Introduction
Pancreatic cancer remains closely correlated with an
unfavorable prognosis, characterized by a 5-year overall survival
(OS) rate of <5% (1). Despite
the implementation of surgical resection coupled with
chemoradiotherapy, the 5-year OS rate was only marginally improved
to 20–25% (2). Of note, a paradigm
shift in pancreatic cancer treatment has emerged with the advent of
immune checkpoint molecule targeting (3). Immune checkpoint inhibitors (ICIs)
have the ability to activate T cells, offering a promising avenue
for tumor immunotherapy. Examples include anti-cytotoxic
T-lymphocyte-associated protein 4 and anti-programmed cell death 1
agents (4). In essence, the
identification of biomarkers capable of serving as targets for
immunotherapy, while also predicting both the prognosis of patients
with pancreatic cancer and their responsiveness to chemotherapy,
holds immense potential. Such biomarkers may substantially enhance
treatment effectiveness and simultaneously mitigate unnecessary
interventions in the realm of pancreatic cancer therapy (5).
Immunogenic cell death (ICD) represents a regulated
form of cell demise triggered by treatments such as chemotherapy
and radiotherapy. This process effectively prompts an immune
response within the tumor microenvironment (TME) (6). ICD achieves this by liberating
tumor-associated antigens and tumor-specific antigens, thereby
setting off critical ‘danger signals’ that serve as triggers for
immune activation (7). Of note, ICD
is characterized by the release and elevation of damage-related
molecular patterns (DAMPs), pro-antigen inflammatory cytokines and
inflammatory mediators. Among the DAMPs, significant players
encompass ATP, calreticulin, high mobility group box protein B1
(HMGB1), heat shock proteins, type I interferon (IFN) and Annexin
1. The orchestration of these molecules culminates in the
activation and recruitment of antigen-presenting cells,
subsequently prompting T-cell activation and an adaptive immune
response directed at tumor antigens. Recognizing the potential of
combining multiple immunotherapeutic strategies rooted in ICD
induction, a novel avenue for advancing tumor immunotherapy
emerges, exemplified by the synergy achieved through combined ICIs
(8). Consequently, the exploration
of therapeutic targets and methodologies associated with ICD holds
a prominent position in contemporary pancreatic cancer
research.
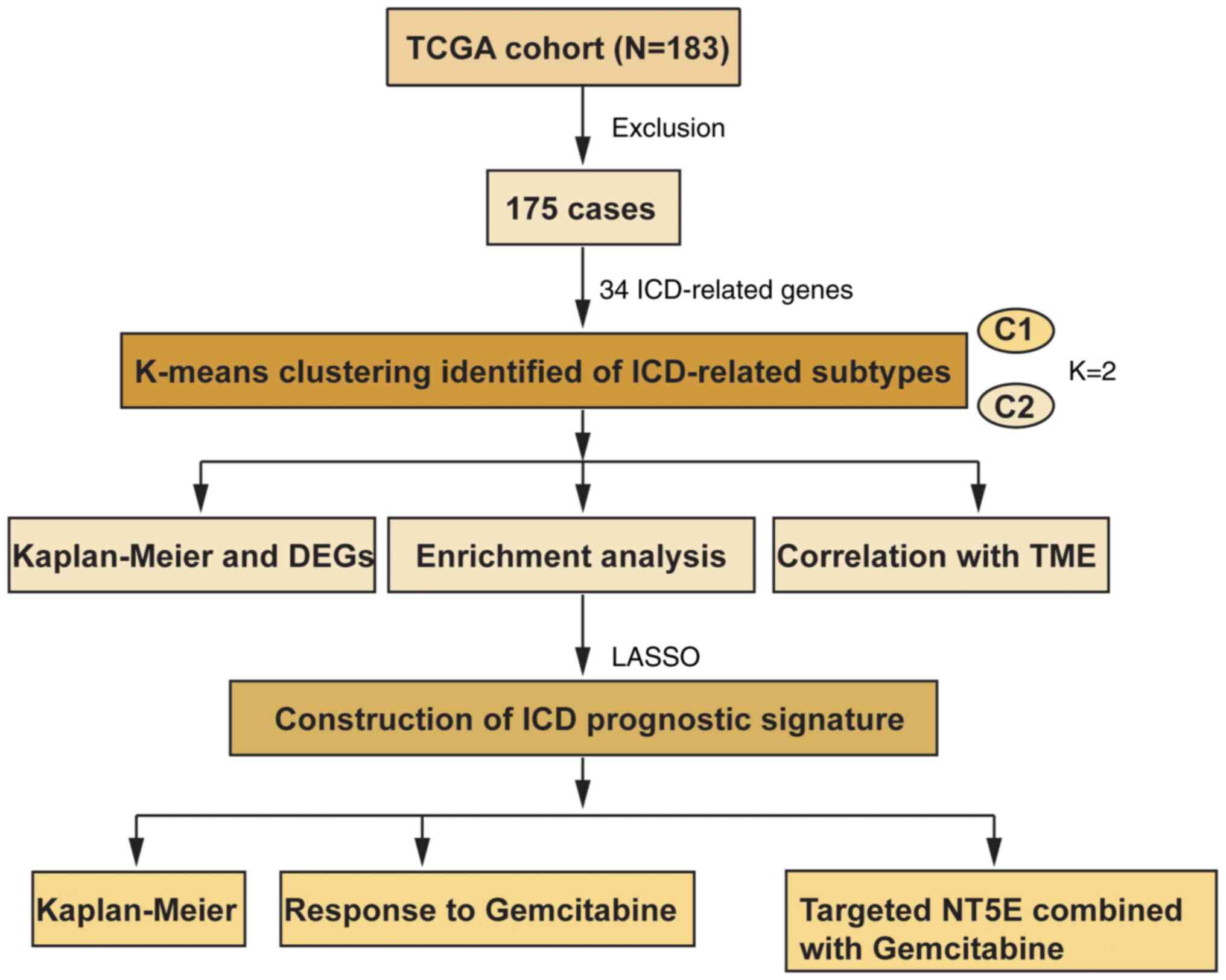
In the present study, a comprehensive analysis was
conducted involving consensus clustering of pancreatic cancer
samples into two distinct subtypes based on the expression patterns
of ICD-associated genes (Fig. 1).
Of note, high ICD levels were found to be associated with
unfavorable prognostic outcomes and compromised immune status in
patients with pancreatic cancer. Furthermore, an innovative
prognostic signature named the ICD-Related Prognostic Signature
(IRPS) was devised and its efficacy in predicting individual
responses to both chemotherapy and immunotherapy interventions was
demonstrated. The present results shed new light on the potential
immunotherapeutic strategies to manage pancreatic cancer.
Materials and methods
Patient data acquisition
Gene expression data, alongside clinical data and
single nucleotide mutation data of 183 patients with pancreatic
cancer were obtained from The Cancer Genome Atlas (TCGA) data
portal (https://cancergenome.nih.gov/).
Subsequent to the exclusion of cases with incomplete clinical
information (included survival status or time, T, N, and M stage),
a comprehensive follow-up dataset comprising 175 patients was
compiled for analysis (female, n=78; male, n=97; age, >60 years,
n=119; age ≤60 years, n=56). The dataset GSE183795 (9), containing 105 samples of normal
pancreatic tissue and 139 samples of pancreatic cancer tissue, was
downloaded from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/).
Consensus clustering
A collection of 34 ICD-related genes, as delineated
in previous studies (10,11), was curated. Protein-protein
interaction network was visulized by STRING (https://cn.string-db.org/). These genes were
subsequently subjected to a consensus clustering analysis through
the utilization of the ‘ConsensusClusterPlus’ package of the R
software. The K-means clustering algorithm was employed to discern
and delineate robust and stable ICD-related subtypes within the
realm of pancreatic cancer.
Functional enrichment analysis
The ‘Limma’ package of R software was used for the
identification of differentially expressed genes (DEGs), with a
screening threshold set at a false discovery rate-corrected
P<0.001 and |log fold change| >2. Subsequently, these DEGs
underwent comprehensive functional elucidation and pathway analysis
through the application of Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG). The enrichment analyses
for GO and KEGG were conducted using the ‘clusterProfiler’
package.
Association between ICD-related
subtypes and somatic mutations or TME
For the visualization of copy number variation (CNV)
data, the ‘ComplexHeatmap’ package of the R software was employed
to generate a waterfall plot. The composition of 22
immune-infiltrating cell types was quantified using the CIBERSORT
methodology. Furthermore, the ESTIMATE algorithm was applied to
calculate both stromal scores and immune scores. To discern
distinctions between the two ICD subtypes, a comparison of the
expression levels of human leukocyte antigen (HLA) gene families
and immune checkpoint molecules was undertaken.
Construction of the IRPS
ICD-related genes were utilized to perform
univariate Cox regression analysis and subsequent Least Absolute
Shrinkage and Selection Operator Cox regression analysis to
calculate coefficient values. The risk score for each patient was
calculated according to the following formula: Risk
score=∑k=0ncoef(k)*x(k),
where coef (k) and × (k) are regresion coefficients. The OS or
progression-free survival (PFS) and the receiver operating
characteristic (ROC) curve were used to determine the prognostic
value of the IRPS.
Relationship between IRPS model and
TME or response to immunotherapy
The tumor mutation burden (TMB) was quantified by
tallying the number of mutations within each sample. Leveraging the
R package ‘pRRophetic’ of the R software, an exploration was
conducted to pinpoint potential drugs with sensitivity for
pancreatic cancer, guided by the ICD-related prognostic signature.
To gauge the anticipated response to immunotherapy, Tumor Immune
Dysfunction and Exclusion (TIDE; http://tide.dfci.harvard.edu/) was applied.
Human Protein Atlas
Immunohistochemical staining images of caspase 1
(CASP1) [Tumor (n=23), Normal (n=3)] and 5′-nucleotidase ecto
(NT5E) [Tumor (n=46), Normal (n=6)] in normal and pancreatic cancer
tissues (poor differentiation; moderate differentiation; highly
differentiation) were obtained from the Human Protein Atlas
(https://www.proteinatlas.org/). The
staining intensity was scored as follows: None, 0; pale yellow, 1;
brownish yellow, 2; deep brown, 3. The percentage of positive
staining was scored as follows: 0–5%, 0; 6–25%, 1; 26–50%, 2;
51–75%, 3; >75%, 4. For scoring a certain type of cells, 5
fields of view were selected and 100 cells of this type per 400×
high-magnification field were counted. The formula for the final
staining score was as follows: Staining intensity per field of view
× Positive cell percentage. The rating was as follows: 0 points,
negative; 1–4 points, weakly positive; 5–8 points, moderately
positive; 9–12 points, strongly positive.
Cell culture
The human pancreatic cancer cell lines ASPC-1,
PANC-1, SW1990 and T3M4 and the normal ductal epithelial cell line
hTERT-HPNE were obtained from the American Type Culture Collection.
Pancreatic cancer cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) + 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) in a cell incubator with 5% CO2
at 37°C. hTERT-HPNE cells were cultured in DMEM with 1 ng/ml
epidermal growth factor (cat. no. HY-P7109; MedChemExpress) and 10%
FBS.
Western blot analysis
Extracted cells were suspended in cell lysis buffer
(cat. no. P0013; Beyotime Institute of Biotechnology). The protein
concentration in each group was determined using a BCA protein
assay kit (cat. no. P0012S; Beyotime Institute of Biotechnology).
Roughly 40 µg of denatured protein was loaded into 10% SDS-PAGE
gels and subjected to electrophoresis at 200V. Subsequently, the
proteins were transferred to a PVDF membrane (Merck Millipore),
followed by blocking with 5% skimmed milk for 1 h at 37°C. The PVDF
membrane was then subjected to an overnight incubation at 4°C with
the following primary antibodies: Anti-NT5E (cat. no. ab133582),
anti-Bcl-2 (cat. no. ab32124), anti-Bax (cat. no. ab32503) and
anti-β-actin (cat. no. ab8226; all from Abcam; 1:1,000 dilution).
On the following day, the PVDF membrane was exposed to secondary
antibody [anti-rabbit (cat. no. 7074) or anti-mouse IgG (cat. no.
7076); both from Cell Signaling Technology, Inc.; 1:2,000 dilution]
for 1 h at room temperaure. Chemiluminescent substrate (cat. no.
WBKLS0500; Merck KGaA) was applied according to the manufacturer's
protocol, and chemiluminescence was captured using a Molecular
Imager ChemiDoc XRS+ (Bio-Rad Laboratories, Inc.). Image Lab
software v5.0 (Bio-Rad Laboratories, Inc.) was employed for
subsequent analysis.
Reverse transcription-quantitative
(RT-q)PCR
RNAiso Plus Regent (Takara Bio, Inc.) was used to
extract total RNA from harvested cells. RNA was reverse transcribed
to cDNA using a High Capacity cDNA kit (cat. no. 4374966; Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Real-time qPCR was performed with TB GREEN
(Takara Bio, Inc.) according to the manufacturer's protocol using
an ABI Prism 7900HT Sequence Detection System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions were
as follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles of denaturation, annealing, elongation (95°C for 20 sec,
55°C for 20 sec and 72°C for 20 sec, respectively), and final
extension (95°C for 10 sec). The primer sequences were as follows:
GAPDH sense, 5′-GCACCGTCAAGGCTGAGAAC-3′ and antisense,
5′-ATGAGGTCCACCACCCTGTTG-3′; NT5E sense, 5′-AGCGAGGACTCCAGCAAGTG-3′
and antisense, 5′-CTTGATCCGACCTTCAACTGCTG-3′. GAPDH was used as an
internal reference, and relative mRNA expression was calculated
using the 2−∆∆Cq method (12).
Lentiviral transfection
Lentiviruses [short hairpin RNA (sh)-NT5E and
negative control] were purchased from Shanghai GenePharma Co., Ltd.
The sequences were as follows: sh-NT5E, 5′-GGATACACTTCCAAAGAAA-3′;
negative control, 5′-GATGGAGAAGCTCGCTGATTT-3′. The pancreatic
cancer cells were transfected with lentivirus according to the
manufacturer's instructions and the stably transfected cells were
selected by puromycin (2 µg/ml; cat. no. A1113803; Thermo Fisher
Scientific, Inc.) for two weeks. Extraction of total protein and
detection of knockdown efficiency were then performed.
Cell viability assay
Roughly 1×104 cells were plated per well
in 96-well plates and allowed to incubate for 24 h. Following
treatment with Gemcitabine (1 µM; cat. no. HY-B0003;
MedChemExpress), the cell viability was determined by treating the
cells with CCK-8 reagent (cat. no. 96992-3000TESTS-F; Merck KGaA)
at 37°C for 2 h, in accordance with the manufacturer's
instructions. Subsequently, the absorbance was recorded at 564 nm
using a microplate reader (Infinite 200 PRO; Tecan Group, Ltd.) to
quantify cell viability.
Apoptosis assessment
Approximately 1×106 cells originating
from each group were subjected to incubation with Annexin V-FITC
(20 µg/ml) and propidium iodide (50 µg/ml; cat. no. C1062M;
Beyotime Institute of Biotechnology) for a duration of 30 min.
Subsequent to incubation, flow cytometry using FC500 (Beckman
Coulter, Inc.) was employed to determine cell apoptosis, and data
were then subjected to analysis using BD CellQuest Pro (v 5.1; BD
Biosciences).
Statistical analysis
Values are expressed as the mean ± standard
deviation. R software (v.4.2.1) and SPSS (v.19.0; IBM Corp.) were
used to perform analysis and visualization. Spearman correlation
analysis was used to determine the correlation between immune
cells. Kaplan-Meier plotter (https://kmplot.com) was used for survival analysis.
DComparisons of two groups were performed by unpaired Student's
t-tests. One-way ANOVA was used to conduct comparisons of multiple
groups, followed by Tukey's post-hoc test. P<0.05 was considered
to indicate statistical significance.
Results
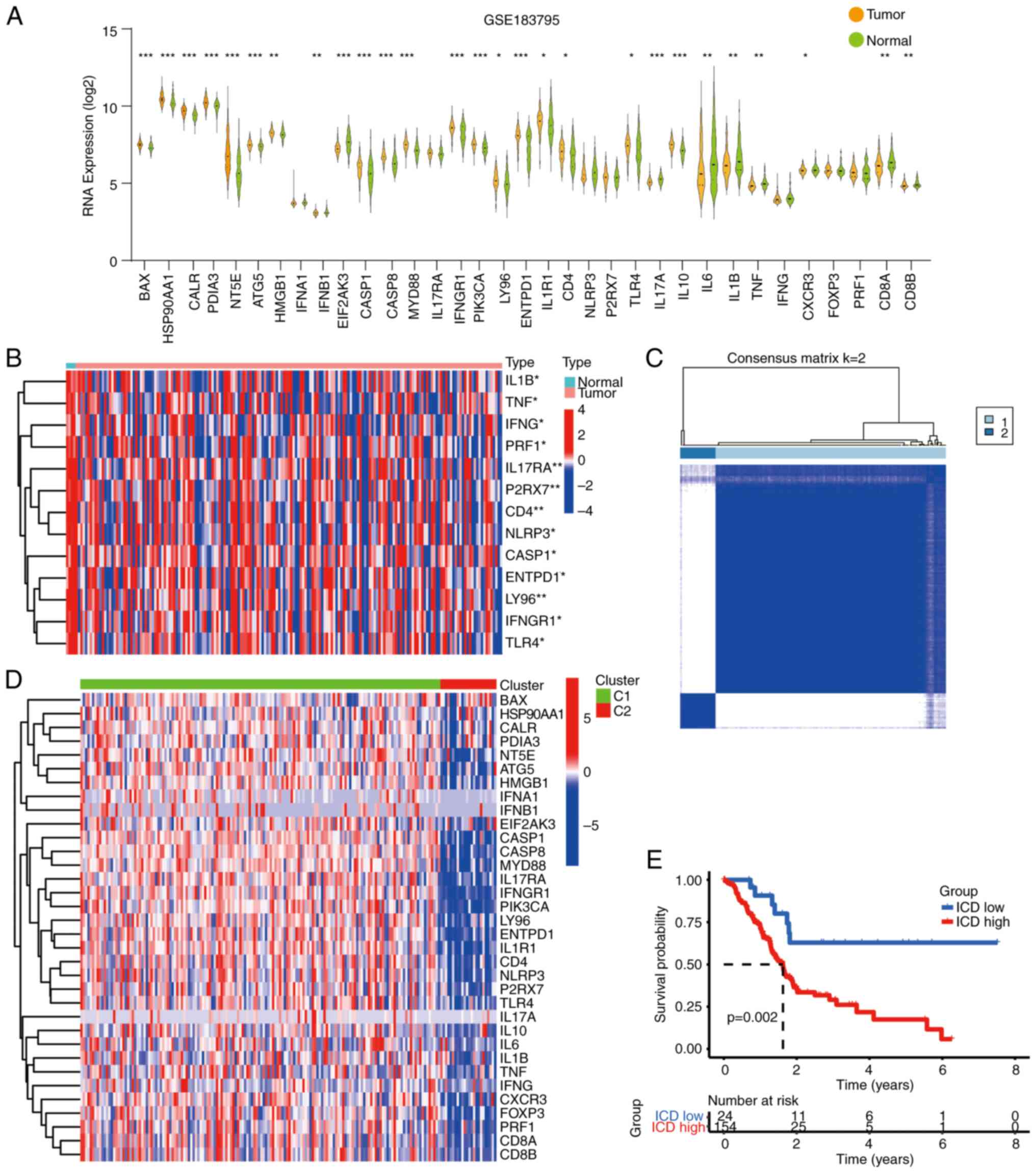
Identification of two ICD-related
subtypes
Utilizing a set of 34 ICD-related genes, a consensus
clustering analysis was conducted on pancreatic cancer samples. Of
note, certain ICD-related genes exhibited abnormal expression in
pancreatic cancer samples from the TCGA and GSE183795 datasets
(Fig. 2A and B). The
protein-protein interaction network of these downregulated genes
was further visualized using the STRING database (Fig. S1). Moving forward, a consensus
clustering analysis was performed, yielding the grouping of
pancreatic cancer samples into two distinct subtypes through the
application of the K-means algorithm (Fig. 2C). Of note, the ICD-related genes
displayed an upregulation trend within cluster C1, corresponding to
the ICD-high subtype (Fig. 2D). In
addition, the OS of patients in the ICD-high group was
significantly shorter (Fig. 2E).
Collectively, these findings underscore the prognostic significance
of ICD-related genes within the context of pancreatic cancer.
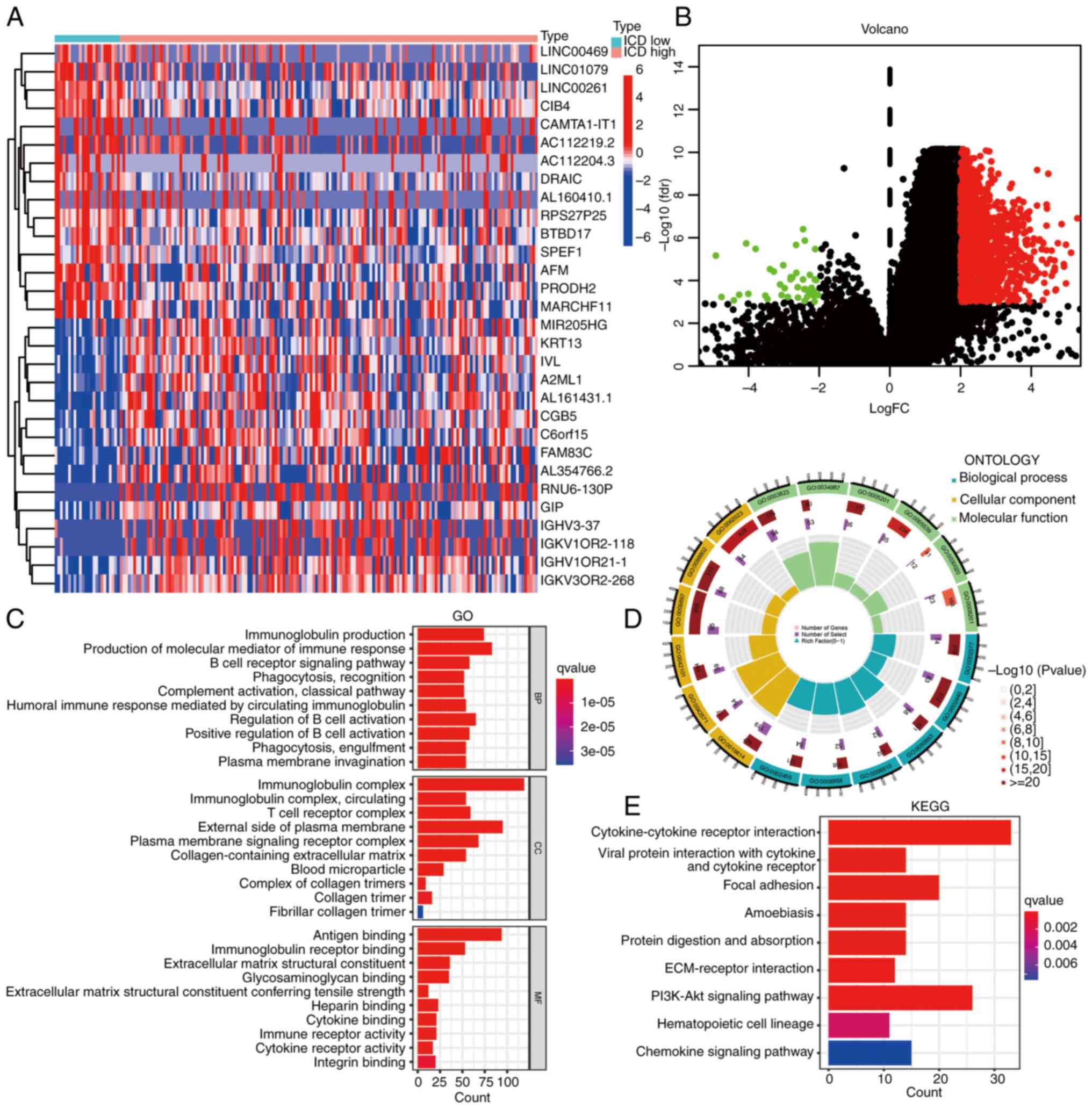
Identification of DEGs between ICD
subtypes
Subsequently, leveraging the prognostic significance
attributed to ICD-related genes, identification of DEGs was
performed for subsequent enrichment analysis. The DEGs were
visually represented through both heatmap and volcano plot
depictions (Fig. 3A and B). As
presented in Figs. 2B and 3B, cluster 2 or downregulated genes were
so few that enrichment analysis could not be performed.
Accordingly, the current enrichment analysis primarily represents
the enrichment results of upregulated genes or cluster 1. Of note,
GO enrichment analysis indicated that DEGs exhibited enrichment in
various immune-related functions, encompassing processes such as
‘immunoglobulin production’, ‘production of molecular mediator of
immune response’, ‘antigen binding’, ‘T-cell receptor complex’ and
‘B-cell receptor signaling pathway’ (Fig. 3C and D). Furthermore, KEGG
enrichment analysis highlighted the involvement of the DEGs with
immune-related pathways, including ‘cytokine-cytokine receptor
interaction’, ‘focal adhesion’, ‘ECM-receptor interaction’ and
‘PI3K-AKT signaling pathway’ (Fig.
3E). These results emphasize the substantial involvement of the
DEGs in various immune processes, ultimately shedding light on
their roles within the realm of immunity.
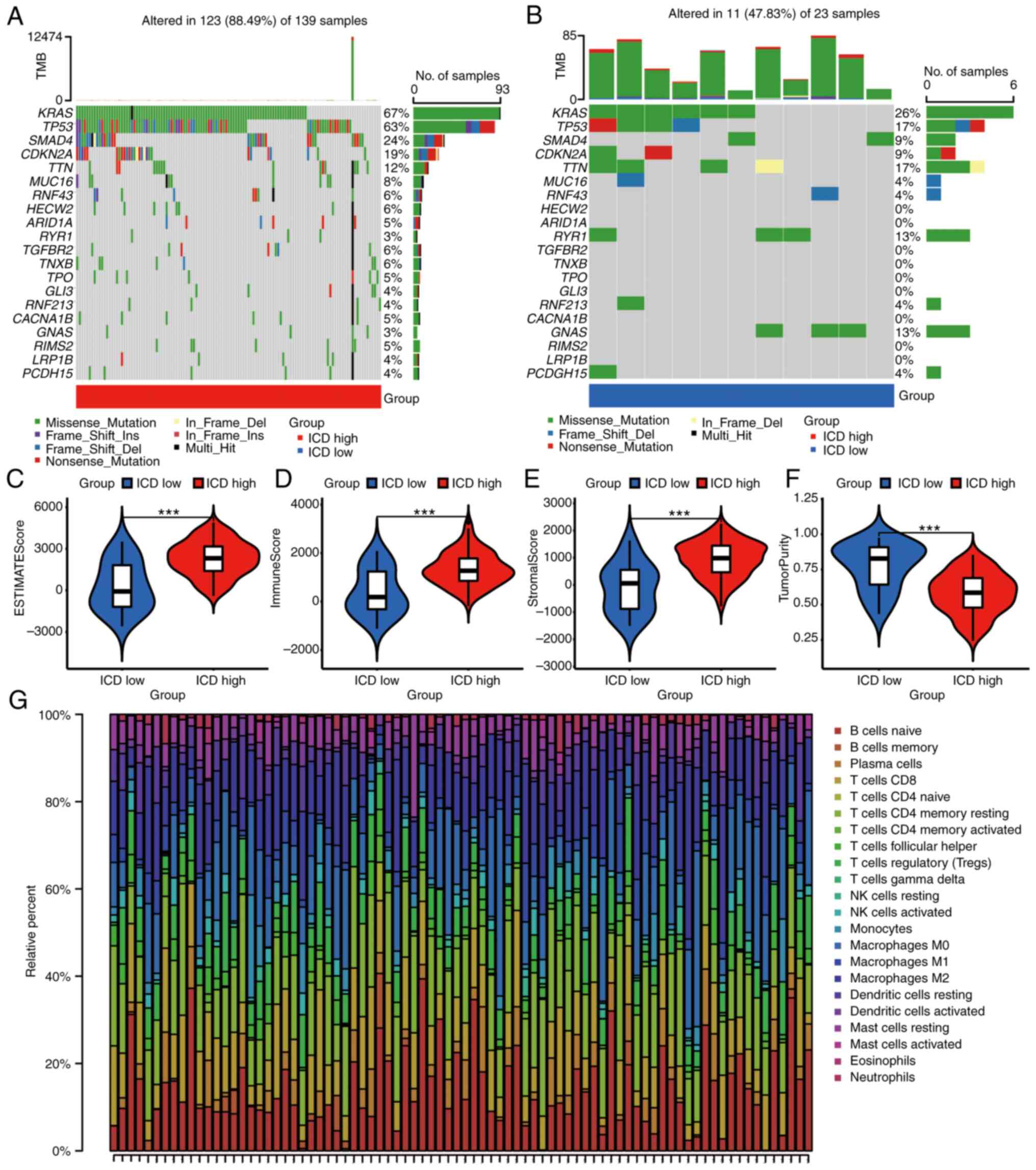
ICD-related subtypes are associated
with TME and mutations
To shed light on the connection between somatic
mutations and ICD-related subtypes, the somatic mutation profiles
within the two subtypes were visualized. Of note, the ICD-high
subtype exhibited a higher frequency of somatic mutations (Fig. 4A and B). These mutations included
KRAS (67%), TP53 (63%), SMAD4 (24%), CDKN2A (19%) and TTN (12%),
among others. Moving forward, a comparative analysis of TME
composition revealed that the ICD-high subtype displayed elevated
values in terms of the estimate score, immune score and stromal
score (Fig. 4C-E). Conversely, the
ICD-low subtype exhibited lower tumor purity (Fig. 4F).
Further exploration encompassed the assessment of
immune cell infiltration between two subtypes. Utilizing CIBERSORT,
the composition of 22 distinct immune cell types within the 175
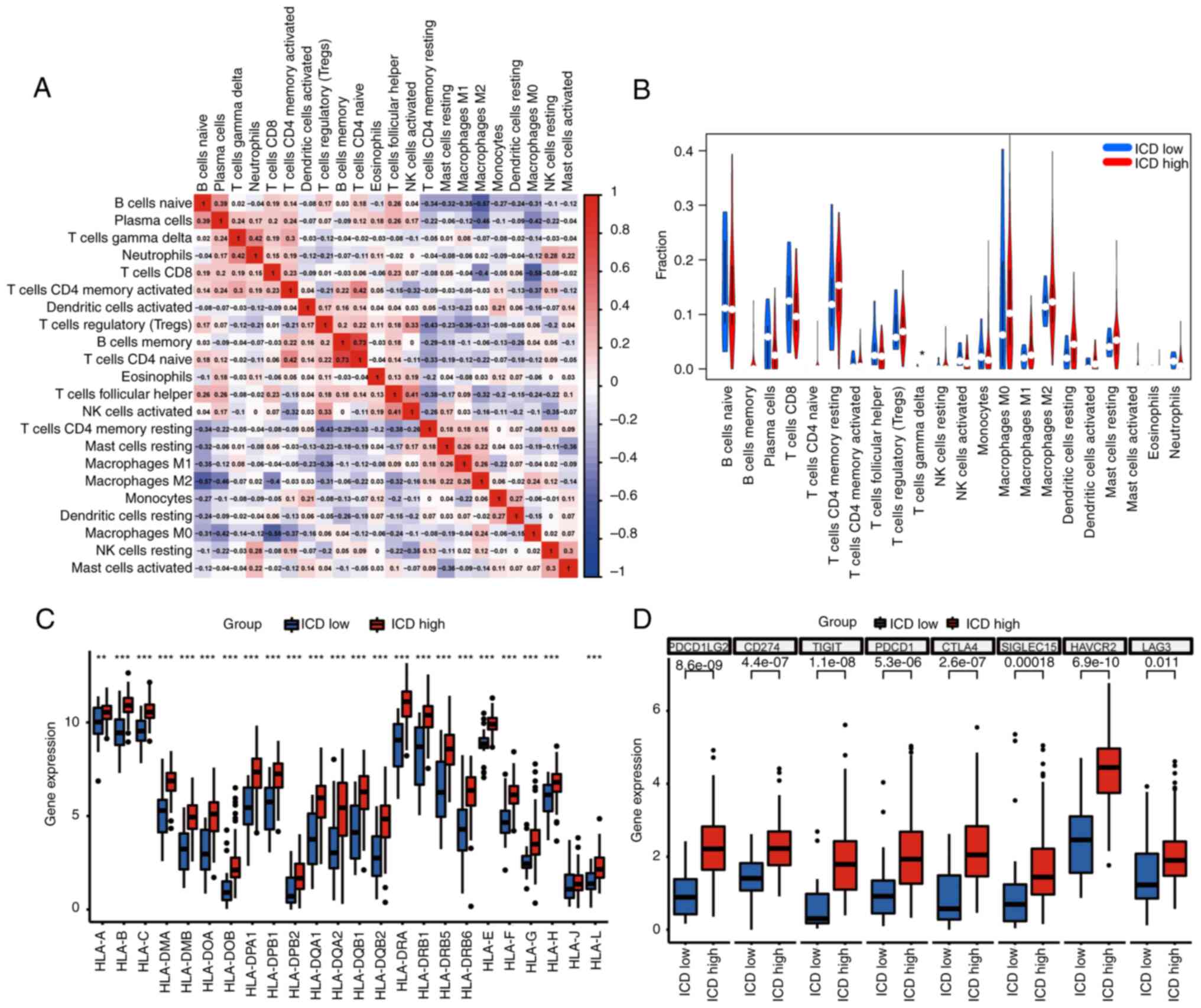
pancreatic cancer samples was visualized (Fig. 4G). Of note, a positive correlation
was observed between CD4-naive T cells and CD4 memory-activated T
cells, while macrophages M0 demonstrated a negative correlation
with T cells CD8 (Fig. 5A).
Specifically, the fraction of T cells gamma delta was noted to be
upregulated among patients in the ICD high group (Fig. 5B). Furthermore, a substantial
portion of HLA gene families and immune checkpoint molecules
displayed upregulation in the ICD-high group (Fig. 5C and D). These collective findings
firmly establish the association between ICD-related subtypes and
the intricate status of the tumor immune milieu.
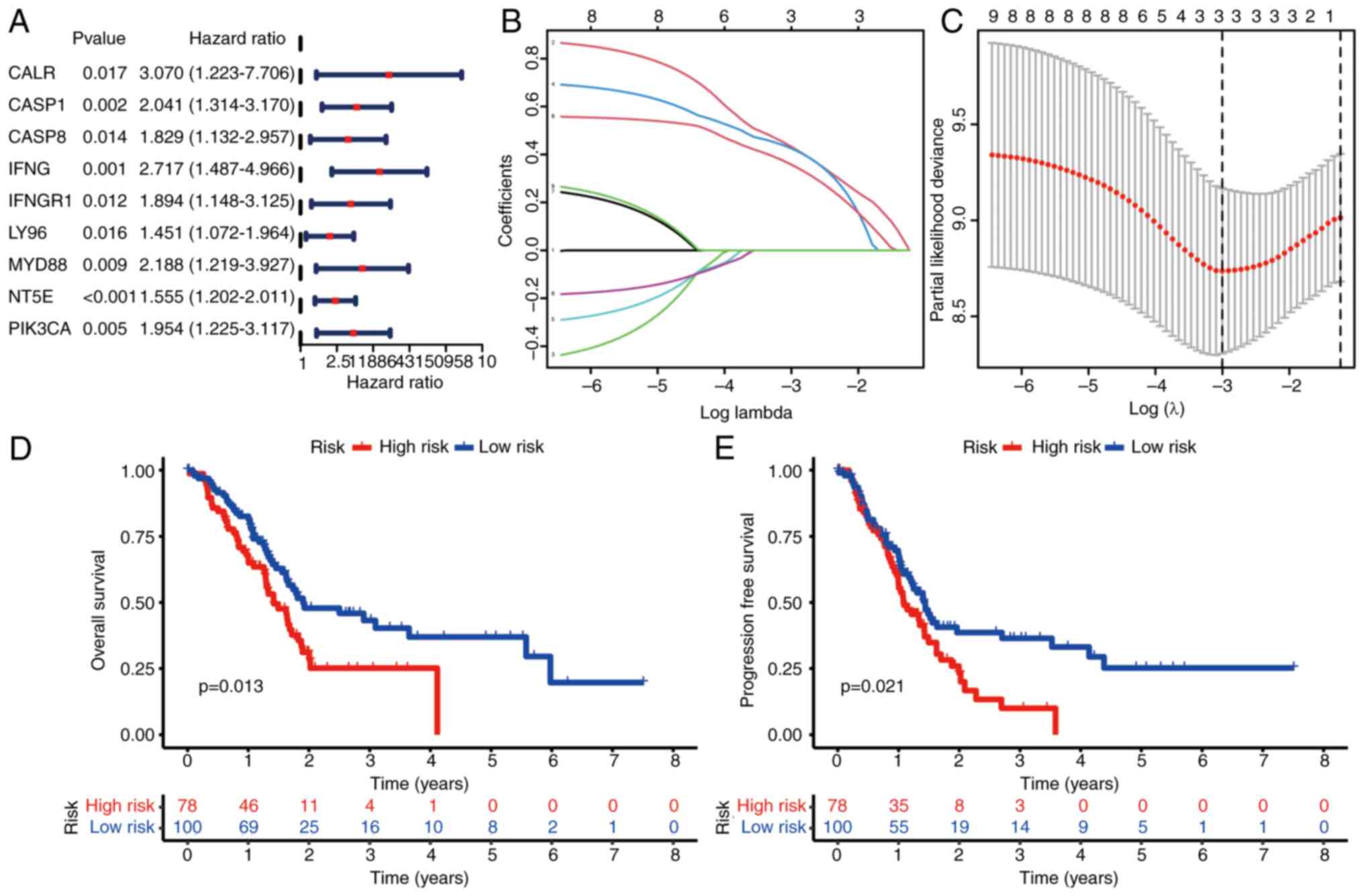
Construction of IRPS model
Subsequently, the ICD-related genes were harnessed
to construct a prognostic signature. Employing univariate Cox
regression analysis, nine ICD-related genes were identified that
exhibited associations with OS (Fig.
6A). Out of these, two ICD-related genes were meticulously
chosen for the construction of the IRPS model (Fig. 6B and C). The OS and PFS of
individuals categorized as high-risk were notably shorter in
comparison to those classified as low-risk (Fig. 6D and E). This trend was mirrored in
the higher count of deceased patients within the high-risk group
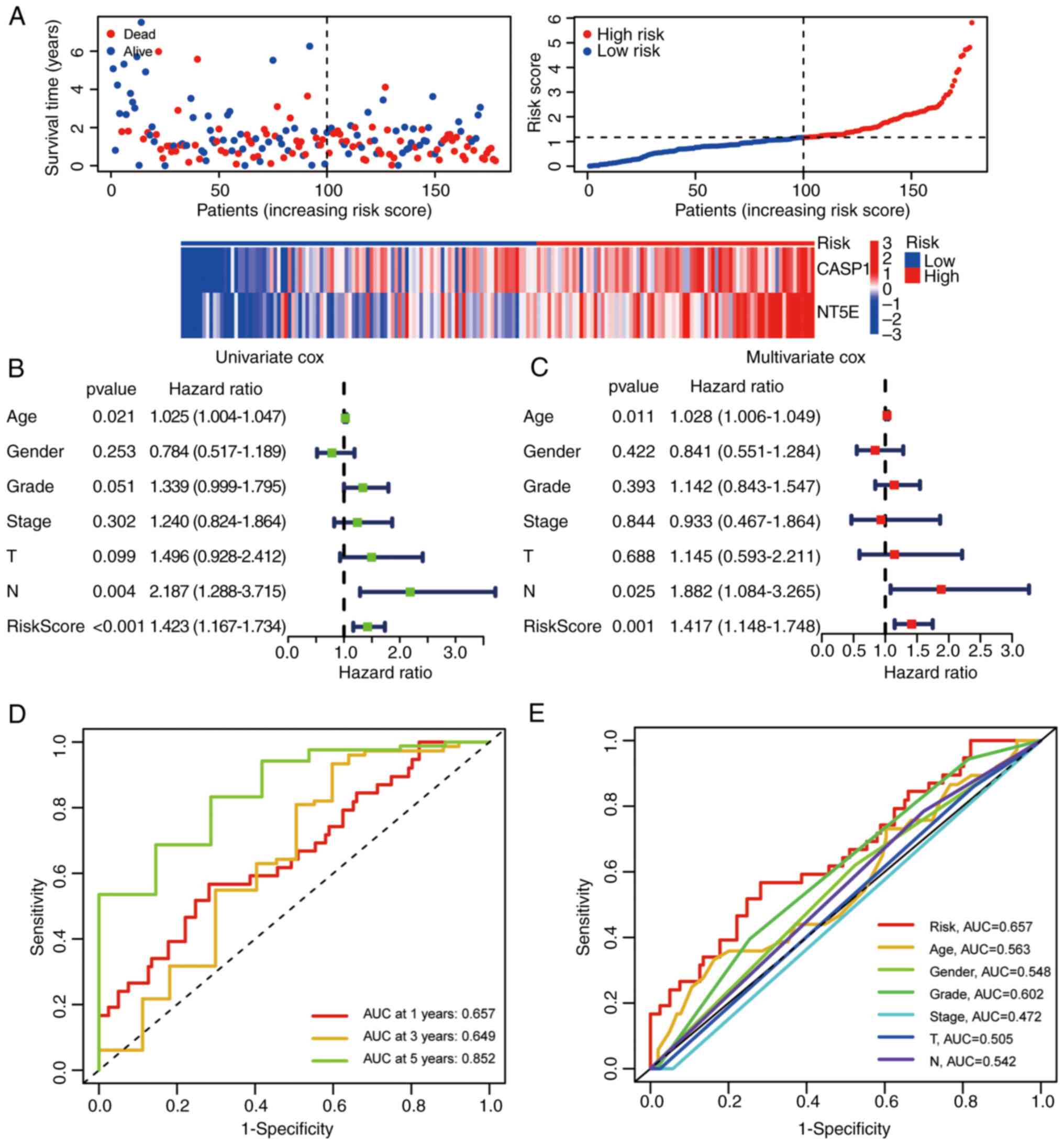
(Fig. 7A). Both univariate and
multivariate Cox regression analyses demonstrated that the IRPS
model stood as an independent prognostic factor in pancreatic
cancer (Fig. 7B and C). Finally,
the IRPS model's predictive capacity for OS was evaluated through
ROC curve analysis. The AUC values for 1, 3 and 5 years were 0.657,
0.649 and 0.852, respectively (Fig.
7D). The OS prediction potential of the risk score was further
compared against other clinical features at 1 year, yielding the
following AUC values: IRPS risk score, 0.657; age, 0.563; gender,
0.548; grade, 0.602; stage, 0.472; T, 0.505; and N, 0.542 (Fig. 7E).
Relationship between IRPS model and
TME
Recognizing the pivotal role of ICD in response to
immunotherapy, an exploration into the association between the IRPS
model and the TME was undertaken. The results of this analysis
demonstrated a notable association between patients categorized as
high-risk and factors such as HLA, T-cell inhibition and checkpoint
molecules (Fig. 8A). Furthermore,
individuals classified as high-risk were characterized by an
elevated frequency of somatic mutations (Fig. 8B and C). Although the TMB score in
high-risk patients did not rank extremely high (Fig. 8D), the OS among high-risk patients
with an elevated TMB was comparatively shorter compared to other
types (Fig. 8E and F). Despite a
moderate TIDE score for high-risk patients (Fig. 8G), their OS remained diminished.
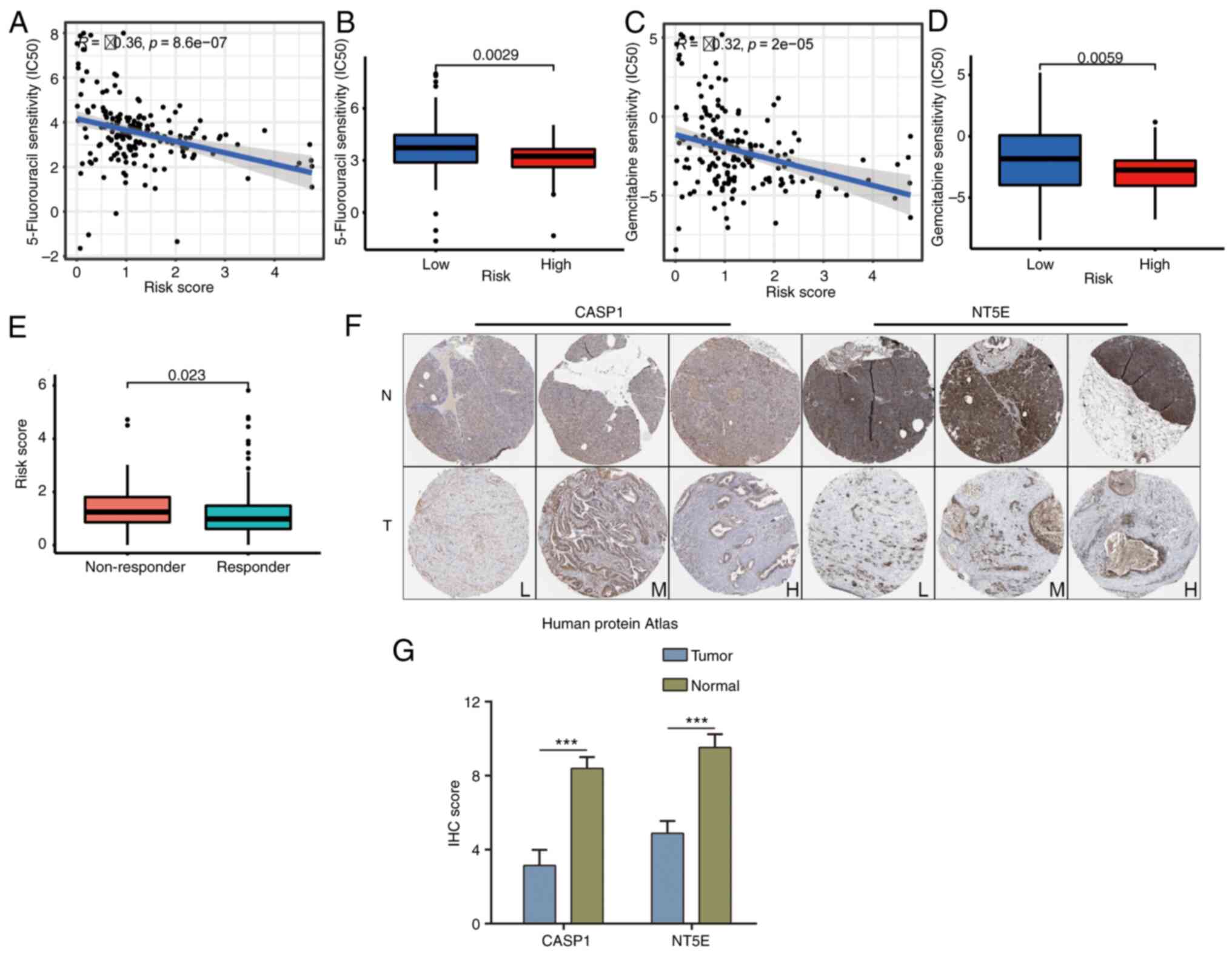
Further insight revealed that individuals categorized as low-risk
exhibited limited sensitivity to 5-Fluorouracil and Gemcitabine
treatments (Fig. 9A-D). Of note,
these low-risk patients displayed a greater propensity for positive
responses to immunotherapy (Fig.
9E). Remarkably, CASP1 and NT5E were also identified as
upregulated in pancreatic cancer tissues (Fig. 9F and G). These findings collectively
underscore the intricate interplay between the IRPS model, TME,
therapeutic responses and the expression of key genes within
pancreatic cancer.
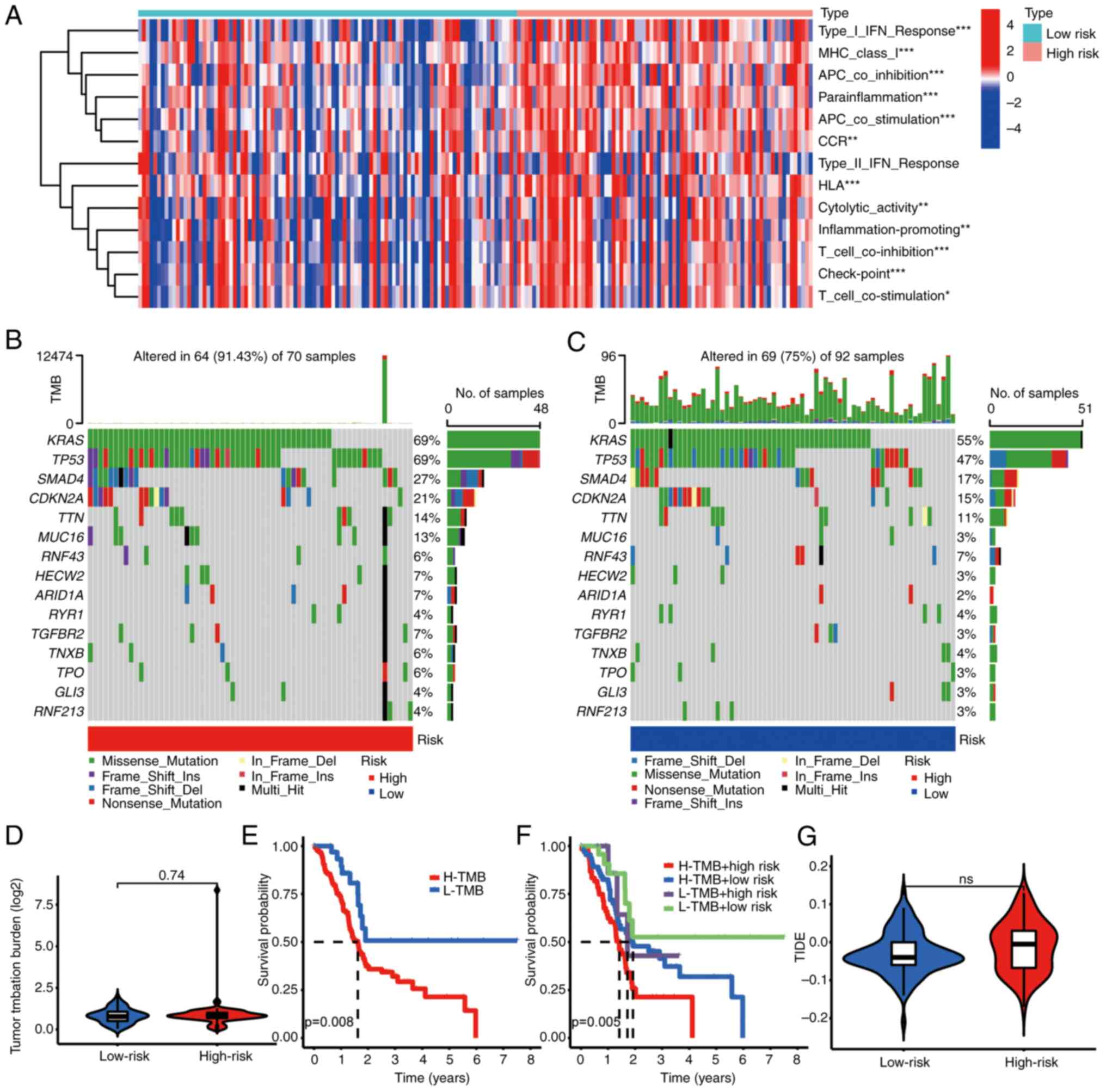 | Figure 8.Association between IRPS and tumor
mutation. (A) Patients with a high-risk IRPS were correlated with
immune function (type I IFN response, MHC class I, APC
co-inhibition, etc.). (B and C) Patients with (B) a high-risk IRPS
had a higher mutation frequency than (C) those with a low-risk
IRPS. (D) The TMB score in the high-risk group exhibited a trend to
be higher. (E) Patients with a high TMB score had poor prognosis.
(F) Patients with a high-risk IRPS had poor prognosis. (G)
Association between IRPS risk score and TIDE score. *P<0.05,
**P<0.01, ***P<0.001. IRPS, immunogenic cell death-related
prognostic signature; Del, deletion; Ins, insertion; ns, no
significance; APC, antigen-presenting cell; IFN, interferon;
H/L-TMB, high/low tumor mutation burden; TIDE, Tumor Immune
Dysfunction and Exclusion; TMB, tumor mutation burden. |
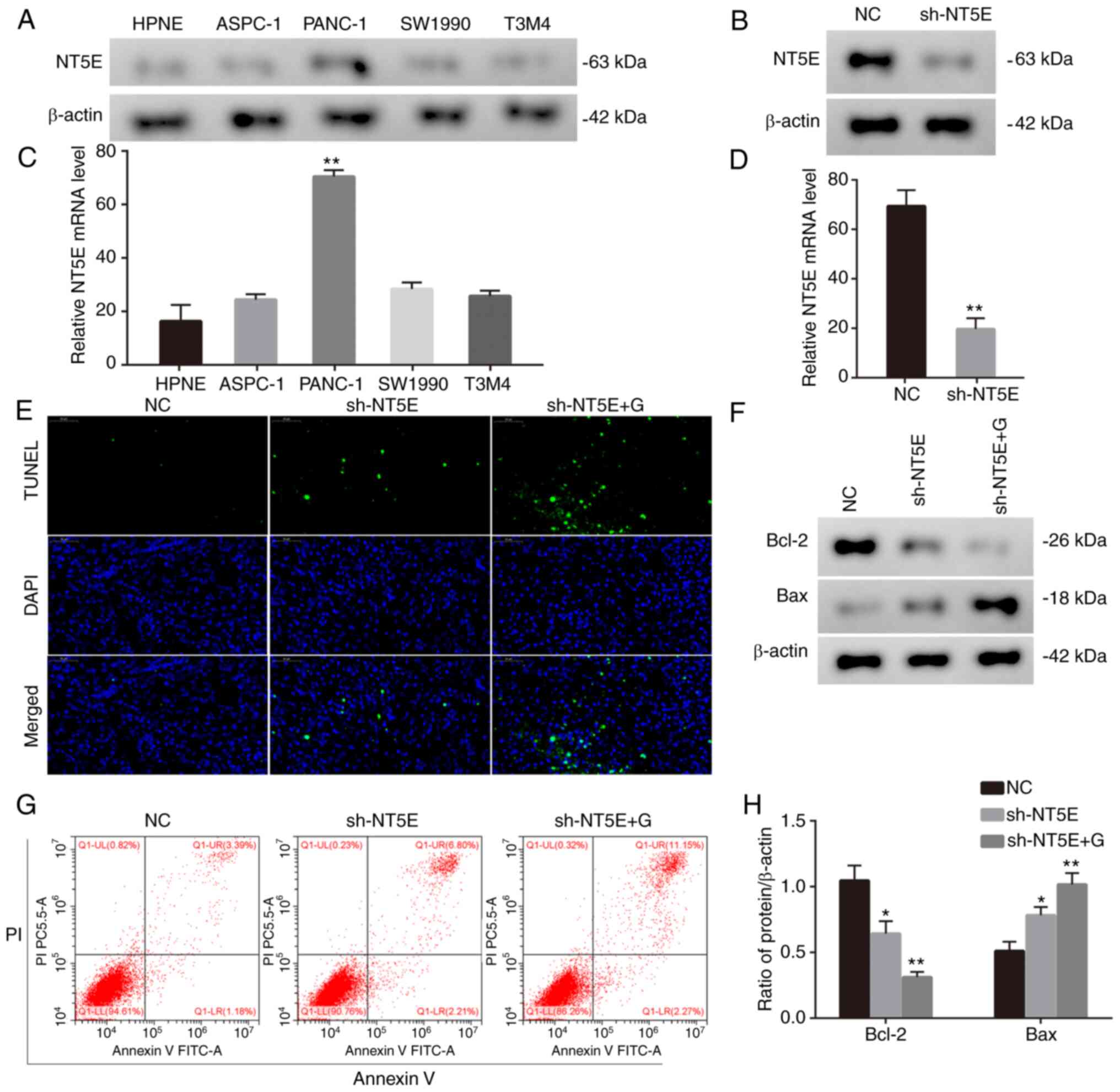
NT5E promotes progression of
pancreatic cancer
Given that CASP1 is a member of the caspase family,
its role in tumors has been explored. However, in comparison to
NT5E, its significance in research is relatively lower. NT5E has a
comprehensive role in the initiation and progression of diverse
tumors, exhibiting intricate and multifaceted mechanisms. This
complexity renders it immensely valuable for further investigation.
Thus, our focus has shifted to developing a cell line in which NT5E
expression is suppressed, allowing us to conduct in-depth
functional validation. Finally, the biological function of NT5E was
also investigated. Given the observed upregulation of NT5E in
pancreatic cancer cells (Fig. 10A and
B), lentivirus-mediated knockdown was employed to attenuate
NT5E expression in PANC-1 cells (Fig.
10C and D). Seeking to examine the utility of NT5E in the
treatment of pancreatic cancer, the effects of downregulated NT5E
in conjunction with gemcitabine treatment on pancreatic cancer
cells were explored. Of note, the results indicated that the
combination of NT5E downregulation and gemcitabine treatment
significantly promoted cell apoptosis (Fig. 10E and F). Furthermore, this
treatment led to the downregulation of Bcl-2 and upregulation of
Bax in vitro (Fig. 10G and
H). These results collectively underscore the potential role of
NT5E in influencing pancreatic cancer cell responses to treatment,
particularly in combination with gemcitabine.
Discussion
Recent research has accumulated a wealth of evidence
underscoring the pivotal role played by the tumor immune
microenvironment in the evolution of pancreatic cancer (13,14).
Despite numerous clinical trials aimed at evaluating the
effectiveness of diverse immunotherapeutic strategies in managing
pancreatic cancer, such as ICIs, cancer vaccines and chemotherapy,
the outcomes from a majority of these trials have been
underwhelming (15–17). Of note, the quest to identify
reliable biomarkers capable of categorizing patients based on their
responsiveness to immunotherapy (18), particularly ICD immunotherapy, has
emerged as a significant pursuit. In the present study, the
segmentation of pancreatic cancer samples into two distinctive
subtypes was achieved through the application of ICD-related genes.
These findings demonstrated the potential of ICD-related genes to
provide a benefit for patients undergoing chemotherapy or
immunotherapy.
The present study revealed that patients classified
under the ICD high subtype exhibited a compromised immune status
and prognosis. During the process of ICD induction, deteriorating
tumor cells release DAMPs. Subsequent to ICD, these DAMPs activate
pattern recognition receptors present on macrophages, dendritic
cells and natural killer cells, thereby triggering T-cell
activation and the initiation of immune responses (19). Furthermore, the utilization of ICIs
was observed to bolster the activity of effector T cells,
subsequently augmenting the anti-tumor efficacy (20). In line with these findings, the
present results demonstrated that the DEGs were closely linked to
immune function. Of note, the expression of genes belonging to the
HLA families and checkpoint molecules was markedly upregulated in
patients from the ICD-high group. These results strongly suggest
the potential of ICD-related genes to have a role in enhancing the
effectiveness of ICI treatment for patients with pancreatic
cancer.
Moving forward, the present study unveiled a
noteworthy trend: Patients characterized by a high IRPS risk score
exhibited a higher likelihood of responding positively to
chemotherapy or immunotherapy within the context of pancreatic
cancer treatment. The TME in pancreatic cancer comprises a diverse
array of immune cells with varying functions. Among these are
CD4+/CD8+ T cells, natural killer cells and dendritic cells, all of
which exert anti-tumor effects. Conversely, the TME is abundant in
immunosuppressive elements, such as regulatory T cells,
myeloid-derived suppressor cells and tumor-associated macrophages
(21). These cells secrete an array
of immunosuppressive factors, including IL-10, IL-23, TGF-β and
indoleamine 2,3-dioxygenase 1, contributing to the formation of an
immunosuppressive milieu conducive to pancreatic cancer progression
(22–24). Consequently, this immunosuppressive
TME curtails the immune response, leading to immune evasion and
thereby influencing the efficacy of immunotherapeutic approaches in
treating pancreatic cancer. In light of these observations, it
becomes apparent that the IRPS model, in conjunction with ICIs,
holds promise as an effective strategy to enhance the outcomes of
immunotherapy for pancreatic cancer.
Furthermore, it is worth highlighting that both NT5E
and CASP1, key components in the construction of the IRPS model,
were upregulated in pancreatic cancer tissues. In line with this,
Angelova et al (25)
reported that the infection of pancreatic cancer cells by oncolytic
parvovirus H-1 triggers signaling via the secretion of the alarmin
HMGB1 and activation of an inflammasome/CASP1 platform. Of note,
the CASP1/4/5 genes have a role in regulating innate immunity and
T-cell responses, suggesting their potential to enhance tumor
checkpoint inhibition (26). While
CASP1′s role as part of the caspase family has prompted scrutiny,
its significance as a downstream factor within the apoptotic
pathway has somewhat dampened its research appeal, particularly
when compared to the multifaceted and prominent NT5E. The intricate
involvement of NT5E across diverse aspects of tumor initiation and
progression enhances its research relevance substantially.
Consequently, our focus remains on establishing a cell line
characterized by suppressed NT5E expression for rigorous functional
validation. Importantly, previous investigations have hinted at
NT5E's involvement in inducing resistance to gemcitabine. Thus, our
strategy involves combining targeted intervention against NT5E with
gemcitabine treatment, aiming to determine whether NT5E targeting
may potentially counteract gemcitabine resistance within pancreatic
cancer scenarios. Furthermore, the present study revealed that NT5E
displayed upregulation in the pancreatic cell line PANC-1 and
patients with a lower IRPS risk score exhibited increased
sensitivity to 5-Fluorouracil and Gemcitabine. Intriguingly, the
combination of NT5E downregulation and Gemcitabine treatment was
found to facilitate cell apoptosis. These findings are in
accordance with the observations made by Chen et al
(27), who proposed a correlation
between higher NT5E expression and elevated programmed death-ligand
1 expression and TMB in patients with pancreatic cancer. In
addition, King et al (28)
noted an increase in IFN-γ expression by intratumoral CD4+ and CD8+
cells upon NT5E knockdown in pancreatic cancer. In light of these
findings, inhibition of NT5E in conjunction with Gemcitabine holds
promise as a potentially impactful therapeutic approach for
pancreatic cancer.
However, it is important to acknowledge that the
current study is not without its limitations. First, the scarcity
of available GEO datasets pertaining to pancreatic cancer posed
challenges in the external validation of the IRPS model.
Furthermore, the relatively limited sample size within the TCGA
pancreatic cancer dataset may potentially compromise the
statistical robustness of the findings. Finally, it becomes evident
that further experimentation is required to substantiate the
underlying molecular mechanisms associated with NT5E in the context
of pancreatic cancer. In future research, in vivo and in
vitro experiments will be designed to validate the functions of
NT5E. In addition, RNA sequencing should be conducted to identify
the pathways and functions influenced by it.
In conclusion, the present study has provided
valuable insight into the intricate interplay between ICD clusters
and the tumor immune microenvironment within the realm of
pancreatic cancer. Furthermore, an IRPS model that holds the
capability to predict OS and the potential response to
immunotherapy among individuals with pancreatic cancer was
successfully developed.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX was responsible for conceptualization, ML for
data collection and analysis, and supervision, and WY for the
formal analysis. WY and JX helped review and edit the manuscript.
JZ, ML and WY confirm the authenticity of all the raw data. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Declaration of interest
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ICD
|
immunogenic cell death
|
|
DAMPs
|
damage-associated molecular
patterns
|
|
TME
|
tumor microenvironment
|
|
ICI
|
immune checkpoint inhibitor
|
|
DEG
|
differentially expressed gene
|
|
IRPS
|
ICD-related prognostic signature
|
|
OS
|
overall survival
|
|
HMGB1
|
high mobility group box protein B1
|
|
IFN I
|
type I interferon
|
|
APCs
|
antigen-presenting cells
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GO
|
Gene Ontology
|
|
LASSO
|
Least Absolute Shrinkage and Selection
Operator
|
|
PFS
|
progression-free survival
|
|
ROC
|
receiver operating characteristic
|
|
TMB
|
tumor mutation burden
|
|
TIDE
|
Tumor Immune Dysfunction and
Exclusion
|
|
FBS
|
fetal bovine serum
|
References
|
1
|
Versteijne E, van Dam JL, Suker M, Janssen
QP, Groothuis K, Akkermans-Vogelaar JM, Besselink MG, Bonsing BA,
Buijsen J, Busch OR, et al: Neoadjuvant chemoradiotherapy versus
upfront surgery for resectable and borderline resectable pancreatic
cancer: Long-term results of the Dutch randomized PREOPANC trial. J
Clin Oncol. 40:1220–1230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Truty MJ, Kendrick ML, Nagorney DM, Smoot
RL, Cleary SP, Graham RP, Goenka AH, Hallemeier CL, Haddock MG,
Harmsen WS, et al: Factors predicting response, perioperative
outcomes, and survival following total neoadjuvant therapy for
borderline/locally advanced pancreatic cancer. Ann Surg.
273:341–349. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu J and Cai J: Dilemma and challenge of
immunotherapy for pancreatic cancer. Dig Dis Sci. 66:359–368. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zong L, Mo S, Sun Z, Lu Z, Yu S, Chen J
and Xiang Y: Analysis of the immune checkpoint V-domain
Ig-containing suppressor of T-cell activation (VISTA) in
endometrial cancer. Mod Pathol. 35:266–273. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ortega MA, Fraile-Martinez O, Pekarek L,
Garcia-Montero C, Alvarez-Mon MA, Castellanos AJ, García-Honduvilla
N, Buján J, Alvarez-Mon M, Sáez MA, et al: Oxidative stress markers
are associated with a poor prognosis in patients with pancreatic
cancer. Antioxidants (Basel). 11:7592022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fucikova J, Kepp O, Kasikova L, Petroni G,
Yamazaki T, Liu P, Zhao L, Spisek R, Kroemer G and Galluzzi L:
Detection of immunogenic cell death and its relevance for cancer
therapy. Cell Death Dis. 11:10132020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim R and Kin T: Current and future
therapies for immunogenic cell death and related molecules to
potentially cure primary breast cancer. Cancers (Basel).
13:47562021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hayashi K, Nikolos F and Chan KS:
Inhibitory DAMPs in immunogenic cell death and its clinical
implications. Cell Stress. 5:52–54. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang S, Tang W, Azizian A, Gaedcke J,
Ströbel P, Wang L, Cawley H, Ohara Y, Valenzuela P, Zhang L, et al:
Dysregulation of HNF1B/Clusterin axis enhances disease progression
in a highly aggressive subset of pancreatic cancer patients.
Carcinogenesis. 43:1198–1210. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Wu S, Liu F, Ke D, Wang X, Pan D,
Xu W, Zhou L and He W: An immunogenic cell death-related
classification predicts prognosis and response to immunotherapy in
head and neck squamous cell carcinoma. Front Immunol.
12:7814662021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garg AD, De Ruysscher D and Agostinis P:
Immunological metagene signatures derived from immunogenic cancer
cell death associate with improved survival of patients with lung,
breast or ovarian malignancies: A large-scale meta-analysis.
Oncoimmunology. 5:e10699382015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ahn HR, Baek GO, Yoon MG, Son JA, You D,
Yoon JH, Cho HJ, Kim SS, Cheong JY and Eun JW: HMBS is the most
suitable reference gene for RT-qPCR in human HCC tissues and blood
samples. Oncol Lett. 22:7912021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grünwald BT, Devisme A, Andrieux G, Vyas
F, Aliar K, McCloskey CW, Macklin A, Jang GH, Denroche R, Romero
JM, et al: Spatially confined sub-tumor microenvironments in
pancreatic cancer. Cell. 184:5577–5592.e18. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pekarek L, Fraile-Martinez O,
Garcia-Montero C, Alvarez-Mon MA, Acero J, Ruiz-Llorente L,
García-Honduvilla N, Albillos A, Buján J, Alvarez-Mon M, et al:
Towards an updated view on the clinical management of pancreatic
adenocarcinoma: Current and future perspectives. Oncol Lett.
22:8092021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan JQ, Wang MF, Chen HL, Shang D, Das JK
and Song J: Current advances and outlooks in immunotherapy for
pancreatic ductal adenocarcinoma. Mol Cancer. 19:322020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reiss KA, Mick R, O'Hara MH, Teitelbaum U,
Karasic TB, Schneider C, Cowden S, Southwell T, Romeo J, Izgur N,
et al: Phase II study of maintenance rucaparib in patients with
platinum-sensitive advanced pancreatic cancer and a pathogenic
germline or somatic variant in BRCA1, BRCA2, or PALB2. J Clin
Oncol. 39:2497–2505. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Parikh AR, Szabolcs A, Allen JN, Clark JW,
Wo JY, Raabe M, Thel H, Hoyos D, Mehta A, Arshad S, et al:
Radiation therapy enhances immunotherapy response in microsatellite
stable colorectal and pancreatic adenocarcinoma in a phase II
trial. Nat Cancer. 2:1124–1135. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pekarek L, Fraile-Martinez O,
Garcia-Montero C, Saez MA, Barquero-Pozanco I, Del Hierro-Marlasca
L, de Castro Martinez P, Romero-Bazán A, Alvarez-Mon MA, Monserrat
J, et al: Clinical applications of classical and novel biological
markers of pancreatic cancer. Cancers (Basel). 14:18662022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jentho E and Weis S: DAMPs and innate
immune training. Front Immunol. 12:6995632021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jardim DL, Goodman A, de Melo Gagliato D
and Kurzrock R: The challenges of tumor mutational burden as an
immunotherapy biomarker. Cancer Cell. 39:154–173. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu YT and Sun ZJ: Turning cold tumors
into hot tumors by improving T-cell infiltration. Theranostics.
11:5365–5386. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Lao M, Xu J, Duan Y, Yang H, Li
M, Ying H, He L, Sun K, Guo C, et al: Combination cancer
immunotherapy targeting TNFR2 and PD-1/PD-L1 signaling reduces
immunosuppressive effects in the microenvironment of pancreatic
tumors. J Immunother Cancer. 10:e0039822022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Wang L, Mo Q, Dong Y, Wang G and
Ji A: Changes of Th17/Treg cell and related cytokines in pancreatic
cancer patients. Int J Clin Exp Pathol. 8:5702–5708.
2015.PubMed/NCBI
|
|
24
|
Gabitova-Cornell L, Surumbayeva A, Peri S,
Franco-Barraza J, Restifo D, Weitz N, Ogier C, Goldman AR, Hartman
TR, Francescone R, et al: Cholesterol pathway inhibition induces
TGF-β signaling to promote basal differentiation in pancreatic
cancer. Cancer Cell. 38:567–583.e11. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Angelova AL, Grekova SP, Heller A,
Kuhlmann O, Soyka E, Giese T, Aprahamian M, Bour G, Rüffer S,
Cziepluch C, et al: Complementary induction of immunogenic cell
death by oncolytic parvovirus H-1PV and gemcitabine in pancreatic
cancer. J Virol. 88:5263–5276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hong W, Gu Y, Guan R, Xie D, Zhou H and Yu
M: Pan-cancer analysis of the CASP gene family in relation to
survival, tumor-infiltrating immune cells and therapeutic targets.
Genomics. 112:4304–4315. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Q, Pu N, Yin H, Zhang J, Zhao G, Lou
W and Wu W: CD73 acts as a prognostic biomarker and promotes
progression and immune escape in pancreatic cancer. J Cell Mol Med.
24:8674–8686. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
King RJ, Shukla SK, He C, Vernucci E,
Thakur R, Attri KS, Dasgupta A, Chaika NV, Mulder SE, Abrego J, et
al: CD73 induces GM-CSF/MDSC-mediated suppression of T cells to
accelerate pancreatic cancer pathogenesis. Oncogene. 41:971–982.
2022. View Article : Google Scholar : PubMed/NCBI
|