Pancreatic adenocarcinoma (PDAC) is a
life-threatening condition, the incidence of which has been
increasing, and is now predicted to be the second leading cause of
cancer-associated death in certain regions of the world (1). Given that patients have no apparent
symptoms specific to PDAC and that PDAC usually shows aggressive
invasion, distant metastasis, and chemotherapeutic resistance even
in the initial stages of carcinogenesis (2), a lack of early diagnosis is considered
a leading challenge in the management of PDAC (3). Since PDAC is commonly diagnosed in the
first instance at an advanced stage, when most treatment regimens
are ineffective, investigating PDAC and understanding the mechanism
of how it is biologically aggressive is indispensable for the
development of innovative therapeutic approaches against PDAC
(4).
Previous efforts in understanding the therapeutic
resistance of tumors led to the identification of cancer stem cells
(CSCs), and there is overwhelming evidence that virtually all
cancers are clonal and represent a single-cell progeny (5–7). A
recent study demonstrated that the identification of pre-leukemic
hematopoietic stem cells in acute leukemia and mapping of the
evolutionary trajectory derived from the first somatic mutation to
the eventual development of cancer resulted in an increased
understanding of the mechanistic underpinnings of CSC lineages
(8). Given that CSCs are
biologically malignant in nature, exhibiting high invasive and
metastatic potential, increased survival, and possessing enhanced
therapeutic resistance (9),
understanding the stemness mechanism may highlight novel avenues
for overcoming PDAC, a life-threatening disease. Here, the
intracellular and intercellular signaling pathways employed by CSCs
and their surrounding cells in the tumor microenvironment of the
pancreas are discussed.
The study of gastrointestinal cancer identified the
presence of CSCs in colon cancer (10) and hepatocellular cancer (11,12).
The study of PDAC indicated that CSC-specific markers include
prominin 1 (CD133), small cell lung carcinoma cluster 4 antigen
(CD24), hyaluronate receptor (CD44), leukocyte-derived seven
transmembrane domain receptor (CXCR4), epithelial cell adhesion
molecule (EpCAM), ATP-binding cassette subfamily G member 2
(ABCG2), MET proto-oncogene, receptor tyrosine kinase (c-Met),
aldehyde dehydrogenase 1 family member A1 (ALDH1), and nestin
(Table I) (13).
Although the role of RNA methylation in the control
of CSC markers of PDAC is under investigation, previous reports
demonstrated that METTL3 improves oxaliplatin resistance of CD133+
CSCs in the stomach by promoting mRNA stability of poly(ADP-ribose)
polymerase 1 (PARP1) (14).
Alteration of the m6A modification reportedly reduces the efficacy
of drugs by regulating the expression of several drug efflux
transporters, including ABCG2, and altering the m6A modification
may prevent drug-mediated cell death through regulation of DNA
damage repair in the tumor microenvironment (15). m6A methylation-regulated AF4/FMR2
family member 4 (AFF4) was reported to enhance the self-renewal of
bladder CSCs as identified by ALDH activity (16). Therefore, m6A modifications are
proposed to be intricately associated with the functions of
CSCs.
Recent technological advancements have allowed the
study of RNA expression profiles in each cell, highlighting the
true heterogeneity of a cancer (17,18). A
recent study indicated that single-cell RNA sequencing analysis of
PDAC from patients and control pancreatic tissues revealed the
transformation process of CSC-like ductal cells into ductal cells
with invasive potential and determined CSC-related prognostic genes
associated with significantly worse overall survival, suggesting an
insight into the invasive trajectory for the treatment of PDAC
(19). Moreover, a recent study of
RNA transcription mechanisms revealed that polymerase II-associated
factor 1 (PAF1), an RNA PAF, forms a complex to maintain CSC
pluripotency by interacting with DEAD (Asp-Glu-Ala-Asp) box
helicase 3, X-linked (DDX3), and PHD-finger protein 5A (PHF5A, a
subunit of the splicing factor 3b protein complex) to regulate the
expression of self-renewal markers (homeobox transcription factor
NANOG, SRY-box transcription factor 9 (SOX9), and β-catenin), in
therapeutic-resistant phenotypes (CD44v6 and ALDH1), and other
metastasis-associated gene signatures (20). The results of a knockdown experiment
of PAF1 in mice suggested that strategies targeting the
PAF1-PHF5A-DDX3 complex may reduce or inhibit PDAC progression
(20).
During the early elongation stage of RNA
transcription, the PAF1 complex is required to determine its
association with compass-orientational RNA polymerase II (21). Given that monoubiquitination of
histone H2B, catalyzed by ubiquitin-conjugating enzyme E2 (Rad6)-E3
Ubiquitin-Protein Ligase BRE1, is required for histone H3
methylation on lysine residues 4 and 79, catalyzed by the
Set1-containing complex, the Paf1 complex is required for Rad6-Bre1
catalytic activity (21).
Dysregulation of the human RNA polymerase-II-associated factor
complex was observed in the cancer cells (22). Although the significance of the PAF1
complex in CSCs remains to be completely understood, its biological
function may be observed by binding with partners such as PHF5A in
CSCs.
Although the significance of the relationship
between pattern recognition against pathogenic organisms to
cellular RNA modification by PHF5A requires further elucidation, a
recent study suggested that the data in colorectal cancer (CRC) may
support this theory regarding roles of PHF5A in CSCs. A study on
CRC reported that PHF5A trans-activated superoxide dismutase 2
(SOD2) by regulating lysine acetyltransferase 2A [with an
alternative name of GCN5, a histone acetyltransferase (HAT) that
primarily functions as a transcriptional activator] messenger RNA
alternative splicing after being exposed to enterotoxigenic
Bacteroides fragilis (ETBF) was strongly associated with the
occurrence of inflammatory bowel disease (IBD), colitis-associated
colorectal cancer, and CRC (23).
Interestingly, PHF5A upregulation was associated with miR-149-3p
downregulation, and this was dependent on
N6-adenosine-methyltransferase subunit METTL14-mediated
N6-methyladenosine methylation (23). Targeting the ETBF/miR-149-3p pathway
presents a promising approach for treating patients with IBD and
CRC with higher ETBF expression levels (23), suggesting that pattern recognition
against pathogenic organisms may be associated with cellular RNA
modification (24).
Studies of RNA modifications demonstrate that RNA
modification is performed by writers [addition of CH3-
to RNA by the enzymatic reactions of methyltransferase 3,
N6-adenosine-methyltransferase complex catalytic subunit (METTL3),
METTL14, and Wilms tumor 1 associated protein (WTAP)], erasers
[removal of CH3- from RNA by the enzymatic reactions of
fat mass and obesity-associated protein (FTO) and
α-ketoglutarate-dependent dioxygenase AlkB Homolog (ALKBH5)], and
readers [recognition of methylated RNA by RNA binding proteins,
such as heterogeneous nuclear ribonucleoprotein (hnRNP) and
N6-methyladenosine RNA binding, YTH domain family protein (YTHDF)]
(25). In cancer, cell-cell
communication in the tumor microenvironment, such as that between
epithelial and mesenchymal cells induces the signaling pathways
that result in RNA modifications (25). In 1974, m6A was detected in poly(A)
RNA fractions (26,27), and recent sequencing methods
revealed that m6A levels in mRNA appeared to be dynamic, with
levels varying in terms of development and response to cellular
stresses (28,29). Furthermore, the enzyme FTO, which is
associated with human obesity (30), could reportedly demethylate m6A.
Therefore, aside from methylation, m6A can be dynamically regulated
through removal (31). The
biochemical process of RNA modification mediated by methylation,
demethylation, and recognition of the methylation status has been
studied, whereas the mechanisms of up or downregulation of gene
expression of the enzymes which are involved in the RNA
modification process remain to be fully understood (25). In gastrointestinal cancer, the MYC
proto-oncogene and BHLH transcription factor oncogene are shown to
promote the expression of RNA modification readers at m6A, which
can contribute to the imbalance of the epitranscriptome system in
cancer (25). Considering that
methylation, demethylation, and recognition of changes are involved
in the regulation of the cellular process, recent studies of PDAC
have emerged in recent years and indicated that METTL3 largely
plays a role in promoting PDAC, although it is also involved in the
counterbalance via a complex mechanism, summarized in Table II.
Importantly, quantification of the m6A RNA
methylation modulator pattern has been demonstrated to allow
precise evaluation of the tumor microenvironment of PDAC, including
immune response, suggesting the usefulness of a potential biomarker
for prognosis (32). RNA
modification is associated with the progression of tumor
heterogeneity (25). A study of
data obtained from TCGA (185 samples) indicated that m6A regulatory
genes played an important role in the prognosis, progression, and
regulation of the immune microenvironment in PDAC (33). In the tumor microenvironment,
glutamate from the nerve cells upregulated the expression of
hexokinase 2 (HK2) through METTL3-mediated mRNA m6A modification,
N-methyl-d-aspartate receptor (NMDAR2B), and downstream
Ca2+-dependent calcium/calmodulin-dependent protein
kinase (CaM Kinase) II/mitogen-activated protein kinase 1
(ERK)-mitogen-activated protein kinase (MAPK) pathway in PDAC
(34).
Recent studies have suggested that m6A RNA
alterations play essential physiological and pathological roles,
particularly in the initiation and progression of various types of
cancer, such as those of hematopoietic malignancies, central
nervous tumors, and reproductive cancers (35). Particularly in PDAC, the formation,
and evolution of the disease show both common and unique pathways
when compared with other malignancies (36). Although the common pathways are
responsible for regulating the balance of methylation and
demethylation, the unique pathways include the mechanism through
which ALKBH5 suppresses period circadian regulator 1-ataxia
telangiectasia mutated-checkpoint kinase 2-tumor protein P53
(TP53)-cell division cycle 25C signaling in an m6A-YTHDF2-dependent
manner, and TP53-induced ALKBH5 activation acts as a feedback loop
regulating m6A modification in PDAC (37). These studies suggest that RNA
alterations are potential therapeutic targets and early diagnostic
and prognostic cancer biomarkers (35–37).
Previous studies have shown the significant role of RNA
modifications in whole cancer tumor tissues, but not in the CSC
fraction, in several types of cancer (35), such as PDAC (36), through RNA processing (37). However, the RNA processing-dependent
mechanism in a CSC fraction in PDAC remains to be fully understood.
Here, an update on the current body of knowledge regarding recent
advances in studies on RNA methylation in the CSC fraction in tumor
tissues of PDAC is provided.
Previous studies demonstrated that RNA modification
plays a role in the initiation and maintenance of CSCs in several
types of cancer (38), including
hematopoietic malignancies (39,40)
such as breast cancer (41,42) and brain tumors (43–45).
m6A modification affects the properties of CSC, including tumor
progression and treatment responses (38). m6A modification is involved in the
function of protein-coding mRNAs and non-coding RNAs in CSCs
(46). A recent study revealed that
m6A methylation is associated with cancer, contributing to the
self-renewal of CSC, promotion and initiation of cancer
progression, and resistance to radiotherapy or chemotherapy
(47).
Nevertheless, the significance and implication of
RNA modifications in CSCs of PDAC remain to be fully investigated,
although several reports have provided critical insights into the
multifaceted roles of CSCs in PDAC (48,49).
This may be associated with the insufficient power of single-cell
analysis technology for RNA modification to investigate tumor
heterogeneity in PDAC. Given that mRNA profiling was successfully
performed with single-cell analysis technology (50), the significance and implications of
RNA modification in CSCs of PDAC may be elucidated in the near
future.
To understand the intractability of PDAC and create
innovative therapeutic approaches to overcome this disease, the
molecular and cellular biological properties of pancreatic CSCs
should be elucidated. As mentioned before, CSCs have self-renewal,
pluripotent, and tumorigenic properties under the control of the
epigenome, and this is dependent on the metabolic characteristics
of cancer (such as the presence of a hypoxic environment) in the
tumor microenvironment (9,39), and this allows a cancer to exhibit
resistance to anticancer drugs and radiation therapy (51).
The findings of previous studies have emphasized the
importance of RNA methylation in the pathogenesis of several types
of cancer, including pancreatic cancer (14,15,25).
In particular, a recent study showed the role of RNA methylation in
CSCs, and its importance in pancreatic CSCs is expected to be
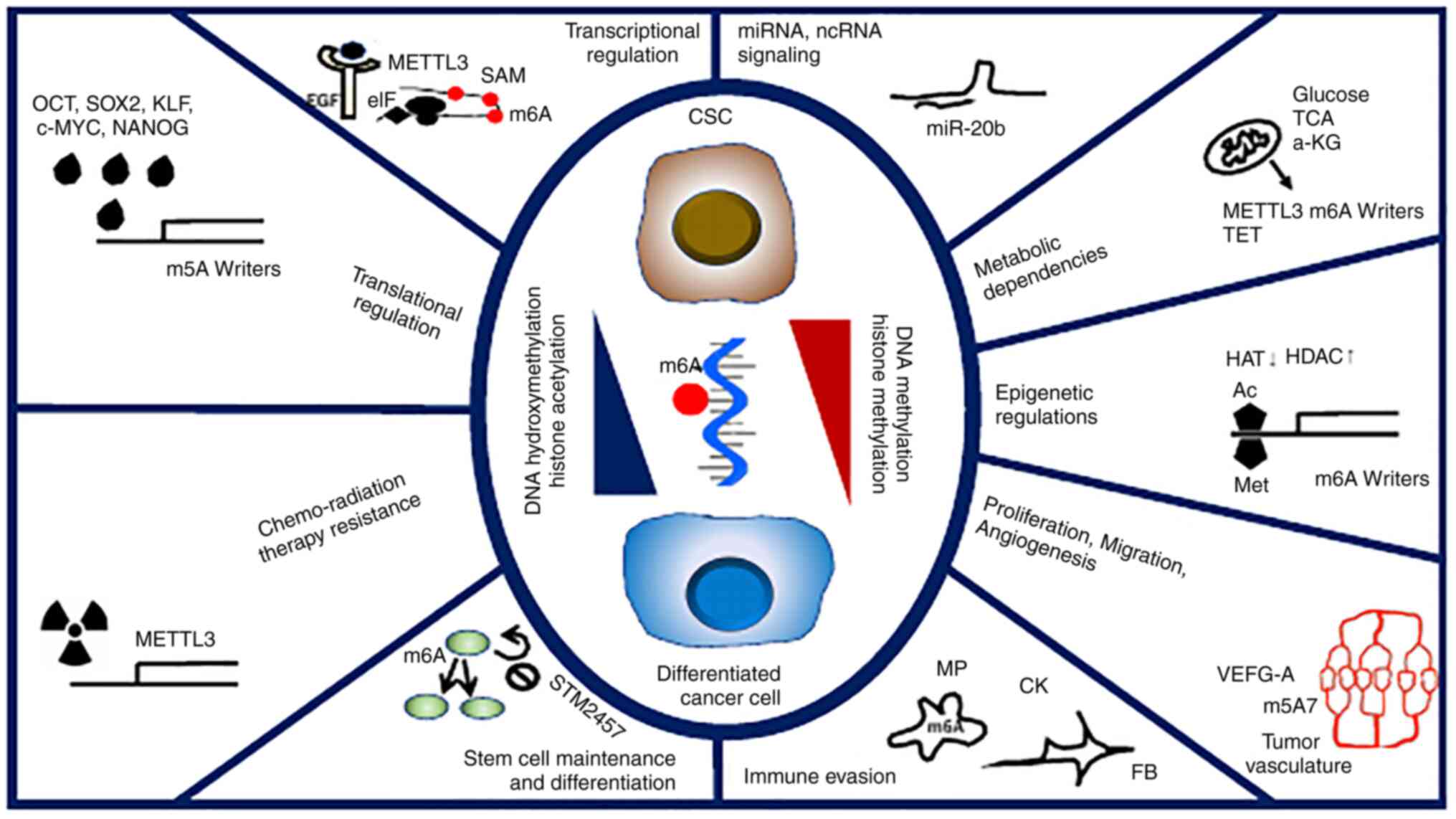
elucidated (25) (Fig. 1).
Furthermore, RNA methylation is hypothesized to play
an important role in pancreatic CSCs for the following reasons: i)
The epigenome reflects the extracellular environment and is
destined for intracellular metabolism. RNA methylation enzymes
convert the target RNA into a methylation donor using
S-adenosylmethionine (SAM) produced under inflammatory stimuli
(52). RNA demethylase is a
dioxygenase working in the nucleus with mitochondrial
α-ketoglutarate, a cofactor involved in cancer aggressiveness, and
could be a therapeutic target (52–54).
The RNA methylation process, (that is, the methylosystem) may be a
target of several cancers as a cancer-sieging strategy (53). DNA demethylase is also catalyzed by
dioxygenases that belong to the ten-eleven translocation (TET)
family (55). The TET proteins are
important players in dioxygenase for DNA hydroxymethylation, which
regulates gene expression in tumor cells, though its roles in PDAC
remain to be fully investigated (55). ii) RNA methylation is involved in
splicing and translation and plays an essential role in cell
differentiation (56,57). The hierarchy of differentiation in
CSCs is hypothesized to develop through abnormal RNA processing
rather than through the inclusion of new DNA mutations (53,56,57);
this is due to the fact that cellular reprogramming occurs during
the process of epigenetic mechanism in iPS cells (58,59).
Beyond the occurrence of DNA mutations in cancer, RNA methylation
is involved in splicing and translation, but also in RNA editing
via posttranscriptional regulation, although the significance of
this in PDAC remains to be fully understood (Fig. 1). iii) By clarifying RNA methylation
at the single-cell level, precision medicine can be established
(50). Although RNA-methylation
writer proteins are generally oncogenic in all pancreatic cancers,
the role of erasers in cancer varies considerably (25). The significance and role of RNA
methylation at the molecular level are expected to be clarified by
examining information at the cellular and gene levels. Although
certain studies have already been conducted to assess the whole
status of RNA methylation in tumors (25), the development and practical
application of a single-cell technique for RNA methylation analysis
is warranted. Although standard single-cell analysis allows for the
identification of sequence information of mRNAs with a poly(A)
tail, recent studies indicate the possible involvement of
non-coding RNAs (Fig. 1). iv)
Treatments targeting RNA methylation and the associated pathways
using low-molecular-weight drugs and related strategies have been
assessed in clinical studies (60–62).
It has already been shown that pharmacological inhibition of METTL3
in vivo leads to impaired engraftment and prolonged survival
in animal models of hematopoietic malignancies, specifically
targeting key stem cell subpopulations of leukemia (60). If the targets for diagnosis and
therapy can be clarified and similarly narrowed down, it can
potentially reveal hitherto unknown combinations of CSC-targeting
strategies for the management of PDAC. Whether targeting RNA
methylation in PDAC is a suitable approach remains to be
determined, the beneficial effects can be expected in the
therapeutic targeting of RNA methylation for hematopoietic
malignancies as shown in the studies of STM2457, which exhibited
pronounced anti-tumor efficacy in animal models (60). In the case of possible application
for the management of PDAC or other types of cancers, adverse
effects may serve to limit the viability of this approach. To the
best of our knowledge, there have been no reports on adverse events
from small animal tests, that have led to the discontinuation of
the entire development process. However, careful considerations and
precautions should be taken such as testing medium-sized animals,
before moving on to human studies. The efficacy and cytotoxicity of
these treatments remain to be investigated fully in humans.
Therefore, extra experimental and clinical data are warranted for
the development of this approach as novel anti-tumor therapeutic
reagents. However, further investigation regarding RNA
modifications may highlight novel frontiers for the field of cancer
research and treatment, whilst also providing further insight into
the molecular mechanisms underlying the development and progression
of PDAC.
Finally, the mechanisms underlying the development
and the functions of CSCs in PDACs are not fully understood. For
example, the involvement of long non-coding RNAs, such as NEAT1,
which is regulated by short non-coding RNAs has been hypothesized,
though its significance in a CSC population of PDAC remains to be
elucidated fully (63). Further
mechanistic studies will contribute to the elucidation of the
molecular mechanisms, drug discovery, and the promotion of
precision medicine of PDAC, by targeting a CSC population in PDAC.
Strategies designed to elucidate the mechanism and the profile of
CSCs in PDAC, both in the lab and in clinical practice will be
required to ensure they allow for more personalized treatments
based on a patient's specific profile.
This review article summarizes the available body of
knowledge on targeting RNA methylation in the management of PDAC
and discusses potential future approaches, with the aim of
improving the survival and the quality of life of patients with
PDAC.
The present study reviewed the functional roles and
mechanisms of RNA methylation in the regulation of CSCs in PDAC.
Given that the existence of CSCs in PDAC tissues and CSCs plays a
role in response to chemotherapy and radiation therapy, and cancer
invasion and metastasis, the understanding of the metastatic
cascade of CTCs will give rise to tremendous potential for the
identification of therapeutic targets and the development of novel
approaches. Further investigation will be necessary for precision
medicine against the deleterious cancer PDAC.
Not applicable.
This work was supported in part by a Grant-in-Aid for Scientific
Research from the Ministry of Education, Culture, Sports, Science
and Technology [grant nos. 17cm0106414h0002, JP21lm0203007,
18KK0251, 19K22658, 20H00541, 21K19526, 22H03146, 22K19559, and
16H06279 (PAGS)) and in part by funding from the Mitsubishi
Foundation (2021).
Not applicable.
HI conceived the study. YT, TH, SM, HS, YA, KO, and
HI contributed to creating the figure, reviewing the literature,
and writing the manuscript. TH and HI were involved in drafting in
the manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
Partial institutional endowments were received from
Hirotsu Bio Science Inc. (Tokyo, Japan); Kinshu-kai Medical
Corporation (Osaka, Japan); Kyowa-kai Medical Corporation (Osaka,
Japan); IDEA Consultants Inc. (Tokyo, Japan); and Unitech Co. Ltd.
(Chiba, Japan). KO is an employee of IDEA Consultants Inc. (Tokyo,
Japan). However, these funders had no role in the procurement of
the main experimental equipment, supply expenses, study design,
data collection and analysis, decision to publish, or preparation
of this manuscript.
|
1
|
McGuigan A, Kelly P, Turkington RC, Jones
C, Coleman HG and McCain RS: Pancreatic cancer: A review of
clinical diagnosis, epidemiology, treatment and outcomes. World J
Gastroenterol. 24:4846–4861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng S, Pöttler M, Lan B, Grützmann R,
Pilarsky C and Yang H: Chemoresistance in pancreatic cancer. Int J
Mol Sci. 20:45042019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang L, Sanagapalli S and Stoita A:
Challenges in diagnosis of pancreatic cancer. World J
Gastroenterol. 24:2047–2060. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ansari D, Tingstedt B, Andersson B,
Holmquist F, Sturesson C, Williamsson C, Sasor A, Borg D, Bauden M
and Andersson R: Pancreatic cancer: Yesterday, today and tomorrow.
Future Oncol. 12:1929–1946. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fialkow PJ, Singer JW, Raskind WH, Adamson
JW, Jacobson RJ, Bernstein ID, Dow LW, Najfeld V and Veith R:
Clonal development, stem-cell differentiation, and clinical
remissions in acute nonlymphocytic leukemia. N Engl J Med.
317:468–473. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McCulloch EA, Howatson AF, Buick RN,
Minden MD and Izaguirre CA: Acute myeloblastic leukemia considered
as a clonal hemopathy. Blood Cells. 5:261–282. 1979.PubMed/NCBI
|
|
7
|
Vogelstein B, Fearon ER, Hamilton SR and
Feinberg AP: Use of restriction fragment length polymorphisms to
determine the clonal origin of human tumors. Science. 227:642–645.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shlush LI, Zandi S, Mitchell A, Chen WC,
Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW,
et al: Identification of pre-leukaemic haematopoietic stem cells in
acute leukaemia. Nature. 506:328–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haraguchi N, Ohkuma M, Sakashita H,
Matsuzaki S, Tanaka F, Mimori K, Kamohara Y, Inoue H and Mori M:
CD133+CD44+ population efficiently enriches colon cancer initiating
cells. Ann Surg Oncol. 15:2927–2933. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Haraguchi N, Ishii H, Mimori K, Tanaka F,
Ohkuma M, Kim HM, Akita H, Takiuchi D, Hatano H, Nagano H, et al:
CD13 is a therapeutic target in human liver cancer stem cells. J
Clin Invest. 120:3326–3339. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishiwata T, Matsuda Y, Yoshimura H, Sasaki
N, Ishiwata S, Ishikawa N, Takubo K, Arai T and Aida J: Pancreatic
cancer stem cells: Features and detection methods. Pathol Oncol
Res. 24:797–805. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Wang C, Lan L, Yan L, Li W, Evans I,
Ruiz EJ, Su Q, Zhao G, Wu W, et al: METTL3 promotes oxaliplatin
resistance of gastric cancer CD133+ stem cells by promoting PARP1
mRNA stability. Cell Mol Life Sci. 79:1352022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS
and Huang C: Surmounting cancer drug resistance: New insights from
the perspective of N6-methyladenosine RNA modification. Drug Resist
Updat. 53:1007202020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao Q, Zheng J, Ni Z, Sun P, Yang C, Cheng
M, Wu M, Zhang X, Yuan L, Zhang Y and Li Y: The m6A
methylation-regulated AFF4 promotes self-renewal of bladder cancer
stem cells. Stem Cells Int. 2020:88492182020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ziegenhain C, Vieth B, Parekh S, Reinius
B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I
and Enard W: Comparative analysis of single-cell RNA sequencing
methods. Mol Cell. 65:631–643.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu
J, Yu X and Shi S: Applications of single-cell sequencing in cancer
research: Progress and perspectives. J Hematol Oncol. 14:912021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ren X, Zhou C, Lu Y, Ma F, Fan Y and Wang
C: Single-cell RNA-seq reveals invasive trajectory and determines
cancer stem cell-related prognostic genes in pancreatic cancer.
Bioengineered. 12:5056–5068. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karmakar S, Rauth S, Nallasamy P, Perumal
N, Nimmakayala RK, Leon F, Gupta R, Barkeer S, Venkata RC, Raman V,
et al: RNA polymerase II-associated factor 1 regulates stem cell
features of pancreatic cancer cells, independently of the PAF1
complex, via interactions with PHF5A and DDX3. Gastroenterology.
159:1898–1915.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wood A, Schneider J, Dover J, Johnston M
and Shilatifard A: The Paf1 complex is essential for histone
monoubiquitination by the Rad6-Bre1 complex, which signals for
histone methylation by COMPASS and Dot1p. J Biol Chem.
278:34739–34742. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chaudhary K, Deb S, Moniaux N, Ponnusamy
MP and Batra SK: Human RNA polymerase II-associated factor complex:
Dysregulation in cancer. Oncogene. 26:7499–7507. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B,
Shen C, Ma Y, Jiang S, Ma D, et al: Enterotoxigenic
bacteroidesfragilis promotes intestinal inflammation and malignancy
by inhibiting exosome-packaged miR-149-3p. Gastroenterology.
161:1552–1566.e12. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sato H, Sasaki K, Hara T, Kobayashi S,
Doki Y, Eguchi H, Satoh T and Ishii H: Targeting the regulation of
aberrant protein production pathway in gastrointestinal cancer
treatment. Front Oncol. 12:10183332022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Konno M, Taniguchi M and Ishii H:
Significant epitranscriptomes in heterogeneous cancer. Cancer Sci.
110:2318–2327. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Porc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Perry RP and Kelley DE: Existence of
methylated messenger RNA in mouse L cells. Cell. 1:37–42. 1974.
View Article : Google Scholar
|
|
28
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3´ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-Methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meyer KD and Jaffrey SR: Rethinking m6A
readers, writers, and erasers. Annu Rev Cell Dev Biol. 33:319–342.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L, Zhang S, Li H and Xu Y, Wu Q, Shen
J, Li T and Xu Y: Quantification of m6A RNA methylation modulators
pattern was a potential biomarker for prognosis and associated with
tumor immune microenvironment of pancreatic adenocarcinoma. BMC
Cancer. 21:8762021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu F, Zhang Z, Yuan M, Zhao Y, Zhou Y, Pei
H and Bai L: M6A regulatory genes play an important role in the
prognosis, progression and immune microenvironment of pancreatic
adenocarcinoma. Cancer Invest. 39:39–54. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li F, He C, Yao H, Zhao Y, Ye X, Zhou S,
Zou J, Li Y, Li J, Chen S, et al: Glutamate from nerve cells
promotes perineural invasion in pancreatic cancer by regulating
tumor glycolysis through HK2 mRNA-m6A modification. Pharmacol Res.
187:1065552023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Wang F, Liu Y, Wang H and Ni B:
N6-methyladenosine (m6A) in pancreatic cancer: Regulatory
mechanisms and future direction. Int J Biol Sci. 17:2323–2335.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang
Y, Feng Y, Pan Q and Wan R: RNA demethylase ALKBH5 prevents
pancreatic cancer progression by posttranscriptional activation of
PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 19:912020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma Z and Ji J: N6-methyladenosine (m6A)
RNA modification in cancer stem cells. Stem Cells. 38:1511–1519.
2020. View Article : Google Scholar
|
|
39
|
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang
X, Dong L, Chen H, Su R, Yin Z, Li W, et al: RNA demethylase ALKBH5
selectively promotes tumorigenesis and cancer stem cell
self-renewal in acute myeloid leukemia. Cell Stem Cell.
27:64–80.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Paris J, Morgan M, Campos J, Spencer GJ,
Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA,
Sepulveda C, et al: Targeting the RNA m6A reader YTHDF2 selectively
compromises cancer stem cells in acute myeloid leukemia. Cell Stem
Cell. 25:137–148.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang C, Samanta D, Lu H, Bullen JW, Zhang
H, Chen I, He X and Semenza GL: Hypoxia induces the breast cancer
stem cell phenotype by HIF-dependent and ALKBH5-mediated
m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA.
113:E2047–E2056. 2016.PubMed/NCBI
|
|
42
|
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T,
Zeng H, Qin Y, Wan X, Qiao Y, et al: A novel hypoxic long noncoding
RNA KB-1980E6.3 maintains breast cancer stem cell stemness via
interacting with IGF2BP1 to facilitate c-Myc mRNA stability.
Oncogene. 40:1609–1627. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dixit D, Prager BC, Gimple RC, Poh HX,
Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, et al: The RNA m6A
reader YTHDF2 maintains oncogene expression and is a targetable
dependency in glioblastoma stem cells. Cancer Discov. 11:480–499.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A Demethylase
ALKBH5 maintains tumorigenicity of glioblastoma Stem-like Cells by
sustaining FOXM1 expression and cell proliferation program. Cancer
Cell. 31:591–606.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation regulates the
self-renewal and tumorigenesis of glioblastoma stem cells. Cell
Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G,
Yuan W, Kan Q and Sun Z: The interplay between m6A RNA methylation
and noncoding RNA in cancer. J Hematol Oncol. 12:1212019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dai D, Wang H, Zhu L, Jin H and Wang X:
N6-methyladenosine links RNA metabolism to cancer progression. Cell
Death Dis. 9:1242018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma X, Cao J, Zhou Z, Lu Y, Li Q, Jin Y,
Chen G, Wang W, Ge W, Chen X, et al: N6-methyladenosine
modification-mediated mRNA metabolism is essential for human
pancreatic lineage specification and islet organogenesis. Nat
Commun. 13:41482022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Garg R, Melstrom L, Chen J, He C and Goel
A: Targeting FTO suppresses pancreatic carcinogenesis via
regulating stem cell maintenance and EMT pathway. Cancers (Basel).
14:59192022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chijimatsu R, Kobayashi S, Takeda Y,
Kitakaze M, Tatekawa S, Arao Y, Nakayama M, Tachibana N, Saito T,
Ennishi D, et al: Establishment of a reference single-cell RNA
sequencing dataset for human pancreatic adenocarcinoma. iScience.
25:1046592022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ishii H, Iwatsuki M, Ieta K, Ohta D,
Haraguchi N, Mimori K and Mori M: Cancer stem cells and
chemoradiation resistance. Cancer Science. 99:1871–1877. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mehdi A and Rabbani SA: Role of
methylation in pro- and anti-cancer immunity. Cancers (Basel).
13:5452021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tatekawa S, Ofusa K, Chijimatsu R,
Vecchione A, Tamari K, Ogawa K and Ishii H: Methylosystem for
cancer sieging strategy. Cancers (Basel). 13:50882021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Monné M, Marobbio CMT, Agrimi G, Palmieri
L and Palmieri F: Mitochondrial transport and metabolism of the
major methyl donor and versatile cofactor S-adenosylmethionine, and
related diseases: A review. IUBMB Life. 74:573–591. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu X and Zhang Y: TET-mediated active DNA
demethylation: Mechanism, function and beyond. Nat Rev Genet.
18:517–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G:
Functions of N6-methyladenosine and its role in cancer. Mol Cancer.
18:1762019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lan Q, Liu PY, Haase J, Bell JL,
Hüttelmaier S and Liu T: The critical role of RNA m6A methylation
in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ryall JG, Cliff T, Dalton S and Sartorelli
V: Metabolic reprogramming of stem cell epigenetics. Cell Stem
Cell. 17:651–662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nuñez JK, Chen J, Pommier GC, Cogan JZ,
Replogle JM, Adriaens C, Ramadoss GN, Shi Q, Hung KL, Samelson AJ,
et al: Genome-wide programmable transcriptional memory by
CRISPR-based epigenome editing. Cell. 184:2503–2519.e17. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yankova E, Blackaby W, Albertella M, Rak
J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D,
Hendrick AG, et al: Small molecule inhibition of METTL3 as a
strategy against myeloid leukaemia. Nature. 593:597–601. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Huff S, Tiwari SK, Gonzalez GM, Wang Y and
Rana TM: m6A-RNA demethylase FTO inhibitors impair self-renewal in
glioblastoma stem cells. ACS Chem Biol. 16:324–333. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang JN, Wang F, Ke J, Li Z, Xu CH, Yang
Q, Chen X, He XY, He Y, Suo XG, et al: Inhibition of METTL3
attenuates renal injury and inflammation by alleviating TAB3 m6A
modifications via IGF2BP2-dependent mechanisms. Sci Transl Med.
14:eabk27092022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huang B, Liu C, Wu Q, Zhang J, Min Q,
Sheng T, Wang X and Zou Y: Long non-coding RNA NEAT1 facilitates
pancreatic cancer progression through negative modulation of
miR-506-3p. Biochem Biophys Res Commun. 482:828–834. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gupta VK and Banerjee S: Isolation of
lipid raft proteins from CD133+ cancer stem cells. Methods Mol
Biol. 1609:25–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhou T, Liu J, Xie Y, Yuan S, Guo Y, Bai
W, Zhao K, Jiang W, Wang H, Wang H, et al: ESE3/EHF, a promising
target of rosiglitazone, suppresses pancreatic cancer stemness by
downregulating CXCR4. Gut. 71:357–371. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hamada S, Satoh K, Hirota M, Kanno A,
Umino J, Ito H, Masamune A, Kikuta K, Kume K and Shimosegawa T: The
homeobox gene MSX2 determines chemosensitivity of pancreatic cancer
cells via the regulation of transporter gene ABCG2. J Cell Physiol.
227:729–738. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ling X, Wu W, Fan C, Xu C, Liao J, Rich
LJ, Huang RY, Repasky EA, Wang X and Li F: An ABCG2 non-substrate
anticancer agent FL118 targets drug-resistant cancer stem-like
cells and overcomes treatment resistance of human pancreatic
cancer. J Exp Clin Cancer Res. 37:2402018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li C, Wu JJ, Hynes M, Dosch J, Sarkar B,
Welling TH, Pasca di Magliano M and Simeone DM: c-Met is a marker
of pancreatic cancer stem cells and therapeutic target.
Gastroenterology. 141:2218–2227.e5. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Aliebrahimi S, Kouhsari SM, Arab SS,
Shadboorestan A and Ostad SN: Phytochemicals, withaferin A and
carnosol, overcome pancreatic cancer stem cells as c-Met
inhibitors. Biomed Pharmacother. 106:1527–1536. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Nimmakayala RK, Leon F, Rachagani S, Rauth
S, Nallasamy P, Marimuthu S, Shailendra GK, Chhonker YS, Chugh S,
Chirravuri R, et al: Metabolic programming of distinct cancer stem
cells promotes metastasis of pancreatic ductal adenocarcinoma.
Oncogene. 40:215–231. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Matsuda Y, Tanaka M, Sawabe M, Mori S,
Muramatsu M, Mieno MN, Ishiwata T and Arai T: The stem
cell-specific intermediate filament nestin missense variation
p.A1199P is associated with pancreatic cancer. Oncol Lett.
17:4647–4654. 2019.PubMed/NCBI
|
|
73
|
Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang
J, Lu Z, Wu P, Cai B, Miao Y and Jiang K: The RNA m6A
methyltransferase METTL3 promotes pancreatic cancer cell
proliferation and invasion. Pathol Res Pract. 215:1526662019.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C,
Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via
N6-methyladenosine stimulated by cigarette smoke promotes
pancreatic cancer progression. Nat Commun. 10:18582019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Tang Y, Gao G, Xia WW and Wang JB: METTL3
promotes the growth and metastasis of pancreatic cancer by
regulating the m6A modification and stability of E2F5. Cell Signal.
99:1104402022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Guo Z, Zhang X, Lin C, Huang Y, Zhong Y,
Guo H, Zheng Z and Weng S: METTL3-IGF2BP3-axis mediates the
proliferation and migration of pancreatic cancer by regulating
spermine synthase m6A modification. Front Oncol. 12:9622042022.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Y, Huang H, Zhu Y, Xu B, Chen J, Liu Y,
Zheng X and Chen L: Increased expression of METTL3 in pancreatic
cancer tissues associates with poor survival of the patients. World
J Surg Oncol. 20:2832022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Song Z, Wang X, Chen F, Chen Q, Liu W,
Yang X, Zhu X, Liu X and Wang P: LncRNA MALAT1 regulates
METTL3-mediated PD-L1 expression and immune infiltrates in
pancreatic cancer. Front Oncol. 12:10042122022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Taketo K, Konno M, Asai A, Koseki J,
Toratani M, Satoh T, Doki Y, Mori M, Ishii H and Ogawa K: The
epitranscriptome m6A writer METTL3 promotes chemo- and
radioresistance in pancreatic cancer cells. Int J Oncol.
52:621–629. 2018.PubMed/NCBI
|
|
80
|
Jiang Z, Song X, Wei Y, Li Y, Kong D and
Sun J: N(6)-methyladenosine-mediated miR-380-3p maturation and
upregulation promotes cancer aggressiveness in pancreatic cancer.
Bioengineered. 13:14460–14471. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen JQ, Tao YP, Hong YG, Li HF, Huang ZP,
Xu XF, Zheng H and Hu LK: M6A-mediated up-regulation of LncRNA
LIFR-AS1 enhances the progression of pancreatic cancer via
miRNA-150-5p/VEGFA/Akt signaling. Cell Cycle. 20:2507–2518. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
He Y, Liu Y, Wu D, Chen L, Luo Z, Shi X,
Li K, Hu H, Qu G, Zhao Q and Lian C: Linc-UROD stabilizes ENO1 and
PKM to strengthen glycolysis, proliferation and migration of
pancreatic cancer cells. Transl Oncol. 27:1015832023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Tatekawa S, Tamari K, Chijimatsu R, Konno
M, Motooka D, Mitsufuji S, Akita H, Kobayashi S, Murakumo Y, Doki
Y, et al: N(6)-methyladenosine methylation-regulated polo-like
kinase 1 cell cycle homeostasis as a potential target of
radiotherapy in pancreatic adenocarcinoma. Sci Rep. 12:110742022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ye X, Wang LP, Han C, Hu H, Ni CM, Qiao
GL, Ouyang L and Ni JS: Increased m6A modification of lncRNA
DBH-AS1 suppresses pancreatic cancer growth and gemcitabine
resistance via the miR-3163/USP44 axis. Ann Transl Med. 10:3042022.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Huang C, Zhou S, Zhang C, Jin Y, Xu G,
Zhou L, Ding G, Pang T, Jia S and Cao L: ZC3H13-mediated
N6-methyladenosine modification of PHF10 is impaired by fisetin
which inhibits the DNA damage response in pancreatic cancer. Cancer
Lett. 530:16–28. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hou J, Wang Z, Li H, Zhang H and Luo L:
Gene signature and identification of clinical trait-related m6 A
regulators in pancreatic cancer. Front Genet. 11:5222020.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang W, He Y, Zhai LL, Chen LJ, Yao LC, Wu
L, Tang ZG and Ning JZ: m6A RNA demethylase FTO promotes the
growth, migration and invasion of pancreatic cancer cells through
inhibiting TFPI-2. Epigenetics. 17:1738–1752. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Huang R, Yang L, Zhang Z, Liu X, Fei Y,
Tong WM, Niu Y and Liang Z: RNA m6A demethylase ALKBH5 protects
against pancreatic ductal adenocarcinoma via targeting regulators
of iron metabolism. Front Cell Dev Biol. 9:7242822021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cui L, Ma R, Cai J, Guo C, Chen Z, Yao L,
Wang Y, Fan R, Wang X and Shi Y: RNA modifications: Importance in
immune cell biology and related diseases. Signal Transduct Target
Ther. 7:3342022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Sato H, Hara T, Tatekawa S, Sasaki K,
Kobayashi S, Kitagawa T, Doki Y, Eguchi H, Ogawa K, Uchida S and
Ishii H: Emerging roles of long noncoding and circular RNAs in
pancreatic ductal adenocarcinoma. Front Physiol. 13:10259232022.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Takeda Y, Chijimatsu R, Vecchione A, Arai
T, Kitagawa T, Ofusa K, Yabumoto M, Hirotsu T, Eguchi H, Doki Y and
Ishii H: Impact of one-carbon metabolism-driving epitranscriptome
as a therapeutic target for gastrointestinal cancer. Int J Mol Sci.
22:72782021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Takeda Y, Chijimatsu R, Ofusa K, Kobayashi
S, Doki Y, Eguchi H and Ishii H: Cancer metabolism challenges
genomic instability and clonal evolution as therapeutic targets.
Cancer Sci. 113:1097–1104. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zagorac S, Garcia-Bermejo L and Sainz B
Jr: The epigenetic landscape of pancreatic cancer stem cells.
Epigenomes. 2:102018. View Article : Google Scholar
|
|
94
|
Liu Y, Tang G and Li J: Long non-coding
RNA NEAT1 participates in ventilator-induced lung injury by
regulating miR-20b expression. Mol Med Rep. 25:662022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Xia L, Li F, Qiu J, Feng Z, Xu Z, Chen Z
and Sun J: Oncogenic miR-20b-5p contributes to malignant behaviors
of breast cancer stem cells by bidirectionally regulating CCND1 and
E2F1. BMC Cancer. 20:9492020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kroeze LI, van der Reijden BA and Jansen
JH: 5-Hydroxymethylcytosine: An epigenetic mark frequently
deregulated in cancer. Biochim Biophys Acta. 1855:144–154.
2015.PubMed/NCBI
|