Introduction
Nasopharyngeal carcinoma (NPC) is one of the most
common head and neck malignancies in Southeast Asia (1), and radiation therapy (RT) is the main
treatment approach (2).
Intensity-modulated RT (IMRT) is a conformal RT technique that has
been widely used in patients with NPC (3). IMRT accurately controls the radiation
dose locally in the lesion, reduces damage to surrounding normal
tissues, decreases the incidence of complications (such as temporal
lobe necrosis, cranial neuropathy and/or hypothyroidism), and
improves the local area control rate of the tumor, while increasing
the 5-year overall survival rate of patients with NPC to ~80%
(4–6). However, ~85% of patients treated with
IMRT experience different degrees of acute and late
radiation-induced injuries such as oral mucositis, dermatitis,
pharyngitis, temporal lobe neuropathy, late xerostomia and trismus
(7,8). More specifically, ~20% of these
patients suffer from acute severe radiation-induced injuries, which
often cause uncontrollable pain, resulting in treatment
interruptions (8,9). These interruptions eventually lead to
failure of tumor control, thereby shortening the overall survival
time (10,11). However, the lack of sensitive and
specific biomarkers makes it difficult to predict the occurrence of
acute severe radiation-induced injury in patients.
To understand the mechanism by which
radiation-induced injury occurs and to find useful markers,
numerous risk factors have been analyzed over the past few decades,
including age, sex, smoking status, radiation dose, RT technique,
chemotherapy and genetics-related factors such as genetic variants
and epigenetics (12,13). With the rapid advancement of
radio-genomics, numerous studies have presented associations
between genetic variants of candidate genes and the toxicity and
efficacy of radiotherapy in patients with NPC (14,15).
Since radiation exposure destroys cells by inducing
DNA damage, the ability for DNA damage repair is a central factor
that influences tissue radiation sensitivity or damage (16). X-ray repair cross-complementing
group-1 (XRCC1) is a pivotal DNA repair gene. The XRCC1 protein
encoded by this gene serves a critical role in repairing base
excision and single-strand breaks induced by radiation (17,18).
Prior research has indicated that minor sequence variations within
these DNA repair genes, such as single-nucleotide polymorphisms
(SNPs), have the potential to disrupt the function of these genes,
subsequently altering protein function and the ability of
individuals to effectively repair damaged DNA (19). Ultimately, these genetic variations
can influence susceptibility to radiation-induced injuries
(20).
Through the analysis of the human XRCC1 gene
sequence, ~16 SNP sites have been found to be located in exons or
promoter regions, with the three most important functional SNP
sites being Arg194Trp (rs1799782), Arg280His (rs25489) and
Arg399Gln (rs25487) (19). A
previous meta-analysis of the existing literature showed that, in
breast, prostate and other cancer types, the Arg399Gln SNP was
notably associated with the risk of acute adverse reactions induced
by RT (21). These results suggest
that this SNP in the XRCC1 gene is likely to be a predictor of
individual response to radiation.
However, in NPC, most of the studies on XRCC1 SNPs
investigated their relationship with tumor susceptibility (22,23),
whereas the associations with RT response and prognosis have not
been extensively analyzed (24,25).
Therefore, relevant observational studies are urgently needed to
extend the current understanding of the relationship between SNPs
in the XRCC1 gene and the therapeutic effect of RT in NPC. The
purpose of the present study was to investigate whether the
aforementioned three major SNPs in the XRCC1 gene are associated
with the severity of acute radiation-induced injury and prognosis
of patients with NPC treated with IMRT.
Materials and methods
Patient selection
All patients with primary NPC who were first
diagnosed and treated with IMRT at Fujian Cancer Hospital (Fuzhou,
China) between July 2012 and October 2013 were considered for
inclusion in the present retrospective study.
Patients with NPC who met any of the following
criteria on admission were excluded: i) A Karnofsky Performance
Status (26) score <80; ii)
severe dysfunction of the heart, lungs, liver and/or kidneys; iii)
history of any other malignancies; and iv) prior clinical
interventions such as surgery, radiotherapy and chemotherapy.
The following inclusion criteria were applied: i)
Availability of comprehensive diagnostic information, including
general clinical characteristics, pathology reports, radiological
findings, as well as routine laboratory test results, with a
particular focus on plasma EBV-DNA concentration; ii) peripheral
blood was collected before treatment, genotype analysis of XRCC1
SNP was performed, and relevant results were recorded; iii)
definitive post-admission IMRT treatment was received,
radiation-induced injuries were examined and the severity of damage
was recorded; and iv) after the end of treatment, prognostic
follow-up examinations of the patients were regularly performed,
and complete case data, including recurrence or death, were
recorded.
The research protocol used in the present study was
approved by the Ethics Committee of Fujian Cancer Hospital
(approval no. K2021-046-01; Fuzhou, China). Written informed
consent was obtained from all adult participants, as well as from
the parents or guardians of minors included in the study.
SNP genotyping
EDTA-K2 anticoagulated peripheral blood (10 ml) was
collected from patients before treatment. Subsequently, 2 ml whole
blood was utilized for the genotype analysis of three SNP loci
XRCC1-Arg194Trp, -Arg280His and -Arg399Gln, which was completed
within 3 months. The remaining blood samples were kept at −80°C for
re-examination.
For SNP genotyping, genomic DNA was extracted from
whole-blood samples with the DNeasy Blood and Tissue Kit (Qiagen,
Inc.) following the manufacturer's instructions. Relevant segments
of DNA were amplified by PCR using HotStar Taq DNA polymerase
(Qiagen, Inc.) under the following conditions: An initial
denaturation step at 95°C for 2 min, followed by 45 cycles of 15
sec at 94°C and 35 sec at 60°C, concluding with a final cooling
step at 25°C for 1 min. The primer sequences for the SNP sites were
as follows: Arg194Trp forward, 5′-GCCAGGGCCCCTCCTTCAA-3′ and
reverse, 5′-TACCCTCAGACCCACGAGT-3′; Arg280His forward,
5′-CCAGTGGTGCTAACCTAATC-3′ and reverse, 5′-CACTCAGCACCACTACCACA-3′;
and Arg399Gln forward, 5′-TTGTGCTTTCTCTGTGTCCA-3′ and reverse,
5′-TCCTCCAGCCTTTTCTGATA-3′. The quality of the PCR product was
tested by running 5 µl PCR product on a 1% agarose gel containing
SYBR Green I Dye (Biosharp Life Sciences). Next, the PCR products
underwent Sanger sequencing, employing the BigDye Terminator v3.1
Cyclic Sequencing Kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
sequencing was performed on the ABI 3100 sequencer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Sequencing results
were aligned to the corresponding reference sequence (NG_033799.1),
and the SNPs were genotyped using SeqManII sequence analysis
software (version 6.3; DNASTAR, Inc.). Finally, the variants of the
three candidate SNPs in the XRCC1 gene were classified into
wild-type and polymorphic variants: Arg194Trp, CC and CT/TT;
Arg280His, GG and GA/AA; and Arg399Gln, GG and GA/AA,
respectively.
Grading of acute radiation-induced
injuries
The detailed RT regimen was as previously described
(27). Briefly, patients were
examined by their radiation therapists weekly during IMRT and at
4–6 weeks post-treatment for radiation-induced injury. The severity
of radiation-induced injury was assessed using the radiation
toxicity grading criteria of the Radiation Therapy Oncology Group
(28). Using this scale, grades 0–4
were designated, where 0 represented the absence of change over the
baseline and 4 indicated ulceration, hemorrhage or necrosis of the
mucosa or skin. All scores were confirmed by the same senior
consultant physician to eliminate observer bias.
Follow-up frequency and content
The first post-treatment visit of all patients was
scheduled at 4 weeks post-IMRT treatment. The follow-up frequency
was as follows: Once a month for the first 3 months, every 2 months
for the next 6 months, every 3 months for the next 2 years, and
every 6 months thereafter. The clinical follow-up included medical
history collection, physical examination, direct laryngoscopy and
routine laboratory tests such as liver function, complete blood
count and plasma EBV-DNA concentration measurement, as well as MRI
of the nasopharynx, neck and skull base every 6–12 months.
Additional chest CT, abdominal ultrasound and bone scans were
performed if necessary to detect local or distant recurrence.
Outpatient visits, sending text messages, telephone inquiries and
reviewing medical records were implemented as follow-up methods.
The follow-up period ended in August 2020.
Data collection
Basic information and general clinicopathological
characteristics such as name, age, sex, pathological type, tumor
size, lymph node status and distant metastasis were collected from
patient admission to the time of diagnosis. Clinical staging of
patients was performed according to the American Joint Commission
on Cancer guidelines (7th edition; 2010) (29). The grade of the most serious acute
radiation-induced injuries that occurred in patients during the
observation period was recorded. Acute radiation-induced injuries
of less than grade 2 were generally considered to represent an
acceptable level of injury that did not further affect RT outcomes.
Therefore, grades 0–1 were defined as none or mild, and grades 2+
were defined as moderate to severe. Throughout the follow-up
period, the progression-free survival (PFS), instances of disease
recurrence, metastasis, development of second primary tumor and
various survival endpoints were documented. If follow-up continued
to the end of the study in August 2020, the survival time was
calculated from the beginning of treatment to the end of the study
in August 2020. Incomplete data from patients who were lost to
follow-up were treated as censored data.
Finally, the basic patient information and general
clinicopathological characteristics, tumor RT regimen and dose,
chemotherapy information, SNP genotyping, laboratory test results,
type and severity of acute radiation-induced injury, as well as
clinical follow-up time and outcomes were documented.
Statistical analysis
The data are presented as the mean or as number (%).
Every SNP was tested for deviation from the Hardy-Weinberg
equilibrium (HWE) using the Pearson χ2 test with two
degrees of freedom.
The following univariate tests were employed to
compare the frequency of oral mucositis, dermatitis and pharyngitis
among patients with different genotypes, adhering to the following
criteria: When the sample size (n) was ≥40 and all cells had a
theoretical frequency (T) of ≥5, Pearson χ2 test was
utilized; when n≥40 but at least one cell had a T of 1–5, the
Yates's correction for continuity was applied; and when n<40 or
at least one cell had T<1, Fisher's exact test was used
(30,31). Subsequently, multivariate logistic
regression analysis was performed to evaluate the association
between every factor and the occurrence of acute severe
radiation-induced injuries by computing the odds ratio (OR) and the
corresponding 95% CI.
Survival rates were estimated using a univariate
Kaplan-Meier survival curve, which determined whether the risk of
PFS varied depending on the patient genotype. A log-rank test was
implemented to compare the differences in survival time. In
addition, multivariate Cox regression analysis was performed,
incorporating other variables that had previously been reported as
significant in the literature, to identify the parameters having an
independent, significant influence on PFS, and to calculate hazard
ratios (HRs).
All statistical analyses were performed using SPSS
(version 22.0; IBM Corp.), and P≤0.05 was considered to indicate a
statistically significant difference. Graphs were created with
GraphPad Prism (version 8.3.0; Dotmatics).
Results
Clinicopathological characteristics of
patients with NPC
A total of 158 patients with NPC were included in
the present study, and their general clinicopathological
characteristics are presented in Table
I. The majority of patients with NPC were male, and the mean
age was 43.6 years (range, 11–74 years). The predominant
pathological type was non-keratinizing undifferentiated NPC. Before
treatment, 72 cases (46.5%) were plasma EBV-DNA-positive, whereas
83 cases (53.5%) were negative; no results were obtained for 3
cases. In general, 140 cases (88.6%) received the intended
radiation dose ranging from 66 to 70.95 Gy for the gross tumor
volume of the primary focus. Additionally, 10 cases (6.3%) received
a dose <66 Gy, while 8 cases (5.1%) received a dose >70.95
Gy. The total incidence of acute radiation-induced injuries,
including oral mucositis, dermatitis and pharyngitis, in the
patient cohort was 98.1, 84.2 and 88.0%, respectively.
 | Table I.Clinicopathological characteristics
of 158 patients with nasopharyngeal carcinoma. |
Table I.
Clinicopathological characteristics
of 158 patients with nasopharyngeal carcinoma.
| Clinicopathological
characteristics | No. (%) |
|---|
| Sex |
|
|
Male | 113 (71.5) |
|
Female | 45
(28.5) |
| Age at diagnosis,
years |
|
|
<60 | 145 (91.8) |
|
≥60 | 13
(8.2) |
| Pathological
types |
|
|
NKU | 150 (94.9) |
|
Others | 8
(5.1) |
| T stage |
|
|
T1-T2 | 69
(43.7) |
|
T3-T4 | 89
(56.3) |
| N stage |
|
|
N0-N1 | 45
(28.5) |
|
N2-N3 | 113 (71.5) |
| Distant
metastasis |
|
| M0 | 152 (96.2) |
| M1 | 6
(3.8) |
| Clinical stage |
|
|
I–II | 15
(9.5) |
|
III–IV | 143 (90.5) |
| EBV-DNA
(n=155)a |
|
|
Positive | 72
(46.5) |
|
Negative | 83
(53.5) |
| Radiation dose,
Gy |
|
|
<66 | 10
(6.3) |
|
66-70.95 | 140 (88.6) |
|
>70.95 | 8
(5.1) |
| Acute radiation
toxicityb |
|
| Oral
mucositis | 155 (98.1) |
|
Dermatitis | 133 (84.2) |
|
Pharyngitis | 139 (88.0) |
Allele frequencies and genotype
distribution
The representative gel images and Sanger sequencing
traces of PCR products containing XRCC1 gene SNPs are shown in
Fig. S1, and the characteristics
of the three candidate SNPs of the XRCC1 gene are presented in
Table II. Overall, all the
genotype distributions were in HWE (P>0.05 based on
χ2 test for each allele).
 | Table II.Genotype distribution of the three
candidate SNPs in the X-ray repair cross-complementing group-1
gene. |
Table II.
Genotype distribution of the three
candidate SNPs in the X-ray repair cross-complementing group-1
gene.
| SNP
(genotypes) | NCBI dbSNP ID | Alleles (amino
acids) | Genotype
distributiona, n | HWE (P-value) |
|---|
| Arg194Trp
(CC/CT/TT) | rs1799782 | C>T
(Arg>Trp) | 80/67/11 | 0.547 |
| Arg280His
(GG/GA/AA) | rs25489 | G>A
(Arg>His) | 133/22/3 | 0.083 |
| Arg399Gln
(GG/GA/AA) | rs25487 | G>A
(Arg>Gln) | 85/63/10 | 0.712 |
Association between candidate SNPs and
the severity of acute radiation-induced injuries
Acute radiation-induced injuries that are most
frequently observed in patients with NPC during IMRT treatment
include oral mucositis, dermatitis and pharyngitis or dysphagia
(3). In the present study, 24.7%
(n=39) and 75.3% (n=119) of patients experienced grade 0–1 and 2+
radioactive oral mucositis, respectively. A total of 84.2% (n=133)
and 15.8% (n=25) of patients experienced grade 0–1 and 2+
radioactive dermatitis, respectively. A total of 88.6% (n=140) and
11.4% (n=18) of the patients developed grade 0–1 and 2+ radioactive
pharyngitis, respectively. No grade 4 adverse reactions were
observed in the current study (Table
III).
 | Table III.Relationship between candidate SNPs
and the severity of acute radiation-induced injuries. |
Table III.
Relationship between candidate SNPs
and the severity of acute radiation-induced injuries.
|
| Oral mucositis
grade, n (%) |
| Dermatitis grade, n
(%) |
| Pharyngitis grade,
n (%) |
|
|---|
|
|
|
|
|
|
|
|
|---|
|
|
G0-1 | G2+ |
|
G0-1 | G2+ |
|
G0-1 | G2+ |
|
|---|
| SNP/genotypes | (n=39) | (n=119) | P-value | (n=133) | (n=25) | P-value | (n=140) | (n=18) | P-value |
|---|
|
XRCC1-Arg194Trp |
|
|
|
|
|
|
|
|
|
| CC-wild
(n=80) | 17 (10.8) | 63 (39.9) | 0.311 | 67 (42.4) | 13 (8.2) | 0.882 | 69 (43.7) | 11 (7.0) | 0.345 |
| CT + TT
(n=78) | 22 (13.9) | 56 (35.4) |
| 66 (41.8) | 12 (7.6) |
| 71 (44.9) | 7 (4.4) |
|
|
TT-mutant (n=11) | 4 (2.5) | 7 (4.4) | 0.569 | 10 (6.3) | 1 (0.6) | 0.837 | 11 (6.9) | 0 (0.0) | 0.616 |
| CT + CC
(n=147) | 35 (22.2) | 112 (70.9) |
| 123 (77.9) | 24 (15.2) |
| 129 (81.7) | 18 (11.4) |
|
| XRCC1-
Arg280His |
|
|
|
|
|
|
|
|
|
| GG-wild
(n=133) | 32 (20.3) | 101 (63.9) | 0.675 | 113 (71.5) | 20 (12.7) | 0.533 | 119 (75.3) | 14 (8.9) | 0.655 |
| GA + AA
(n=25) | 7 (4.4) | 18 (11.4) |
| 20 (12.7) | 5 (3.1) |
| 21 (13.3) | 4 (2.5) |
|
|
AA-mutant (n=3) | 0 (0.0) | 3 (1.9) | >0.999 | 3 (1.9) | 0 (0.0) | >0.999 | 3 (1.9) | 0 (0.0) | >0.999 |
| GA + GG
(n=155) | 39 (24.7) | 116 (73.4) |
| 130 (82.3) | 25 (15.8) |
| 137 (86.7) | 18 (11.4) |
|
| XRCC1-
Arg399Gln |
|
|
|
|
|
|
|
|
|
| GG-wild
(n=85) | 21 (13.3) | 64 (40.5) | 0.994 | 73 (46.2) | 12 (7.6) | 0.526 | 78 (49.4) | 7 (4.4) | 0.178 |
| GA + AA
(n=73) | 18 (11.4) | 55 (34.8) |
| 60 (38.0) | 13 (8.2) |
| 62 (39.2) | 11 (7.0) |
|
|
AA-mutant (n=10) | 1 (0.6) | 9 (5.7) | 0.463 | 8 (5.1) | 2 (1.3) | >0.999 | 8 (5.1) | 2 (1.3) | 0.711 |
| GA + GG
(n=148) | 38 (24.1) | 110 (69.6) |
| 125 (79.1) | 23 (14.5) |
| 132 (83.5) | 16 (10.1) |
|
The data indicated no statistically significant
association between Arg194Trp, Arg280His and Arg399Gln SNPs in the
XRCC1 gene and the severity of acute radiation-induced injuries in
patients with NPC treated with IMRT (P>0.05).
Risk factors for moderate to severe
radiation-induced injuries
Multivariate logistic regression analysis was used
to evaluate the effects of risk factors that could have affected
the occurrence of acute severe oral mucositis, dermatitis and
pharyngitis. Sex, age, pathological type, tumor stage, nodal stage,
distant metastasis, clinical stage, plasma EBV-DNA concentration,
radiation dose, and XRCC1-Arg194Trp, XRCC1-Arg280His and
XRCC1-Arg399Gln were included as independent variables (Table IV). Of these, only nodal stage was
significantly associated with the occurrence of acute severe
radiation-induced oral mucositis (OR, 2.213; 95% CI, 1.149–4.263;
P=0.018). Nodal stage also showed a trend towards an association
with moderate to the incidence of acute severe radiation-induced
pharyngitis, although this was not statistically significant (OR,
4.796; 95% CI, 0.930–24.736; P=0.061). No association was observed
between acute severe radiation-induced injuries and the three
candidate SNPs (P>0.05).
 | Table IV.Multivariate logistic regression
analyses of the risk factors for acute severe radiation
injuries. |
Table IV.
Multivariate logistic regression
analyses of the risk factors for acute severe radiation
injuries.
|
| Oral mucositis
(G2+) | Dermatitis
(G2+) | Pharyngitis
(G2+) |
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Adjusted OR | 95% CI | P-value | Adjusted OR | 95% CI | P-value | Adjusted OR | 95% CI | P-value |
|---|
| Sex | 0.60 | 0.257–1.413 | 0.244 | 0.60 | 0.185–1.963 | 0.400 | 0.67 | 0.164–2.715 | 0.573 |
| Age | 0.99 | 0.951–1.021 | 0.415 | 1.02 | 0.979–1.069 | 0.314 | 1.04 | 0.982–1.107 | 0.168 |
| Pathological
type | 2.44 | 0.269–22.207 | 0.428 | 0.00a |
<0.001->999.999a | 0.999a | 0.00a |
<0.001->999.999a | 0.999a |
| Tumor stage | 1.40 | 0.826–2.352 | 0.214 | 1.08 | 0.600–1.934 | 0.802 | 1.04 | 0.531–2.062 | 0.896 |
| Nodal stage | 2.21 | 1.149–4.263 | 0.018 | 1.18 | 0.517–2.694 | 0.695 | 4.80 | 0.930–24.736 | 0.061 |
| Distant
metastasis |
>999.99a |
<0.001->999.999a | 0.999a | 2.76 | 0.375–20.216 | 0.319 | 1.81 | 0.140–23.407 | 0.651 |
| Clinical stage | 0.71 | 0.295–1.727 | 0.454 | 1.09 | 0.357–3.320 | 0.881 | 0.31 | 0.053–1.824 | 0.195 |
| Plasma EBV-DNA
concentration | 0.99 | 0.999–1.000 | 0.176 | 0.99 | 0.992–1.003 | 0.441 | 0.99 | 0.968–1.009 | 0.267 |
| Radiation dose | 0.97 | 0.876–1.079 | 0.595 | 1.06 | 0.910–1.225 | 0.475 | 0.98 | 0.915–1.039 | 0.433 |
|
XRCC1-Arg194Trp |
|
|
|
|
|
|
|
|
|
| CC vs.
CT + TT | 0.68 | 0.288–1.596 | 0.374 | 1.25 | 0.452–3.443 | 0.669 | 1.44 | 0.414–4.989 | 0.568 |
| TT vs.
CT + CC | 2.39 | 0.602–9.453 | 0.216 | 2.07 | 0.237–18.063 | 0.511 |
>999.99a |
<0.001->999.999a | 0.999a |
|
XRCC1-Arg280His |
|
|
|
|
|
|
|
|
|
| GG vs.
GA + AA | 0.57 | 0.203–1.626 | 0.296 | 1.53 | 0.471–4.966 | 0.479 | 2.20 | 0.576–8.417 | 0.249 |
| AA vs.
GA + GG | 0.00a |
<0.001->999.999a | 0.999a |
>999.99a |
<0.001->999.999a | 0.999a |
>999.99a |
<0.001->999.999a | 0.999a |
|
XRCC1-Arg399Gln |
|
|
|
|
|
|
|
|
|
| GG vs.
GA + AA | 0.59 | 0.246–1.414 | 0.237 | 1.28 | 0.457–3.587 | 0.638 | 2.28 | 0.650–8.024 | 0.198 |
| AA vs.
GA + GG | 0.52 | 0.058–4.777 | 0.567 | 0.82 | 0.133–5.058 | 0.830 | 0.83 | 0.132–5.164 | 0.837 |
Associations between candidate SNPs
and PFS
The present study followed up patients from July
2012 to August 2020. During the follow-up period, primary lymph
node recurrence occurred in 6 patients, a local relapse in 7
patients and metastasis in 29 patients. Additionally, a second
primary tumor developed in 6 patients. The mean PFS time was 69.8
months (range, 0–97 months).
Patients with the AA genotype at the XRCC1-Arg399Gln
SNP had a worse PFS compared with patients with the GA + GG
genotype; however, this was not statistically significant (P=0.069;
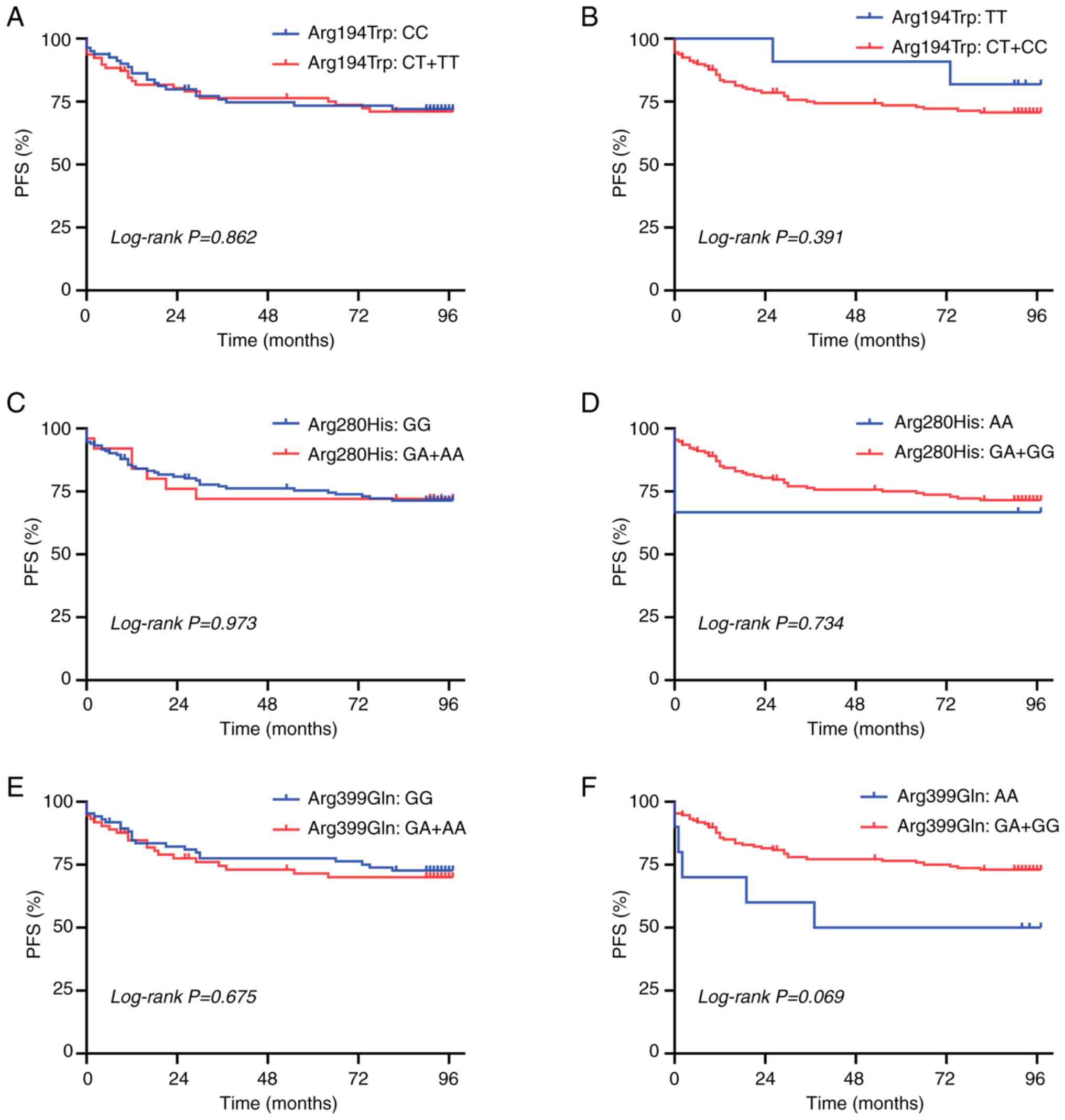
Fig. 1F). None of the other SNPs
examined in the present study was associated with PFS (all
P>0.05; Fig. 1A-E).
Survival analysis
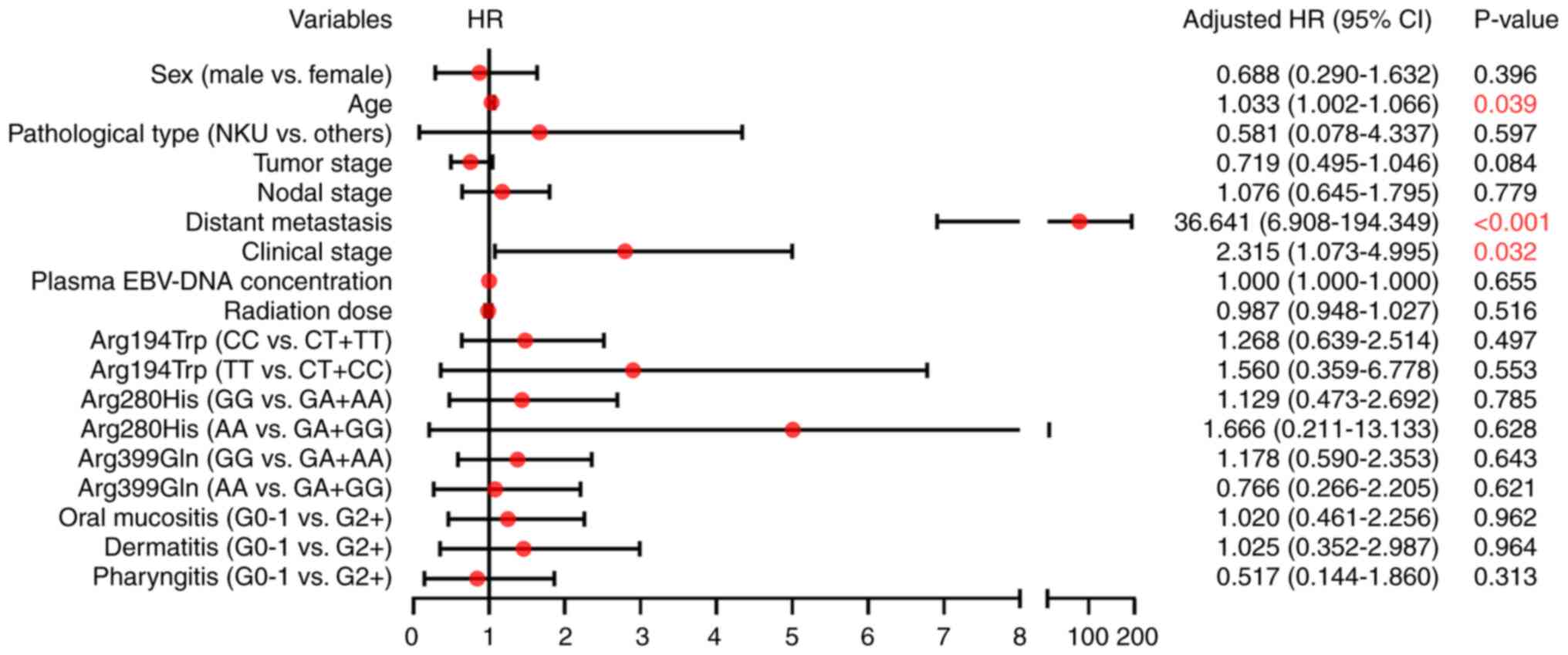
Multivariate Cox regression analysis was performed
to identify the factors associated with PFS. The results suggested
that age (HR, 1.033; 95% CI, 1.002–1.066; P=0.039), distant
metastasis (HR, 36.641; 95% CI, 6.908–194.349; P<0.001) and
clinical stage (HR, 2.315; 95% CI, 1.073–4.995; P=0.032) were
independent risk factors for PFS. No statistically significant
association was observed between the investigated SNPs and PFS
(P>0.05; Fig. 2).
Discussion
In the present study, the relationship between SNPs
in the XRCC1 gene, which is a key factor in the DNA damage repair
pathway (17,18), and the severity of acute
radiation-induced injuries as well as prognosis in patients with
NPC who received IMRT treatment, were retrospectively analyzed. The
analyses of the current study aimed to explore biological
indicators that predict the response of patients with NPC to RT,
which would provide guidance to clinicians in the choice of
personalized treatment plans based on the specific tissue damage
and tumor progression risk.
The results of the present study indicated no
significant association between the three candidate SNPs in the
XRCC1 gene and acute radiation-induced injuries during IMRT
treatment in patients with NPC. This is consistent with the
research findings of Wang et al (32) in that the SNPs of two loci in the
XRCC1 gene, Arg280His and Arg399Gln, were not associated with the
severity of acute radiation mucositis and dermatitis in patients
with NPC who received IMRT combined with chemotherapy. Similarly,
Zhai et al (33) did not
observe an association between XRCC1-Arg399Gln and acute
radiation-induced injuries to the skin, mucous membranes and
salivary glands of 60 patients with stage III–IVA NPC. Chen et
al (34) found no notable
difference in the severity of acute radiation-induced oral
mucositis among patients with NPC treated with IMRT with different
genotypes of XRCC1-Arg399Gln; however, the risk of acute
radiation-induced dermatitis of grade 2 or more in patients with
the GG genotype was notably higher than that of patients with the
other two genotypes. Li et al (35) also observed that the GA genotype of
XRCC1-Arg399Gln was significantly associated with the occurrence of
grade 3 dermatitis (P=0.037) and also showed a trend towards an
association with the incidence of grade 3 mucositis (P=0.065) in
patients with NPC. The discrepancies between the findings of Li
et al (35) and the present
study may be due to analysis biases in their cases, which used not
only IMRT but also three-dimensional (3D) conformal RT.
The multivariate logistic regression analysis
performed in the present study demonstrated a significant
association between nodal stage and the occurrence of acute severe
radiation-induced oral mucositis, and there was a trend towards an
association with the incidence of acute severe radiation-induced
pharyngitis; however, this was not statistically significant.
Similar results were also obtained in the study by Chen et
al (34), which established
that nodal stage was significantly associated with grade 2 or
greater acute radiation-induced dermatitis (P<0.001). These
findings might suggest that for improved tumor control, as the
nodal stage increases, the local irradiation dose and area of the
oropharynx and/or neck near the lymph nodes should be accordingly
increased, resulting in intensification of related radiation
damage. Chen et al (34)
also demonstrated that there was no significant association between
the severity of acute radiation-induced injuries and radiation dose
in patients with NPC treated with IMRT, which is consistent with
the findings in the studies by Wang et al (32) and Li et al (35). This lack of association might have
been caused by higher uniformity of the irradiation dose
administered to local lesions, as IMRT can be programmed with a
target volume consistent with the lesion parameters by 3D conformal
technology, thereby concentrating the effective dose on the
lesion.
Prognostic analysis in the present study revealed
that the AA genotype of the XRCC1-Arg399Gln SNP tended to be
associated with a worse PFS compared with the GA + GG genotype
(P=0.069); however, this was not statistically significant, and
there was no evidence that other candidate SNPs in the XRCC1 gene
were associated with poor prognosis in patients with NPC. These
results are consistent with the findings in the study by Zhai et
al (33). However, the study by
Jin et al (36) showed that
heavy smokers (>20 packs/year) with the XRCC1-Arg399Gln GG
genotype had significantly higher PFS times than smokers with other
genotypes (P=0.047). This discrepancy could potentially be
attributed to their stratification of patients based on smoking
status, as well as their detection of XRCC1 Arg399Gln from
paraffin-embedded biopsy specimens. Wang et al (32) found that the GG genotype of
XRCC1-Arg280His was positively associated with primary tumor
response at the end of RT in patients with NPC, which is
inconsistent with the findings of the present study. Nevertheless,
the main difference in the methodologies was that the study by Wang
et al (32) only explored
short-term effects 3 months after RT, which is not comparable with
long-term effects at the 7- to 8-year follow-up assessed in the
current study.
The multivariate Cox regression analysis conducted
in the present study showed that older age, the presence of distant
metastasis and a higher clinical stage were independent risk
factors for poor PFS in patients with NPC, which is consistent with
previously published results (37).
However, the findings of the current study showed that PFS time in
patients with NPC was not associated with pretreatment plasma
EBV-DNA results, which is inconsistent with the results of other
studies (38,39). We hypothesized that the observed
discrepancy may have been caused by differences in primer fragments
used at the Clinical Laboratory of Fujian Cancer Hospital (Fuzhou,
China) to detect plasma EBV-DNA between 2012 and 2013, and those
used after 2016 (40,41).
The main strengths of the current study include the
following: i) To the best of our knowledge, the study was the first
to simultaneously analyze the three most important functional SNPs
in the XRCC1 gene in patients with NPC, which provides a more
comprehensive understanding of the impact that the XRCC1 gene
exerts in these patients; ii) prognostic follow-up examinations
were carried out over a period of 7–8 years, which is notably
longer than the follow-up duration adopted in a number of previous
studies (36,42), thus the present study provided
strong evidence for clarifying the relationship between these SNPs
and prognosis; and iii) the choice of radiotherapy modality and the
specific types of acute radiation-induced injuries were
deliberately restricted to minimize potential sources of bias in
the study design.
Nonetheless, several limitations of the present
study should be acknowledged: i) The study was a single-center,
retrospective study, and selection biases could not be avoided; ii)
the size of the patient cohort and the total number of included
events are relatively small. This limitation is particularly
noticeable within specific subgroups, such as patients with distant
metastases, where the total number of cases is limited. This may
result in ‘sparse data bias’ in some subgroups, consequently
limiting statistical power (43,44);
iii) in view of the limited clinical data collected ≥10 years ago,
not all patients had complete records of relevant
clinicopathological, including the absence of descriptions of
smoking status, drinking habits and oral hygiene (45), which might have influenced the
accuracy of the multivariate analysis results; and iv) due to the
limited number of cases, the present study did not stratify
patients with cancer who received RT alone from those undergoing
combined radiochemotherapy regimens. The use of chemotherapeutic
drugs often exacerbates the severity of acute radiation-induced
injuries (46,47), which also might have introduced bias
into the analysis.
Previous studies have demonstrated that, in addition
to the SNPs in the DNA damage repair gene mentioned in the present
study, numerous other gene SNPs are also involved in the process of
radiation damage and repair, including SNPs in genes associated
with angiogenesis (48), autophagy
(49), the Wnt/β-catenin pathway
(50), and the cell cycle and NF-κB
pathway (51), suggesting that the
mechanism of acute radiation-induced injury may be influenced by
the combined effect of multiple SNP pathways. In radiogenomics, an
essential focal point revolves around the creation of risk scores
based on combinations of SNPs within genes associated with
radiation-induced signaling pathways. These risk scores are
primarily aimed at predicting the likelihood of experiencing acute
or delayed radiation-induced injuries, serving as valuable guidance
for informing clinical decision-making (52). Within these risk scores, each SNP is
assigned individualized weights, which are determined by their
respective significance. Therefore, an in-depth elucidation of the
relationship between each SNP and radiation damage is important for
overall risk assessment.
In summary, the current study suggested that the
Arg194Trp, Arg280His and Arg399Gln SNPs in the XRCC1 gene of the
DNA damage repair pathway cannot independently predict the severity
of acute radiation-induced injury and prognosis in patients with
NPC treated with IMRT. It may be necessary to expand the study
across multiple medical centers, and increase the sample size
through a prospective cohort study, or combine data with that of
other related gene SNPs to build a prediction model, so as to
explore the relationship between gene-related factors and RT
efficacy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fujian Provincial Health
Technology Project (grant no. 2020GGA016) and the National Natural
Science Foundation of China (grant no. 81972717).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YHZ and YC conceived the study idea and designed the
experiments. YHZ, JYG and XQX performed the experiments. JFZ, YSC
and TZL collected and analyzed the data. YHZ wrote the first draft
of the manuscript. YC supervised the study and commented on
previous versions of the manuscript. YHZ and YC confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in the present study
involving biological samples/medical record information/data were
approved by the Ethics Committee of Fujian Cancer Hospital
(approval no. K2021-046-01; Fuzhou, China). Written informed
consent was obtained from all adult participants, as well as from
the parents or guardians of minors included in the study. All
methods were performed in accordance with the relevant guidelines
and regulations (Declaration of Helsinki).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EBV
|
Epstein-Barr virus
|
|
HR
|
hazard ratio
|
|
IMRT
|
intensity-modulated radiation
therapy
|
|
NPC
|
nasopharyngeal carcinoma
|
|
OR
|
odds ratio
|
|
PFS
|
progression-free survival
|
|
RT
|
radiation therapy
|
|
XRCC1
|
X-ray repair cross-complementing
group-1
|
References
|
1
|
World Health Organization:
4-Nasopharynx-fact-sheet. Global Cancer Observatory. 2020.
|
|
2
|
Lee AW, Ma BB, Ng WT and Chan AT:
Management of nasopharyngeal carcinoma: Current practice and future
perspective. J Clin Oncol. 33:3356–3364. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen YP, Chan ATC, Le QT, Blanchard P, Sun
Y and Ma J: Nasopharyngeal carcinoma. Lancet. 394:64–80. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma Z, Umezawa R, Yamamoto T, Ishikawa Y,
Takahashi N, Takeda K, Suzuki Y, Tang L, Ito K, Kadoya N and Jingu
K: IMRT improves local control in patients with NPC compared with
conventional radiotherapy: Propensity score-matched analysis. Jpn J
Clin Oncol. 51:1444–1451. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yeh SA, Hwang TZ, Wang CC, Yang CC, Lien
CF, Wang CC, Hsu TY, Hsu RF, Shih YC, Huang YC, et al: Outcomes of
patients with nasopharyngeal carcinoma treated with
intensity-modulated radiotherapy. J Radiat Res. 62:438–447. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen L, Zhang Y, Lai SZ, Li WF, Hu WH, Sun
R, Liu LZ, Zhang F, Peng H, Du XJ, et al: 10-Year results of
therapeutic ratio by Intensity-Modulated radiotherapy versus
Two-Dimensional radiotherapy in patients with Nasopharyngeal
Carcinoma. Oncologist. 24:e38–e45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kouloulias V, Thalassinou S, Platoni K,
Zygogianni A, Kouvaris J, Antypas C, Efstathopoulos E and Nikolaos
K: The treatment outcome and radiation-induced toxicity for
patients with head and neck carcinoma in the IMRT era: A systematic
review with dosimetric and clinical parameters. Biomed Res Int.
2013:4012612013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao Z and Cheng B: Morbidity in patients
with nasopharyngeal carcinoma and Radiation-induced skin lesions:
Cause, risk factors, and dermatitis evolution and severity. Adv
Skin Wound Care. 34:1–8. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bossi P, Cossu Rocca M, Corvo R, Depenni
R, Guardamagna V, Marinangeli F, Micciche F and Trippa F: The
vicious circle of treatment-induced toxicities in locally advanced
head and neck cancer and the impact on treatment intensity. Crit
Rev Oncol Hematol. 116:82–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang XL, Zhou GQ, Lin L, Zhang LL, Chen
FP, Lv JW, Kou J, Wen DW, Ma J, Sun Y, et al: Prognostic value of
radiation interruption in different periods for nasopharyngeal
carcinoma patients in the intensity-modulated RT era. Cancer Med.
10:143–155. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yao JJ, Zhang F, Gao TS, Zhang WJ,
Lawrence WR, Zhu BT, Zhou GQ, Ma J, Wang SY and Sun Y: Survival
impact of radiotherapy interruption in nasopharyngeal carcinoma in
the intensity-modulated radiotherapy era: A big-data intelligence
platform-based analysis. Radiother Oncol. 132:178–187. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Applegate KE, Ruhm W, Wojcik A,
Bourguignon M, Brenner A, Hamasaki K, Imai T, Imaizumi M, Imaoka T,
Kakinuma S, et al: Individual response of humans to ionising
radiation: Governing factors and importance for radiological
protection. Radiat Environ Biophys. 59:185–209. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wardill HR, Sonis ST, Blijlevens NMA, Van
Sebille YZA, Ciorba MA, Loeffen EAH, Cheng KKF, Bossi P, Porcello
L, Castillo DA, et al: Prediction of mucositis risk secondary to
cancer therapy: A systematic review of current evidence and call to
action. Support Care Cancer. 28:5059–5073. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang DW, Wang TM, Zhang JB, Li XZ, He YQ,
Xiao R, Xue WQ, Zheng XH, Zhang PF, Zhang SD, et al: Genome-wide
association study identifies genetic susceptibility loci and
pathways of radiation-induced acute oral mucositis. J Transl Med.
18:2242020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brothwell MRS, West CM, Dunning AM, Burnet
NG and Barnett GC: Radiogenomics in the era of advanced
radiotherapy. Clin Oncol (R Coll Radiol). 31:319–325. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu J, Bi K, Yang R, Li H, Nikitaki Z and
Chang L: Role of DNA damage and repair in radiation cancer therapy:
A current update and a look to the future. Int J Radiat Biol.
96:1329–1338. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
London RE: XRCC1-Strategies for
coordinating and assembling a versatile DNA damage response. DNA
Repair (Amst). 93:1029172020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo Z, Shu Y, Zhou H, Zhang W and Wang H:
Radiogenomics helps to achieve personalized therapy by evaluating
patient responses to radiation treatment. Carcinogenesis.
36:307–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xi T, Jones IM and Mohrenweiser HW: Many
amino acid substitution variants identified in DNA repair genes
during human population screenings are predicted to impact protein
function. Genomics. 83:970–979. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rosen EM, Fan S, Goldberg ID and Rockwell
S: Biological basis of radiation sensitivity. Part 2: Cellular and
molecular determinants of radiosensitivity. Oncology. 14:741–757.
2000.PubMed/NCBI
|
|
21
|
Zhao J, Zhi Z, Zhang M, Li Q, Li J, Wang X
and Ma C: Predictive value of single nucleotide polymorphisms in
XRCC1 for radiation-induced normal tissue toxicity. Onco Targets
Ther. 11:3901–3918. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin J, Ye Q, Wang Y, Wang Y and Zeng Y:
Association between XRCC1 single-nucleotide polymorphisms and
susceptibility to nasopharyngeal carcinoma: An update
meta-analysis. Medicine (Baltimore). 97:e118522018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang J, Li L, Yin X, Wu F, Shen J, Peng Y,
Liu Y, Sun Y, Lu H and Zhang Y: The association between gene
polymorphisms and risk of nasopharyngeal carcinoma. Med Oncol.
32:3982015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang SQ, Pan SM, Liang SX, Han YS, Chen
HB and Li JC: Research status and prospects of biomarkers for
nasopharyngeal carcinoma in the era of highthroughput omics
(Review). Int J Oncol. 58:92021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siak PY, Khoo AS, Leong CO, Hoh BP and
Cheah SC: Current status and future perspectives about molecular
biomarkers of nasopharyngeal carcinoma. Cancers (Basel).
13:34902021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mor V, Laliberte L, Morris JN and Wiemann
M: The karnofsky performance status scale. An examination of its
reliability and validity in a research setting. Cancer.
53:2002–2007. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin S, Pan J, Han L, Guo Q, Hu C, Zong J,
Zhang X and Lu JJ: Update report of nasopharyngeal carcinoma
treated with reduced-volume intensity-modulated RT and hypothesis
of the optimal margin. Radiother Oncol. 110:385–389. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cox JD, Stetz J and Pajak TF: Toxicity
criteria of the radiation therapy oncology group (RTOG) and the
european organization for research and treatment of cancer (EORTC).
Int J Radiat Oncol Biol Phys. 31:1341–1346. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. Seventh
Edition. New York; Springer: 2010
|
|
30
|
Jiqian Fang: Health Statistics. People's
Medical Publishing House; Beijing: pp. 150–171. 2012
|
|
31
|
Prescott RJ: Two-tailed significance tests
for 2×2 contingency tables: What is the alternative? Stat Med.
38:4264–4269. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Guo C, Gong X, Ao F, Huang Y,
Huang L, Tang Y, Jiang C, Xie X, Dong Q, et al: The impacts of
genetic polymorphisms in genes of base excision repair pathway on
the efficacy and acute toxicities of (chemo)radiotherapy in
patients with nasopharyngeal carcinoma. Oncotarget. 8:78633–78641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhai XM, Hu QC, Gu K, Wang JP, Zhang JN
and Wu YW: Significance of XRCC1 Codon399 polymorphisms in Chinese
patients with locally advanced nasopharyngeal carcinoma treated
with radiation therapy. Asia Pac J Clin Oncol. 12:e125–e132. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen H, Wu M, Li G, Hua L, Chen S and
Huang H: Association between XRCC1 single-nucleotide polymorphism
and acute radiation reaction in patients with nasopharyngeal
carcinoma: A cohort study. Medicine (Baltimore). 96:e82022017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li H, You Y, Lin C, Zheng M, Hong C, Chen
J, Li D, Au WW and Chen Z: XRCC1 codon 399Gln polymorphism is
associated with radiotherapy-induced acute dermatitis and mucositis
in nasopharyngeal carcinoma patients. Radiat Oncol. 8:312013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin H, Xie X, Wang H, Hu J, Liu F, Liu Z,
Zhou J, Zhang Y, Xi X, Hu B, et al: ERCC1 Cys8092Ala and XRCC1
Arg399Gln polymorphisms predict progression-free survival after
curative radiotherapy for nasopharyngeal carcinoma. PLoS One.
9:e1012562014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chiang CL, Guo Q, Ng WT, Lin S, Ma TSW, Xu
Z, Xiao Y, Li J, Lu T, Choi HCW, et al: Prognostic factors for
overall survival in nasopharyngeal cancer and implication for TNM
staging by UICC: A systematic review of the literature. Front
Oncol. 11:7039952021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alami IE, Gihbid A, Charoute H, Khaali W,
Brahim SM, Tawfiq N, Cadi R, Belghmi K, El Mzibri M and Khyatti M:
Prognostic value of Epstein-Barr virus DNA load in nasopharyngeal
carcinoma: A meta-analysis. Pan Afr Med J. 41:62022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qu H, Huang Y, Zhao S, Zhou Y and Lv W:
Prognostic value of Epstein-Barr virus DNA level for nasopharyngeal
carcinoma: A meta-analysis of 8128 cases. Eur Arch
Otorhinolaryngol. 277:9–18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zong J, Zheng Y, Lin C, Chen Y, Chen C,
Pan J and Lin S: The value of plasma EBV DNA in monitoring the
therapeutic effect of nasopharyngeal carcinoma. Chin J Radiat
Oncol. 28:881–885. 2019.
|
|
41
|
Zong JF, Zheng YH, Weng YI, Chen LS, Yun
XU, Chen Y, Pan JJ and Lin SJ: Prognostic Value of Pre-treatment
plasma EBV DNA in nasopharyngeal carcinoma patients treated with
intensity-modulated radiation therapy. J Chin Oncol. 22:810–815.
2016.
|
|
42
|
Nanda SS, Gandhi AK, Rastogi M, Khurana R,
Hadi R, Sahni K, Mishra SP, Srivastava AK, Bhatt MLB and Parmar D:
Evaluation of XRCC1 gene polymorphism as a biomarker in head and
neck cancer patients undergoing chemoradiation therapy. Int J
Radiat Oncol Biol Phys. 101:593–601. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Leopold SS and Porcher R: Editorial:
Sparse-data Bias-What the savvy reader needs to know. Clin Orthop
Relat Res. 476:657–659. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Greenland S, Mansournia MA and Altman DG:
Sparse-data bias: A problem hiding in plain sight. BMJ.
352:i19812016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vera-Llonch M, Oster G, Hagiwara M and
Sonis S: Oral mucositis in patients undergoing radiation treatment
for head and neck carcinoma. Cancer. 106:329–336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang C, Wang F, Min X, Zhang Q, Shen LJ,
Jiang Y and Yan J: Toxicities of chemoradiotherapy and radiotherapy
in nasopharyngeal carcinoma: An updated meta-analysis. J Int Med
Res. 47:2832–2847. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu YC, Chen KH, Liang ZG and Zhu XD: A
systematic review and Meta-Analysis of studies comparing concurrent
chemoradiotherapy with radiotherapy alone in the treatment of stage
II Nasopharyngeal carcinoma. Front Oncol. 12:8436752022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma WL, Liu R, Huang LH, Zou C, Huang J,
Wang J, Chen SJ, Meng XG, Yang JK, Li H, et al: Impact of
polymorphisms in angiogenesis-related genes on clinical outcomes of
radiotherapy in patients with nasopharyngeal carcinoma. Clin Exp
Pharmacol Physiol. 44:539–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang Z and Liu Z: Potentially functional
variants of autophagy-related genes are associated with the
efficacy and toxicity of radiotherapy in patients with
Nasopharyngeal carcinoma. Mol Genet Genomic Med. 7:e10302019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu J, Huang Y, Liu L, Wang J, Yin J, Huang
L, Chen S, Li J, Yuan H, Yang G, et al: Genetic polymorphisms of
Wnt/β-catenin pathway genes are associated with the efficacy and
toxicities of radiotherapy in patients with nasopharyngeal
carcinoma. Oncotarget. 7:82528–82537. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Guo C, Huang Y, Yu J, Liu L, Gong X, Huang
M, Jiang C, Liao Y, Huang L, Yang G and Li J: The impacts of single
nucleotide polymorphisms in genes of cell cycle and NF-kB pathways
on the efficacy and acute toxicities of radiotherapy in patients
with nasopharyngeal carcinoma. Oncotarget. 8:25334–25344. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Andreassen CN, Schack LM, Laursen LV and
Alsner J: Radiogenomics-current status, challenges and future
directions. Cancer Lett. 382:127–136. 2016. View Article : Google Scholar : PubMed/NCBI
|