Introduction
Breast cancer is one of the most common invasive
cancers among women and has a high mortality rate. Globally, 2.3
million women were diagnosed with breast cancer in 2020, and
685,000 deaths were reported due to breast cancer (1). Breast cancer is classified into four
subtypes based on the expression of the estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor
receptor 2 (HER2). These four subtypes include luminal A
(ER+/PR+/HER2−), luminal B
(ER+/PR+/HER2+ or
ER+/PR+/HER2−), HER2
(ER−/PR−/HER2+), and triple
negative breast cancer (TNBC) or basal-like
(ER−/PR−/HER2−) breast cancer.
These subtypes possess different abilities to metastasize to the
distal organs, with luminal A having the longest survival time
followed by luminal B, HER2, and TNBC, respectively (2). Remarkably, TNBC accounts for 10–20% of
all cases (3) and is associated
with high metastasis, which leads to poor prognosis and high
mortality (4). Patients with the
TNBC subtype do not respond well to hormonal therapies, such as
HER2-targeted therapy (3), which is
a significant challenge for the successful treatment of patients
with TNBC. Therefore, the development of novel biomarkers for the
identification of the TNBC subtype is crucial.
The two breast cancer cell lines, MCF-7 and
MDA-MB-231 have been well characterized for their low and high
metastatic phenotypes, respectively. The MCF-7 cell line represents
the luminal A subtype, while the MDA-MB-231 cell line represents
the TNBC subtype (5). The MCF-7
cell line can form only the primary tumor and can be suppressed
following treatment by tamoxifen, an ER antagonist, while the
MDA-MB-231 cell line can induce metastasis and is resistant to
tamoxifen (5).
MicroRNAs (miRs/miRNAs) are small non-coding RNAs
(snRNA) 18–22 nucleotides in length, which regulate several
biological processes in tumorigenesis including proliferation,
stress response, cell adhesion, motility and apoptosis (6). Previous studies have reported aberrant
expression of certain miRNAs in TNBC. For example, Fang et
al (7) reported that miR-21 was
upregulated in MDA-MB-468 cells and that it supported proliferation
and invasion. Similarly, miR-25-3p has been reported to be
overexpressed in TNBC where it promotes proliferation both in
vitro and in vivo (8).
Previous studies have also reported that elevated expression of
miR-93 (9) and miR-455-3p (10) induced proliferation, invasion and
metastasis in TNBC. Downregulation of certain miRNAs has also been
reported in TNBC; they have been demonstrated to regulate
proliferation, migration and invasion in TNBC (6). These data support the roles of miRNAs
in maintaining the invasive phenotype of TNBC and therefore hold
promise for their potential application as diagnostic and
prognostic biomarkers.
A previous study indicated that miR-222 expression
was elevated in breast cancer tissues compared with that of normal
tissues, which suggested that it could be a promising biomarker for
breast cancer diagnosis (11).
miR-222 expression in breast cancer tissues exhibited prognostic
significance in lymph node (LN) negative breast cancer, which
suggested that it could be used as a differential biomarker between
LN negative and positive patients (12). Although miR-222 was highly expressed
in breast cancer tissues, it remains unclear whether this miRNA is
highly expressed in TNBC. The present study aimed to study miRNAs
associated with breast cancer metastasis using two breast cancer
cell lines, the low-metastatic MCF-7 cell line and the highly
metastatic MDA-MB-231 cell line. The expression of one candidate
miRNA, miR-222-3p, was validated. Suppression of miR-222-3p
expression was performed to study its roles in MDA-MB-213 cells.
The data indicated the crucial roles of miR-222-3p in supporting
growth and migration in TNBC.
Materials and methods
Cell culture
The breast cancer MCF-7 (HTB22; American Type
Culture Collection) and MDA-MB-231 (HTB26; American Type Culture
Collection) cell lines were cultured in DMEM high glucose
(HyClone™; Cytiva) supplemented with 10% (v/v) FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin at 37°C, in
the presence of 5% CO2. For the anoikis resistance
assay, 700 µl of 20 mg/ml poly-2-hydroxyethyl methacrylate
(polyHEMA) diluted in 95% ethanol was applied to a 6-well plate and
dried for 2 days. Breast cancer cells were plated (5×105
cells/well for MCF-7 and 1×106 cells/well for
MDA-MB-231) in a polyHEMA-coated 6-well plate.
RNA extraction, complementary DNA
(cDNA) synthesis and miRNA array
RNA was extracted from the adherent and
anoikis-resistant MCF-7 and MDA-MB-231 cells using
TRIzol® reagent according to the manufacturer's protocol
(Invitrogen; Thermo Fisher Scientific, Inc.). A total of 200 ng RNA
from each cell line was used to synthesize cDNA using the miRCURY
LNA RT Kit (cat. no. 339340; Qiagen, Inc.) according to the
manufacturer's protocol. To determine the miRNA expression
profiles, an miRNA array was performed using miRCURY LNA miRNA
focus panel (cat. no. 339325; Qiagen, Inc.) coated with specific
primers for 84 miRNAs associated with cancer. The miRNA target
sequences are presented in Table
SI. The miRCURY LNA SYBR Green PCR Kit(cat no. 339345; Qiagen,
Inc.) was used for qPCR reactions. The 96-well plate of the miRNA
array was subjected to PCR using a real-time PCR machine. The PCR
cycling conditions were: Initial heat activation at 95°C for 2 min,
and 40 cycles of denaturation at 95°C for 10 sec and
annealing/extension at 56°C for 60 sec. The relative expression was
determined using the 2−∆∆Cq method (13).
RT-qPCR
A candidate miRNA, miR-222-3p, from the miRNA array
was assessed using RT-qPCR. RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. A total of 100 ng
RNA extracted from adherent or anoikis-resistant MCF-7 and
MDA-MB-231 cultures was converted to cDNA using a
TaqMan® Advanced MicroRNA Synthesis Kit (cat. no.
A25576; Applied Biosystems; Thermo Fisher Scientific, Inc.). cDNA
synthesis was performed using four steps, including Poly (A)
tailing, adaptor ligation, RT and cDNA amplification reactions,
according to the manufacturer's protocol. The expression levels of
miR-222-3p were determined by qPCR using TaqMan Advanced MicroRNA
Assays (miR-222-3p, cat. no. A25576; Applied Biosystems; Thermo
Fisher Scientific, Inc.) containing forward and reverse primers and
probes in a single tube and TaqMan® Fast Advanced Master
Mix (cat. no. 4440038; Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
qPCR thermal conditions were: Uracil-N-glycosylase incubation at
50°C for 2 min and polymerase activation at 95°C for 10 min,
followed by 40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 60 sec. The relative expression was
compared using the 2−ΔΔCq method (13) and U6 snRNA (cat. no. 4427975;
Applied Biosystems; Thermo Fisher Scientific, Inc.) was used as the
internal control. The miRNA sequences used in this experiment were
as follows: Mature miRNA sequence of miR-222-3p,
5′-AGCUACAUCUGGCUACUGGGU-3′ (cat. no. A25576; Applied Biosystems;
Thermo Fisher Scientific, Inc.); and U6 snRNA control sequence,
5′-GTGCTCGCTTCGGCAGCACATATACTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTT-3′
(cat. no. 4427975; Applied Biosystems; Thermo Fisher Scientific,
Inc.). The forward and reverse primer sequences used in this
experiment are proprietary information of Applied Biosystems;
Thermo Fisher Scientific, Inc.
Transfection of cells with the
miR-222-3p inhibitor
A total of 2.5×105 MDA-MB-231 cells were
plated in a 12-well plate and incubated at 37°C overnight, and 50
nM miR-222-3p inhibitor (cat. no. 339121; Qiagen, Inc.) or 50 nM
negative control (cat. no. 339126; Qiagen, Inc.) was transfected
into the cells using HiPerFect Transfection Reagent (cat. no.
301704, Qiagen, Inc.). The transfection was performed according to
the manufacturer's protocol (Qiagen, Inc.). miR-222-3p inhibitor or
negative control was diluted with 100 µl culture medium without
serum. HiperFect transfection reagent (3 µl) was added, followed by
mixing by vortexing. The mixtures were incubated at room
temperature for 10 min to allow the formation of transfection
complexes before addition to the cells. The plate was gently
swirled to ensure uniform distribution of the transfection
complexes. Then, the cells were incubated with the transfection
complexes at 37°C for 48 h before use in further experiments. The
negative control (cat. no. 339126; Qiagen, Inc.) used in this
experiment was scramble-miRNA control (negative control A miRCURY
LNA miRNA inhibitor control), which demonstrated no hits of >70%
homology for any sequence in any organism in the NCBI (https://www.ncbi.nlm.nih.gov/) and miRBase (https://www.mirbase.org/) databases. The sequences of
negative control and miR-222-3p inhibitor were
5′-TAACACGTCTATACGCCCA-3′ and 5′-CCCAGTAGCCAGATGTAGC-3′,
respectively. To confirm miR-222-3p inhibition, qPCR was performed
at 48 h post-transfection. The relative expression levels of
miR-222-3p were compared between the miR-222-3p inhibitor and
scramble control transfected cells. The expression levels of
miR-222-3p of miRNA inhibitor-transfected cells were relative to
those of the scramble control cells. The expression levels of the
scramble control cells were set as 100%.
Trypan blue exclusion assay
The transfected cells were collected at 48 h
post-transfection and re-plated (100,000 cells/well) into 12-well
plates for 5 days. The cells were incubated at 37°C in a 5%
CO2 incubator and counted at days 1, 2, 3, 4 and 5. At
each time point, the cells were trypsinized using 0.25%
Trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 3
min and resuspended with 900 µl culture media. Cell suspension (100
µl) was mixed with 100 µl 0.4% trypan blue (Gibco; Thermo Fisher
Scientific, Inc.) to obtain a 1:2 dilution. The mixture was
incubated at room temperature for 5 min to allow the trypan blue to
stain the dead cells. Subsequently, the cells were counted using a
hemocytometer under a light microscope. The results are presented
as the mean cell density (cells/ml) ± SD.
Migration assay
The migration assay was performed using Transwell
inserts (6.5 mm diameter, polyvinylpyrrolidone-free polycarbonate
filters with 8 µm pore size; Corning, Inc.). A total of
1.2×105 cells were re-suspended in 200 µl serum-free
DMEM and plated into the upper chamber of the Transwell insert. A
total of 600 µl DMEM supplemented with 10% (v/v) FBS (Gibco; Thermo
Fisher Scientific, Inc.) was added to the lower chamber to induce
cell migration. Following 6 h of incubation at 37°C, the migrated
cells were fixed using 4% paraformaldehyde in 1X PBS at room
temperature for 20 min and stained using 0.5% crystal violet in 25%
methanol as previously described (14). The number of migrated cells was
counted in 5 different randomly selected fields of view under a
light microscope, and the numbers in the miR-222-3p knockdown and
scrambled control groups were compared. The data are presented as
the mean ± standard deviation (miR-222-3p knockdown cells vs.
scrambled control cells).
miRNA target prediction
The mRNA targets of miR-222-3p were predicted using
three programs, including TargetScan 8.0 (15), miRDB (16) and PicTar (17). The total context++ score was
calculated using TargetScan 8.0 (15,18),
and the mRNA targets were ranked from the lowest to the highest
total context++ score (Table
SII).
Statistical analysis
The results were obtained from two independent
experiments, each in duplicate. All values are presented as mean ±
standard deviation. The statistical analysis was performed using
unpaired Student's t-test, Mann-Whitney test and one-way ANOVA
followed by Tukey's multiple comparison test. Statistical analysis
was performed using GraphPad Prism version 5.0 software
(Dotmatics). P<0.05 was considered to indicate a statistically
significant difference.
Results
Differential miRNA expression profiles
in MCF-7 and MDA-MB-231 cell lines
To determine the miRNA expression profiles
associated with the invasive phenotype, an miRNA array was used to
assess the MCF-7 and MDA-MB-231 breast cancer cell lines. Due to
the different characteristics of the two cell lines, notably the
degree of metastatic potential, it was hypothesized that miRNAs
involved in metastasis would be highly expressed in MDA-MB-231
cells. A total of 46 miRNAs were demonstrated to be differentially
expressed between these two cell lines. A total of 16 miRNAs were
upregulated and 30 miRNAs were downregulated in MDA-MB-231 cells
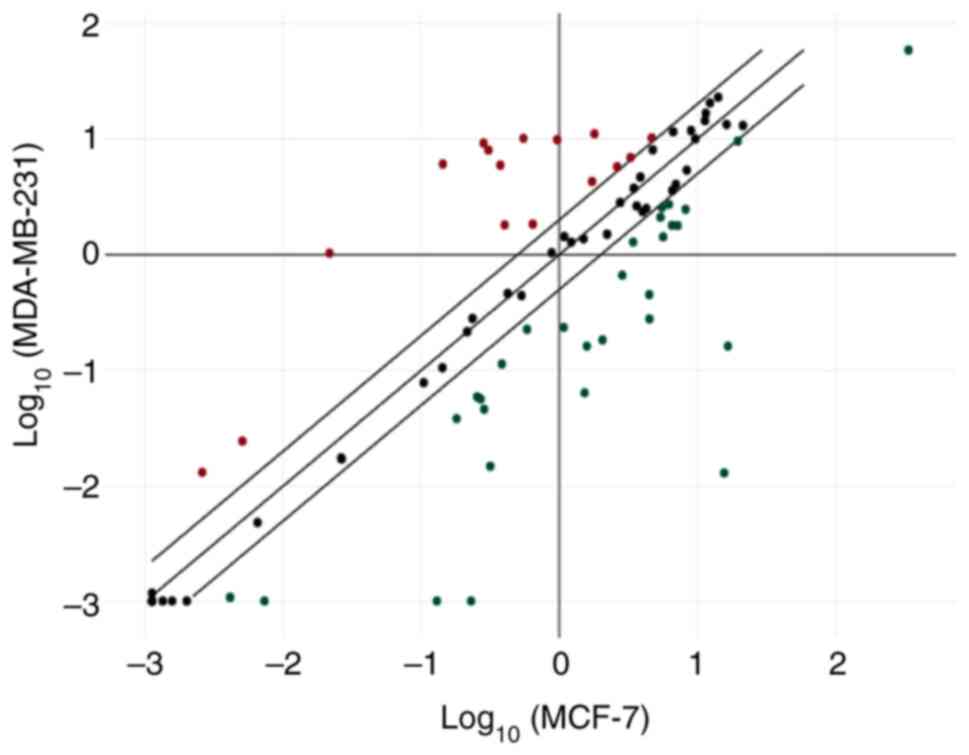
compared with MCF-7 cells (Fig. 1).
Details of the upregulated and downregulated miRNAs are presented
in Tables I and II, respectively.
 | Table I.Up-regulated miRNAs in the highly
invasive MDA-MB-231 breast cancer cell line compared with the
non-invasive MCF-7 breast cancer cell line. |
Table I.
Up-regulated miRNAs in the highly
invasive MDA-MB-231 breast cancer cell line compared with the
non-invasive MCF-7 breast cancer cell line.
| miRNA name | Fold change |
|---|
| hsa-miR-100-5p | 32.51 |
|
hsa-miR-106a-5p | 2.19 |
|
hsa-miR-125b-5p | 18.4 |
|
hsa-miR-130a-3p | 47.45 |
| hsa-miR-10b-5p | 5.06 |
|
hsa-miR-146a-5p | 42.31 |
| hsa-miR-17-5p | 2.11 |
| hsa-miR-18a-5p | 2.86 |
| hsa-miR-10a-5p | 4.83 |
| hsa-miR-20a-5p | 2.19 |
| hsa-miR-221-3p | 26.09 |
| hsa-miR-222-3p | 15.85 |
| hsa-miR-29a-3p | 10.22 |
| hsa-miR-29b-3p | 4.50 |
| hsa-miR-29c-3p | 6.16 |
| hsa-miR-30c-5p | 2.48 |
 | Table II.Down-regulated miRNAs in the highly
invasive MDA-MB-231 breast cancer cell line compared with the
non-invasive MCF-7 breast cancer cell line. |
Table II.
Down-regulated miRNAs in the highly
invasive MDA-MB-231 breast cancer cell line compared with the
non-invasive MCF-7 breast cancer cell line.
| miRNA name | Fold change |
|---|
|
hsa-miR-200a-3p | −23.78 |
| hsa-let-7f-5p | −2.29 |
| hsa-miR-101-3p | −4.57 |
|
hsa-miR-106b-5p | −4.02 |
| hsa-miR-126-3p | −3.38 |
| hsa-miR-141-3p | −1,201.53 |
| hsa-miR-145-5p | −3.78 |
| hsa-miR-26a-5p | −2.66 |
| hsa-miR-15a-5p | −2.17 |
| hsa-miR-149-3p | −7.22 |
| hsa-miR-16-5p | −2.04 |
| hsa-miR-182-5p | −11.20 |
| hsa-miR-191-5p | −3.95 |
| hsa-miR-192-5p | −4.31 |
|
hsa-miR-148a-3p | −2.58 |
| hsa-miR-194-5p | −6.19 |
| hsa-miR-195-5p | −21.28 |
|
hsa-miR-196a-5p | −127.68 |
|
hsa-miR-200b-3p | −16.14 |
|
hsa-miR-200c-3p | −102.39 |
| hsa-miR-205-5p | −225.92 |
| hsa-miR-21-5p | −5.75 |
| hsa-miR-215-5p | −4.70 |
| hsa-miR-25-3p | −3.66 |
| hsa-miR-26b-5p | −4.28 |
| hsa-miR-34a-5p | −9.75 |
| hsa-miR-7-5p | −9.92 |
| hsa-miR-9-5p | −4.77 |
| hsa-let-7e-5p | −2.56 |
| hsa-miR-93-5p | −3.32 |
Expression levels of miR-222-3p are
upregulated in MDA-MB-231 breast cancer cells
Based on the miRNA array, there were 16 upregulated
miRNAs in highly metastatic MDA-MB-231 cells compared with
low-metastatic MCF-7 cells, indicating that these miRNAs may be
associated with breast cancer metastasis (Table I). One candidate miRNA, miR-222-3p,
was validated in the present study. The miRNA array results
indicated that the miRNA expression levels of miR-222-3p were
upregulated in the MDA-MB-231 cell line; the levels were ~15-fold
higher than those noted in the MCF-7 cell line. Therefore,
miR-222-3p may be associated with the invasive phenotype of
MDA-MB-231 cells. The miRNA expression levels of miR-222-3p were
further assessed in MDA-MB-231 and MCF-7 cell lines using RT-qPCR.
The miR-222-3p expression levels in MDA-MB-231 cells were 250-fold
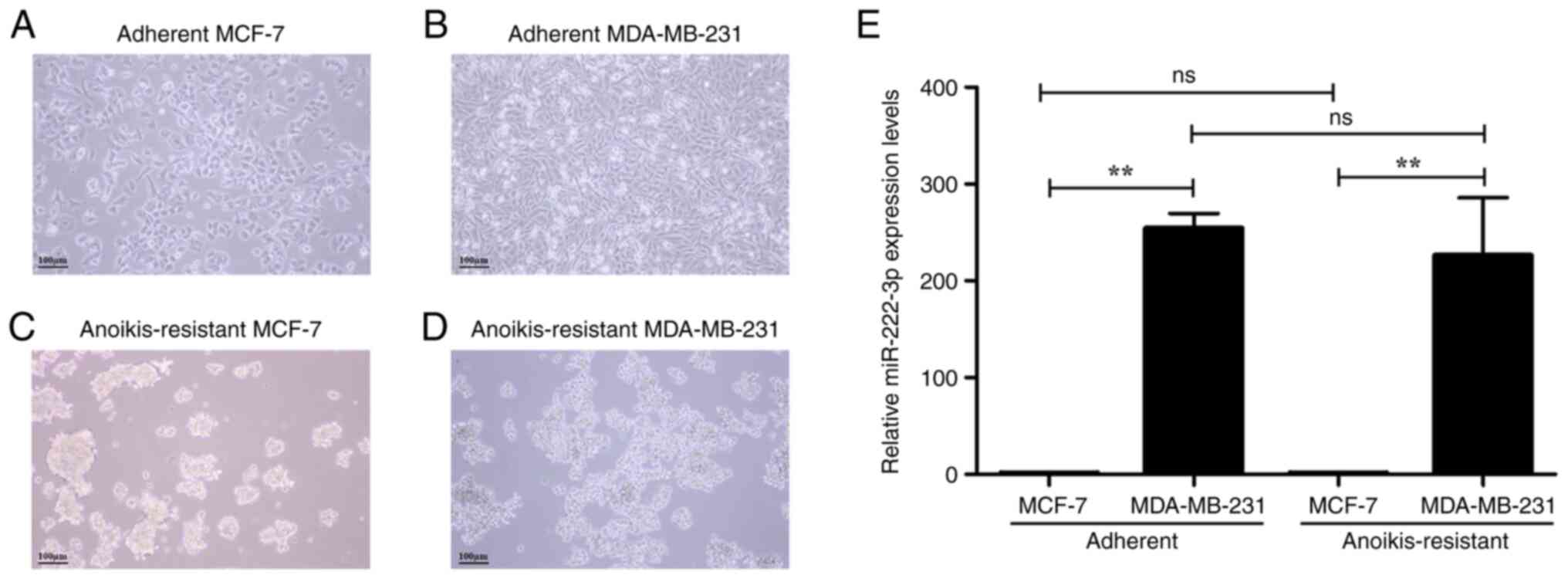
higher than those demonstrated in MCF-7 cells (Fig. 2E). The expression levels of
miR-222-3p were also determined under anoikis resistance
conditions. The anoikis-resistant cells were generated to mimic the
phenotype of those cancer cells which survive following detachment
from the primary site and remain in the circulation. MCF-7 and
MDA-MB-231 cells were cultured in an anti-adhesive polymer
(polyHEMA)-coated plate, which resulted in loss of cell attachment.
The cells which survived and proliferated in an
anchorage-independent manner were considered to be
anoikis-resistant cells (19). The
adherent MCF-7 and MDA-MB-231 cells had epithelial-like
morphologies. MCF-7 cells formed a monolayer and their shape was
dome-like (Fig. 2A) and MDA-MB-231
cells were spindle-shaped (long and thin) (Fig. 2B). The morphologies of MCF-7 and
MDA-MB-231 anoikis-resistant cells were round with spheroid shape
compared with their parental cell lines (Fig. 2C and D). Following two days of cell
culture under anoikis conditions, the anoikis-resistant cells were
collected to determine the expression levels of miR-222-3p. Under
anoikis-resistance conditions, MDA-MB-231 cells expressed
miR-222-3p at a similar level as that noted following cell culture
under the adherent conditions (Fig.
2E). The mRNA targets of miR-222-3p were predicted using
TargetScan 8.0 (15), miRDB
(16) and PicTar (17). TargetScan 8.0, miRDB and PicTar
predicted 254, 619, and 177 mRNA targets, respectively. A total of
25 mRNA targets were commonly identified by all three programs
(Fig. S1). Certain of these 25
mRNA targets were associated with key proteins involved in cell
proliferation and metastasis, such as cyclin-dependent kinase
inhibitor 1B (CDKN1B), eukaryotic translation initiation factor 5A2
(EIF5A2), iroquois homeobox 5 (IRX5), ADP-ribosylation factor 4
(ARF4), Bcl2 modifying factor (BMF), and estrogen receptor 1
(ESR1). The 25 common mRNA targets ranked from the lowest to the
highest total context++ score calculated using TargetScan 8.0
(15,18) are presented in Table SII.
Suppression of miR-222-3p reduces
proliferation in the MDA-MB-231 breast cancer cell line
The miRNA expression levels of miR-222-3p were
elevated in the MDA-MB-231 cell line compared with those of the
MCF-7 cell line (Fig. 2E). This
miRNA may contribute to the proliferative ability of MDA-MB-231
cells. Therefore, the present study assessed whether the
suppression of miR-222-3p expression in MDA-MB-231 cells affected
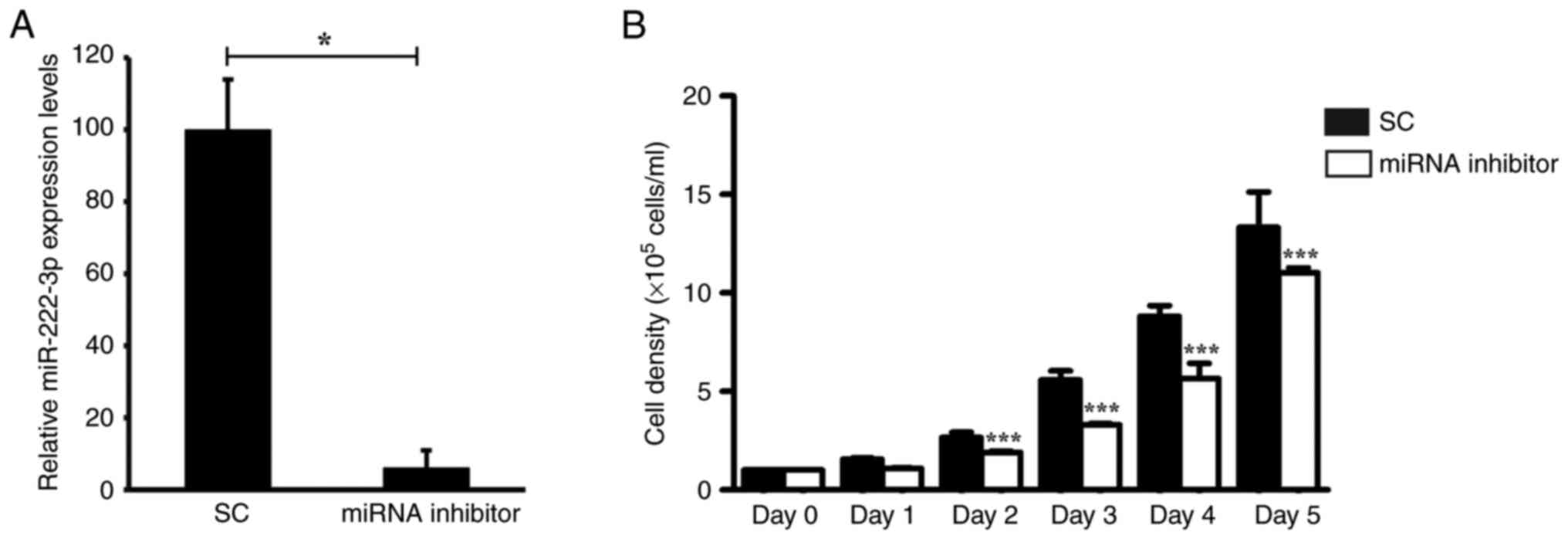
their proliferation. Transfection of the cells with the miR-222-3p
inhibitor markedly reduced the expression levels of miR-222-3p by
94% (Fig. 3A). At 48 h
post-transfection, the proliferative rate of miR-222-3p-knockdown
MDA-MB-231 cells was assessed for 5 days. MDA-MB-231 cells
transfected with the miR-222-3p inhibitor indicated a significant,
20–40%, reduction in cell viability from day 2 onward (Fig. 3B).
Suppression of miR-222-3p reduces
migration in highly metastatic breast cancer MDA-MB-231 cells
As the miRNA expression levels of miR-222-3p were
elevated in MDA-MB-231 cells, it was hypothesized that this miRNA
may be involved in the migratory ability of this cell line.
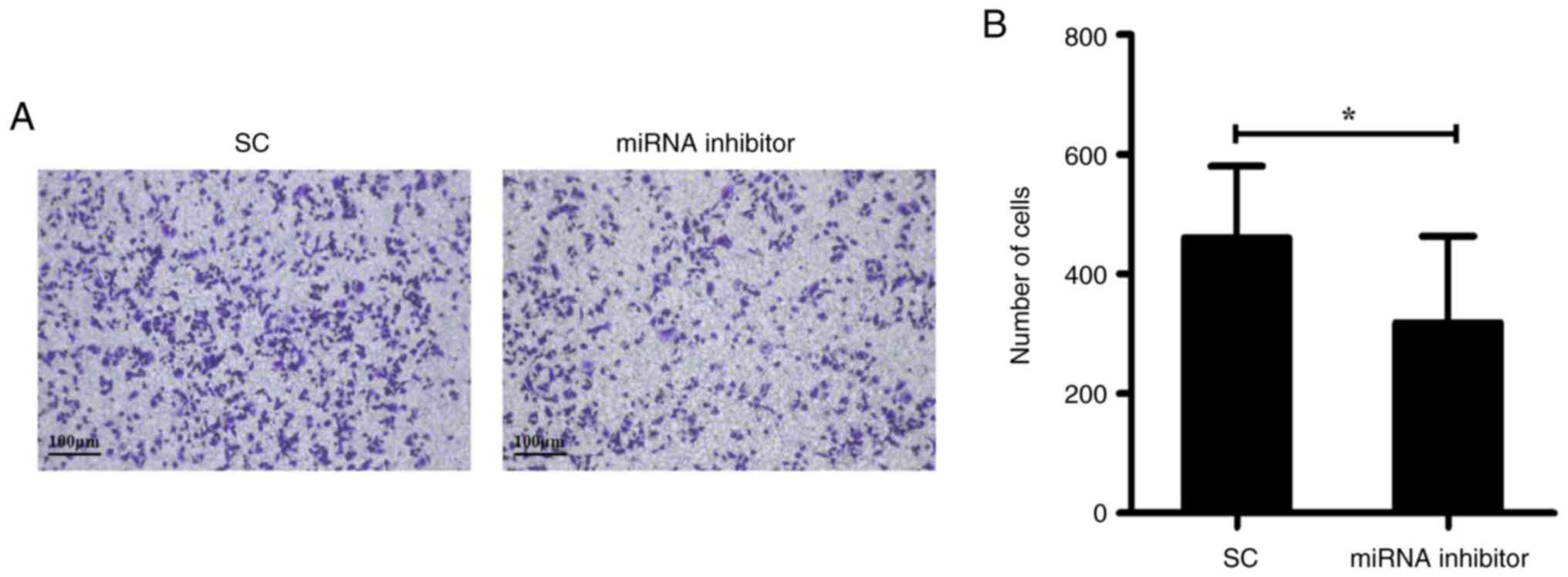
Endogenous miR-222-3p expression in MDA-MB-231 cells was suppressed
using a miR-222-3p inhibitor. At 48 h post-transfection,
miR-222-3p-knockdown-MDA-MB-231 cells demonstrated a significant,
30% reduction in migration (Fig.
4).
Discussion
miRNAs serve a crucial role in tumorigenesis
including proliferation, stress responses, cell adhesion, motility
and apoptosis (6). numerous miRNAs
have been reported to regulate metastasis in certain types of human
cancer (20). In the present study,
miRNA expression profiles were assessed in the highly metastatic
MDA-MB-231 cells and compared with those in the low-metastatic
MCF-7 cells. A total of 16 miRNAs were highly expressed in the
highly metastatic MDA-MB-231 cell line; however, their expression
levels were low in the MCF-7 cell line, which suggested that these
miRNAs may be associated with the aggressive breast cancer
phenotype. A total of 30 miRNAs were downregulated in MDA-MB-231
cells compared with MCF-7 cells. From the miRNA expression
profiles, these miRNAs could be considered as biomarkers for
metastatic breast cancer in the future. However, further studies
are required to validate the expression of these candidate miRNAs
in breast cancer cell lines and samples from patients with
different stages of breast cancer.
miR-221/222 has been previously reported to be an
onco-miR or a tumor suppressor miRNA, depending on the cellular
context (21). miR-222 has been
reported as an onco-miR in gastric cancer, bladder cancer, lung
cancer, colorectal cancer, cervical cancer and ovarian cancer;
whereas, it has been reported to act as a tumor suppressor miRNA in
tongue squamous cell carcinoma (TSCC), colorectal cancer, and
ovarian cancer (21). The onco-miRs
regulate tumor suppressor genes to promote cancer development,
while the tumor suppressor miRNAs target oncogenes to inhibit
cancer progression (22). In oral
TSCC (OTSCC), miR-222 serves a role as a tumor suppressor miRNA,
which inhibits OTSCC cell invasion by regulating matrix
metalloproteinase 1 (MMP1) expression. This is mediated by the
targeting of MMP1 mRNA and indirectly controlling its gene
expression via the targeting of manganese superoxide dismutase 2
(23). miR-222 serves oncogenic
roles in gastric cancer. Upregulation of miR-222 induces the cell
proliferation and invasion of the gastric cancer cell line SGC7901,
whereas suppression of miR-222 reverses these phenotypes via
induction of PTEN, a direct target of miR-222 (24). In colorectal cancer, miR-222
directly targets a disintegrin and metalloprotease 17, which is
downregulated in multidrug-resistant colorectal cancer cells and
increases cancer cell apoptosis (25). In contrast to these reports, miR-222
acts as an onco-miRNA in colorectal cancer cells by the direct
targeting of mammalian Ste20-like protein kinase 3 (26), and MIA SH3 domain ER export factor 3
(27) to promote cancer cell
migration and invasion.
The association of miR-222 and breast cancer has not
been clearly elucidated. A limited number of studies that examined
the expression of miR-222 in breast cancer have reported the roles
of miR-222 to be coupled with miR-221. Li et al (28) reported that the miRNA expression
levels of miR-221/222 were elevated in cisplatin-resistant
MDA-MB-231 cells and in patients with cisplatin-resistant breast
cancer. Suppression of miR-221/222 in MDA-MB-231 cells increased
their sensitivity to cisplatin in vitro and induced
apoptosis, which suggested that the combination of anti-miR-221 and
anti-miR-222 produced the synergistic effects noted following
cisplatin treatment (28).
Therefore, the present study further evaluated the expression and
functions of miR-222.
The miRNA array results indicated that miR-222-3p
expression was upregulated in MDA-MB-231 compared with MCF-7 cells.
The results from RT-qPCR analysis were consistent with the miRNA
array results, which indicated that miR-222 was expressed at higher
levels in MDA-MB-231 compared with MCF-7 cells. A previous study
also reported that miR-222 expression was upregulated in breast
cancer tissues compared with those in non-cancerous breast tissues
(11). Moreover, miR-222-3p
expression was previously reported to be elevated in the sera of
patients with breast cancer, which suggested that it could be used
as a non-invasive biomarker for human breast cancer. However, this
miRNA cannot be used as a biomarker to differentiate between early
stage and advanced stage breast cancer because the subjects of the
previous study were only from stages II and III of the disease; in
addition, the expression level of miR-222-3p was not significantly
different between stage II and stage III tumors (29). The results of the present study,
that a higher expression level of miR-222-3p was detected in
invasive breast cancer cells, were in line with a previous report
that the miR-222 level was elevated in patients with breast cancer
with lymphatic metastasis (30). It
should be noted that the present study indicated that miR-222-3p
expression was significantly higher in MDA-MB-231 cells, which
represented the highly metastatic model of breast cancer, compared
with the corresponding levels demonstrated in the non-metastatic
MCF-7 cells. This indicated the potential of miR-222-3p as a
prognostic biomarker for breast cancer metastasis. The present
study further examined other functions of this miRNA, including in
anoikis-resistance, proliferation and migration in the highly
metastatic MDA-MB-231 breast cancer cell line. Inhibition of
miR-222-3p in MDA-MB-231 cells suppressed the high proliferation
and migration, which are related to breast cancer metastasis.
Numerous miRNAs have been reported to promote or suppress anoikis
in certain types of cancer. miR-31, miR-220b and miR-200c have been
previously reported to enhance anoikis, while miR-181a promotes
anoikis-resistance in breast cancer (31). miR-141 has been reported to promote
anoikis resistance in ovarian cancer cells and was highly expressed
in anchorage-independent ovarian cancer cell lines compared with
anchorage-dependent cells (32).
However, the results of the present study demonstrated that the
expression levels of miR-222-3p in MDA-MB-231 cells under
anoikis-resistance condition were not higher than those in the
adherent MDA-MB-231 cells. Similarly, the levels of miR-222-3p in
anoikis-resistant MCF-7 cells were not higher than those in the
adherent MCF-7 cells. These results indicated that miR-222-3p was
not associated with the anoikis resistance of breast cancer.
Due to the high miRNA expression level of miR-222 in
patients with breast cancer and lymphatic metastasis and in the
MDA-MB-231 cell line, this miRNA may serve crucial roles in cell
proliferation and motility, which are the pre-requisites for cancer
progression. The present study demonstrated that proliferation of
miR-222-3p knockdown cells was suppressed by 20–40%, which
indicated that this miRNA supported the proliferative ability of
MDA-MB-231 cells. Moreover, suppression of miR-222-3p inhibited the
migratory ability of MDA-MB-231 cells by ~30%, which indicated that
this miRNA induced the migration of MDA-MB-231 cells. The migratory
results correlated with those of a previous study on the function
of exosomal miR-222, which reported that suppression of miR-222
reduced the migratory ability of MDA-MB-231 cells (30). The study also reported the decreased
invasive ability of miR-222 knockdown MDA-MB-231 cells (30). The present study demonstrated the
role of miR-222-3p in supporting cancer cell proliferation and to a
lesser extent migration. The increased expression of miR-222-3p in
the highly metastatic breast cancer cell line may also support
other breast cancer phenotypes, such as chemoresistance. According
to the functions of this miRNA in cell proliferation and migration
in breast cancer, the mRNA targets of miR-222-3p were predicted
using bioinformatics analysis (Fig.
S1); however, these require validation in future studies. Some
of the 25 mRNA targets were reported to be involved in cell
proliferation and metastasis in cancer. For example, CDKN1B/p27 has
been reported as the target of miR-221 which functions as an
oncogenic miRNA in hepatocarcinogenesis by promoting cell
proliferation and regulating the expression of cell-cycle
inhibitors (33). In addition, the
EIF5A2 gene has been reported as a direct target of miR-221-3p and
its expression level was decreased in medulloblastoma cell lines
(34). Overexpression of miR-221-3p
in these cell lines reduced their proliferation (34). The suppressive effect of miR-221-3p
on cell proliferation was reported to be alleviated by the
restoration of EIF5A2 in miR-221-3p-overexpressing DAOY cells
(34). Aberrant expression of IRX5
was previously reported in TSCC tissues and cell lines.
Overexpression of IRX5 promoted proliferation, migration, and
invasion of TSCC cells, whereas knockdown of IRX5 expression caused
the opposite effects (35). BMF was
reported as a target regulated by miR-221 using the dual-luciferase
reporter gene assay. The inhibition of miR-221 expression
significantly increased BMF expression in ovarian cancer SKOV3
cells, accompanied by decreased cell proliferation and increased
cell apoptosis (36). ESR1 has been
reported as the target of miR-222/221 and its expression levels
were reported to be markedly higher in ESR1 negative breast cancer
cells. In clinical samples, miR-222 expression was reported only in
TNBC, whereas miR-222 was absent in luminal A breast cancer, which
indicated that this miRNA acted cooperatively to decrease ERα
expression (37). ARF4 is one of
the putative targets of miR-221-3p. Overexpression of miR-221-3p
inhibited the proliferation and migration of epithelial ovarian
cancer (EOC) cells in vitro. The negative correlation
between ARF4 and miR-221-3p levels was previously reported in EOC
specimens, which suggested that the tumor suppressive role of
miR-221-3p in EOC may be via the direct targeting of ARF4 (38).
The analysis of additional upregulated miRNAs
demonstrated upregulation of miR-100-5p and 146a-5p in MDA-MB-231
cells compared with the corresponding miRNA expression levels
demonstrated in MCF-7 cells. Elevated miRNA expression levels of
miR-100-5p in post-neoadjuvant chemotherapy samples was reported to
be significantly correlated with improved event-free survival and
overall survival compared with the effects noted in normal samples
or samples with lower miRNA expression levels (39), which suggested its potential
application as a biomarker for predicting the outcome in patients
with early breast cancer. High miRNA expression levels of
miR-100-5p in insulin-like growth factor binding protein
6-knockdown MDA-MB-231 cells were reported to be correlated with
the decrease of insulin receptor and cyclin D1 genes which were
associated with the insulin-like growth factor signaling pathway
and the proliferative and migratory activity during the metastatic
cascade in breast cancer (40).
Upregulation of miR-146a-5p expression modulated by the
methyltransferase 14, N6-adenosine-methyltransferase subunit
promoted cell migration and invasion by breast cancer cells
(41). Downregulation of
miR-130a-3p was previously evaluated in TNBC cells and was compared
with the expression noted in normal cells (42), while the present study showed that
miR-130a-3p expression was upregulated in TNBC MDA-MB-231 cells
compared with the MCF-7 luminal A subtype cells
(ER+/PR+/HER2−). Overexpression of
miR-130a-3p was reported to reduce the proliferation,
anchorage-independent growth and migratory activity by
downregulating the Wnt signaling cascade in TNBC cells (42). Therefore, the function of
miR-130a-3p warrants further evaluation.
With regard to the downregulated miRNAs, the present
study identified certain downregulated miRNAs, including miR-141-3p
and miR-205-5p, which have been reported to be associated with
pathogenic roles in previous studies. The expression levels of
miR-141-3p were downregulated in breast cancer compared with the
corresponding levels noted in adjacent non-tumor tissues (43). Overexpression of miR-141-3p has been
reported to inhibit cell proliferation, migration and invasion in
MCF-7 and MDA-MB-231 cells (43).
The expression of miR-205-5p was also reported to be downregulated
in breast cancer compared with normal breast tissues. miR-205-5p
exhibited tumor suppressive functions by inhibiting tumor growth
and metastasis in breast cancer (44). miR-205-5p was expressed at the
lowest level in TNBCs compared with other subtypes (44). A limitation of the present study was
the limited number of miRNAs detected on the miRNA array,
additional miRNAs that are involved in cancer metastasis may not
have been detected by the miRNA array used in the present
study.
In summary, the present study indicated that
miR-222-3p expression was upregulated in the highly metastatic
breast cancer MDA-MB-231 cell line, suggesting that this miRNA may
be considered as a potential biomarker of metastatic breast cancer
in the future. Suppression of miR-222-3p lowered proliferation and
the migratory ability of MDA-MB-231 cells, which highlighted the
possible roles of miR-222-3p in supporting these processes during
cancer progression in high metastatic breast cancer. The present
study provided miRNA profiles that require further evaluation for
the determination of their functions in MDA-MB-231 cells.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Shaun R.
McColl (University of Adelaide, Adelaide, South Australia) for
providing the cell lines used in the present study. The authors
would also like to thank Professor Saovaros Svasti (Head of
Thalassemia Research Center, Institute of Molecular Biosciences,
Mahidol University, Nakhon Pathom, Thailand)for her general support
for this project.
Funding
This research project was supported by Mahidol University (grant
no. A42/2562) and the Institute of Molecular Biosciences (matching
fund).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PP designed and performed the experiments and
drafted the manuscript. CA provided suggestions regarding the
experimental design and data interpretation, discussed the results
and edited the manuscript. SJ contributed to the conception, design
of the works, provided advice and suggestions, and edited the
manuscript for important intellectual content. PP and CA confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miR/miRNA
|
microRNA
|
|
MCF-7
|
Michigan Cancer Foundation-7
|
|
MDA-MB-231
|
M.D. Anderson-Metastatic Breast
231
|
|
TNBC
|
triple-negative breast cancer
|
|
PR
|
progesterone receptor
|
|
ER
|
estrogen receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
polyHEMA
|
poly-2-hydroxyethyl methacrylate
|
|
LN
|
lymph node
|
|
cDNA
|
complementary DNA
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
CDKN1B
|
cyclin-dependent kinase inhibitor
1B
|
|
EIF5A2
|
eukaryotic translation initiation
factor 5A2
|
|
IRX5
|
iroquois homeobox 5
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
BMF
|
Bcl2 modifying factor
|
|
ESR1
|
estrogen receptor 1
|
|
has
|
Homo sapiens
|
|
TSCC
|
tongue squamous cell carcinoma
|
|
OTSCC
|
oral tongue squamous cell
carcinoma
|
|
MMP1
|
matrix metallo-proteinase 1
|
|
EOC
|
epithelial ovarian cancer
|
References
|
1
|
Breast cancer, . World Health
Organization. 2021.[updated 26 March 2021]. Available from:.
https://www.who.int/news-room/fact-sheets/detail/breast-cancer
|
|
2
|
Kennecke H, Yerushalmi R, Woods R, Cheang
MCU, Voduc D, Speers CH, Nielsen TO and Gelmon K: Metastatic
behavior of breast cancer subtypes. J Clin Oncol. 28:3271–3277.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al-Mahmood S, Sapiezynski J, Garbuzenko OB
and Minko T: Metastatic and triple-negative breast cancer:
Challenges and treatment options. Drug Deliv Transl Res.
8:1483–1507. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mayer IA, Abramson VG, Lehmann BD and
Pietenpol JA: New strategies for triple-negative breast
cancer-deciphering the heterogeneity. Clin Cancer Res. 20:782–790.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13:2152011. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu J, Wu KJ, Jia QJ and Ding XF: Roles of
miRNA and lncRNA in triple-negative breast cancer. J Zhejiang Univ
Sci B. 21:673–689. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang H, Xie J, Zhang M, Zhao Z, Wan Y and
Yao Y: miRNA-21 promotes proliferation and invasion of
triple-negative breast cancer cells through targeting PTEN. Am J
Transl Res. 9:953–961. 2017.PubMed/NCBI
|
|
8
|
Chen H, Pan H, Qian Y, Zhou W and Liu X:
MiR-25-3p promotes the proliferation of triple negative breast
cancer by targeting BTG2. Mol Cancer. 17:42018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu J, Xu J, Wu Y, Chen Q, Zheng W, Lu X,
Zhou C and Jiao D: Identification of microRNA-93 as a functional
dysregulated miRNA in triple-negative breast cancer. Tumour Biol.
36:251–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Meng Q, Pan A, Wu X, Cui J, Wang Y
and Li L: MicroRNA-455-3p promotes invasion and migration in triple
negative breast cancer by targeting tumor suppressor EI24.
Oncotarget. 8:19455–19466. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Amini S, Abak A, Estiar MA, Montazeri V,
Abhari A and Sakhinia E: Expression analysis of MicroRNA-222 in
breast cancer. Clin Lab. 64:491–496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Falkenberg N, Anastasov N, Rappl K,
Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler
H, et al: MiR-221/-222 differentiate prognostic groups in advanced
breast cancers and influence cell invasion. Br J Cancer.
109:2714–2723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Phannasil P, Thuwajit C, Warnnissorn M,
Wallace JC, MacDonald MJ and Jitrapakdee S: Pyruvate carboxylase is
up-regulated in breast cancer and essential to support growth and
invasion of MDA-MB-231 cells. PLoS One. 10:e01298482015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McGeary SE, Lin KS, Shi CY, Pham TM,
Bisaria N, Kelley GM and Bartel DP: The biochemical basis of
microRNA targeting efficacy. Science. 366:eaav17412019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48(D1): D127–D131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, Da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akekawatchai C, Roytrakul S, Kittisenachai
S, Isarankura-Na-Ayudhya P and Jitrapakdee S: Protein profiles
associated with anoikis resistance of metastatic MDA-MB-231 breast
cancer cells. Asian Pac J Cancer Prev. 17:581–590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McGuire A, Brown JA and Kerin MJ:
Metastatic breast cancer: The potential of miRNA for diagnosis and
treatment monitoring. Cancer Metastasis Rev. 34:145–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song Q, An Q, Niu B, Lu X, Zhang N and Cao
X: Role of miR-221/222 in tumor development and the underlying
mechanism. J Oncol. 2019:72520132019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H
and Zhou X: MicroRNA-222 regulates cell invasion by targeting
matrix metalloproteinase 1 (MMP1) and manganese superoxide
dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines.
Cancer Genomics Proteomics. 6:131–139. 2009.PubMed/NCBI
|
|
24
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu K, Liang X, Shen K, Sun L, Cui D, Zhao
Y, Tian J, Ni L and Liu J: MiR-222 modulates multidrug resistance
in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell
Res. 318:2168–2177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo F, Zhou J, Wang S, Sun Z, Han Q and
Bai C: microRNA-222 promotes colorectal cancer cell migration and
invasion by targeting MST3. FEBS Open Bio. 9:901–913. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao H, Cong X, Zhou J and Guan M:
MicroRNA-222 influences migration and invasion through MIA3 in
colorectal cancer. Cancer Cell Int. 17:782017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Li Q, Lü J, Zhao Q, Li D, Shen L,
Wang Z, Liu J, Xie D, Cho WC, et al: Targeted inhibition of
miR-221/222 promotes cell sensitivity to cisplatin in
triple-negative breast cancer MDA-MB-231 cells. Front Genet.
10:12782020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Said MN, Muawia S, Helal A, Fawzy A, Allam
RM and Shafik NF: Regulation of CDK inhibitor p27 by microRNA 222
in breast cancer patients. Exp Mol Pathol. 123:1047182021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding J, Xu Z, Zhang Y, Tan C, Hu W, Wang
M, Xu Y and Tang J: Exosome-mediated miR-222 transferring: An
insight into NF-κB-mediated breast cancer metastasis. Exp Cell Res.
369:129–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Malagobadan S and Nagoor NH: Evaluation of
MicroRNAs regulating anoikis pathways and its therapeutic
potential. Biomed Res Int. 2015:7168162015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mak CS, Yung MM, Hui LM, Leung LL, Liang
R, Chen K, Liu SS, Qin Y, Leung TH, Lee KF, et al: MicroRNA-141
enhances anoikis resistance in metastatic progression of ovarian
cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer.
16:112017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fornari F, Gramantieri L, Ferracin M,
Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM,
Bolondi L and Negrini M: MiR-221 controls CDKN1C/p57 and CDKN1B/p27
expression in human hepatocellular carcinoma. Oncogene.
27:5651–5661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Cui H and Wang X: Downregulation
of EIF5A2 by miR-221-3p inhibits cell proliferation, promotes cell
cycle arrest and apoptosis in medulloblastoma cells. Biosci
Biotechnol Biochem. 83:400–408. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang L, Song F, Sun H, Zhang L and Huang
C: IRX5 promotes NF-κB signalling to increase proliferation,
migration and invasion via OPN in tongue squamous cell carcinoma. J
Cell Mol Med. 22:3899–3910. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie X, Huang Y, Chen L and Wang J: miR-221
regulates proliferation and apoptosis of ovarian cancer cells by
targeting BMF. Oncol Lett. 16:6697–6704. 2018.PubMed/NCBI
|
|
37
|
Cochrane DR, Cittelly DM, Howe EN,
Spoelstra NS, McKinsey EL, LaPara K, Elias A, Yee D and Richer JK:
MicroRNAs link estrogen receptor alpha status and Dicer levels in
breast cancer. Horm Cancer. 1:306–319. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y,
Xiao Q, Li T, Ouyang C, Hu Y, et al: MiR-221-3p targets ARF4 and
inhibits the proliferation and migration of epithelial ovarian
cancer cells. Biochem Biophys Res Commun. 497:1162–1170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fuso P, Di Salvatore M, Santonocito C,
Guarino D, Autilio C, Mulè A, Arciuolo D, Rinninella A, Mignone F,
Ramundo M, et al: Let-7a-5p, miR-100-5p, miR-101-3p, and
miR-199a-3p hyperexpression as potential predictive biomarkers in
early breast cancer patients. J Pers Med. 11:8162021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Poloznikov AA, Nikulin SV, Raigorodskaya
MP, Fomicheva KA, Zakharova GS, Makarova YA and Alekseev BY:
Changes in the metastatic properties of MDA-MB-231 cells after
IGFBP6 gene knockdown is associated with increased expression of
miRNA genes controlling INSR, IGF1R, and CCND1 genes. Bull Exp Biol
Med. 166:641–645. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yi D, Wang R, Shi X, Xu L, Yilihamu Y and
Sang J: METTL14 promotes the migration and invasion of breast
cancer cells by modulating N6-methyladenosine and has-miR-146a-5p
expression. Oncol Rep. 43:1375–1386. 2020.PubMed/NCBI
|
|
42
|
Poodineh J, Sirati-Sabet M, Rajabibazl M
and Mohammadi-Yeganeh S: MiR-130a-3p blocks Wnt signaling cascade
in the triple-negative breast cancer by targeting the key players
at multiple points. Heliyon. 6:e054342020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun S, Ma J, Xie P, Wu Z and Tian X:
Hypoxia-responsive miR-141-3p is involved in the progression of
breast cancer via mediating the HMGB1/HIF-1α signaling pathway. J
Gene Med. 22:e32302020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xiao Y, Humphries B, Yang C and Wang Z:
MiR-205 dysregulations in breast cancer: The complexity and
opportunities. Noncoding RNA. 5:532019.PubMed/NCBI
|