Introduction
The latest genome-wide transcriptome analyses
revealed that ~2/3 of the genomic DNA of mammalian species is
transcribed but >98% of the transcripts lack protein-coding
capacity (1), indicating that most
mammalian genes are non-protein coding genes. Long non-coding RNAs
(lncRNAs) are a class of transcripts with pivotal biological
functions in many processes (2),
such as cell growth, embryonic development, and disease progression
by regulating downstream genes at post-transcriptional and
translational levels (3,4). Extensive studies have shown that
lncRNAs play critical roles in cancer biology (5), and regulation of lncRNAs has shown
potentials in inhibiting cancer development (6). However, functions of most lncRNAs are
still unknown.
Although the mortality rate of breast cancer has
dropped significantly during the past decades, it still accounts
for >1/5 deaths among females (7). As a major subtype of breast cancer,
triple-negative breast cancer (TNBC) is characterized by inhibited
expression of estrogen receptor α, progesterone receptor, and human
epidermal growth factor receptor 2 (8). The development and progression of TNBC
involve the regulation of multiple lncRNAs (9,10).
lncRNA PTCSC3 is characterized as a tumor suppressor in glioma and
thyroid cancer, while its role in the recurrence of TNBC is unknown
(11–13). It was reported that lncRNA MIR100HG
promotes cell viability in TNBC (14). The interaction between different
lncRNAs play important roles in tumor growth. For example, lncRNA
PTCSC3 alleviates gastric cancer by regulating lncRNA HOXA11-AS
(15). The present study
investigated the involvement of PTCSC3 in TNBC and explored its
potential interaction with MIR100HG. lncRNA PTCSC3 may serve as a
target for the treatment and prognosis of gastric cancer.
Materials and methods
Research subjects
A total of 82 patients diagnosed with TNBC admitted
at Changle People's Hospital between January 2010 and January 2013
were enrolled in the present study (Table I). The inclusion criteria included:
i) Patients diagnosed by pathological biopsy; ii) patients
diagnosed and treated for the first time; iii) patients who signed
the informed consent. The exclusion criteria were as follows: i)
Patients treated before admission; ii) patients diagnosed with
multiple diseases, such as cardiovascular diseases, diabetes and
gastroenteritis. The age range of the 82 patients with TNBC was
between 30 and 68 years old, and the mean age was 47.7±5.0 years
old. Based on AJCC staging, there were 25 cases at stage I, 24
cases at stage II, 19 cases at stage III and 14 cases at stage IV.
The present study was approved by the Ethics Committee of Changle
People's Hospital before the admission of patients. The ethics
committee approval number is Changle-20160049.
 | Table I.Clinical characteristics of patients
with triple-negative breast cancer. |
Table I.
Clinical characteristics of patients
with triple-negative breast cancer.
| Characteristics | Number | Percentage |
|---|
| Age, years |
|
|
|
≤50 | 45 | 54.9 |
|
≥50 | 37 | 45.1 |
| Stage |
|
|
| I | 25 | 30.5 |
| II | 24 | 29.3 |
|
III | 19 | 23.2 |
| IV | 14 | 17.0 |
| Patients not
treated | 73 | 89.0 |
| Patients treated
for the first time (arubicin + paclitaxel) | 9 | 11.0 |
Specimens and cell lines
Tumor tissues and adjacent non-cancerous tissues (3
cm away from the tumor tissue) were obtained from each patient
through biopsy. All tissue specimens were confirmed by 3
experienced pathologists. Specimens were stored in liquid nitrogen
before use.
BT-549 and HCC70 cell lines purchased from American
Type Culture Collection were used for all in vitro cell
experiments in the present study. Cells were cultivated in Roswell
Park Memorial Institute (RPMI)-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 0.023 IU/ml insulin at 37°C with 5%
CO2.
Follow-up
All patients were followed-up for 5 years from the
day of admission to record their overall survival conditions.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Tissue specimens were ground in liquid nitrogen.
RNAzol reagent (Sigma-Aldrich; Merck KGaA) was used to extract
total RNAs from tissue specimens and in vitro cultivated
cells. SuperScript IV Reverse Transcriptase (Thermo Fisher
Scientific, Inc.) was used to synthesize cDNA through reverse
transcription (gentle mixing and incubation at 37°C for 2 min,
followed by incubation of the sample for 15 min at 55°C in the
presence of 10 mM dithiothreitol to inactivate the enzyme). To
detect the expression levels of PTCSC3 and MIR100HG,
SYBR® Green Quantitative RT-qPCR (Sigma-Aldrich; Merck
KGaA) was used to prepare all PCR reactions. The PCR reaction
conditions were: 55 sec at 95°C, followed by 12 sec at 95°C and 32
sec at 58.5°C for 40 cycles. PCRs were performed on an ABI 7500
System. 18S RNA was used as the endogenous control. Primers of
PTCSC3 and MIR100HG as well as 18S RNA were designed and
synthesized by Sangon Biotech Co., Ltd. The expression levels of
PTCSC3 and MIR100HG were normalized to 18S RNA using
2−ΔΔCq method (16). The
primer sequences were listed as follows: lncRNA PTCSC3 forward,
5′-CCAGGGGGATCGCATTTTTC-3′; lncRNA PTCSC3 reverse,
5′-CTTCTGCTTGGCCTTTGACC-3′; lncRNA MIR100HG forward,
5′-TCGAACTTTGGAGTGTGGCA-3′; lncRNA MIR100HG reverse,
5′-TCCCTGGTTACTGCCCAGAT-3′; 18S RNA forward,
5′-CGGCTACCACATCCAAGGAA-3′; 18S RNA reverse,
5′-TGTCACTACCTCCCCGTGTCA-3′.
Vectors and cell transfection
The vector expressing PTCSC3 or MIR100HG were
designed using pcDNA3.1 vector as backbone that was synthesized by
Sangon Biotech Co., Ltd. BT-549 and HCC70 cells were seeded in
6-well plates (1×105 cells/well) and transfected with 10
nM vector using Lipofectamine® 2000 reagent (cat. no.
11668-019; Invitrogen; Thermo Fisher Scientific, Inc.). Cells
transfected with the empty vectors were used as negative control
cells. Non-transfected cells treated with Lipofectamine®
2000 reagent only were used as the control cells. Cells were
harvested at 24 h after transfection for subsequent
experiments.
Western blotting
Total protein was isolated from cells using RIPA
buffer reagent (Thermo Fisher Scientific, Inc.) containing protease
inhibitor (Cocktail, Roche Diagnostics). The protein concentration
was measured using a BCA assay kit (Thermo Fisher Scientific,
Inc.). The same amount of protein samples (30 µg) were separated by
10% SDS-PAGE (Shanghai Enzyme Link Biotechnology Co., Ltd.) and
transferred onto nitrocellulose membranes (GE Healthcare). Blocking
was performed using 5% BSA at room temperature for 2 h. GAPDH was
selected as the housekeeping protein for general western blotting.
The protein-membrane was incubated with the primary antibodies of
anti-phosphorylated (p)-LATS1 (1:1,000; cat. no. ab111344; Abcam),
anti-LATS1 (1:1,000; cat. no. ab243656; Abcam), anti-p-YAP
(1:1,000; cat. no. ab76252; Abcam), anti-Yes-associated protein
(YAP; 1:1,000; cat. no. ab52771; Abcam) and anti-GAPDH (1:1,000;
cat. no. ab8245; Abcam) at 4°C overnight. For the detection of
nuclear YAP, laminB was selected as the internal control, and the
membranes were also incubated with anti-nuclear YAP antibody
(1:1,000; cat. no. ab52771; Abcam) and anti-laminB (1:1,000; cat.
no. ab16048; Abcam) at 4°C overnight. TBST buffer was used to elute
the membrane twice. The membranes were then incubated with the
secondary antibody [Goat Anti-Rabbit IgG H&L (HRP) preadsorbed;
cat. no. ab7090; dilution, 1:5,000; Abcam; or Goat Anti-Mouse
IgG+IgM H&L (HRP) preadsorbed; cat. no. ab47827; dilution,
1:5,000; Abcam] at 37°C for 2 h. The ECL detection system (Thermo
Fisher Scientific, Inc.) was used to detect the protein bands.
ImageJ software v.1.51j8 (National Institutes of Health) was used
for densitometry analyses.
In vitro cell viability assay
In vitro cell viability assay was carried out
using Cell Counting Kit-8 kit (CCK-8; cat. no. ab228554, Abcam) to
detect cell viability at 24 h post transfection. Briefly, single
cell suspensions were prepared using RPMI-1640 medium and cell
density was adjusted to 5×104 cells/ml. Cells were
cultivated in a 96-well plate with 100 µl cell suspension per well
at 37°C with 5% CO2. CCK-8 solution (10 µl) was added at
24, 48, 72 and 96 h after the beginning of cell culture.
Subsequently, cells were cultivated for an additional 4 h and 10 µl
dimethyl sulfoxide (DMSO) was added. OD values at 450 nm were used
to reflect cell viability abilities.
Cell invasion and migration
assays
BT-549 and HCC70 cells were seeded onto the upper
chamber of Transwell inserts (8 µm pore; Corning, Inc.) with 3,000
cells in 0.1 ml non-serum medium per well. The lower chamber of the
Transwell was filled with medium supplemented with 20% FBS.
Matrigel-coated membrane was used in invasion assay. Cells were
cultured at 37°C with 5% CO2 for 12 h. Subsequently, the
upper surface of membrane was cleaned, and 0.5% crystal violet
(Sigma-Aldrich; Merck KGaA) was used to stain the cells in the
lower surface at 25°C for 30 min. The stained cells were observed
under an optical microscope (magnification, ×100; Olympus DX51;
Olympus Corporation). All cell groups were normalized to the
control (C) group.
Cell apoptosis assay
Cell apoptosis was determined by flow cytometry
using Annexin V-FITC Apoptosis Detection kit (BD Biosciences).
Briefly, BT-549 and HCC70 cells were seeded in 6-well plates
(1×105 cells/well) and incubated for 24 h at 37°C with
5% CO2. Cells were collected and washed by PBS. Then,
cells were re-suspended in 200 µl binding buffer containing 5 µl
Annexin V-FITC and 10 µl propidium iodide (PI) and further
incubated for 30 min at room temperature. Cell apoptosis was
detected by a flow cytometer (BD Bioscience).
BrdU staining
BrdU staining was used to monitor cell viability.
Briefly, BT-549 and HCC70 cells were inoculated into 24-well plates
(10,000 cells/well) and cultured at 37°C overnight. Following 48 h,
10 µg/ml BrdU was added to the medium and incubated for 2 h at room
temperature, followed by cell fixation using 4% paraformaldehyde
for 15 min at room temperature. Following treatment with 2 M HCL
and 0.3% Triton X100, cells were blocked with 10% goat serum
(ZSGB-Bio). Cells were then incubated with BrdU primary antibody
(cat. no. A1482; mouse anti-BrdU primary antibody; dilution, 1:50;
lot. 347580; BD Biosciences) and then secondary antibody
(FITC-conjugated rabbit anti-mouse secondary antibody; dilution,
1:25; cat. no. FI-2000; Vector Laboratories, Inc.). Cells were then
stained with DAPI for 30 min at room temperature. BrdU-positive
cells were counted in three random areas under a Leica confocal
microscope (TCS SP8; Leica Microsystems, Inc.).
Immunofluorescence staining
BT-549 and HCC70 cells were fixed in 4%
paraformaldehyde for 15 min at room temperature and then
permeabilized in 0.1% Triton X-100 at room temperature for 20 min.
Cardiomyocytes were then blocked in PBS containing 0.5% bovine
serum albumin at 37°C for 60 min and incubated with primary rabbit
antibody against YAP (cat. no. ab52771; 1:200, Abcam) at 4°C
overnight, followed by incubation with Alexa Fluor 488-conjugated
anti-rabbit IgG secondary antibody (dilution, 1:3,000; cat. no.
A-11034; Invitrogen; Thermo Fisher Scientific, Inc.). Finally, the
nuclei were stained with DAPI for 30 min at room temperature.
Images were visualized and captured with a Leica SP8 confocal
microscopy (magnification, ×200; Leica Microsystems, Inc.).
Nuclear/cytoplasmic fractionation
For nuclear YAP accumulation assay, BT-549 and HCC70
cells were harvested and lysed to obtain cytoplasmic and nuclear
lysates using the Keygen Nuclear-Cytosol Protein Extraction kit
(Nanjing KeyGen Biotech. Co., Ltd.). Western blotting was performed
to detect the cytoplasmic and nuclear lysates.
Statistical analyses
All experiments were repeated for 3 times and data
of 3 replicates were used to calculate the mean ± standard
deviation (SD). Statistical analyses were performed using GraphPad
Prism 6 software (GraphPad Software Inc.). Comparison of the
expression levels of PTCSC3 and MIR100HG between tumor and
non-cancerous tissues was performed by paired Student's t-test.
Comparison of the expression levels of PTCSC3 and MIR100HG in cells
with different treatments, as well as cell viability, apoptosis and
migration data, were performed by one-way ANOVA and Tukey's test.
Correlations between the expression levels of PTCSC3 and MIR100HG
were analyzed by Pearson's correlation coefficient. According to
Youden's index, patients were divided into high (n=38) and low
(n=44) PTCSC3 groups, as well as high (n=36) and low (n=46)
MIR100HG level groups. Kaplan-Meier method was used to plot
survival curves, which were compared by log-rank test. P<0.05 as
considered to indicate a statistically significant difference.
Results
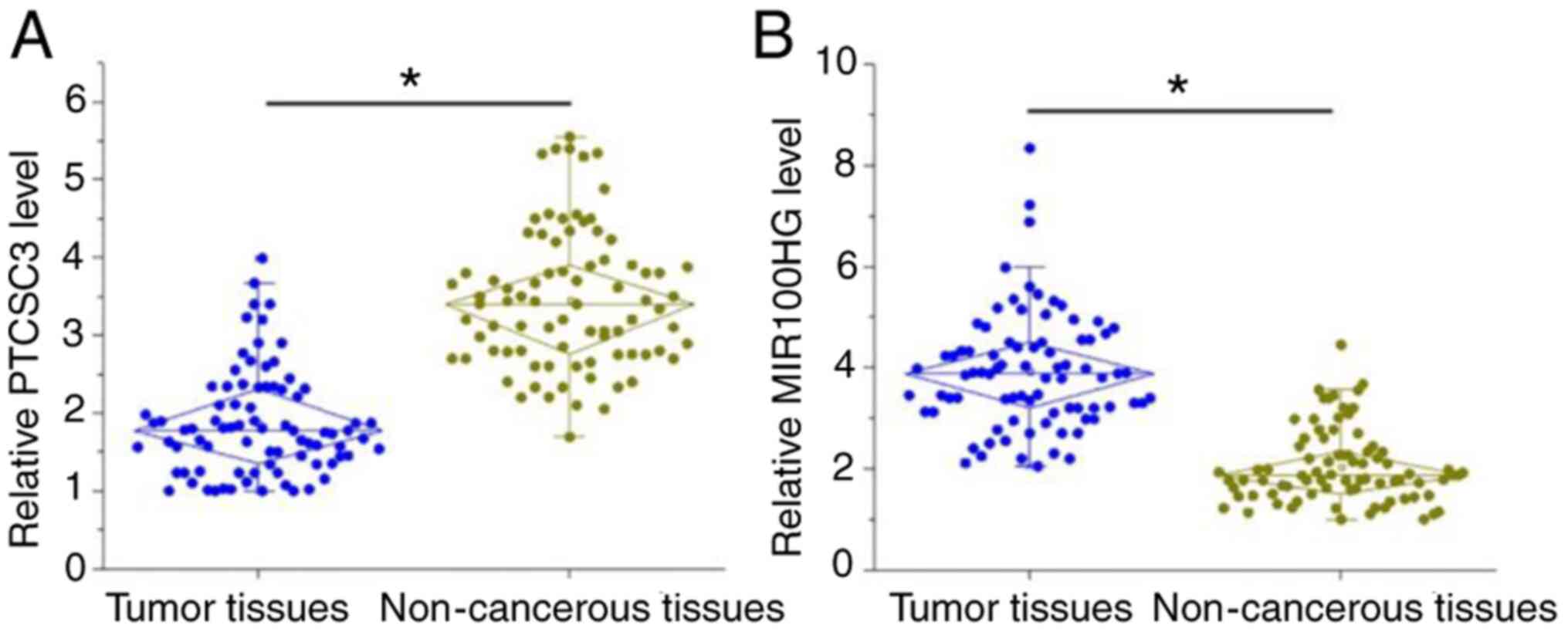
Expression levels of PTCSC3 and
MIR100HG are altered in TNBC tissues
The expression levels of PTCSC3 and MIR100HG in both
tumor tissues and adjacent non-cancerous tissues of the 82 patients
with TNBC were detected by RT-qPCR. Comparison of the expression
levels of PTCSC3 and MIR100HG between tumor and non-cancerous
tissues were performed by paired Student's t-test. PTCSC3 was
significantly downregulated (Fig.
1A), while lncRNA MIR100HG was significantly upregulated
(Fig. 1B) in tumor tissues compared
with adjacent non-cancerous tissues of patients with TNBC
(P<0.05).
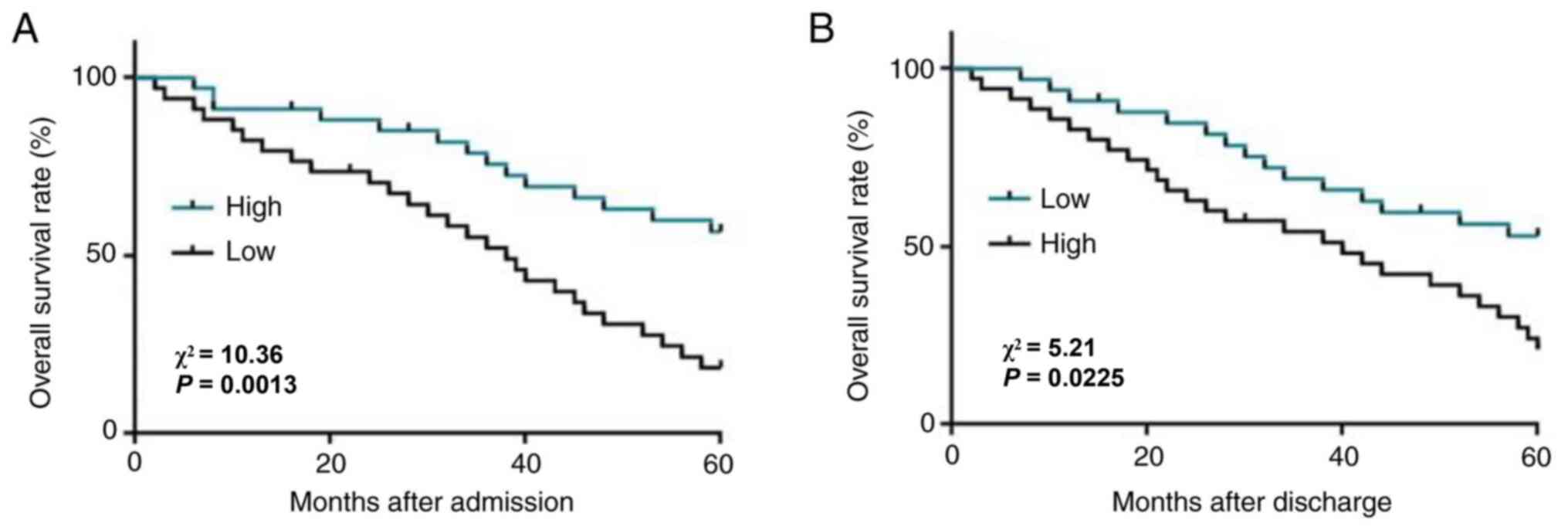
Expression levels of PTCSC3 and
MIR100HG in tumor tissues predicts survival of patients with
TNBC
According to Youden's index, patients were divided
into high (n=38) and low (n=44) PTCSC3 groups, as well as high
(n=36) and low (n=46) MIR100HG level groups. As shown in Fig. 2A, the survival rate of patients in
the low level PTCSC3 group was significantly lower compared with
that of patients in the high level PTCSC3 group (P=0.0013). In
contrast, the survival rate of patients in the high level MIR100HG
group was significantly lower compared with that of patients in the
low level MIR100HG group (Fig. 2B;
P=0.0225). Based on the role of PTSCS3 and MIR100HG in the survival
of TNBC, Cox proportional hazard model was used to compare the
importance of lncRNA PTCSC3 and MIR100HG with SNHG22 and H19
(17,18). PTCSC3 and MIR100HG had independent
significance for the survival rate of patients with TBNC (Table II).
 | Table II.Cox analysis of prognosis of patients
with triple-negative breast cancer. |
Table II.
Cox analysis of prognosis of patients
with triple-negative breast cancer.
| Factors | β | SE | P-value | RR | 95% Cl |
|---|
| PTCSC3 | 1.909 | 0.371 | <0.001 | 0.148 | 0.072-0.307 |
| MIR100HG | 1.688 | 0.584 | 0.004 | 0.185 | 0.059-0.581 |
| SNHG22 | 2.198 | 0.637 | 0.001 | 0.111 | 0.032-0.387 |
| H19 | 1.004 | 0.344 | <0.001 | 0.366 | 0.213-0.629 |
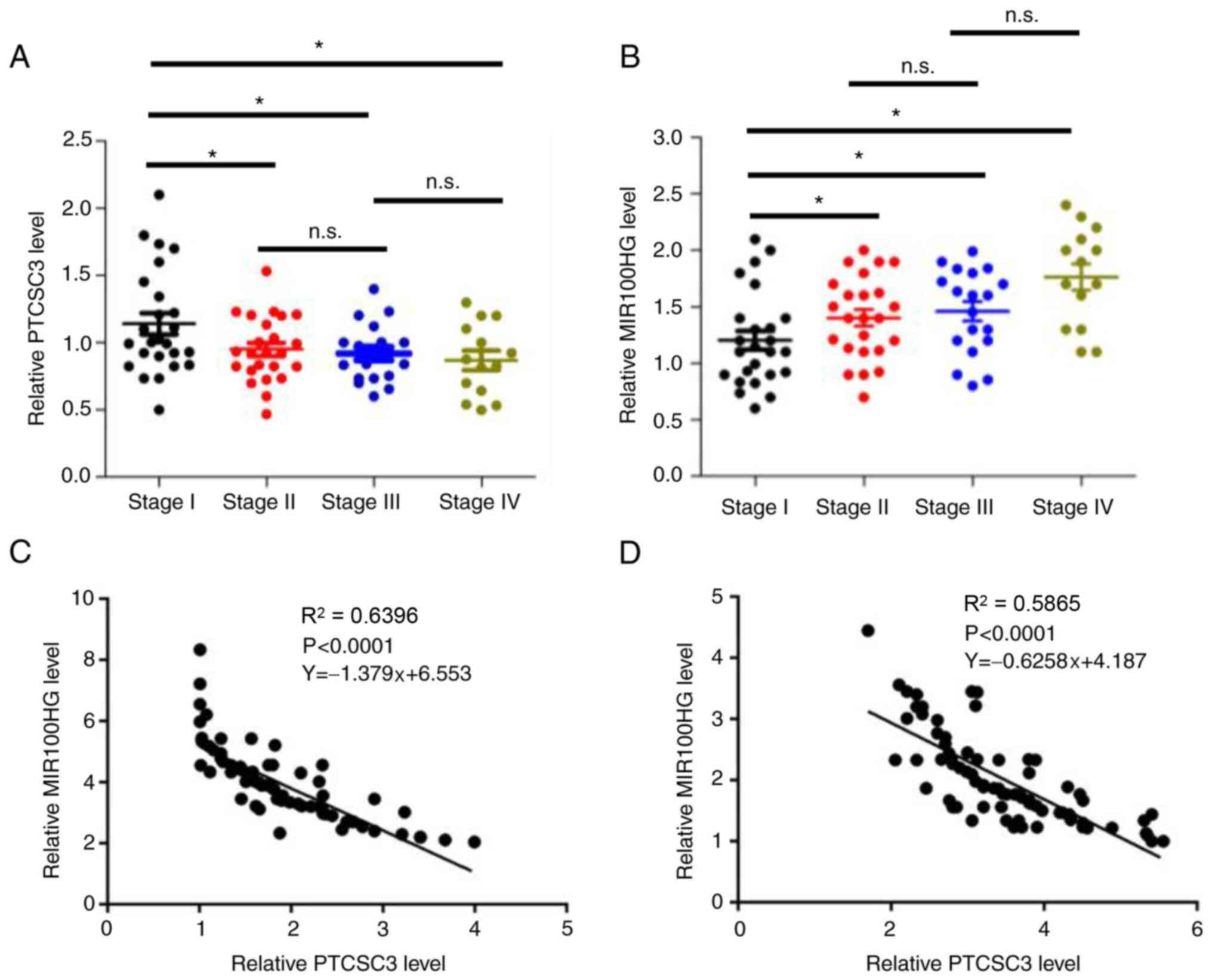
Expression levels of PTCSC3 and
MIR100HG are inversely correlated
The expression levels of PTCSC3 and MIR100HG in
tumor tissues were detected according to the different stages of 82
patients with TNBC. The expression levels of PTCSC3 was
significantly decreased in stage II compared with that in stage I,
and there was no significant difference in expression between
stages II and III or between stages III and IV (Fig. 3A; P<0.05). However, the
expression levels of MIR100HG significantly increased in stage II,
and remained unchanged at stages III and IV (Fig. 3B; P<0.05). Correlations between
the expression levels of PTCSC3 and MIR100HG were then analyzed by
Pearson's correlation coefficient and a significantly inverse
correlation was found between tumor tissues (Fig. 3C) and adjacent non-cancerous tissues
(Fig. 3D).
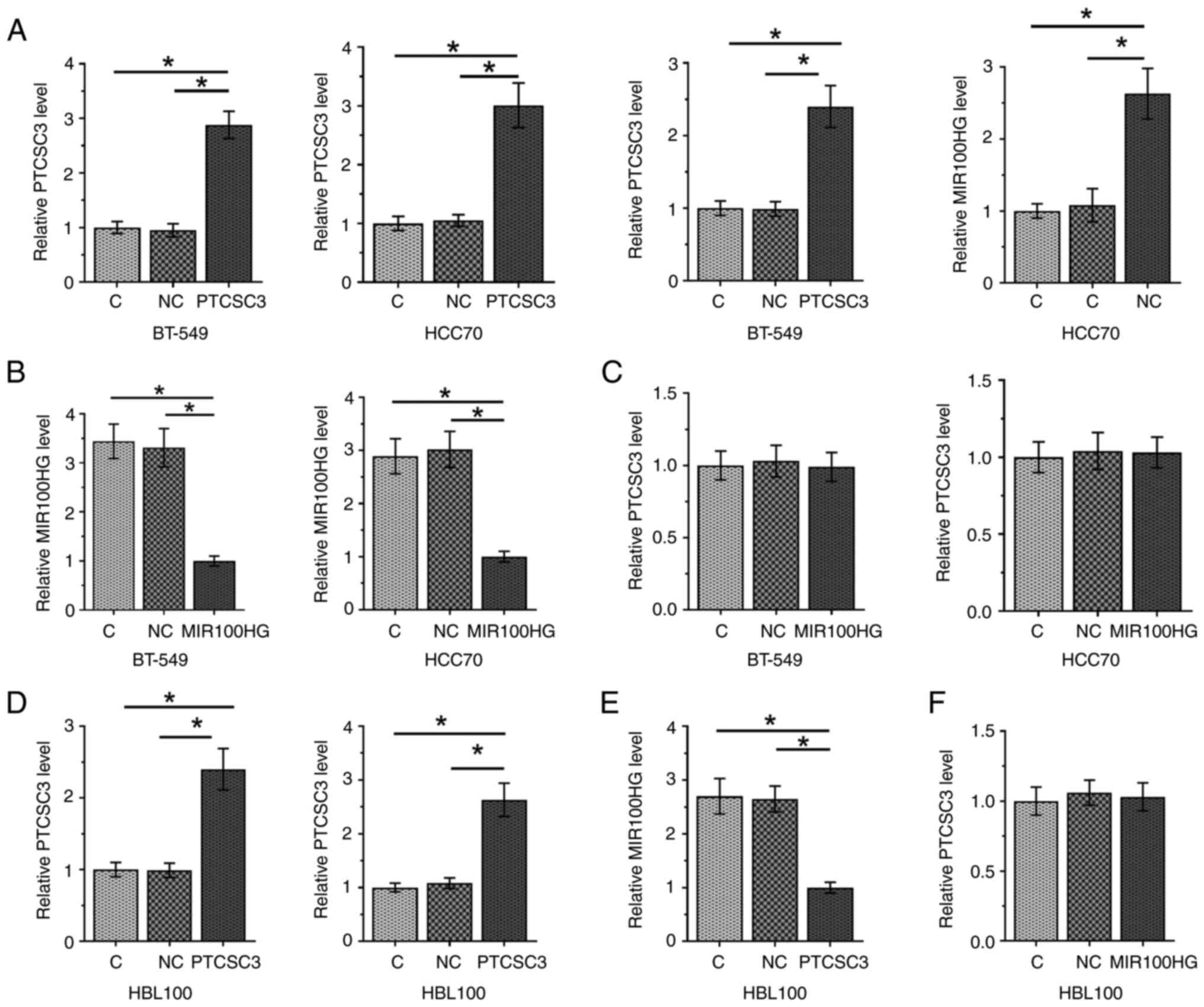
PTCSC3 is an upstream inhibitor of
MIR100HG in TNBC cells
To further assess the interaction between PTCSC3 and
MIR100HG, PTCSC3 and MIR100HG overexpression vectors were
transfected into BT-549 and HCC70 cells; overexpression was
confirmed at 24 h after transfection (Fig. 4A; P<0.05). Compared with the C
and NC groups, overexpression of PTCSC3 inhibited the expression
level of MIR100HG in cells of both cell lines (Fig. 4B; P<0.05). In contrast,
overexpression of MIR100HG did not affect the expression level of
PTCSC3 (Fig. 4C).
Overexpression of PTCSC3 inhibits
viability and promotes apoptosis of TNBC cells through
MIR100HG
Compared with the C and NC groups, overexpression of
PTCSC3 inhibited, while overexpression of MIR100HG promoted the
viability of TNBC BT-549 (Fig. 5A)
and HCC70 (Fig. 5B) cells
(P<0.05). In addition, rescue experiments showed that
overexpression of MIR100HG attenuated the effects of overexpression
of PTCSC3 on cancer cell viability (P<0.05). Cell apoptosis
assay showed increased apoptosis rate in cancer cells with the
overexpression of PTCSC3 compared with that in the C group
(P<0.01), while overexpression of PTCSC3 and MIR100HG reversed
the effect of overexpression of PTCSC3 in BT-549 (Fig. 5C) and HCC70 (Fig. 5D) (P<0.05). In addition, BrdU
staining assay results showed that PTCSC3 increased cell viability
and MIR100HG had the opposite effect. Also, overexpression of
PTCSC3 and MIR100HG reversed the effect of PTCSC3 overexpression in
BT-549 (Fig. 5E) and HCC70
(Fig. 5F) (P<0.05). However, the
overexpression of PTCSC3 and MIR100HG did not affect cancer cell
migration and invasion in BT-549 (Fig.
5G) and HCC70 (Fig. 5H)
cells.
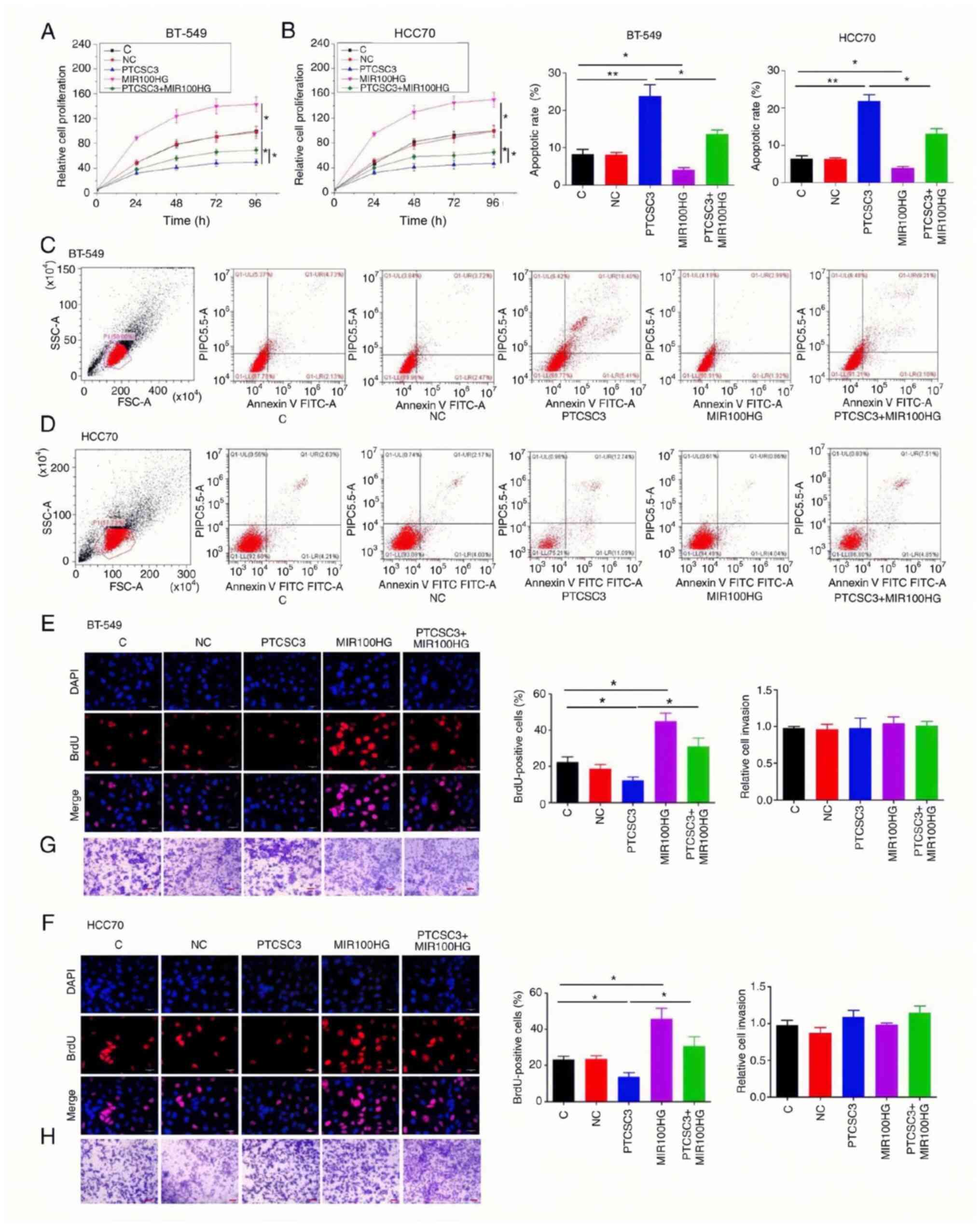 | Figure 5.Overexpression of PTCSC3 inhibits
viability and promotes apoptosis of TNBC cells through MIR100HG.
Overexpression of PTCSC3 led to inhibited, while overexpression of
MIR100HG led to promoted viability of TNBC BT-549 (A) and HCC70 (B)
cells. In addition, rescue experiments showed that overexpression
of MIR100HG attenuated the effects of PTCSC3 overexpression on
cancer cell viability. Overexpression of PTCSC3 led to promoted,
while overexpression of MIR100HG led to inhibited viability of TNBC
BT-549 (C) and HCC70 (D) cells. Overexpression of MIR100HG
attenuated the effects of PTCSC3 overexpression on cancer cell
apoptosis. Moreover, BrdU was used for staining to further detect
the TNBC cell viability. Overexpression of PTCSC3 inhibited, while
overexpression of MIR100HG promoted the viability of TNBC BT-549
(E) and HCC70 (F) cells by BrdU staining. In addition, rescue
experiments showed that overexpression of MIR100HG attenuated the
effects of PTCSC3 overexpression on cancer cell viability. Scale
bars, 30 µm. Overexpression of PTCSC3 and MIR100HG did not affect
the migration of TNBC BT-549 (G) and HCC70 (H) cells. Scale bars,
100 µm. **P<0.01; *P<0.05. TNBC, triple-negative breast
cancer; C, control; NC, negative control. |
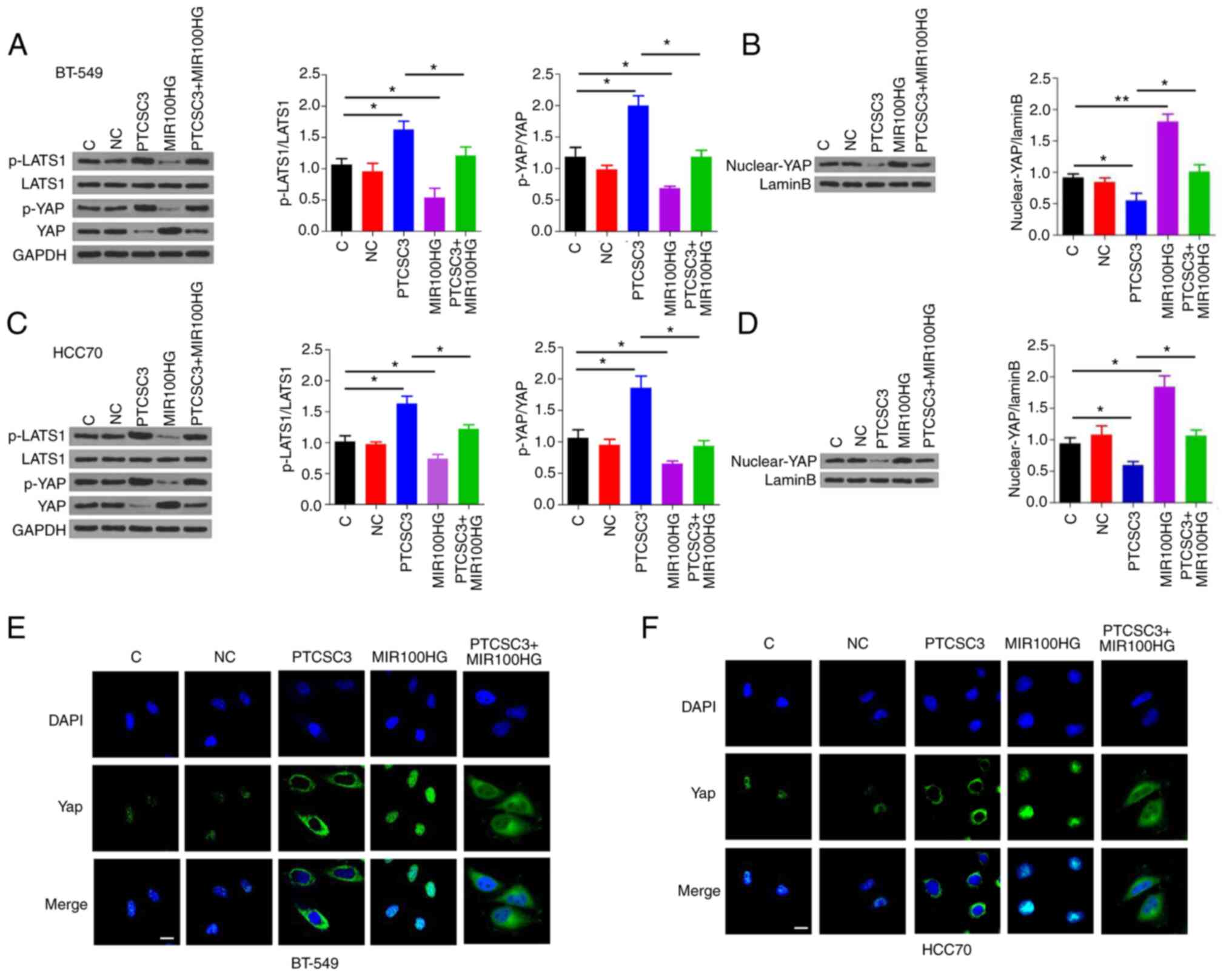
PTCSC3 suppresses viability and
promotes apoptosis of TNBC cells through the Hippo signaling
pathway
Hippo signaling pathway plays a key role in cancer
progression. Western blot analysis demonstrated that overexpression
of PTCSC3 was associated with increased p-YAP/YAP and
p-LATS1/LATS1, while overexpression of MIR100HG was associated with
decreased p-YAP/YAP and p-LATS1/LATS1 in TNBC BT-549 (Fig. 6A) and HCC70 (Fig. 6C) cells (P<0.05). In addition,
overexpression of MIR100HG attenuated the effect of PTCSC3 on
p-YAP/YAP and p-LATS1/LATS1 (Fig. 6A
and C; P<0.05). Nucleus/cytoplasm fractionation and western
blot analysis revealed that the nuclear expression levels of YAP
were decreased following overexpression of PTCSC3, while these were
increased following overexpression of MIR100HG in BT-549 (Fig. 6B) and HCC70 (Fig. 6D) cells (P<0.05). In addition,
the overexpression of MIR100HG attenuated the effect of PTCSC3 on
the nuclear expression levels of YAP (Fig. 6B and D; P<0.05). The effects of
PTCSC3 and MIR100HG on the nuclear expression levels of YAP were
confirmed by immunofluorescence assays in BT-549 (Fig. 6E) and HCC70 (Fig. 6F) cells.
Discussion
lncRNA PTCSC3 is characterized as a tumor suppressor
in thyroid cancer and glioma, while its involvement in other types
of cancer is unknown. The key finding of the present study is that
PTCSC3 is downregulated in TNBC and may play a tumor suppressive
role in TNBC by downregulating MIR100HG, which is an oncogenic
lncRNA in TNBC. A recent study reported that MIR100HG is
upregulated in TNBC and is correlated with increased viability of
cancer cells (19). Consistent with
previous study (20), the present
study also observed upregulated expression of MIR100HG in TNBC
tissues compared with that in adjacent non-cancerous tissues; and
upregulation of MIR100HG significantly promoted TNBC cell
viability. Besides, the follow-up study also showed that the high
expression levels of MIR100HG in tumor tissues is significantly
correlated with the poor survival of patients with TNBC. Therefore,
MIR100HG may serve as a prognostic marker for TNBC.
lncRNAs exert their biological functions by
regulating downstream oncogenic and tumor suppression pathways,
such as the MTDH-Twist1 signaling, Akt signaling and other
non-coding RNAs such as miRNAs (21,22).
However, interactions between lncRNAs have not been well-studied.
One study demonstrated the crosstalk of lncRNA PTCSC3 and lncRNA
H19 in TNBC, proved the interactions between different lncRNAs,
while the association of PTCSC3 and MIR100HG and their downstream
pathways are unknow (23). In the
present study, it was demonstrated that PTCSC3 is likely an
upstream inhibitor of MIR100HG in TNBC cells, and downregulation of
MIR100HG by PTCSC3 is shown to participate in the regulation of
cancer cell viability and apoptosis. MIR100HG was a
well-characterized oncogenic lncRNA in different types of cancer.
The inhibition of MIR100HG could serve as a therapeutic target for
cancer treatment by inhibiting cancer cell viability, migration and
invasion (24,25). In the development of TNBC, MIR100HG
was upregulated and resulted in promoted progression of TNBC by
sponging miR-5590-3p or regulating P27 (20). In the present study, significantly
upregulated expression of MIR100HG in tumor tissues of patients
with TNBC was observed. In addition, the in vitro cell
viability and apoptosis assays also suggested the regulatory role
of MIR100HG in TNBC cells. The present study further confirmed the
oncogenic role of MIR100HG in TNBC.
lncRNA PTCSC3 is known as a tumor suppressor in
thyroid cancer and gastric cancer. For instance, in thyroid cancer,
PTCSC3 is downregulated in the plasma of patients with gastric
cancer and can regulate multiple pathways, such as the Wnt and
STAT3 signaling pathway, to impact cancer cellular behaviors
including viability, migration and invasion (12,26,27).
In addition, PTCSC3 is downregulated in gastric cancer and can
interact with lncRNA HULC, HOXA11-AS and Linc-pint to suppress
invasion and viability of cancer cells (28,29).
Moreover, recent studies suggested that PTCSC3 is a potential
biomarker for the treatment and prognosis of gastric cancer
(29,30), whereas its role in TNBC is unknown.
The present study found that lncRNA PTCSC3 could negatively
regulate the expression of MIR100HG and inhibit TNBC cells.
Notably, the interaction between MIR100HG and PTCSC3 was also found
in breast cancer cells that are non-triple-negative, indicating
that they might also play important roles in other cancer types.
However, to date, the functions of MIR100HG and PTCSC3 in cancer
biology have not been fully elucidated and absence of non-cancerous
control cells is a limitation in the present study.
It is worth noting that overexpression of MIR100HG
only partially rescued the inhibitory effects of PTCSC3
overexpression on cancer cell viability. Therefore, it is
speculated that PTCSC3 may also interact with other downstream
targets to regulate TNBC cell viability. PTCSC3 inhibits cancer
cell migration and invasion in thyroid cancer and glioma (13). However, in the present study PTCSC3
did not affect the migration and invasion of TNBC cells. Therefore,
PTCSC3 may have different functions in different types of cancer.
In addition, PTCSC3 inhibited TNBC cell viability and promoted cell
apoptosis without affecting cell migration, which can be used to
control disease development at early stages. However, the absence
of experiments in other tumors is the big limitation of the present
study and will be further investigated in other tumors in future
studies.
Yes-associated protein (YAP), a downstream regulator
of the Hippo pathway, is upregulated in human cancer and is
associated with viability, apoptosis, migration, invasion and
resistance to chemotherapy drugs in breast cancer (31). It was reported that lncRNA FOXD2-AS1
regulates the tumorigenesis and progression of breast cancer
through the S100 calcium binding protein A1/Hippo signaling pathway
(32). The present study found that
PTCSC3 regulates TNBC cell viability through the Hippo pathway,
which downregulated the phosphorylation level of LATS1 and YAP, and
improved the translocation of YAP. MIR100HG has the opposite effect
on PTCSC3 and abolished the effect of PTCSC3. The aforementioned
data demonstrated that the Hippo signaling pathway is required for
the protective effects of PTCSC3.
In conclusion, PTCSC3 is downregulated in TNBC, and
overexpression of PTCSC3 may inhibit TNBC cell viability and
promote TNBC cell apoptosis by downregulating MIR100HG. The present
study findings might provide a potential target for the prevention
and treatment of TNBC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GZ and XW conceived and designed the experiments.
LG, JZ, RW and XW performed the experiments and analyzed the data.
GZ contributed reagents and materials. GZ and XW wrote the
manuscript. GZ and XW confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Changle People's Hospital before the admission of
patients. The ethics committee approval number is Changle-20160049.
All patients signed the informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao XB and Ren GS: lncRNA
Taurine-Upregulated Gene 1 promotes cell proliferation by
inhibiting microRNA-9 in MCF-7 cells. J Breast Cancer. 19:349–357.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Engreitz JM, Ollikainen N and Guttman M:
Long non-coding RNAs: Spatial amplifiers that control nuclear
structure and gene expression. Nat Rev Mol Cell Biol. 17:756–770.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peto R, Boreham J, Clarke M, Davies C and
Beral V: UK and USA breast cancer deaths down 25% in year 2000 at
ages 20–69 years. Lancet. 355:18222000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signaling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan M, Li X, Jiang W, Huang Y, Li J and
Wang Z: A long non-coding RNA, PTCSC3, as a tumor suppressor and a
target of miRNAs in thyroid cancer cells. Exp Ther Med.
5:1143–1146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang X, Lu X, Geng Z, Yang G and Shi Y:
lncRNA PTCSC3/miR-574-5p governs cell proliferation and migration
of papillary thyroid carcinoma via Wnt/β-catenin signaling. J Cell
Biochem. 118:4745–4752. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia S, Ji R and Zhan W: Long noncoding RNA
papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3)
inhibits proliferation and invasion of glioma cells by suppressing
the Wnt/β-catenin signaling pathway. BMC Neurol. 17:302017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang S, Ke H, Zhang H, Ma Y, Ao L, Zou L,
Yang Q, Zhu H, Nie J, Wu C and Jiao B: lncRNA MIR100HG promotes
cell proliferation in triple-negative breast cancer through triplex
formation with p27 loci. Cell Death Dis. 9:8052018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu J, Zhang Y, You Q, Fu H, Zhao X, Lu K,
Yan R and Yang D: lncRNA PTCSC3 Alleviates the postoperative
distant recurrence of gastric cancer by suppression of lncRNA
HOXA11-AS. Cancer Manag Res. 12:2623–2629. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang X, Zhang J, Li C, Liu J, Shi Z and
Zhou P: Long non-coding RNA SNHG22 facilitates the malignant
phenotypes in triple-negative breast cancer via sponging miR-324-3p
and upregulating SUDS3. Cancer Cell Int. 20:2522020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Ma HY, Hu XW, Qu YY, Wen X, Zhang Y
and Xu QY: lncRNA H19 promotes triple-negative breast cancer cells
invasion and metastasis through the p53/TNFAIP8 pathway. Cancer
Cell Int. 20:2002020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo L, Tang H, Ling L, Li N, Jia X, Zhang
Z, Wang X, Shi L, Yin J, Qiu N, et al: LINC01638 lncRNA activates
MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation
in triple-negative breast cancer. Ocogene. 37:6166–6179. 2018.
|
|
20
|
Chen FY, Zhou ZY, Zhang KJ, Pang J and
Wang SM: Long non-coding RNA MIR100HG promotes the migration,
invasion and proliferation of triple-negative breast cancer cells
by targeting the miR-5590-3p/OTX1 axis. Cancer Cell Int.
20:5082020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han J, Han B, Wu X, Hao J, Dong X, Shen Q
and Pang H: Knockdown of lncRNA H19 restores chemo-sensitivity in
paclitaxel-resistant triple-negative breast cancer through
triggering apoptosis and regulating Akt signaling pathway. Toxicol
Appl Pharmacol. 359:55–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zuo Y, Li Y, Zhou Z, Ma M and Fu K: Long
non-coding RNA MALAT1 promotes proliferation and invasion via
targeting miR-129-5p in triple-negative breast cancer. Biomed
Pharmacother. 95:922–928. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang N, Hou M, Zhan Y and Sheng X: lncRNA
PTCSC3 inhibits triple-negative breast cancer cell proliferation by
downregulating lncRNA H19. J Cell Biochem. 120:15083–15088. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li P, Ge D, Li P, Hu F, Chu J, Chen X,
Song W, Wang A, Tian G and Gu X: CXXC finger protein 4 inhibits the
CDK18-ERK1/2 axis to suppress the immune escape of gastric cancer
cells with involvement of ELK1/MIR100HG pathway. J Cell Mol Med.
24:10151–10165. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Y, Zhang C and Zhou Y: lncRNA
MIR100HG promotes cancer cell proliferation, migration and invasion
in laryngeal squamous cell carcinoma through the downregulation of
miR-204-5p. Onco Targets Ther. 12:2967–2973. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang M, Song Y and Yu L: lncRNA PTCSC3
suppressed cervical carcinoma cell invasion and proliferation via
regulating miR-574-5p. Am J Transl Res. 11:7186–7194.
2019.PubMed/NCBI
|
|
27
|
Wang XM, Liu Y, Fan YX, Liu Z, Yuan QL,
Jia M, Geng ZS, Gu L and Lu XB: lncRNA PTCSC3 affects drug
resistance of anaplastic thyroid cancer through STAT3/INO80
pathway. Cancer Biol Ther. 19:590–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cai Y, Li Y, Sun B, Wang H, Zhang W, Zhao
Y, Zhao H, Zhang J, Xu J and Wang Y: lncRNA PTCSC3 and lncRNA HULC
negatively affect each other to regulate cancer cell invasion and
migration in gastric cancer. Cancer Manag Res. 12:8535–8543. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hong L, Wang H, Wang J, Wei S, Zhang F,
Han J, Liu Y, Ma M, Liu C, Xu Y and Jiang D: lncRNA PTCSC3 inhibits
tumor growth and cancer cell stemness in gastric cancer by
interacting with lncRNA Linc-pint. Cancer Manag Res.
11:10393–10399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang G, Chi N, Lu Q, Zhu D and Zhuang Y:
lncRNA PTCSC3 Is a biomarker for the treatment and prognosis of
gastric cancer. Cancer Biother Radiopharm. 35:77–81.
2020.PubMed/NCBI
|
|
31
|
Ma J, Fan Z, Tang Q, Xia H, Zhang T and Bi
F: Aspirin attenuates YAP and β-catenin expression by promoting
β-TrCP to overcome docetaxel and vinorelbine resistance in
triple-negative breast cancer. Cell Death Dis. 11:5302020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang K, Zhang M, Yao Q, Han X, Zhao Y,
Zheng L, Li G, Liu Q, Chang Y, Zhang P, et al: The
hepatocyte-specifically expressed lnc-HSER alleviates hepatic
fibrosis by inhibiting hepatocyte apoptosis and
epithelial-mesenchymal transition. Theranostics. 9:7566–7582. 2019.
View Article : Google Scholar : PubMed/NCBI
|