Introduction
Solute carrier family 17 member 9 (SLC17A9) is
localized in lysosomes and its gene encodes for a vesicular
nucleotide transporter protein, a member of the transmembrane
protein family (1). It is involved
in small molecule transportation in cells, specifically the active
transport of ATP to lysosomes. Therefore, SLC17A9 dysfunction
reduces ATP accumulation in the lysosomes, leading to cell death
(1). Furthermore, studies have
demonstrated that SLC17A9 is critically involved in cell viability
and the physiology of lysosomes (1,2).
In colorectal cancer, a correlation was observed
between enhanced SLC17A9 expression levels and a number of
clinical as well as pathological characteristics of patients. In
addition, the overall survival (OS) and disease-free survival of
patients with colorectal cancer expressing high SLC17A9
levels in tumor tissues were poor (3). Studies have demonstrated that the
survival of patients with gastric cancer expressing high
SLC17A9 levels was poor (4,5).
SLC17A9 affects liver hepatocellular carcinoma (LIHC)
progression. It is involved in the infiltration of immune cells
into tumors and ferroptosis. Furthermore, a decrease in
SLC17A9 expression levels inhibited the proliferation,
migration and colony formation of HepG2 cells (6). By contrast, low SLC17A9
expression promotes prostate cancer cell proliferation, migration
and invasion, and inhibits cell apoptosis (7). Taken together, this indicates the
involvement of SLC17A9 in cancer; however, its influence on
the prognosis of patients has remained elusive.
Therefore, in the present study, the SLC17A9
expression levels were examined in 34 types of cancer. Next, the
association between SLC17A9 expression levels and the
prognosis of patients, stemness indices, immunity and drug
sensitivity in these types of cancer were determined. Finally, Cell
Counting Kit-8 (CCK-8) and colony-formation assays were performed
to determine the effect of SLC17A9 expression levels on
osteosarcoma (OSS) cells. The results of the present study
indicated that SLC17A9 may serve as a prognostic marker and
a therapeutic target for OSS.
Materials and methods
SLC17A9 expression levels in a
pan-cancer panel
Standardized pan-cancer data were retrieved from
databases including The Cancer Genome Atlas (TCGA), Therapeutically
Applicable Research to Generate Effective Treatments (TARGET) and
Genotype-Tissue Expression using the University of California Santa
Cruz (UCSC) genome browser database (https://xenabrowser.net/datapages/) (8,9). The
abbreviations used for 34 types of cancer are presented in Table SI. The data on the SLC17A9
expression pattern in all samples were extracted and plotted using
the R software. However, the data on SLC17A9 expression levels in
corresponding healthy tissues for certain types of cancer from TCGA
were missing. Thus, data on the SLC17A9 expression levels in
these types of cancer and corresponding healthy tissues, including
adrenocortical carcinoma (ACC), lymphoid neoplasm diffuse large
B-cell lymphoma (DLBC), acute myeloid leukemia (LAML), low grade
glioma (LGG), ovarian serous cystadenocarcinoma, skin cutaneous
melanoma (SKCM), testicular germ cell tumors (TGCT), thymoma
(THYM), and uterine carcinosarcoma (UCS), were obtained from the
Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) database (10).
Analysis of the association between
SLC17A9 expression levels and survival of patients with cancer
To determine the association between SLC17A9
expression and survival outcomes of various types of cancer, the
OS, progression-free survival (PFS) and disease-specific survival
(DSS) data of patients were obtained from the TCGA and TARGET
cohorts. TCGA data including expression data and clinical data were
obtained from the University of California, Santa Cruz (UCSC)
database, which is an open, pubic database (https://xenabrowser.net/datapages/). In the UCSC
database, not all tumor types contained the three survival times
(OS, PFS, and DSS), some cancer types only contained OS or PFS.
Survival analysis was performed using the Kaplan-Meier (KM) method
with the median of SLC17A9 expression as the cut-off value and the
survival curves were plotted using the survival package in R. In
addition, only when P was less than 0.05 can the survival curves be
graphed. Next, a univariate Cox regression analysis was performed
using the forestplot R package to determine the association between
SLC17A9 expression levels and survival. In addition, the
association between SLC17A9 expression levels and the
survival of patients in the immunotherapy cohort was determined
using KM plotter (https://kmplot.com/analysis/) (11). The detailed steps were as follows:
i) Enter the KM plotter website and select the ‘start KM plotter
for immunotherapy’ button; ii) enter the gene symbol ‘SLC17A9’ and
select the cut-off value ‘median’; and iii) select survival ‘OS or
PFS’, select follow-up threshold ‘all’, select Anti-PD-L1 treatment
‘all’, select tumor type ‘all, bladder...’.
Analysis of the stemness index
Stemness refers to the self-renewal and
dedifferentiation properties of cells, which aid in the progression
and invasion of cancer cells, thus resulting in poor prognosis of
the patient (12). The two
independent stemness indices are the mRNA expression-based stemness
index (RNAss), which demonstrates gene expression, and the DNA
methylation-based stemness index (DNAss), which reflects epigenetic
features. The stemness scores of the patients were obtained from
the TCGA cohort based on the UCSC database. The correlation between
SLC17A9 expression levels and the stemness indices was
determined by Spearman correlation analysis.
Analysis of tumor immunity
The tumor microenvironment (TME) serves a crucial
role in cancer progression. The proportion of stromal and immune
cells in the TME of patients was determined from the TCGA cohort by
calculating the estimation of stromal and immune cells in malignant
tumor tissues using expression data (ESTIMATE) score (13) determined with the ESTIMATE algorithm
(https://bioinformatics.mdanderson.org/public-software/estimate/).
In addition, the cell-type identification by estimating relative
subsets of RNA transcripts (CIBERSORT) tool (https://cibersortx.stanford.edu/) was used to
determine the infiltration status of tumor-infiltrating immune
cells (TIICs) in 34 types of cancer (14). Subsequently, the effect of
SLC17A9 expression on the TME and TIICs was determined.
Drug activity analysis
To predict potential drugs targeting SLC17A9,
the correlation between SLC17A9 expression levels and drug
sensitivity was determined by Pearson's correlation coefficient.
Furthermore, the CellMiner database (https://discover.nci.nih.gov/cellminer/) was examined
to obtain data on drug activity (15).
Gene Ontology (GO), Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway enrichment and gene set
variation analyses (GSVA)
The clusterProfiler R package (16) was employed to perform the GO and
KEGG pathway enrichment analyses to determine the functions and
pathways enriched by SLC17A9. Next, the GSVA R package
(17) was used to estimate the
variations in key gene sets in patients with OSS.
Cell culture and transfection
OSS cells were obtained from The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences. U2OS, Saos2,
MG63 and HOS cells were cultured in Dulbecco's Modified Eagle's
Medium supplemented with 10% fetal bovine serum (both from
Biological Industries), and 143 B cells were cultured in Modified
Eagle's Medium (Biological Industries) supplemented with 10% fetal
bovine serum. All cells were incubated at 37°C in a humidified
atmosphere containing 5% CO2. GV492 was used as the vector for
overexpression and empty plasmid, and SLC17A9 overexpression (OE)
and empty (NC) vectors were constructed and provided by Shanghai
GeneChem Co., Ltd. Small interfering RNAs (siRNAs) targeting
SLC17A9 (si-1, stB0012921A and si-2, stB0012921B) and
negative control siRNA (si-nc, siN0000001-1-5) were designed and
synthesized by Guangzhou RiboBio Co., Ltd. The si-1 and si-2 target
sequences are presented in Table
SII. These vectors or siRNAs were separately transfected into
cells with the aid of Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) or riboFECT CP Transfection Kit (cat. no.
C10511-05; Guangzhou RiboBio Co., Ltd). HOS and MG63
(1×106/well) cells were seeded into a 6-well plate.
After 24 h, vectors and Lipofectamine 3000 (or siRNAs and riboFECT
CP reagent) were mixed and left at room temperature for 15 min,
then the mixture was added to each well of the 6-well plate. MG63
cells were transfected with 5 µg vector in each well. HOS cells
were transfected with 100 nM siRNA per well. The transfected cells
were incubated for 48 h at 37°C in a humidified atmosphere
containing 5% CO2, and then used in subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR) and western blotting (WB)
RT-qPCR and WB were performed as described
previously (18). RT-qPCR reagents
such as AG RNAex Pro Reagent, SYBR Green Premix Pro Taq HS qPCR kit
and the Evo M-MLV RT Premix kit were purchased from Accurate
Biology. The sequences of the primers used are presented in
Table SII. The reagents used to
perform WB, including RIPA buffer and BeyoECL Plus kit, were
purchased from Beyotime Institute of Biotechnology. Furthermore,
anti-GAPDH (1:1,000; cat. no. 10494-1-AP) and anti-SLC17A9 (1:500;
cat. no. 26731-1-AP) antibodies were purchased from Wuhan Sanying
Biotechnology, and the HRP-conjugated Affinipure Goat Anti-Rabbit
IgG antibody (1:1,000; cat. no. SA00001-2) was purchased from
Proteintech Group, Inc (Wuhan Sanying Biotechnology).
Cell-proliferation and
colony-formation assays
The proliferation of cells was determined using the
CCK-8 assay (Beyotime Institute of Biotechnology) following the
manufacturer's protocols. For this assay, 2×103
cells/well were seeded and cultured in a 96-well plate at 37°C for
0, 24, 48 and 72 h. For the colony-formation assay, 1,000
cells/well were seeded and incubated in a 6-well plate and
incubated for 14 days, with the media being changed every 2 days.
Next, the colonies were stained at room temperature with 1% crystal
violet for 20 min. The colonies that had formed (colonies with ≥50
cells) were manually counted under a microscope.
Statistical analysis
Bioinformatics and statistical analyses, such as the
normalization and transformation of RNA sequencing data, and the
correlation, survival, CIBERSORT, ESTIMATE, GSVA, GO and KEGG
analyses, were performed using the R software (version 4.0.0;
http://www.r-project.org/). Furthermore,
the coxph algorithm of the survival R package was used to perform
univariate and multivariate Cox regression analyses. Pearson
correlation analysis was performed to determine the correlation
between SLC17A9 expression levels and the activity of the
drugs from the CellMiner database. All quantitative data from in
vitro experiments are expressed as the mean ± standard
deviation of three independent experiments. One-way analysis of
variance followed by Tukey's post hoc test was performed using the
GraphPad Prism 8.0 Software (Dotmatics) to determine differences
across groups. A two-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
SLC17A9 expression levels in
pan-cancer panel
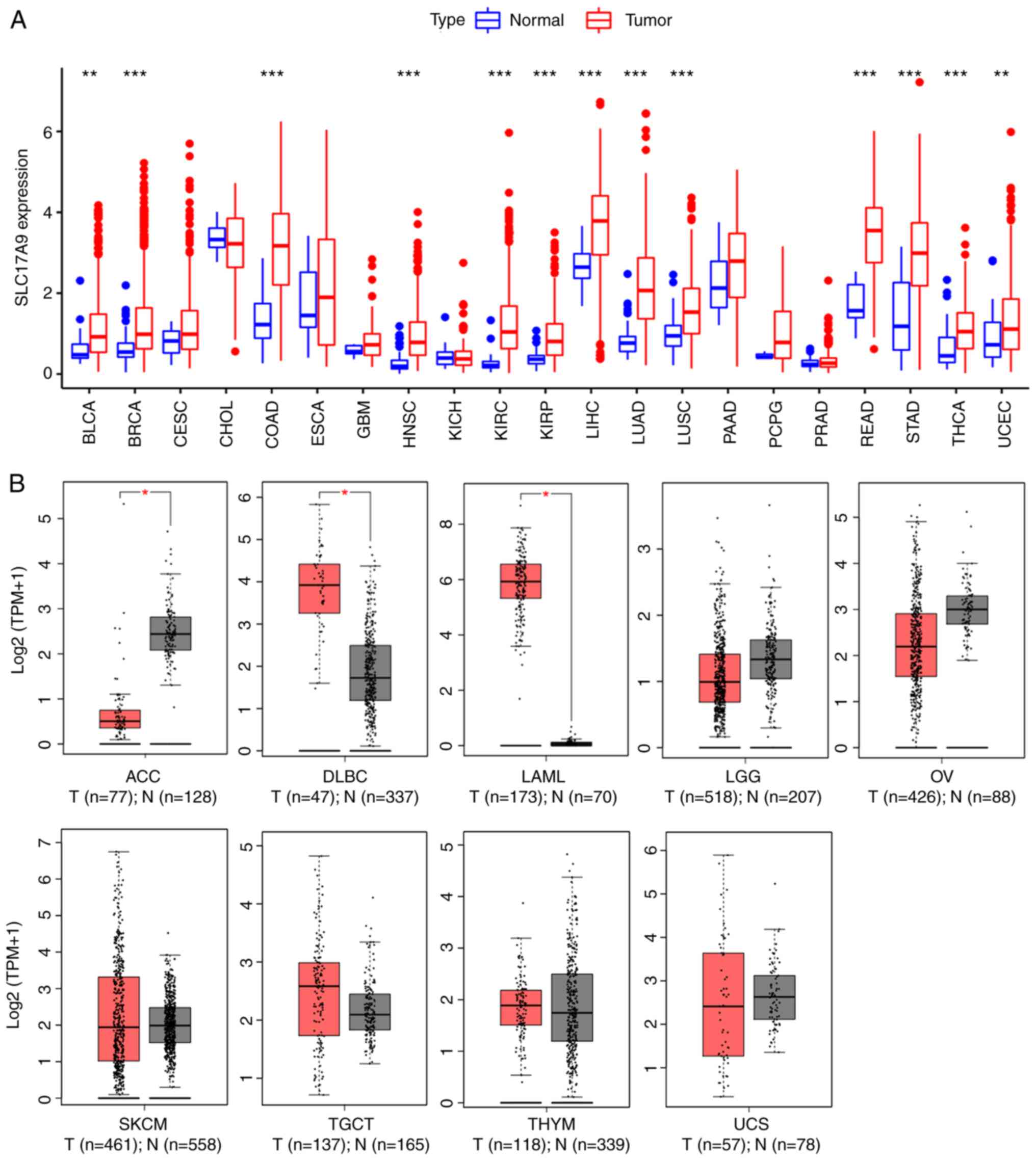
The differences in SLC17A9 expression
profiles between pan-cancer tumor and normal samples were analyzed
after obtaining data of the SLC17A9 expression profiles for
pan-cancer using publicly available data. A significant increase in
SLC17A9 expression levels was observed in 15 types of cancer
(Fig. 1A and B), including LUAD,
COAD, BRCA, KIRP, DLBC, head and neck squamous cell carcinoma
(HNSC), LAML, KIRC, LIHC, BLCA, READ, STAD, LUSC, THCA and UCEC.
However, a significant decrease in SLC17A9 expression levels
was observed in the tissues of patients with ACC (Fig. 1B).
Significance of SLC17A9 expression
levels in predicting the prognosis of patients
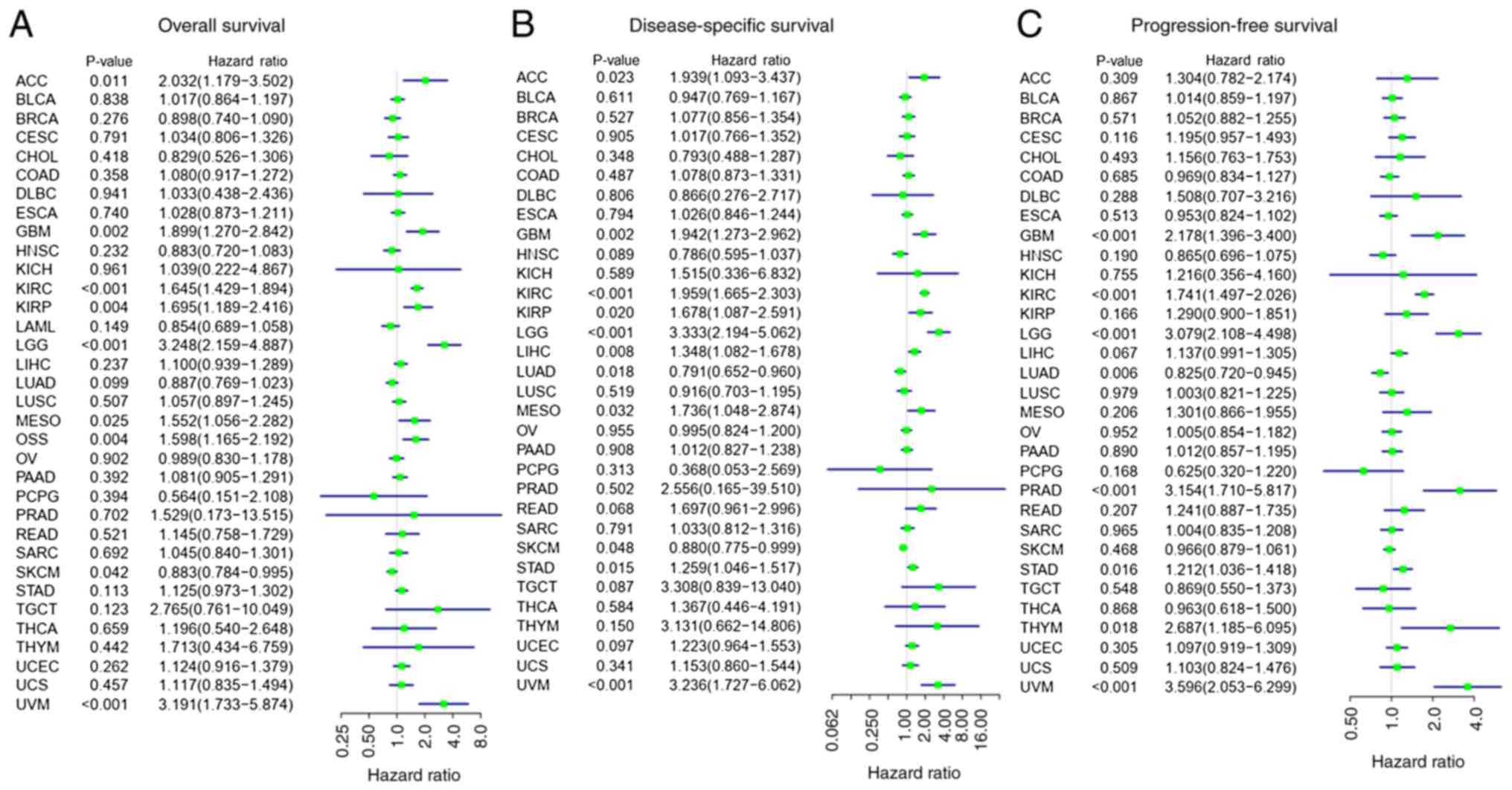
Univariate Cox regression and survival analyses were
performed to determine the association between SLC17A9
expression levels and the prognosis of patients in 34 types of
cancer. The univariate Cox regression analysis demonstrated that
SLC17A9 expression was an independent prognostic factor
associated with the OS of patients with various types of cancer,
such as OSS, ACC, LGG, KIRC, MESO, KIRP, SKCM and UVM (Fig. 2A). Furthermore, the KM analysis
demonstrated that patients with high SLC17A9 expression and
types of cancer such as KIRC, LGG, OSS or UVM had a reduced OS
duration compared with patients with low SLC17A9 expression.
On the contrary, the OS duration of patients with high
SLC17A9 expression levels and BRCA was increased compared
with that of patients with low SLC17A9 expression levels
(Fig. S1).
The SLC17A9 expression levels were an
independent risk factor for DSS in patients with types of cancer
such as KIRC, ACC, KIRP, LGG, MESO, LIHC, SKCM, LUAD, STAD and UVM
(Fig. 2B). KM analysis demonstrated
that the DSS duration of patients with high SLC17A9
expression levels and KIRC, LGG or UVM was reduced compared with
that of patients with low SLC17A9 expression levels.
However, the DSS duration of patients with high SLC17A9
expression levels and LUAD was increased compared with that of
patients with low SLC17A9 expression levels (Fig. S1). Forest plots demonstrated a
significant association between SLC17A9 expression levels
and the PFS of patients with glioblastoma (GBM), LGG, KIRC, LUAD,
PRAD, THYM, STAD and UVM (Fig. 2C).
In addition, KM analysis demonstrated that the PFS duration of
patients with high SLC17A9 expression levels and KIRC, LGG,
PRAD or UVM was reduced compared with that of patients with low
SLC17A9 expression levels (Fig.
S1). The DSS and PFS of patients with high SLC17A9
expression levels and LUAD were increased compared with those of
patients with low SLC17A9 expression levels.
The association between the SLC17A9
expression levels and the prognosis of patients from the
immunotherapy cohort was also investigated using the KM plotter
web-based tool. The results revealed that in several types of
cancer, including bladder cancer, esophaseal adenocarcinoma, GBM,
hepatocellular carcinoma, melanoma, HNSC, non-small cell lung
cancer, non-squamous lung carcinoma and urothelial cancer, the OS
of patients with high SLC17A9 expression levels in the
antibody directed against programmed cell death-1 ligand 1
(anti-PD-L1) therapy cohort was increased (Fig. S2A). Furthermore, a significant
association was observed between the SLC17A9 expression
levels and the OS of patients with bladder carcinoma (Fig. S2B) in the anti-PD-L1 therapy
cohort. Thus, SLC17A9 may be a potential prognostic marker
for various types of cancer.
Analysis of stemness indices and tumor
immunity
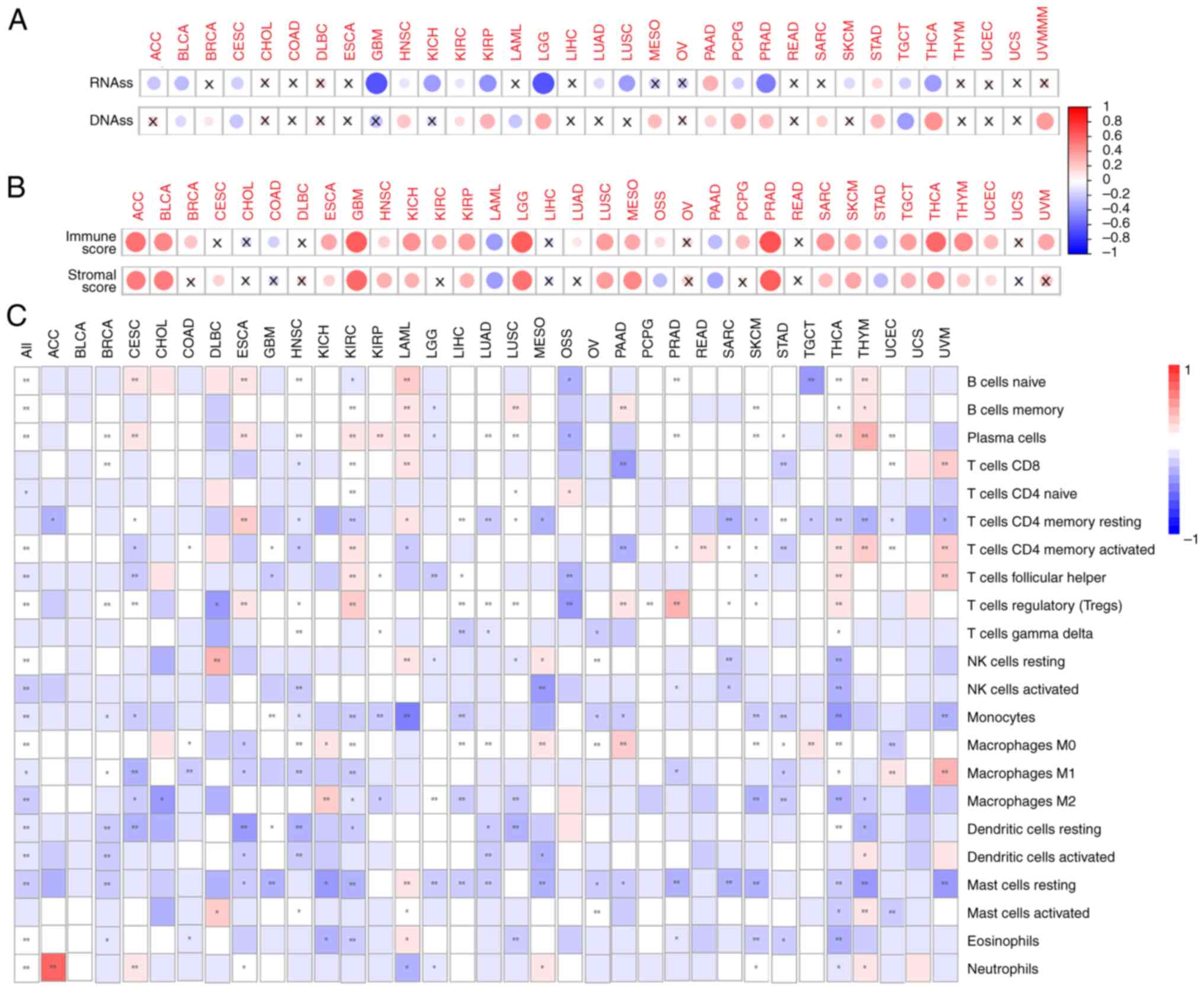
The correlation between the stemness indices and the
SLC17A9 expression levels exhibited differences among
various cancer types. A negative correlation was revealed between
SLC17A9 expression levels and the RNAss stemness index in
various types of cancer, such as ACC, CESC, GBM, BLCA, HNSC, KICH,
LUAD, KIRP, KIRC, LUSC, PCPG, LGG, PRAD, TGCT, SKCM and THCA.
However, a positive correlation was observed between SLC17A9
expression levels and the RNAss stemness index in other types of
cancer, such as PAAD and STAD (Fig.
3A). In addition, the SLC17A9 expression level was
negatively correlated with the DNAss stemness index in BLCA, CESC,
LAML and TGCT, and positively correlated in other types of cancer,
such as HNSC, BRCA, KIRC, LGG, MESO, KIRP, PAAD, THCA, PRAD, SARC,
PCPG, STAD and UVM (Fig. 3A).
The ESTIMATE algorithm was used to calculate the
stromal and immune scores of patients. A positive correlation was
observed between the SLC17A9 expression levels and the
immune score of patients with BRCA, KIRC, GBM, ACC, HNSC, ESCA,
UCEC, KIRP, LGG, KICH, SKCM, LUAD, BLCA, MESO, OSS, PCPG, SARC,
LUSC, TGCT, PRAD, THYM, THCA and UVM. However, a negative
correlation was observed between the SLC17A9 expression
level and the immune score of patients with COAD, LAML, PAAD and
STAD (Fig. 3B). Furthermore, the
results revealed a positive correlation between the SLC17A9
expression level and the stromal scores of patients with ACC, BLCA,
ESCA, ESCA, KICH, GBM, HNSC, KIRP, LGG, LUSC, MESO, PRAD, SKCM,
TGCT, THYM, SARC, THCA and UCEC. However, a negative correlation
was observed between the SLC17A9 expression level and the
stromal scores of patients with LAML, OSS, PAAD and STAD (Fig. 3B). Taken together, these results
suggest a significant correlation between the SLC17A9
expression level and stemness indices such as RNAss and DNAss, as
well as the immune and stromal scores of patients with BLCA, HNSC,
LGG, PAAD, TGCT, PRAD, KIRP, STAD and THCA.
Next, whether there was a correlation between
SLC17A9 expression and the levels of 22 types of TIIC was
determined. The results demonstrated a positive correlation between
SLC17A9 expression and nine TIICs, including plasma cells,
naïve and memory B cells, memory-activated CD4 T cells, resting
natural killer (NK) cells, regulatory T (Treg) cells, M0
macrophages, eosinophils and neutrophils. On the contrary, a
negative correlation was observed between SLC17A9 expression
levels and the levels of other types of immune cell, such as naïve
CD4 T cells, activated NK cells, T follicular helper (Tfh) cells,
monocytes, M1 and M2 macrophages, resting and activated dendritic
cells, and activated mast cells (Fig.
3C). Furthermore, differences in TIIC levels were observed
among patients with different types of cancer. In patients with ACC
or DLBC, there was a strong positive correlation between the
SLC17A9 expression levels and immune cells such as
neutrophils and resting NK cells, respectively. Furthermore,
SLC17A9 expression levels were revealed to be positively
correlated with resting memory CD4 T cells in ESCA and to be
negatively correlated with monocytes in LAML. In patients with
PRAD, THYM or UVM, there was a positive correlation between the
SLC17A9 expression levels and immune cells such as Treg
cells, plasma cells and M1 macrophages, respectively (Fig. 3C). However, no significant
correlation was observed between SLC17A9 expression levels
and the infiltration of 22 immune cell types in patients with BLCA
or UCS (Fig. 3C). These results
indicate a correlation between the SLC17A9 expression level
and the degree of immune-cell infiltration in different types of
cancer.
SLC17A9 enhances OSS cell
proliferation and viability
In the dataset GSE16088, an increase in
SLC17A9 expression levels was observed in the tissues of
patients with OSS compared with those in healthy bone tissues
(Fig. 4A). Furthermore, in the
TCGA-TARGET dataset, the OS (Figs.
4B and S1) and relapse-free
survival (Fig. 4B) of patients with
high SLC17A9 expression levels were poor compared with those
of patients with low SLC17A9 expression levels. Univariate
and multivariate Cox regression analyses of the TCGA-TARGET dataset
demonstrated that SLC17A9 may be an independent risk factor
for the OS of patients with OSS (Fig.
4C).
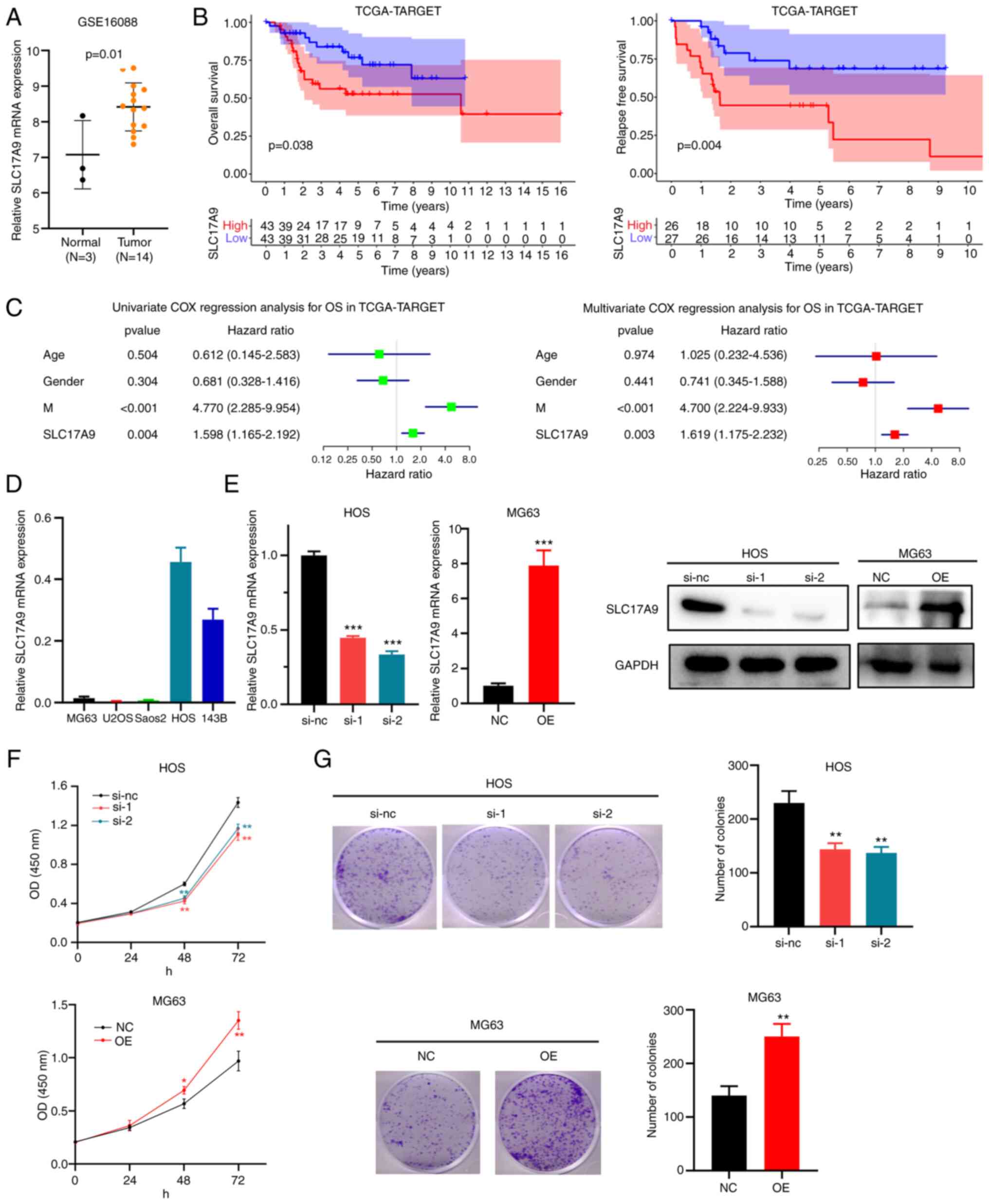 | Figure 4.Cellular functions of SLC17A9
in OSS. (A) Differential expression of SLC17A9 in tumor
tissues compared with normal tissues in OSS (accession no.
GSE16088). (B) Association between high SLC17A9 expression
levels and poor OS and relapse-free survival of patients from the
TCGA-TARGET cohort. (C) Univariate and multivariate Cox regression
analysis of the OS of patients from the TCGA-TARGET cohort. (D)
SLC17A9 expression levels in OSS cells. (E)
SLC17A9-knockdown HOS cells and SLC17A9-OE MG63 cells were
constructed. Reverse transcription-PCR and western blot analysis
were used to determine SLC17A9 expression levels in these cells.
(F) Effect of SLC17A9 on the proliferation of HOS and MG63
cells. (G) Effect of SLC17A9 on the viability of HOS and MG63
cells. **P<0.01 and ***P<0.001 vs. NC. SLC17A9, solute
carrier family 17 member 9; M, metastasis; OSS, osteosarcoma; OS,
overall survival; TCGA, The Cancer Genome Atlas; TARGET,
Therapeutically Applicable Research to Generate Effective
Treatments; si-nc, negative control small interfering RNA; si-1/2,
small interfering RNA targeting SLC17A9; NC, empty
overexpression vector; OE, overexpression; OD, optical density. |
Next, RT-qPCR was performed to determine
SLC17A9 expression levels in several OSS cell lines
(Fig. 4D). HOS and MG63 cells were
used for the subsequent experiments since SLC17A9 levels were high
in HOS and low in MG63 cells. HOS cells were transfected with
siRNAs (si-nc, si-1 and si-2) and MG63 cells were transfected with
SLC17A9-NC or SLC17A9-OE vectors. Finally, RT-qPCR and WB analyses
were performed to confirm the knockdown and overexpression of
SLC17A9 in HOS and MG63 cells, respectively (Fig. 4E). The results of the CCK-8 assay
revealed a decrease in the proliferation capacity of
SLC17A9-knockdown HOS cells (si-1 or si-2) compared with cells
transfected with si-NC. In addition, there was an increase in the
proliferation of MG63 cells transfected with the SLC17A9-OE vector
compared with the cells transfected with SLC17A9-NC (Fig. 4F). The colony-formation assays
indicated a considerable reduction in the colony number of HOS
cells with SLC17A9 knockdown, while there was a substantial
increase in the colony numbers of MG63 cells overexpressing SLC17A9
(Fig. 4G). These results indicate
that SLC17A9 has a role to enhance the proliferation and
viability of OSS cells. Thus, SLC17A9 may be significantly
involved in enhancing OSS progression.
GSVA and GO and KEGG pathway
enrichment analyses
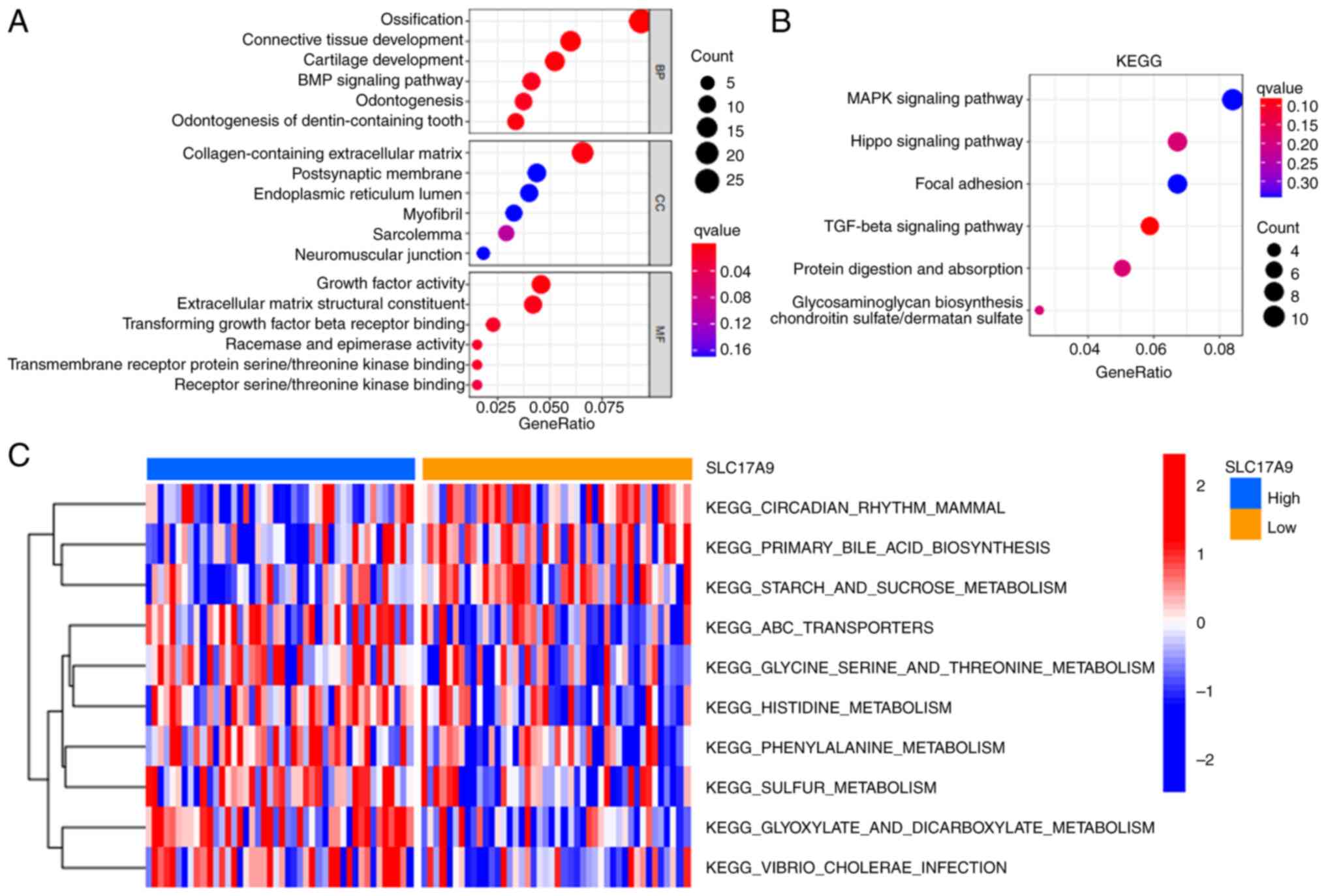
Next, GO and KEGG pathway enrichment analyses were
performed to determine the functions and pathways enriched by
SLC17A9 in OSS. Fig. 5A
reveals that SLC17A9 was enriched in biological processes
such as ossification, connective tissue and cartilage development,
the bone morphogenetic protein signaling pathway and odontogenesis.
In addition, SLC17A9 was enriched in cellular component
terms such as the collagen-containing extracellular matrix. The
molecular function terms enriched by SLC17A9 were
extracellular matrix structural constituents, receptor
serine/threonine kinase and TGF-β receptor binding, growth factors
activity, racemase and epimerase activity. The KEGG pathway
enrichment analysis demonstrated that SLC17A9 was enriched in the
MAPK, Hippo, focal adhesion and TGF-β signaling pathways (Fig. 5B). Finally, GSVA was performed to
compare the pathways enriched in patients with OSS with high or low
SLC17A9 expression levels. The GSVA results revealed significant
differences in pathways such as circadian rhythm in mammals, starch
and sucrose metabolism, ABC transporters, glycine serine/threonine,
histidine, phenylalanine, sulfur, glyoxylate and dicarboxylate
metabolism and Vibrio cholera infection between the high and
low expression groups (Fig.
5C).
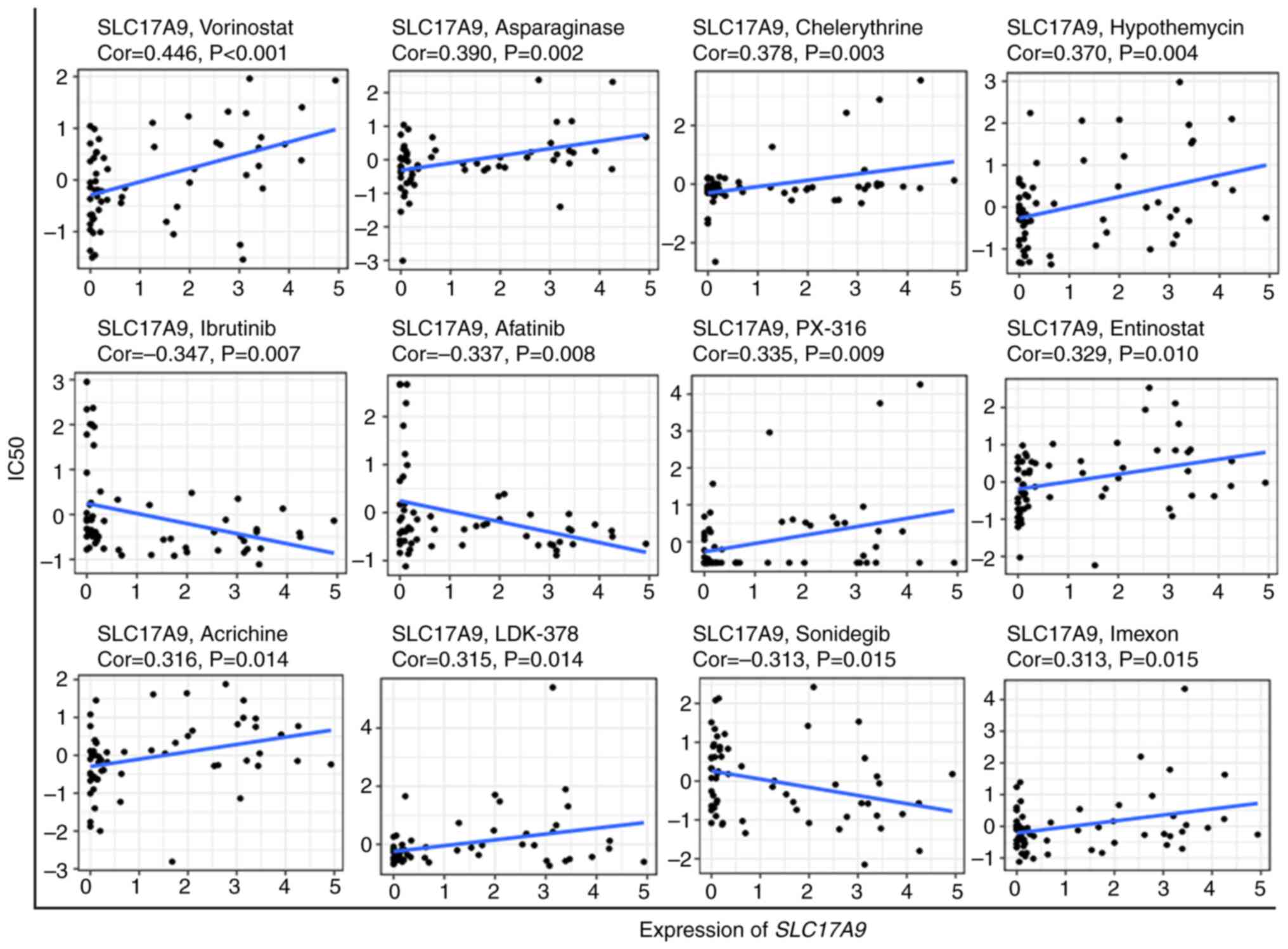
Drug sensitivity analysis
Data on 263 anticancer drugs were retrieved from the
CellMiner database. Next, Pearson correlation analysis was
performed to determine the correlation between SLC17A9
expression levels and the activity of these drugs in NCI-60 cells
(15). The scatterplot in Fig. 6 revealed the correlation analysis
results: A positive correlation was observed between SLC17A9
expression levels and the sensitivity to drugs such as vorinostat,
asparaginase, chelerythrine, hypothemycin, PX-316, entinostat,
acrichine, LDK-378 and imexon. A negative correlation was observed
between SLC17A9 expression levels and the sensitivity to
drugs including ibrutinib, afatinib and sonidegib.
Discussion
A previous study has demonstrated the involvement of
SLC17A9, a transmembrane protein, in small-molecule transportation
(1). Furthermore, SLC17A9 controls
ATP accumulation in lysosomes, thus altering cell survival
(1). Recent studies have
demonstrated that SLC17A9 has a role in cancer development
(6,7). In LIHC, SLC17A9 acts as a tumor
promoter gene that influences tumor progression (6). In PRAD, SLC17A9 acts as a tumor
suppressor that attenuates proliferation, apoptosis and metastasis
of cancer cells (7). However, the
role of SLC17A9 in other types of cancer has remained elusive.
Therefore, in the present study, SLC17A9 expression levels
were comprehensively analyzed in a pan-cancer panel. Next, the
impact of SLC17A9 on patient prognosis, tumor immunity and
stemness was determined in various types of cancer. Finally, the
functions of SLC17A9 were verified in OSS cells.
The results of the present study revealed an
increase in SLC17A9 expression levels in the tumor tissues
of 15 types of cancer, including COAD (3), STAD (4) and LIHC (6,19).
However, a decrease in SLC17A9 expression levels was
observed in ACC tissues. In addition, a significant difference in
the status of methylation of the SLC17A9 promoter was observed in
tumor tissues compared with normal tissues in 14 types of cancer.
Extensive perturbations in DNA methylation alter the expression
levels of genes regulating tumorigenesis, thus affecting the
progression of cancer and the prognosis of the patient (20). These results indicate the
involvement of SLC17A9 in the onset and progression of
different types of cancer.
Survival analysis for multiple types of cancer was
performed using univariate Cox regression analysis and the KM
method. The results revealed an association between SLC17A9
expression levels and the prognosis of patients with various types
of cancer, including COAD (3), STAD
(4,5), LIHC (6,19) and
KIRC (21), consistent with
previous studies. A correlation was observed between high
SLC17A9 expression levels and longer PFS and DSS of patients
with LUAD. SLC17A9 was able to suppress tumorigenesis in LUAD and
PRAD (7). In the anti-PD-L1 therapy
cohort, high SLC17A9 expression levels were associated with
an increase in the OS of patients. Therefore, the potential
mechanisms by which SLC17A9 affects anti-PD-L1 treatment are worth
investigating. These results suggest that SLC17A9 may be a
promising prognostic marker in various tumor types.
A previous study has demonstrated a close
association between stemness, the status of cancer progression and
metastasis and poor prognosis of patients in a pan-cancer panel
(12). Therefore, the association
between SLC17A9 expression levels and the two stemness
indices was investigated. A significant association was observed
between SLC17A9 expression levels and RNAss in 18 types of
cancer, including GBM, LGG and PRAD. In addition, a significant
association was observed between SLC17A9 expression levels
and DNAss in 17 types of cancer, including TGCT, THCA and UVM.
Furthermore, these stemness indices were associated with intratumor
heterogeneity, immune microenvironment and immune response
(12). Therefore, the role of
SLC17A9 in different types of cancer should be further
investigated.
Previous studies have demonstrated that the TME
serves a significant role in tumor onset and progression; thus,
targeting the TME appears to be a promising approach in the
treatment of cancer (22,23). First, the results of the present
study indicated a significant difference in the SLC17A9
expression levels in six immune subtypes of 13 types of cancer.
Next, the stromal and immune scores were calculated to determine
the status of the TME of patients. The results of the present study
indicated a positive correlation between the SLC17A9
expression levels and the stromal scores in 18 types of cancer, as
well as the immune scores in 23 types of cancer. However, a
significant negative correlation was observed between the
SLC17A9 expression levels and the stromal as well as the
immune scores of patients with LAML, PAAD and STAD.
In addition, the results of the present study
demonstrated a significant correlation between the SLC17A9
expression levels and the infiltration of immune cells in 32 types
of cancer. The results of the present study indicated a negative
correlation between the SLC17A9 expression levels and the
immune cells such as Tfh and Treg cells in patients with OSS. Tfh
cells express high PD1 levels and are closely associated with
antitumor immunity. In addition, Tfh-like cells are present in
several types of cancer (24). A
previous study has revealed the synergistic effect of combining
therapies targeting Treg cells and other treatment strategies
(25). These results suggest
synergistic immunotherapy may be used to target SLC17A9 for
treating patients with OSS.
However, the role of SLC17A9 in cancer remains
controversial. Therefore, in vitro experiments were
performed using OSS cells to determine the functions of
SLC17A9. The results suggested that SLC17A9 enhances
OSS cell proliferation and viability. Next, the underlying
mechanisms of SLC17A9 were investigated. The KEGG pathway
enrichment analysis indicated that SLC17A9 was enriched in
the MAPK, Hippo, focal adhesion and TGF-β signaling pathways.
Furthermore, several known drugs targeting SLC17A9 were
identified.
Of note, the present study has certain limitations.
First, data were obtained, analyzed and integrated from several
publicly available databases, which primarily included patients of
Caucasian ethnicity. This may cause bias due to ethnicity.
Furthermore, the SLC17A9 protein significantly influences the
occurrence and progression of cancer; therefore, it is necessary to
determine the effect of SLC17A9 protein expression in multiple
types of cancer. In addition, the underlying mechanisms of the role
of SCL17A9 remain to be elucidated. Thus, additional studies should
be performed to design new SLC17A9-based therapeutic strategies for
improving patient survival.
In conclusion, the results of the present study
demonstrate that SCL17A9 was differentially expressed and
significantly associated with tumor immunity as well as the
prognosis of patients with various types of cancer. It was
demonstrated that SLC17A9 functions as a tumor promoter in OSS.
SLC17A9 may be a novel prognostic biomarker and a potential
target for synergistic immunotherapy of OSS.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MZ and JY conceptualized and supervised the study.
JL performed the bioinformatic analysis and statistical analysis.
MZ performed the in vitro experiments. FW, LS and HZ
performed the data curation and interpretation. JL wrote the
original manuscript. MZ and JY confirm the authenticity of all the
raw data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cao Q, Zhao K, Zhong XZ, Zou Y, Yu H,
Huang P, Xu TL and Dong XP: SLC17A9 protein functions as a
lysosomal ATP transporter and regulates cell viability. J Biol
Chem. 289:23189–23199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang P, Cao Q, Xu M and Dong XP:
Lysosomal ATP Transporter SLC17A9 controls cell viability via
regulating cathepsin D. Cells. 11:8872022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L, Chen Z, Xiong W, Ren H, Zhai E, Xu
K, Yang H, Zhang Z, Ding L, He Y, et al: High expression of SLC17A9
correlates with poor prognosis in colorectal cancer. Hum Pathol.
84:62–70. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li J, Su T, Yang L, Deng L, Zhang C and He
Y: High SLC17A9 expression correlates with poor survival in gastric
carcinoma. Future Oncol. 15:4155–4166. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun M, Qiu J, Zhai H, Wang Y, Ma P, Li M
and Chen B: Prognostic implications of novel gene signatures in
gastric cancer microenvironment. Med Sci Monit. 26:e9246042020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kui XY, Gao Y, Liu XS, Zeng J, Yang JW,
Zhou LM, Liu XY, Zhang Y, Zhang YH and Pei ZJ: Comprehensive
analysis of SLC17A9 and its prognostic value in hepatocellular
carcinoma. Front Oncol. 12:8098472022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mi YY, Sun CY, Zhang LF, Wang J, Shao HB,
Qin F, Xia GW and Zhu LJ: Long Non-coding RNAs LINC01679 as a
competitive endogenous RNAs inhibits the development and
progression of prostate cancer via regulating the
miR-3150a-3p/SLC17A9 axis. Front Cell Dev Biol. 9:7378122021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blum A, Wang P and Zenklusen JC: SnapShot:
TCGA-Analyzed tumors. Cell. 173:5302018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lánczky A and Győrffy B: Web-Based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res. 23:e276332021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Malta TM, Sokolov A, Gentles AJ,
Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J,
Omberg L, Gevaert O, et al: Machine learning identifies stemness
features associated with oncogenic dedifferentiation. Cell.
173:338–354.e15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reinhold WC, Sunshine M, Liu H, Varma S,
Kohn KW, Morris J, Doroshow J and Pommier Y: CellMiner: A web-based
suite of genomic and pharmacologic tools to explore transcript and
drug patterns in the NCI-60 cell line set. Cancer Res.
72:3499–3511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Su L, Xiao X, Wu F, Du G, Guo X,
Kong F, Yao J and Zhu H: Development and validation of novel
prognostic models for immune-related genes in osteosarcoma. Front
Mol Biosci. 9:8288862022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu J, Yang Y and Song J: Expression of
SLC17A9 in hepatocellular carcinoma and its clinical significance.
Oncol Lett. 20:1822020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hao X, Luo H, Krawczyk M, Wei W, Wang W,
Wang J, Flagg K, Hou J, Zhang H, Yi S, et al: DNA methylation
markers for diagnosis and prognosis of common cancers. Proc Natl
Acad Sci USA. 114:7414–7419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng M, Lan T, Tian D, Qin Z, Li Y, Li J
and Cao H: Integrative bioinformatics analysis demonstrates the
prognostic value of chromatin accessibility biomarkers in clear
cell renal cell carcinoma. Front Oncol. 11:8143962021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao Y and Yu D: Tumor microenvironment as
a therapeutic target in cancer. Pharmacol Ther. 221:1077532021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crotty S: T follicular helper cell
biology: A decade of discovery and diseases. Immunity.
50:1132–1148. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C, Jiang P, Wei S, Xu X and Wang J:
Regulatory T cells in tumor microenvironment: New mechanisms,
potential therapeutic strategies and future prospects. Mol Cancer.
19:1162020. View Article : Google Scholar : PubMed/NCBI
|