Introduction
Breast cancer is one of the three most common
cancers in the world. The World Health Organization's International
Agency for Research on Cancer has released the latest global cancer
statistics for 2020, revealing that breast cancer has surpassed
lung cancer as the world's most prevalent cancer, with 2.26 million
new cases reported globally (1).
Based on the type of hormone receptor and the tumor proliferation
status, breast cancer can be divided into either luminal A or B,
HER2-positive or triple-negative breast cancer (2,3).
Current studies show that early screening is still key to
preventing breast cancer. Ultrasonography (USG) is a commonly used
and convenient method for early screening of breast cancer, but it
has disadvantages of low specificity (4). Magnetic Resonance Imaging (MRI) is the
precise imaging of soft tissues (5). Treating with antiestrogen drugs, such
as tamoxifen or raloxifene, may reduce the risk of an individual
developing breast cancer (6). For
patients with diagnosed breast cancer, different treatment
strategies can be adopted such as targeted therapy, hormone
therapy, radiation therapy, surgery and chemotherapy. For patients
with distant metastasis, the goal of treatment is usually to
improve their quality of life and survival rate (7). However, although an increasing number
of treatment methods has been discovered for the treatment of
breast cancer, and the 5-year survival rate of breast cancer
increasing year by year, there are still various obstacles in
treating breast cancer. For example, numerous chemotherapy drugs
have serious side effects, and the development of resistance has
always been an issue in the treatment of breast cancer. The
treatment of most patients with distant metastases fails due to
intolerance to multiple chemotherapy drugs (8,9).
Therefore, identifying additional effective treatment methods with
less side effects remains urgent.
Vincristine (VCR) is a biologically-derived alkaloid
extracted from the periwinkle plant, often used in combination with
other chemotherapy drugs to treat various types of cancer,
including breast cancer (10). VCR
disrupts the formation of microtubules in the mitotic spindle,
leading to the arrest of cells undergoing mitosis. Resistance to
VCR often occurs during the treatment of breast cancer (11). There are several ways that
resistance develops during treatment with chemotherapy, including
enhanced drug metabolism, reduced cellular drug uptake, altered
expression of the drug target, intracellular drug sequestration and
altered expression of genes involved in either cell death, cell
cycle or DNA repair (12).
Additionally, changes in autophagy and inflammatory pathways are
often associated with resistance (13,14).
However, current research has not yet provided a reasonable
explanation for the development of resistance to VCR during
treatment of breast cancer.
Sequencing technology has propelled the development
of scientific research. With the rapid development of
second-generation sequencing technologies, an increasing number of
scientists are relying on it to solve biological issues. Whole
transcriptome sequencing has become increasingly advanced, allowing
for the analysis of alternative splicing, gene expression and other
research (15).
RNA alternative splicing refers to the process by
which different mature mRNAs can be produced from the same pre-mRNA
through different splicing patterns, which can then be translated
into proteins with multiple functions (16). Alternative splicing plays an
important role in regulating mRNA and protein diversity (17). It is noteworthy that RNA splicing
processes are strictly regulated in different tissues and
developmental stages, and abnormal RNA splicing regulation is
closely related to various human diseases, including breast cancer
(18,19). High-throughput sequencing and
functional enrichment analysis of genes with altered expression are
routinely used in biological research.
Based on the background of the development of
transcriptome sequencing technology, the present study investigated
the development of resistance to VCR in breast cancer cells using
transcriptome sequencing technology. A breast cancer cell line
resistant to VCR was established by gradually increasing the
concentration of VCR. We established this cell line to assist in
addressing the issue of VCR resistance that arises during the
clinical treatment of breast cancer. In order to explore the
mechanism behind this resistance, transcriptome sequencing was
performed. Gene expression analysis showed that multiple genes were
deregulated in the resistant cell line. Additionally, alternative
splicing events were also found to be altered in the resistant cell
line. The present study focused on two genes, IL1B and VEGFA, with
the aim of finding the key to drug resistance in patients with
breast. The present results provide mechanistic insight into VCR
resistance and a foundation for developing clinical interventions
to overcome this resistance.
Materials and methods
Cell culture
The cell lines used in the present study were
obtained from the American Type Culture Collection (ATCC) and were
maintained in the culture medium recommended by ATCC under standard
culture conditions. MCF7 cells were cultured in MEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (cat. no.
FBS-Superior-L; Sinsagetech Co. Ltd.), at 37°C in a humidified
incubator with 5% CO2.
Inhibitory concentration 50 (IC50)
measurement
The IC50 value was assessed using Cell Counting
Kit-8 (CCK-8; APExBIO Technology LLC). MCF7 cells were seeded into
96-well plates and treated with 1 nM VCR in 100 µl medium for 5
days, followed by 5, 10, 50, 100, 500, 1,000, 5,000 and 10,000 nM
VCR for 5 days each. Eventually, the surviving MCF7 cells were
expanded and screened to obtain the VCR-resistant MCF7 cell line.
After 48 h of incubation, the media were removed and replaced with
100 µl of culture media containing 10 µl CCK-8 reagent, and
incubated at 37°C for 2 h. Spectrophotometric absorbance was
measured at 450 nm to determine the dose-response curves, and the
IC50 value of VCR was calculated using GraphPad Prism (version 8;
Dotmatics).
Cell proliferation assay
Cell proliferation was assessed using a CCK-8 assay.
Cells were initially counted and 2,000 cells were seeded onto
96-well plates, followed by incubation in a CO2
incubator at 37°C for 24 h. Cells were then treated with VCR at
various concentrations in 100 µl of medium. After a 48-h incubation
period, the media were replaced with 100 µl of culture media
containing 10 µl of CCK-8 reagent, and incubated at 37°C for 2 h.
Absorbance was measured at 450 nm using a microplate reader
(DNM-9602; Perlong Medical Equipment Co., Ltd.), and the
dose-response curve was plotted using GraphPad Prism (version 8;
Dotmatics).
Colony formation assay
The relevant cells (MCF7 or VCR-resistant MCF7
cells) were initially counted and 1,000 cells were seeded into a
6-cm dish and cultured in an incubator at 37°C. Over a 2-week
period, the growth media were refreshed every 4 days. The cell
colonies were fixed with paraformaldehyde in room temperature for
15 min, followed by washing with PBS 3 times and staining with 0.1%
crystal violet for 20 min. A gel imaging system (Tanon-2500R; Tanon
Science and Technology Co., Ltd.) was used to capture images of the
cell colonies, which were subsequently quantified using ImageJ
v.1.50i (National Institutes of Health). Deep color visible to the
naked eye was counted as clones.
RNA-sequencing (RNA-seq) analysis
The RNA of both MCF7-wild type (WT) and
VCR-resistant VCR/MCF7 cells was extracted using the
TRIzol™ Reagent (Ambion; Thermo Fisher Scientific,
Inc.), followed by RNA-seq analysis performed by Haplox Co. Ltd.
[Platform: GPL20795 HiSeq X Ten (Homo sapiens)]. Each cell
line was analyzed once. To identify enriched pathways, Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
analyses were conducted using Metascape (20) and KOBAS-3.0 (http://kobas.cbi.pku.edu.cn/). Additionally,
functional interaction networks were generated by analyzing the
functional association of VCR-induced targets using protein
interaction data from the STRING database (https://cn.string-db.org/).
Reverse transcription-quantitative
(RT-q)PCR
RT was carried out using the Evo M-MLV Plus cDNA
Synthesis Kit (cat. no. AG11705; Accurate Biology) according to the
manufacturer's instructions. Mixed genomic DNA (gDNA) was removed
from the RNA template using gDNA Clean Reagent (included in the
above-mentioned kit) at 42°C for 2 min. The RT solution was added
and incubated at 37°C for 15 min, followed by 85°C for 5 sec. qPCR
was conducted using High-specificity Chemically-colored
Quantitative PCR Premix (Low ROX) (cat. no. MQ00601S; Monad Biotech
Co., Ltd.) according to the manufacturer's instructions. The
thermocycling conditions were as follows: 95°C/10 min for initial
denaturation, followed by 40 cycles of 95°C/10 sec for
denaturation, 60°C/10 sec for primer annealing and 72°C/30 sec for
extension. Dissociation curve was used as the default option of
QuantStudio3 (Thermo Fisher Scientific, Inc.). Expression was
normalized to GAPDH and quantified using the 2−ΔΔCq
method (21). Primer sequences can
be found in Table SI.
Public datasets
The functional association networks of genes related
to VCR resistance were analyzed using the STRING database. TCGA
data analysis of IL1B/VEGFA expression in BRCA and normal tissues
was based on sample types, individual cancer stages and nodal
metastasis status. Kaplan-Meier survival curves showing overall
survival of patients with BRCA bearing either high or low
IL1B/VEGFA were generated.
ELISA
Human interleukin-1β (IL-1β) ELISA Kit (cat. no.
E-EL-H0149c; Elabscience Biotechnology Inc.) was used to study the
protein expression of IL-1β in the VCR-resistant breast cancer cell
line. The experimental procedures were performed according to
manufacturer's instructions.
Western blot analysis
Total protein was extracted from cells using RIPA
lysis buffer (cat. no. R0020; Solarbio) containing 1 mM Cocktail.
The protein concentration was determined using the BCA assay. A
total of 30 µg protein sample was denatured by heating at 95°C for
5 min in 1X SDS sample buffer, and then separated by 12% SDS-PAGE.
The separated proteins were subsequently transferred onto a PVDF
membrane (cat. no. 10600023; GE Healthcare Life Science Co. Ltd.).
10% milk was used for blocking for 1 h in room temperature. The
membrane was then incubated with primary antibody at 4°C overnight.
PBS with 0.05% Tween-20 was used to wash the PVDF membrane 3 times.
Subsequently, membranes were incubated with secondary antibody with
HRP at room temperature for 1 h. Finally, MINICHEMI
(MiniChemi® 580; Sinsagetech Co. Ltd.) was used to
visualization, and NcmECL (NCM Biotech cat. no. P10300B) was used
as visualisation reagent. The following antibodies were used:
vascular endothelial growth factor A (VEGFA; 1:1,000 dilution; cat.
no. 19003-1-AP; Proteintech Group, Inc.) and GAPDH (1:5,000
dilution; cat. no. 10494-1-AP; Proteintech Group, Inc.).
Statistical analysis
The experiments were conducted using three
biologically independent repeats, and the data are presented as
mean ± SD. Statistical significance was determined using a
2-tailed, unpaired Student's t-test, as well as one-way or two-way
ANOVA followed by Tukey's post-hoc test in GraphPad Prism (version
8; Dotmatics). Kaplan-Meier plots were used to investigate patient
survival (http://kmplot.com/analysis/index.php?p=background).
Fisher's exact test was used in KEGG.
Results
VCR-resistant cells have stronger
proliferation and cloning formation abilities
In order to investigate the specific mechanism
underlying the development of VCR resistance in breast cancer
treatment, the VCR-resistant breast cancer cell line VCR/MCF7 was
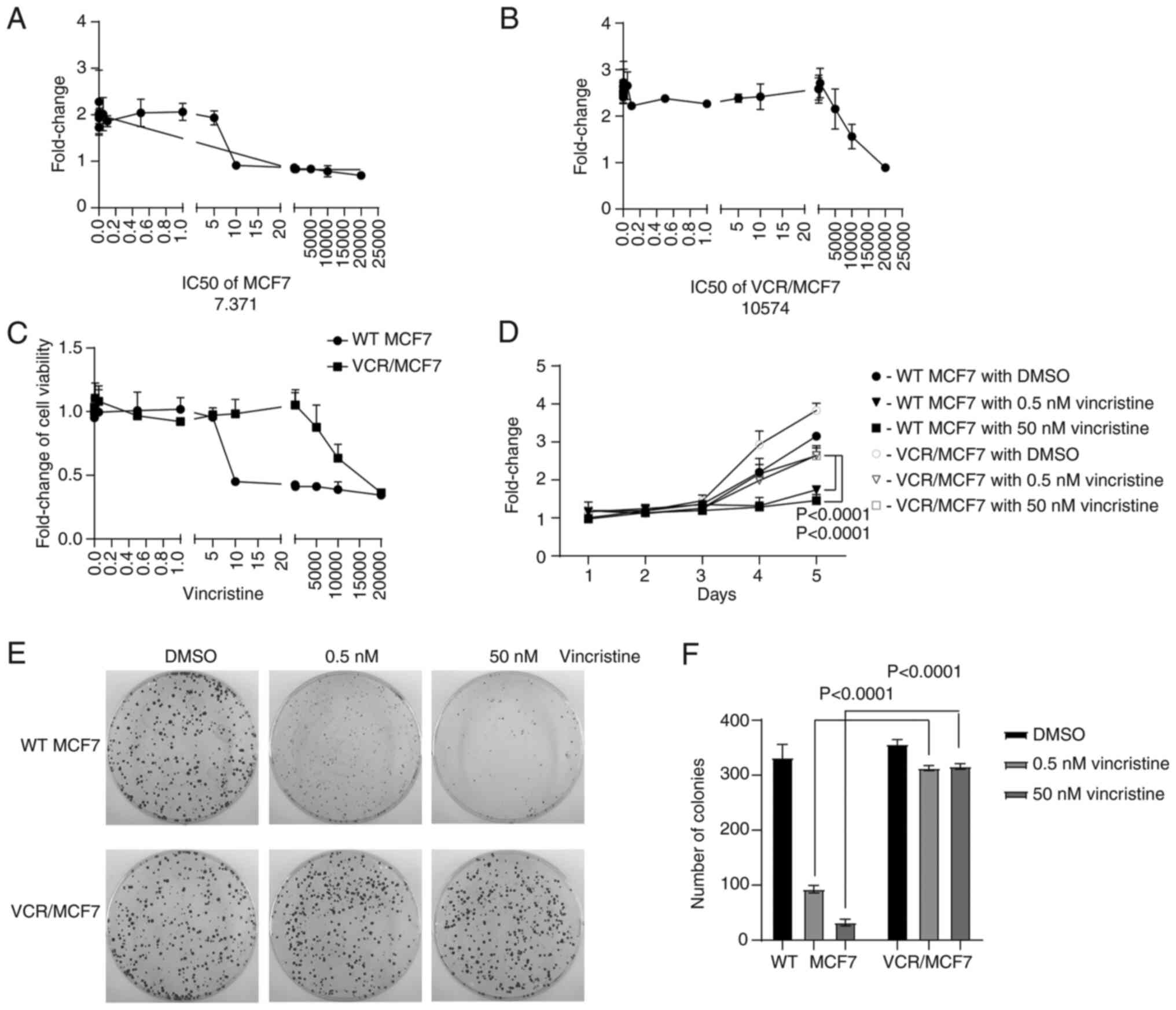
generated. First, the IC50 of MCF7-WT and VCR/MCF7 cells to VCR was
measured using a CCK-8 assay, and it was found to be 7.371 nM
(Fig. 1A) and 10,574 nM (Fig. 1B), respectively. The tolerance of
the two cell lines to VCR treatment was then investigated and it
was shown that the VCR/MCF7 cells were more resistant to VCR
compared with the MCF7-WT cells (Fig.
1C). MCF7-WT and VCR/MCF7 cells were treated with 0.5 and 50 nM
VCR, respectively, while DMSO was used as a control. It was shown
that the number of colonies of VCR/MCF7 cells after VCR treatment
was significantly higher than that of MCF7-WT cells (P<0.0001;
Fig. 1F). These results indicated
that a VCR-resistant cell line was successfully constructed.
The tumor phenotype of the VCR-resistant cells was
further investigated. It was shown that the VCR/MCF7 cells had
stronger proliferation ability than the MCF7-WT cells through a
growth curve experiment (Fig. 1D).
Moreover, by performing a colony-formation assay, it was revealed
that VCR/MCF7 cells had stronger colony formation ability compared
with that of MCF7-WT cells (Fig.
1E). This suggested that VCR-resistant breast cancer cells have
a more aggressive tumor phenotype than MCF7-WT cells.
VCR-resistant breast cancer cells have
a broad change in gene expression levels
To investigate the mechanism of VCR resistance in
breast cancer cells, RNA was extracted from both the VCR/MCF7 and
MCF7-WT cells, and RNA-seq was performed after purification.
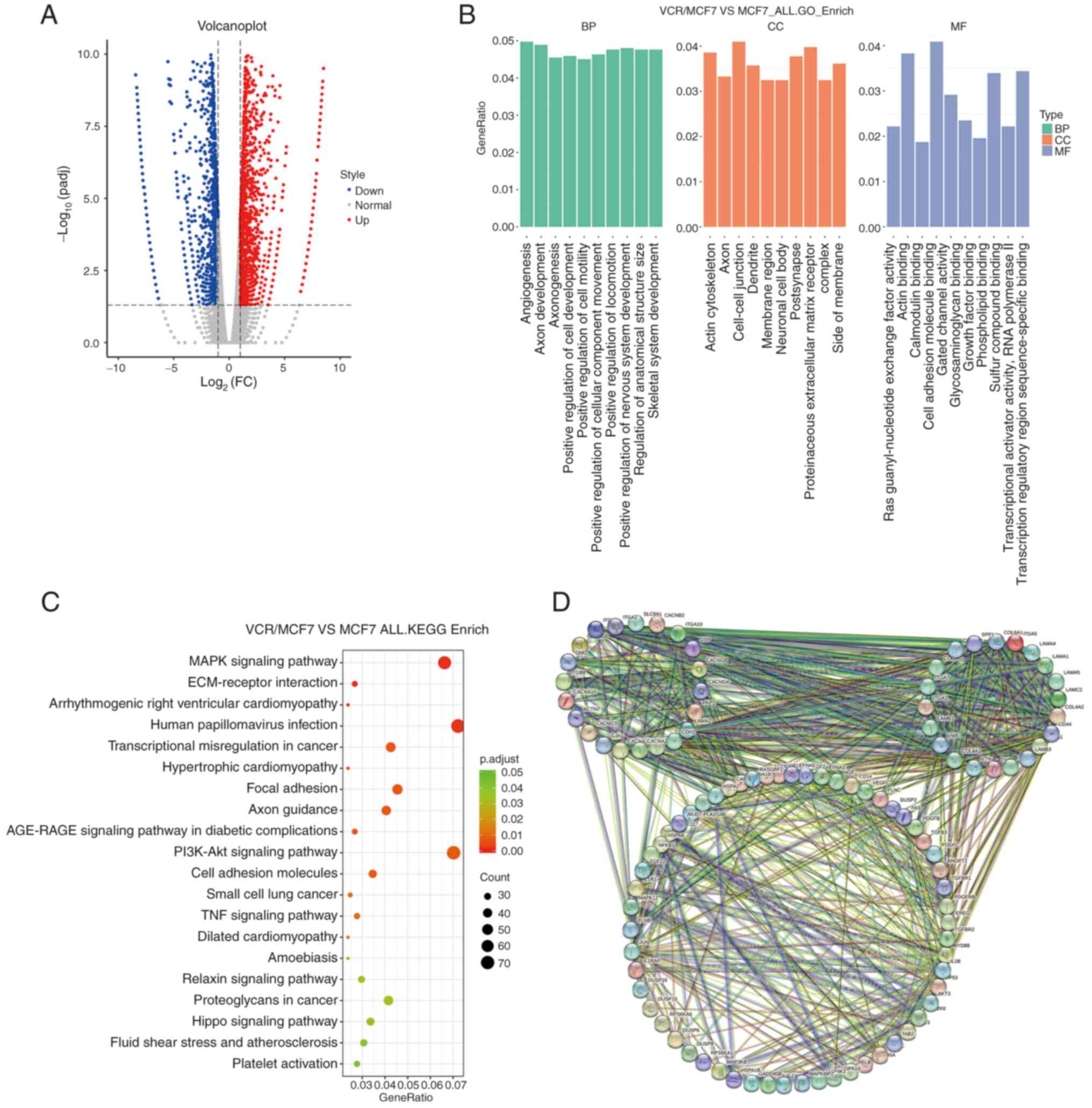
According to the analysis of RNA-seq data, it was revealed that the
expression levels of 263 genes were altered (log2fold change >1;
P<0.05) in the VCR-resistant breast cancer cells compared with
those in the MCF7-WT cells; more specifically, the expression
levels of 94 and 169 genes increased and decreased, respectively
(Fig. 2A). A GO analysis was then
performed on the genes of the sequencing data and it was found that
in terms of biological processes, these genes were mainly
concentrated in ‘angiogenesis’ and ‘positive regulation of cell
motility’, while in terms of cellular component, these genes were
mainly concentrated in ‘actin cytoskeleton’. In terms of molecular
function, these genes mainly concentrated in ‘actin binding’
(Fig. 2B). These results indicated
that the genes the expression of which was altered in the
VCR-resistant breast cancer cell line mainly concentrate on genes
related to microtubules. This suggested that there might be changes
in microtubule-related functions in the VCR-resistant cells.
Furthermore, through KEGG analysis, it was shown that VCR-resistant
is related to the pathways including ‘MAPK signaling pathway’,
‘human papillomavirus infection’ and others (Fig. 2C). Through STRING analysis, it was
shown that the gene expression, which changed in VCR-resistant
breast cancer cells, was functionally related (Fig. 2D). These results revealed that there
were extensive changes in the expression levels of certain genes in
the VCR-resistant breast cancer cells, which may associated with
VCR resistance.
Drug-resistant VCR/MCF7 cells undergo
extensive changes at the level of gene splicing
In order to further explore the possible mechanism
of drug resistance in VCR-resistant breast cancer cells, the
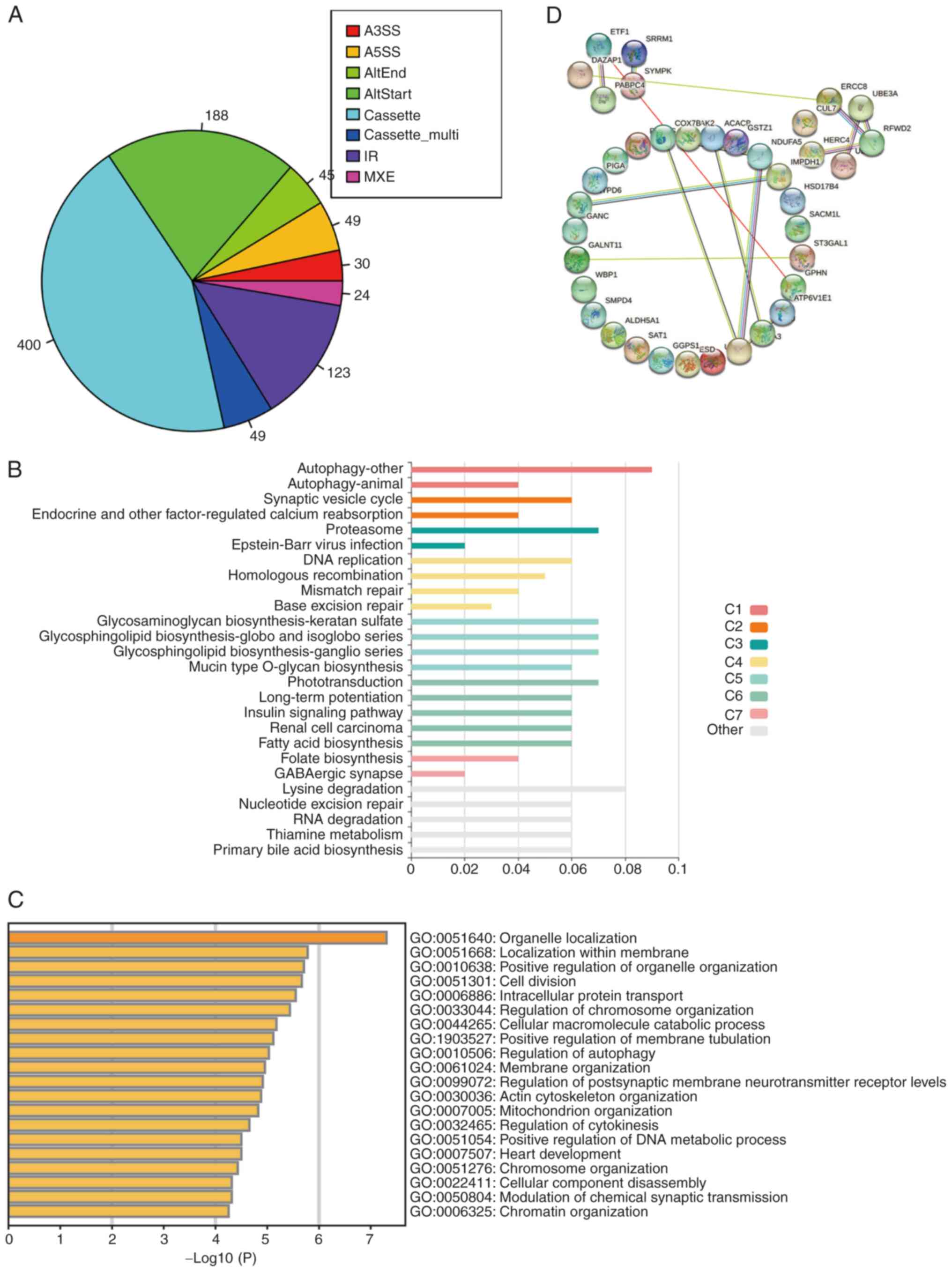
splicing changes in the sequencing data were further analyzed. The
results showed that compared with the MCF7-WT breast cancer cells,
VCR-resistant breast cancer cells undergo various alternative
splicing events. By analyzing the splicing types of these events,
it was found that the cassette type was the main splicing event
type that changed in the VCR-resistant breast cancer cells
(Fig. 3A). KEGG analysis of these
splicing events showed that the genes undergoing variable splicing
changes in the VCR-resistant breast cancer cells were mainly
concentrated on the autophagy signaling pathway (Fig. 3B). This prompted the hypothesis that
the autophagy pathway could act as a possible compensatory pathway
for VCR resistance. Further GO analysis showed that the genes the
splicing of which changed in drug-resistant VCR/MCF7 breast cancer
cells, were mainly focused on ‘organelle localization’ and other
functions (Fig. 3C). Analysis using
the STRING website revealed that the genes the splicing of which
changed in the VCR-resistant breast cancer cells, were functionally
related (Fig. 3D). The
aforementioned results indicated that the VCR-resistant breast
cancer cells had extensive changes at the gene splicing level,
which suggested that the variable splicing process may be involved
in the development of VCR resistance.
The expression levels of genes such as
VEGFA and IL-1β are altered in VCR-resistant breast cancer
cells
In order to validate the genes, the expression of
which changed in the sequencing data, some genes were selected for
validation. qPCR primers were designed for these genes and details
of the qPCR primer sequences are shown in Table SI. The results showed that the
expression levels of several genes in the VCR-resistant breast
cancer cells were consistent with the aforementioned sequencing
results (P<0.01; Fig. 4A-J). The
genes with increased expression levels included VEGFA, the
functions of which include inducing endothelial cell proliferation,
promoting cell migration, inhibiting apoptosis and inducing
permeabilization of blood vessels. The genes with decreased
expression levels included IL1B, which encodes the IL-1β protein
and its main functions include prostaglandin synthesis, neutrophil
influx and activation, T-cell activation and cytokine production,
B-cell activation and antibody production, fibroblast proliferation
and collagen production. Changes in autophagy and inflammatory
pathways are often associated with drug resistance (13,14).
VEGFA can promote the autophagy process (22), while IL1B is related to inflammatory
pathways (23), and thus, the study
focused on these two genes.
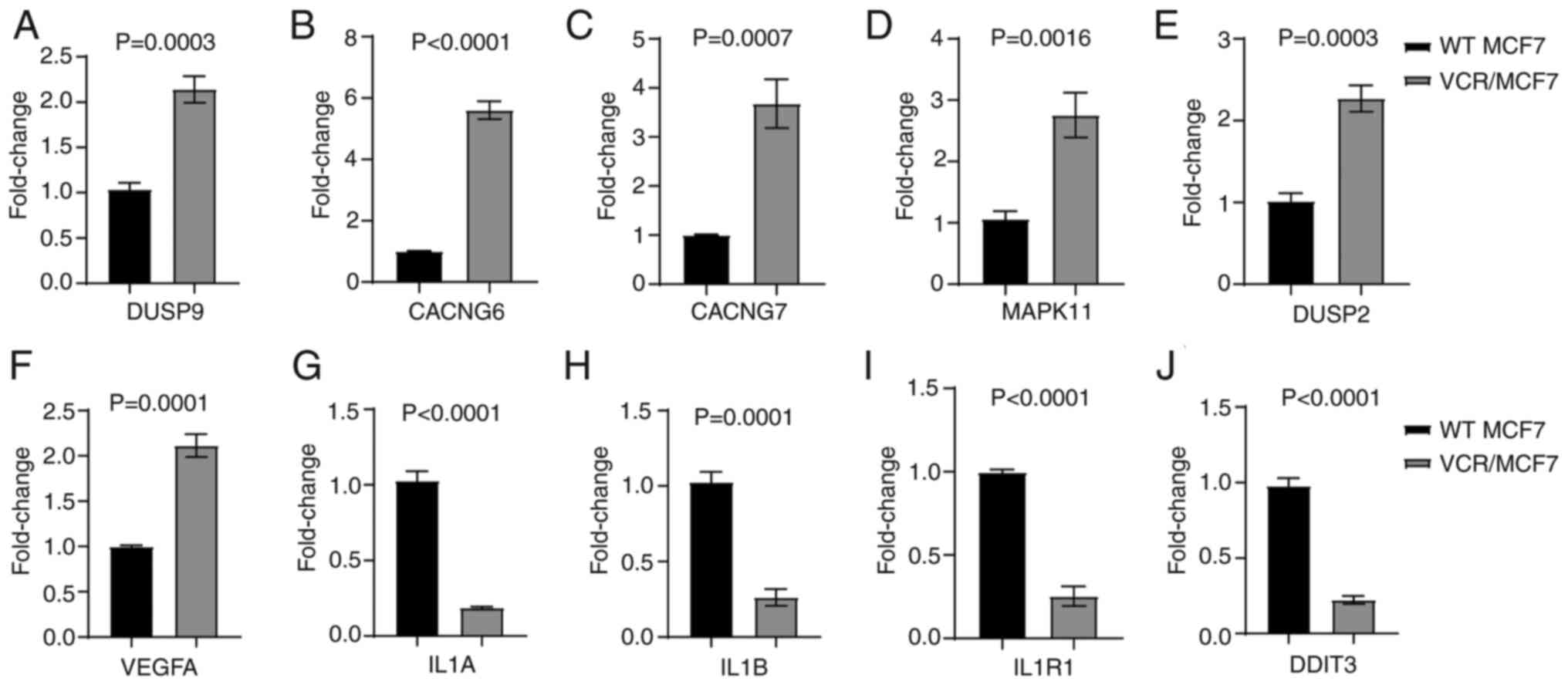 | Figure 4.Expression levels of genes such as
VEGFA and IL-1β are altered in drug-resistant VCR/MCF7 cells. (A-J)
The changes in expression levels of (A) DUSP9, (B) CACNG6, (C)
CACNG7, (D) MAPK11, (E) DUSP2, (F) VEGFA, (G) IL-1A, (H) IL1B, (I)
IL1R1 and (J) DDIT3 confirmed via reverse
transcription-quantitative PCR. The mean ± SD of the relative fold
changes obtained from triplicate experiments were plotted, and
P-values were calculated using an unpaired Student's t-test. VEGFA,
vascular endothelial growth factor A; IL, interleukin; VCR,
vincristine; WT, wild-type; DUSP9, dual specificity protein
phosphatase 9; CACNG6, voltage-dependent calcium channel gamma-6
subunit; MAPK11, mitogen-activated protein kinase 11; VEGFA,
vascular endothelial growth factor A, long form; IL1B:
Interleukin-1β; IL1R1, interleukin-1 receptor type 1; DDIT3, DNA
damage-inducible transcript 3 protein. |
VCR resistance may be due to changes
in the expression of either VEGFA or IL-1β
In order to validate the impact of VEGFA and IL1B on
tumors, TCGA data were analyzed, and all the selected patients were
VCR-resistant. It was shown that the expression of IL1B was
significantly lower in breast cancer tissues than in normal tissues
(Fig. 5A), while the expression of
VEGFA was higher in breast cancer tissues (Fig. 5F). Further analysis showed that the
expression levels of IL1B and VEGFA were abnormally expressed at
various clinical stages and nodal metastasis states, and the
expression of VEGFA was positively correlated with the clinical
stage, and IL1B was negatively correlated with the clinical stage
(Fig. 5B-C and G-H). Next,
Kaplan-Meier analysis was used to analyze the relationship between
VEGFA and IL1B, and breast cancer survival, and Kaplan-Meier
survival curves were generated. The analysis revealed that high
expression of VEGFA was associated with low breast cancer survival
rates (Fig. 5D), while low
expression of IL1B was associated with high breast cancer survival
rates (Fig. 5I). The protein
expression of VEGFA and IL-1β in the VCR-resistant breast cancer
cell line was then verified, and it was found that IL-1β expression
was significantly lower in the VCR-resistant breast cancer cells as
shown by ELISA (P<0.0001; Fig.
5E), while VEGFA protein expression was higher in VCR-resistant
breast cancer cells than MCF7-WT breast cancer cells as shown by
western blotting (Fig. 5J). This
suggested that VCR resistance may be due to changes in the
expression of either VEGFA or IL-1β.
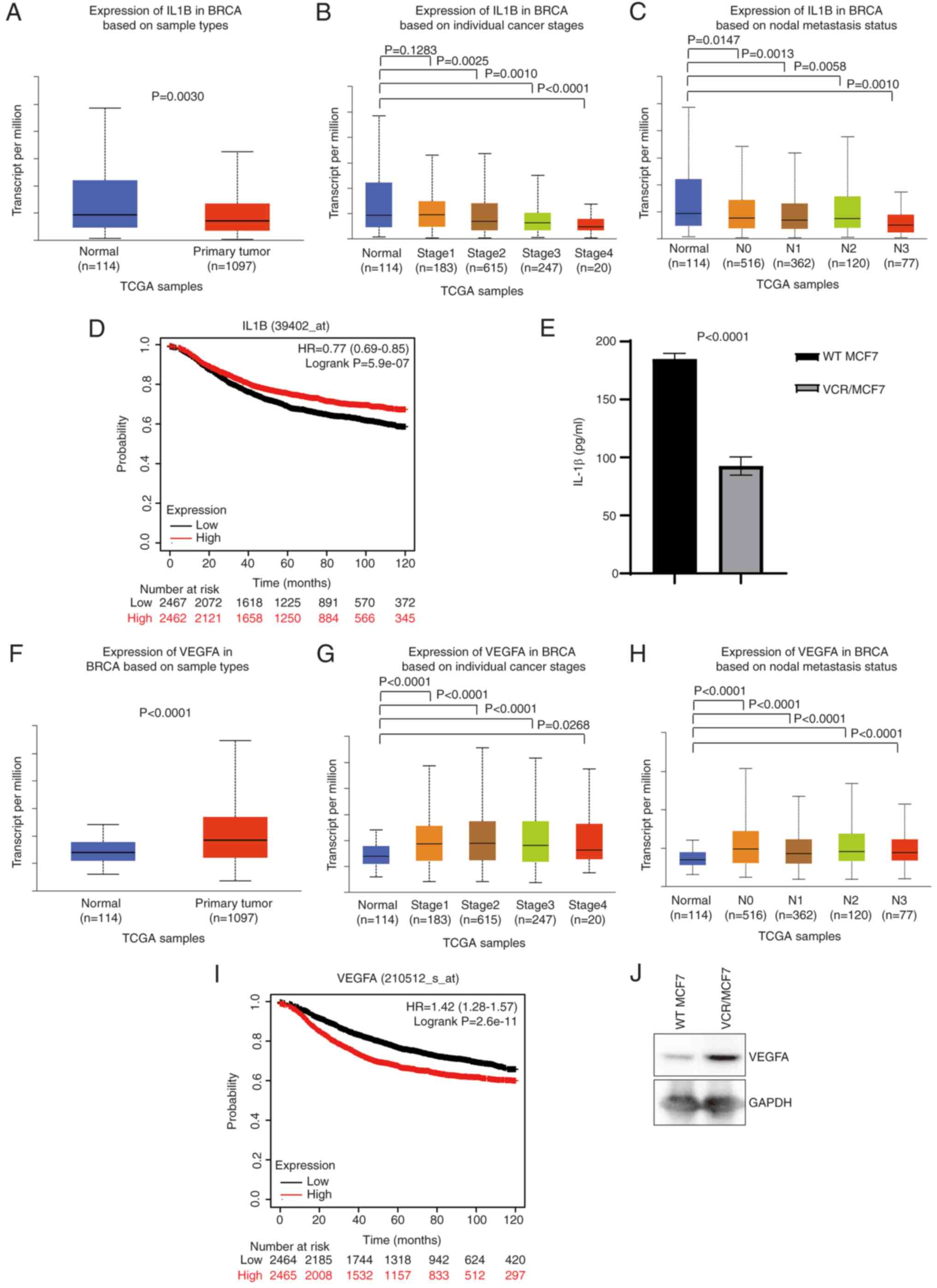 | Figure 5.VCR resistance may be due to changes
in the expression of VEGFA or IL-1β. (A-C) TCGA data analysis of
IL1B expression in BRCA and normal tissues based on (A) sample
types, (B) individual cancer stages and (C) nodal metastasis
status. (D) Kaplan-Meier survival curve showing overall survival of
patients with BRCA bearing either high or low IL1B. (E) ELISA was
used to verify IL-1β level expression. TCGA data analysis of VEGFA
expression in BRCA and normal tissues based on (F) sample types,
(G) individual cancer stages and (H) nodal metastasis status. (I)
Kaplan-Meier survival curves showing overall survival of patients
with BRCA bearing either high or low VEGFA. (J) Western blotting
was used to verify VEGFA protein level expression. HR, hazard
ratio; IL-1β, interleukin-1β; TCGA, The Cancer Genome Atlas; BRCA,
breast cancer; N, nodal; VEGFA, vascular endothelial growth factor
A; WT, wild type; VCR, vincristine. |
Discussion
Breast cancer is the most common cancer in women,
second only to lung cancer in the number of annual deaths (24,25).
Epidemiological studies show that the incidence range of breast
cancer worldwide is 22–26% and the mortality rate is ~18% (26,27).
During the treatment of breast cancer, metastasis and chemotherapy
resistance are often the main reasons for treatment failure.
Chemotherapy resistance is often the result of multiple factors,
including mutations in the tubulin protein (28), alternative tubulin expression
(29,30), changes in other cytoskeletal
proteins, activation of autophagy or intracellular detoxification
systems, and changes in drug transport protein expression, which
lead to a decrease in intracellular drug concentration (31–33).
Changes in apoptosis-related proteins are also often associated
with cell resistance (34,35).
Through growth curve and clone formation
experiments, it was shown that the VCR-resistant breast cancer cell
line became less sensitive to VCR treatment. RNA-seq showed that
multiple gene expression levels, including microtubule assembly,
were altered in the VCR-resistant breast cancer cell line. Analysis
of splicing events also revealed that the variable splicing of
numerous genes, including autophagy-related genes, was altered in
the VCR-resistant breast cancer cell line. Further verification
showed that the expression levels of certain genes, including VEGFA
and IL1B, were altered in the VCR-resistant breast cancer cell
line, and these gene expression changes were related to VCR
resistance. The results of the present study indicated that the
production of the VCR-resistant cell line VCR/MCF7 may be caused by
changes in microtubule proteins or drug metabolism pathways such as
autophagy, providing new avenues for the development of VCR
resistance.
The sequencing data were then validated and it was
found that the expression levels of several genes in the
VCR-resistant breast cancer cells were consistent with the
aforementioned sequencing results. The further analysis focused on
VEGFA and IL1B1.
Most cells in the human body can produce VEGFA and
express it at a higher level under hypoxia (36). In the development of tumors, VEGFA
is mainly produced by tumor cells in low oxygen environments, as
well as by endothelial cells and tumor-associated macrophages
(37). Its function is mainly
related to angiogenesis, and the regulation of VEGFA on endothelial
cell proliferation and invasive properties is strictly controlled.
The ERK and PI3K/Akt pathways are the primary regulators of
endothelial cell proliferation (38,39),
whereas endothelial cell invasion is facilitated by the release of
matrix metalloproteinases that break down the basement membrane and
extracellular matrix, promoting the migration of new endothelial
cells and the development of capillary buds. VEGFA has been shown
to regulate the activity of proteins such as MMP-2 and −9 through
the activation of β-catenin and NF-κB dependent on Akt (39,40).
IL-1β mainly regulates the inflammatory signaling
pathway, and is mainly produced by blood monocytes, tissue
macrophages, skin dendritic cells and brain microglia (41). IL-1β is produced in the form of a
precursor peptide, which is cleaved and activated under the
stimulation of PAMPs and DAMPs, and is a cellular stress response
mechanism induced by invading pathogens and other danger signals,
such as mycobacterium tuberculosis components (42,43).
IL-1β is activated after being cleaved by caspase-1, which is in
turn activated by the NLRP3 inflammatory body complex. Once IL-1β
is activated, it is released into the extracellular space and
causes an inflammatory response by activating the IL-1R1 receptor
(44). The expression of IL-1β in
breast cancer cells is often closely related to the development of
breast cancer (45).
Through TCGA data analysis, it was shown that the
expression of IL-1β in breast cancer tissue was significantly lower
than that in normal tissue, while the expression of VEGFA was
higher in breast cancer tissue. Survival analysis showed that high
expression of VEGFA was related to low survival rates in breast
cancer, while low expression of IL-1β was related to high survival
rates in breast cancer. These results indicate that VEGFA and IL-1β
may participate in the process of drug resistance in breast cancer
cells. VEGFA is known to mediate vasculogenesis and angiogenesis by
regulating the activity of endothelial cells (46). Accumulating evidence suggests that
VEGFA is expressed at high levels in a range of human cancers,
including liver, ovary, kidney and colon cancers, and is associated
with tumor progression and poor prognosis (47–50).
It has been shown that patients with metastatic breast cancer have
higher circulating VEGFA levels than those without metastasis
(51). In addition to regulating
angiogenesis, VEGFA also promotes tumor growth, metastasis and
survival directly (46). In ovarian
cancer, VEGFA upregulation is associated with poor survival, and it
has been proposed as a biomarker for subsets of advanced ovarian
tumors (52). Various therapies
against VEGFA have been used for anticancer treatment (53,54).
However, the role of VEGFA in chemotherapy resistance is still not
clear. As an inflammatory mediator, IL-1β is frequently upregulated
in a variety of cancers, which is different from the experimental
findings of the present study, and its production is associated
with poor prognosis (55,56). Some studies suggest that IL-1β
induces neoangiogenesis and regulates the expression of soluble
mediators in stromal cells to enhance tumor cell survival and
metastasis (56,57).
Activation of autophagy leads to a decrease in
intracellular drug concentration, which often associated with cell
resistance (13). Changes in
apoptosis-related proteins are also often associated with cell
resistance. Based on a study that showed that VEGFA can promote the
autophagy process (22), it was
hypothesized that VEGFA may reduce the intracellular drug
concentration by promoting autophagy, leading to the generation of
drug-resistant cells. The decreased expression of IL-1β may play a
role in the formation of drug-resistant cells by participating in
the process of apoptosis (23).
There are still some deficiencies and limitations in
the current study: It only included cellular experiments without
involving animal models or analysis of clinical specimens;
sequencing of drug-resistant cell lines often has a large number of
genes with expression or splicing changes, but only a small number
of these genes are typically involved in the generation of drug
resistance. Therefore, it is important to identify those genes that
are truly related to drug resistance among others. This requires
the analysis and the validation processes of the present study to
be further optimized.
The present study showed that the development of
resistance to VCR may be related not only to changes in microtubule
proteins or drug metabolism pathways such as autophagy, but also to
changes in the expression of VEGFA and IL-1β, providing a
theoretical basis for exploring the mechanism of VCR resistance
clinically.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Dalian Medical Science
Research Program (grant no. 2021D004).
Availability of data and materials
The data generated in the present study may be found
in the GEO database (accession no. GSE241356).
Authors' contributions
YC and CMW conceived and designed the study. LLY and
CW were involved in data collection. YC performed statistical
analysis and prepared the figures. All authors have read and
approved the final manuscript. YC and CMW confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of the First Affiliated Hospital of Dalian Medical
University in accordance with the Declaration of Helsinki (approval
no. PJ-KS-KY-2021-145). Written informed consent was obtained from
each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B and Senn HJ; Panel members, :
Personalizing the treatment of women with early breast cancer:
Highlights of the St Gallen international expert consensus on the
primary therapy of early breast cancer 2013. Ann Oncol.
24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B and Senn HJ; Panel
Members, : Tailoring therapies-improving the management of early
breast cancer: St Gallen international expert consensus on the
primary therapy of early breast cancer 2015. Ann Oncol.
26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mehnati P and Tirtash MJ: Comparative
efficacy of four imaging instruments for breast cancer screening.
Asian Pac J Cancer Prev. 16:6177–6186. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang SN, Li FJ, Liao YH, Chen YS, Shen WC
and Huang TC: Identification of breast cancer using integrated
information from MRI and mammography. PLoS One. 10:e01284042015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng J, Sengupta S and Jordan VC:
Potential of selective estrogen receptor modulators as treatments
and preventives of breast cancer. Anticancer Agents Med Chem.
9:481–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reeder JG and Vogel VG: Breast cancer
prevention. Cancer Treat Res. 141:149–164. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liscovitch M and Lavie Y: Cancer multidrug
resistance: A review of recent drug discovery research. IDrugs.
5:349–355. 2002.PubMed/NCBI
|
|
10
|
De Lena M, Brambilla C, Morabito A and
Bonadonna G: Adriamycin plus vincristine compared to and combined
with cyclophosphamide, methotrexate, and 5-fluorouracil for
advanced breast cancer. Cancer. 35:1108–1115. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bates D and Eastman A: Microtubule
destabilising agents: Far more than just antimitotic anticancer
drugs. Br J Clin Pharmacol. 83:255–268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mansoori B, Mohammadi A, Davudian S,
Shirjang S and Baradaran B: The different mechanisms of cancer drug
resistance: A brief review. Adv Pharm Bull. 7:339–348. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu F, Song D, Yan Y, Huang C, Shen C, Lan
J, Chen Y, Liu A, Wu Q, Sun L, et al: IL-6 regulates autophagy and
chemotherapy resistance by promoting BECN1 phosphorylation. Nat
Commun. 12:36512021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Novototskaya-Vlasova KA, Neznanov NS,
Molodtsov I, Hall BM, Commane M, Gleiberman AS, Murray J, Haber M,
Norris MD, Leonova KI and Gudkov AV: Inflammatory response to
retrotransposons drives tumor drug resistance that can be prevented
by reverse transcriptase inhibitors. Proc Natl Acad Sci USA.
119:e22131461192022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duncavage EJ, Bagg A, Hasserjian RP,
DiNardo CD, Godley LA, Iacobucci I, Jaiswal S, Malcovati L,
Vannucchi AM, Patel KP, et al: Genomic profiling for clinical
decision making in myeloid neoplasms and acute leukemia. Blood.
140:2228–2247. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marasco LE and Kornblihtt AR: The
physiology of alternative splicing. Nat Rev Mol Cell Biol.
24:242–254. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv Y, Zhang W, Zhao J, Sun B, Qi Y, Ji H,
Chen C, Zhang J, Sheng J, Wang T, et al: SRSF1 inhibits autophagy
through regulating Bcl-x splicing and interacting with PIK3C3 in
lung cancer. Signal Transduct Target Ther. 6:1082021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kitamura K and Nimura K: Regulation of RNA
splicing: Aberrant splicing regulation and therapeutic targets in
cancer. Cells. 10:9232021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Read A and Natrajan R: Splicing
dysregulation as a driver of breast cancer. Endocr Relat Cancer.
25:R467–R478. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li X, Hu Z, Shi H, Wang C, Lei J and Cheng
Y: Inhibition of VEGFA increases the sensitivity of ovarian cancer
cells to chemotherapy by suppressing VEGFA-mediated autophagy. Onco
Targets Ther. 13:8161–8171. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang BW, Jiang Y, Yao ZL, Chen PS, Yu B
and Wang SN: Aucubin protects chondrocytes against IL-1β-induced
apoptosis in vitro and inhibits osteoarthritis in mice model. Drug
Des Devel Ther. 13:3529–3538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wooster R and Weber BL: Breast and ovarian
cancer. N Engl J Med. 348:2339–2347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kavallaris M, Tait AS, Walsh BJ, He L,
Horwitz SB, Norris MD and Haber M: Multiple microtubule alterations
are associated with vinca alkaloid resistance in human leukemia
cells. Cancer Res. 61:5803–5809. 2001.PubMed/NCBI
|
|
29
|
Sirotnak FM, Danenberg KD, Chen J, Fritz F
and Danenberg PV: Markedly decreased binding of vincristine to
tubulin in vinca alkaloid-resistant Chinese hamster cells is
associated with selective overexpression of alpha and beta tubulin
isoforms. Biochem Biophys Res Commun. 269:21–24. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Verrills NM, Walsh BJ, Cobon GS, Hains PG
and Kavallaris M: Proteome analysis of vinca alkaloid response and
resistance in acute lymphoblastic leukemia reveals novel
cytoskeletal alterations. J Biol Chem. 278:45082–45093. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stow MW and Warr JR: Reduced influx is a
factor in accounting for reduced vincristine accumulation in
certain verapamil-hypersensitive multidrug-resistant CHO cell
lines. FEBS Lett. 320:87–91. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yao D, Ding S, Burchell B, Wolf CR and
Friedberg T: Detoxication of vinca alkaloids by human P450
CYP3A4-mediated metabolism: Implications for the development of
drug resistance. J Pharmacol Exp Ther. 294:387–395. 2000.PubMed/NCBI
|
|
34
|
Kontos CK, Christodoulou MI and Scorilas
A: Apoptosis-related BCL2-family members: Key players in
chemotherapy. Anticancer Agents Med Chem. 14:353–374. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Urfali-Mamatoglu C, Kazan HH and Gündüz U:
Dual function of programmed cell death 10 (PDCD10) in drug
resistance. Biomed Pharmacother. 101:129–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ferrara N: Vascular endothelial growth
factor: Basic science and clinical progress. Endocr Rev.
25:581–611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Delforce SJ, Lumbers ER, Morosin SK, Wang
Y and Pringle KG: The Angiotensin II type 1 receptor mediates the
effects of low oxygen on early placental angiogenesis. Placenta.
75:54–61. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Takahashi T, Yamaguchi S, Chida K and
Shibuya M: A single autophosphorylation site on KDR/Flk-1 is
essential for VEGF-A-dependent activation of PLC-gamma and DNA
synthesis in vascular endothelial cells. EMBO J. 20:2768–2778.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang BH and Liu LZ: PI3K/PTEN signaling
in angiogenesis and tumorigenesis. Adv Cancer Res. 102:19–65. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
van Hinsbergh VW and Koolwijk P:
Endothelial sprouting and angiogenesis: Matrix metalloproteinases
in the lead. Cardiovasc Res. 78:203–212. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Garlanda C, Dinarello CA and Mantovani A:
The interleukin-1 family: Back to the future. Immunity.
39:1003–1018. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brough D, Tyrrell PJ and Allan SM:
Regulation of interleukin-1 in acute brain injury. Trends Pharmacol
Sci. 32:617–622. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baroja-Mazo A, Martín-Sánchez F, Gomez AI,
Martínez CM, Amores-Iniesta J, Compan V, Barberà-Cremades M, Yagüe
J, Ruiz-Ortiz E, Antón J, et al: The NLRP3 inflammasome is released
as a particulate danger signal that amplifies the inflammatory
response. Nat Immunol. 15:738–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dinarello CA: Interleukin-1. Cytokine
Growth Factor Rev. 8:253–265. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tulotta C and Ottewell P: The role of
IL-1B in breast cancer bone metastasis. Endocr Relat Cancer.
25:R421–R434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jang K, Kim M, Gilbert CA, Simpkins F,
Ince TA and Slingerland JM: VEGFA activates an epigenetic pathway
upregulating ovarian cancer-initiating cells. EMBO Mol Med.
9:304–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang W, Ren F, Wu Q, Jiang D, Li H and Shi
H: MicroRNA-497 suppresses angiogenesis by targeting vascular
endothelial growth factor A through the PI3K/AKT and MAPK/ERK
pathways in ovarian cancer. Oncol Rep. 32:2127–2133. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yan JJ, Zhang YN, Liao JZ, Ke KP, Chang Y,
Li PY, Wang M, Lin JS and He XX: MiR-497 suppresses angiogenesis
and metastasis of hepatocellular carcinoma by inhibiting VEGFA and
AEG-1. Oncotarget. 6:29527–29542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zeng FC, Zeng MQ, Huang L, Li YL, Gao BM,
Chen JJ, Xue RZ and Tang ZY: Downregulation of VEGFA inhibits
proliferation, promotes apoptosis, and suppresses migration and
invasion of renal clear cell carcinoma. Onco Targets Ther.
9:2131–2141. 2016.PubMed/NCBI
|
|
50
|
Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G,
Hou Y and Jiang P: MicroRNA-140-5p inhibits the progression of
colorectal cancer by targeting VEGFA. Cell Physiol Biochem.
37:1123–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Adams J, Carder PJ, Downey S, Forbes MA,
MacLennan K, Allgar V, Kaufman S, Hallam S, Bicknell R, Walker JJ,
et al: Vascular endothelial growth factor (VEGF) in breast cancer:
Comparison of plasma, serum, and tissue VEGF and microvessel
density and effects of tamoxifen. Cancer Res. 60:2898–2905.
2000.PubMed/NCBI
|
|
52
|
Lawicki S, Będkowska GE, Gacuta-Szumarska
E and Szmitkowski M: The plasma concentration of VEGF, HE4 and
CA125 as a new biomarkers panel in different stages and sub-types
of epithelial ovarian tumors. J Ovarian Res. 6:452013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fleming ND, Coleman RL, Tung C, Westin SN,
Hu W, Sun Y, Bhosale P, Munsell MF and Sood AK: Phase II trial of
bevacizumab with dose-dense paclitaxel as first-line treatment in
patients with advanced ovarian cancer. Gynecol Oncol. 147:41–46.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fahmy K, Gonzalez A, Arafa M, Peixoto P,
Bellahcène A, Turtoi A, Delvenne P, Thiry M, Castronovo V and
Peulen O: Myoferlin plays a key role in VEGFA secretion and impacts
tumor-associated angiogenesis in human pancreas cancer. Int J
Cancer. 138:652–663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bent R, Moll L, Grabbe S and Bros M:
Interleukin-1 beta-A friend or foe in malignancies? Int J Mol Sci.
19:21552018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Apte RN, Dotan S, Elkabets M, White MR,
Reich E, Carmi Y, Song X, Dvozkin T, Krelin Y and Voronov E: The
involvement of IL-1 in tumorigenesis, tumor invasiveness,
metastasis and tumor-host interactions. Cancer Metastasis Rev.
25:387–408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Saijo Y, Tanaka M, Miki M, Usui K, Suzuki
T, Maemondo M, Hong X, Tazawa R, Kikuchi T, Matsushima K and Nukiwa
T: Proinflammatory cytokine IL-1 beta promotes tumor growth of
Lewis lung carcinoma by induction of angiogenic factors: In vivo
analysis of tumor-stromal interaction. J Immunol. 169:469–475.
2002. View Article : Google Scholar : PubMed/NCBI
|