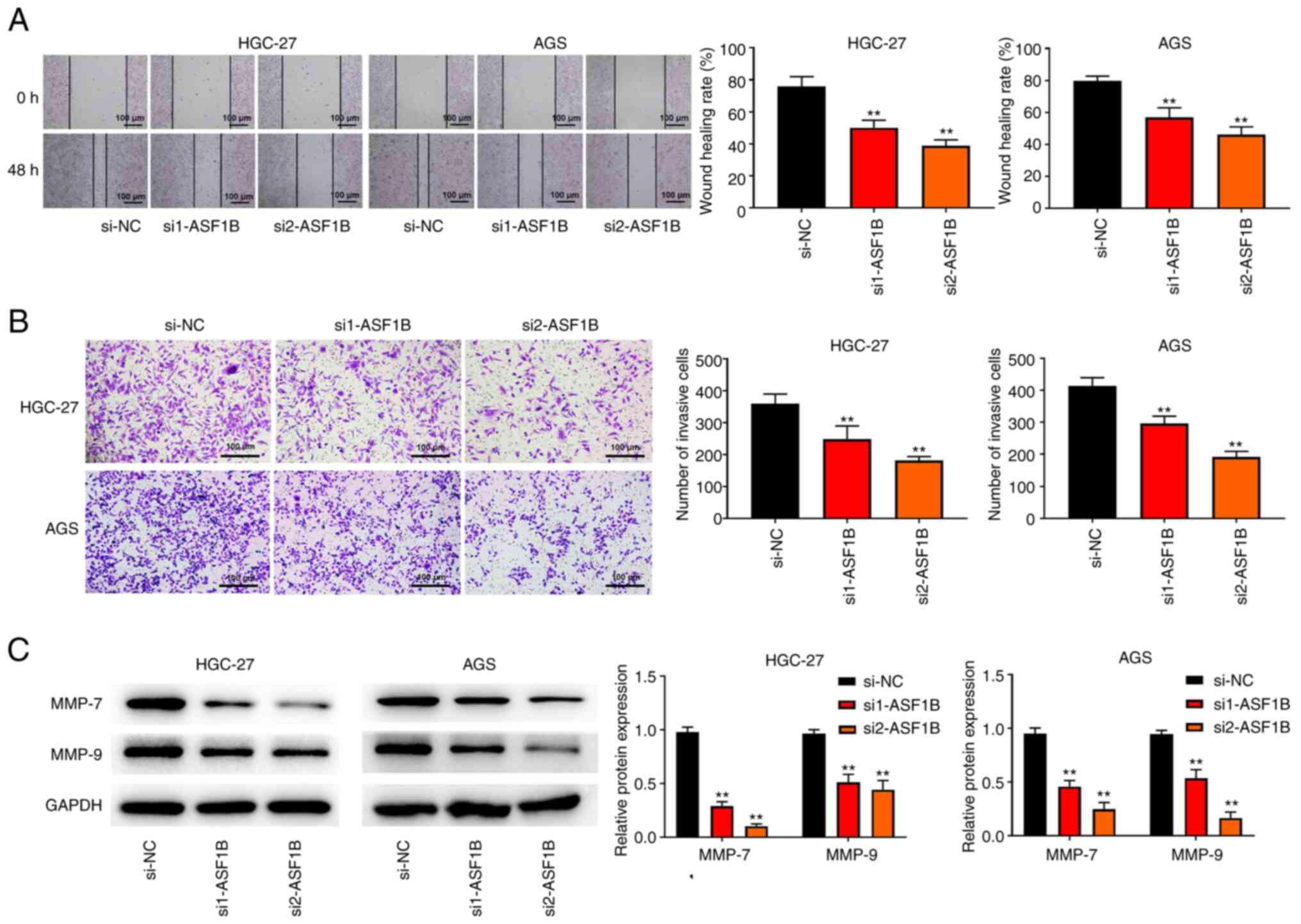
Knockdown of ASF1B inhibits GC cell
migration and invasion
Subsequently, wound healing and Transwell
experiments were conducted to explore the effect of ASF1B on the
migratory and invasive ability of GC cells. Knockdown of ASF1B
inhibited cell migration in both HGC-27 and AGS cells (Fig. 3A). Consistently, the results of
Transwell assay verified that the number of invaded cells was
decreased after ASF1B knockdown, compared with the si-NC group
(Fig. 3B). Furthermore, western
blotting revealed that MMP-7 and MMP-9 protein levels were notably
attenuated in HGC-27 and AGS cells transfected with si-ASF1B
compared with the si-NC group (Fig.
3C).
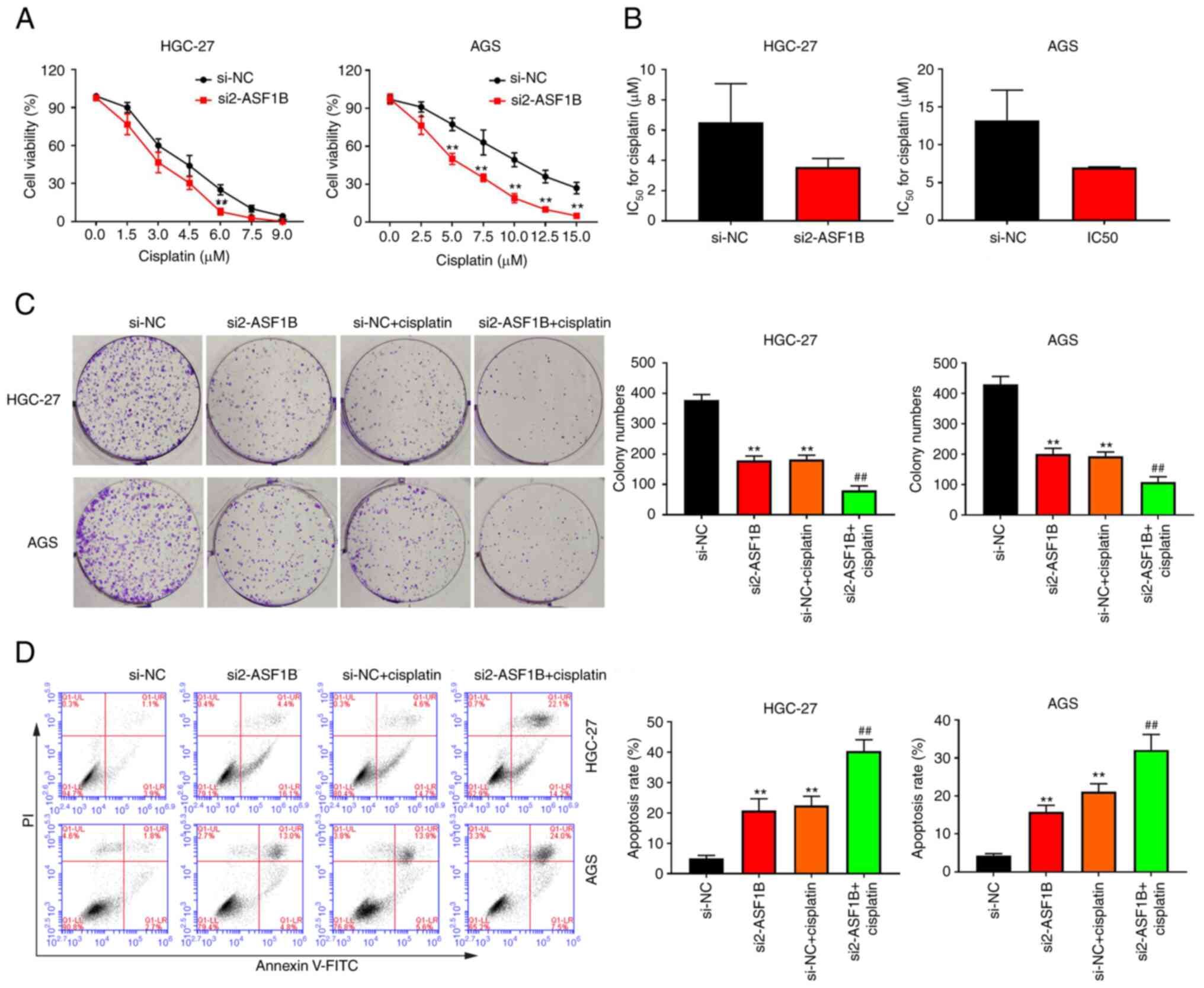
Knockdown of ASF1B inhibits cisplatin
resistance in GC cells
It was hypothesized that ASF1B may affect the
sensitivity of GC cells to cisplatin. Due to the difference in
sensitivity of different tumor cells to cisplatin (23,24),
HGC-27 and AGS cells in the si-NC and si2-ASF1B groups were treated
with different doses of cisplatin. CCK-8 results showed that the
proliferative potential of HGC-27 and AGS cells transfected with
si2-ASF1B was lower than that in the si-NC group under cisplatin
treatment (Fig. 4A). In both HGC-27
and AGS cells, the IC50 for cisplatin was markedly
decreased in the si2-ASF1B group compared with the si-NC group
(Fig. 4B). As shown in Fig. 4C, the addition of cisplatin
increased the inhibitory effect of ASF1B knockdown on colony
formation in GC cells. Apoptosis analysis showed that the apoptotic
rate of GC cells treated with cisplatin markedly increased compared
with the si-NC group, and the combination of si2-ASF1B with
cisplatin treatment further enhanced the apoptotic rate of both
HGC-27 and AGS cells compared with the si-NC + cisplatin group
(Fig. 4D).
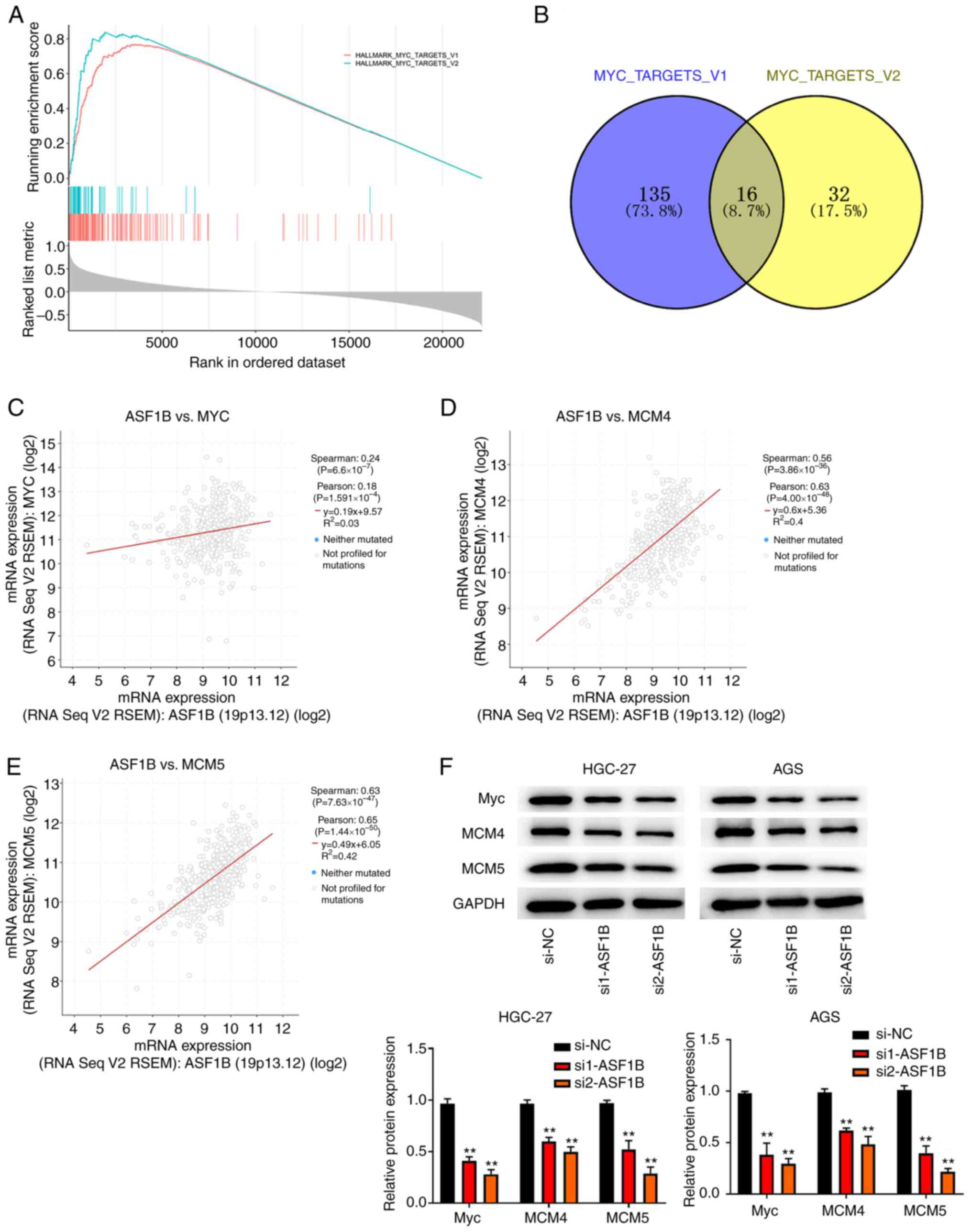
Silencing ASF1B attenuates the Myc
signaling pathway
To detect the underlying mechanism of ASF1B in GC
progression, analysis of expression profiles from TCGA database
indicated that the Myc-targets-v1 and Myc-targets-v2 pathways could
be activated by ASF1B (Fig. 5A).
Subsequently, the interaction of the downstream genes of the
Myc-targets-v1 and Myc-targets-v2 pathways was explored and a total
of 16 overlapping genes were obtained, including Myc, MCM4 and MCM5
(Fig. 5B). Pearson's correlation
analysis revealed that Myc, MCM4 and MCM5 expression were
positively correlated with the expression of ASF1B (Fig. 5C-E). Following ASF1B knockdown, the
protein levels of Myc, MCM4 and MCM5 were all markedly attenuated
in HGC-27 and AGS cells (Fig.
5F).
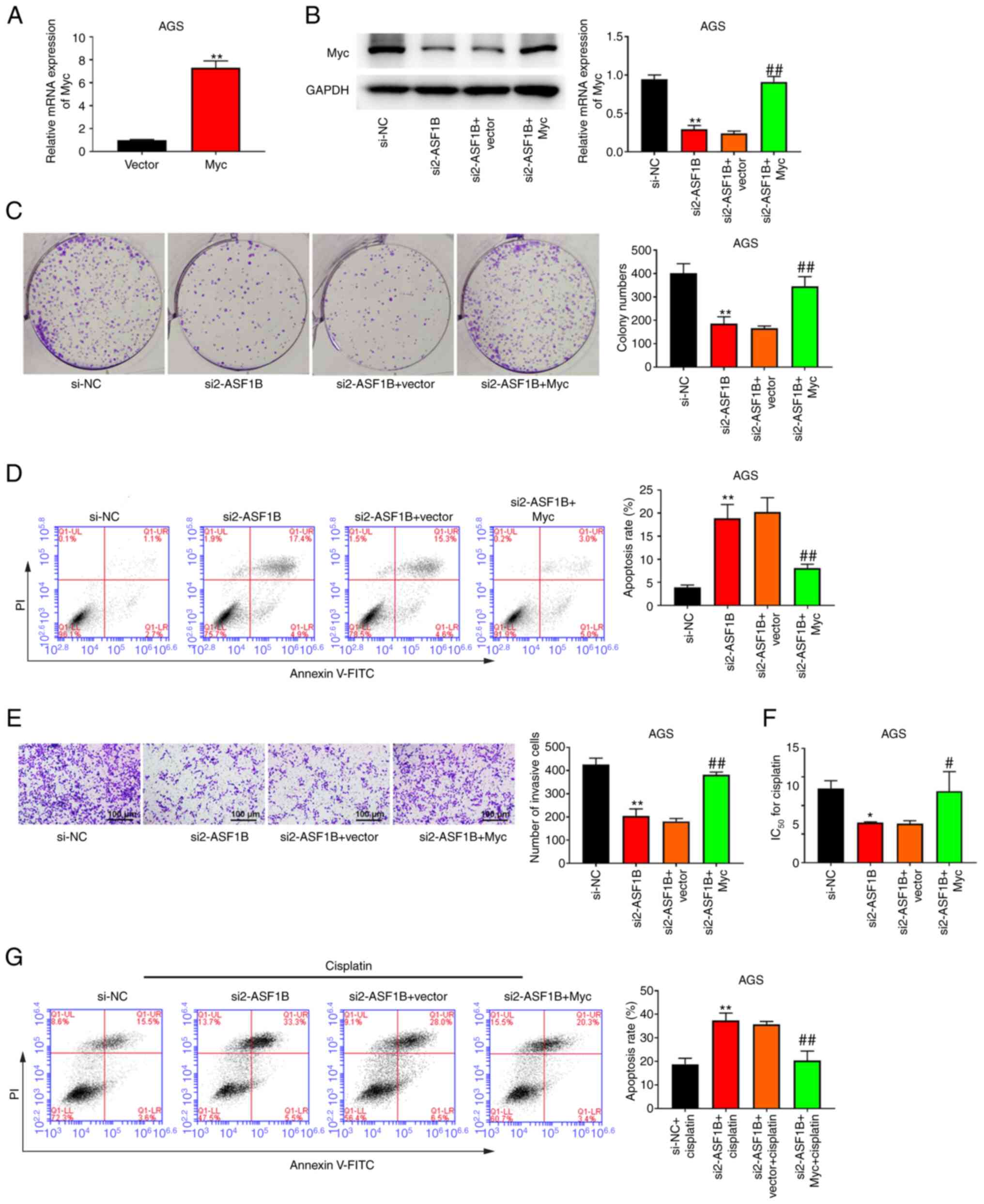
ASF1B regulates cell proliferation,
apoptosis, invasion and the cisplatin resistance of GC cells by
regulating the Myc pathway
Compared with GES-1 cells, AGS cells showed high
ASF1B expression. To further examine the molecular mechanism of
ASF1B in GC, AGS cells were selected for Myc overexpression.
Successful Myc overexpression is shown in Fig. 6A. Transfection of the Myc
overexpression plasmid increased Myc protein expression compared
with the si2-ASF1B + Vector group (Fig.
6B). Colony formation experiments revealed that overexpression
of Myc reversed the suppressive role of si2-ASF1B on cell
proliferation (Fig. 6C). In
addition, overexpression of Myc increased the number of invasive
cells compared with the si2-ASF1B + Vector group (Fig. 6E). Compared with the si2-ASF1B +
Vector group, the si2-ASF1B + Myc group exhibited a higher
IC50 for cisplatin (Fig.
6F). Furthermore, it was observed that Myc overexpression
significantly reversed the promotion of apoptosis caused by ASF1B
silencing in AGS cells, both with or without cisplatin treatment
(Fig. 6D and G).
Discussion
GC is a prevalent malignancy worldwide, and surgical
treatment is considered the fundamental approach to curing this
tumor. For patients diagnosed with early GC, survival rates after
surgery can reach >90% (25).
However, due to the low diagnostic rate of early GC, patients with
advanced GC mainly benefit from chemotherapy (26). The effect of chemotherapy on
improving survival has been previously reported by data analysis
and systematic reviews on individuals with GC (27). Additionally, since GC is an
aggressive disease and systemic metastasis is present in most
patients with GC (28),
chemotherapy has a critical role in GC treatment Nevertheless, drug
resistance often leads to an unfavorable prognosis for patients
with GC. Therefore, it is urgent to explore novel genes to identify
potential therapeutic targets for drug resistance.
The present study revealed that ASF1B was highly
expressed in GC tissues and cells. High expression of ASF1B was
associated with poor outcomes of patients with GC. These results
are similar to those of previous studies. For example, based on a
comprehensive pan-cancer analysis, Hu et al (29) indicated that ASF1B was a prognostic
and immunological biomarker in nermous cancers, such as
adrenocortical cancer, bladder cancer, breast cancer and lung
adenocarcinoma. Furthermore, ASF1B has been determined to be a
biomarker for the prognosis of thyroid cancer (17), and increased levels of ASF1B in
human lung tissues and cells have been associated with poor
prognosis and metastasis (30).
However, there is no evidence for the effect of ASF1B on GC
development, to the best of our knowledge. To detect the functions
of ASF1B, in vitro experiments were performed. Silencing
ASF1B expression suppressed GC cell proliferation, migration and
invasion, and induced cell apoptosis. Furthermore, it was
demonstrated that cisplatin resistance was significantly reduced
following ASF1B knockdown. Cisplatin is known for its strong
anticancer activity and is widely used in chemotherapy (31). However, the emergence of cisplatin
resistance significantly affects therapeutic outcomes, thereby
inducing the tumor recurrence (32,33).
In summary, the present results suggested that ASF1B is an
important therapeutic target for patients with GC.
Moreover, to explore the underlying mechanism of
ASF1B in GC, GSEA was performed. The results suggested that ASF1B
was associated with the activation of the Myc-targets-v1 and
Myc-targets-v2 signaling pathways. Members of the Myc family are
regarded as oncogenes and exert crucial roles in the progression of
multiple malignances (34). Myc is
an important transcription activator, which can regulate numerous
genes and modulate cell viability and apoptosis (35). In addition, the expression of
double-stranded DNA break repair genes, such as poly(ADP-Ribose)
polymerase 1 and DNA ligase 3, have been indicated to be influenced
by Myc (36,37). A number of studies have illustrated
that Myc is able to mediate drug resistance in hepatocellular
carcinoma, prostate cancer and colorectal cancer (38–40).
These findings suggest that Myc may be an alternative therapeutic
target for tumor treatment. Therefore, to verify the relationship
between ASF1B and the Myc pathway, western blotting was used to
measure the protein expression levels of Myc, MCM4 and MCM5 in
HGC-27 and AGS cells after si-ASF1B transfection. The results
suggested that reduction of ASF1B expression inhibited the levels
of Myc, MCM4 and MCM5. Therefore, it was assumed that ASF1B may
regulate the cisplatin resistance of GC through the Myc pathway.
The present results revealed that the suppressive effect of ASF1B
knockdown on cisplatin resistance was reversed by Myc
overexpression. Moreover, Myc overexpression reversed the
inhibitory effect of ASF1B silencing on GC cell proliferation and
invasion.
In conclusion, the present study demonstrated that
downregulation of ASF1B may attenuate cell proliferation, invasion
and cisplatin resistance by regulating the Myc pathway, suggesting
that ASF1B may be a therapeutic target for cisplatin-based
chemotherapy in GC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ, MN and JZ designed the research study. ZZ and MN
performed the experiments. ZZ and JZ contributed essential reagents
or tools. LL, ZL and YW analyzed the data. ZZ and MN confirm the
authenticity of all the raw data. ZZ wrote the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures involving human participants were
performed in accordance with The Declaration of Helsinki. The
research protocol was approved by the Ethics Committee of Cangzhou
Central Hospital (Cangzhou, China; approval no. 2021-018-01).
Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ASF1B
|
anti-silencing function 1B
|
|
GC
|
gastric cancer
|
References
|
1
|
Sexton RE, Al Hallak MN, Diab M and Azmi
AS: Gastric cancer: A comprehensive review of current and future
treatment strategies. Cancer Metastasis Rev. 39:1179–1203. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brodsky AS, Khurana J, Guo KS, Wu EY, Yang
D, Siddique AS, Wong IY, Gamsiz Uzun ED and Resnick MB: Somatic
mutations in collagens are associated with a distinct tumor
environment and overall survival in gastric cancer. BMC Cancer.
22:1392022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tan Z: Recent advances in the surgical
treatment of advanced gastric cancer: A Review. Med Sci Monit.
25:3537–3541. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Joshi SS and Badgwell BD: Current
treatment and recent progress in gastric cancer. CA Cancer J Clin.
71:264–279. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kenmotsu H, Yamamoto N, Yamanaka T,
Yoshiya K, Takahashi T, Ueno T, Goto K, Daga H, Ikeda N, Sugio K,
et al: Randomized phase III study of pemetrexed plus cisplatin
versus vinorelbine plus cisplatin for completely resected stage II
to IIIA nonsquamous non-small-cell lung cancer. J Clin Oncol.
38:2187–2196. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang DM, Gupta S, Kitchlu A, Meraz-Munoz
A, North SA, Alimohamed NS, Blais N and Sridhar SS: Defining
cisplatin eligibility in patients with muscle-invasive bladder
cancer. Nat Rev Urol. 18:104–114. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang
JY, Zhao FY, Qiao HY and Wang HQ: m6A-YTHDF1-mediated TRIM29
upregulation facilitates the stem cell-like phenotype of
cisplatin-resistant ovarian cancer cells. Biochim Biophys Acta Mol
Cell Res. 1868:1188782021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Safi A, Bastami M, Delghir S, Ilkhani K,
Seif F and Alivand MR: miRNAs modulate the dichotomy of cisplatin
resistance or sensitivity in breast cancer: An update of
therapeutic implications. Anticancer Agents Med Chem. 21:1069–1081.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu L, Fan J, Ai G, Liu J, Luo N, Li C and
Cheng Z: Berberine in combination with cisplatin induces
necroptosis and apoptosis in ovarian cancer cells. Biol Res.
52:372019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu L, Yuan Y, Yuan L, Li L, Liu F, Liu J,
Chen Y, Lu Y and Cheng J: Activation of TFEB-mediated autophagy by
trehalose attenuates mitochondrial dysfunction in cisplatin-induced
acute kidney injury. Theranostics. 10:5829–5844. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Raudenska M, Balvan J, Fojtu M, Gumulec J
and Masarik M: Unexpected therapeutic effects of cisplatin.
Metallomics. 11:1182–1199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cocetta V, Ragazzi E and Montopoli M:
Links between cancer metabolism and cisplatin resistance. Int Rev
Cell Mol Biol. 354:107–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ouyang X, Lv L, Zhao Y, Zhang F, Hu Q, Li
Z, Zhu D and Li L: ASF1B serves as a potential therapeutic target
by influencing cell cycle and proliferation in hepatocellular
carcinoma. Front Oncol. 11:8015062021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Song J, Zhang Y, Wang H, Sun H,
Feng X, Hou M, Chen G, Tang Q and Ji M: ASF1B promotes cervical
cancer progression through stabilization of CDK9. Cell Death Dis.
11:7052020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu L and Jie B: Protumor effects of
histone H3-H4 chaperone antisilencing Feature 1B gene on lung
adenocarcinoma: In silico and in vitro analyses. Comput Math
Methods Med. 16:50054592021.PubMed/NCBI
|
|
17
|
Ma J, Han W and Lu K: Comprehensive
pan-cancer analysis and the regulatory mechanism of ASF1B, a gene
associated with thyroid cancer prognosis in the tumor
micro-environment. Front Oncol. 11:7117562021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhan T, Gao X, Wang G, Li F, Shen J, Lu C,
Xu L, Li Y and Zhang J: Construction of novel lncRNA-miRNA-mRNA
network associated with recurrence and identification of
immune-related potential regulatory axis in hepatocellular
carcinoma. Front Oncol. 11:6266632021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiangqiao Z, Tao Q, Zhongbao C, Xiaoxiong
M, Long Z, Jilin Z and Tianyu W: Anti-silencing function 1B histone
chaperone promotes cell proliferation and migration via activation
of the AKT pathway in clear cell renal cell carcinoma. Biochem
Biophys Res Commun. 511:165–172. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Lv W, Liu Y, Fu W, Chen B, Ma Q,
Gao X and Cui X: Knockdown of serum- and glucocorticoid-regulated
Kinase 1 enhances cisplatin sensitivity of gastric cancer through
suppressing the nuclear factor kappa-b signaling pathway. Balkan
Med J. 38:331–340. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang G, Wang X and Han M: Loss of DAB2IP
contributes to cell proliferation and cisplatin resistance in
gastric cancer. Onco Targets Ther. 14:979–988. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song W, Cui Z, Liu H, Xue L and Ju H: The
expression and prognostic value of miR-195-5p in patients with
advanced gastric cancer after chemotherapy. J BUON. 25:2332–2340.
2020.PubMed/NCBI
|
|
26
|
Qiao XL, Zhong ZL, Dong Y and Gao F:
LncRNA HMGA1P4 promotes cisplatin-resistance in gastric cancer. Eur
Rev Med Pharmacol Sci. 24:8830–8836. 2020.PubMed/NCBI
|
|
27
|
Ge L, Hou L, Yang Q, Wu Y, Shi X, Li J and
Yang K: A systematic review and network meta-analysis protocol of
adjuvant chemotherapy regimens for resected gastric cancer.
Medicine (Baltimore). 98:e144782019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo Z, Rong Z and Huang C: Surgery
strategies for gastric cancer with liver metastasis. Front Oncol.
9:13532019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu X, Zhu H, Zhang X, He X and Xu X:
Comprehensive analysis of pan-cancer reveals potential of ASF1B as
a prognostic and immunological biomarker. Cancer Med. 10:6897–6916.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang W, Xiao L, Pan D and Hu L: ASF1B
enhances migration and invasion of lung cancers cell via regulating
the P53-mediated epithelial-mesenchymal transformation (EMT)
signaling pathway. Neoplasma. 69:3692022. View Article : Google Scholar
|
|
31
|
Qi L, Luo Q and Zhang Y: Advances in
toxicological research of the anticancer drug cisplatin. Chem Res
Toxicol. 32:1469–1486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, Wu X, Yang H, Li L, Ye Z and Rao Y:
A nuclear targeted Dox-aptamer loaded liposome delivery platform
for the circumvention of drug resistance in breast cancer. Biomed
Pharmacother. 117:122019. View Article : Google Scholar
|
|
33
|
Liang W, Zheng Y, Zhang J and Sun X:
Multiscale modeling reveals angiogenesis-induced drug resistance in
brain tumors and predicts a synergistic drug combination targeting
EGFR and VEGFR pathways. BMC Bioinformatics. 20 (Suppl 7):S2032019.
View Article : Google Scholar
|
|
34
|
Li Y, Zu X, Hu X, Wang L and He W:
Forkhead Box R2 knockdown decreases chemoresistance to cisplatin
via MYC pathway in bladder cancer. Med Sci Monit. 25:8928–8939.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mei Y, Liu YB, Hu DL and Zhou HH: Effect
of RIF1 on response of non-small-cell lung cancer patients to
platinum-based chemotherapy by regulating MYC signaling pathway.
Int J Biol Sci. 14:1859–1872. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pyndiah S, Tanida S, Ahmed KM, Cassimere
EK, Choe C and Sakamuro D: c-MYC suppresses BIN1 to release
poly(ADP-ribose) polymerase 1: A mechanism by which cancer cells
acquire cisplatin resistance. Sci Signal. 4:20015562011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Muvarak N, Kelley S, Robert C, Baer MR,
Perrotti D, Gambacorti-Passerini C, Civin C, Scheibner K and
Rassool FV: c-MYC Generates repair errors via increased
transcription of alternative-NHEJ factors, LIG3 and PARP1, in
tyrosine kinase-activated leukemias. Mol Cancer Res. 13:699–712.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang
G, Liu Y, Yao Y, Chen X, Ma W, et al: MYC-targeted WDR4 promotes
proliferation, metastasis, and sorafenib resistance by inducing
CCNB1 translation in hepatocellular carcinoma. Cell Death Dis.
12:6912021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li M, Fang L, Kwantwi LB, He G, Luo W,
Yang L, Huang Y, Yin S, Cai Y, Ma W, et al: N-Myc promotes
angiogenesis and therapeutic resistance of prostate cancer by TEM8.
Med Oncol. 38:1272021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun W, Li J, Zhou L, Han J, Liu R, Zhang
H, Ning T, Gao Z, Liu B, Chen X and Ba Y: The
c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in
colorectal cancer. Theranostics. 10:1981–1996. 2020. View Article : Google Scholar : PubMed/NCBI
|