Introduction
Gastric cancer (GC) was the fifth most frequently
diagnosed cancer and the third leading cause of cancer death
worldwide in 2018 (1). The
incidence and mortality of GC has decreased substantially in the
United states and Western Europe over the past several decades;
however, the number of new cases and current mortality rate
contributes to ~50% of the global health problem, especially in
East Asian countries (2). The
5-year survival rate of GC in Japan was ~50% in 2000, but in the
United states, the 5-year survival rate ranges from 5–20%, as
patients with GC are usually diagnosed at an advanced stage of
disease with an increasing risk for tumor metastasis (3). Diagnosing GC at an early stage allows
timely treatment interventions and can improve the overall
prognosis for this type of malignancy (4).
The current recommended standard method for
diagnosing GC is endoscopic biopsy (5). However, due to the discomfort caused,
the invasive nature of the procedure and the high cost to the
general public, the use of endoscopic biopsy for screening early
stage GC is difficult in clinical practice (6). Serum biomarkers for GC, such as cancer
antigen 724 and carcinoembryonic antigen, are associated with poor
sensitivity and specificity for diagnosis (7,8).
Furthermore, gastric precursor lesions, such as intestinal
metaplasia and atypical hyperplasia, in addition to persistent
Helicobacter pylori infection, increase the difficulty of
the screening process for early GC (4). Thus, developing non-invasive and
affordable screening approaches with a high specificity and
sensitivity is important for clinical practice.
Extracellular vesicles (EVs) are secreted by
numerous cell types and are nanostructured lipid bilayer membrane
capsules (9). EVs contain numerous
types of molecules, including nucleic acids such as DNA, mRNA and
non-coding RNA, in addition to proteins, which enable communication
from donor to recipient cells (9,10). EVs
are present in certain biofluids, including plasma, serum, urine,
gastric juice and saliva (10).
Tumor-derived EVs modify tumor microenvironment, promote tumor
progression, angiogenesis, metastasis and immune evasion, and RNAs
contained in tumor-derived EVs are associated with tumor
progression, metastasis and aggressive tumor phenotypes (11,12).
Previous studies reported that molecules contained in EVs,
particularly exosome RNAs that can cause changes in gene
expression, have the potential to serve as non-invasive, robust
biomarkers for cancer screening (10,11,13).
In the present study, the diagnostic performance of EV biomarkers
for GC was summarized and analyzed, and subgroup analysis to
determine the diagnostic accuracy of EV microRNAs (miRNAs/miRs) and
long non-coding RNAs (lncRNAs) for GC was performed.
Materials and methods
Search strategy
The present review was performed according to the
preferred reporting items for systematic reviews and meta-analysis
(14). The PubMed (https://pubmed.ncbi.nlm.nih.gov/), Web of Science
(http://webofknowledge.com) and Medline
(https://www.nlm.nih.gov/medline) online
databases were searched (from May 1983 to September 18, 2022) for
literature using the following key words: (Gastric OR stomach) AND
(cancer OR carcinoma OR neoplasm OR tumor OR malignancy OR
adenocarcinoma OR adenoma) AND (detection OR diagnosis OR biomarker
OR marker OR sensitivity OR specificity OR area under the curve)
AND (exosome OR extracellular vesicles OR exosomal OR membrane
vesicles OR intracellular multivesicular endosomes). Duplicate
studies were removed from the analysis.
Literature selection and data
abstraction
Non-English language, non-human, non-original,
non-related GC studies and articles not relevant to the research
topic were excluded from the analysis. Subsequently, two authors
independently screened all potential studies for inclusion into the
meta-analysis. Inclusion criteria included: i) Studies that
identified EV biomarkers for diagnosis of GC in plasma and serum;
ii) patients with GC diagnosed according to histological
examination; and iii) studies that reported the diagnostic value of
EV biomarkers for GC, such as sensitivity, specificity, area under
the curve (AUC) or receiver operator characteristic (ROC) curve.
Any discrepancy surrounding study screening was resolved through
discussion by the authors. Relevant information in the eligible
studies was extracted using a pre-designed data collection table
and the key information included was as follows: First author, year
of publication, the country the study was performed in, study
design, population characteristics (including sample size, mean age
and sex distribution), type of blood-based specimen, GC stage,
population composition of control group, names or panels of target
biomarkers, detection method of target biomarkers, preparation
approach of EVs, sensitivity, specificity and AUC value.
Quality assessment
The quality of each eligible study was evaluated
using the diagnostic accuracy studies-2 checklist using Review
Manager (v. 5.3; The Cochrane Collaboration) (15). The risk of bias and clinical
application of eligible studies were assessed. Publication bias was
assessed using Egger's test and the symmetry of the funnel plot was
evaluated using R software (v. 3.5.3; R Foundation for Statistical
Computing) (16).
Statistical analysis
If the values of sensitivity and specificity were
not reported in the original study, the present study estimated
these two diagnostic indicators based on ROC curves using OriginPro
software (v. 9.0; OriginLab) according to the maximum Youden's
index. The bivariate meta-analysis model was used to summarize the
diagnostic value. The control groups contained healthy patients
and/or those with benign diseases, and the present study analyzed
the healthy patients; if the control groups contained healthy
people and benign disease, they were analyzed as a whole. The
sensitivity, specificity and AUC values of EV biomarkers were
pooled for subgroup analysis using Meta-DiSc software (v.1.4)
(17) using the random-effect model
(18). Heterogeneity across studies
was assessed using the c2 and I2 statistic.
P<0.05 was considered to indicate a statistically significant
difference and I2>50% indicated a statistically
significant heterogeneity.
Results
Database search results
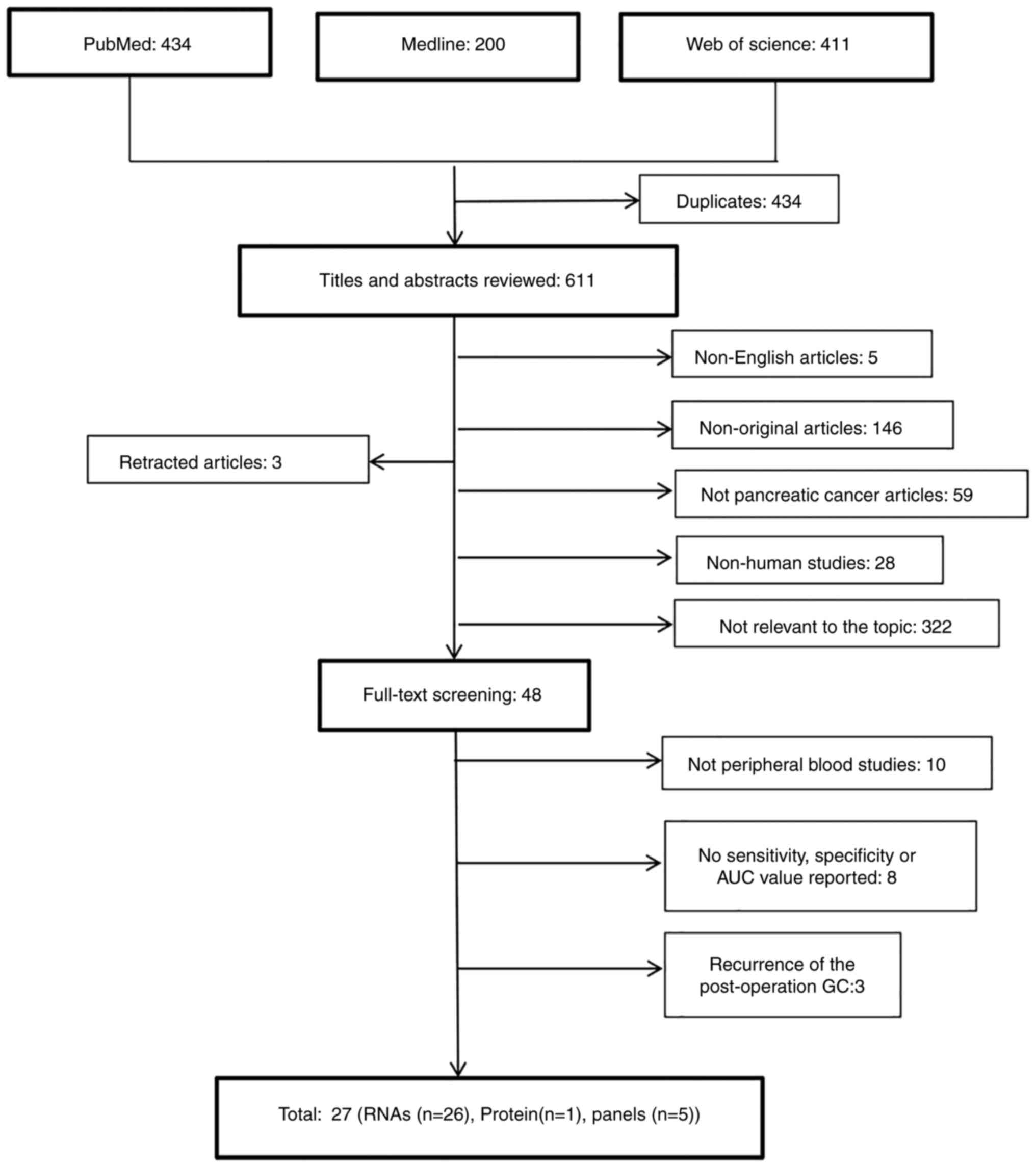
A total of 1,045 studies were found as a result of
the database searches and of these studies, 434 duplicates were
detected and removed from the analysis (Fig. 1). After screening the title and
abstracts of the remaining studies, 48 studies were selected for
full review. Then, 21 studies were excluded due to the following
criteria: i) Sample specimens used in 10 studies were not plasma or
serum; ii) 8 studies reported no sensitivity, specificity or AUC
value; and iii) 3 studies reported the EV biomarkers used to
diagnose the recurrence of post-operation patients with GC. A total
of 27 eligible studies were identified for further analysis.
Study characteristics
All eligible studies were performed in Asia and
reported results from a total of 2,831 cases of GC and 2,117
controls (Tables I and II) (19–45). A
singular study conducted prospective research (19), whereas the remaining studies were
case control studies. The mean sample size of groups of patients
with GC was 98 (range, 23–386 patients), whereas the mean sample
size of the control groups was 62 (range, 12–151 patients). A total
of 26 studies analyzed the diagnostic value of RNAs for GC: MiRNAs
in 13 studies (22,24,25,28–33,37,40,42,44,45),
four of which performed validation tests (25,29,31,44);
lncRNAs in nine studies (19,20,23,26,35,38,39,41,45);
circular RNAs (circRNAs) in three studies (21,27,34);
P-element induced wimpy testis-interacting RNAs (piRNAs) in
one study (42); mRNA in one study
(43); and a single study reported
the diagnostic value of protein (36). A total of nine studies set a
diagnostic cut-off value, which was determined using the Youden
Index (19,23,27,31,33,34,39,41,45). A
total of five studies reported the diagnostic value of RNA panels
(25,27,29,31,40),
two of which performed validation testing (25,29). A
total of six studies reported the diagnostic performance of EV
biomarkers for early stage GC (stage I/II) (31,33,34,36,38,41),
of which one study performed validation testing (31).
 | Table I.Diagnostic performance of biomarkers
in extracellular vesicles for gastric cancer. |
Table I.
Diagnostic performance of biomarkers
in extracellular vesicles for gastric cancer.
| A, miRNA |
|---|
|
|---|
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | Country of
study | Number | Mean age | Sex (% male) | Sample type | GC stage | Health status of
controls | Detection
method | Marker | Sensitivity
(%) | Specificity
(%) | AUC | P-value | Cut-off value | (Refs.) |
|---|
| Wang et al,
2022 | China | 24/24 |
| 58/ | Serum | I–IV | HC | RT-qPCR | miR-10401-3p | 91 | 70 | 0.833 | <0.001 |
| (30) |
|
|
|
|
|
|
|
|
|
| miR-1225-5p | 83 | 86 | 0.832 | <0.001 |
|
|
|
|
|
|
|
|
|
|
|
| miR-6736-5p | 95 | 63 | 0.814 | <0.001 |
|
|
| Tang et
al, | China | 120/60 | 49/ |
| Plasma | I–IV | HC | RT-qPCR | miR-4741 | 77 | 87 | 0.855 |
|
| (31) |
| 2022 |
|
|
|
|
|
|
|
| miR-32 | 79 | 99 | 0.946 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-3149 | 51 | 93 | 0.768 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-6727 | 66 | 96 | 0.892 |
|
|
|
| Kahroba et
al, | Iran | 43/40 |
| 42/43 | Serum |
| HC | RT-qPCR | miR-10a-5p | 76 | 74 | 0.801 |
|
| (40) |
| 2022 |
|
|
|
|
|
|
|
| miR-18a-5p | 72 | 71 | 0.721 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-19b-3p | 74 | 69 | 0.780 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-215-5p | 68 | 67 | 0.736 |
|
|
|
| Zheng et al,
2021 | China | 168/50 | 61/ |
| Serum | I–IV | HC | RT-qPCR | miR-590-5p | 64 | 86 | 0.810 |
| 3.470 | (22) |
| Zhang et al,
2021 | China | 118/70 |
|
| Serum | I–IV | HC | RT-qPCR | miR-215-5p | 69 | 97 | 0.866 |
|
| (24) |
|
|
| 118/60 |
|
|
|
| BGD |
|
| 65 | 95 | 0.808 |
|
|
|
| Yang et al,
2021 | China | 108/108 |
|
| Plasma | I–IV | HC |
| miR-195-5p | 67 | 73 | a0.762 |
|
| (25) |
|
|
|
|
|
|
|
|
|
| miR-211-5p | 59 | 90 | a0.798 |
|
|
|
| Lu et al,
2021 | China | 131/122 |
| 59/ | Serum | I–IV | HC | RT-qPCR | miR-92a-3p | 66 | 88 | 0.829 | 0.026 |
| (37) |
| Wei et al,
2020 | China | 108/108 | 63/61 | 66/66 | Serum |
| HC | RT-qPCR | miR-15b-3p | 74 | 81 | 0.820 |
|
| (28) |
| Ge et al,
2020 | China | 70/60 | 59/59 | 57/67 | Serum | I–IV | HC | RT-qPCR | miR-1307-3p | 81 | 77 | 0.845 | <0.001 |
| (42) |
| Tang et al,
2020 | China | 50/50 | 58/ | 76/ | Serum | Ia-IIb | HC | RT-qPCR | Let-7g-5p | b54 | b88 | a0.756 |
| <4.184 | (31) |
|
|
|
|
|
|
|
|
|
| miR-92b-3p | b58 | b80 | a0.714 |
| <1.690 |
|
|
|
|
|
|
|
|
|
|
| miR-146b-5p | b46 | b83 | a0.674 |
| <0.674 |
|
|
|
|
|
|
|
|
|
|
| miR-9-5p | b50 | b84 | a0.626 |
| <0.626 |
|
|
|
| 36/12 | 57/ | 70/ | Serum | Ia-IIb | HC | RT-qPCR | Let-7c-5p | 80 | 91 | 0.886 | 0.0001 |
|
|
|
|
|
|
|
|
|
|
|
| miR-101-3p | 75 | 91 | 0.805 | 0.0057 |
|
|
|
|
|
|
|
|
|
|
|
| miR-21-5p | 100 | 91 | 1.000 | 0.0005 |
|
|
|
|
|
|
|
|
|
|
|
| miR-26a-5p | 86 | 91 | 0.961 | 0.0013 |
|
|
| Chung et al,
2020 | China | 20/20 |
| 55/65 | Serum | IIb-IIIb | HC | PBP | miR-423-5p |
|
| a0.780 |
|
| (44) |
|
|
|
|
|
|
|
|
|
| miR-484 |
|
| a0.560 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-186-5p |
|
| a0.540 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-142-5p |
|
| a0.750 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-320d |
|
| a0.740 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-320a |
|
| a0.770 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-320b |
|
| a0.720 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-17-5p |
|
| a0.640 |
|
|
|
|
|
| 15/15 |
| 47/67 |
| Ia-IIIb |
|
| miR-629-5p |
|
| 0.750 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-363-3p |
|
| 0.780 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-337-5p |
|
| 0.750 |
|
|
|
|
|
|
|
|
|
|
|
|
| miR-27a-3p |
|
| 0.770 |
|
|
|
| Shi et
al, | China | 85/50 | 60/58 | 66/68 | Serum | I–IV | HC | RT-qPCR | miR-1246 | 82 | 86 | 0.911 | <0.001 | 1.670 | (33) |
| 2019 |
| 28/50 | /58 | /68 |
| I | HC |
|
| 86 | 74 | 0.843 | <0.001 |
|
|
|
|
| 28/30 | /58 | /67 |
|
| BD |
|
| 79 | 80 | 0.811 |
|
|
|
| Wang et al,
2017 | China | 110/110 |
| 55/ | Serum |
| HC | RT-qPCR | miR-106a −5p | c63 | c89 | a0.786 | <0.0001 |
| (29) |
|
|
|
|
|
|
|
|
|
| miR-19b-3p | c84 | c51 | a0.769 | <0.0001 |
|
|
|
| B,
lncRNA |
|
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| First author,
year | Country of
study | Number | Mean
age | Sex (%
male) | Sample
type | GC
stage | Health status of
controls | Detection
method | Marker | Sensitivity
(%) | Specificity
(%) | AUC | P-value | Cut-off
value | (Refs.) |
|
| bZhou et al, 2020 | China | 81/78 | 64/60 | 63/ | Serum | I–IV | HC | RT-qPCR | LncRNA H19 | 74 | 84 | 0.849 | <0.01 | 1.770 | (19) |
| Zheng et al,
2020 | China | 60/60 |
| 63/ | Plasma | I–IV | HC | RT-qPCR | Lnc-SLC2A12- | 69 | 75 | 0.776 | <0.001 |
| (20) |
|
|
|
|
|
|
|
|
|
| 10:1 |
|
|
|
|
|
|
| Xu et al,
2020 | China | 109/50 |
| 74/ | Serum | I–IV | HC | RT-qPCR | LncRNA | c82 | c94 | 0.892 |
|
| (26) |
|
|
| 109/48 |
| 74/ |
|
| GA |
| MIAT | c65 | c89 | 0.787 |
|
|
|
| Guo et al,
2020 | China | 386/151 | 61/ | 59/62 | Serum | I–IV | HC | RT-qPCR | LncRNA-GC1 | 85 | 85 | 0.898 |
| 5.200 | (41) |
|
|
| 386/37 | 61/54 | 59/70 |
|
| CAG |
|
| 89 | 88 | 0.842 |
|
|
|
|
|
| 386/48 | 61/55 | 59/65 |
|
| IM |
|
| 90 | 81 | 0.860 |
|
|
|
|
|
| 179/151 |
| /62 |
| I–II | HC |
|
| 89 | 80 | 0.861 |
|
|
|
|
|
| 179/ 37 | /54 | /70 |
|
| CAG |
|
| 92 | 82 | 0.884 |
|
|
|
|
|
| 179/48 | /55 | /65 |
|
| IM |
|
| 81 | 88 | 0.885 |
|
|
|
| Piao et al,
2020 | China | 281/80 |
|
| Plasma | I–IV | HC | RT-qPCR | CEBPA-ASI | c72 | c87 | 0.824 |
|
| (35) |
| Li et al,
2020 | China | 43/27 |
| 74/ | Plasma | I–IV | HC | RT-qPCR | Lnc-GNAQ-6:1 | 84 | 56 | 0.736 | <0.001 | 1.855 | (39) |
| Cai et al,
2019 | China | 63/29 |
| 71/ | Serum | I–IV | HC | RT-PqCR | Lnc RNA | 84 | 87 | 0.896 |
| 2.390 | (45) |
|
|
|
|
|
|
|
|
|
| PCSK1-2:1 |
|
|
|
|
|
|
| Zhao et al,
2018 | China | 126/120 |
| 55/ | Serum | I–IV | HC |
| Lnc RNA | 70 | 85 | 0.827 |
| 1.720 | (23) |
|
|
|
|
|
|
|
|
|
| HOTTIP |
|
|
|
|
|
|
| Lin et al,
2018 | China | 51/60 | 61/58 | 61/63 | Plasma | Ia-IIb | HC | RT-qPCR | LncUEGC1 | c97 | c96 | 0.876 | <0.0001 |
| (38) |
|
|
|
|
|
|
|
|
|
| lncUEGC2 | c89 | c58 | 0.758 | <0.0001 |
|
|
|
|
| 23/60 |
|
|
| I | HC |
| LncUEGC1 | c96 | c73 | 0.850 | <0.0001 |
|
|
|
|
|
|
|
|
|
|
|
| LncUEGC2 | c74 | c71 | 0.749 | 0.0456 |
|
|
|
|
| 23/18 |
|
|
|
| CAG |
| LncUEGC1 | c74 | c88 | 0.841 | 0.0002 |
|
|
|
| C,
circRNA |
|
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| First author,
year | Country of
study | Number | Mean
age | Sex (%
male) | Sample
type | GC
stage | Health status of
controls | Detection
method | Marker | Sensitivity
(%) | Specificity
(%) | AUC | P-value | Cut-off
value | (Refs.) |
|
| Zheng et al,
2022 | China | 60/60 | 63.7/ | 53/ | Plasma | I–IV | CG, HC | RT-qPCR | circ_0015286 | 82 | 66 | 0.778 | <0.001 |
| (21) |
| Xiao et al,
2022 | China | 112/120 |
|
| Serum |
| CG, TH, | RT-qPCR | circRNA | 77 | 66 | 0.726 |
| 1.330 | (27) |
|
|
|
|
|
|
|
| HC |
| Chr10q11 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| circRNA
Chr1p11 | 82 | 77 | 0.822 |
| 2.000 |
|
|
|
|
|
|
|
|
|
|
| circRNA
Chr7q11 | 80 | 59 | 0.749 |
| 1.070 |
|
| Shao et al,
2020 | China | 41/39 |
|
| Plasma | I–II | HC | RT-qPCR | circ-0065149 | 49 | 90 | 0.640 | 0.031 | 6.430 | (34) |
|
| D,
piRNA |
|
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| First author,
year | Country of
study | Number | Mean
age | Sex (%
male) | Sample
type | GC
stage | Health status of
controls | Detection
method | Marker | Sensitivity
(%) | Specificity
(%) | AUC | P-value | Cut-off
value | (Refs.) |
|
| Ge et al,
2020 | China | 70/60 | 59/59 | 57/67 | Serum | I–IV | HC | RT-qPCR | piR-018569 | 44 | 97 | 0.732 | <0.001 |
| (42) |
|
|
|
|
|
|
|
|
|
| piR-004918 | 43 | 95 | 0.754 | <0.001 |
|
|
|
|
|
|
|
|
|
|
|
| piR-019308 | 57 | 92 | 0.820 | <0.001 |
|
|
|
| E, mRNA |
|
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| First author,
year | Country of
study | Number | Mean
age | Sex (%
male) | Sample
type | GC
stage | Health status of
controls | Detection
method | Marker | Sensitivity
(%) | Specificity
(%) | AUC | P-value | Cut-off
value | (Refs.) |
|
| Dong et al,
2019 | China | 119/31 |
| 75/ | Serum | I–IV | HC | RT-qPCR | MT1-MMP | 64 | 87 | 0.788 |
|
| (43) |
|
|
|
|
|
|
|
|
|
| mRNA |
|
|
|
|
|
|
|
| F,
Protein |
|
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| First author,
year | Country of
study | Number | Mean
age | Sex (%
male) | Sample
type | GC
stage | Health status of
controls | Detection
method | Marker | Sensitivity
(%) | Specificity
(%) | AUC | P-value | Cut-off
value | (Refs.) |
|
| Okuda et al,
2021 | Japan | 93/90 | 72/72 | 69/67 | Serum | I–IV | HC | ELISA | Dicer | c93 | c34 | 0.622 |
|
| (36) |
|
|
| 63/90 | /72 | /67 |
| I |
|
|
| c92 | c37 | 0.623 |
|
|
|
 | Table II.Diagnostic performance of biomarker
panels in extracellular vesicles for gastric cancer. |
Table II.
Diagnostic performance of biomarker
panels in extracellular vesicles for gastric cancer.
|
|
| GC
cases/controls |
|
|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | Country of
study | Number | Mean age | Sex (% male) | Sample type | GC stage | Health status of
controls | Detection
Method | Marker panel | Sensitivity
(%) | Specificity
(%) | AUC | P-value | (Refs.) |
|---|
| Kahroba et
al, 2022 | Iran | 43/40 |
| 42/43 | Serum |
| HC | RT-qPCR | A | 73 | 72 | 0.813 |
| (40) |
| Yang et al,
2021 | China | 108/108 |
|
| Plasma | I–IV | HC |
| B | b68 | b89 | a0.820 |
| (25) |
| Tang et al,
2020 | China | 50/50 | 58/ | 76/ | Serum | Ia-IIb | HC | RT-qPCR | C | 64 | 78 | 0.775 | <0.001 | (31) |
|
|
|
|
|
|
|
|
|
| D | 60 | 82 | 0.736 | <0.001 |
|
|
|
|
|
|
|
|
|
|
| E | 44 | 88 | 0.705 | 0.0004 |
|
|
|
|
|
|
|
|
|
|
| F | 58 | 86 | 0.774 | <0.001 |
|
|
|
|
|
|
|
|
|
|
| G | 60 | 82 | 0.774 | <0.001 |
|
|
|
|
|
|
|
|
|
|
| H | 68 | 74 | 0.750 | <0.001 |
|
|
|
|
|
|
|
|
|
|
| I | 60 | 84 | 0.773 | <0.001 |
|
| Wang et al,
2017 | China | 110/110 |
| 55/ | Serum |
| HC | RT-qPCR | J | b84 | b51 | a0.814 | <0.0001 | (29) |
| Xiao et al,
2022 | China | 112/120 |
|
| Serum |
| CG/TH/HC | RT-qPCR | K | 73 | 84 | 0.839 |
| (27) |
With the development of EV extraction technologies,
commercial exosome isolation kits were also used for the extraction
of exosomes (46). From a total of
27 studies, 23 studies analyzed exosome biomarkers, and almost all
extracted exosomes were reported to have a mean size of 30–200 nm
and were positive for CD9, CD81, CD63 and/or TSG101 markers. The
remaining four studies analyzed EV biomarkers.
Quality assessment of included
studies
Quality assessment of the analyzed studies was
performed (Fig. S1). All 27
studies analyzed had a low risk of bias for the index test, four
studies had unclear risk of reference standard, flow and timing.
Quality assessment analysis also demonstrated that all studies had
a low concern for application regarding the index test and
reference standard. A total of four studies demonstrated an unclear
risk of bias of patient selection and an unclear applicability
concern of patient selection, due to non-random patient selection
and a lack of basic patient information reported. Funnel plot
analysis of the publication bias of studies demonstrated no
statistically significant publication bias (Fig. S2).
Diagnostic performance
A total of 58 RNAs were reported in the 27 eligible
studies. Of these RNAs, 39 were miRNAs, 10 were lncRNAs, five were
circRNAs, three were piRNAs and one was mRNA. miR-19b-3p and
miR-215-5p were reported in two studies and were consistently
upregulated (Table III).
 | Table III.Summary of studies reporting a
significant association of miRNA with development of gastric
cancer. |
Table III.
Summary of studies reporting a
significant association of miRNA with development of gastric
cancer.
|
|
First
author, year (refs.) |
|---|
|
|
|
|---|
| miRNA | Chung et al,
2020 (44) | Ge et al,
2020 (42) | Kahroba et
al, 2022 (40) | Lu et al,
2021 (37) | Shi et al,
2019 (33) | Tang et al,
2022 (32) | Tang et al,
2020 (31) | Wang et al,
2022 (30) | Wang et al,
2017 (29) | Wei et al,
2020 (28) | Yang et al,
2021 (25) | Zhang et al,
2021 (24) | Zheng et al,
2021 (22) | Sum of studies
reporting the mi RNA |
|---|
| miR-19b-3p |
|
| aUpregulated |
|
|
|
|
| aUpregulated |
|
|
|
| 2 |
| miR-215-5p |
|
| aUpregulated |
|
|
|
|
|
|
|
| bUpregulated |
| 2 |
| Let-7c-5p |
|
|
|
|
|
| a |
|
|
|
|
|
| 1 |
| Let-7g-5p |
|
|
|
|
|
| aUpregulated |
|
|
|
|
|
| 1 |
| miR-101-3p |
|
|
|
|
|
| a |
|
|
|
|
|
| 1 |
| miR-10401-3p |
|
|
|
|
|
|
| bDownregulated |
|
|
|
|
| 1 |
| miR-106a-5p |
|
|
|
|
|
|
|
| aUpregulated |
|
|
|
| 1 |
| miR-10a-5p |
|
| aUpregulated |
|
|
|
|
|
|
|
|
|
| 1 |
| miR-1225-5p |
|
|
|
|
|
|
| bDownregulated |
|
|
|
|
| 1 |
| miR-1246 |
|
|
|
| bUpregulated |
|
|
|
|
|
|
|
| 1 |
| miR-1307-3p |
| bUpregulated |
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-142-5p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-146b-5p |
|
|
|
|
|
| aUpregulated |
|
|
|
|
|
| 1 |
| miR-15b-3p |
|
|
|
|
|
|
|
|
| bUpregulated |
|
|
| 1 |
| miR-17-5p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-186-5p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-18a-5p |
|
| aUpregulated |
|
|
|
|
|
|
|
|
|
| 1 |
| miR-195-5p |
|
|
|
|
|
|
|
|
|
| b |
|
| 1 |
| miR-211-5p |
|
|
|
|
|
|
|
|
|
| b |
|
| 1 |
| miR-21-5p |
|
|
|
|
|
| a |
|
|
|
|
|
| 1 |
| miR-26a-5p |
|
|
|
|
|
| a |
|
|
|
|
|
| 1 |
| miR-27a-3p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-3149 |
|
|
|
|
| bDownregulated |
|
|
|
|
|
|
| 1 |
| miR-32 |
|
|
|
|
| bDownregulated |
|
|
|
|
|
|
| 1 |
| miR-320a | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-320b | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-320d | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-337-5p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1. |
| miR-363-3p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-423-5p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-4741 |
|
|
|
|
| bUpregulated |
|
|
|
|
|
|
| 1 |
| miR-484 | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-590-5p |
|
|
|
|
|
|
|
|
|
|
|
| bDownregulated | 1 |
| miR-629-5p | b |
|
|
|
|
|
|
|
|
|
|
|
| 1 |
| miR-6727 |
|
|
|
|
| bDownregulated |
|
|
|
|
|
|
| 1 |
| miR-6736-5p |
|
|
|
|
|
|
| bDownregulated |
|
|
|
|
| 1 |
| miR-92a-3p |
|
|
| bDownregulated |
|
|
|
|
|
|
|
|
| 1 |
| miR-92b-3p |
|
|
|
|
|
| aUpregulated |
|
|
|
|
|
| 1 |
| miR-9-5p |
|
|
|
|
|
| aUpregulated |
|
|
|
|
|
| 1 |
The median sensitivity, specificity and AUC value of
the total RNAs were 74% (range, 43–100%), 86% (range, 51–99%) and
0.800 (range, 0.626–1.000), respectively. The median sensitivity,
specificity and AUC value of miRNAs were 74% (range, 46–100%), 86%
(range, 51–99%) and 0.783 (range, 0.540–1.000), respectively. A
previous study by Tang et al (31) reported that miR-21-5p demonstrates
diagnostic value for distinguishing patients with early stage GC
from healthy controls with a sensitivity of 100% and a specificity
of 91%. The median sensitivity, specificity and AUC value of
lncRNAs were 82% (range, 43–97%), 84% (range, 34–97%) and 0.821
(range, 0.622–0.898), respectively. In the prospective study
analyzed, the expression level of exosome lncRNA H19 in serum was
significantly upregulated in patients with GC and the AUC value was
0.849 (19). The optimal cut-off
value was 1.770, with a sensitivity of 74% and a specificity of
84%. The median sensitivity, specificity and AUC value of circRNAs
were 78% (range, 49–82%), 72% (range, 59–90%) and 0.774 (range,
0.640–0.893), respectively. The median sensitivity, specificity and
AUC value of EV biomarker panels were 64% (range, 44–84%), 82%
(range, 51–89%) and 0.774 (range, 0.705–0.839).
Meta-analysis
Meta-analysis was performed according to the type of
molecule reported in the study. The diagnostic values of all EV
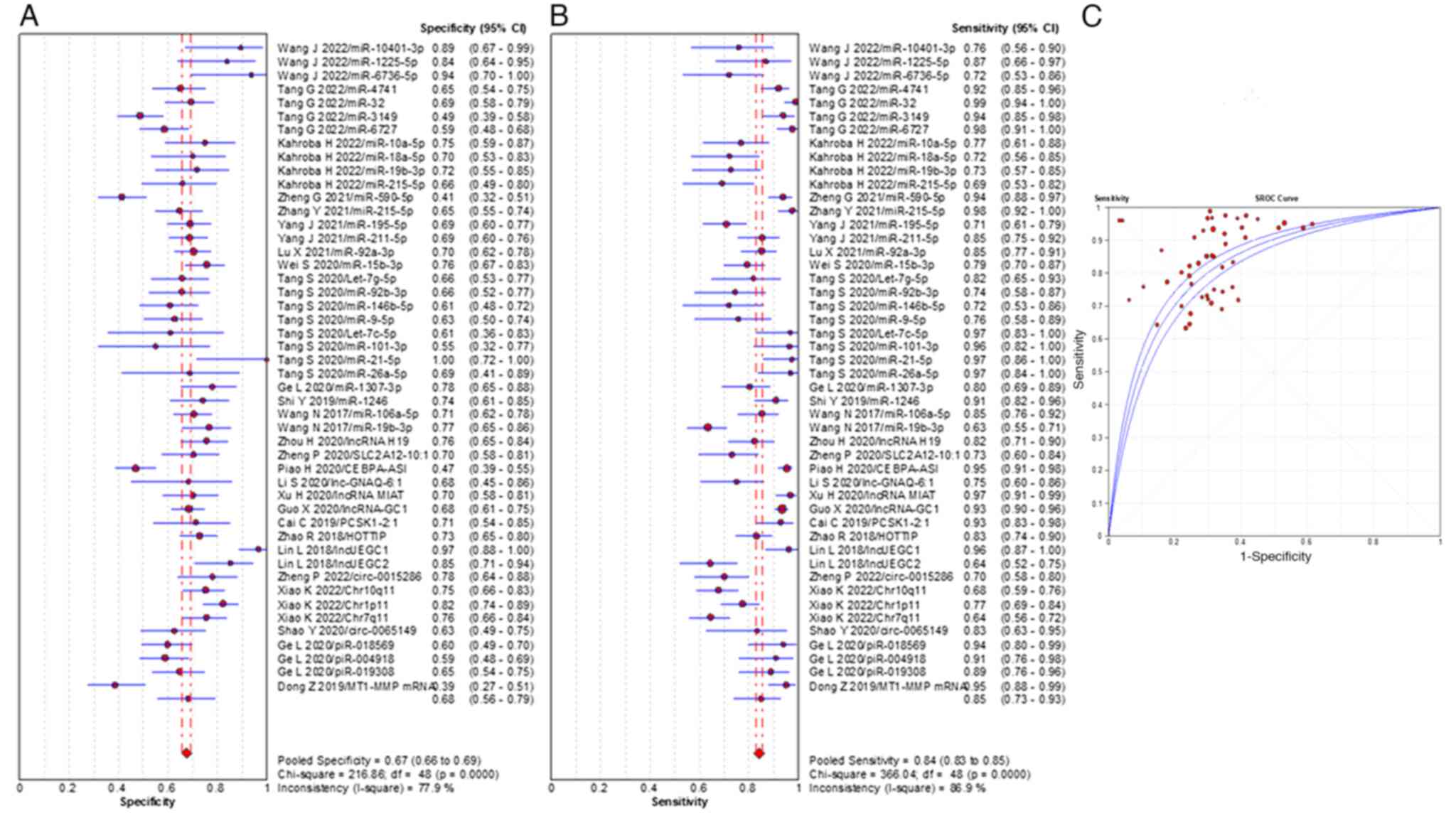
total RNAs were summarized and the meta-analysis demonstrated that
the pooled sensitivity, specificity and the AUC value were 84%
(range, 95% CI 83–85%), 67% (range, 95% CI 66–69%) and 0.822,
respectively (Fig. 2). The pooled
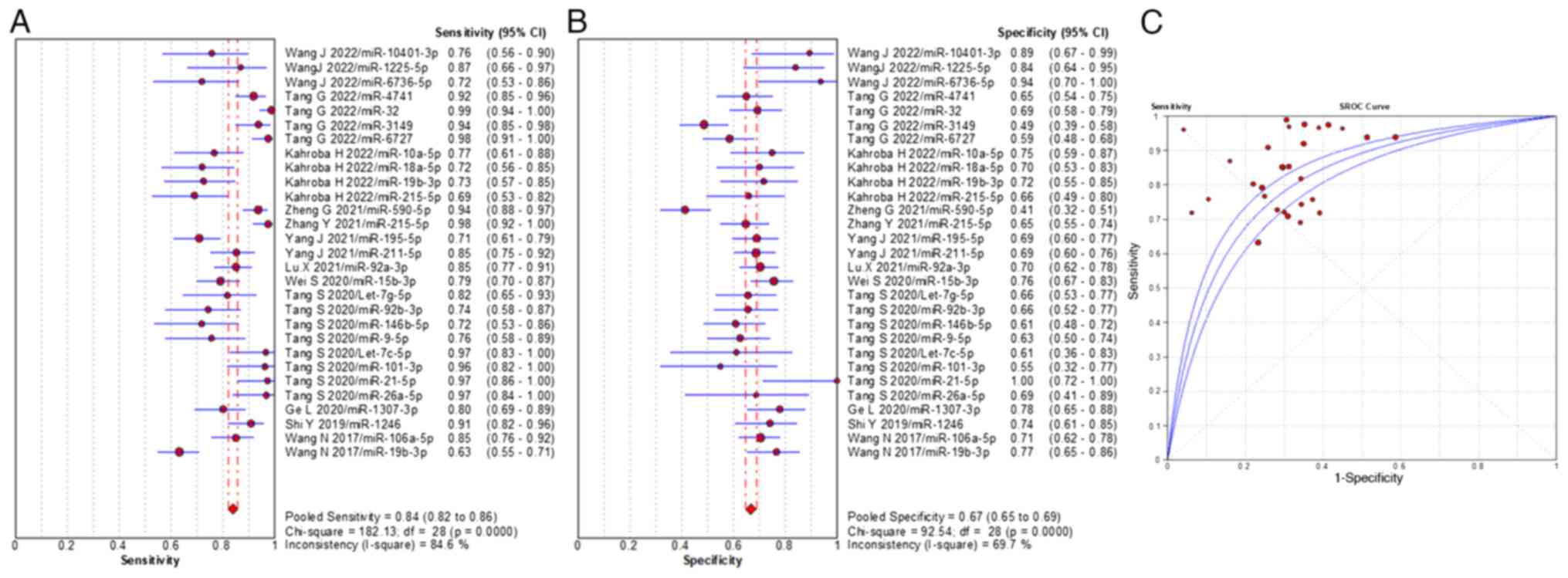
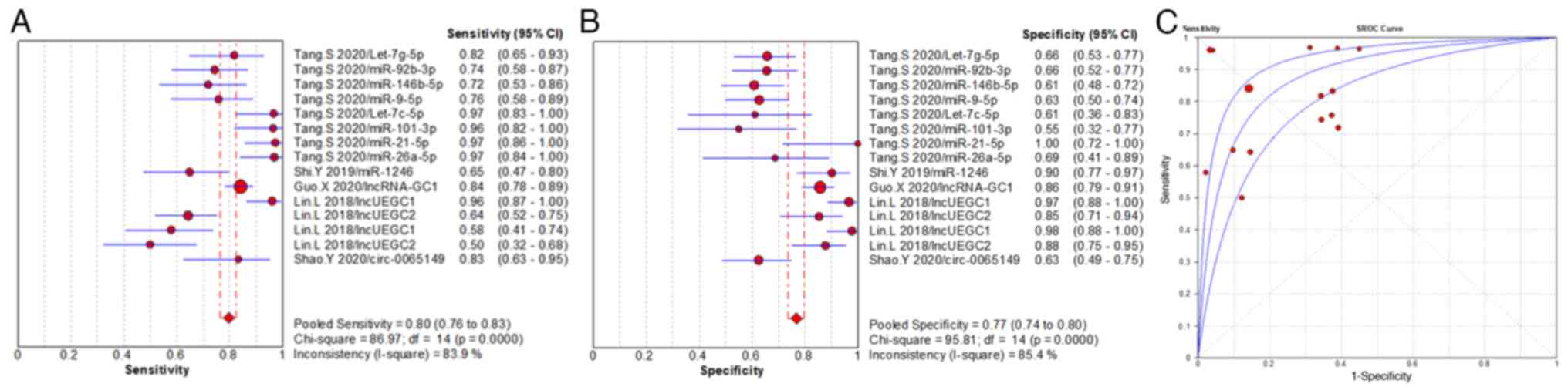
sensitivity, specificity and AUC value of miRNAs were 84% (range,
95% CI 82–86%), 67% (range, 95% CI 65–69%) and 0.808, respectively
(Fig. 3), which demonstrated
consistent diagnostic accuracy with the EV total RNAs. The pooled
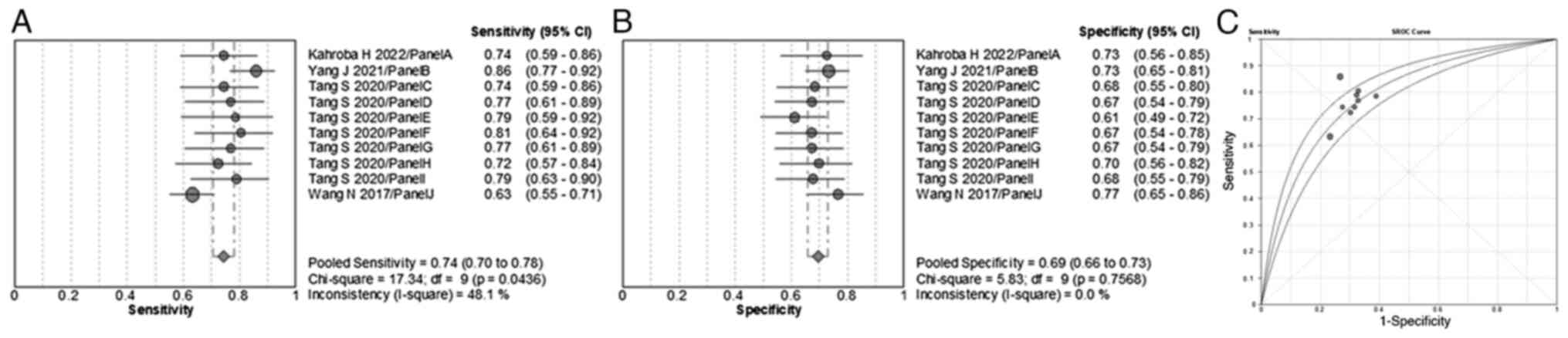
sensitivity, specificity and AUC value of EV miRNA panels were 74%
(range, 95% CI 70–78%), 69% (range, 95% CI 66–73%) and 0.784,
respectively (Fig. 4). The miRNA
panels demonstrated a lower diagnostic efficiency compared with the
individual miRNAs. The pooled sensitivity, specificity and AUC
value of EV lncRNAs were 89% (range, 95% CI 81–91%), 69% (range,
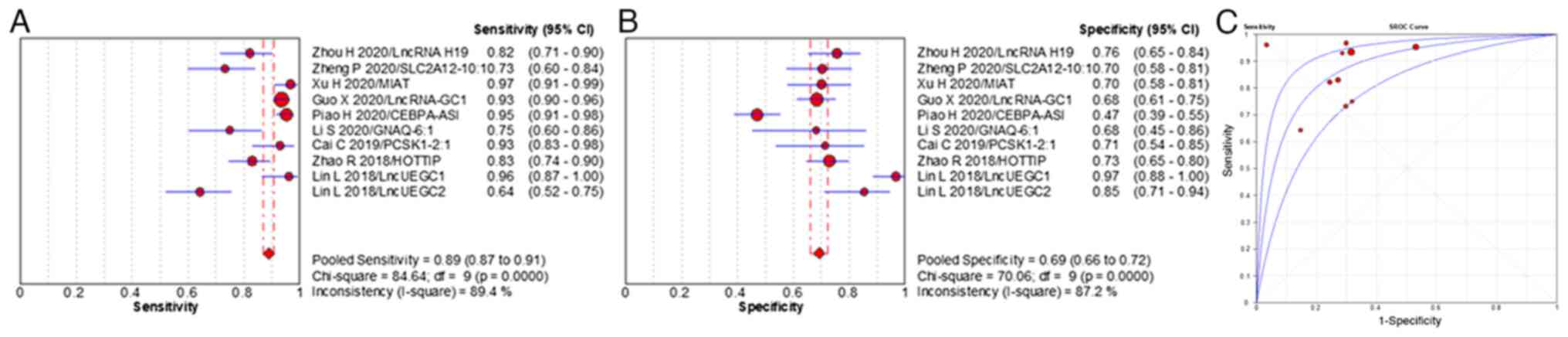
95% CI 66–72%) and 0.872, respectively (Fig. 5). The diagnostic efficiency of EV
lncRNAs was higher compared with that of EV miRNAs. A meta-analysis
of early stage GC cases with 13 individual EV RNAs was performed.
The pooled sensitivity, specificity and AUC value of individual EV
RNAs were 80% (range, 95% CI 76–83%), 77% (range, 95% CI 74–80%)
and 0.879, respectively (Fig. 6).
Therefore, EV RNAs demonstrated a promising diagnostic efficiency
for cases of early stage GC.
Discussion
In the present study, the diagnostic performance of
EV biomarkers in plasma and serum for GC was analyzed. A total of
27 studies that assessed 58 EV RNAs and one EV protein for the
diagnosis of GC were selected for meta-analysis. These studies
reported results from 2,831 patients with GC and 2,117 healthy
controls from 2017-2022. The meta-analysis demonstrated that out of
the total number of miRNAs reported, miR-19b-3p and miR-215-5p were
the only two miRNAs reported twice in the literature, therefore
further studies validating the diagnostic value of these miRNAs are
required. The diagnostic efficiency of EV miRNAs and lncRNAs were
analyzed and EV lncRNAs demonstrated a higher diagnostic
performance compared with EV miRNAs. When compared with the EV
total RNAs, EV miRNAs demonstrated a similar diagnostic
performance. Analysis of the studies that reported the diagnostic
efficiency of EV biomarkers for early stage GC demonstrated that EV
biomarkers showed promise for the diagnosis of early stage GC.
However, in the present review, the majority of the studies
analyzed were case controls; therefore, well-designed prospective
studies are needed to improve the diagnostic accuracy of EV
biomarkers for GC.
Late diagnosis is a major reason for the poor
survival rate of GC patients (2).
In China, the proportion of GC patients diagnosed at an early stage
of disease was 9% in 2008 (47).
The survival rate of patients with early stage GC ranges from
60–80% compared with 15–24% of patients with advanced stage GC
(48). Therefore, it is crucial to
find a novel, non-invasive and efficient diagnostic strategy of
screening for early stage GC. In the present study, the diagnostic
performance of EV RNAs for early stage GC was analyzed and it was
demonstrated that EV RNAs demonstrated an AUC value of 0.879 and
showed a high diagnostic efficiency for early stage GC. Lin et
al (38) reported that in
patients with stage I GC, EV lncUEGC1 effectively distinguished 23
patients with GC from 60 healthy controls with an AUC value of
0.850. In a Chinese population, the presence of EV lncRNA-GC1 is
reported to be sufficient for discriminating between patients with
early stage GC and healthy controls, with a sensitivity of 89% and
a specificity of 80% (41).
Moreover, detection of lncRNA-GC1 is sufficient for discriminating
patients with early stage GC from those with precancerous lesions,
with a sensitivity of 92% and a specificity of 82% (41). Nevertheless, as there was no
repetition study to report the same EV RNAs for early stage GC, it
is essential to perform repetitive researches on the same RNAs for
early stage GC.
In previous years, EV-derived RNAs as novel,
effective, non-invasive biomarkers for the diagnosis of GC have
attracted increasing attention (49). RNAs are one of the most abundant
types of molecule present in EVs (50). EV RNAs are reported to have a high
stability in the blood due to the ability of EVs to protect RNA
from degradation by RNases (51).
EV RNAs can regulate gene expression at post-transcriptional,
transcriptional and translational levels by modulating relevant
signaling pathways in the tumor microenvironment, effecting both
angiogenesis and metastasis (52).
Previous studies have reported that EV-derived RNAs serve critical
roles in the tumorigenesis and metastasis of GC (53), and the most promising EV RNAs used
as diagnostic biomarkers are miRNAs, lncRNAs and circRNAs (52). In the present study, EV miRNAs and
lncRNAs were the most frequently reported type of biomarker, and
the diagnostic performance of lncRNAs was higher compared with the
diagnostic performance of miRNAs. All lncRNAs were reported once in
the literature and no replicated studies were found; therefore,
further studies demonstrating the diagnostic value of these lncRNAs
are needed to verify these results. In the present study,
miR-19b-3p and miR-215-5p were reported twice in the literature,
were both consistently upregulated and miR-19b-3p was also tested
in a validation study. This result suggested that miRNAs are more
promising diagnostic biomarkers for GC, comparing to lncRNAs. A
total of three studies reported five EV circRNAs that had a
powerful diagnostic efficiency for GC (20,27,34).
circRNAs are a class of RNA with a unique closed loop-structure
structure without 5′ and 3′ ends, which increases RNase R
resistance compared with other non-coding RNAs (ncRNAs) (54,55).
Based on the unique structure of circRNAs, EV circRNA could be a
more efficient non-invasive diagnostic marker for GC compared with
other EV ncRNAs. circRNAs are an endogenous RNAs with a covalently
closed cyclic structure, and owing to this structure, circRNAs are
more resistant to RNA exonuclease than linear RNAs (56). However, as the research on the use
of EV circRNAs as a biomarker for GC tumors is currently limited,
further studies are needed to validate this hypothesis.
In previous studies, compared with individual EV
biomarkers, EV biomarker panels have been reported to show a
greater efficacy for the diagnosis of lung and pancreatic cancer
(57,58). Previous studies reported that EV
miRNA panels demonstrate a higher efficiency for distinguishing
patients with GC from healthy controls, with an AUC value of
>0.800, while the AUC value is <0.800 for the corresponding
individual EV miRNAs (25,29). By contrast, previous studies
reported that the diagnostic value of EV miRNA panels are similar
to the corresponding individual EV miRNAs (31,40).
In present study, EV miRNA panels did not demonstrate a higher
diagnostic value compared with individual miRNAs, consistent with
the previous reports, which could be due to fewer studies focused
on EV miRNA panels being included in the meta-analysis. In the
present study, two miRNAs (miR-19b-3p and miR-215-5p) were reported
twice in the literature and were both included in panels A and J.
miR-19b-3p inhibits GC cell proliferation, migration and invasion
by negatively regulating neuropilin-1 (NRP1), and the
miR-19b-3p/NRP1 axis can regulate the epithelial-to-mesenchymal
transition and focal adhesions that occur in GC, which could
contribute to the development and progression of GC (59). Previous studies reported that
miR-215-5p expression is significantly upregulated in GC tissues
and cell lines, and that the aberrant expression of miR-215-5p
promotes the malignancy of GC cells, which results in enhanced
carcinogenesis (60,61). Overexpression of miR-215-5p
stimulates the migration and invasion of cancer cells via the
degradation of Forkhead Box Protein O1 (62). Therefore, miRNAs that have been
repeatedly verified were deemed more suitable than other RNAs to
construct a biomarker panel to improve the robustness and
diagnostic accuracy of these panels. Previous studies reported that
both EV proteins alone and EV proteins combined with miRNA
demonstrate a powerful diagnostic efficiency for certain types of
lung and pancreatic cancer (58,63).
In the present study, only one EV protein was reported, for which
the diagnostic performance was not promising; however, the protein
demonstrated a high sensitivity for the diagnosis of GC (36). Therefore, EV proteins should be
studied to further analyze the diagnostic efficiency of EV
biomarker panels for GC.
Currently, circulating tumor DNAs (ctDNAs),
circulating tumor cells (CTCs) and EVs, particularly exosomes, are
the main components that have been mostly analyzed in liquid biopsy
samples (64,65). A previous study reported that
109 exosome particles can be detected in 1 ml of blood,
while only a few CTCs are detected in the same sample volume
(66). The expression level of
exosomes in biofluids is higher compared with that of CTCs or
ctDNAs and exosomes are more stable than CTCs and ctDNAs due to the
presence of lipid bilayers (66,67).
Therefore, compared with CTCs and ctDNAs, exosomes may potentially
be a more promising non-invasive biomarker tested for in liquid
biopsy.
Currently, ultralcentrifugation (UC) is the
recommended and most widely used extraction method for EV isolation
and separation (68). However,
there is presently no standardized protocol for the centrifugation
time, centrifugal force, or rotor type, which can influence the
purity and yield of isolated EVs (69,70).
Of the studies included for meta-analysis in the present study, one
study reported the use of UC to isolate EVs and no uniform
centrifugal time or number of centrifugations were reported, which
could affect purity and concentration of the target EVs isolated.
Furthermore, due to the high time consumed, high cost, potential
for structural damage of EVs, aggregation into blocks and
lipoprotein co-separation associated with UC, this EV isolation
method is not conducive to clinical applications (71,72).
With the advent of advanced sequencing techniques, the development
of commercial exosome isolation kits occurred, which can be used in
the extraction of exosomes from plasma and serum (46). EV isolation methods in the majority
of studies included in the present meta-analysis used commercial
exosome isolation kits, with transmission electron microscopy and
western blotting used to further verify exosome identity (42,43,45).
These results suggest that commercial exosome isolation kits can be
used to efficiently extract exosomes from both plasma and serum
samples. Additional techniques used to isolate EVs from human
bodily fluids include size-based isolation techniques,
immunoaffinity chromatography and other new isolation techniques
(such as immunomagnetic beads conjugated with combined antibodies)
can also be used for the extraction of EVs, which might be suitable
for extractions from plasma and serum; however, there are currently
a limited number of studies that report using these techniques
(73–75). Thus, it is necessary to develop a
unified, convenient and effective method for the extraction of EVs
from plasma and serum samples.
There were a number of limitations in the present
study. Firstly, all studies selected for meta-analysis performed
analysis on samples obtained from Asian populations, therefore,
there was an absence of samples taken from other ethnicities.
Secondly, plasma and serum were both used as potential sources of
circulating EVs; however, further verification is required to
determine if one is a more suitable source of EVs compared with the
other. There was no standardized method reported for EV extraction
and the cost related to EV detection was also not reported. Thus,
further research is required to determine an effective standard
method for extraction and detection of EVs. Thirdly, from a total
of 27 studies selected for meta-analysis, just nine studies
reported the cut-off values used, no studies reported the cut-off
value of the same biomarker, thus there was no uniform cut-off
value used as a standard reference. Finally, all studies selected
for meta-analysis were case studies, with the exception of a single
prospective study. Therefore, further prospective research should
be conducted to analyze the diagnostic efficiency of EV biomarkers
for GC.
The detection of EV RNAs in plasma and serum
demonstrated promise for use as novel noninvasive biomarkers in the
early diagnosis of GC in Asian populations. Future studies are
required to further research the diagnostic efficacy of EV RNAs and
EV RNA panels.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors read and approved the final version of
the manuscript. EJ contributed to the conception and design of the
study. JX and SQ analyzed data, performed the statistical analysis
and drafted the manuscript. NR, BG and XS acquired data and revised
the manuscript critically for important intellectual content. JX,
SQ, NR, BG, XS and EJ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hochwald SN, Kim S, Klimstra DS, Brennan
MF and Karpeh MS: Analysis of 154 actual five-year survivors of
gastric cancer. J Gastrointest Surg. 4:520–525. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Young E, Philpott H and Singh R:
Endoscopic diagnosis and treatment of gastric dysplasia and early
cancer: Current evidence and what the future may hold. World J
Gastroenterol. 27:5126–5151. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mabe K, Inoue K, Kamada T, Kato K, Kato M
and Haruma K: Endoscopic screening for gastric cancer in Japan:
Current status and future perspectives. Dig Endosc. 34:412–419.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen C, Chen Q, Zhao Q, Liu M and Guo J:
Value of combined detection of serum CEA, CA72-4, CA19-9, CA15-3
and CA12-5 in the diagnosis of gastric cancer. Ann Clin Lab Sci.
47:260–263. 2017.PubMed/NCBI
|
|
8
|
Jelski W and Mroczko B: Molecular and
circulating biomarkers of gastric cancer. Int J Mol Sci.
23:75882022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu R, Rai A, Chen M, Suwakulsiri W,
Greening DW and Simpson RJ: Extracellular vesicles in
cancer-implications for future improvements in cancer care. Nat Rev
Clin Oncol. 15:617–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kinoshita T, Yip KW, Spence T and Liu FF:
MicroRNAs in extracellular vesicles: potential cancer biomarkers. J
Hum Genet. 62:67–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qian Z, Shen Q, Yang X, Qiu Y and Zhang W:
The role of extracellular vesicles: An epigenetic view of the
cancer microenvironment. Biomed Res Int. 2015:6491612015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu YX, Pu SD, Li X, Yu ZW, Zhang YT, Tong
XW, Shan YY and Gao XY: Exosomal ncRNAs: Novel therapeutic target
and biomarker for diabetic complications. Pharmacol Res.
178:1061352022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liberati A, Altman DG, Tetzlaff J, Mulrow
C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J
and Moher D: The PRISMA statement for reporting systematic reviews
and meta-analyses of studies that evaluate health care
interventions: Explanation and elaboration. PLoS Med.
6:e10001002009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM; QUADAS-2 Group, : QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zamora J, Abraira V, Muriel A, Khan KS and
Coomarasamy A: Meta-DiSc: A software for meta-analysis of test
accuracy data. BMC Med Res Methodol. 6:312006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dias S, Welton NJ, Sutton AJ and Ades AE:
NICE DSU technical support document 2: A generalised linear
modelling framework for pairwise and network meta-analysis of
randomised controlled trials [Internet]. NICE Decision Support Unit
Technical Support Documents London: National Institute for Health
and Care Excellence (NICE); 2014
|
|
19
|
Zhou H, Shen W, Zou H, Lv Q and Shao P:
Circulating exosomal long non-coding RNA H19 as a potential novel
diagnostic and prognostic biomarker for gastric cancer. J Int Med
Res. 48:3000605209342972020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng P, Zhang H, Gao H, Sun J, Li J,
Zhang X, Gao L, Ma P and Li S: Plasma exosomal long noncoding RNA
lnc-SLC2A12-10:1 as a novel diagnostic biomarker for gastric
cancer. Onco Targets Ther. 13:4009–4018. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng P, Gao H, Xie X and Lu P: Plasma
exosomal hsa_circ_0015286 as a potential diagnostic and prognostic
biomarker for gastric cancer. Pathol Oncol Res. 28:16104462022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng GD, Xu ZY, Hu C, Lv H, Xie HX, Huang
T, Zhang YQ, Chen GP, Fu YF and Cheng XD: Exosomal miR-590-5p in
serum as a biomarker for the diagnosis and prognosis of gastric
cancer. Front Mol Biosci. 8:6365662021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao R and Zhang Y, Zhang X, Yang Y, Zheng
X, Li X, Liu Y and Zhang Y: Exosomal long noncoding RNA HOTTIP as
potential novel diagnostic and prognostic biomarker test for
gastric cancer. Mol Cancer. 17:682018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Huang F, Xu N, Wang J, Li D and
Yin L: Overexpression of serum extracellular vesicle
microRNA-215-5p is associated with early tumor recurrence and poor
prognosis of gastric cancer. Clinics (Sao Paulo). 76:e20812021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang J, Li X, Wei S, Peng L, Sang H, Jin
D, Chen M, Dang Y and Zhang G: Evaluation of the diagnostic
potential of a plasma exosomal miRNAs panel for gastric cancer.
Front Oncol. 11:6834652021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu H, Zhou J, Tang J, Min X, Yi T, Zhao J
and Ren Y: Identification of serum exosomal lncRNA MIAT as a novel
diagnostic and prognostic biomarker for gastric cancer. J Clin Lab
Anal. 34:e233232020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao K, Li S, Ding J, Wang Z, Wang D, Cao
X, Zhang Y and Dong Z: Expression and clinical value of circRNAs in
serum extracellular vesicles for gastric cancer. Front Oncol.
12:9628312022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei S, Peng L, Yang J, Sang H, Jin D, Li
X, Chen M, Zhang W, Dang Y and Zhang G: Exosomal transfer of
miR-15b-3p enhances tumorigenesis and malignant transformation
through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric
cancer. J Exp Clin Cancer Res. 39:322020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang N, Wang L, Yang Y, Gong L, Xiao B and
Liu X: A serum exosomal microRNA panel as a potential biomarker
test for gastric cancer. Biochem Biophys Res Commun. 493:1322–1328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang JF, Jiang YM, Zhan WH, Ye SP, Li TY
and Zhang JN: Screening of serum exosomal miRNAs as diagnostic
biomarkers for gastric cancer using small RNA sequencing. J Oncol.
2022:53465632022.PubMed/NCBI
|
|
31
|
Tang S, Cheng J, Yao Y, Lou C, Wang L,
Huang X and Zhang Y: Combination of four serum exosomal MiRNAs as
novel diagnostic biomarkers for early-stage gastric cancer. Front
Genet. 11:2372022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang G, Wang J, Dong W, Dai K and Du J:
Exosomal miRNA expression profiling and the roles of exosomal
miR-4741, miR-32, miR-3149, and miR-6727 on gastric cancer
progression. Biomed Res Int. 2022:12638122022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang
J, Yang X and Liu Z: Exosomal miR-1246 in serum as a potential
biomarker for early diagnosis of gastric cancer. Int J Clin Oncol.
25:89–99. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao
B, Ye G and Guo J: Hsa_circ_0065149 is an indicator for early
gastric cancer screening and prognosis prediction. Pathol Oncol
Res. 26:1475–1482. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Piao HY, Guo S, Wang Y and Zhang J:
Exosomal long non-coding RNA CEBPA-AS1 inhibits tumor apoptosis and
functions as a non-invasive biomarker for diagnosis of gastric
cancer. Onco Targets Ther. 13:1365–1374. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Okuda Y, Shimura T, Iwasaki H, Katano T,
Kitagawa M, Nishigaki R, Fukusada S, Natsume M, Tanaka M, Nishie H,
et al: Serum exosomal dicer is a useful biomarker for early
detection of differentiated gastric adenocarcinoma. Digestion.
102:640–649. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu X, Lu J, Wang S, Zhang Y, Ding Y, Shen
X, Jing R, Ju S, Chen H and Cong H: Circulating serum exosomal
miR-92a-3p as a novel biomarker for early diagnosis of gastric
cancer. Future Oncol. 17:907–919. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin LY, Yang L, Zeng Q, Wang L, Chen ML,
Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, et al: Tumor-originated
exosomal lncUEGC1 as a circulating biomarker for early-stage
gastric cancer. Mol Cancer. 17:842018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li S, Zhang M, Zhang H, Hu K, Cai C, Wang
J, Shi L, Ma P, Xu Y and Zheng P: Exosomal long noncoding RNA
lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer.
Clin Chim Acta. 501:252–257. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kahroba H, Samadi N, Mostafazadeh M,
Hejazi MS, Sadeghi MR, Hashemzadeh S, Eftekhar Sadat AT and Karimi
A: Evaluating the presence of deregulated tumoral onco-microRNAs in
serum-derived exosomes of gastric cancer patients as noninvasive
diagnostic biomarkers. Bioimpacts. 12:127–138. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H,
Zhang K, Li J, Chang R, Xie T, et al: Circulating exosomal gastric
cancer-associated long noncoding RNA1 as a biomarker for early
detection and monitoring progression of gastric cancer: A
multiphase study. JAMA Surg. 155:572–579. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ge L, Zhang N, Li D, Wu Y, Wang H and Wang
J: Circulating exosomal small RNAs are promising non-invasive
diagnostic biomarkers for gastric cancer. J Cell Mol Med.
24:14502–14513. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dong Z, Sun X, Xu J, Han X, Xing Z, Wang
D, Ge J, Meng L and Xu X: Serum membrane type 1-matrix
metalloproteinase (MT1-MMP) mRNA protected by exosomes as a
potential biomarker for gastric cancer. Med Sci Monit.
25:7770–7783. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chung KY, Quek JM, Neo SH and Too HP:
Polymer-based precipitation of extracellular vesicular miRNAs from
serum improve gastric cancer miRNA biomarker performance. J Mol
Diagn. 22:610–618. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun
J, Zhang M, Lan T, Gu B, Li S and Ma P: Serum exosomal long
noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker
for gastric cancer. Onco Targets Ther. 12:10035–10041. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Helwa I, Cai J, Drewry MD, Zimmerman A,
Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer
WD, et al: A Comparative study of serum exosome isolation using
differential ultracentrifugation and three commercial reagents.
PLoS One. 12:e01706282017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hu Y, Fang JY and Xiao SD: Can the
incidence of gastric cancer be reduced in the new century? J Dig
Dis. 14:11–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang J, Yu JC, Kang WM and Ma ZQ:
Treatment strategy for early gastric cancer. Surg Oncol.
21:119–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji
JF and Li ZY: Exosome-derived noncoding RNAs in gastric cancer:
Functions and clinical applications. Mol Cancer. 20:992021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D,
Xu X, Zuo Y, Zhao Y, Wei YQ, et al: Exosomal tRNA-derived small RNA
as a promising biomarker for cancer diagnosis. Mol Cancer.
18:742019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li C, Ni YQ, Xu H, Xiang QY, Zhao Y, Zhan
JK, He JY, Li S and Liu YS: Roles and mechanisms of exosomal
non-coding RNAs in human health and diseases. Signal Transduct
Target Ther. 6:3832021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Saaoud F, Drummer IVC, Shao Y, Sun Y, Lu
Y, Xu K, Ni D, Jiang X, Wang H and Yang X: Circular RNAs are a
novel type of non-coding RNAs in ROS regulation, cardiovascular
metabolic inflammations and cancers. Pharmacol Ther.
220:1077152021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Long F, Lin Z, Li L, Ma M, Lu Z, Jing L,
Li X and Lin C: Comprehensive landscape and future perspectives of
circular RNAs in colorectal cancer. Mol Cancer. 20:262021.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang Y, Liang W, Zhang P, Chen J, Qian H,
Zhang X and Xu W: Circular RNAs: Emerging cancer biomarkers and
targets. J Exp Clin Cancer Res. 36:1522017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu H, Guan Z, Cuk K, Brenner H and Zhang
Y: Circulating microRNA biomarkers for lung cancer detection in
Western populations. Cancer Med. 7:4849–4862. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jia E, Ren N, Shi X, Zhang R, Yu H, Yu F,
Qin S and Xue J: Extracellular vesicle biomarkers for pancreatic
cancer diagnosis: A systematic review and meta-analysis. BMC
Cancer. 22:5732022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wei Y, Guo S, Tang J, Wen J, Wang H, Hu X
and Gu Q: MicroRNA-19b-3p suppresses gastric cancer development by
negatively regulating neuropilin-1. Cancer Cell Int. 20:1932020.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen Z, Liu K, Li L, Chen Y and Du S:
miR-215 promotes cell migration and invasion of gastric cancer by
targeting Retinoblastoma tumor suppressor gene 1. Pathol Res Pract.
213:889–894. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li N, Zhang QY, Zou JL, Li ZW, Tian TT,
Dong B, Liu XJ, Ge S, Zhu Y, Gao J and Shen L: miR-215 promotes
malignant progression of gastric cancer by targeting RUNX1.
Oncotarget. 7:4817–4828. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zang Y, Wang T, Pan J and Gao F: miR-215
promotes cell migration and invasion of gastric cancer cell lines
by targeting FOXO1. Neoplasma. 64:579–587. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vaidyanathan R, Soon RH, Zhang P, Jiang K
and Lim CT: Cancer diagnosis: From tumor to liquid biopsy and
beyond. Lab Chip. 19:11–34. 2018.PubMed/NCBI
|
|
65
|
Ye Q, Ling S, Zheng S and Xu X: Liquid
biopsy in hepatocellular carcinoma: Circulating tumor cells and
circulating tumor DNA. Mol Cancer. 18:1142019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cai X, Janku F, Zhan Q and Fan JB:
Accessing genetic information with liquid biopsies. Trends Genet.
31:564–575. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yu W, Hurley J, Roberts D, Chakrabortty
SK, Enderle D, Noerholm M, Breakefield XO and Skog JK:
Exosome-based liquid biopsies in cancer: Opportunities and
challenges. Ann Oncol. 32:466–477. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sidhom K, Obi PO and Saleem A: A review of
exosomal isolation methods: Is size exclusion chromatography the
best option? Int J Mol Sci. 21:64662020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Livshits MA, Khomyakova E, Evtushenko EG,
Lazarev VN, Kulemin NA, Semina SE, Generozov EV and Govorun VM:
Corrigendum: Isolation of exosomes by differential centrifugation:
Theoretical analysis of a commonly used protocol. Sci Rep.
6:214472016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cvjetkovic A, Lötvall J and Lässer C: The
influence of rotor type and centrifugation time on the yield and
purity of extracellular vesicles. J Extracell Vesicles. 3:2014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang XX, Sun C, Wang L and Guo XL: New
insight into isolation, identification techniques and medical
applications of exosomes. J Control Release. 308:119–129. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Böing AN, van der Pol E, Grootemaat AE,
Coumans FA, Sturk A and Nieuwland R: Single-step isolation of
extracellular vesicles by size-exclusion chromatography. J
Extracell Vesicles. 3:2014. View Article : Google Scholar
|
|
73
|
Cha BS, Park KS and Park JS: Signature
mRNA markers in extracellular vesicles for the accurate diagnosis
of colorectal cancer. J Biol Eng. 14:42020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fu F, Jiang W, Zhou L and Chen Z:
Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic
stage and grade of colorectal cancer. Transl Oncol. 11:221–232.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Nazarova I, Slyusarenko M, Sidina E,
Nikiforova N, Semiglazov V, Semiglazova T, Aigner A, Rybakov E and
Malek A: Evaluation of colon-specific plasma nanovesicles as new
markers of colorectal cancer. Cancers (Basel). 13:39052021.
View Article : Google Scholar : PubMed/NCBI
|