Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer globally (10.0%), and the second most important
cause of cancer death (9.4%). More than 1.9 million new CRC cases
and 935,000 deaths were estimated to occur in 2020 (1). The overall incidence increased in most
European countries over the last decade, especially in low- and
middle-income countries (2). It is
therefore clear that new and improved treatment options,
prevention, and early detection are essential. The most important
prognostic factors of CRC known today are the anatomical spread of
the disease (determined by TNM classification), and the status of
resection margins. However, it has been shown that using solely
anatomical spread is an insufficient prognostic factor (3). As a result, a considerable amount of
research has been done at the molecular level, and four consensus
molecular subtypes (CMS) of CRC were defined (4), with distinct molecular and biological
characteristics. CMS classification has led to better understanding
of CRC heterogeneity, progression, and response to treatment
(5). However, this has not
significantly impacted CRC timely diagnosis and prognosis. Fecal
occult blood test and colonoscopy are the two main screening tools
currently in use, but with suboptimal efficacy, in terms of
insufficient diagnostic accuracy, and invasiveness and high costs,
respectively. This further supports the need for new, sensitive,
and specific diagnostic and prognostic biomarkers, especially
considering that more than half of patients with CRC have
metastases at the time of diagnosis, and the average survival time
of patients with metastases is 24 months (6). In metastatic disease, KRAS
mutations status is an indispensable biomarker used to select
patients who are good candidates for anti-EGFR treatment (7). KRAS mutations are present in
approximately 50% of patients with metastatic disease and are
related to anti-EGFR biological therapy resistance (8).
Research on microRNAs (miRNAs) as new reliable
biomarkers of tumor development and progression is emerging. MiRNAs
are short, non-coding RNA sequences, 21–25 nucleotides in length
that negatively regulate the expression of protein-coding genes
(9). MiRNAs inhibit their targets
by acting as translational inhibitors of gene expression or by
degrading mRNA transcripts (10).
The expression levels of most protein coding genes in human cells
are subject to some degree of regulation by miRNAs, meaning that
miRNAs influence nearly all the developmental processes and
diseases in metazoans (11).
Deregulation of miRNAs is involved in the initiation and
progression of cancer, and aberrant miRNA expression is found in
most, if not all cancers (12). It
has been shown that miRNA profiling in patients can be used to
classify tumors with accuracy and predict outcome (13). Functional studies revealed that
miRNAs can act as both oncogenes and tumor suppressors (14); therefore, understanding miRNAs
expression levels in cancer patients could provide clues about the
affected molecular pathways and help identification of new targets
for treatment. MiRNAs high stability in biological fluids, easy
extraction and quantification, proven sensitivity and specificity,
makes them particularly suitable for biomarkers research (15).
In CRC, a current literature review identified over
230 potential candidate miRNAs in various biological fluids, with
some level of association to the pathology (16). However, more detailed mechanistic
studies on the significance of these associations are lacking for
most deregulated miRNAs. Thus, in order to give further insights
into the use of miRNA for diagnosis and treatment of CRC we
conducted a pilot study on small, but well-characterized patient
cohort in Montenegro for which both FFPE tissue and blood samples
were available. We selected a panel of five candidate miRNAs,
miR-29a, miR-101, miR-125b, miR-146a, and miR-155, known to be
involved in all essential cancer molecular and cellular pathways,
with both tumor suppressor and oncogenic roles. More precisely,
quite complex gene networks regulated by these miRNAs were shown to
be involved in proliferation, pro-inflammatory signaling,
apoptosis, angiogenesis, invasion, metastasis, and drug resistance
(17–20). Deregulation of these miRNAs with
various levels of correlation with clinical parameters has already
been shown in other populations (21,22),
however, these findings were often inconsistent and even
contradictory. Population differences in miRNA expression were
shown to have significant biological and pharmacological
implications (23). In addition,
miRNAs expression is also influenced by external factors, such as
physical activity and lifestyle (24). All of the previous statements
prompted us to perform this pilot study and analyze expression
levels of the five selected miRNAs in Montenegrin patients for the
first time. We assessed this panel of miRNAs in both pre-operative
plasma and FFPE tissue samples of patients with CRC and healthy
control subjects and correlated the results with demographic,
clinical and pathological features of the study participants. This
experimental setup enabled us to detect whether any of the studied
circulatory miRNAs potentially originated from the tumor. We also
performed bioinformatics analysis of the putative target genes of
these miRNAs, and their joint regulatory pathways.
Materials and methods
Patient recruitment and study
design
The study protocol was approved by the Ethical
Committee of the Clinical Center of Montenegro (approval no.
03/01-11417/1) and by the Committee for Medical Ethics and
Bioethics of the Faculty of Medicine of the University of
Montenegro (approval no. 3824/4). All the procedures were conducted
in accordance with the Declaration of Helsinki and all participants
signed an informed consent before any study procedures were
performed. All participants completed a standardized questionnaire,
in order to obtain demographic (age, gender), and basic health
information, including eventual comorbidities and frequency of
habits associated with an increased risk for CRC (BMI,
hypertension, hyperlipidemia, diabetes mellitus, history of
smoking, coffee and alcohol consumption, and physical activity).
Peripheral blood samples were also collected from all
participants.
CRC patients were recruited at the Center for
Digestive Surgery of the Clinical Center of Montenegro, between
November 2019 and November 2021. All patients were subjected to
standard clinical practice. Peripheral blood sampling was performed
during preoperative preparation on 24 patients who were admitted
for surgical resection of colon tumors previously diagnosed by
colonoscopy. Patients who received preoperative adjuvant therapy,
those who have clinically diagnosed hereditary adenomatous
polyposis or hereditary non-polyposis CRC, with a previous medical
history of malignancy, poorly controlled systemic disease, and/or
current acute disease, were not included in the study. In addition,
paired CRC and surrounding normal colon tissue samples were
obtained from CRC patients during the surgical treatment.
Healthy volunteers were recruited at the Faculty of
Medicine, University of Montenegro. We have recruited 34 age- and
sex-matched healthy volunteers. Inclusion criteria for the healthy
group of participants were negative history of cancer, uncontrolled
chronic systemic disease, including inflammatory bowel disease
(IBD), and/or current acute disease.
Plasma samples processing and miRNA
quantification by reverse transcription-quantitative PCR
(RT-qPCR)
MicroRNAs were extracted from plasma using Qiagen
miRNeasy Serum/Plasma Advanced kit (cat. No. 217204, Qiagen,
Hilden, Germany) essentially as described in (25). Briefly, peripheral venous blood
samples were collected in the EDTA-containing Vacutainer tubes,
kept on ice, and processed within 1 h from the collection. Plasma
was separated from the whole blood by centrifugation at 1,900 × g
for 10 min at 4°C, followed by an additional centrifugation step at
3,000 × g for 15 min at 4°C. All samples were aliquoted and stored
at −80°C until further analysis.
The miRNA concentration was determined using Qubit
microRNA Assay kit (Q32880, Invitrogen, Thermo Fisher Scientific)
on a Qubit 3.0 fluorimeter (Q33216, Invitrogen, Thermo Fisher
Scientific, USA). Two µl miRNA from each sample were reversely
transcribed to cDNA using TaqMan Advanced miRNA cDNA Synthesis kit
(A28007, Applied Biosystems, USA) and analyzed with TaqMan Advanced
microRNA Assays (A25576, Applied Biosystems, USA) for miR-29a,
miR-101, miR-125b, miR-146a, and miR-155. Context sequences of
TaqMan probes were as follows: ACUGAUUUCUUUUGGUGUUCAG for miR-29a;
CAGUUAUCACAGUGCUGAUGCU for miR-101; UCCCUGAGACCCUAACUUGUGA for
miR-125b; UGAGAACUGAAUUCCAUGGGUU for miR-146a; and
UUAAUGCUAAUCGUGAUAGGGGUU for miR-155. RT-qPCR was run on an Applied
Biosystems 7300 Real Time PCR system (Applied Biosystems, USA),
with the following thermocycling conditions: enzyme activation:
95°C, 20 sec; denaturation: 95°C, 3 sec; annealing: 60°C, 30 sec,
for 40 cycles. The expression levels of target miRNAs were
normalized by using the mean expression levels of miR-361-5p gene
for plasma samples, and miR-186-5p for FFPE tissue samples,
selected as the most stable internal control miRNA by the
NormFinder algorithm (26). These
miRNAs are recommended as endogenous controls due to their
relatively constant expression levels across many different sample
types. Expression of every target gene was calculated using the
2−ΔΔCq method (27).
Every sample was retrotranscribed twice and run in triplicate each
time.
FFPE tissue sample processing and
pathohistological analysis
Biopsy samples were fixed in 10% buffered formalin
and embedded in paraffin. Serial sections, 5 µm thick, were cut
using microtome (Leica SM 200R, Austria). After deparaffinization
in xylene and hydration in descending order of alcohol, sections
were stained with Mayer's hematoxylin and 1% eosin solution, then
illuminated and mounted on slides using dibutylphtalate polystyrene
xylene (DPX).
Morphological analysis and reporting were conducted
by two independent pathologists using CAP protocols (Cancer
Reporting Protocols-College of American Pathologists). For each
sample tumor site and size, histological type and grade, tumor
extent, presence of lymphovascular and perineural invasion,
necrosis, mucus production, inflammatory infiltrate density and
composition, status of margins, lymph nodes status, and disease
stage were estimated. Representative FFPE tissue samples were
selected for miRNA and DNA extraction. Representative image of
hematoxylin and eosin stained FFPE tissue sample of the patient
with CRC is given in Fig. S1A. Red
square in the image denotes the area of the tumor, while the rest
is healthy colonic mucosa. FFPE tissue samples chosen for miRNA and
DNA extraction were selected by pathologists, so that FFPE tissue
sections containing no less than 80% of tumor cells were chosen for
the analysis as tumor samples, and healthy surrounding tissue of
the same patient was used as a control sample (Fig. S1B).
miRNA extraction from FFPE
tissues
MiRNA purification from FFPE tissues was performed
using miRNeasy FFPE kit (cat. no. 217504, Qiagen, Hilden, Germany),
according to the manufacturer's instructions. Two to three 10 µm
inner sections were aseptically collected in nuclease-free
microcentrifuge tube and deparaffinization solution (cat. no.
19093) was used to remove all paraffin. The hematoxylin and
eosin-stained slides were evaluated by pathologist who designated
control and tumor samples, ensuring no cancer cells were present in
the controls (Fig. S1B).
DNA extraction from FFPE tissues and
KRAS mutation analysis
DNA extraction from the FFPE tissue samples was
performed using QIAmp DNA FFPE Tissue kit (cat. No. 56404) from
Qiagen. Two 10 µm sections of FFPE tissue samples were cut, treated
with deparaffinization solution (cat. no. 19093), and DNA was
extracted following the manufacturer's instructions. KRAS
gene mutations were analyzed using RealLine KRAS Detect kit (REF
MED20401) (Bioron diagnostics GmbH). Seven mutations of codons 12
and 13 of the KRAS gene that are associated with resistance
to anti-EGFR therapy were detected with allele specific qPCR.
Target prediction and enrichment
analysis
Potential targets of five studied miRNAs were
predicted by miRTarBase database (https://mirtarbase.cuhk.edu.cn) (28), and only targets with strong evidence
(reporter assay, Western blot, and qPCR) were selected for further
analysis. Subsequently, only genes targeted by more than one miRNA
were selected and Gene Ontology (GO) functional annotation and KEGG
pathway analyses were made using the STRING online resource
(https://string-db.org/) (29).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 9.3.1 (GraphPad Software, San Diego, CA, USA). The
results were considered statistically significant when P<0.05.
Continuous variables were first tested for normality of
distribution by D'Agostino-Pearson and Shapiro-Wilk tests and
analyzed with the unpaired t-test or an appropriate non-parametric
test (Mann-Whitney). Normally distributed variables of tumor FFPE
tissue samples were analyzed with the paired Student's t-test.
Categorical variables were analyzed with the Fisher's exact test.
Receiver operating characteristic (ROC) curve analysis was
performed to evaluate potential diagnostic performance of studied
miRNAs. The area under the curve (AUC) was estimated, along with
the 95% CI. Correlations between miRNA expression and clinical
variables were explored using the Spearman correlation coefficient.
Logistic regression analysis was also performed to evaluate the
association of the investigated variables with the CRC.
Results
Patient characterization
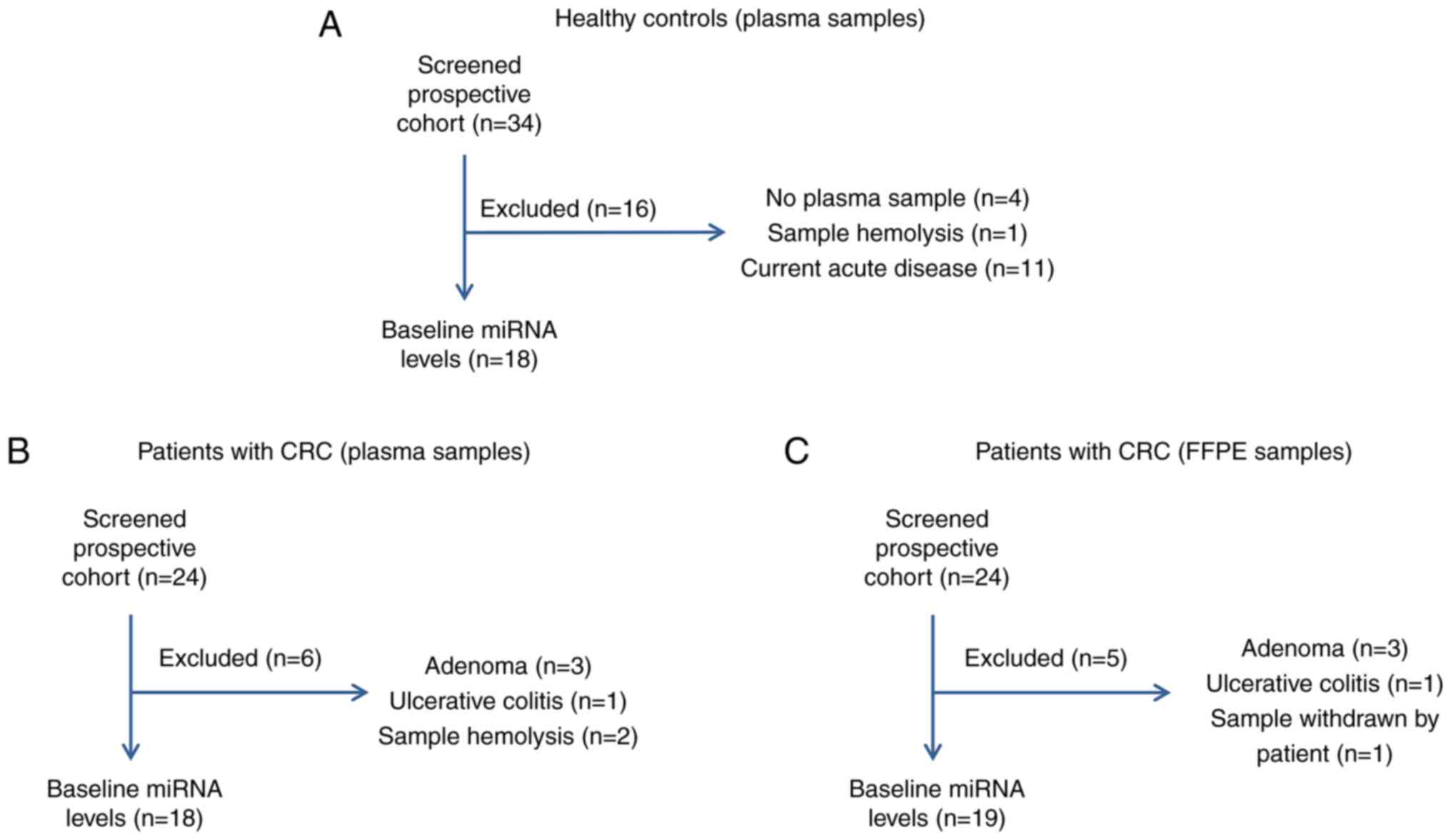
Out of a total of 24 CRC patients who were initially
recruited for this prospective study, six were excluded due to
postoperative pathohistological diagnosis of adenoma (n=3),
ulcerative colitis (n=1), and two due to sample hemolysis (Fig. 1B), thus 18 plasma samples in total
were further analyzed. From the group of 34 healthy volunteers
recruited, 18 were further analyzed, as presented in the scheme of
the recruitment in the Fig. 1A.
Demographic and clinical characteristics of study participants are
given in Table I.
 | Table I.Demographic and clinical
characteristics of the study cohorts (plasma samples). |
Table I.
Demographic and clinical
characteristics of the study cohorts (plasma samples).
| Characteristic | Colon cancer
(n=18) | Healthy controls
(n=18) | P-value |
|---|
| Sex |
|
| >0.999 |
| Female
(%) | 7 (38.9) | 7 (38.9) |
|
| Male
(%) | 11 (61.1) | 11 (61.1) |
|
| Age at diagnosis,
years |
|
|
|
| Mean ±
SD | 66.89±9.64 | 65.44±8.12 | 0.599 |
| Median
(range) | 66.0
(55.0-87.0) | 65.0
(55.0-77.0) |
|
| Mean ± SD BMI,
kg/m2 | 26.88±3.67 | 27.17±3.96 | 0.7842 |
| Hypertension
(%) | 9 (50) | 8 (44.4) | >0.999 |
| Hyperlipidemia
(%) | 2 (11.1) | 7 (38.9) | 0.1212 |
| Diabetes mellitus
(%) | 3 (16.7) | 3 (16.7) | >0.999 |
| Physical activity
(%) | 16 (88.9) | 11 (61.1) | 0.1212 |
| History of smoking
(%) | 10 (55.6) | 10 (55.6) | >0.999 |
| Coffee consumption
(%) | 0 (0) | 7 (38.9) | 0.0076a |
| Alcohol consumption
(%) | 5 (27.8) | 9 (50) | 0.3053 |
There were no significant differences in prevalence
of either gender. The range of ages of the participants enrolled in
our study is given in the Table I
(55.0-87.0 for patients with CRC, and 55.0-77.0 for healthy
controls). The median age of patients with CRC was 66 years, and
for healthy control subjects it was 65, and this difference was not
statistically significant. Body mass index (BMI) was within the
overweight range (>25-30) for both groups, with no statistically
significant difference between the two. Hypertension was the most
prevalent comorbidity in both CRC patients and healthy group,
followed by hyperlipidemia and diabetes mellitus. However, their
prevalence did not differ significantly among the two groups
(Table I). The same applied for
physical activity, history of smoking, and alcohol consumption,
whereas coffee consumption was significantly higher in the healthy
group, since none of the patients with CRC consumed 3 or more cups
of coffee daily. Therefore, patients with CRC and healthy
participants proved to have a homogeneous distribution of the
demographic and clinical variables included in the
questionnaire.
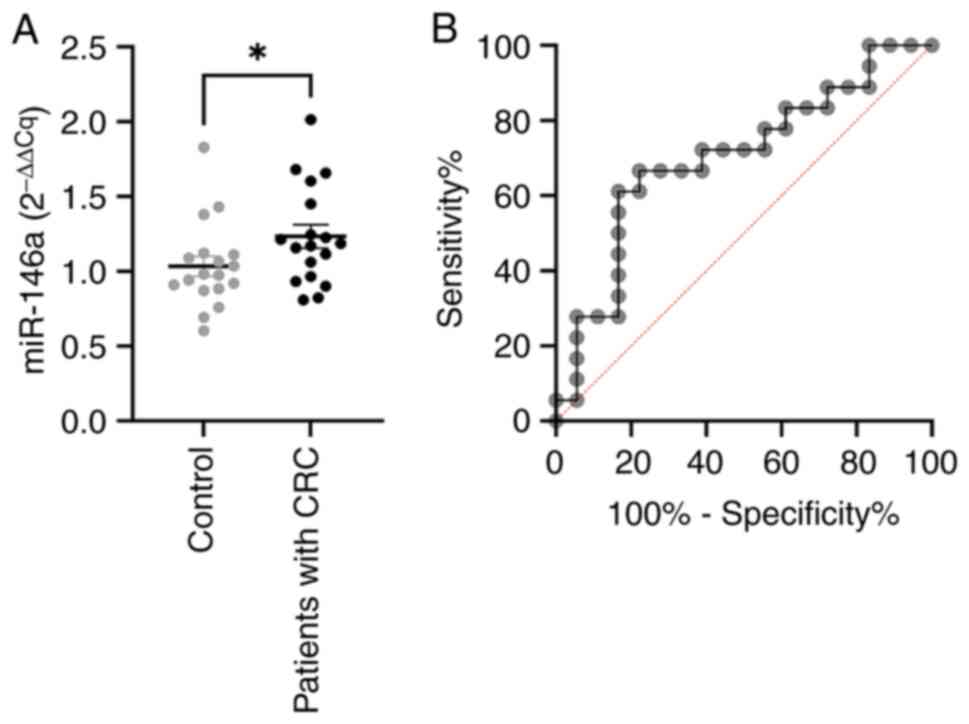
miR-146a is elevated in the plasma of
patients with CRC
In order to assess their role in the development of
CRC, relative expression of five selected miRNAs: miR-29a, miR-101,
miR-125b, miR-146a, and miR-155, was analyzed in plasma samples of
patients with CRC and healthy individuals. No statistically
significant differences were observed between groups for miR-29a,
miR-101, miR-125b, and miR-155 (data not shown). However,
expression levels of miR-146a were significantly higher (P=0.0402)
in the plasma of patients with CRC, compared to healthy
individuals, as shown in Fig.
2A.
miR-146a is one of the most important inflammatory
miRNAs, shown to mediate both the innate and adaptive immune
response (30). In order to assess
the diagnostic potential of miR-146a, we performed ROC analysis
(Fig. 2B), which showed AUC of
0,7006 (CI 95%, 0.5252-0.8761), with 66.7% sensitivity and 77.8%
specificity at a cutoff value of 1.114.
Correlation of plasma miRNA levels
with the demographic and basic health features of the
participants
The associations between all five investigated
miRNAs levels and patient characteristics are presented in the
Table SI. Correlation between
expression levels of miR-29a, miR-101, miR-125b, miR-146a, and
miR-155, and the following variables was investigated: gender, age,
BMI, hypertension, hyperlipidemia, diabetes mellitus, physical
activity, history of smoking, and alcohol consumption.
Only two associations were found to be statistically
significant, namely, miR-146a expression was significantly higher
in male in comparison to female CRC patients (P=0.0441); and lower
levels of miR-101 expression were found in patients with a history
of smoking (P=0.0307) (Table SI).
Given that miR-146a levels were significantly up-regulated in the
plasma of CRC patients, we performed a logistic regression analysis
on all participants using miR-146a expression, and clinical
parameters studied, but the results were not significant (data not
shown). This is probably due to both the small sample size and the
homogeneity of the clinical parameters investigated between the
patients with CRC and healthy participants.
miRNAs deregulated in tissue samples
and their correlation with pathological features
Clinical characteristics of the CRC group of
patients are given in Table II.
After one participant withdrew their tissue samples, there were 19
patients with CRC who were recruited (Fig. 1C).
 | Table II.Demographic and clinical
characteristics of the study cohort (FFPE samples). |
Table II.
Demographic and clinical
characteristics of the study cohort (FFPE samples).
| Characteristic | Colon cancer
(n=19) |
|---|
| Sex |
|
| Female
(%) | 7 (36.8) |
| Male
(%) | 12 (63.2) |
| Age at
diagnosis |
|
| Mean ±
SD | 66.84±9.37 |
| Median
(range) | 66.0
(55.0-87.0) |
| TNM stage |
|
| I
(%) | 0 (0) |
| II
(%) | 1 (5.3) |
| III
(%) | 17 (89.5) |
| IV
(%) | 1 (5.3) |
| Nodal status |
|
|
Positive (%) | 11 (57.9) |
|
Negative (%) | 8 (42.1) |
| Tumor location |
|
| Left
(%) | 6 (31.6) |
| Right
(%) | 13 (68.4) |
| KRAS
mutation statusa |
|
|
Positive (%) | 5 (45.45%) |
|
Negative (%) | 6 (54.54%) |
A majority of patients (89.5%) were TNM stage III;
57.9% had positive nodal status, and tumors were predominantly
located in the right colon (68.4%). KRAS mutation status was
analyzed in all patients with lymph nodes metastasis (n=11), and
five of those patients (45.45%) had KRAS mutations. The
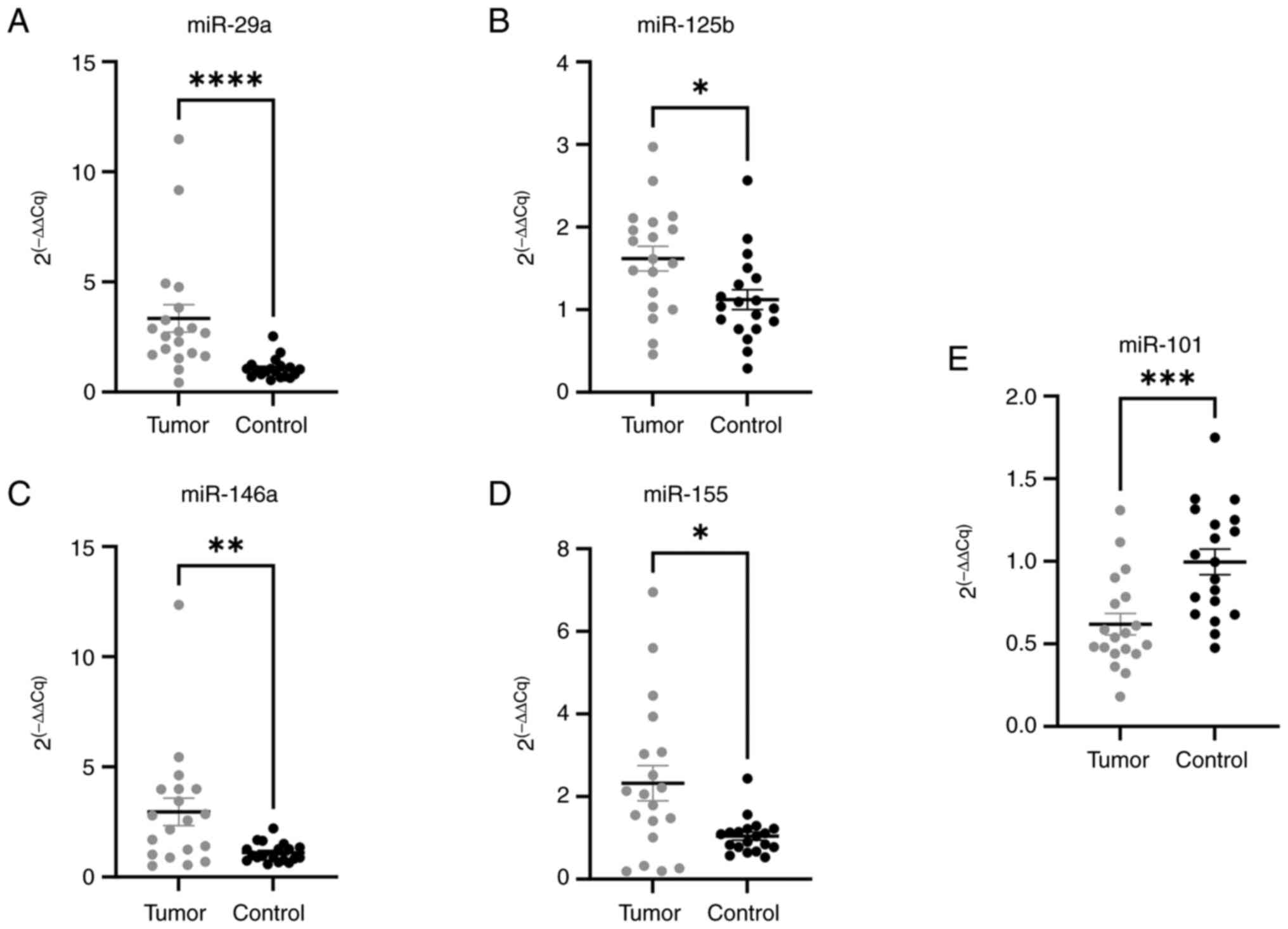
expression levels of miR-29a, miR-101, miR-125b, miR-146a, and
miR-155 were analyzed in FFPE tissue samples from the patients with
CRC by RT-qPCR and correlated with pathological features, in order
to assess their clinical relevance. As shown in the Fig. 3A-E, miR-29a (P<0.0001), miR-125b
(P=0.0141), miR-146a (P=0.0068), and miR-155 (P=0.0130) were
up-regulated in the tumor tissue, whereas miR-101 was found to be
down-regulated (P=0.0007) in the tumor, with respect to the
surrounding healthy colon tissue.
Diagnostic performance of the investigated miRNAs
for CRC was evaluated by computing AUC values of the ROC curves for
each miRNA. The results presented in the Table III show that miR-29a had highest
AUC value (0.8921), with 84.21% sensitivity and 89.47% specificity,
while both miR-101 and miR-146a had AUC levels above 0.75.
 | Table III.Diagnostic performance of all
investigated miRNAs in CRC tissue samples. |
Table III.
Diagnostic performance of all
investigated miRNAs in CRC tissue samples.
| FFPE miRNA | AUC | Cutoff point | Sensitivity, % | Specificity, % |
|---|
| miR-29a | 0.8921 | >1.582 | 84.21 | 89.47 |
| miR-101 | 0.8172 | <0.7511 | 73.68 | 73.68 |
| miR-125b | 0.7313 | >1.421 | 68.42 | 78.95 |
| miR-146a | 0.7535 | >1.683 | 63.16 | 94.74 |
| miR-155 | 0.7341 | >1.347 | 73.68 | 89.47 |
To investigate whether there is any correlation
between CRC tissue and plasma levels, we performed an analysis for
each individual miRNA, but no significant correlation was found
(data not shown). We have also compared miRNA expression levels in
the FFPE samples of the healthy surrounding tissue with miRNA
expression levels in CRC plasma samples, and we did not find any
significant correlation.
Next, we analyzed miRNA expression levels in
correlation with clinical and pathological features of patients
with CRC (Table SII). In
particular, we compared expression levels of all five investigated
miRNAs and the following characteristics: gender, age, histological
grade, nodal status, tumor localization, KRAS mutation
status, number of intratumoral lymphocytes, number of lymphocytes
around tumor, tumor stroma quantity, quantity and composition of
inflammatory infiltrate, necrosis, mucus, lympho-vascular invasion,
and lymph node metastases.
We found that there is a negative correlation
between miR-101 levels in tumor tissue and the lymphovascular
invasion (r=−0.4901, P=0.0389). We have also analyzed each
individual miRNA in two different groups for all investigated
demographic and clinical variables, as shown in the Table SI. Significant differences were
found for miR-146a, whose expression was significantly higher
(P=0.0435) in FFPE CRC tissue samples with higher degree of
necrosis; higher expression of miR-155 (P=0.0339) was found in
samples with no production of mucus; higher miR-125b expression
(P=0.0103) was found in samples without lympho-vascular invasion,
and lower miR-101 expression (P=0.0202) was found in samples with
positive nodal status. No significant differences were found in
expression values of any of the studied miRNA with respect to the
KRAS mutation status (Table
SII).
Putative joint regulatory pathways
identified by bioinformatics analysis
In order to better understand the consequences of
this identified five-miRNA deregulation, we performed a
bioinformatic analysis on the genes regulated by miR-29a, miR-101,
miR-125b, miR-146a, and miR-155. A total of 174 genes were found to
be regulated by at least two out of five studied miRNAs (Table SIII), and the most strongly
enriched GO processes and KEGG pathways analyzed by the STRING
database (31) are presented in the
Table IV.
 | Table IV.The most strongly enriched GO
processes and KEGG pathways-joint analysis. |
Table IV.
The most strongly enriched GO
processes and KEGG pathways-joint analysis.
| A, Biological
process (Gene Ontology) |
|---|
|
|---|
| Term or
pathway | Description | Count in
network | Strength | False discovery
rate |
|---|
| GO:0006211 | 5-methylcytosine
catabolic process | 3 of 3 | 2.05 | 0.00019 |
| GO:1905460 | Negative regulation
of vascular associated smooth muscle cell apoptotic process | 2 of 2 | 2.05 | 0.0044 |
| GO:1905075 | Positive regulation
of tight junction disassembly | 2 of 2 | 2.05 | 0.0044 |
| GO:1904466 | Positive regulation
of matrix metallopeptidase secretion | 2 of 2 | 2.05 | 0.0044 |
| GO:0035622 | Intrahepatic bile
duct development | 2 of 2 | 2.05 | 0.0044 |
| GO:0032707 | Negative regulation
of interleukin-23 production | 2 of 2 | 2.05 | 0.0044 |
| GO:0014740 | Negative regulation
of muscle hyperplasia | 2 of 2 | 2.05 | 0.0044 |
| GO:0003169 | Coronary vein
morphogenesis | 2 of 2 | 2.05 | 0.0044 |
| GO:0002384 | Hepatic immune
response | 2 of 2 | 2.05 | 0.0044 |
| GO:0061419 | Positive regulation
of transcription from RNA polymerase II promoter in response to
hypoxia | 4 of 6 | 1.87 |
2.15×10−5 |
|
| B, Molecular
function (Gene Ontology) |
|
| Term or
pathway |
Description | Count in
network |
Strength | False discovery
rate |
|
| GO:0004517 | Nitric-oxide
synthase activity | 3 of 3 | 2.05 | 0.00062 |
| GO:0003886 | DNA
(cytosine-5-)-methyltransferase activity | 3 of 3 | 2.05 | 0.00062 |
| GO:0070579 | Methylcytosine
dioxygenase activity | 3 of 4 | 1.93 | 0.00098 |
| GO:0034617 | Tetrahydrobiopterin
binding | 3 of 4 | 1.93 | 0.00098 |
| GO:0043125 | Erb-3 class
receptor binding | 3 of 4 | 1.93 | 0.00098 |
| GO:0003958 | NADPH-hemoprotein
reductase activity | 3 of 6 | 1.75 | 0.0021 |
| GO:003958 | Type III
transforming growth factor beta receptor binding | 2 of 4 | 1.75 | 0.0282 |
| GO:0070878 | Primary miRNA
binding | 4 of 10 | 1.65 | 0.00030 |
| GO:0051525 | NFAT protein
binding | 2 of 5 | 1.65 | 0.0375 |
| GO:0046870 | Cadmium ion
binding | 2 of 5 | 1.65 | 0.0375 |
|
| C, KEGG
pathways |
|
| Term or
pathway |
Description | Count in
network |
Strength | False discovery
rate |
|
| hsa05212 | Pancreatic
cancer | 26 of 73 | 1.6 |
1.37×10−29 |
| hsa05219 | Bladder cancer | 13 of 41 | 1.55 |
7.98×10−15 |
| hsa04933 | AGE-RAGE signaling
pathway in diabetic complications | 28 of 98 | 1.51 |
1.37×10−29 |
| hsa05220 | Chronic myeloid
leukemia | 20 of 75 | 1.48 | 5.34×
0−21 |
| hsa05218 | Melanoma | 19 of 72 | 1.47 |
5.88×10−20 |
| hsa05223 | Non-small cell lung
cancer | 17 of 68 | 1.45 |
1.09×10−17 |
| hsa01521 | EGFR tyrosine
kinase inhibitorresistance | 19 of 78 | 1.44 |
1.80×10−19 |
| hsa05213 | Endometrial
cancer | 14 of 57 | 1.44 |
1.22×10−14 |
| hsa05214 | Glioma | 17 of 72 | 1.42 |
2.29×10−17 |
| hsa05222 | Small cell lung
cancer | 21 of 92 | 1.41 |
7.11×10−21 |
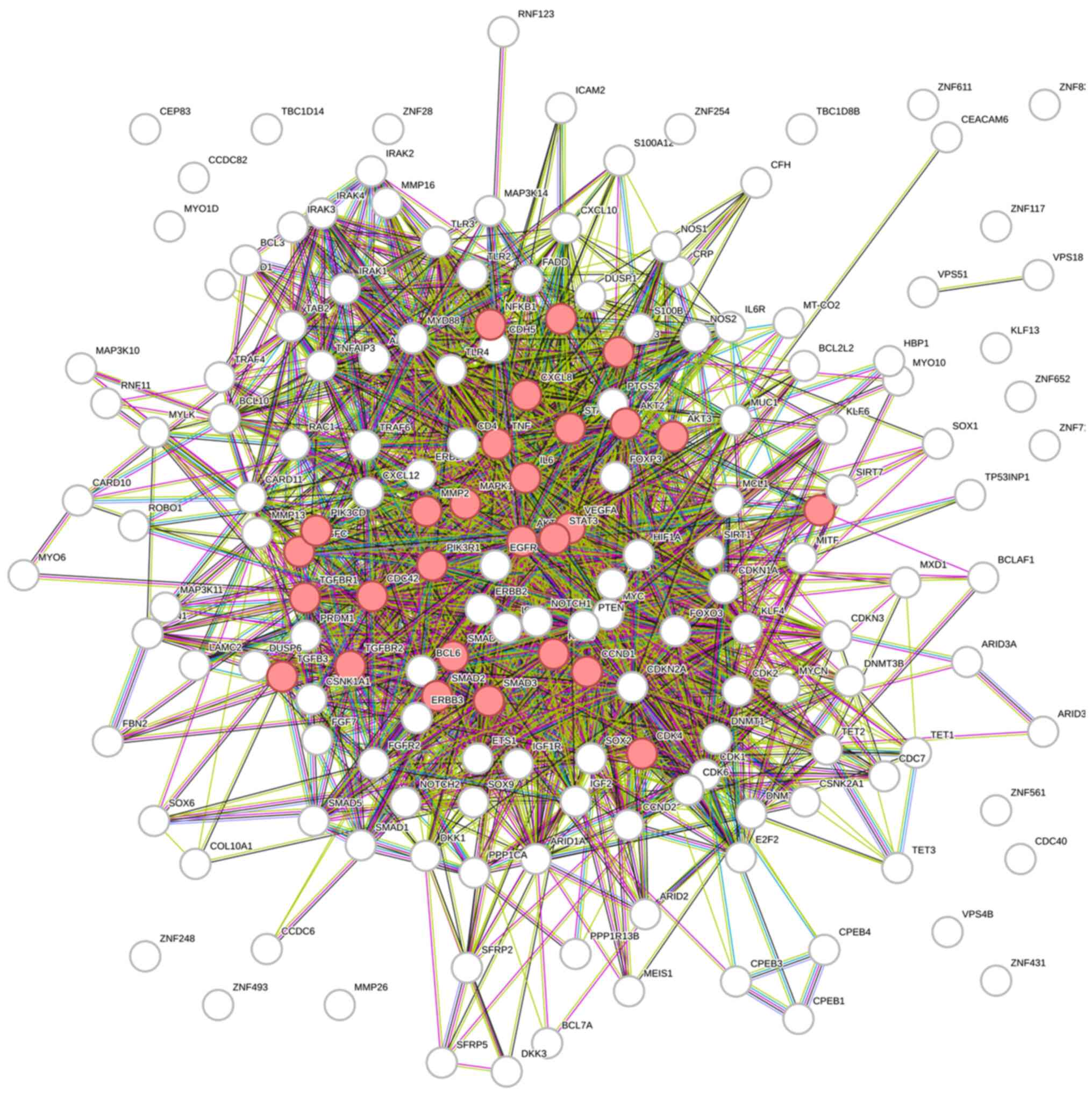
The enrichment analysis revealed that the targets of
candidate miRNAs were involved in pathways of several different
cancers, including pancreatic, bladder, chronic myeloid leukemia,
melanoma, and NSCLC, but also in EGFR tyrosine kinase inhibitor
resistance and interestingly enough, the AGE-RAGE signaling pathway
(Fig. 4).
Binding of advanced glycation end products (AGEs) to
their receptors (RAGEs) activate several different signaling
pathways, such as MAPK, p53, PI3K/Akt/mTOR, JAK-STAT, and NF-κB,
involved in the proliferation of cancer cells, angiogenesis, and
invasion (32), contributing to the
progression of cancer.
Discussion
The present pilot study determined the pattern of
miRNAs deregulation in CRC patients in Montenegro for the first
time, with the aim of contributing new information to the current
knowledge of which specific miRNA signature could be used in
clinical setting. We have shown that all five of the investigated
candidate miRNAs were deregulated in FFPE tissue samples of
patients with CRC, compared to the healthy surrounding colonic
mucosa (Fig. 3A-E), however, only
miR-146a levels were found to be up-regulated in both FFPE tissue
and plasma samples of patients with CRC (Figs. 2A and 3C). This finding suggests that circulatory
miR-146a probably originate from the tumor, further emphasizing its
potential role as a diagnostic biomarker. The discriminatory
potential of miR-146a in the plasma was shown to be modest
(AUC=0.7006), with a sensitivity and specificity of 66.7 and 77.8%,
respectively, at a cutoff value of 1.114. An ideal screening tool
should be highly sensitive and specific, safe, affordable, and
minimally invasive. The RT-qPCR methodology we used is not
expensive, requires venous blood sampling, which is minimally
invasive, and is easily repeatable. Our results on deregulation of
candidate miRNAs are in agreement with previous studies on their
diagnostic ability (21,22,33,34),
and the roles of each studied miRNA in CRC, together with previous
findings on clinical significance of their differential expression
in different populations in comparison to those presented in this
study are discussed.
Our bioinformatic analysis identified the AGE-RAGE
signaling pathway as one of the putative jointly affected pathways
by all five of the investigated miRNAs. AGEs are predominantly
formed as a result of chronic hyperglycemic conditions/diabetes or
aging (35). The binding of AGEs
and activation of their receptor RAGE triggers upregulation of ROS
production, and activation of several signaling cascades via
phosphatidylinositol-3 kinase (PI3K), MAPK and K-Ras to activate
NF-κB (36), ultimately leading to
the exacerbation of inflammation and cellular damage (32).
miR-146a is one of the most prominent inflammatory
miRNAs, with a key modulatory role both in the innate and adaptive
immune response (30). Notably,
miR-146a was found to be a NF-κB-dependent gene, whose direct
molecular targets are tumor necrosis factor receptor-associated
factor 6 (TRAF6), and interleukin 1 receptor associated kinase
(IRAK1), the two main adapter proteins downstream of Toll-like
receptors (TLR) and cytokine receptors (37), crucial for pro-inflammatory
signaling. In this model, miR-146a was proposed to act as a
negative feedback regulator of the innate immune response. High
miR-146a levels found both in tissue and plasma samples of our
cohort of patients with CRC are in line with the role of miR-146a
as a suppressor of the immune response. Furthermore, miR-146a is a
NF-κB-dependent gene, and NF-κB activation is triggered by the
AGE-RAGE signaling pathway, which is one of the jointly regulated
pathways of the five miRNAs investigated in this study. The
anti-inflammatory and anti-tumorigenic roles of miR-146a in CRC
were also demonstrated to occur via modulation of IL-17 signaling
in vivo (38). miR-146a was
shown to have both a tumor suppressive (39), and oncogenic role (40). Finally, miR-146a polymorphisms were
shown to be associated with susceptibility to CRC (41). Conflicting findings on the role of
miR-146a in not only CRC, but all malignancies, warrants the need
for further investigation on its exact function, correlation to
clinical parameters and therapeutic potential.
Serum miR-146a was shown to have a significant
diagnostic ability in CRC, as a member of a three-miRNA panel,
together with miR-30e-3p, and miR-148a-3p (22). A comprehensive meta-analysis
regarding the prognostic utility of its expression levels was
performed by Li et al, who showed that higher miR-146a
expression correlated with better survival in solid cancers,
especially of the digestive system (42). Our finding of elevated miR-146a
expression both in tissue and plasma samples of CRC patients is in
agreement with these previous findings. Population-specific genetic
variation of miRNA expression and/or function (23) might have also influenced miR-146a
levels in the Montenegrin population and should be evaluated. We
will continue to monitor our study group over time, to evaluate
whether miR-146a plays a role in delaying tumor progression and
prolonging overall survival.
miR-29a expression was found to be up-regulated in
FFPE tissue samples of the patients with CRC (Fig. 3A), which is in accordance with
several studies reporting significantly increased miR-29a
expression levels in CRC malignant tissue, plasma, and serum
samples (21,43). miR-29a was found to promote both
inflammation and CRC development (44), but also, contrary to these findings,
to inhibit in vivo tumor growth (45). A recent review by Mo and Cao
highlights this discrepancy regarding the role of miR-29a in CRC
and gives an excellent overview on tissue, circulatory, fecal, and
salivary miR-29a as a biomarker for CRC, as well as on the
underlying molecular mechanisms of miR-29a, and its role in
chemoradiotherapy resistance (17).
Taken together, the clinical significance of miR-29a expression
levels is still debatable. Conflicting findings could be attributed
to the inadequate sample size or analytical procedures used
(17). Therefore, more research is
needed in order to clarify its exact role, underlying mechanisms,
and clinical utility.
We found significantly lower miR-101 expression in
FFPE tissue samples of patients with CRC, compared to the normal
surrounding colonic mucosa (Fig.
3E), which is in agreement with Chandramouli et al
(33). Intricate gene networks
regulated by miR-101 were shown to be involved in proliferation,
apoptosis, angiogenesis, invasion, metastasis, and drug resistance
(18). Serum levels of miR-101 were
shown to have prognostic significance - low miR-101 levels
correlated with advanced cancer stage and a worse overall survival
(OS) (46). Also, miR-101
up-regulation is synergistic with chemotherapeutic drugs and
promotes their inhibitory effect (47). In our study, lower levels of miR-101
expression were found in patients with history of smoking, compared
to those who do not smoke (Table
SI), which is in agreement with the study by Gong et al
(48). This study showed that low
doses of cigarette smoke extract induced miR-101 down-regulation in
esophageal epithelial and cancer cells (48), targeting cyclooxygenase-2
(COX-2), and promoted cancer cell proliferation.
miR-125b was found to be upregulated in both CRC
tumors and metastatic sites, compared to adjacent normal tissues,
and miR-125b overexpression enhanced CRC cells migration and
invasion (49). Also, miR-125b was
found to mediate CRC cancer invasion and resistance to 5-FU through
enhancing cell autophagy (50).
miR-125b expression, together with miR-99a and Let-7c expression
correlated with progression-free survival (PFS) and OS of
metastatic patients with CRC and could potentially have a role in
sensitivity to anti-EGFR monoclonal antibodies therapy (51). miR-125b was identified as one of the
hub miRNAs related to CRC prognosis (52), and as one of the combination
biomarkers, together with miR-21, miR-29a, and miR-92a showed a
good diagnostic accuracy (53). In
the present study, miR-125b was upregulated in the CRC tumor
tissues with respect to the surrounding healthy tissue, in
agreement with (54). High miR-125b
expression was shown to correlate with advanced CRC tumor size,
tumor invasion, and poor prognosis (55). Other studies reported lower miR-125b
levels in CRC tissues, possibly due to different ethnic origin of
the investigated populations (56,57),
and its tumor suppressor functions (58). The dual role of miR-125b in
malignancies could at least partially be explained by its
dependence on the TP53 mutational status. Namely, miR-125b
had anti-cancer effect only in TP53 mutated colon cancer
cells, and not in WT cells (59).
miR-125b undoubtedly plays an important role in CRC tumorigenesis
and further investigations on its precise role will help to
establish its clinical utility.
miR-155 was found to be over-expressed in CRC
(60), and to have strong
tumorigenic role by promoting CRC cells proliferation, invasion,
and metastasis (61). miR-155 is
also an important regulator of homeostasis and function of the
immune system (62). miR-155 was
shown to be involved in mismatch repair and genomic stability in
CRC (63), and interestingly
enough, in promoting the Warburg phenotype by increasing glucose
consumption, lactate production, and HIF-1α levels in human CRC
cell lines (64). However, there
are also opposing findings showing an oncosuppressive role of
miR-155; for example, miR-155 expression was decreased in CRC
tissue and cell lines, and inhibited CRC progression and metastasis
via silencing collagen triple helix repeat containing 1 (65). Regarding its clinical significance,
miR-155 expression was shown to have a potential predictive value
for survival of stage II patients with CRC, together with other two
lncRNAs and miR-200a (66).
Furthermore, an increase in both circulating, and/or tissue miR-155
levels is associated with shorter PFS and OS (67). In addition, re-elevation or
sustained elevation of serum miR-155 in postoperative CRC patients
treated with chemotherapy is a sign of chemoresistance, and
together with miR-200c, and miR-210 levels, a sign of poor
prognosis (68). In our study,
miR-155 was shown to be upregulated in CRC tissue samples, compared
to the adjacent normal tissue (Fig.
3D), which is in agreement with other reports (34,69).
Serum miR-155 levels were also higher in patients with CRC compared
to the healthy controls, and were independent prognostic factor for
PFS, and OS (70). We also observed
higher miR-155 expression in CRC tissue samples without the
presence of mucus, but failed to find a correlation with TNM
staging, invasion, metastasis, and differentiation as in (34,71).
A major limitation of this study that needs to be
considered when interpreting its results is a small sample size,
thus a follow-up study on a larger prospective cohort of patients
is warranted. In general, many potential miRNA biomarkers are still
not utilized in the clinical setting, partially due to inconsistent
findings. The discovery and validation of a novel miRNA signature
in a specific population could potentially aid in the timely
diagnosis of patients at risk. Since the development and
progression of CRC is a multi-step process which involves mutations
in many genes, future combination biomarkers will better reflect
the complexity and heterogeneity of CRC. Further analysis of miRNAs
in various CRC stages, and the various targets of selected miRNAs
is necessary. Circulating miRNAs could potentially be a novel,
noninvasive CRC screening tool, which could be utilized along with
fecal occult blood testing and colonoscopy. Analysis of specific
miRNAs identified in this study, in combination with other CRC
diagnostic biomarkers, such as carcinoembryonic antigen, and
carbohydrate antigen 19-9 could further increase their sensitivity
and specificity.
In conclusion, our study identified statistically
significant up-regulation of circulating miR-146a in patients with
CRC in the Montenegrin population for the first time. miR-146a was
shown to have the potential to be used as a diagnostic biomarker
for CRC. This was further corroborated by the finding that its
expression was altered in both FFPE tissues and in plasma of
patients with CRC. We have also shown for the first time that the
distinct five-miRNA (miR-29a, miR-101, miR-125b, miR-146a, and
miR-155) pattern of expression is highly involved in CRC
carcinogenesis, contributing to the current understanding of which
miRNA signature can be potentially used in a clinical setting.
Bioinformatics analysis identified the AGE-RAGE signaling axis as a
common potentially deregulated pathway. Better understanding of the
interaction between the oncogenic effect of the AGE-RAGE axis and
the inflammatory tumor microenvironment, and their regulation,
could lead to the development of new cancer prevention and
treatment methods.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Katarina Popović
(University of Montenegro, Faculty of Medicine) for English
language editing.
Funding
This research was funded by the Ministry of Science, Montenegro
(grant no. 01-781).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MŽ, NP, JR, LV, IRD, AT, NG, and SG conceived the
study. MŽ, JR, MR, FM, SG, and AT designed the methodology. MŽ, NP,
BV, and IRD recruited patients. MŽ, JR, BV, VT, FV and LV performed
formal analysis and data curation. MŽ prepared the original draft
and data visualization. All authors reviewed and edited the
manuscript. MR, VT, and FV performed project administration. MŽ and
JR confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and was approved by the Ethical
Committee of the Clinical Center of Montenegro (approval no.
03/01-11417/1 on 24.06.2019.) and by the Committee for Medical
Ethics and Bioethics of the Faculty of Medicine of the University
of Montenegro (approval no. 3824/4 on 13.12.2018.). Written
informed consent was obtained from all study subjects.
Patient consent for publication
Written informed consent has been obtained from all
the study subjects to publish this paper.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kocarnik JM, Shiovitz S and Phipps AI:
Molecular phenotypes of colorectal cancer and potential clinical
applications. Gastroenterol Rep (Oxf). 3:269–276. 2015.PubMed/NCBI
|
|
4
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sawayama H, Miyamoto Y, Ogawa K, Yoshida N
and Baba H: Investigation of colorectal cancer in accordance with
consensus molecular subtype classification. Ann Gastroenterol Surg.
4:528–539. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong
QH, Wang L and Wang JP: The prognostic role of BRAF mutation in
metastatic colorectal cancer receiving anti-EGFR monoclonal
antibodies: A meta-analysis. PLoS One. 8:e659952013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Afrăsânie VA, Marinca MV, Alexa-Stratulat
T, Gafton B, Păduraru M, Adavidoaiei AM, Miron L and Rusu C: KRAS,
NRAS, BRAF, HER2 and microsatellite instability in metastatic
colorectal cancer-practical implications for the clinician. Radiol
Oncol. 53:265–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong J, Cho M, Sy M, Salgia R and Fakih M:
Molecular profiling of metastatic colorectal tumors using
next-generation sequencing: A single-institution experience.
Oncotarget. 8:42198–42213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ullah S, John P and Bhatti A: MicroRNAs
with a role in gene regulation and in human diseases. Mol Biol Rep.
41:225–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bagga S, Bracht J, Hunter S, Massirer K,
Holtz J, Eachus R and Pasquinelli AE: Regulation by let-7 and lin-4
miRNAs results in target mRNA degradation. Cell. 122:553–563. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Sun C, Zhao Y, Wang Q, Guo J, Ye
B and Yu G: Overview of MicroRNAs as diagnostic and prognostic
biomarkers for high-incidence cancers in 2021. Int J Mol Sci.
23:113892022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Backes C, Meese E and Keller A: Specific
miRNA disease biomarkers in blood, serum and plasma: Challenges and
prospects. Mol Diagn Ther. 20:509–518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jorgensen BG and Ro S: MicroRNAs and
‘sponging’ competitive endogenous RNAs dysregulated in colorectal
cancer: Potential as noninvasive biomarkers and therapeutic
targets. Int J Mol Sci. 23:21662022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mo WY and Cao SQ: MiR-29a-3p: A potential
biomarker and therapeutic target in colorectal cancer. Clin Transl
Oncol. 25:563–577. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang CZ, Deng F, Li H, Wang DD, Zhang W,
Ding L and Tang JH: MiR-101: A potential therapeutic target of
cancers. Am J Transl Res. 10:3310–3321. 2018.PubMed/NCBI
|
|
19
|
Yin H, Sun Y, Wang X, Park J, Zhang Y, Li
M, Yin J, Liu Q and Wei M: Progress on the relationship between
miR-125 family and tumorigenesis. Exp Cell Res. 339:252–260. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang D, Wang X, Song Y, Si M, Sun Y, Liu
X, Cui S, Qu X and Yu X: Exosomal miR-146a-5p and miR-155-5p
promote CXCL12/CXCR7-induced metastasis of colorectal cancer by
crosstalk with cancer-associated fibroblasts. Cell Death Dis.
13:3802022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamada A, Horimatsu T, Okugawa Y, Nishida
N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, et al: Serum
miR-21, miR-29a, and miR-125b Are promising biomarkers for the
early detection of colorectal neoplasia. Clin Cancer Res.
21:4234–4242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng X, Wang J, Zhang C, Liu K, Zhao L,
Chen X, Huang G and Lai Y: A three-miRNA panel in serum as a
noninvasive biomarker for colorectal cancer detection. Int J Biol
Markers. 35:74–82. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rawlings-Goss RA, Campbell MC and Tishkoff
SA: Global population-specific variation in miRNA associated with
cancer risk and clinical biomarkers. BMC Med Genomics. 7:532014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Panico A, Tumolo MR, Leo CG, Donno A,
Grassi T, Bagordo F, Serio F, Idolo A, Masi R, Mincarone P and
Sabina S: The influence of lifestyle factors on miRNA expression
and signal pathways: A review. Epigenomics. 13:145–164. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rovčanin Dragović I, Popović N, Ždralević
M, Radulović L, Vuković T, Marzano F, Tullo A and Radunović M:
Inflammation-related microRNAs-146a and −155 are upregulated in
mild cognitive impairment subjects among older age population in
montenegro. J Alzheimers Dis. 90:625–638. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang HY, Lin YCD, Cui S, Huang Y, Tang Y,
Xu J, Bao J, Li Y, Wen J, Zuo H, et al: miRTarBase update 2022: An
informative resource for experimentally validated miRNA-target
interactions. Nucleic Acids Res. 50(D1): D222–D230. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang H, Li X, Li T, Wang L, Wu X, Liu J,
Xu Y and Wei W: Multiple roles of microRNA-146a in immune responses
and hepatocellular carcinoma (review). Oncol Lett. 18:5033–5042.
2019.PubMed/NCBI
|
|
31
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49(D1): D605–D612. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
El-Far AH, Sroga G, Jaouni SKA and Mousa
SA: Role and mechanisms of RAGE-ligand complexes and
RAGE-inhibitors in cancer progression. Int J Mol Sci. 21:36132020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chandramouli A, Onyeagucha BC,
Mercado-Pimentel ME, Stankova L, Shahin NA, LaFleur BJ, Heimark RL,
Bhattacharyya AK and Nelson MA: MicroRNA-101 (miR-101)
post-transcriptionally regulates the expression of EP4 receptor in
colon cancers. Cancer Biol Ther. 13:175–183. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao H, Huang S, Liu A and Chen Z:
Up-regulated expression of miR-155 in human colonic cancer. J
Cancer Res Ther. 14:604–607. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Waghela BN, Vaidya FU, Ranjan K, Chhipa
AS, Tiwari BS and Pathak C: AGE-RAGE synergy influences programmed
cell death signaling to promote cancer. Mol Cell Biochem.
476:585–598. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Snelson M, Lucut E and Coughlan MT: The
role of AGE-RAGE signalling as a modulator of gut permeability in
diabetes. Int J Mol Sci. 23:17662022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Tan S, Shen Y and Guo L: miR-146a-5p
negatively regulates the IL-1β-stimulated inflammatory response via
downregulation of the IRAK1/TRAF6 signaling pathway in human
intestinal epithelial cells. Exp Ther Med. 24:6152022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Garo LP, Ajay AK, Fujiwara M, Gabriely G,
Raheja R, Kuhn C, Kenyon B, Skillin N, Kadowaki-Saga R, Saxena S
and Murugaiyan G: MicroRNA-146a limits tumorigenic inflammation in
colorectal cancer. Nat Commun. 12:24192021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sathyanarayanan A, Chandrasekaran KS and
Karunagaran D: microRNA-146a inhibits proliferation, migration and
invasion of human cervical and colorectal cancer cells. Biochem
Biophys Res Commun. 480:528–533. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lu D, Yao Q, Zhan C, Le-Meng Z, Liu H, Cai
Y, Tu C, Li X, Zou Y and Zhang S: MicroRNA-146a promote cell
migration and invasion in human colorectal cancer via
carboxypeptidase M/src-FAK pathway. Oncotarget. 8:22674–22684.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ,
Park JY, Jeon HS, Park JS and Choi GS: A miR-146a polymorphism
(rs2910164) predicts risk of and survival from colorectal cancer.
Anticancer Res. 33:3233–3239. 2013.PubMed/NCBI
|
|
42
|
Li MW, Gao L, Dang YW, Li P, Li ZY, Chen G
and Luo DZ: Protective potential of miR-146a-5p and its underlying
molecular mechanism in diverse cancers: A comprehensive
meta-analysis and bioinformatics analysis. Cancer Cell Int.
19:1672019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fellizar A, Refuerzo V, Ramos JD and
Albano PM: Expression of specific microRNAs in tissue and plasma in
colorectal cancer. J Pathol Transl Med. May 3–2022.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang A, Deng S, Chen X, Yu C, Du Q, Wu Y,
Chen G, Hu L, Hu C and Li Y: miR-29a-5p/STAT3 positive feedback
loop regulates TETs in colitis-associated colorectal cancer.
Inflamm Bowel Dis. 26:524–533. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang J, Chen X, Xie C, Sun M, Hu C, Zhang
Z, Luan L, Zhou J, Zhou J, Zhu X, et al: MicroRNA miR-29a inhibits
colon cancer progression by downregulating B7-H3 expression:
Potential molecular targets for colon cancer therapy. Mol
Biotechnol. 63:849–861. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He D, Yue Z, Li G, Chen L, Feng H and Sun
J: Low serum levels of miR-101 are associated with poor prognosis
of colorectal cancer patients after curative resection. Med Sci
Monit. 24:7475–7481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen LG, Xia YJ and Cui Y: Upregulation of
miR-101 enhances the cytotoxic effect of anticancer drugs through
inhibition of colon cancer cell proliferation. Oncol Rep.
38:100–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gong J, Chu Y, Xu M, Huo J and Lv L:
Esophageal squamous cell carcinoma cell proliferation induced by
exposure to low concentration of cigarette smoke extract is
mediated via targeting miR-101-3p/COX-2 pathway. Oncol Rep.
35:463–471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang X, Li T, Han YN, Ge M, Wang P, Sun
L, Liu H, Cao T, Nie Y, Fan D, et al: miR-125b promotes colorectal
cancer migration and invasion by dual-targeting CFTR and CGN.
Cancers (Basel). 13:57102021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu X, Shi W, Zhang Y, Wang X, Sun S, Song
Z, Liu M, Zeng Q, Cui S and Qu X: CXCL12/CXCR4 axis induced
miR-125b promotes invasion and confers 5-fluorouracil resistance
through enhancing autophagy in colorectal cancer. Sci Rep.
7:422262017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cappuzzo F, Sacconi A, Landi L, Ludovini
V, Biagioni F, D'Incecco A, Capodanno A, Salvini J, Corgna E,
Cupini S, et al: MicroRNA signature in metastatic colorectal cancer
patients treated with anti-EGFR monoclonal antibodies. Clin
Colorectal Cancer. 13:37–45.e4. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou XG, Huang XL, Liang SY, Tang SM, Wu
SK, Huang TT, Mo ZN and Wang QY: Identifying miRNA and gene modules
of colon cancer associated with pathological stage by weighted gene
co-expression network analysis. Onco Targets Ther. 11:2815–2830.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu HN, Liu TT, Wu H, Chen YJ, Tseng YJ,
Yao C, Weng SQ, Dong L and Shen XZ: Serum microRNA signatures and
metabolomics have high diagnostic value in colorectal cancer using
two novel methods. Cancer Sci. 109:1185–1194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yagi T, Iinuma H, Hayama T, Matsuda K,
Nozawa K, Tsukamoto M, Shimada R, Akahane T, Tsuchiya T, Ozawa T
and Hashiguchi Y: Plasma exosomal microRNA-125b as a monitoring
biomarker of resistance to mFOLFOX6-based chemotherapy in advanced
and recurrent colorectal cancer patients. Mol Clin Oncol.
11:416–424. 2019.PubMed/NCBI
|
|
55
|
Nishida N, Yokobori T, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M: MicroRNA
miR-125b is a prognostic marker in human colorectal cancer. Int J
Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
56
|
Ak S, Tunca B, Tezcan G, Cecener G, Egeli
U, Yilmazlar T, Ozturk E and Yerci O: MicroRNA expression patterns
of tumors in early-onset colorectal cancer patients. J Surg Res.
191:113–122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zeng ZL, Lu JH, Wang Y, Sheng H, Wang YN,
Chen ZH, Wu QN, Zheng JB, Chen YX, Yang DD, et al: The lncRNA
XIST/miR-125b-2-3p axis modulates cell proliferation and
chemotherapeutic sensitivity via targeting Wee1 in colorectal
cancer. Cancer Med. 10:2423–2441. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu M, Yi Z, Chen S, Chen X and Xie X:
MIR4435-2HG, miR-125b-5p, and Sema4D axis affects the
aggressiveness of colorectal cancer cells. Folia Histochem
Cytobiol. 60:191–202. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cenariu D, Zimta AA, Munteanu R, Onaciu A,
Moldovan CS, Jurj A, Raduly L, Moldovan A, Florea A, Budisan L, et
al: Hsa-miR-125b therapeutic role in colon cancer is dependent on
the mutation status of the TP53 gene. Pharmaceutics. 13:6642021.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qu YL, Wang HF, Sun ZQ, Tang Y, Han XN, Yu
XB and Liu K: Up-regulated miR-155-5p promotes cell proliferation,
invasion and metastasis in colorectal carcinoma. Int J Clin Exp
Pathol. 8:6988–6994. 2015.PubMed/NCBI
|
|
62
|
Rodriguez A, Vigorito E, Clare S, Warren
MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska
EA, et al: Requirement of bic/microRNA-155 for normal immune
function. Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Valeri N, Gasparini P, Fabbri M, Braconi
C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, et
al: Modulation of mismatch repair and genomic stability by miR-155.
Proc Natl Acad Sci USA. 107:6982–6987. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xu Y, Han S, Lei K, Chang X, Wang K, Li Z
and Liu J: Anti-Warburg effect of rosmarinic acid via miR-155 in
colorectal carcinoma cells. Eur J Cancer Prev. 25:481–489. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu J, Chen Z, Xiang J and Gu X:
MicroRNA-155 acts as a tumor suppressor in colorectal cancer by
targeting CTHRC1 in vitro. Oncol Lett. 15:5561–5568.
2018.PubMed/NCBI
|
|
66
|
Wang XJ, Zeng B, Lin S, Chen M and Chi P:
An integrated miRNA-lncRNA signature predicts the survival of stage
II colon cancer. Ann Clin Lab Sci. 49:730–739. 2019.PubMed/NCBI
|
|
67
|
Ulivi P, Canale M, Passardi A, Marisi G,
Valgiusti M, Frassineti GL, Calistri D, Amadori D and Scarpi E:
Circulating plasma levels of miR-20b, miR-29b and miR-155 as
predictors of bevacizumab efficacy in patients with metastatic
colorectal cancer. Int J Mol Sci. 19:3072018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen J, Wang W, Zhang Y, Chen Y and Hu T:
Predicting distant metastasis and chemoresistance using plasma
miRNAs. Med Oncol. 31:7992014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hongliang C, Shaojun H, Aihua L and Hua J:
Correlation between expression of miR-155 in colon cancer and serum
carcinoembryonic antigen level and its contribution to recurrence
and metastasis forecast. Saudi Med J. 35:547–553. 2014.PubMed/NCBI
|
|
70
|
Lv ZC, Fan YS, Chen HB and Zhao DW:
Investigation of microRNA-155 as a serum diagnostic and prognostic
biomarker for colorectal cancer. Tumor Biol. 36:1619–1625. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS
and Zhou T: Upregulation of microRNA-155 promotes the migration and
invasion of colorectal cancer cells through the regulation of
claudin-1 expression. Int J Mol Med. 31:1375–1380. 2013. View Article : Google Scholar : PubMed/NCBI
|