Introduction
As the malignant tumor with fast growth in incidence
(28.3 per 100,000) and mortality rate (23.0 per 100,000), lung
cancer poses a threat to human health (1,2).
According to the histological origin, lung cancer is usually
divided into small cell lung cancer (SCLC) and non-SCLC (NSCLC)
(3). NSCLC is the most common
pathological type of lung cancer, accounting for ~85%, including
lung adenocarcinoma (40%), lung squamous cell carcinoma (25%),
large cell carcinoma and other subtypes (4,5). Lung
cancer prognosis is improved if it is detected and treated at an
early stage. However, patients often miss the optimal treatment
period due to late diagnosis and the advanced stage of the disease.
Although patients receive a series of treatments, including
radiotherapy and chemotherapy, targeted therapy, and immune
checkpoint therapy, the prognosis is poor (6). Therefore, effective
biomarkers/signatures for the early diagnosis of lung cancer are
important for patients with lung cancer.
Endocytosis is an important physiological function
in cell activity that engulfs extracellular substances into cells
in a membrane-dependent manner, maintaining cell-cell interactions
via molecule exchange (7).
Endocytosis mainly includes the ingestion of two types of
molecules, namely large particles and small vesicles. In cancer
cells, receptor-mediated and vesicle-dependent endocytosis not only
provides sufficient energy and substances for promoting the
malignant progression of tumors and activates important signaling
pathways for tumorigenesis, but also affects the activation state
of the intracellular signaling pathway for cell-cell communication
and acquired chemoresistance (8,9).
Emerging evidence indicated that endocytosis contributed to
signaling via its ‘canonical’ and ‘noncanonical’ mechanisms
(10). For example, inhibition of
endocytosis blocked H-Ras-mediated cell differentiation and Raf-1
activation (11). In addition,
tumor cells can drive a microenvironment suitable for tumorigenesis
and metastasis via interaction with surrounding cells including
stromal, immune and vascular cells. For example, clathrin light
chain b, the essential isoform of the clathrin receptor in
clathrin-mediated endocytosis, enhances the epidermal growth factor
receptor (EGFR)/AKT/glycogen synthase kinase 3β (GSK-3β) signal
transduction, contributing to tumor progression and metastasis in
lung cancer (12). Therapeutic
strategies targeting endocytosis have been demonstrated to have the
potential to inhibit tumor growth and sensitize lung cancer to
conventional chemotherapeutic drugs (12–14).
For example, the clathrin-mediated endocytosis inhibitor PAO
combined with gefitinib resulted in tumor regression and increased
apoptosis in NSCLC (12). The
aforementioned in vitro studies, carried out using lung
cancer cell lines such as H358, Calu-3, SNU-1327, H1299, HCC4017,
H441 and H522, and in vivo experiments involving xenografts
established in nude mice, revealed that inhibition of
clathrin-mediated endocytosis decreased the malignant progression
and enhanced the anti-tumor effect of tyrosine kinase inhibitors
(TKIs) against lung cancer (15–18).
However, studies focusing on lung cancer prognostic prediction
models based on endocytosis-associated genes have not been
reported.
In the present study, the differentially expressed
genes (DEGs) in tumor samples compared with paracancerous tissues
from the lung adenocarcinoma (LUAD) cohort in The Cancer Genome
Atlas (TCGA) database were screened, and these DEGs were integrated
with endocytosis-associated genes. To investigate the effect of
these endocytosis-associated genes on the prognosis prediction of
patients with LUAD, a novel endocytosis-associated gene signature
was constructed, and its prognostic value was examined using TCGA
and Gene Expression Omnibus (GEO) databases. Based on the
experimental verification of the expression patterns of these genes
in vitro, the endocytosis inhibitor chloroquine (CQ) was
used to investigate changes in the expression of the aforementioned
genes and their anti-tumor effect.
Materials and methods
Acquisition of LUAD transcriptome data
in TCGA and GEO databases
The RNA sequencing data of LUAD in TCGA database
were downloaded from the UCSC Xena online server (https://xenabrowser.net/). The gene expression
matrices of GSE30219 and GSE31210 were obtained from the GEO
database (https://www.ncbi.nlm.nih.gov/geo/). Data were
transformed as follows: i=2i−1 for subsequent
analysis.
Collection of endocytosis-associated
genes
Endocytosis-associated genes were collected from the
Gene Ontology (GO) (https://geneontology.org/), Kyoto Encyclopedia of
Genes and Genomes (https://www.kegg.jp/) database and the Reactome
Knowledgebase (www.reactome.org). The search term ‘endocytosis’ was
used. In total, 24 gene sets were obtained, and were intersected
for endocytosis-associated genes.
DEG analysis in TCGA-LUAD cohort
Based on the clinical information of the LUAD cohort
from TCGA, the RNA sequencing data of 60 tumor and paired
paracancerous tissues were used for differential expression
analysis via the DESeq2 R package (version 1.30.0) (19). DEGs were identified using the
criteria of absolute value of Log fold-change (FC) >1.2,
adjusted P≤0.05 and visualized as volcano plots. The intersection
between DEGs and endocytosis-associated genes was used as a
candidate for univariant Cox regression analysis.
Cox regression and Kaplan-Meier
survival analyses
Cox regression and Kaplan-Meier survival analyses
were performed against candidate genes using the Survival R package
(version 3.5–5; http://github.com/therneau/survival) to measure the
association between expression level and overall survival in
TCGA-LUAD, GSE30219 and GSE31210 cohorts. Genes with significant
P-values were selected for Lasso Cox regression analysis using the
glmnet R package (version 4.1–3; http://glmnet.stanford.edu/), and the refined
prognostic model was constructed. The risk scores of each patient
were calculated using the following formula:
RiskScore=Σexpi × βi, where expi
was the expression of gene i and βi referred to the
coefficient calculated by univariate Cox regression analysis of
gene i.
GO analysis
Metascape (https://metascape.org) is an online tool for gene
function annotation analysis, and it was used to analyze the GO
enrichment of the 18 genes in the novel signature. Results were
visualized as network plots.
Detection of tumor purity and immune
cell composition
The tumor purities of TCGA-LUAD, GSE30219 and
GSE31210 cohorts were calculated using the ESTIMATE R package
(version 1.0.13; http://r-forge.r-project.org/projects/estimate/)
(20). Briefly, gene symbols and
their corresponding expression levels were used as input data.
After transformation and calculation by the ‘filterCommonGenes’ and
‘estimateScore’ functions, purity data were obtained. The Pearson's
correlation coefficient was used for investigating the correlation
between the riskScore and tumor purity was visualized as scatter
plots.
Immune cell compositions were acquired via the
CIBERSORTx (https://cibersortx.stanford.edu/) online analytical
tool. By performing a series of steps to input the signature matrix
file and expression file with default settings, the results were
downloaded after the analysis was finished. The risk score
distributions of each patient in the high and low groups were
visualized as boxplots.
Cell culture
The human lung epithelial cell line BEAS-2B, and the
NSCLC cell lines A549, H1299 and H1975 were obtained from the
American Type Culture Collection. The NSCLC cell line PC9 was
purchased from the National Collection of Authenticated Cell
Cultures. The BEAS-2B and A549 cell lines were cultured in
Dulbecco's Modified Eagle Medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo
Fisher Scientific, Inc.). The H1299, H1975 and PC9 cell lines were
cultured in Roswell Park Memorial Institute-1640 (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS. All cells were
cultured at 37°C and 5% CO2.
Cell viability assay
Cells were trypsinized to a single-cell suspension
and counted. A total of 3,000 cells per well were seeded into
96-well culture plates. CQ (cat. no. S6999; Selleck Chemicals) was
dissolved in DMSO to a storage concentration of 10 mM and diluted
in culture media to a working concentration of 20 µM. After either
24, 48, 72, 96 or 120 h of CQ treatment, Cell Counting Kit-8 (cat.
no. CK04; Dojindo Laboratories, Inc.) reagent was added, followed
by a 2 h-incubation. The OD450 absorbance was measured using a
microplate reader.
5-ethynyl-2′-deoxyuridine (EdU)
assay
Cultured cells were seeded into glass bottom culture
dishes. After 48 h of treatment with 20 µM CQ, EdU (cat. no.
C0078S; Beyotime Institute of Biotechnology.) incorporation and
staining were performed according to the manufacturer's protocol.
Briefly, after pretreatment with 10 µM EdU for 2 h, cells were
fixed with 4% paraformaldehyde solution for 10 min at room
temperature and permeated using PBS with 0.2% Triton X-100 for 10
min at room temperature. Subsequently, Azid Alexa Fluor 594 was
used to label EdU, and Hoechst 33342 was employed to stain nuclei
for 10 min at room temperature protected from light. Samples were
imaged using a laser confocal microscope (Olympus Corporation). The
proportion of EdU-positive cells was calculated by counting the
EdU-positive cells and the total number of cells using ImageJ
(version 1.53; National Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
After 48 h of 20 µM CQ treatment, total RNA was
isolated from cells using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.) reagent according to the manufacturer's
protocol, and reverse-transcribed to cDNA by using the RevertAid
First Strand cDNA Synthesis Kit (cat. no. K1622; Thermo Fisher
Scientific, Inc.) at 42°C for 60 min, 70°C for 5 min, and then
stored at 4°C. The SYBR Green Mix (cat. no. Q711; Vazyme Biotech
Co., Ltd.) was used in a thermocycler instrument (QuantStudio 3;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions as follows: Predenaturation at 95°C for 5 min,
followed by 40 cycles of denaturation at 95°C for 15 sec, and
annealing and extension at 60°C for 30 sec. GAPDH served as the
internal control. Primer sequences are summarized in Table SI. The mRNA levels were quantified
using the 2−ΔΔCq method (21).
Apoptosis assay
Cell apoptosis was stained using Annexin V-FITC
apoptosis detection kit (cat. no. C1062M; Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Briefly,
after 48 h of 20 µM CQ treatment, total cells in the supernatant
medium and on the plate, were harvested and centrifuged at 500 × g
for 10 min at room temperature. Cell pellets were resuspended in
PBS buffer and counted. A total of 1×105 cells were
centrifuged at 500 × g for 10 min at room temperature, and
resuspended in binding buffer (cat. no. C1062M; Beyotime Institute
of Biotechnology) containing Annexin V-FITC and propidium iodide
and incubated for 15 min at room temperature in the dark.
Subsequently, cell apoptosis was detected using an Agilent NovoCyte
flow cytometer (Agilent Technologies, Inc.). The data were analyzed
using FlowJo (version 10.6.2; FlowJo LLC).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism (version 8, Dotmatics) and R (version 4.0.2;
http://www.r-project.org/about.html).
A paired Student's t-test was used for the analysis of two paired
groups, and an unpaired independent-sample Student's t-test was
used for the comparison of two experimental groups. One-way
analysis of variance was performed to compare ≥3 groups, and
Dunnett's post hoc test was used for multiple comparisons. The
Pearson's correlation coefficient was used to investigate the
correlation between risk score and either stromal or immune score,
or tumor purity. Statistical results are presented as mean ±
standard deviation. Univariate and multivariate Cox regression
analysis were used to analyze the effect of risk factors on
survival. Kaplan-Meier plots were used to assess the correlation
between risk score and survival. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of differentially
expressed endocytosis-associated genes correlated with the survival
rate of patients with LUAD
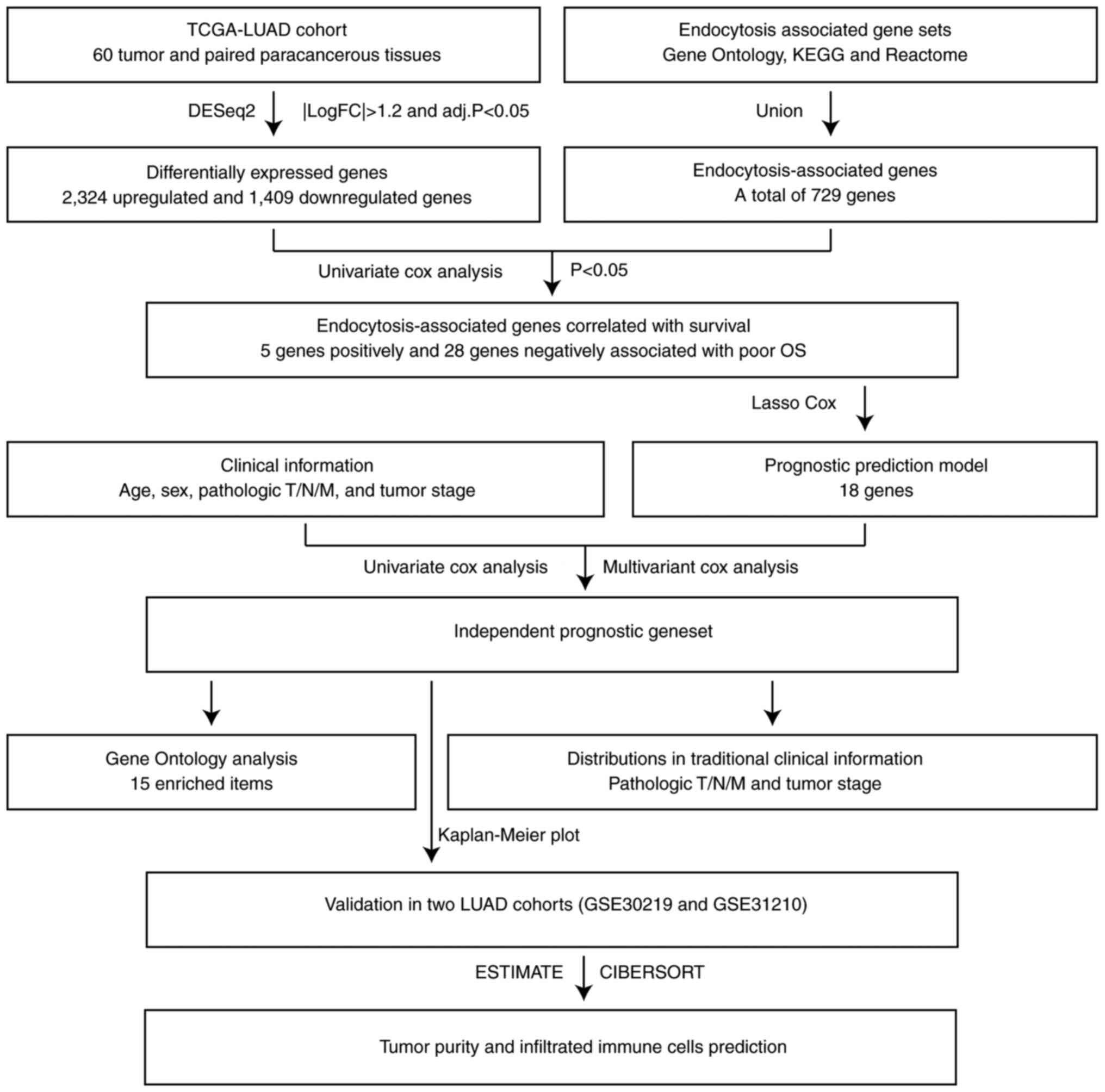
The analysis flow chart of the present study is
presented in Fig. 1. First, LUAD
data were downloaded from TCGA database via the UCSC XENA online
server, and the transcriptome sequencing data of paired cancerous
and adjacent normal samples were selected for the identification of
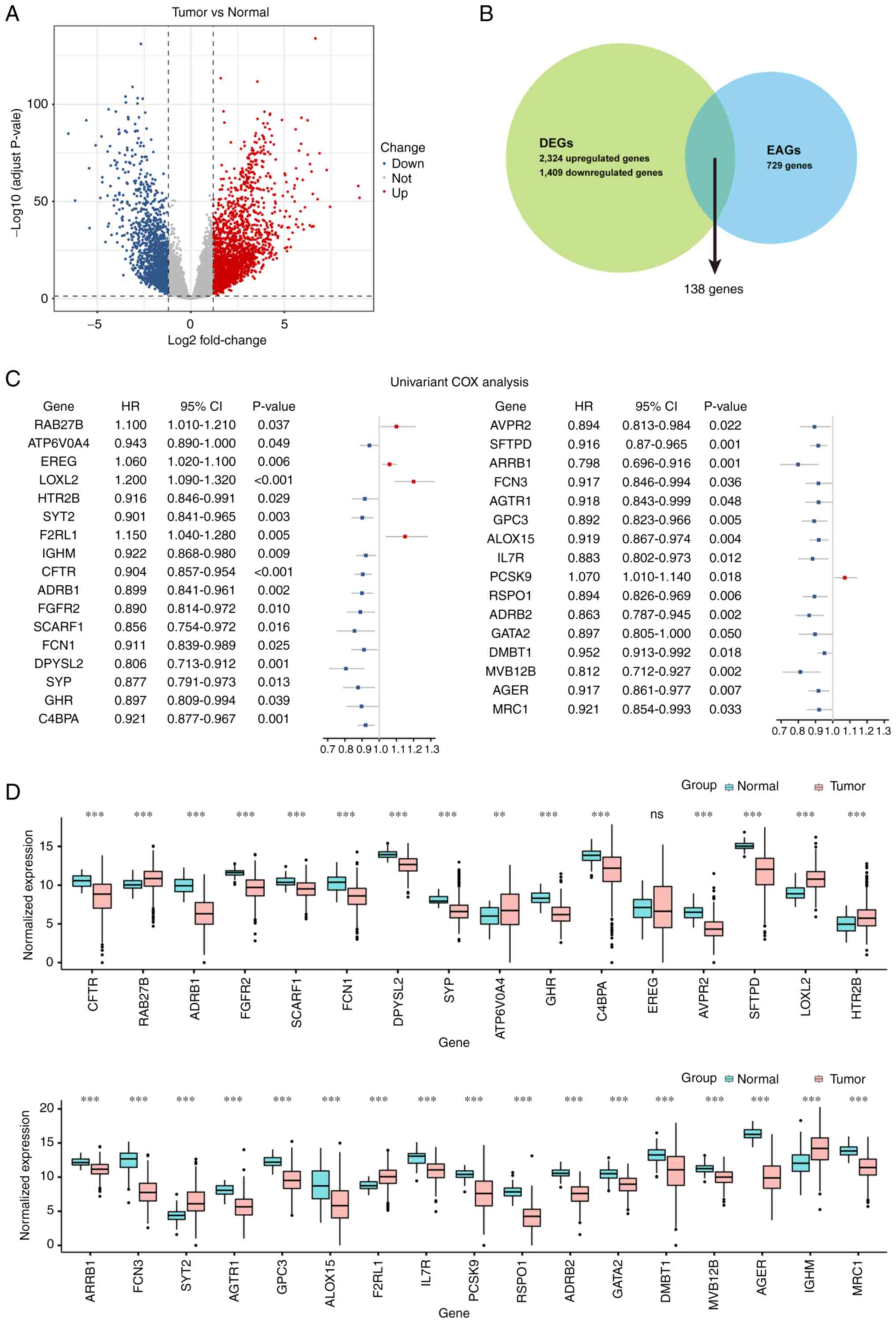
DEGs using the DESeq2 R package. A total of 2,324 upregulated and
1,409 downregulated DEGs were identified using the filtering
criteria of absolute value of LogFC >1.2 and P-adjust ≤0.05
(Table SII), and were visualized
as a volcano plot (Fig. 2A). To
select the DEGs correlated with the function of endocytosis, the
list of DEGs and the endocytosis-associated gene set were
intersected (Table SIII), and 138
candidate genes were obtained (Fig.
2B). Subsequently, a univariate Cox regression analysis was
performed to evaluate the association between the expression levels
of these genes and the overall survival of patients. A total of 33
genes were indicated to be significantly correlated with survival
times, of which the expression levels of five genes were associated
with poor prognosis, and the others were associated with a
favorable prognosis (Fig. 2C and
Table SIV). The expression levels
of these genes in tumor tissues (n=525) and normal tissues (n=60)
in the LUAD cohort from TCGA containing a total of 585 samples were
compared and visualized as boxplots (Fig. 2D). Compared with the normal tissues,
the expression levels of RAB27B, ATP6V0A4, LOXL2, HTR2B, SYT2,
F2RL1 and IGHM were significantly increased in tumor tissues,
whereas the expression levels of the other genes, excluding EREG,
were significantly decreased in the tumor tissues.
Construction of a novel gene signature
to predict prognosis in TCGA-LUAD cohort
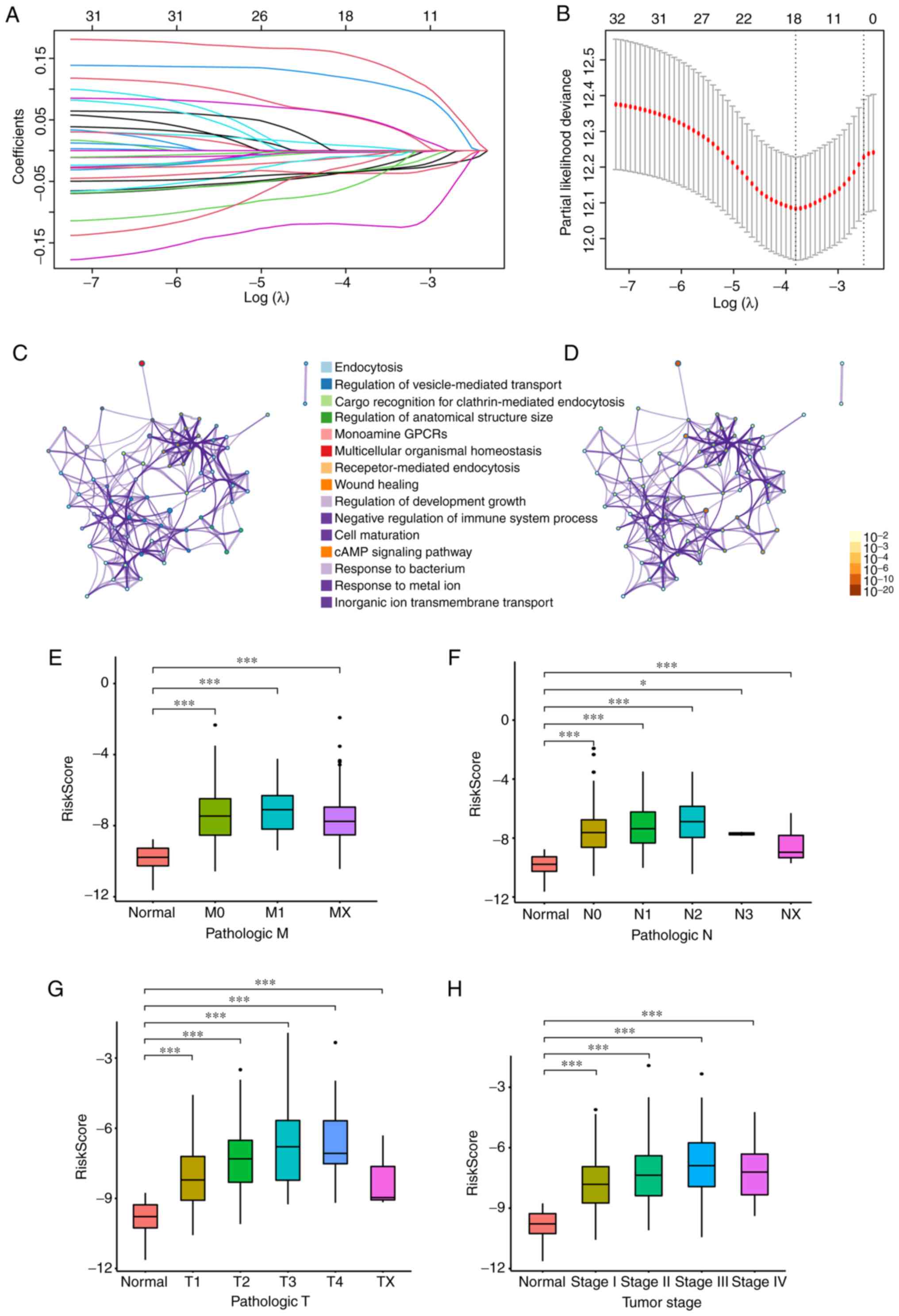
Based on the aforementioned identified candidate
genes, Lasso Cox regression analysis was used to screen the optimal
genes to construct the refined prognostic model. The coefficient
results calculated by Lasso Cox regression analysis are presented
in Fig. 3A. According to the
calculation of risk factors using Cox regression, and Kaplan-Meier
survival analyses, a novel gene signature of 18 genes, including
CFTR, RAB27B, ADRB1, DPYSL2, ATP6V0A4, EREG, SFTPD, LOXL2, HTR2B,
SYT2, ALOX15, F2RL1, IL7R, PCSK9, ADRB2, GATA2, IGHM and MRC1, was
involved (Fig. 3B and Table SV). The GO analysis results
revealed the enriched signaling pathways and molecular events,
among which the functions associated with ‘endocytosis’ and
‘regulation of vesicle-mediated transport’ were the most
significantly enriched (Fig. 3C and
D).
The risk score of every patient in the LUAD cohort
from TCGA was calculated based on the expression of 18 genes and
the distributions in the clinical information were examined. As
indicated in Fig. 3E-H, the risk
score of the novel signature in different pathologic tumor
(T)/nodal (N)/metastasis (M) grades and tumor stages in LUAD was
significantly increased compared with that in normal tissues. As
the grades or stages increased, the risk score also had an upward
trend. Furthermore, univariate and multivariate Cox regression
analyses were performed to evaluate the prognostic value of the
novel gene signature. As presented in Table I, the pathologic M [M1 vs. M0;
hazard ratio (HR), 2.112; 95% confidence interval (CI),
1.235–3.612], pathologic N (N1 vs. N0; HR, 2.392; 95% CI,
1.700–3.365; N2 vs. N0; HR, 3.046; 95% CI, 2.084–4.452), pathologic
T (T2 vs. T1; HR, 1.491; 95% CI, 1.046–2.125; T3 vs. T1; HR, 2.971;
95% CI, 1.765–4.999; T4 vs. T1; HR, 3.004; 95% CI, 1.547–5.834; TX
vs. T1; HR, 4.794; 95% CI, 1.153–19.939) and tumor stage (stage II
vs. stage I; HR, 2.451; 95% CI, 1.710–3.513; stage III vs. stage I;
HR, 3.492; 95% CI, 2.390–5.102; stage IV vs. stage I; HR, 3.813;
95% CI, 2.203–6.599) indicated statistical significances in the
univariant Cox regression analysis, whereas these were not
statistically significant in the multivariant Cox regression
analysis. Both univariant and multivariant Cox regression analyses
of the risk score of the novel gene signature exhibited poor
survival for patient prognosis (univariant; HR, 1.476; 95% CI,
1.336–1.630; multivariant; HR, 1.418, 95% CI, 1.272–1.580),
demonstrating that this novel gene signature could be an
independent prognostic marker.
 | Table I.Cox regression analysis in The Cancer
Genome Atlas-lung adenocarcinoma cohort. |
Table I.
Cox regression analysis in The Cancer
Genome Atlas-lung adenocarcinoma cohort.
|
|
| Univariant | Multivariant |
|---|
|
|
|
|
|
|---|
|
Characteristics | Total, n | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years | 526 | 1.007 | 0.992–1.022 |
3.521×10−1a | 1.014 | 0.998–1.030 |
8.802×10−2a |
| Sex |
|
|
|
|
|
|
|
|
Male | 244 | Reference |
|
| Reference |
|
|
|
Female | 282 | 0.958 | 0.717–1.279 |
7.694×10−1a | 1.101 | 0.808–1.500 |
5.412×10−1a |
| Pathologic M |
|
|
|
|
|
|
|
| M0 | 354 | Reference |
|
| Reference |
|
|
| M1 | 25 | 2.112 | 1.235–3.612 |
6.305×10−3b | 0.233 | 0.014–3.910 |
3.111×10−1a |
| MX | 142 | 0.854 | 0.596–1.222 |
3.879×10−1a | 0.923 | 0.637–1.340 |
6.726×10−1a |
| Pathologic N |
|
|
|
|
|
|
|
| N0 | 341 | Reference |
|
| Reference |
|
|
| N1 | 95 | 2.392 | 1.700–3.365 |
5.556×10−7c | 1.583 | 0.878–2.850 |
1.263×10−1a |
| N2 | 74 | 3.046 | 2.084–4.452 |
8.752×10−9c | 1.212 | 0.510–2.880 |
6.635×10−1a |
| N3 | 2 | 0.000 | 0.000-Infinite |
9.943×10−1a | 0.000 | 0.000-Infinite |
9.920×10−1a |
| NX | 13 | 1.417 | 0.519–3.868 |
4.959×10−1a | 1.392 | 0.337–5.750 |
6.474×10−1a |
| Pathologic T |
|
|
|
|
|
|
|
| T1 | 172 | Reference |
|
| Reference |
|
|
| T2 | 284 | 1.491 | 1.046–2.125 |
2.705×10−2b | 1.001 | 0.683–1.470 |
9.973×10−1a |
| T3 | 48 | 2.971 | 1.765–4.999 |
4.124×10−5c | 1.397 | 0.723–2.700 |
3.198×10−1a |
| T4 | 19 | 3.004 | 1.547–5.834 |
1.163×10−3b | 0.844 | 0.378–1.880 |
6.789×10−1a |
| TX | 3 | 4.794 | 1.153–19.939 |
3.113×10−2b | 0.936 | 0.076–11.560 |
9.587×10−1a |
| Tumor stage |
|
|
|
|
|
|
|
| Stage
I | 286 | Reference |
|
| Reference |
|
|
| Stage
II | 122 | 2.451 | 1.710–3.513 |
1.050×10−6c | 1.372 | 0.734–2.560 |
3.217×10−1a |
| Stage
III | 84 | 3.492 | 2.390–5.102 |
1.030×10−0c | 2.291 | 0.884–5.940 |
8.810×10−2a |
| Stage
IV | 26 | 3.813 | 2.203–6.599 |
1.738×10−6c | 11.619 | 0.632–213.470 |
9.865×10−2a |
| RiskScore | 526 | 1.476 | 1.336–1.630 |
2.072×10−14c | 1.418 | 1.272–1.580 |
3.394×10−10c |
Estimation of the novel gene signature
for independent prognostic prediction in training and validation
sets
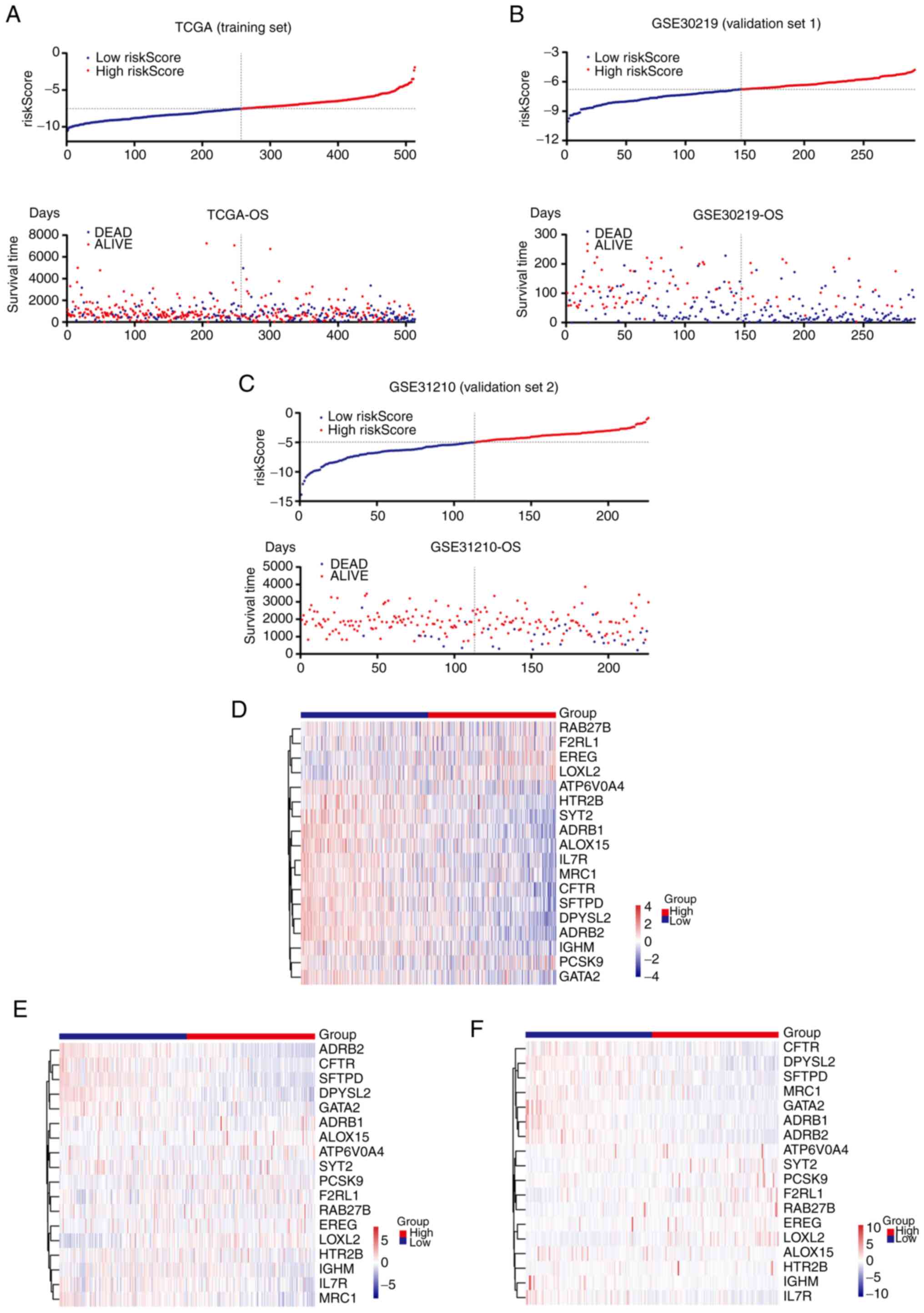
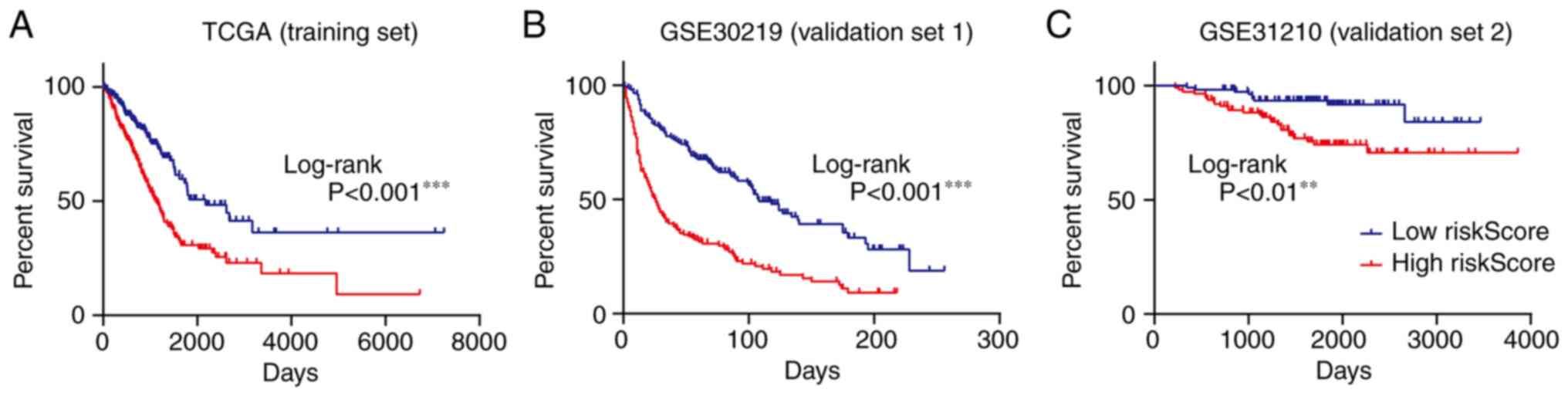
To further validate the efficacy of the risk score
in the prognostic prediction of patients with LUAD, TCGA-LUAD
cohort (training set) was divided into two groups according to the
median value of the risk score. The patients in the high-risk score
group had a shorter survival time and an increased ratio of ‘DEAD’
status compared with the patients in the low-risk score group
(Fig. 4A). The same results were
illustrated in the two independent validation sets, GSE30219 and
GSE31210, obtained from the GEO database (Fig. 4B and C). Heatmaps of the 18 genes in
the training set and validation sets are presented in Fig. 4D-F.
Furthermore, Kaplan-Meier plots were used to examine
the ability of the novel signature to predict prognosis. As
presented in Fig. 5A-C, an
increased probability of a longer survival time was positively
correlated with a low-risk score of the novel gene signature in the
training set and the two validation sets.
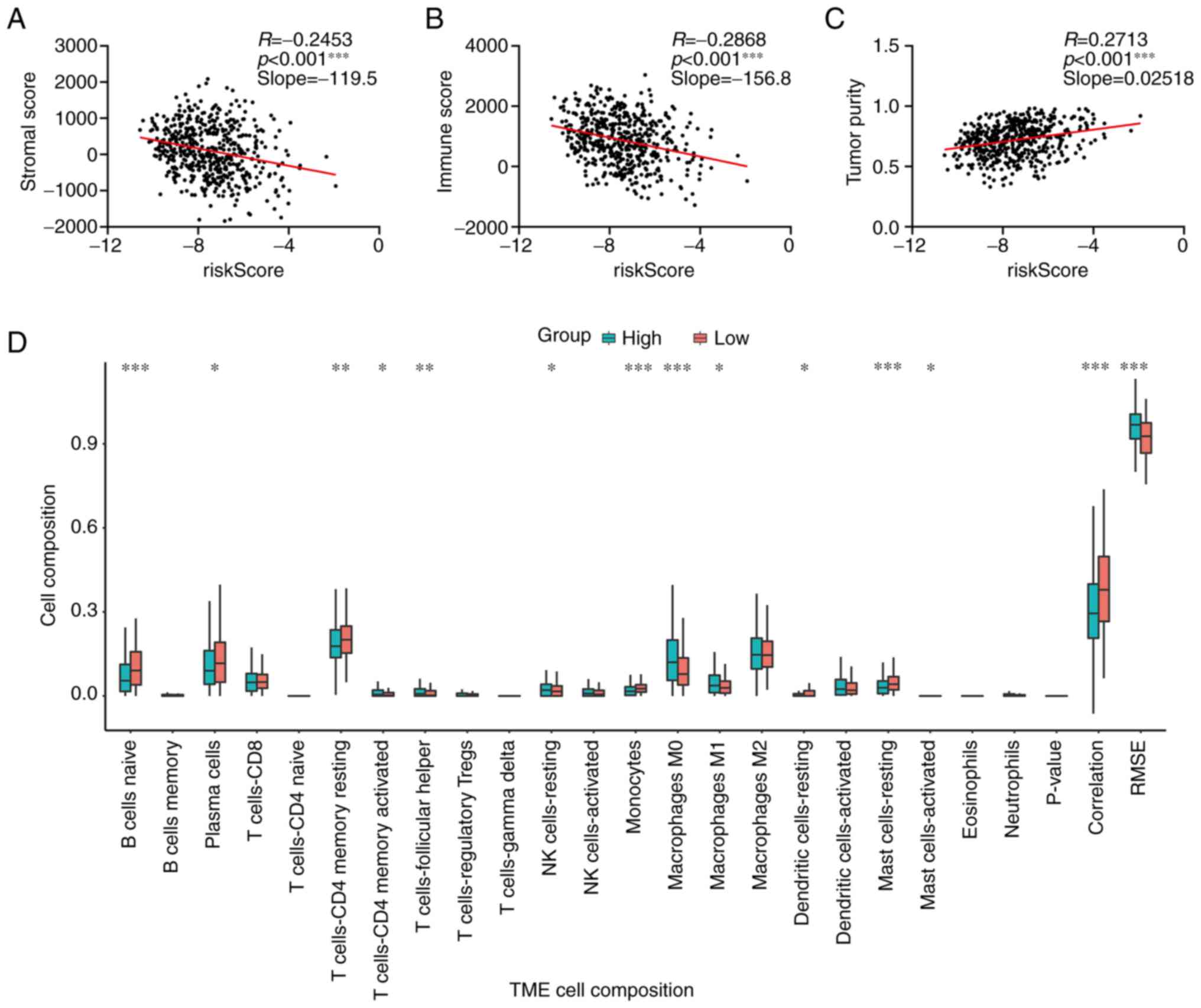
Correlation between the novel gene
signature and tumor-infiltrating immune cells
Tumor cells interact with tumor-infiltrating immune
cells such as macrophages to induce the immunoinhibitory phenotype
(22,23). Considering that these genes were
endocytosis-associated genes, the present study attempted to
investigate the association between the novel signature and
tumor-infiltrating immune cells of LUAD. The ESTIMATE algorithm was
used to calculate tumor purity scores, and these were compared with
the risk scores of the novel signature in TCGA-LUAD cohort. As
presented in Fig. 6A-C, the risk
score was negatively correlated with the stromal and immune scores,
but positively correlated with tumor purity, indicating that
patients with a low-risk score had a more complicated immune
microenvironment. Immune cell compositions in different groups with
high or low risk scores were further examined using the CIBERSORTx
algorithm. B cells, plasma cells, CD4+ T cells,
monocytes, dendritic and mast cells had an increased composition in
the low-risk score group compared with the high-risk score group;
however, the NK cells and M0 and M1 macrophages were increased in
the high-risk score group compared with the low-risk score group
(Fig. 6D). In addition, the
correlation between the risk score and marker genes of different
immune cells was calculated. As presented in Table II, the majority of genes were
negatively associated with these genes. These findings indicated
that the novel gene signature could hint at the complexity of the
tumor microenvironment.
 | Table II.Correlation analysis between
riskScore and marker genes of immune cells in The Cancer Genome
Atlas-lung adenocarcinoma cohort. |
Table II.
Correlation analysis between
riskScore and marker genes of immune cells in The Cancer Genome
Atlas-lung adenocarcinoma cohort.
| Immune cell | Marker gene | Correlation | P-value |
|---|
| T cell | CD3G | −0.372 |
1.19×10−16a |
|
| CD3D | −0.296 |
2.80×10−10a |
|
| CD3E | −0.362 |
1.05×10−15a |
|
| CD2 | −0.362 |
1.17×10−15a |
| Monocyte | CD86 | −0.342 |
6.57×10−14a |
|
| CSF1R | −0.393 |
1.08×10−18a |
| Tumor-associated
macrophage | CCL2 | −0.211 |
4.02×10−5a |
|
| CD68 | −0.233 |
3.13×10−6a |
|
| IL10 | −0.338 |
1.44×10−13a |
| M1 macrophage | INOS | −0.200 |
1.34×10−4a |
|
| IRF5 | −0.255 |
1.53×10−7a |
|
| COX2 | −0.109 |
2.80×10−1b |
| M2 macrophage | CD163 | −0.314 |
1.41×10−11a |
|
| VSIG4 | −0.308 |
3.88×10−11a |
|
| MS4A4A | −0.355 |
4.68×10−15a |
|
| MRC1 | −0.439 |
6.14×10−24a |
| Dendritic cell | HLA-DPB1 | −0.492 |
4.07×10−31a |
|
| HLA-DQB1 | −0.414 |
5.30×10−21a |
|
| HLA-DRA | −0.442 |
2.83×10−24a |
|
| CD1C | −0.531 |
3.13×10−37a |
|
| NRP1 | −0.238 |
1.58×10−6a |
|
| ITGAX | −0.424 |
3.25×10−22a |
| Regulatory T
cell | FOXP3 | −0.295 |
3.52×10−10a |
|
| CCR8 | −0.315 |
1.16×10−11a |
|
| STAT5B | −0.406 |
3.50×10−20a |
|
| TGFB1 | −0.247 |
4.66×10−7a |
| T cell
exhaustion | PD-1 | −0.196 |
2.00×10−4a |
|
| CTLA4 | −0.315 |
1.24×10−11a |
|
| LAG3 | −0.151 |
1.40×10−2c |
|
| TIM-3 | −0.303 |
9.39×10−11a |
|
| GZMB | 0.027 |
5.41×10−1b |
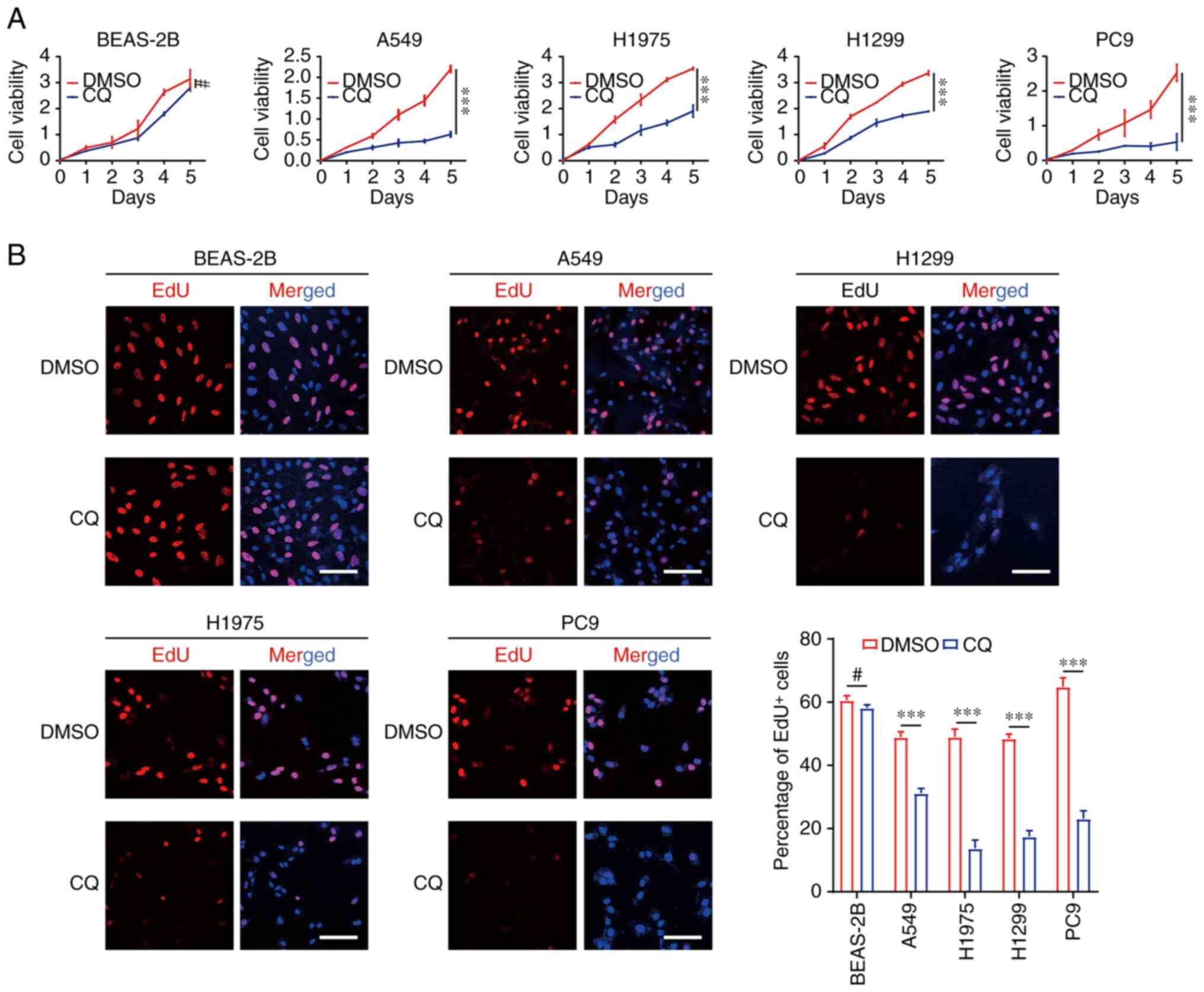
Inhibition of endocytosis by CQ
reduced proliferation and increased apoptosis in lung cancer
To further investigate the expression levels of
endocytosis-associated genes in the novel signature, total RNA was
isolated from the lung epithelial cell line BEAS-2B and the lung
cancer cell lines A549, H1975, H1299 and PC9, and then reverse
transcribed for qPCR assays. The result was visualized as a heatmap
in Fig. 7A. Compared with the
levels in BEAS-2B cells, the expression levels of RAB27B, ATP6V0A4,
LOXL2, HTR2B, SYT2, F2RL1 and IGHM were increased in lung cancer
cells, and the other genes had decreased expression levels in tumor
cells, which was consistent with the expression patterns in
TCGA-LUAD dataset (Fig. 2D).
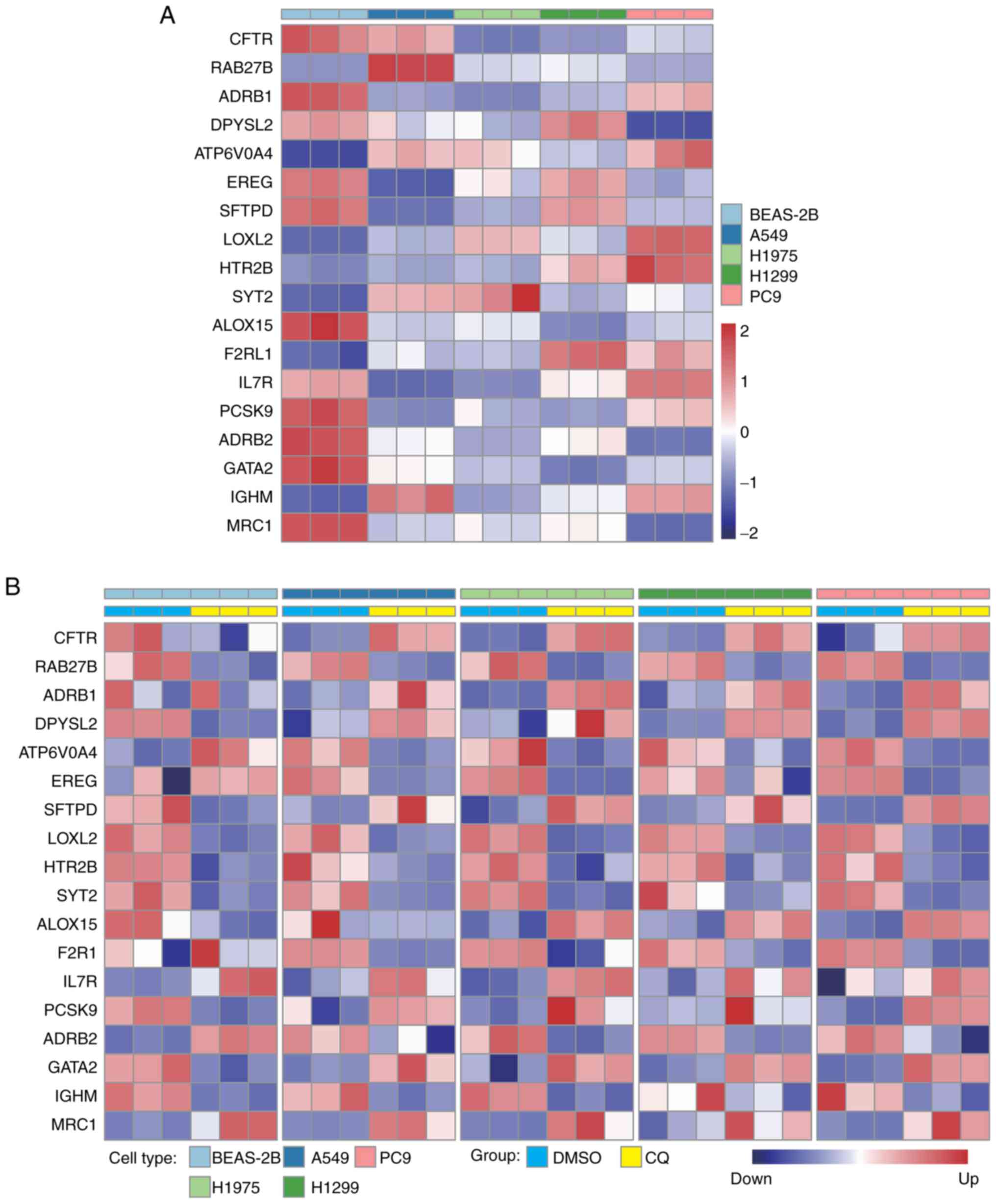 | Figure 7.CQ treatment altered the expression
profiles of signature genes. (A) Relative expression levels of
CFTR, RAB27B, ADRB1, DPYSL2, ATP6V0A4, EREG, SFTPD, LOXL2, HTR2B,
SYT2, ALOX15, F2RL1, IL7R, PCSK9, ADRB2, GATA2, IGHM and MRC1 in
BEAS-2B, A549, H1975, H1299 and PC9 cells. (B) Relative expression
levels of the aforementioned genes were detected after CQ
treatment. CQ, chloroquine. |
CQ is a conventional drug for treating amebiasis. CQ
has been reported to induce a blockade of endocytosis via pH
changes in lysosomes in cells (24). Given the roles of endocytosis in
LUAD progression and the value of the endocytosis-associated
signature in the prognostic prediction of patients with LUAD, CQ
was used to treat lung cancer cell lines as well as normal lung
epithelial cell lines. CQ treatment decreased the expression levels
of the genes that were positively correlated with poor prognosis,
and increased the expression levels of genes that were negatively
associated with short survival time in lung cancer cells such as
A549, H1299, H1975 and PC9 (Fig.
7B). However, the CQ-induced expression level changes in these
genes were not consistently observed in BEAS-2B (Fig. 7B), indicating that CQ had a
preferential effect on tumor cells. In addition, CQ treatment
decreased the viability of A549, H1975, H1299 and PC9 cells, but no
such effect was observed in BEAS-2B cells (Fig. 8A). EdU staining indicated reduced
proliferation of lung cancer cell lines in the CQ treatment group
compared with that in the DMSO group (Fig. 8B). Additionally, the level of
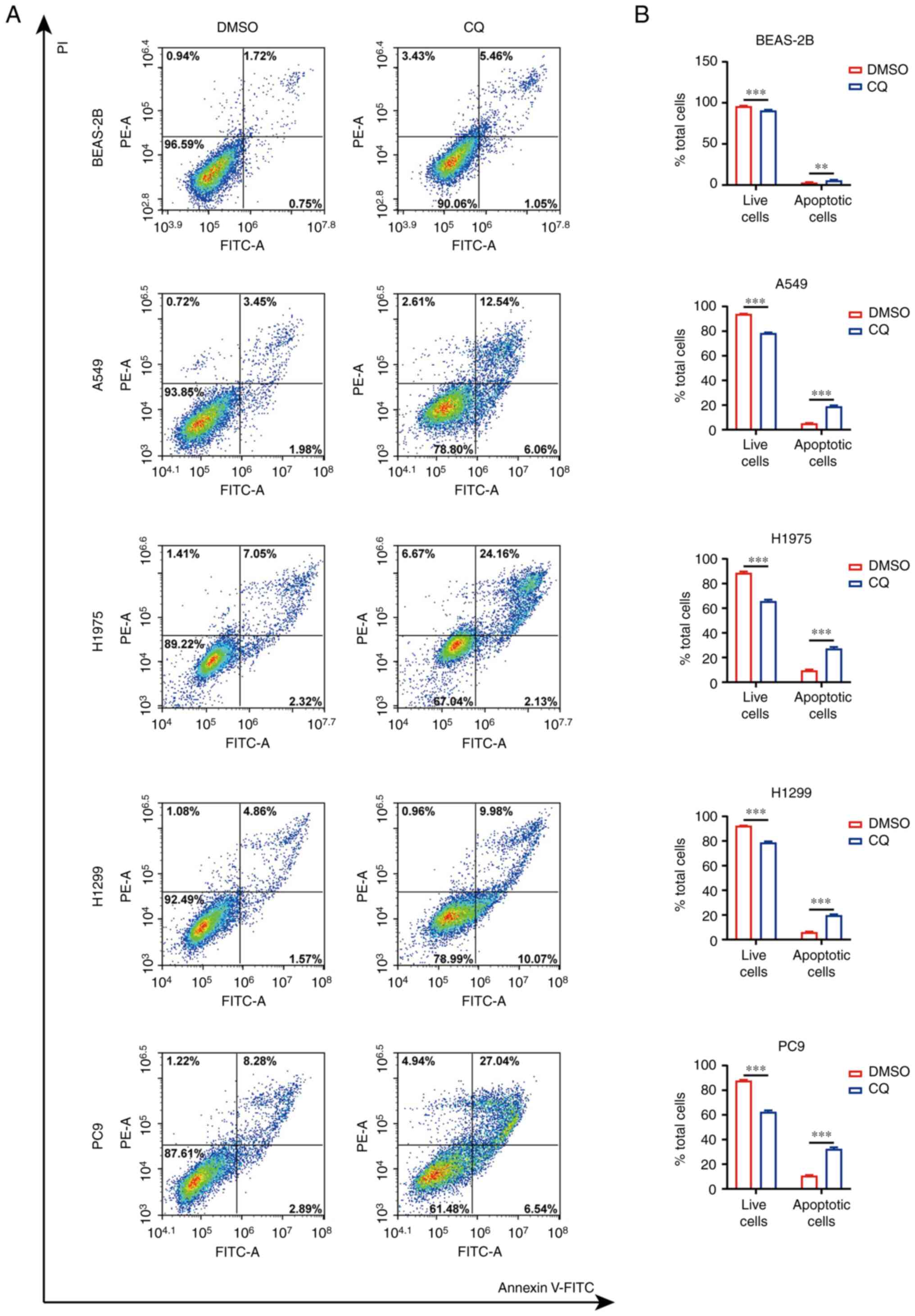
apoptosis after stimulation with CQ was examined in these cells.
Flow cytometry results revealed that the percentage of apoptotic
cells was increased, and that of live cells was decreased after CQ
treatment (Fig. 9). Taken together,
these results suggest that CQ treatment resulted in an inhibited
proliferation and increased apoptosis of lung cancer cells,
presumably due to the reversal of gene expression patterns in
tumors, demonstrating that targeting endocytosis could benefit the
clinical treatment of patients with lung cancer, including patients
with a high-risk score.
Discussion
Endocytosis is one of the essential functions of
cell biology and serves an extensive role in the development of
tumors (25). Uptake and excretion
are mediated by endocytosis and exocytosis maintains the integrity
and kinetics of the cell membrane, supporting the rapid
proliferation of tumor cells (26).
Furthermore, endocytosis can seize the majority of nutrients from
the surroundings to promote energy metabolism and lead to nutrient
deficiency of ambient stromal cells and infiltrated immune cells,
thus regulating the immunosuppressive microenvironment construction
(27). In the present study, the
focus was on an endocytosis-associated gene set used to screen
candidate genes that not only promote the malignant progression of
LUAD, but are also associated with a poor prognosis. Subsequently,
a novel prognostic gene signature consisting of 18 genes was
further refined and established, and the association between this
signature and tumor grade and stage was examined. Finally, it was
also confirmed that a high score of the gene signature was
positively correlated with lower tumor purity and more complex
immune cell infiltration. Since no endocytosis-associated gene
signature has been reported in LUAD for prognosis prediction, the
findings of the present study are expected to further confirm the
key roles of endocytosis in tumorigenesis, and provide a powerful
strategy for early diagnosis and prognosis determination of
LUAD.
Receptor tyrosine kinases (RTKs) are a type of
transmembrane protein that transduce stimulatory signals to
downstream proteins in a ligand-dependent manner (28). Aberrant activation of the RTK
pathway usually occurs in tumorigenesis and malignant progression
(29). EGFR is an important RTK,
and its oncogenic mutation accounts for ~20% of patients with LUAD
(30). TKIs target kinase
receptors, such as EGFR, fibroblast growth factor receptor,
platelet-derived growth factor receptor and vascular endothelial
growth factor receptor, which block activation of the downstream
cascade. Currently, first-generation (gefitinib and erlotinib),
second-generation (afatinib) and third-generation (osimertinib)
TKIs have been approved for clinical LUAD treatment (31–33). A
number of patients can benefit from TKI treatment, while others
will still suffer from recurrence (34). It has been reported that
internalization of EGFR mediated by endocytosis is one of the
direct reasons for TKI treatment failure (35). In addition, tumors can also become
drug resistant through a bypass activation mechanism, leading to
poor prognosis (36). The findings
of the present study classified patients with LUAD using an
endocytosis-associated signature. CQ treatment could reduce the
proliferation of lung cancer cells. Considering the correlation
between endocytosis and RTK activation, the combined treatment
strategy of TKIs and endocytosis blockers is expected to result in
a better prognosis for patients with high-score LUAD.
In recent years, tumor immunotherapy has
demonstrated strong antitumor effects, but a number of patients
have developed drug resistance to this therapy (37). Chew et al (38) revealed that inhibition of
endocytosis can effectively improve the killing effect of
monoclonal antibodies against tumor immunotherapy. Through
retrospective analysis, it was confirmed that the score based on
the endocytosis-associated signature was correlated with immune
infiltration. Therefore, patients with high scores might benefit
from improved therapeutic effects through treatment with
immunotherapy affiliated with endocytosis inhibitors.
Currently, various studies have explored prognostic
prediction of lung cancer from different perspectives, the majority
of which focus on the area of tumor immunity. Sun et al
(39) established a long non-coding
RNA signature associated with the immune infiltration of LUAD.
Another study identified 10 genes associated with immune
infiltration and constructed a prediction model for SCLC (40). Li et al (41) integrated multiple cohorts to develop
and validate an immune signature composed of 25 genes predicting
early-stage NSCLC. A study used Lasso Cox regression analysis to
identify four immune-associated gene signatures that could
adequately predict the prognosis of patients with LUAD (42). In addition, a number of studies have
also established models for crucial biological processes of tumors,
such as anoikic (43) and
glycolysis (44), which were
reported to be associated with the prognosis of patients. In the
present study, a univariate/multivariate Cox regression analysis
combined with Lasso Cox regression analysis was used to identify 18
genes that were significantly associated with patient outcomes, and
a novel signature in terms of tumor endocytosis was established.
Subsequently, treatment with the endocytosis inhibitor CQ
significantly inhibited proliferation, and increased apoptosis
levels of LUAD cells. The present study not only provided a
prognostic prediction model for clinical patients with LUAD, but
also an alternative treatment for patients with high scores.
In the present study, the novel
endocytosis-associated gene signature established by the LUAD
cohort of TCGA database and verified by two GEO datasets included
the following 18 genes: CFTR, RAB27B, ADRB1, DPYSL2, ATP6V0A4,
EREG, SFTPD, LOXL2, HTR2B, SYT2, ALOX15, F2R1, IL7R, PCSK9, ADRB2,
GATA2, IGMH and MRC1. Considering that these genes belong to the
gene set of endocytosis, a number of them are membrane localization
proteins, including CFTR, ADRB1, ATP6V0A4, EREG, SFTPD, HTR2B,
ALOX15, F2RL1, IL7R, ADRB2, IGMH and MRC1. Notably, the functional
importance of a number of the genes in LUAD has been reported. CFTR
mutations are closely associated with cystic fibrosis and
tumorigenesis of lung cancer (45,46).
EREG induces EGFR/ErbB2 heterodimer formation to phosphorylate AKT
and block TKI-mediated apoptosis in NSCLC (47), whereas SFTPD inhibits the
dimerization and activation of EGFR (48). LOXL2 induced by the
microRNA-200/ZEB1 axis mediates extracellular matrix reprogramming
to promote the invasion and metastasis of lung cancer (49). IL7R not only activates the JAK/STAT5
signaling pathway to sensitize NSCLC to chemotherapeutics (50), but also acts as a transmitter for
IL-7-induced sensitization to cisplatin by activating the PI3K/AKT
pathway and enhancing ABCG2 expression (51). GATA2 was proven to be essential for
RAS-driven NSCLC (52). The
expression levels of RAB27B (53),
DPYSL2 (54), ALOX15 (55) and PCSK9 (56) were reported to be closely associated
with the prognosis of patients with lung cancer. In addition, the
functional roles of HTR2B, F2RL1 and SYT2 in lung cancer remain
unclear, suggesting that a number of mechanisms require further
investigation.
The present study has certain limitations which need
to be carefully addressed. The endocytosis-associated signature
established was based on data from TCGA and two other cohorts
originating from RNA sequencing. The expression profiles of
signature genes under the treatment of CQ were validated by qPCR in
LUAD cell lines. However, the levels of proteins involved in this
signature remained unclear after CQ treatment, and a number of
proteins might serve important roles in malignant behavior of lung
cancer, the mechanism of which requires further investigation. In
addition, it is hypothesized that if verified using a larger
dataset, a PCR-based diagnostic kit according to this signature
could be developed for accurate prognostic prediction of patients
with LUAD.
In conclusion, a novel endocytosis-associated
prognostic signature was depicted using TCGA and GEO datasets. High
risk scores of patients with LUAD, calculated according to the
signature, indicated poor prognosis and short survival time. The
gene signature helped to identify the immunosuppressive tumor
microenvironment, suggesting that anti-endocytosis therapy could
significantly improve the prognosis of patients with LUAD.
Additionally, targeting endocytosis via CQ could repress tumor
proliferation in vitro. Based on the aforementioned
findings, personalized diagnosis and targeted combinational
therapeutic strategies for patients with LUAD could be
provided.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
TCGA data used in the present study were downloaded
from the UCSC Xena online server (https://xenabrowser.net/). The GSE30219 and GSE31210
datasets were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
YiZ, ML and KZ conceptualized and designed the
study. YiZ, SL, YaZ and ML carried out the experiments. YiZ and SL
analyzed the data and generated the figures. YiZ, ML and KZ drafted
the manuscript. ML and KZ confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li C, Lei S, Ding L, Xu Y, Wu X, Wang H,
Zhang Z, Gao T, Zhang Y and Li L: Global burden and trends of lung
cancer incidence and mortality. Chin Med J (Engl). 136:1583–1590.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duma N, Santana-Davila R and Molina JR:
Non-small cell lung cancer: epidemiology, screening, diagnosis, and
treatment. Mayo Clin Proc. 94:1623–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doherty GJ and McMahon HT: Mechanisms of
endocytosis. Annu Rev Biochem. 78:857–902. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lanzetti L and Di Fiore PP: Endocytosis
and cancer: An ‘insider’ network with dangerous liaisons. Traffic.
9:2011–2021. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Théry C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen PH, Bendris N, Hsiao YJ, Reis CR,
Mettlen M, Chen HY, Yu SL and Schmid SL: Crosstalk between
CLCb/Dyn1-mediated adaptive clathrin-mediated endocytosis and
epidermal growth factor receptor signaling increases metastasis.
Dev Cell. 40:278–288.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ketteler J and Klein D: Caveolin-1, cancer
and therapy resistance. Int J Cancer. 143:2092–2104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roy S, Wyse B and Hancock JF: H-Ras
signaling and K-Ras signaling are differentially dependent on
endocytosis. Mol Cell Biol. 22:5128–5140. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim B, Park YS, Sung JS, Lee JW, Lee SB
and Kim YH: Clathrin-mediated EGFR endocytosis as a potential
therapeutic strategy for overcoming primary resistance of EGFR TKI
in wild-type EGFR non-small cell lung cancer. Cancer Med.
10:372–385. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao GY, Mohanakrishnan A and Schmid SL:
Role for ERK1/2-dependent activation of FCHSD2 in cancer
cell-selective regulation of clathrin-mediated endocytosis. Proc
Natl Acad Sci USA. 115:E9570–E9579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jo U, Park KH, Whang YM, Sung JS, Won NH,
Park JK and Kim YH: EGFR endocytosis is a novel therapeutic target
in lung cancer with wild-type EGFR. Oncotarget. 5:1265–1278. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nishimura Y, Bereczky B and Ono M: The
EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis
of EGFR via the early/late endocytic pathway in non-small cell lung
cancer cell lines. Histochem Cell Biol. 127:541–553. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nishimura Y, Yoshioka K, Bereczky B and
Itoh K: Evidence for efficient phosphorylation of EGFR and rapid
endocytosis of phosphorylated EGFR via the early/late endocytic
pathway in a gefitinib-sensitive non-small cell lung cancer cell
line. Mol Cancer. 7:422008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tanaka T, Ozawa T, Oga E, Muraguchi A and
Sakurai H: Cisplatin-induced non-canonical endocytosis of EGFR via
p38 phosphorylation of the C-terminal region containing Ser-1015 in
non-small cell lung cancer cells. Oncol Lett. 15:9251–9256.
2018.PubMed/NCBI
|
|
19
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park JV, Chandra R, Cai L, Ganguly D, Li
H, Toombs JE, Girard L, Brekken RA and Minna JD: Tumor cells
modulate macrophage phenotype in a novel in vitro co-culture model
of the NSCLC tumor microenvironment. J Thorac Oncol. 17:1178–1191.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park JE, Dutta B, Tse SW, Gupta N, Tan CF,
Low JK, Yeoh KW, Kon OL, Tam JP and Sze SK: Hypoxia-induced tumor
exosomes promote M2-like macrophage polarization of infiltrating
myeloid cells and microRNA-mediated metabolic shift. Oncogene.
38:5158–5173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tripathy S, Dassarma B, Roy S, Chabalala H
and Matsabisa MG: A review on possible modes of action of
chloroquine/hydroxychloroquine: Repurposing against SAR-CoV-2
(COVID-19) pandemic. Int J Antimicrob Agents. 56:1060282020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Johannes L and Billet A: Glycosylation and
raft endocytosis in cancer. Cancer Metastasis Rev. 39:375–396.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cooper ST and McNeil PL: Membrane repair:
Mechanisms and pathophysiology. Physiol Rev. 95:1205–1240. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao Y, Rabien A, Buschow R, Amtislavskiy
V, Busch J, Kilic E, Villegas SL, Timmermann B, Schütte M, Mielke
T, et al: Endocytosis-mediated replenishment of amino acids favors
cancer cell proliferation and survival in chromophobe renal cell
carcinoma. Cancer Res. 80:5491–5501. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Azad T, Rezaei R, Surendran A, Singaravelu
R, Boulton S, Dave J, Bell JC and Ilkow CS: Hippo signaling pathway
as a central mediator of receptors tyrosine kinases (RTKs) in
tumorigenesis. Cancers (Basel). 12:20422020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du Z and Lovly CM: Mechanisms of receptor
tyrosine kinase activation in cancer. Mol Cancer. 17:582018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
da Cunha Santos G, Shepherd FA and Tsao
MS: EGFR mutations and lung cancer. Annu Rev Pathol. 6:49–69. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sim EH, Yang IA, Wood-Baker R, Bowman RV
and Fong KM: Gefitinib for advanced non-small cell lung cancer.
Cochrane Database Syst Rev. 1:CD0068472018.PubMed/NCBI
|
|
32
|
Sartori G, Belluomini L, Lombardo F,
Avancini A, Trestini I, Vita E, Tregnago D, Menis J, Bria E,
Milella M and Pilotto S: Efficacy and safety of afatinib for
non-small-cell lung cancer: State-of-the-art and future
perspectives. Expert Rev Anticancer Ther. 20:531–542. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Remon J, Steuer CE, Ramalingam SS and
Felip E: Osimertinib and other third-generation EGFR TKI in
EGFR-mutant NSCLC patients. Ann Oncol. 29 (Suppl 1):i20–i27. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu SG and Shih JY: Management of acquired
resistance to EGFR TKI-targeted therapy in advanced non-small cell
lung cancer. Mol Cancer. 17:382018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cruz Da Silva E, Choulier L,
Thevenard-Devy J, Schneider C, Carl P, Ronde P, Dedieu S and
Lehmann M: Role of endocytosis proteins in gefitinib-mediated EGFR
internalisation in glioma cells. Cells. 10:32582021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McCoach CE, Le AT, Gowan K, Jones K,
Schubert L, Doak A, Estrada-Bernal A, Davies KD, Merrick DT, Bunn
PA Jr, et al: Resistance mechanisms to targeted therapies in
ROS1+ and ALK+ non-small cell lung cancer.
Clin Cancer Res. 24:3334–3347. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mamdani H, Matosevic S, Khalid AB, Durm G
and Jalal SI: Immunotherapy in lung cancer: Current landscape and
future directions. Front Immunol. 13:8236182022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chew HY, De Lima PO, Gonzalez Cruz JL,
Banushi B, Echejoh G, Hu L, Joseph SR, Lum B, Rae J, O'Donnell JS,
et al: Endocytosis inhibition in humans to improve responses to
ADCC-mediating antibodies. Cell. 180:895–914.e27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun J, Zhang Z, Bao S, Yan C, Hou P, Wu N,
Su J, Xu L and Zhou M: Identification of tumor immune
infiltration-associated lncRNAs for improving prognosis and
immunotherapy response of patients with non-small cell lung cancer.
J Immunother Cancer. 8:e0001102020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xie Q, Chu H, Yi J, Yu H, Gu T, Guan Y,
Liu X, Liang J, Li Y and Wang J: Identification of a prognostic
immune-related signature for small cell lung cancer. Cancer Med.
10:9115–9128. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li B, Cui Y, Diehn M and Li R: Development
and validation of an individualized immune prognostic signature in
early-stage nonsquamous non-small cell lung cancer. JAMA Oncol.
3:1529–1537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun S, Guo W, Wang Z, Wang X, Zhang G,
Zhang H, Li R, Gao Y, Qiu B, Tan F, et al: Development and
validation of an immune-related prognostic signature in lung
adenocarcinoma. Cancer Med. 9:5960–5975. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Diao X, Guo C and Li S: Identification of
a novel anoikis-related gene signature to predict prognosis and
tumor microenvironment in lung adenocarcinoma. Thorac Cancer.
14:320–330. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Zhang Z and Yu Z: Identification
of a novel glycolysis-related gene signature for predicting
metastasis and survival in patients with lung adenocarcinoma. J
Transl Med. 17:4232019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Govindan R, Ding L, Griffith M,
Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L,
Wallis J, et al: Genomic landscape of non-small cell lung cancer in
smokers and never-smokers. Cell. 150:1121–1134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi X, Kou M, Dong X, Zhai J, Liu X, Lu D,
Ni Z, Jiang J and Cai K: Integrative pan cancer analysis reveals
the importance of CFTR in lung adenocarcinoma prognosis. Genomics.
114:1102792022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma S, Zhang L, Ren Y, Dai W, Chen T, Luo
L, Zeng J, Mi K, Lang J and Cao B: Epiregulin confers EGFR-TKI
resistance via EGFR/ErbB2 heterodimer in non-small cell lung
cancer. Oncogene. 40:2596–2609. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Umeda Y, Hasegawa Y, Otsuka M, Ariki S,
Takamiya R, Saito A, Uehara Y, Saijo H, Kuronuma K, Chiba H, et al:
Surfactant protein D inhibits activation of non-small cell lung
cancer-associated mutant EGFR and affects clinical outcomes of
patients. Oncogene. 36:6432–6445. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Peng DH, Ungewiss C, Tong P, Byers LA,
Wang J, Canales JR, Villalobos PA, Uraoka N, Mino B, Behrens C, et
al: ZEB1 induces LOXL2-mediated collagen stabilization and
deposition in the extracellular matrix to drive lung cancer
invasion and metastasis. Oncogene. 36:1925–1938. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi L, Xu Z, Yang Q, Huang Y, Gong Y, Wang
F and Ke B: IL-7-Mediated IL-7R-JAK3/STAT5 signalling pathway
contributes to chemotherapeutic sensitivity in non-small-cell lung
cancer. Cell Prolif. 52:e126992019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ke B, Wei T, Huang Y, Gong Y, Wu G, Liu J,
Chen X and Shi L: Interleukin-7 resensitizes non-small-cell lung
cancer to cisplatin via inhibition of ABCG2. Mediators Inflamm.
2019:72414182019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kumar MS, Hancock DC, Molina-Arcas M,
Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E,
Lassailly F, Matthews N, Nye E, et al: The GATA2 transcriptional
network is requisite for RAS oncogene-driven non-small cell lung
cancer. Cell. 149:642–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Koh HM and Song DH: Prognostic role of
Rab27A and Rab27B expression in patients with non-small cell lung
carcinoma. Thorac Cancer. 10:143–149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu YJ, Nai AT, He GC, Xiao F, Li ZM, Tang
SY, Liu YP and Ai XH: DPYSL2 as potential diagnostic and prognostic
biomarker linked to immune infiltration in lung adenocarcinoma.
World J Surg Oncol. 19:2742021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ren Z, Hu M, Wang Z, Ge J, Zhou X, Zhang G
and Zheng H: Ferroptosis-related genes in lung adenocarcinoma:
Prognostic signature and immune, drug resistance, mutation
analysis. Front Genet. 12:6729042021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xie M, Yu X, Chu X, Xie H, Zhou J, Zhao J
and Su C: Low baseline plasma PCSK9 level is associated with good
clinical outcomes of immune checkpoint inhibitors in advanced
non-small cell lung cancer. Thorac Cancer. 13:353–360. 2022.
View Article : Google Scholar : PubMed/NCBI
|