Introduction
Ovarian cancer (OC) is the fifth leading cause of
cancer-associated mortality among women worldwide (1). Among the gynecological malignancies,
OC is the leading cause of mortality due to its late diagnosis and
recurrence (2). In general, ~75% of
patients with OC are initially diagnosed with intra-abdominal
disease and a 5-year survival is encountered in <40% of patients
with stage III disease (3,4). Ovarian tumors are prone to becoming
diffuse, resulting in peritoneal and intraperitoneal metastasis;
these tumors also relatively resistant to conventional
chemotherapeutics (4). All of the
aforementioned factors contribute to a strong frequency of
therapeutic recurrence and resistance.
MicroRNAs (miRNAs/miRs) are defined as short
non-coding RNA molecules of 19 to 22 nucleotides in length. They
function as tumor suppressors or oncogenes (oncomiRs) in the
initiation and progression of human cancer. miRNA mimics or
molecules targeted at miRNAs (antimiRs) have been reported to be
promising as therapeutic drugs in pre-clinical development
(5), including a mimic of the tumor
suppressor miR-34 (6), which
reached phase I clinical trials for hepatocellular carcinoma, and
anti-miRs targeting miR-122, which reached phase II trials for
hepatitis (7). Several miRNAs have
been reported to be involved in OC tumorigenesis, invasion or
metastasis, the function of extracellular matrix and angiogenesis
(8). For example, circulating
levels of different panels of miRNAs may be used as promising
classifiers for OC (8–10), greatly enhancing the feasibility of
circulating miRNAs as non-invasive prognostic markers (11). Therefore, examining the expression
profile and underlying mechanisms of miRNAs in OC may be
crucial.
Recently, miR-4732-5p expression in plasma-derived
exosomes was identified as a potential diagnosis biomarker for
distinguishing patients with epithelial OC (EOC) from healthy
subjects with 85.7% sensitivity and 82.4% specificity, and as a
monitor for EOC progression from the early to the late stage
(12). However, the role of
miR-4732-5p and its underlying molecular mechanisms in OC remain
unclear. In the present study, the expression of the miR-4732 was
analyzed using The Cancer Genome Atlas (TCGA) database and the
functional effects of miR-4732-5p on OC cell proliferation,
migration and invasion were defined by gain-of-function and
loss-of-function experiments in vitro. Finally,
mitochondrial calcium uniporter regulator 1 (MCUR1) was validated
as one of the direct targets of miR-4732-5p, potentially involved
in the promotion role of miR-4732-5p in OC, at least partially.
Materials and methods
TCGA database
OC RNA expression, miRNA expression and clinical
data were downloaded from TCGA database (https://xenabrowser.net/datapages/). Firstly, the link
for ‘GDC TCGA Ovarian Cancer (TCGA-OV) (15 datasets)’ was selected
using the following web link: https://xenabrowser.net/datapages/. Subsequently, the
links ‘gene expression RNAseq/HTSeq-FPKM (n=379) GDC Hub’ for RNA
expression (https://xenabrowser.net/datapages/?dataset=TCGA-OV.htseq_fpkm.tsv&host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443),
‘stem loop expression/miRNA Expression Quantification (n=498) GDC
Hub’ for miRNA expression (https://xenabrowser.net/datapages/?dataset=TCGA.OV.sampleMap%2FmiRNA_HiSeq_gene&host=https%3A%2F%2Ftcga.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443),
‘phenotype/Phenotype (n=758) GDC Hub (https://xenabrowser.net/datapages/?dataset=TCGA-OV.GDC_phenotype.tsv&host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443)
and survival data (n=731) GDC Hub’ (https://xenabrowser.net/datapages/?dataset=TCGA-OV.survival.tsv&host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443)
were selected, in order to acquire expression level information for
RNA, miRNAs and clinical information. The clinical phenotype and
miR-4732 expression were matched according to the sample ID. In
addition, the normalized expression data (level 3) of miR-4732 were
analyzed according to TNM stage, lymph node metastasis and survival
status following surgery. Finally, receiver operating
characteristic (ROC) curve was performed to predict the mortality
of patients. The expression value of miR-4732 at the maximum
Youden's Index (Sensitivity + Specificity-1) was used as the
cut-off value to divide the high and low miR-4732 expression
groups. Kaplan-Meier (K-M) survival curve analysis was used to
analyze the overall survival (OS) percentage of patients with a
high or low miR-4732 expression.
Cells and cell culture
The IGROV1 (cat. no. CTCC-009-0048) human OC cells
were obtained from the Cell Center of Meisen CTCC (https://www.ctcc.online/). The cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 mg/ml streptomycin at 37°C with 5%
CO2. 293FT cells were purchased from Invitrogen; Thermo
Fisher Scientific, Inc. (cat. no. R70007) and cultured according to
manufacturer's manuals.
Cell transfection with miR-4732-5p
mimics or inhibitors
miRNA mimics or inhibitors for miRabse accession no.
MIMAT0019855 were synthesized by Guangzhou RiboBio Co., Ltd., and
transiently transfected into 1×105 IGROV1 cells at a 50
nmol/l concentration using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol cultured in an incubator at 37°C with 5%
CO2 for 24 h to subsequent experimentation. The
synthesis of mimic sequences (micrON™ miRNA mimics) was designed as
double-strand 5′-UGUAGAGCAGGGAGCAGGAAGCU-3′; the inhibitor sequence
(micrOFF™ miRNA inhibitor) was chemically modified as single-strand
5′-AGCUUCCUGCUCCCUGCUCUACA-3′; the control sequences were: Mimics
control [micrON™ mimic negative control (NC) #24; cat. no.
miR1N0000002-1; https://www.ribobio.com/product_detail/?sku=miR1N0000002-1-1]
and inhibitor control (micrOFF inhibitor NC #24; cat. no.
miR2N0000002-1; http://www.ribobio.com/product_detail/?sku=miR2N0000002-1-1);
all sequences were designed and purchased from RiboBio Co. Ltd.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA extraction and cDNA synthesis for miRNAs
by adding polyA tail followed by onligodT-adaptor primer revers
transcription were performed according to a previous study
(13). Briefly, total RNA was
isolated from the cells using miRNeasy Mini kits (Qiagen, Inc.)
according to the manufacturer's protocols. Poly(A) Polymerase (New
England BioLabs, Inc.) was used to add Poly(A) tails to 100 ng of
total RNA 3′-ends according to the manufacturer's protocols.
Subsequently, RNA was reverse transcribed using an oligodT-adaptor
primer (5′-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3′) and
Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.). The following primers were used to
amplify the miRNA or U6: miR-4732-5p forward,
5′-TGTAGAGCAGGGAGCAGGAAG-3′ and reverse,
5′-GCGAGCACAGAATTAATACGAC-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The primer pairs were designed using Primer-blast. (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The expression levels of miRNAs were evaluated using SYBR™ Select
Master Mix (Thermo Fisher Scientific, Inc.) and an ABI7500 fast
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
cycling conditions were as follows: 10 min at 95°C followed by 40
cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 42 sec. U6
was used as the reference gene. The expression level of miR-4732-5p
(fold change relative to the control group) was calculated using
the 2−∆∆Cq method, whereby ∆Cq=Cq (miR-4732-5p)-Cq (U6)
(14).
Cell viability assay
A total of 1×103 cells/well were seeded
into 96-well plates in triplicate and cultured for 1, 2, 3 and 4
days. Cell viability was evaluated using a Cell Counting Kit-8
(CCK-8; Dojindo Molecular Technologies, Inc.), according to the
manufacturer instructions. Briefly, a total of 10 µl CCK-8 solution
was added to each well at the 0, 24, 48, 72 and 96 h time points.
Following incubation at 37°C with 5% CO2 for 1 h, the
absorbance at a wavelength of 450 nm was then measured using a
microplate reader (iMark; Bio-Rad Laboratories, Inc.). The
experiments were performed in three independent experiments in
triplicate.
Transwell migration and invasion
assays
The cells (1×104) were seeded into the
upper chambers of a Transwell plate (cat. no. 3422, 8.0 µM;
Corning, Inc.) pre-coated in triplicate without or with Matrigel
(1:30 dilution with RPMI-1640 medium at 37°C for 30 min), in 100 µl
RPMI-1640 medium containing 1% FBS. A total of 500 µl RPMI-1640
medium containing 10% FBS as a chemoattractant was placed in each
lower chamber. Following a 24-h incubation at 37°C in a cell
incubator (Heracell VIOS 160i; Thermo Fisher Scientific, Inc.), the
migrated or invaded cells were fixed with 4% formaldehyde for 5 min
at room temperature and stained with 1% crystal violet (cat. no.
G1062; Beijing Solarbio Science & Technology Co., Ltd.) for 5
min at room temperature. The number of migrated or invasive cells
was counted using a light microscope (Leica Dim8; Leica
Microsystems GmbH; magnification, ×20) in four randomly selected
microscopic fields. The experiments were performed in three
independent experiments in triplicate.
Western blotting
The 106 cells were harvested and protein
extracts were obtained with RIPA lysis buffer (Beijing Solarbio
Science & Technology Co., Ltd.). The concentration of protein
was determined by BCA BCA assay kit (Beijing Solarbio Science &
Technology Co., Ltd.). Equal amounts (30 µg) of protein were
electrophoresed on 10% SDS-PAGE gels, and then transferred to PVDF
membrane (MilliporeSigma). After blocking with 5% non-fat milk
solution at room temperature for 1 h, the membranes were incubated
with anti-MCUR1 (1:5,000; cat. no. 13706; Cell Signaling
Technology, Inc.) and anti-actin (1:10,000; cat. no. 3700; Cell
Signaling Technology, Inc.) antibodies at 4°C overnight, followed
by incubation with horseradish peroxidase-conjugated secondary
antibodies (1:50,000; cat. no. 111-035-003; Jackson ImmunoResearch
Laboratories, Inc.) at room temperature for 1 h. ECL solution
(Millipore) was used to visualize specific bands and the
immunoblots were scanned on an Amersham Imager 680 (AI680; GE
Healthcare).
Prediction of the candidate targets of
miR-4732-5p using bioinformatics analysis
Firstly, TargetScan version 7.1 (http://www.targetscan.org/vert_71/) was used to
predict the direct candidate targets. After selecting ‘Human’ in
the species option inputting miR-4732-5p as the microRNA name,
candidate genes for hsa-miR-4732-5p were predicted. Secondly, Gene
Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) was used to identify the
OS-related genes (15). The most
differential survival genes were acquired by selecting
‘Survival/Most Differential Survival Genes’ function, ‘OC’ for
dataset selection, ‘overall survival’ for method and ‘Median’ value
of the gene expression in the cancer tissues for group cut-off.
Finally, the predicted targets using TargetScan and
survival-related genes (P<0.05) were intersected using a Venn
plot and the potential candidate targets of miR-4732-5p were
obtained. In addition, a literature search using PubMed (https://pubmed.ncbi.nlm.nih.gov/) for these
candidate targets was performed to define the oncogene or
suppressor role of these genes in the cancer.
Dual luciferase report assay
The wild-type (WT) MCUR1 3′-UTR fragments or the
mutant (mutation at the predicted miR-4732-5p binding sites), were
inserted downstream of the Firefly luciferase gene in the
pGL3-control (cat. no. E1741, Promega Corporation) plasmid. A total
of 0.5 µg of the Firefly reporter plasmid pGL3-MCUR1 WT/Mutation
(containing position 612–618 of MCUR1 3′UTR at sequences of
“CUCUACA” and corresponding mutation with sequences of “AGAGCAG”,
26 ng of pRL-TK plasmids (cat. no. E2241, Promega Corporation)
containing Renilla luciferase, along with 10 pmol/ml
miR-4732-5p mimics or negative control, were transfected into 293FT
cells seeded into 24-well plate in triplicate. Following a 48-h
transfection using Lipofectamine 2000® (cat. no.
11668019; Invitrogen; Thermo Fisher Scientific, Inc.), cell lysates
were collected, and luciferase activities were measured using the
Dual Luciferase Reporter System (Promega Corporation) according to
manufacturer's protocol with a Tecan Infinite M200 Pro instrument
(Tecan Group, Ltd.). The luminescence intensity of Firefly
luciferase was normalized to that of Renilla luciferase.
Statistical analysis
Data were analyzed using SPSS 13.0 software (SPSS,
Inc.). The continuous variables with normal distribution
(Shapiro-Wilk test) are expressed as the mean ± standard deviation
from three independent experiments. Statistical significance was
determined using an unpaired two-tailed Student's t-test for two
groups, and using one-way ANOVA with a Bonferroni post hoc test for
multiple comparisons. OS was analyzed using the K-M method
(log-rank test). The cut-off value for miR-4732-5p expression was
based on the ROC corresponding to the maximum the Youden's index
(Sensitivity + Specificity-1). Pearson's correlation analysis was
used to investigate the expression correlation between miR-4732 and
MCUR1 expression. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-4732 in OC
tissues
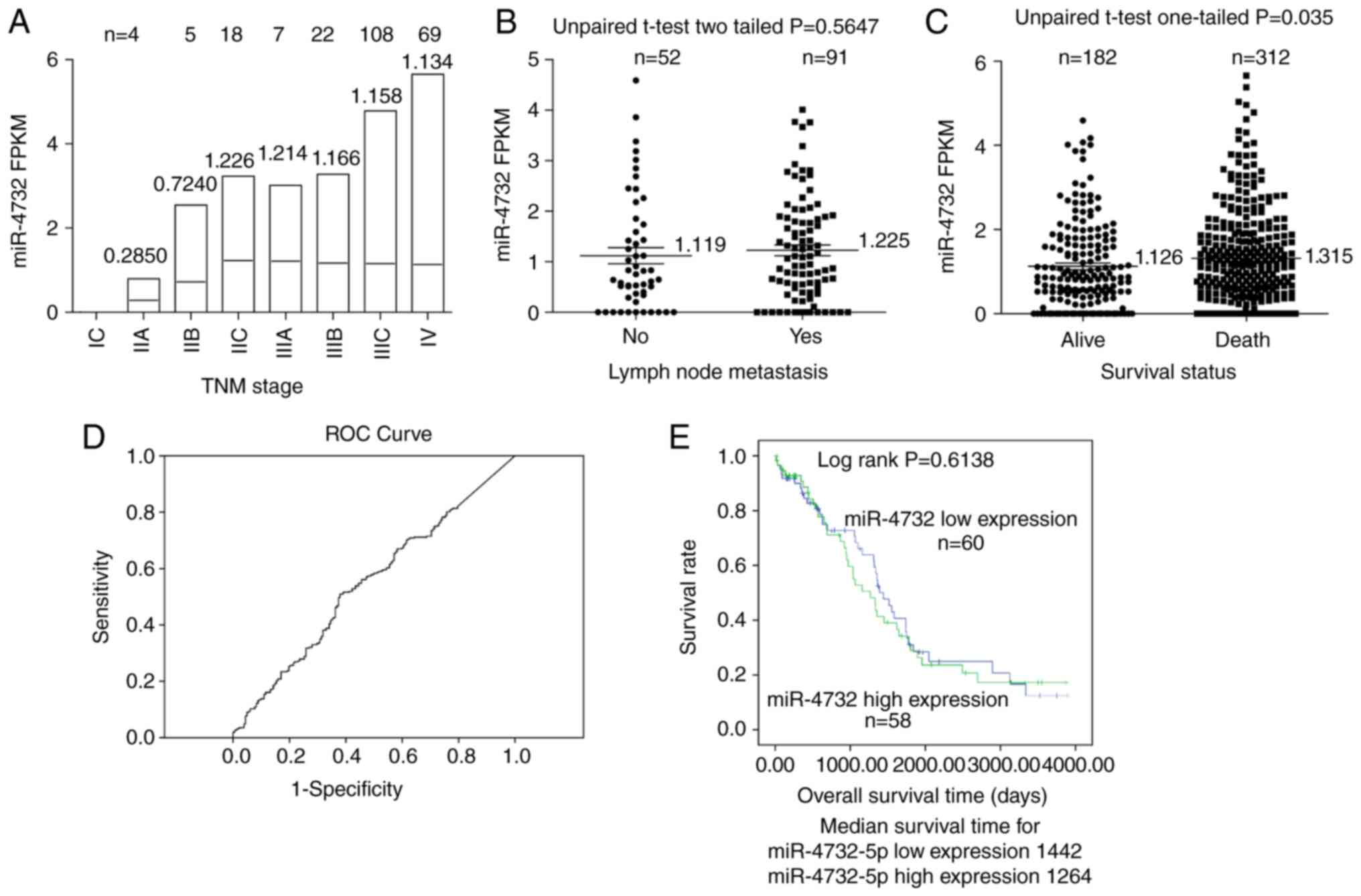
The miR-4732 expression levels in TCGA-OV database
were downloaded and analyzed. Although there was no significant
difference among the TNM subgroups according to the results of
one-way ANOVA, there was a trend towards an increased miR-4732
expression in the early TNM stage (IIA, IIB and IIC; Fig. 1A). In addition, in the late TNM
stage (IIIA, IIIB, IIIC and IV), the expression levels of miR-4732
appeared to reach a specific threshold, being similar to those in
the IIC stage (Fig. 1A), indicating
that the miR-4732 may play a role in the early stages of
tumorigenesis. No statistically significant differences were
observed when comparing the expression levels of miR-4732 in tissue
groups with and without lymph node metastasis (Fig. 1B). However, the expression levels of
miR-4732 in the patients with the ‘death’ OS status option (OS=1 in
the survival analysis acquired from TCGA-OV) following surgery were
significantly higher than those with the ‘alive’ OS status option
(OS=0 in the survival analysis acquired from TCGA-OV; Fig. 1C; unpaired student's t-test,
one-tailed; P=0.035). Herein, a one-tailed test was used, since the
present study was only concerned about one direction, not both,
indicating that the increased level of miR-4732 expression
predicted a poor prognosis. According to the ROC curve analysis
(Fig. 1D), the cut-off value for
miR-4732 expression was determined by the point corresponding to
the maximum the Youden's index (Sensitivity + Specificity-1).
According to this cut-off value, the patients were classified into
the high or low miR-4732 expression groups. The K-M survival curve
suggested that the OS of patients with a high miR-4732 expression
exhibited no significant difference as compared with that of those
with a low miR-4732 expression (Fig,
1E; log-rank test; P=0.6138). These results indicate that
miR-4732 expression may be used for defining a poor prognosis or
mortality, although not for defining the OS time using the K-M
survival curve for deceased patients after surgery.
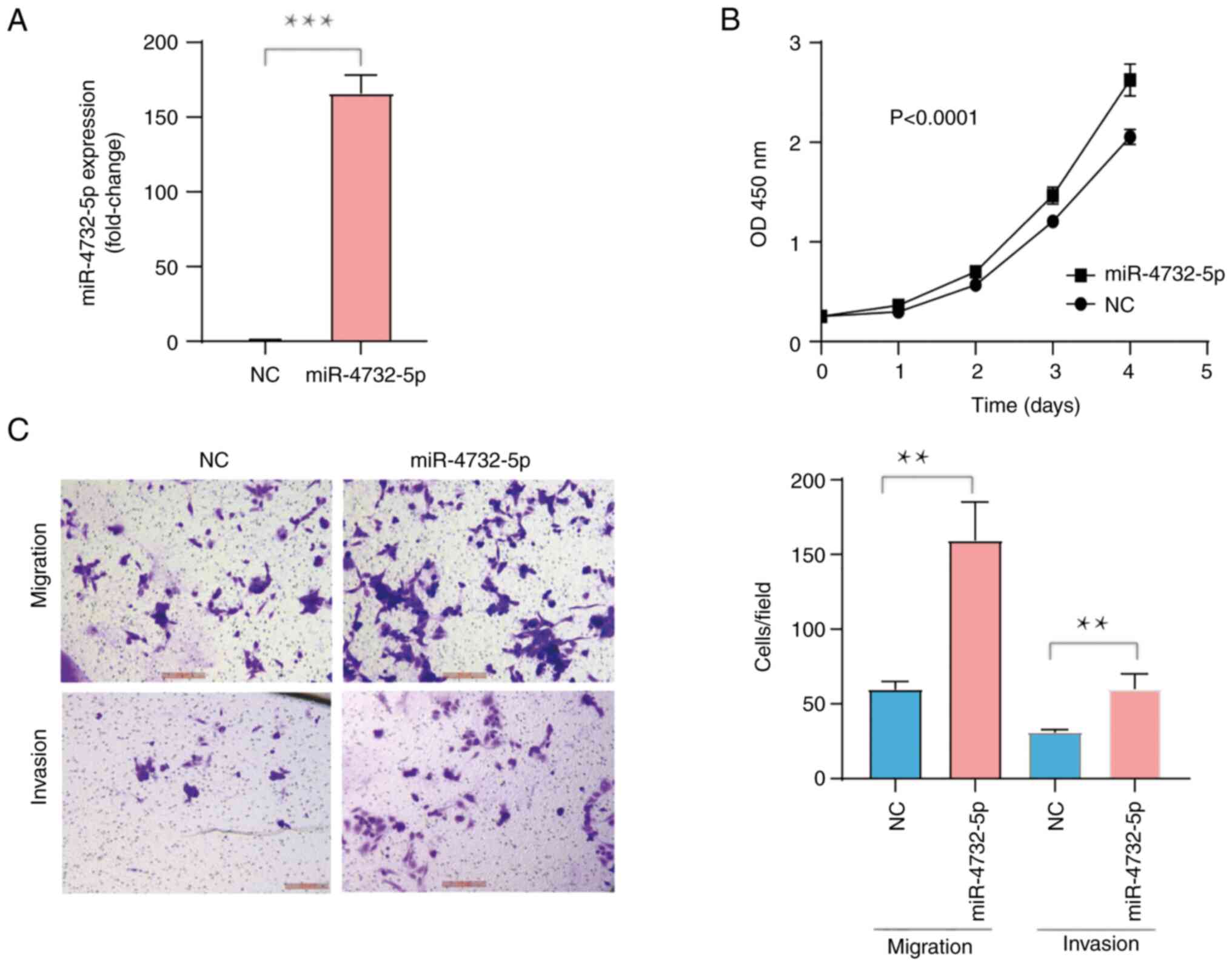
Ectopic overexpression of miR-4732
promotes the viability, migration and invasion of IGROV1 cells in
vitro
miR-4732 includes miR-4732-5p and miR-4732-3p.
Recently, miR-4732-5p expression in plasma-derived exosomes from
patients with EOC was confirmed to be increased, as compared with
that of healthy subjects, and that it could be used as a diagnostic
marker or as a monitor for EOC progression from the early to the
late stage (12). Herein, since it
was hypothesized that miR-4732-5p may function as an oncogene in
OC, the phenotypic alterations were evaluated following the induced
overexpression of miR-4732 using miRNA mimics in IGROV1 cells.
RT-qPCR analysis demonstrated miR-4732-5p expression was increased
166-fold, as compared with the cells transfected with miRNA NC
(Fig. 2A; P<0.0001).
Concurrently, IGROV1 cell viability significantly increased
following transfection with miRNA mimics in comparison with the
cells transfected with mimics NC, as evaluated using CCK-8 assay
(Fig. 2B; P<0.0001).
Furthermore, the migratory or invasive ability of the IGROV1 cells
transfected with miRNA mimics was markedly enhanced, as compared
with the cells transfected with NC (Fig. 3C; P<0.001), as shown by Transwell
assay. These results thus suggested that miR-4732-5p overexpression
promoted IGROV1 cell proliferation, migration and invasion in
vitro.
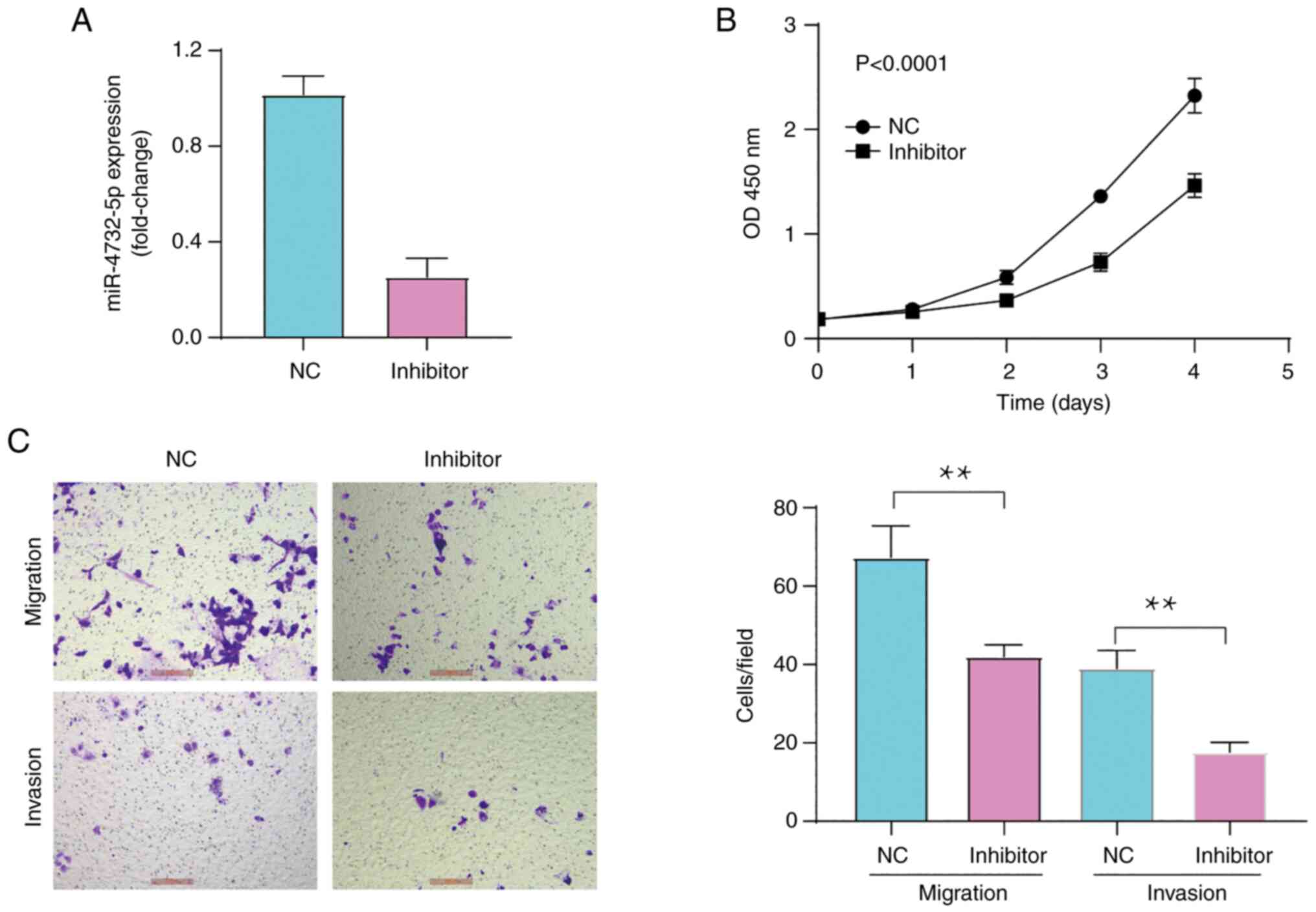
Inhibition of miR-4732-5p suppresses
cell viability, migration and invasion in vitro
To further confirm the aforementioned results on the
viability and the migratory and invasive ability of OC cells, the
IGROV1 cells were then transfected with miR-4732-5p inhibitors.
RT-qPCR analysis demonstrated that miR-125a-5p expression was
inhibited by 68% in the cells transfected with miR-4732-5p
inhibitors (Fig. 3A; P<0.001).
Consistent with the aforementioned results, the viability (Fig. 3B; P<0.0001), as well as the
migratory or invasive ability (Fig.
3C; P<0.001) of the IGROV1 cells transfected with
miR-4732-5p inhibitors was significantly decreased compared to
cells transfected with inhibitor NC. Therefore, miR-4732-5p
silencing exerted an inhibitory effect on IGROV1 cell
proliferation, migration and invasion in vitro.
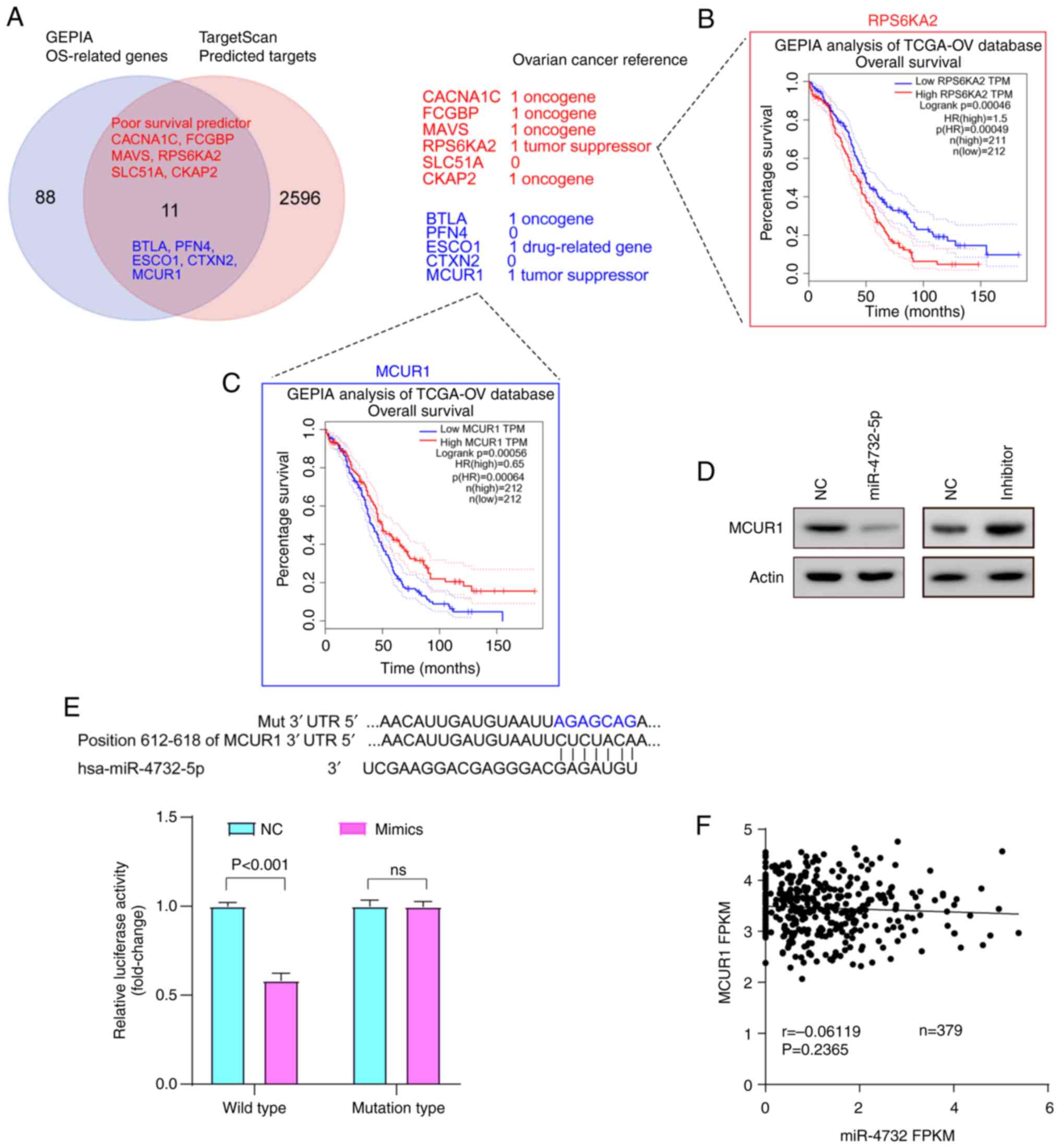
Tumor suppressor MCUR1 may be a direct
target of miR-4732-5p
TargetScan version 7.1 predicted a total of 99
direct targets, and GEPIA identified 2,607 OS-related genes. In
total, 11 candidate targets were obtained through the intersection
genes of above 2 tables according to the presented Venn plot
(Fig. 4A). According to the
hypothesis of miR-4732-5p being an oncogene in OC, it was suggested
that targets should act as tumor suppressor in OC. Thus, a
literature search using PubMed (https://pubmed.ncbi.nlm.nih.gov/) was performed. Two
out of the 11 genes [ribosomal protein S6 kinase A2 (RPS6KA2)
(16) and MCUR1 (17)] had been reported as tumor
suppressors, five genes [CACNA1C (18), FCGBP (19), MAVS (20), CKAP2 (21) and BTLA (22)] as oncogenes, ESCO1 was reported as a
drug-related gene (23) and three
genes (SLC51A, PFN4 and CTXN2) had no reference reported in OC.
According to the OS of patients analyzed using GEPIA K-M survival
curve analysis, the increased expression of RPS6KA2 predicted a
poor OS (Fig. 4B), which was
contrary to the suppressive role of RPS6KA2 reported in OC
(16), while the increased levels
of MCUR1 expression predicted an improved OS (Fig. 4C), which was consistent with the
tumor suppressor role of MCUR1 reported in OC (17). Thus, MCUR1 was selected to be
validated in further experiments. The expression of MCUR1 in the
cells was then validated after altering the miR-4732-5p expression
levels. Consistent with the prediction, MCUR1 expression was
decreased in the cells following transfection with miR-4732-5p
mimics, whereas it was increased in the cells following
transfection with miR-4732-5p inhibitors (Fig. 4D). Usually, miRNA inhibited
downstream targets by complementary binding to the seed region of
mRNA 3′UTR. There was one binding region at the position 612–618 WT
fragment of MCUR1 3′UTR for miR-4732-5p predicted by TargetScan
(Fig. 4E, binding site). The WT
MCUR1 3′UTR fragments or the mutant type at the predicted
miR-4732-5p binding sites were inserted pGL3-control plasmid
downstream of the Firefly luciferase gene. Dual-luciferase reporter
assay suggested that the relative luciferase activity was
significantly suppressed in miR-4732-5p mimics group for the WT
MCUR1 3′UTR (Fig. 4E bar graph;
P<0.001); however, the contrary was observed for the mutant type
of MCUR1 3′UTR (Fig. 4E), as
compared with the negative control, indicating that miR-4732-5p may
inhibit MCUR1 by binding to the predicted binding region. These
results indicated that miR-4732-5p may function as an oncogene by
directly targeting a tumor suppressor, MCUR1. However, it was not
possible to identify a negative correlation between the miR-4732-5p
and MCUR1 expression in the GEPIA correlation analysis (Fig. 4F).
Discussion
The hsa-miR-4732-5p has been reported either as an
oncogene or tumor suppressor in various types of cancer. Firstly,
miR-4732-5p has been reported to promote breast cancer progression
by targeting tetraspanin 13 (TSPAN13) (24). In addition, a high level of
miR-4732-5p has also been reported as a predictor for relapse
following S-1 adjuvant chemotherapy in gastric cancer (25). By contrast, miR-4732-5p has been
shown to inhibit tumor cell proliferation, migration and invasion
in non-small cell lung cancer through the modulation of TSPAN13
(26) and in lung adenocarcinoma
via the PI3K/Akt/GSK3β/Snail pathway (27). In OC, the increased expression of
miR-4732-5p in plasma-derived exosomes is a promising non-invasive
diagnostic biomarker, being able to distinguish cancer patients
from healthy individuals and also monitor cancer progression from
the early to the late stage (12).
These findings may suggest the critical role of miR-4732-5p and its
diverse functions in cancer progression. However, the effect of
miR-4732-5p on the malignant phenotype of OC cells remains
undefined. In the present study, the oncogenic role of miR-4732-5p
in OC was determined by gain-function and loss-function experiments
in order to elucidate an underlying potential mechanism.
In accordance with the aforementioned studies
(12,25), the results of the present study
demonstrated that the expression of miR-4732 in the patients with
the ‘death’ OS status following surgery in the survival analysis
acquired from TCGA-OV was significantly higher than in those with
the ‘alive’ OS status, suggesting that it may be a poor prognostic
predictor for patients with OC.
The expression levels of miR-4732 reported in TGCA
include both miR-4732-3p and miR-4732-5p expression. Since the high
expression of miR-4732-5p in plasma-derived exosomes is a promising
non-invasive diagnostic biomarker in OC (12), only the effects of miR-4732-5p on OC
cells were analyzed in the present study. The role of miR-4732-3p
in OC needs to be investigated in future studies. Previously, it
was reported that miR-4732-5p can function as either a tumor
suppressor or oncogene (oncomiRs) in various type of cancer.
Consistent with the findings of previous studies on breast and
gastric cancer supporting the oncomiR role of miR-4732-5p (24,25),
the results from present study demonstrated that miR-4732-5p also
functioned as an oncogene in OC. The results from previous research
(26,27) mentioning miR-4732-5p as a tumor
suppressor gene, which are inconsistent with the results of the
present study may be attributed to the differential background of
various cancer types.
In total, 11 candidate direct target genes were
obtained by intersecting 99 genes predicted using TargetScan and
2,607 OS-related genes from GEPIA. Through further reference
analysis, the analysis was limited to two possible target genes of
miR-4732-5p, including RPS6KA2 and MCUR1. Previously, it was
reported that RPS6KA2 reduced the proliferation, induced G1 arrest,
increased apoptosis, reduced the levels of phosphorylated
extracellular signal-regulated kinase and altered the levels of
other cell cycle proteins in UCI101 cells (16), and small interfering RNA against
RPS6KA2 demonstrated the opposite effect in 41M cells (16). In a previous study on the MCUR1 gene
in OC (17), the expression of
MCUR1 was acquired from TCGA database and it was revealed that the
decreased expression of MCUR1 was associated with a poor prognosis
of patients with OC. In the present study, the K-M survival curves
in relation to two candidate genes, RPS6KA2 and MCUR1, were
investigated using GEPIA analysis. The results indicated that the
increased expression of the tumor suppressor RPS6KA2 was associated
with a poorer prognosis, which was inconsistent with the tumor
suppressor role of RPS6KA2 in OC. This may be explained by the fact
that one gene can play exactly the opposite role in two different
contexts, similarly to the roles of the well-known tumor suppressor
‘wild-type’ version of p53 (WTp53), which has been reported to
promote tumors by stimulating the cancer metabolic switch of the
oxidative phosphorylation in glycolysis by promoting the
PUMA-mediated disruption of mitochondrial pyruvate uptake in
hepatocellular carcinoma, rather than exerting a suppressive effect
(28). MCUR1 was selected for
further study due to the available consistent results from the
GEPIA analysis. Additionally, MCUR1 was validated as the direct
target of miR-4732-5p, which is consistent with a previous study on
OC (17). According to OS of
patients examined using the GIPIA K-M survival curve analysis, the
high level expression of RPS6KA2 predicted a poor OS (Fig. 4B), which was contrary to the
suppressive role of RPS6KA2 reported in OC (16), while a high level of MCUR1
expression predicted a good OS (Fig.
4C), which was consistent with the tumor suppressive role of
MCUR1 reported in OC (17). In the
present study, it was hypothesized based on previously available
data (12) that miR-4732-5p, as an
oncogene, targeted tumor suppressors and the target genes were
narrowed down into two genes, RPS6KA2 and MCUR1, based on previous
reference (16,17). However, there is only one study
available in OC which suggests RPS6KA2 (16) and MCUR1 (17), respectively, as tumor suppressors.
Thus, additional further experimental data are required in order to
validate any possible suppressive role. In a study on the MCUR1
gene in OC (17), the expression of
MCUR1 was acquired from TCGA database and it was found that the
decreased expression of MCUR1 was associated with a poor prognosis
of patients with OC.
Of note, a limitation of the present study is the
lack of MCUR1 and RPS6KA2 overexpression experiments. Thus, the
gene function of MCUR1 in OC warrants further confirmation.
Furthermore, the expression correlations between MCUR1 and
miR-4732-5p in both OC and non-cancerous cell lines require further
investigation. Biologically, MCUR1 protein plays a crucial role in
mitochondrial calcium uptake (29).
Mitochondrial Ca2+ uptake occurs via the mitochondrial
Ca2+ uniporter (MCU) complex and MCUR1 acting as a
scaffold factor for MCU channel function is an essential MCU
regulators. Thus, it was suggested that MCUR1 may regulate cancer
progression by affecting the cellular Ca2+ concentration
and Ca2+-related signaling. It was determined that
miR-4732-5p may bind to the MCUR1-3′UTR and inhibit its expression
though the UTR binding site, as presented in Fig. 4E. However, it was not possible to
identify the negative correlation between miR-4732-5p and MCUR1
expression in the GEPIA correlation analysis (Fig. 4F). As regards RPS6KA2, it has been
reported that RPS6KA2 suppresses proliferation, induces G1 arrest,
increases apoptosis, reduces the levels of phosphorylated
extracellular signal-regulated kinase and alters the levels of
other cell cycle proteins in OC cell lines (16). Thus, it can be hypothesized that
RPS6KA2 may reduce cancer progression through a similar
mechanism.
In conclusion, the results of the present study
supported the conclusions of a previous study (12) that miR-4732-5p may function as an
oncogene in OC by possibly targeting MCUR1.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from Medical Health
Science and Technology Project of Zhejiang Province Health
Commission (grant no. 2022KY1160).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL performed the majority of the experiments. XW and
JW participated in the in vitro study and acquired cell
viability data. XL designed the experiments, coordinated the study
and wrote the manuscript. XL, XW and JW confirm the authenticity of
all the raw data. All authors have read and approved the final
version of this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Ningbo Women and Children's Hospital (Approval
No.:EC2022-024).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer Statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rojas V, Hirshfield KM, Ganesan S and
Rodriguez-Rodriguez L: Molecular characterization of epithelial
ovarian cancer: Implications for diagnosis and treatment. Int J Mol
Sci. 17:21132016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giampaolino P, Della Corte L, Foreste V,
Vitale SG, Chiofalo B, Cianci S, Zullo F and Bifulco G: Unraveling
a difficult diagnosis: The tricks for early recognition of ovarian
cancer. Minerva Med. 110:279–291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stewart C, Ralyea C and Lockwood S:
Ovarian cancer: An integrated review. Semin Oncol Nurs. 35:151–156.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bader AG: miR-34-a microRNA replacement
therapy is headed to the clinic. Front Genet. 3:1202012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ottosen S, Parsley TB, Yang L, Zeh K, van
Doorn LJ, van der Veer E, Raney AK, Hodges MR and Patick AK: In
vitro antiviral activity and preclinical and clinical resistance
profile of miravirsen, a novel anti-hepatitis C virus therapeutic
targeting the human factor miR-122. Antimicrob Agents Chemother.
59:599–608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen SN, Chang R, Lin LT, Chern CU, Tsai
HW, Wen ZH, Li YH, Li CJ and Tsui KH: MicroRNA in Ovarian cancer:
Biology, pathogenesis, and therapeutic opportunities. Int J Environ
Res Public Health. 16:15102019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng H, Liu JY, Song FJ and Chen KX:
Advances in circulating microRNAs as diagnostic and prognostic
markers for ovarian cancer. Cancer Biol Med. 10:123–130.
2013.PubMed/NCBI
|
|
10
|
Resnick KE, Alder H, Hagan JP, Richardson
DL, Croce CM and Cohn DE: The detection of differentially expressed
microRNAs from the serum of ovarian cancer patients using a novel
real-time PCR platform. Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Llaurado M, Majem B, Altadill T, Lanau L,
Castellví J, Sánchez-Iglesias JL, Cabrera S, De la Torre J,
Díaz-Feijoo B, Pérez-Benavente A, et al: MicroRNAs as prognostic
markers in ovarian cancer. Mol Cell Endocrinol. 390:73–84. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu J, Yoo J, Ho JY, Jung Y, Lee S, Hur SY
and Choi YJ: Plasma-derived exosomal miR-4732-5p is a promising
noninvasive diagnostic biomarker for epithelial ovarian cancer. J
Ovarian Res. 14:592021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li G, Ao S, Hou J and Lyu G: Low
expression of miR-125a-5p is associated with poor prognosis in
patients with gastric cancer. Oncol Lett. 18:1483–1490.
2019.PubMed/NCBI
|
|
14
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bignone PA, Lee KY, Liu Y, Emilion G,
Finch J, Soosay AE, Charnock FM, Beck S, Dunham I, Mungall AJ and
Ganesan TS: RPS6KA2, a putative tumour suppressor gene at 6q27 in
sporadic epithelial ovarian cancer. Oncogene. 26:683–700. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan L, Yang H, Zhang B and Ding H: MCUR1
is a prognostic biomarker for ovarian cancer patients. Cancer
Biomark. 33:311–316. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang X and Dong Y: CACNA1C is a
prognostic predictor for patients with ovarian cancer. J Ovarian
Res. 14:882021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang K, Guan C, Shang X, Ying X, Mei S,
Zhu H, Xia L and Chai Z: A bioinformatic analysis: The
overexpression and clinical significance of FCGBP in ovarian
cancer. Aging (Albany NY). 13:7416–7429. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen L, Hou J, You B, Song F, Tu X and
Cheng X: An analysis regarding the prognostic significance of mavs
and its underlying biological mechanism in ovarian cancer. Front
Cell Dev Biol. 9:7280612021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang M and Zhao L: CKAP2 promotes ovarian
cancer proliferation and tumorigenesis through the FAK-ERK pathway.
DNA Cell Biol. 36:983–990. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang RR, Wang LM and Shen JJ:
Overexpression of miR-32 inhibits the proliferation and metastasis
of ovarian cancer cells by targeting BTLA. Eur Rev Med Pharmacol
Sci. 24:4671–4678. 2020.PubMed/NCBI
|
|
23
|
Zhai DK, Liu B, Bai XF and Wen JA:
Identification of biomarkers and pathway-related modules involved
in ovarian cancer based on topological centralities. J BUON.
21:208–220. 2016.PubMed/NCBI
|
|
24
|
Wang YW, Zhao S, Yuan XY, Liu Y, Zhang K,
Wang J, Zhu J and Ma R: miR-4732-5p promotes breast cancer
progression by targeting TSPAN13. J Cell Mol Med. 23:2549–2557.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Omura T, Shimada Y, Nagata T, Okumura T,
Fukuoka J, Yamagishi F, Tajika S, Nakajima S, Kawabe A and Tsukada
K: Relapse-associated microRNA in gastric cancer patients after S-1
adjuvant chemotherapy. Oncol Rep. 31:613–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang X, Liu S, Cui Y and Zhao Y:
MicroRNA-4732 is downregulated in non-small cell lung cancer and
inhibits tumor cell proliferation, migration, and invasion. Respir
Med Res. 80:1008652021.PubMed/NCBI
|
|
27
|
Hu Y, Bai J, Zhou D, Zhang L, Chen X, Chen
L, Liu Y, Zhang B, Li H and Yin C: The miR-4732-5p/XPR1 axis
suppresses the invasion, metastasis, and epithelial-mesenchymal
transition of lung adenocarcinoma via the PI3K/Akt/GSK3β/Snail
pathway. Mol Omics. 18:417–429. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim J, Yu L, Chen W, Xu Y, Wu M, Todorova
D, Tang Q, Feng B, Jiang L, He J, et al: Wild-Type p53 promotes
cancer metabolic switch by inducing PUMA-Dependent suppression of
oxidative phosphorylation. Cancer Cell. 35:191–203. e82019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alevriadou BR, Patel A, Noble M, Ghosh S,
Gohil VM, Stathopulos PB and Madesh M: Molecular nature and
physiological role of the mitochondrial calcium uniporter channel.
Am J Physiol Cell Physiol. 320:C465–C482. 2021. View Article : Google Scholar : PubMed/NCBI
|