The precise control of cell activity in response to
systemic or local signals is largely governed by the regulation of
homoeostasis (1). Proteins are
known to execute a diverse range of intracellular processes
(2). In order to accurately perform
various physiological functions, the intracellular protein quality
must be precisely and strictly controlled, and this level of
control depends on the balance between protein degradation and
synthesis (3). The lysosomal
systems and the ubiquitin-proteasome system (UPS) form an
interconnected protein quality control network for lysosomal and
proteasomal protein degradation, respectively (4,5). In
the UPS, ubiquitin is conjugated to targeted proteins, which are
subsequently recognized by the proteasome for degradation (6,7).
Ubiquitinases and deubiquitinases (DUBs) in the ubiquitin
proteasome pathway are involved in various pathological and
physiological processes, such as DNA repair (8), apoptosis (9) and cancer metabolism (10). DUBs are classified into seven
families on the basis of their sequences and structural homology,
namely ubiquitin-specific proteases (USPs), ovarian tumor
proteases, the JAB1/MPN/MOV34 family of metalloenzymes, ubiquitin
carboxy-terminal hydrolases (UCHs), the motif interacting with
ubiquitin-containing novel DUB family, Machado-Josephin
domain-containing proteases and the zinc finger-containing
ubiquitin peptidase 1. The structural or functional abnormalities
of these enzymes can lead to numerous diseases, including cancer
(11,12). The USP family accounts for the
largest proportion of DUBs that have been identified, and share a
catalytic domain that comprises 300–800 amino acids (13).
Ubiquitin-specific peptidase 9X (USP9X), a member of
the USP family, regulates numerous signaling pathways by
deubiquitinating essential proteins, such as myeloid leukemia cell
differentiation 1 (MCL-1) (14,15).
USP9X is closely associated with neurological disorders (16–18),
atherosclerosis (19) and cancers
(20). However, to date, USP9X and
its precise roles in different types of cancer have not been widely
investigated or specifically and systematically reviewed. In
addition, the value of USP9X in terms of its clinical and potential
application as a cancer target may be underestimated. Therefore, in
the present review, the current evidence in support of the cellular
functions and underlying regulatory mechanisms of USP9X in
tumorigenesis is summarized, as well as the biological mechanisms
associated with USP9X in different types of cancer.
The highly conserved DUB USP9X is located on
chromosome Xp11.4 and was first identified as a human homologue of
the Drosophila fat facets (faf) gene, subsequently
named DFFRX, which escapes X-chromosome inactivation and serves
vital roles in embryo development (21). The protein sequence of USP9X and its
molecular functions are evolutionarily conserved across species
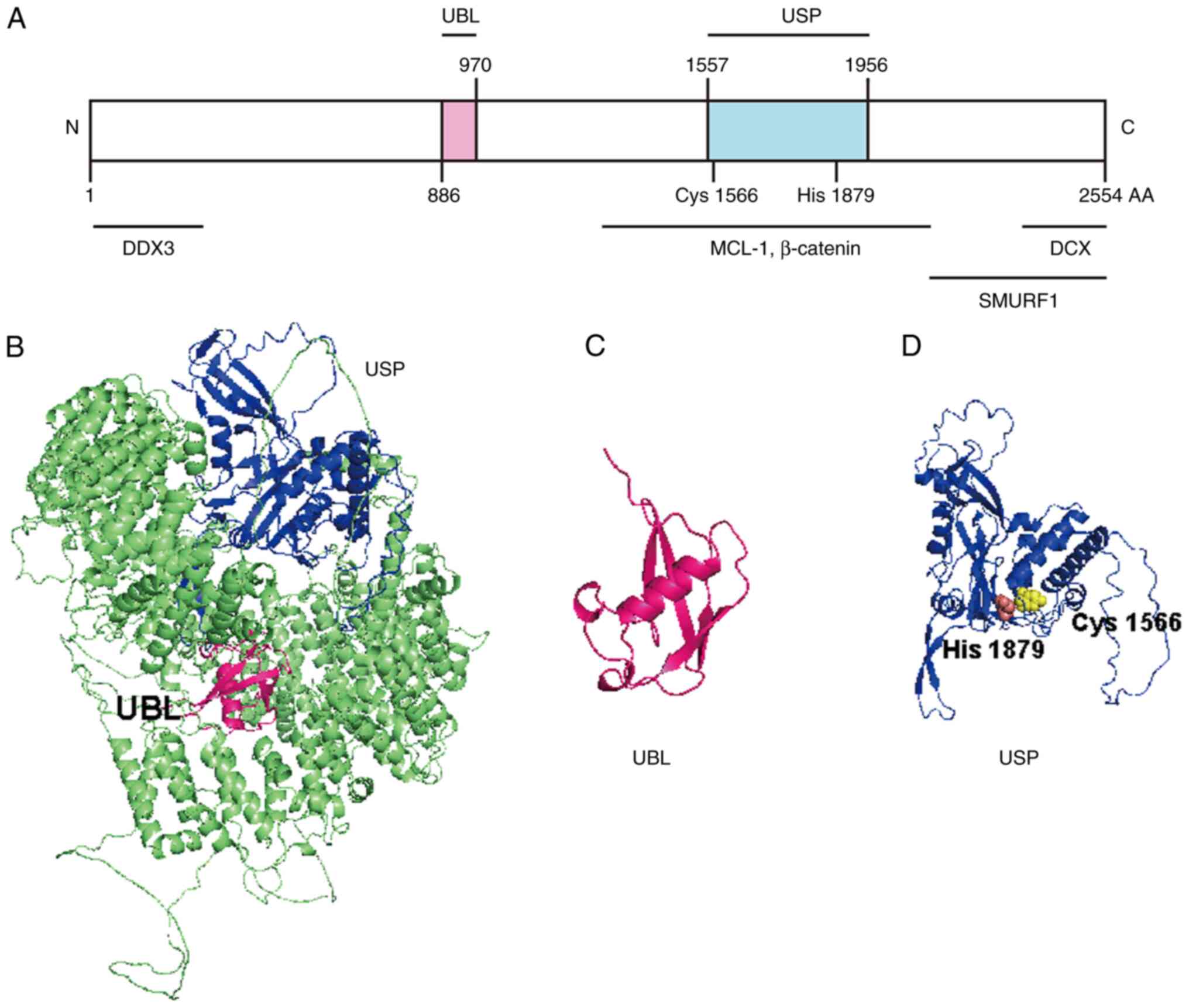
(22–25). The two recognizable domains of USP9X
are the ubiquitin-like module (UBL) domain (amino acids 886–970)
and the catalytic domain with USP-definitive cysteine (amino acid
1,566) and histidine (amino acid 1,879) box catalytic motifs
(Fig. 1A) (15). The long non-conserved N- and
C-terminal extensions flank the catalytic domains of USP9X
(26,27). The homology model of USP9X and its
functional domains were constructed from the Alphafold Database
(https://alphafold.com/) with PyMOL (The PyMOL
Molecular Graphics System; version 2.3; Schrödinger, LLC; Fig. 1B-D). The crystal structure of USP9X
has been solved and reported as a conserved and canonical USP-fold
consisting of palm, finger and thumb subdomains, and a β-hairpin
insertion (28). The catalytic
domain of USP9X harbors a zinc finger motif and three ubiquitin
binding sites in the fingers subdomain, and the β-hairpin
insertion, contributing to polyubiquitin chain processing and
cleavage of lysine (Lys)11-, Lys63-, Lys48- and Lys6-linkages,
enabling the protein to perform a variety of cellular functions
(29–34).
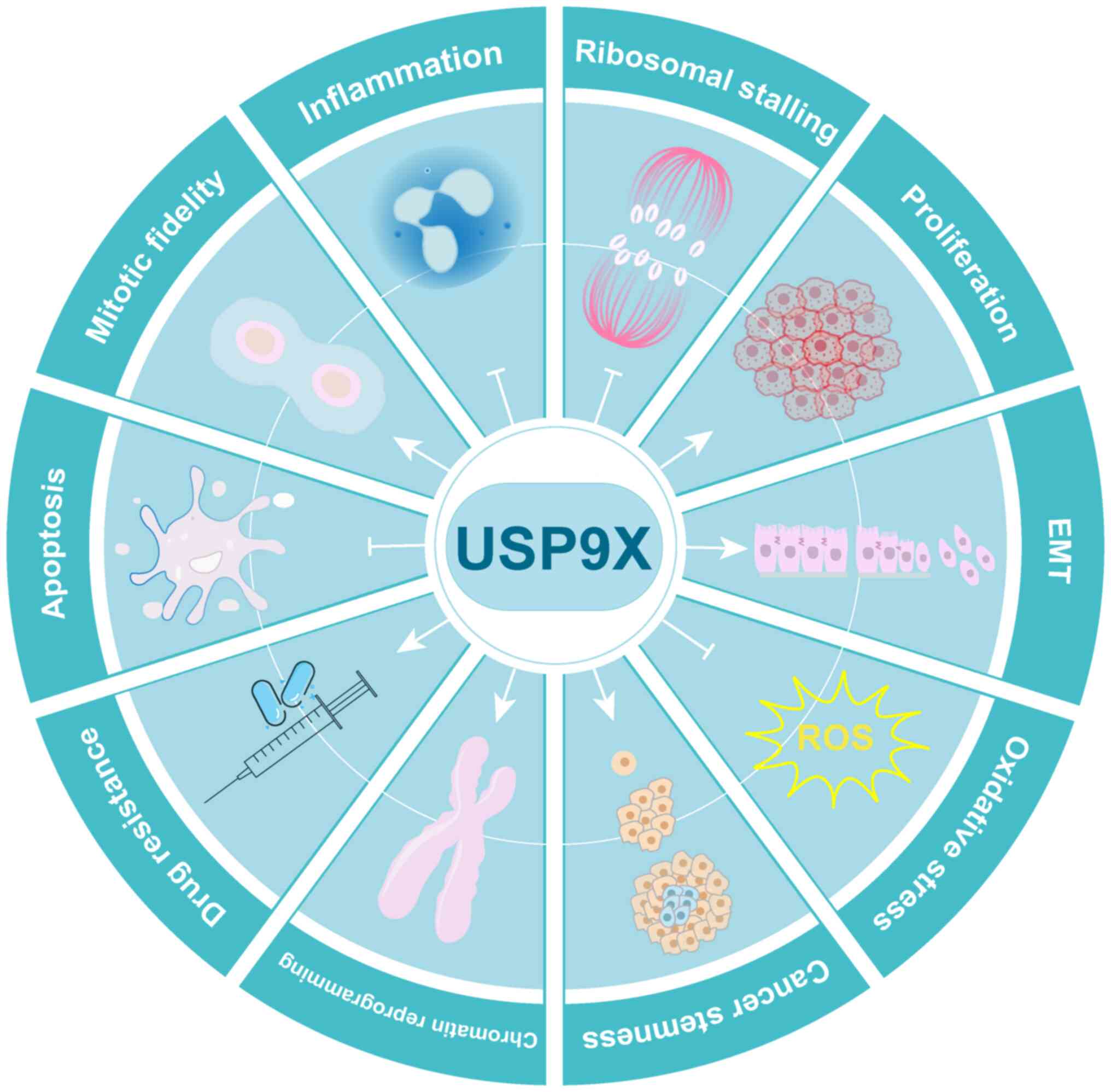
Regulation of both the expression level of USP9X and
its functions could lead to diverse and dynamic biological
behavioral changes under a variety of cellular conditions. USP9X
has been identified to have a series of substrates that enable the
DUB to regulate cell apoptosis and survival, mitotic fidelity and
the cell cycle, cell migration and invasion, and DNA damage repair.
USP9X exhibits considerable control over these cellular functions
and also in the development of a number of diseases.
The execution of apoptosis is countered by the
action of anti-apoptotic proteins (58). Since certain substrates of USP9X,
such as MCL-1 and XIAP, are key factors in cellular apoptosis
signaling pathways that drive cell apoptosis, targeting these
substrates via USP9X might lead to the regulation of cell apoptosis
(41–45). USP9X displays both pro- and
anti-apoptotic functions, mediated by the deubiquitination of
critical components of the apoptotic signaling networks (53,59).
Previous studies have reported that the expression of
stress-sensing, pro-apoptotic kinases is regulated by USP9X to
initiate the apoptotic JNK signaling cascade (59,60).
The pro-apoptotic kinase, apoptosis signal-regulating kinase 1
(ASK-1), is activated under conditions of oxidative stress, leading
to the selective activation of the JNK signaling pathways (59). USP9X also interacts with ASK-1,
protecting it from proteasomal degradation to mediate oxidative
stress-induced cell death (59).
USP9X also activates and stabilizes the dual leucine zipper kinase
in response to extracellular and intracellular stress in neurons,
thereby enabling the activation of pro-apoptotic JNK signaling
(60).
By contrast, USP9X enhances the activities of a
large spectrum of anti-apoptotic factors for cell survival. In the
classical mitochondrial apoptosis pathway, the pro-survival B-cell
lymphoma-2 family proteins, including MCL-1, preserve mitochondrial
integrity and indirectly inhibit the activation of caspase-3 and
−7, ultimately limiting the rate of cell apoptosis (17). USP9X stabilizes MCL-1 through its
interaction with the protein and the removal of the Lys48-linked
polyubiquitin chains that mark a protein for proteasomal
degradation (53). A recent study
reported that the RNA helicase Asp-Glu-Ala-Asp-box polypeptide 3
interacted with the N-terminus of USP9X and participated in
deubiquitination of MCL-1 (61).
Therefore, human tumors with a high level of MCL-1 expression may
be accompanied by the overexpression of USP9X (53,62).
Upregulated expression of USP9X promotes tumorigenicity and cell
survival through stabilizing cell death regulators, including
X-linked inhibitor of apoptosis protein (XIAP), and inhibiting the
induction of apoptosis by specifically stabilizing MCL-1 (53). WP1130 is a small molecule that
directly decreases the DUB activity of USP9X, which leads to the
downregulation of MCL-1, ultimately facilitating apoptosis
(63,64). A previous study reported that
apoptosis was induced by the inhibition of USP9X at least partly
through oxidative stress, which activated DNA damage responses and
stress-associated mitogen-activated protein kinase (MAPK) signaling
pathways (43). Therefore, USP9X,
when co-expressed with multiple apoptosis-associated proteins,
exerts an anti-apoptotic role in cancers, such as oral cancer,
prostate cancer, chronic myeloid leukemia (CML) and acute myeloid
leukemia (AML) (65–67). Overexpression or depletion of USP9X
is therefore an important factor in cell apoptosis and
survival.
Regulation of mitosis safeguards cellular integrity
and its failure contributes to the progression, maintenance and
drug resistance of cancer (68–70).
In the cell cycle, the mitotic checkpoint complex (MCC) senses the
orientation of sister chromatids on the mitotic spindle and
restricts the activity of anaphase-promoting complex/cyclosome
(APC/C) ubiquitin ligase to initiate mitotic exit, consequently
ensuring that chromosome segregation and anaphase are able to occur
(71). In this way, the spindle
assembly checkpoint is strengthened and chromosomal stability is
made more secure through restricting APC/C-mediated MCC turnover
(71). By contrast, downregulation
of USP9X contributes to a reduction in the efficacy of the spindle
assembly checkpoint, an increase in chromosome segregation defects
and chromosomal instability, which leads to the subsequent
promotion of cancer (71).
Activating the mitotic phosphorylation of USP9X promotes cell
survival through counteracting mitotic ubiquitination and the
ensuing proteasomal degradation of Wilms' tumor protein 1, the
latter of which modulates the transcription and secretion of CXC
motif chemokine ligand 8/interleukin-8 in mitosis (72). In addition, the dephosphorylation of
USP9X mediated by cell division cycle 14B (CDC14B) has previously
been shown to promote mitotic apoptosis (72). USP9X is an integral component of the
centrosome, where it functions to stabilize certain centrosome
proteins, centrosomal protein 55 and pericentriolar material 1,
thereby promoting centrosome biogenesis (73). Activation of USP9X enhances
centrosome amplification and chromosome instability, whereas
inactivation of USP9X leads to an impairment of centrosome
duplication (40). It has been
previously reported that USP9X may stabilize cell cycle-associated
proteins to control ribosomal stalling (74), regulate centrosome duplication
(40), and antagonize mitotic cell
death and chemoresistance (41).
Proteins, such as USP9X, that coordinate the normal functioning of
the cell cycle may also trigger inappropriate cell divisions,
thereby taking on dysfunctional roles in certain pathological
disorders, including X-linked intellectual disability (16,18)
and malignancies (40).
USP9X also serves pivotal roles in cell migration
and invasion. Doublecortin (DCX) is a microtubule-associated
protein involved in vesicle transport and microtubule dynamics
(75). The C-terminus of USP9X
binds DCX, which acts as a regulator of neuronal cell migration
(76). Loss of USP9X has been shown
not only to reduce axon growth, but also to cause a reduction in
neuronal cell migration both in vivo and in vitro via
USP9X-mediated disruption of the neuronal cytoskeleton (18). Accordingly, USP9X is required for
normal neuronal cell migration. In addition, USP9X-mediated
deubiquitination of integrin a5b1 serves a role in a5b1-dependent
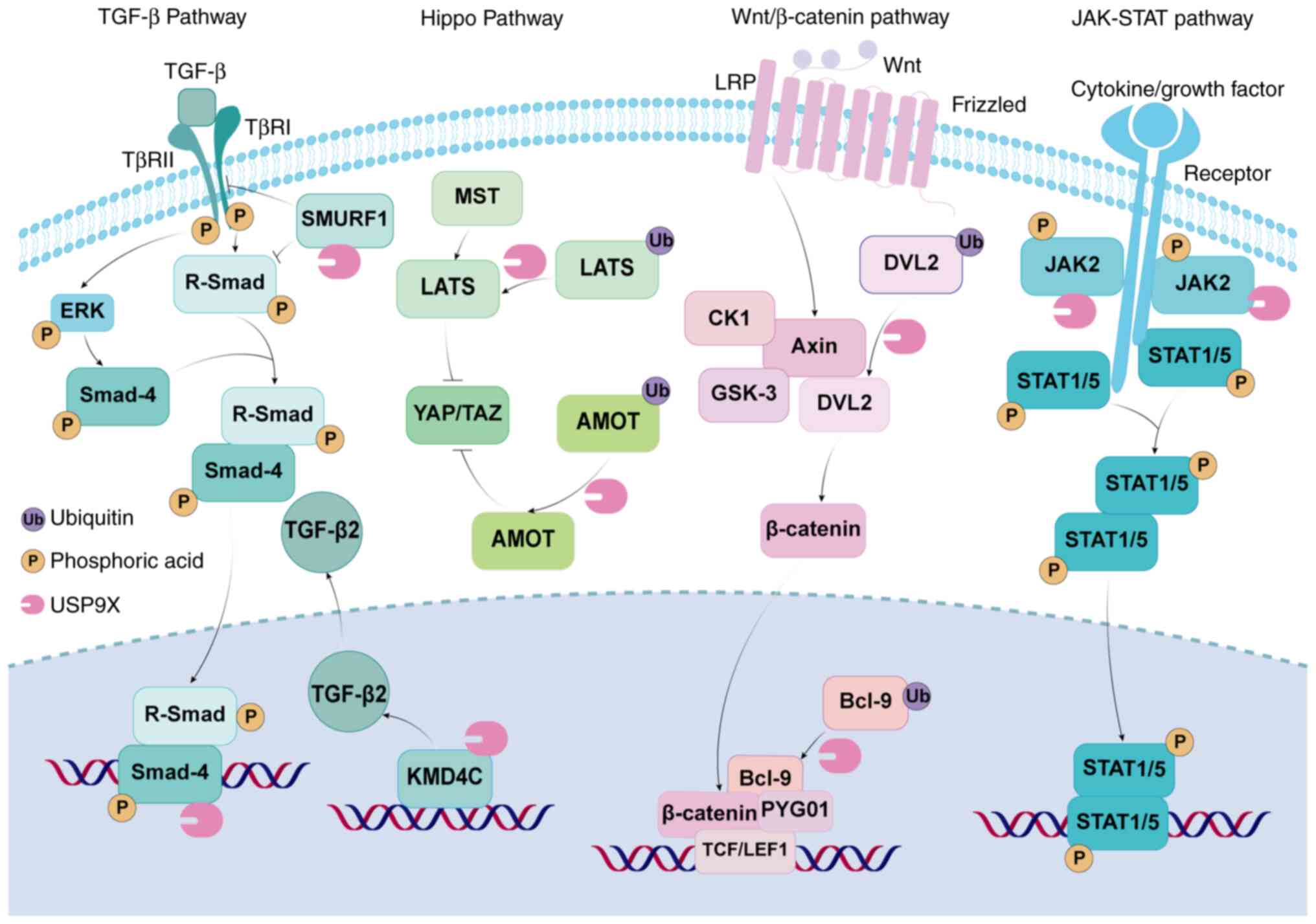
cell migration (77). A previous
study reported that USP9X promoted TGF-β-dependent cancer
progression and metastasis through interaction with SMAD4 in the
TGF-β signaling pathway, thereby inhibiting the E3
ubiquitin-protein ligase TIF1γ-mediated ubiquitination of SMAD4
(46). SMAD-specific E3 ubiquitin
protein ligase 1 (SMURF1) was originally identified to block the
TGF/bone morphogenetic protein (BMP) signaling pathway by
specifically degrading SMAD1 and SMAD5, as well as TGF/BMP
receptors (78,79). The negative regulation of USP9X
destabilizes SMURF1 and inhibits SMURF1-dependent cell migration in
breast cancer cells (80). USP9X
has also been shown to activate the prostaglandin E synthase
(PTGES)/prostaglandin E2 (PGE2) pathway, thereby promoting
metastatic features of non-small cell lung cancer (NSCLC) cells
through the deubiquitination and stabilization of PTGES, which
functions as a key enzyme for the process of PGE2 synthesis in the
arachidonic acid pathway (81). It
has previously been demonstrated that USP9X is able to disrupt
neuronal cell migration and growth, which is associated with
X-linked intellectual disability; therefore, USP9X may serve roles
in both neurodevelopment and the modulation of neural apoptosis
(18). Taken together, these
findings demonstrate the importance of UPS9X in regulating cell
migration and invasion.
The available evidence demonstrates that the
USP9X-induced aggressive and metastatic phenotypes of cancer are
regulated by the abnormal expression of non-coding RNAs (91–97)
(Table II). The long non-coding
RNA LINC01433 has been shown to enhance the interaction between
USP9X and Yes-associated protein (YAP), thereby stabilizing YAP and
leading to the promotion of tumor progression and chemotherapy
resistance in gastric cancer (GC) cells (91). In addition, the circular RNA
hsa_circ_0008434 is highly expressed in GC and serves the role of
upregulating the expression of USP9X, thereby promoting the
malignant phenotypes of GC cells by serving as a microRNA (miR)
sponge for miR-6838-5p (93).
Moreover, downregulation of the cellular FADD-like
interleukin-1β-converting enzyme-inhibitory protein, which is
induced by WP1130, a type of selective USP9X inhibitor that
decreases the activity of USP9X, was shown to be regulated by
miR-708 through the inhibition of USP9X, which induces the
apoptosis of CaSki cells (98). In
addition, miR-132 was previously shown to target USP9X-induced EMT,
which led to inhibition of the migration and invasive capabilities
of NSCLC cells (97).
USP9X has been shown to exert opposing effects in
hematological malignancies. It has been reported that USP9X can
promote tumor cell survival in human diffuse large B-cell lymphoma,
multiple myeloma and follicular lymphoma through deubiquitinating
MCL-1, which protects it from degradation, resulting in poor
clinical outcomes (38,45,99,100).
In addition, a high level of USP9X in patients with aggressive
B-cell lymphoma has been shown to be associated with excessive
B-cell proliferation, resulting in an adverse prognosis and
resistance to treatment therapies through the deubiquitination and
stabilization of XIAP, independent of MCL-1 (41). A BCR-ABL-positive diagnosis is a
typical characteristic of patients with CML. The WP1130-mediated
inhibition of USP9X is associated with a reduction in MCL-1 levels,
followed by blockade of BCR-ABL kinase signaling, which leads to
the rapid onset of CML cell apoptosis (21). However, USP9X is not involved in
BCR-ABL ubiquitination or cellular localization (101). Moreover, USP9X silencing was shown
to lead to lymphoma growth suppression, leading to decreased
chemotherapy resistance in B-cell lymphoma (63). Internal tandem duplications of
FMS-like tyrosine kinase 3 (FLT3-ITD) occur frequently in AML, and
is associated with poor outcomes in patients with AML. USP9X
interacts with FLT3-ITD to inhibit its Lys63-linked
polyubiquitination (43,102). Furthermore, FLT3-ITD reversely
induces the ubiquitination and tyrosine-phosphorylation of USP9X,
thereby promoting its proteasomal degradation (102). USP9X also stabilizes RNA m6A
demethylase ALKBH5 by removing the K48-linked polyubiquitin chain
at K57 and promotes AML cell survival (44). Taken together, these findings
identify USP9X as an oncogenic gene that promotes the development
of hematological malignancies, and the therapeutic targeting of
USP9X may lead to preferential inhibition of certain types of
leukemia.
USP9X also exerts negative effects in hematological
malignancies by acting as a tumor suppressor gene. USP9X has been
identified as a novel leukemia susceptibility gene associated with
B-cell acute lymphoblastic leukemia (B-ALL) and multiple
neurodevelopmental and congenital abnormalities (103). Low USP9X expression is associated
with decreased survival in patients with high-risk B-ALL (103). Genetic or pharmacological
inhibition of USP9X can restrict JAK signaling to enhance the
survival of cytokine receptor-like factor 2-positive B-ALL in
patients with Down syndrome (104), which suggests that USP9X may exert
differential effects in different types of leukemia.
The underlying mechanisms of USP9X in breast cancer
are complex. YAP is an important oncogene that drives cancer
progression, and dysregulation of the Hippo/YAP1 signaling pathway
is involved in breast cancer development (105). YAP1 is deubiquitinated and
stabilized by USP9X, thereby promoting breast cancer cell survival
and resulting in chemotherapy resistance (47). USP9X stabilizes Snail1, a key factor
regulating the EMT process, contributing to metastasis and
chemoresistance in triple-negative breast cancer (48). Furthermore, downregulation of USP9X
renders estrogen receptor α-positive breast cancer resistant to
tamoxifen, leading to a poor outcome for patients following
adjuvant tamoxifen treatment (89).
USP9X can modulate centrosome biogenesis and enhance breast
carcinogenesis by deubiquitinating and stabilizing the centriolar
satellite protein, CEP131 (40). In
a previous study, the arginine methylation of USP9X was shown to
improve its interaction with Tudor domain-containing protein 3
(TDRD3), which led to a subsequent enhancement of its
anti-apoptotic activities in breast cancer cells (35). The aforementioned study also
reported that downregulation of TDRD3 improved the sensitivity of
chemotherapeutic drug-induced apoptosis in breast cancer cells,
which is likely due to its regulation of USP9X DUB activity on the
anti-apoptotic protein MCL-1 and stress granule localization
(35).
Obesity may be a risk factor for the development of
breast cancer, as it is associated with reduced survival and
increases the risk of distant metastasis for female patients
diagnosed with breast cancer (106). Plasma free fatty acids further
facilitate this biological progression of breast cancer in obese
patients (46). USP9X can also be
recruited by the transcription factor Nanog to stabilize the
hypoxia-inducible factor-1α protein, which leads to an enhancement
of self-renewal in breast cancer stem cells (49). Moreover, USP9X selectively promotes
activation of the Notch developmental signaling pathway in
triple-negative breast cancer, and small molecule EOAI3402143
(G9)-mediated USP9X inhibition specifically inhibits the Notch
pathway, which leads to a remodeling of the tumor immune landscape
and suppresses tumor growth (107). Taken together, these studies
suggest that a diverse range of USP9X-associated mechanisms are
involved in breast cancer.
Histone lysine demethylase 4C (KDM4C), a substrate
of USP9X, is associated with poor clinical outcomes in patients
with lung cancer (50). A recent
study reported that USP9X activated the TGF-β/SMAD signaling
pathway, thereby inducing radioresistance by deubiquitinating and
stabilizing KDM4C in lung cancer (50). USP9X is also positively correlated
with the dual-specificity protein kinase known as threonine
tyrosine kinase, which modulates cell proliferation, migration and
tumorigenesis in NSCLC (51).
Furthermore, the inhibition of USP9X has been shown to contribute
towards MCL-1-mediated proteasomal degradation, radiosensitivity
and apoptosis in NSCLC cells (52).
Moreover, the chemotherapeutic agent pemetrexed has been shown to
induce apoptosis by increasing the expression of the pro-apoptotic
protein Noxa, thereby activating the Noxa-USP9X-MCL-1 axis, which
demonstrates that USP9X serves a critical role in human lung cancer
cells (108). Collectively, these
studies have shown that USP9X exerts a range of different functions
in lung cancer.
Pancreatic cancer is a lethal malignancy, and
effective targeted therapies are urgently required to combat its
high morbidity rates (2.6%) and mortality (4.7%) rates worldwide in
2020 (109). To date, USP9X has
been reported to show tumor-suppressing potential in pancreatic
cancer (110–112). USP9X suppressed tumorigenesis in a
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog mouse model of
pancreatic ductal adenocarcinoma (PDAC) and its downregulation
enhanced malignant transformation, protecting PDAC cells from
anoikis (110). However, these
properties of USP9X in pancreatic cancer predominantly depend on
its intrinsic deubiquitinase activity. USP9X mediates acute MCL-1
stabilization and protection from apoptosis in response to MAPK
suppression caused by MEK inhibitors (113), which may provide a promising
therapeutic strategy for pancreatic cancer.
The differential expression or genetic alterations
of USP9X have been identified in multiple types of human cancer,
which suggests that it potentially exerts different roles in tumor
progression. Higher expression levels of USP9X are observed in oral
squamous cell carcinoma (OSCC) cells, where it drives oral
tumorigenesis by deubiquitinating and stabilizing the
anti-apoptotic protein MCL-1, which was shown to correlate with
poor outcomes in patients with OSCC (53). In addition, aldehyde dehydrogenase 1
family member A3 (ALDH1A3) functions as a key enzyme for
maintaining the self-renewal and mesenchymal (MES) features of
glioblastoma stem cells (54).
Depletion of USP9X leads to a marked downregulation of ALDH1A3,
which leads to a loss of the tumorigenic and self-renewal
capabilities of MES glioblastoma stem cells. By contrast, a high
expression level of USP9X is indicative of potent tumorigenic
capability in MES glioblastoma stem cells with enrichment of
ALDH1A3 (114). A previous study
reported that USP9X-mediated deubiquitination and stabilization of
Ets-1 promoted the expression of the N-RAS oncogene and
carcinogenesis in melanoma (56).
Although USP9X has been reported to exert oncogenic
functions in several different types of cancer, a number of
previous studies have also reported the tumor-suppressive
properties of USP9X in carcinogenesis. In the murine intestine,
USP9X is necessary for tissue homeostasis and regeneration
following acute colitis (55); it
regulates the function and protein expression levels of the tumor
suppressor FBW7, which consequently protects it from proteasomal
degradation (55). Therefore, the
restricted level of c-Myc that is regulated by USP9X via
stabilization of FBW7 reduces the risk of colitis-mediated
colorectal cancer (CRC) in mice, whereas the silencing of USP9X is
associated with a poor prognosis in human CRC (55). USP9X expression is downregulated in
CRC and CHOL (89). Moreover, USP9X
is involved in the activation of apoptosis in CHOL, which also
suppresses tumor cell proliferation (57). USP9X exerts its tumor-suppressive
functions through deubiquitinating Egl-9 family hypoxia inducible
factor 3, thereby activating the apoptosis signaling pathway
components, kinesin KIF1Bβ and cleaved caspase-3 (57). USP9X also targets and regulates the
stability of angiomotin to indirectly inhibit YAP/transcriptional
coactivator with PDZ-binding motif activity, and a low level of
USP9X is correlated with poor clinical outcome in renal clear cell
carcinoma (115). Taken together,
these findings suggest that there may be potential for USP9X to be
used as a therapeutic target for the treatment of certain types of
cancer.
Small molecule inhibitors that target DUBs are
emerging as a novel form of anticancer therapeutic strategy
(82). A previous study reported
that USP9X overexpression inhibited apoptosis and promoted tumor
cell survival (41,57). Moreover, previous studies have
identified USP9X as an oncogene in various types of cancer
(47,53), suggesting that it may be a future
potential target for therapeutic development. However, no small
molecule inhibitors specific to USP9X are currently available for
use in a clinical setting. WP1130, previously known as Degrasyn
(116), is the best-characterized
USP9X inhibitor capable of inhibiting USP9X activity, in addition
to the activity of several other DUBs (USP5, USP14, USP24, UCH37
and UCH-L1) (117), thereby
highlighting its role as a type of partially selective DUB
inhibitor. The proliferation, inhibition and anti-apoptotic effects
of WP1130 against diverse tumors have been reported in B-cell
malignancies (118), AML (43), NSCLC (51) and glioblastoma (114), mainly through the inhibition of
USP9X with the subsequent accumulation of polyubiquitinated
proteins and downregulation of antiapoptotic proteins, including
MCL-1 and p53. WP1130 has also been shown to increase tumor cell
sensitivity to chemotherapy (43).
BIX-01294 is an inhibitor developed to inhibit the activity of
euchromatic histone-lysine N-methyltransferase 2 and has been shown
to promote USP9X downregulation by stimulating both endoplasmic
reticulum stress and the expression of
phorbol-12-myristate-13-acetate-induced protein 1, resulting in a
downregulation of the level of MCL-1, which promotes apoptosis in
bladder cancer cells (119).
Peterson et al (118)
reported on a small molecule inhibitor of USP9X/USP24,
EOAI3402143(G9), which inhibited the DUB activity of USP9X and
USP24 in a dose-dependent manner, increased apoptosis in myeloma
and fully regressed or blocked myeloma tumors in mice. G9 was shown
to induce apoptosis in lymphoma and myeloma cell lines in
vitro, inhibiting tumor progression with little overt toxicity
(120). BRD0476 selectively
inhibits USP9X activity to suppress the JAK-STAT pathway, which
protects human pancreatic β-cells and cancer cells from
cytokine-induced apoptosis (121).
It was also demonstrated that the disruption of USP9X by the
CRISPR/Cas9 system and small interfering RNA intervention brought
about similar protective effects resulting from BRD0476 treatment,
which suggests that BRD0476 may function as a modulator of USP9X
(121). Isothiocyanates, such as
phenethyl isothiocyanate and benzyl isothiocyanate, have also been
shown to exert anticancer activity by inhibiting USP9X and other
DUBs in physiologically relevant time scales and concentrations,
particularly in hematological malignancies (122). Taken together, these study results
highlight the potential of pharmacological inhibitors targeting
USP9X, which may be an effective therapeutic target courtesy of its
DUB activity. Although inhibitors of USP9X are not at present
available for clinical use, novel therapeutic approaches targeting
USP9X in human cancers may be developed further in the future.
USP9X interacts with an extensive range of
substrates and has the potential to regulate multiple signaling and
survival pathways with cellular responses. USP9X controls a wide
variety of cellular pathological and physiological processes
through its DUB activity. Previous studies have reported on the
significance of USP9X in the cell cycle, apoptosis and survival,
and its crucial functions in cell migration and invasion, and DNA
damage repair are mediated predominantly via
deubiquitination-mediated regulation of the turnover of certain
substrates. However, the roles of USP9X in tumor development are
complex, and contradictory functions have been identified in
different developmental contexts, wherein USP9X displays both
oncogenic activity and tumor suppressor functions. In addition to
the substrates and downstream signaling pathways regulated by USP9X
in tumor progression, the upstream regulatory factors involved in
modulating USP9X protein activation in carcinogenesis also merit
further exploration. In the future, the development of specific
novel inhibitors against USP9X may be a successful strategy for
tumor therapy.
Not applicable.
This study was supported by grants from the National Natural
Science Foundation of China (grant no. 82102816), the Zhejiang
Provincial Natural Science Foundation of China (grant no.
LQ22H160014) and Zhejiang Provincial People's Hospital Scientific
Research Foundation for The Excellent Youth (grant no.
ZRY2020B003).
Not applicable.
YH conceived and designed the study. YM, YH, ZQ, CH
and SY prepared the figures and tables. YH, YM and LY wrote and
edited the manuscript. ZQ, CH and SY revised the manuscript. All
authors read and approved the final version of the manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Ermolaeva M, Neri F, Ori A and Rudolph KL:
Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol
Cell Biol. 19:594–610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holguin-Cruz JA, Foster LJ and Gsponer J:
Where protein structure and cell diversity meet. Trends Cell Biol.
32:996–1007. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Toyama BH and Hetzer MW: Protein
homeostasis: Live long, won't prosper. Nat Rev Mol Cell Biol.
14:55–61. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beynon RJ and Bond JS: Catabolism of
intracellular protein: Molecular aspects. Am J Physiol.
251:141–152. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Komander D and Rape M: The ubiquitin code.
Annu Rev Biochem. 81:203–229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X, Elmira E, Rohondia S, Wang J, Liu J
and Dou QP: A patent review of the ubiquitin ligase system:
2015–2018. Expert Opin Ther Pat. 28:919–937. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jang HH: Regulation of protein degradation
by proteasomes in cancer. J Cancer Prev. 23:153–161. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Becker JR, Clifford G, Bonnet C, Groth A,
Wilson MD and Chapman JR: BARD1 reads H2A lysine 15 ubiquitination
to direct homologous recombination. Nature. 596:433–437. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roberts JZ, Crawford N and Longley DB: The
role of ubiquitination in apoptosis and necroptosis. Cell Death
Differ. 29:272–284. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun T, Liu Z and Yang Q: The role of
ubiquitination and deubiquitination in cancer metabolism. Mol
Cancer. 19:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schmidt MF, Gan ZY, Komander D and Dewson
G: Ubiquitin signalling in neurodegeneration: Mechanisms and
therapeutic opportunities. Cell Death Differ. 28:570–590. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fhu CW and Ali A: Dysregulation of the
ubiquitin proteasome system in human malignancies: A window for
therapeutic intervention. Cancers (Basel). 13:15132021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grabbe C, Husnjak K and Dikic I: The
spatial and temporal organization of ubiquitin networks. Nat Rev
Mol Cell Biol. 12:295–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shimizu K, Gi M, Suzuki S, North BJ,
Watahiki A, Fukumoto S, Asara JM, Tokunaga F, Wei W and Inuzuka H:
Interplay between protein acetylation and ubiquitination controls
MCL1 protein stability. Cell Rep. 37:1099882021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murtaza M, Jolly LA, Gecz J and Wood SA:
La FAM fatale: USP9X in development and disease. Cell Mol Life Sci.
72:2075–2089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johnson BV, Kumar R, Oishi S, Alexander S,
Kasherman M, Vega MS, Ivancevic A, Gardner A, Domingo D, Corbett M,
et al: Partial loss of USP9X function leads to a male
neurodevelopmental and behavioral disorder converging on
transforming growth factor beta signaling. Biol Psychiatry.
87:100–112. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Delbridge ARD, Kueh AJ, Ke F, Zamudio NM,
El-Saafin F, Jansz N, Wang GY, Iminitoff M, Beck T, Haupt S, et al:
Loss of p53 causes stochastic aberrant x-chromosome inactivation
and female-specific neural tube defects. Cell Rep. 27:442–454.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Homan CC, Kumar R, Nguyen LS, Haan E,
Raymond FL, Abidi F, Raynaud M, Schwartz CE, Wood SA, Gecz J, et
al: Mutations in USP9X are associated with X-linked intellectual
disability and disrupt neuronal cell migration and growth. Am J Hum
Genet. 94:470–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang B, Tang X, Yao L, Wang Y, Chen Z, Li
M, Wu N, Wu D, Dai X, Jiang H and Ai D: Disruption of USP9X in
macrophages promotes foam cell formation and atherosclerosis. J
Clin Invest. 132:e1542172022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Greenhill C: Pancreatic cancer: USP9X can
be used to predict pancreatic cancer outcomes. Nat Rev
Gastroenterol Hepatol. 9:3022012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones MH, Furlong RA, Burkin H, Chalmers
IJ, Brown GM, Khwaja O and Affara NA: The Drosophila developmental
gene fat facets has a human homologue in Xp11.4 which escapes
X-inactivation and has related sequences on Yq11.2. Hum Mol Gene.
5:1695–1701. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wood SA, Pascoe WS, Ru K, Yamada T,
Hirchenhain J, Kemler R and Mattick JS: Cloning and expression
analysis of a novel mouse gene with sequence similarity to the
Drosophila fat facets gene. Mech Dev. 63:29–38. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Overstreet E, Wood SA and Fischer
JA: On the conservation of function of the Drosophila fat facets
deubiquitinating enzyme and Fam, its mouse homolog. Dev Genes Evol.
210:603–610. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fischer-Vize JA, Rubin GM and Lehmann R:
The fat facets gene is required for Drosophila eye and embryo
development. Development. 116:985–1000. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khut PY, Tucker B, Lardelli M and Wood SA:
Evolutionary and expression analysis of the zebrafish
deubiquitylating enzyme, usp9. Zebrafish. 4:95–101. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nijman SM, Luna-Vargas MP, Velds A,
Brummelkamp TR, Dirac AM, Sixma TK and Bernards R: A genomic and
functional inventory of deubiquitinating enzymes. Cell.
123:773–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amerik AY and Hochstrasser M: Mechanism
and function of deubiquitinating enzymes. Biochim Biophys Acta.
1695:189–207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Paudel P, Zhang Q, Leung C, Greenberg HC,
Guo Y, Chern YH, Dong A, Li Y, Vedadi M, Zhuang Z and Tong Y:
Crystal structure and activity-based labeling reveal the mechanisms
for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc Natl Acad Sci USA. 116:7288–7297. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marx C, Held JM, Gibson BW and Benz CC:
ErbB2 trafficking and degradation associated with K48 and K63
polyubiquitination. Cancer Res. 70:3709–3717. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al-Hakim AK, Zagorska A, Chapman L, Deak
M, Peggie M and Alessi DR: Control of AMPK-related kinases by USP9X
and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem
J. 411:249–260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dupont S, Mamidi A, Cordenonsi M,
Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ,
Soligo S, Morsut L, et al: FAM/USP9×, a deubiquitinating enzyme
essential for TGFbeta signaling, controls Smad4 monoubiquitination.
Cell. 136:123–135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mouchantaf R, Azakir BA, McPherson PS,
Millard SM, Wood SA and Angers A: The ubiquitin ligase itch is
auto-ubiquitylated in vivo and in vitro but is protected from
degradation by interacting with the deubiquitylating enzyme
FAM/USP9X. J Biol Chem. 281:38738–38747. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Taya S, Yamamoto T, Kano K, Kawano Y,
Iwamatsu A, Tsuchiya T, Tanaka K, Kanai-Azuma M, Wood SA, Mattick
JS and Kaibuchi K: The Ras target AF-6 is a substrate of the fam
deubiquitinating enzyme. J Cell Biol. 142:1053–1062. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vong QP, Cao K, Li HY, Iglesias PA and
Zheng Y: Chromosome alignment and segregation regulated by
ubiquitination of survivin. Science. 310:1499–1504. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Narayanan N, Wang Z, Li L and Yang Y:
Arginine methylation of USP9X promotes its interaction with TDRD3
and its anti-apoptotic activities in breast cancer cells. Cell
Discov. 3:160482017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murray RZ, Jolly LA and Wood SA: The FAM
deubiquitylating enzyme localizes to multiple points of protein
trafficking in epithelia, where it associates with E-cadherin and
beta-catenin. Mol Biol Cell. 15:1591–1599. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Theard D, Labarrade F, Partisani M,
Milanini J, Sakagami H, Fon EA, Wood SA, Franco M and Luton F:
USP9×-mediated deubiquitination of EFA6 regulates de novo tight
junction assembly. EMBO J. 29:1499–1509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schwickart M, Huang X, Lill JR, Liu J,
Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F,
Eastham-Anderson J, et al: Deubiquitinase USP9X stabilizes MCL1 and
promotes tumour cell survival. Nature. 463:103–107. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia
R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U,
Leonhardt H and Lamond A: Identifying specific protein interaction
partners using quantitative mass spectrometry and bead proteomes. J
Cell Biol. 183:223–239. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Song N, Liu L, Liu X, Ding X, Song
X, Yang S, Shan L, Zhou X, Su D, et al: USP9X regulates centrosome
duplication and promotes breast carcinogenesis. Nat Commun.
8:148662017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Engel K, Rudelius M, Slawska J, Jacobs L,
Abhari BA, Altmann B, Kurutz J, Rathakrishnan A, Fernandez-Saiz V,
Brunner A, et al: USP9X stabilizes XIAP to regulate mitotic cell
death and chemoresistance in aggressive B-cell lymphoma. EMBO Mol
Med. 8:851–862. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grou CP, Francisco T, Rodrigues TA,
Freitas MO, Pinto MP, Carvalho AF, Domingues P, Wood SA,
Rodriguez-Borges JE, Sa-Miranda C, et al: Identification of
ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting
on ubiquitin-peroxin 5 (PEX5) thioester conjugate. J Biol Chem.
287:12815–12827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Akiyama H, Umezawa Y, Ishida S, Okada K,
Nogami A and Miura O: Inhibition of USP9X induces apoptosis in
FLT3-ITD-positive AML cells cooperatively by inhibiting the mutant
kinase through aggresomal translocation and inducing oxidative
stress. Cancer Lett. 453:84–94. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang P, Wang J, Yao S, Cui M, Cheng Y, Liu
W, Gao Z, Hu J, Zhang J and Zhang H: Deubiquitinase USP9X
stabilizes RNA m(6)A demethylase ALKBH5 and promotes acute myeloid
leukemia cell survival. J Biol Chem. 299:1050552023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Akiyama H, Umezawa Y, Watanabe D, Okada K,
Ishida S, Nogami A and Miura O: Inhibition of USP9X downregulates
JAK2-V617F and induces apoptosis synergistically with BH3 mimetics
preferentially in ruxolitinib-persistent JAK2-V617F-positive
leukemic cells. Cancers (Basel). 12:4062020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu Y, Yu X, Yi X, Wu K, Dwabe S, Atefi M,
Elshimali Y, Kemp KT II, Bhat K, Haro J, et al: Aberrant
phosphorylation of SMAD4 Thr277-mediated USP9×-SMAD4 interaction by
free fatty acids promotes breast cancer metastasis. Cancer Res.
77:1383–1394. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li L, Liu T, Li Y, Wu C, Luo K, Yin Y,
Chen Y, Nowsheen S, Wu J, Lou Z and Yuan J: The deubiquitinase
USP9X promotes tumor cell survival and confers chemoresistance
through YAP1 stabilization. Oncogene. 37:2422–2431. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guan T, Yang X, Liang H, Chen J, Chen Y,
Zhu Y and Liu T: Deubiquitinating enzyme USP9X regulates metastasis
and chemoresistance in triple-negative breast cancer by stabilizing
Snail1. J Cell Physiol. 237:2992–3000. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu H, Lyu Y, Tran L, Lan J, Xie Y, Yang Y,
Murugan NL, Wang YJ and Semenza GL: HIF-1 recruits NANOG as a
coactivator for TERT gene transcription in hypoxic breast cancer
stem cells. Cell Rep. 36:1097572021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jie X, Fong WP, Zhou R, Zhao Y, Zhao Y,
Meng R, Zhang S, Dong X, Zhang T, Yang K, et al: USP9X-mediated
KDM4C deubiquitination promotes lung cancer radioresistance by
epigenetically inducing TGF-β2 transcription. Cell Death Differ.
28:2095–2111. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen X, Yu C, Gao J, Zhu H, Cui B, Zhang
T, Zhou Y, Liu Q, He H, Xiao R, et al: A novel USP9X substrate TTK
contributes to tumorigenesis in non-small-cell lung cancer.
Theranostics. 8:2348–2360. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kushwaha D, O'Leary C, Cron KR, Deraska P,
Zhu K, D'Andrea AD and Kozono D: USP9X inhibition promotes
radiation-induced apoptosis in non-small cell lung cancer cells
expressing mid-to-high MCL1. Cancer Biol Ther. 16:392–401. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sulkshane P, Pawar SN, Waghole R, Pawar
SS, Rajput P, Uthale A, Oak S, Kalkar P, Wani H, Patil R, et al:
Elevated USP9X drives early-to-late-stage oral tumorigenesis via
stabilisation of anti-apoptotic MCL-1 protein and impacts outcome
in oral cancers. Br J Cancer. 125:547–560. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cheng P, Wang J, Waghmare I, Sartini S,
Coviello V, Zhang Z, Kim SH, Mohyeldin A, Pavlyukov MS, Minata M,
et al: FOXD1-ALDH1A3 signaling is a determinant for the
self-renewal and tumorigenicity of mesenchymal glioma stem cells.
Cancer Res. 76:7219–7230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Khan OM, Carvalho J, Spencer-Dene B,
Mitter R, Frith D, Snijders AP, Wood SA and Behrens A: The
deubiquitinase USP9X regulates FBW7 stability and suppresses
colorectal cancer. J Clin Invest. 128:1326–1337. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Potu H, Peterson LF, Kandarpa M, Pal A,
Sun H, Durham A, Harms PW, Hollenhorst PC, Eskiocak U, Talpaz M and
Donato NJ: Usp9× regulates Ets-1 ubiquitination and stability to
control NRAS expression and tumorigenicity in melanoma. Nat Commun.
8:144492017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen W, Song J, Liu S, Tang B, Shen L, Zhu
J, Fang S, Wu F, Zheng L, Qiu R, et al: USP9X promotes apoptosis in
cholangiocarcinoma by modulation expression of KIF1Bbeta via
deubiquitinating EGLN3. J Biomed Sci. 28:442021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Czabotar PE, Lessene G, Strasser A and
Adams JM: Control of apoptosis by the BCL-2 protein family:
Implications for physiology and therapy. Nat Rev Mol Cell Biol.
15:49–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nagai H, Noguchi T, Homma K, Katagiri K,
Takeda K, Matsuzawa A and Ichijo H: Ubiquitin-like sequence in ASK1
plays critical roles in the recognition and stabilization by USP9X
and oxidative stress-induced cell death. Mol Cell. 36:805–818.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huntwork-Rodriguez S, Wang B, Watkins T,
Ghosh AS, Pozniak CD, Bustos D, Newton K, Kirkpatrick DS and
Lewcock JW: JNK-mediated phosphorylation of DLK suppresses its
ubiquitination to promote neuronal apoptosis. J Cell Biol.
202:747–763. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lai MC, Chen YP, Li DA, Yu JS, Hung HY and
Tarn WY: DDX3 interacts with USP9X and participates in
deubiquitination of the anti-apoptotic protein MCL1. FEBS J.
289:1043–1061. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hogh-Binder SA, Klein D, Wolfsperger F,
Huber SM, Hennenlotter J, Stenzl A and Rudner J: Protein levels of
anti-apoptotic Mcl-1 and the deubiquitinase USP9× are cooperatively
upregulated during prostate cancer progression and limit response
of prostate cancer cells to radiotherapy. Cancers (Basel).
15:24962023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun H, Kapuria V, Peterson LF, Fang D,
Bornmann WG, Bartholomeusz G, Talpaz M and Donato NJ: Bcr-Abl
ubiquitination and Usp9× inhibition block kinase signaling and
promote CML cell apoptosis. Blood. 117:3151–3162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kapuria V, Peterson LF, Fang D, Bornmann
WG, Talpaz M and Donato NJ: Deubiquitinase inhibition by
small-molecule WP1130 triggers aggresome formation and tumor cell
apoptosis. Cancer Res. 70:9265–9276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Karpel-Massler G, Ishida CT, Bianchetti E,
Shu C, Perez-Lorenzo R, Horst B, Banu M, Roth KA, Bruce JN, Canoll
P, et al: Inhibition of mitochondrial matrix chaperones and
antiapoptotic Bcl-2 family proteins empower antitumor therapeutic
responses. Cancer Res. 77:3513–3526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kim S, Woo SM, Min KJ, Seo SU, Lee TJ,
Kubatka P, Kim DE and Kwon TK: WP1130 enhances TRAIL-induced
apoptosis through USP9X-dependent miR-708-mediated downregulation
of c-FLIP. Cancers (Basel). 11:3442019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu Y, Xu X, Lin P, He Y, Zhang Y, Cao B,
Zhang Z, Sethi G, Liu J, Zhou X and Mao X: Inhibition of the
deubiquitinase USP9× induces pre-B cell homeobox 1 (PBX1)
degradation and thereby stimulates prostate cancer cell apoptosis.
J Biol Chem. 294:4572–4582. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Dominguez-Brauer C, Thu KL, Mason JM,
Blaser H, Bray MR and Mak TW: Targeting mitosis in cancer: Emerging
strategies. Mol Cell. 60:524–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Motofei IG: Biology of cancer:
Understanding the supracellular control of mitosis in physiological
processes and malignancy. Semin Cancer Biol. 92:42–44. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Schoonen PM, Talens F, Stok C, Gogola E,
Heijink AM, Bouwman P, Foijer F, Tarsounas M, Blatter S, Jonkers J,
et al: Progression through mitosis promotes PARP inhibitor-induced
cytotoxicity in homologous recombination-deficient cancer cells.
Nat Commun. 8:159812017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Skowyra A, Allan LA, Saurin AT and Clarke
PR: USP9X limits mitotic checkpoint complex turnover to strengthen
the spindle assembly checkpoint and guard against chromosomal
instability. Cell Rep. 23:852–865. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Dietachmayr M, Rathakrishnan A, Karpiuk O,
von Zweydorf F, Engleitner T, Fernandez-Saiz V, Schenk P, Ueffing
M, Rad R, Eilers M, et al: Antagonistic activities of CDC14B and
CDK1 on USP9X regulate WT1-dependent mitotic transcription and
survival. Nat Commun. 11:12682020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang Q, Tang Y, Xu Y, Xu S, Jiang Y, Dong
Q, Zhou Y and Ge W: The X-linked deubiquitinase USP9X is an
integral component of centrosome. J Biol Chem. 292:12874–12884.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Clancy A, Heride C, Pinto-Fernandez A,
Elcocks H, Kallinos A, Kayser-Bricker KJ, Wang W, Smith V, Davis S,
Fessler S, et al: The deubiquitylase USP9X controls ribosomal
stalling. J Cell Biol. 220:2020042112021. View Article : Google Scholar
|
|
75
|
Weimer JM and Anton ES: Doubling up on
microtubule stabilizers: Synergistic functions of doublecortin-like
kinase and doublecortin in the developing cerebral cortex. Neuron.
49:3–4. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gleeson JG, Lin PT, Flanagan LA and Walsh
CA: Doublecortin is a microtubule-associated protein and is
expressed widely by migrating neurons. Neuron. 23:257–271. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kharitidi D, Apaja PM, Manteghi S, Suzuki
K, Malitskaya E, Roldan A, Gingras MC, Takagi J, Lukacs GL and
Pause A: Interplay of endosomal pH and ligand occupancy in integrin
alpha5beta1 ubiquitination, endocytic sorting, and cell migration.
Cell Rep. 13:599–609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhu H, Kavsak P, Abdollah S, Wrana JL and
Thomsen GH: A SMAD ubiquitin ligase targets the BMP pathway and
affects embryonic pattern formation. Nature. 400:687–693. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ebisawa T, Fukuchi M, Murakami G, Chiba T,
Tanaka K, Imamura T and Miyazono K: Smurf1 interacts with
transforming growth factor-beta type I receptor through Smad7 and
induces receptor degradation. J Biol Chem. 276:12477–12480. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xie Y, Avello M, Schirle M, McWhinnie E,
Feng Y, Bric-Furlong E, Wilson C, Nathans R, Zhang J, Kirschner MW,
et al: Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin
ligase SMURF1 protein and protects it from ligase
activity-dependent self-degradation. J Biol Chem. 288:2976–2985.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang T, Jing B, Sun B, Liao Y, Song H, Xu
D, Guo W, Li K, Hu M, Liu S, et al: Stabilization of PTGES by
deubiquitinase USP9X promotes metastatic features of lung cancer
via PGE(2) signaling. Am J Cancer Res. 9:1145–1160. 2019.PubMed/NCBI
|
|
82
|
Harrigan JA, Jacq X, Martin NM and Jackson
SP: Deubiquitylating enzymes and drug discovery: Emerging
opportunities. Nat Rev Drug Discov. 17:57–78. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability-an evolving hallmark of cancer. Nat Rev Mol
Cell Biol. 11:220–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Kang JW, Zhan Z, Ji G, Sang Y, Zhou D, Li
Y, Feng H and Cheng T: PUMA facilitates EMI1-promoted cytoplasmic
Rad51 ubiquitination and inhibits DNA repair in stem and progenitor
cells. Signal Transduct Target Ther. 6:1292021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
McGarry E, Gaboriau D, Rainey MD,
Restuccia U, Bachi A and Santocanale C: The deubiquitinase USP9X
maintains DNA replication fork stability and DNA damage checkpoint
responses by regulating CLASPIN during S-Phase. Cancer Res.
76:2384–2393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wolfsperger F, Hogh-Binder SA,
Schittenhelm J, Psaras T, Ritter V, Bornes L, Huber SM, Jendrossek
V and Rudner J: Deubiquitylating enzyme USP9× regulates
radiosensitivity in glioblastoma cells by Mcl-1-dependent and
-independent mechanisms. Cell Death Dis. 7:20392016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lautenbacher L, Samaras P, Muller J,
Grafberger A, Shraideh M, Rank J, Fuchs ST, Schmidt TK, The M,
Dallago C, et al: ProteomicsDB: Toward a FAIR open-source resource
for life-science research. Nucleic Acids Res. 50:D1541–D1552. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang C, Cai TY, Zhu H, Yang LQ, Jiang H,
Dong XW, Hu YZ, Lin NM, He QJ and Yang B: Synergistic antitumor
activity of gemcitabine and ABT-737 in vitro and in vivo through
disrupting the interaction of USP9X and Mcl-1. Mol Cancer Ther.
10:1264–1275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Oosterkamp HM, Hijmans EM, Brummelkamp TR,
Canisius S, Wessels LF, Zwart W and Bernards R: USP9X
downregulation renders breast cancer cells resistant to tamoxifen.
Cancer Res. 74:3810–3820. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang C, Qian H, Liu K, Zhao W and Wang L:
A feedback loop regulation of LINC01433 and YAP promotes malignant
behavior in gastric cancer cells. Onco Targets Ther. 12:7949–7962.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chen H, Yang F, Li X, Gong ZJ and Wang LW:
Long noncoding RNA LNC473 inhibits the ubiquitination of survivin
via association with USP9X and enhances cell proliferation and
invasion in hepatocellular carcinoma cells. Biochem Biophys Res
Commun. 499:702–710. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xu X, Wang S, Wang H, Pan C, Yang W and Yu
J: Hsa_circ_0008434 regulates USP9X expression by sponging
miR-6838-5p to promote gastric cancer growth, migration and
invasion. BMC Cancer. 21:12892021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shen G, Lin Y, Yang X, Zhang J, Xu Z and
Jia H: MicroRNA-26b inhibits epithelial-mesenchymal transition in
hepatocellular carcinoma by targeting USP9X. BMC Cancer.
14:3932014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chen W, Huang Y, Zhang S, Zheng X, Xie S,
Mao J, Cai Y, Lu X, Hu L, Shen J, et al: MicroRNA-212 suppresses
nonsmall lung cancer invasion and migration by regulating
ubiquitin-specific protease-9. J Cell Biochem. 120:6482–6489. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen W, Zhou Y, Zhi X, Ma T, Liu H, Chen
BW, Zheng X, Xie S, Zhao B, Feng X, et al: Delivery of miR-212 by
chimeric peptide-condensed supramolecular nanoparticles enhances
the sensitivity of pancreatic ductal adenocarcinoma to doxorubicin.
Biomaterials. 192:590–600. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Guo H, Zhang X, Chen Q, Bao Y, Dong C and
Wang X: miR-132 suppresses the migration and invasion of lung
cancer cells by blocking USP9X-induced epithelial-mesenchymal
transition. Am J Transl Res. 10:224–234. 2018.PubMed/NCBI
|
|
98
|
Chen E, Li E, Liu H, Zhou Y, Wen L, Wang
J, Wang Y, Ye L and Liang T: miR-26b enhances the sensitivity of
hepatocellular carcinoma to Doxorubicin via USP9X-dependent
degradation of p53 and regulation of autophagy. Int J Biol Sci.
17:781–795. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Madan V, Li J, Zhou S, Teoh WW, Han L,
Meggendorfer M, Malcovati L, Cazzola M, Ogawa S, Haferlach T, et
al: Distinct and convergent consequences of splice factor mutations
in myelodysplastic syndromes. Am J Hematol. 95:133–143. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Aqaqe N, Yassin M, Yassin AA, Ershaid N,
Katz-Even C, Zipin-Roitman A, Kugler E, Lechman ER, Gan OI,
Mitchell A, et al: An ERG enhancer-based reporter identifies
leukemia cells with elevated leukemogenic potential driven by
ERG-USP9X feed-forward regulation. Cancer Res. 79:3862–3876. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shen SM, Zhang C, Ge MK, Dong SS, Xia L,
He P, Zhang N, Ji Y, Yang S, Yu Y, et al: PTENalpha and PTENbeta
promote carcinogenesis through WDR5 and H3K4 trimethylation. Nat
Cell Biol. 21:1436–1448. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kapoor S, Natarajan K, Baldwin PR, Doshi
KA, Lapidus RG, Mathias TJ, Scarpa M, Trotta R, Davila E, Kraus M,
et al: Concurrent inhibition of pim and FLT3 kinases enhances
apoptosis of FLT3-ITD acute myeloid leukemia cells through
increased Mcl-1 proteasomal degradation. Clin Cancer Res.
24:234–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Sisoudiya SD, Mishra P, Li H, Schraw JM,
Scheurer ME, Salvi S, Doddapaneni H, Muzny D, Mitchell D, Taylor O,
et al: Identification of USP9X as a leukemia susceptibility gene.
Blood Adv. 7:4563–4575. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Schwartzman O, Savino AM, Gombert M, Palmi
C, Cario G, Schrappe M, Eckert C, von Stackelberg A, Huang JY,
Hameiri-Grossman M, et al: Suppressors and activators of JAK-STAT
signaling at diagnosis and relapse of acute lymphoblastic leukemia
in Down syndrome. Proc Natl Acad Sci USA. 114:4030–4039. 2017.
View Article : Google Scholar
|
|
105
|
Moroishi T, Hansen CG and Guan KL: The
emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 15:73–79.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Demark-Wahnefried W, Platz EA, Ligibel JA,
Blair CK, Courneya KS, Meyerhardt JA, Ganz PA, Rock CL, Schmitz KH,
Wadden T, et al: The role of obesity in cancer survival and
recurrence. Cancer Epidemiol Biomarkers Prev. 21:1244–1259. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Jaiswal A, Murakami K, Elia A, Shibahara
Y, Done SJ, Wood SA, Donato NJ, Ohashi PS and Reedijk M:
Therapeutic inhibition of USP9×-mediated Notch signaling in
triple-negative breast cancer. Proc Natl Acad Sci USA.
118:e21015921182021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Yan J, Zhong N, Liu G, Chen K, Liu X, Su L
and Singhal S: Usp9×- and Noxa-mediated Mcl-1 downregulation
contributes to pemetrexed-induced apoptosis in human non-small-cell
lung cancer cells. Cell Death Dis. 5:e13162014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Perez-Mancera PA, Rust AG, van der Weyden
L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grutzmann R,
Aust D, Rummele P, et al: The deubiquitinase USP9X suppresses
pancreatic ductal adenocarcinoma. Nature. 486:266–270. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Pal A, Dziubinski M, Di Magliano MP,
Simeone DM, Owens S, Thomas D, Peterson L, Potu H, Talpaz M and
Donato NJ: Usp9× promotes survival in human pancreatic cancer and
its inhibition suppresses pancreatic ductal adenocarcinoma in vivo
tumor growth. Neoplasia. 20:152–164. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Guo M, Luo G, Jin K, Long J, Cheng H, Lu
Y, Wang Z, Yang C, Xu J, Ni Q, et al: Somatic genetic variation in
solid pseudopapillary tumor of the pancreas by whole exome
sequencing. Int J Mol Sci. 18:812017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Perurena N, Lock R, Davis RA, Raghavan S,
Pilla NF, Ng R, Loi P, Guild CJ, Miller AL, Sicinska E, et al:
USP9X mediates an acute adaptive response to MAPK suppression in
pancreatic cancer but creates multiple actionable therapeutic
vulnerabilities. Cell Rep Med. 4:1010072023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chen Z, Wang HW, Wang S, Fan L, Feng S,
Cai X, Peng C, Wu X, Lu J, Chen D, et al: USP9X deubiquitinates
ALDH1A3 and maintains mesenchymal identity in glioblastoma stem
cells. J Clin Invest. 129:2043–2055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nguyen HT, Andrejeva D, Gupta R, Choudhary
C, Hong X, Eichhorn PJ, Loya AC and Cohen SM: Deubiquitylating
enzyme USP9× regulates hippo pathway activity by controlling
angiomotin protein turnover. Cell Discov. 2:160012016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Bartholomeusz G, Talpaz M, Bornmann W,
Kong LY and Donato NJ: Degrasyn activates proteasomal-dependent
degradation of c-Myc. Cancer Res. 67:3912–3918. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Luo H, Jing B, Xia Y, Zhang Y, Hu M, Cai
H, Tong Y, Zhou L, Yang L, Yang J, et al: WP1130 reveals USP24 as a
novel target in T-cell acute lymphoblastic leukemia. Cancer Cell
Int. 19:562019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Peterson LF, Sun H, Liu Y, Potu H,
Kandarpa M, Ermann M, Courtney SM, Young M, Showalter HD, Sun D, et
al: Targeting deubiquitinase activity with a novel small-molecule
inhibitor as therapy for B-cell malignancies. Blood. 125:3588–3597.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Cui J, Sun W, Hao X, Wei M, Su X, Zhang Y,
Su L and Liu X: EHMT2 inhibitor BIX-01294 induces apoptosis through
PMAIP1-USP9X-MCL1 axis in human bladder cancer cells. Cancer Cell
Int. 15:42015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Boise LH: DUB-ling down on B-cell
malignancies. Blood. 125:3522–3523. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chou DH, Vetere A, Choudhary A, Scully SS,
Schenone M, Tang A, Gomez R, Burns SM, Lundh M, Vital T, et al:
Kinase-independent small-molecule inhibition of JAK-STAT signaling.
J Am Chem Soc. 137:7929–7934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lawson AP, Long MJC, Coffey RT, Qian Y,
Weerapana E, El Oualid F and Hedstrom L: Naturally occurring
isothiocyanates exert anticancer effects by inhibiting
deubiquitinating enzymes. Cancer Res. 75:5130–5142. 2015.
View Article : Google Scholar : PubMed/NCBI
|