Introduction
Corticotropin-releasing factor (CRF) is a 41-amino
acid peptide (1), which plays an
essential role in maintaining homeostasis (2). The CRF family of peptides consists of
four members, including CRF and urocortins (UCN1,2,3). The latter
have their amino acid sequence homologous to CRF and also play a
role in the process of homeostasis by modulating, for example, the
cardiovascular, endocrine, immune, and reproductive systems
(3). CRF and UCNs exert their
actions on target cells through the activation of CRF receptors
[Corticotropin-Releasing Factor Receptors (CRFRs) type 1 (CRFR1)
and type 2 (CRFR2)] (3). CRFRs
belong to the G-protein-coupled receptor family, have seven
transmembrane domains, are encoded by two different genes and
exhibit distinct pharmacological properties and selectivity
compared to agonists (3). CRF
family peptides and their receptors are expressed in the central
nervous system and other systems, such as the urogenital system
(4). Activation of CRFRs appears to
have different biological effects on each tissue (5). For example, in the gastrointestinal
system, CRFR1 activation appears to increase gut motility, whereas
CRFR2 activation decreases it (6).
While UCN2 and UCN3 have a selective affinity for CRFR2, UCN1 has a
high affinity for both receptors (7). In addition to its central expression,
UCN1 has also been found in peripheral tissues such as adipose
tissue, heart, thymus, spleen, skin, testis, kidney, adrenal gland
and gastrointestinal tract (8). The
CRF system plays a significant role in the physiology and
pathophysiology of various systems (esophagus, intestine, skin,
etc.). However, very little is known about its role in the
urogenital system, particularly in the bladder (4). CRF, UCN1 and CRFRs have been
identified in the bladder of experimental animals (4,9).
Studies have shown that CRF, UCNs and CRFRs are expressed in both
normal and inflammatory (cystitis) feline bladder mucosa
(urothelium), showing functional differences (4). CRF and CRFR2 also show increased
expression in the bladder of adult mice with
cyclophosphamide-induced chemical cystitis. (9). CRFR1 has also been found in the
bladder of adult mice and even more strongly in various
inflammatory conditions (10).
Recently, a study has shown for the first time the presence of
CRFR1 and CRFR2 peptides in the human bladder, using western blot
and immunochemistry (11). However,
it is not clear what occurs with the expression of UCN1.
Moreover, the expression of CRF family peptides and
their receptors has also been associated with different types of
cancer in humans, playing a possible role in their progression
(12). Indeed, in the urogenital
system, the expression of CRF family genes has been studied in
prostate and renal cancer, where it has been shown that the degree
of growth or apoptosis of cancer cells can be modified by an
agonist or antagonist of CRFRs (13–15).
It is well known that most bladder tumours are of
epithelial origin, up to 99.5% of cases (16), highlighting the importance of
developing cancer therapies, especially local treatments (e.g.
intravesical instillations), in the early stages of the disease.
Theoretically, intravesical administration of an antagonist of a
receptor whose upregulation is associated with bladder cancer could
be applicable. Similarly, administering an agonist of a receptor
whose downregulation was associated with carcinogenesis could
provide a field of investigation. To our knowledge, no reports
about the mRNA expression levels of CRF family genes in bladder
cancer are available. This research aimed to investigate the mRNA
expression of CRF family members in the normal human bladder and
urothelial carcinoma, using quantitative real-time polymerase chain
reaction (qPCR) analysis and compare the expression of the genes
between healthy individuals and cancer patients.
Materials and methods
Study population
Our study protocol was approved by the ethical
committee of the University General Hospital of Heraklion, Greece
(Protocol no. 20/25-07-2018, 804). All patient candidates signed
the informed consent before any intervention under the Helsinki
Declaration (17). From August 2018
to July 2021, we enrolled 43 patients according to our research
protocol. We divided the patients into three groups. Group A
included 14 healthy subjects (control group), group B included 20
patients with bladder cancer, and group C included nine patients
with a history of bladder cancer, from whom we obtained tissue
samples from normal mucosa. Samples from healthy individuals were
obtained during prostatectomy or endoscopic ureterolithotripsy
using cold cup biopsy forceps. Samples were taken only from the
dome and not from the bladder triangle. In group B, tissues were
taken from the surface of the tumour with cold biopsy forceps and
in group C, from normal mucosa away from the neoplasm site before
transurethral resection of the bladder tumour (TURBT). The
exclusion criteria of the study are summarized in Table I. We recorded the patients'
demographic and clinical data, including age, sex, type of surgery,
grade of tumour, t stage of the tumour, and risk of progression in
patients with cancer. The grade was classified according to the
world health organization (WHO) 2004/2016 system, and the risk of
tumour progression was based on the study by Sylvester et al
(18). We excluded two patients
with muscle-invasive bladder cancer and one with a clear cell type
variant from group B.
 | Table I.Exclusion criteria for the study
protocol. |
Table I.
Exclusion criteria for the study
protocol.
| Patients with
cancer | Patients without
cancer (controls) |
|---|
| Age <18 years | Age <18 years |
| History of neurogenic
bladder | History of neurogenic
bladder |
| Urinary tract
infection (sterile urine culture) required preoperatively | Urinary tract
infection (sterile urine culture) required preoperatively |
| History of bladder
catheterization, lithiasis and intravesical treatment (BCG or
chemotherapy) | History of bladder
catheterization or lithiasis |
| History or suspicion
of prostate cancer, renal cancer and other malignancy | History or suspicion
of prostate, renal, bladder cancer and other malignancy |
| Immunosuppression,
immunodepression | Immunosuppression,
immunodepression |
| Pregnancy or
breastfeeding | Pregnancy or
breastfeeding |
| Muscle-invasive
bladder cancer, metastatic disease, second malignancy and other
than typical papillary bladder cancer | Endoscopic findings:
Detrusor hypertrophy, ulcers and inflammatory lesions |
| Participation in
another research protocol | Participation in
another research protocol |
RNA extraction
After the surgical procedure, the samples were
immediately stored at −80°C until used. All the tissues had 2–3 mm
thickness. Total RNA was extracted from the tissues with TRIzol
reagent (Invitrogen Life Technologies), and cDNAs were synthesized
using TAKARA PrimeScript 1st strand cDNA synthesis kit (Takara Bio)
(19). Expression of each gene of
interest was determined using SYBR Green master mix (Kapa
Biosystems) containing a specific set of primers in a final volume
of 10 µl. Amplification conditions included denaturation at 95°C
for 2 min followed by 40 cycles at 95°C for 30 secs and at 60°C for
30 sec. To verify the accuracy of qPCR (melting curves and PCR
products), we ran 1.5–2% agarose gel. All samples were initially
tested for the housekeeping gene glyceraldehyde 3-phosphate
dehydrogenase (GAPDH). Additionally, we used the ribosomal protein
S23 (RPS23) gene, which seemed to be the most suitable and stably
expressed housekeeping gene in bladder samples (20). Therefore, the calculations are done
according to the formula RQ=E^-(min Cq-sample Cq) (21), (where Cq is the cycle threshold and
E the primer's efficiency) based on the geometric mean between the
relative quantities (RQ) of GAPDH and RPS23. Using two reference
genes also balanced the problems of expression variation between
pathological and normal tissue. We performed the experiments in
triplicate to allow for statistical assessment. We also created a
standard curve from 5-point cDNA dilution series finding the slope
and the primer's efficiency.
Primers
Primers for housekeeping and CRF family genes are
shown in Table II.
 | Table II.PCR primers used in the present
study. |
Table II.
PCR primers used in the present
study.
| Gene | Sense primer
(5′-3′) | Antisense primer
(5′-3′) | Size (bp) |
|---|
| CRF |
CAC-CCT-CAG-CCC-TTG-GAT-TTC |
GCC-CTG-GCC-ATT-TCC-AAG-AC | 413 |
| UCN1 |
CAG-GCG-AGC-GGC-CGC-G |
CTT-GCC-CAC-CGA-GTC-GAA-T | 146 |
| UCN2 |
AGA-CCA-CAG-GAC-AGT-AGT-GC |
GTG-AGG-TCA-GGC-GCC-AC | 90 |
| UCN3 |
TGC-TGC-TCC-TGC-TGC-TGC-TC-3 |
GTG-TCC-TGG-CGT-GGC-TTT-CCC-3′ | 310 |
| CRFR1 |
GGC-AGC-AGC-TAG-TGG-TTC-GGC-C |
TCG-CAG-GCA-CCG-GAT-GCT-C | 272 |
| CRFR2 |
ATG-GAC-GCG-GCA-CTG-CTC-CA |
CAC-GGC-CTC-TCC-ACG-AGG-G | 342 |
| RPS23 |
TGG-AGG-TGC-TTC-TCA-TGC-AA |
AAT-GGC-AGA-ATT-TGG-CTG-TTT-G | 76 |
| GAPDH |
CTG-CAC-CAC-CAA-CTG-CTT-AG |
GGG-CCA-TCC-ACA-GTC-TTC | 120 |
Statistical analysis
We used the Kolmogorov-Smirnov test to check the
normality of the data. We compared the relative gene expression
between groups using the Kruskal-Wallis and ANOVA tests followed by
Dunn's-Bonferroni and Tukey HSD post hoc analysis, respectively
(SPSS for Linux, version 23). In addition, we examined the
correlation of patients' data (tumour grade, tumour stage, tumour
progression risk and age) with gene expression using Spearman
analysis. Finally, we performed an ANOVA test regarding patients'
age between the groups. Thus, to the extent possible, we have
excluded potential confounding factors affecting the exposure and
the outcomes. P<0.05 was considered to indicate a statistically
significant difference.
Results
Initially, all tissues were investigated for the
presence of the GAPDH reference gene. Of the 14 normal tissues,
GAPDH expression was observed in 11. Of the remaining 17 cancerous
tissues, GAPDH expression was observed in 13. Finally, of the nine
patients with a history of bladder cancer, the expression was
observed in 8. In addition, we decided to study the more suitable
and stably expressed housekeeping gene, RPS23 (20). The main characteristics of the
patients per group who were studied are presented in Table III. UCN1 mRNA expression was
identified in 10 of 11 samples in group A, 12 of 13 in group B and
in all samples (n=8) from group C. The average Cq value ± standard
deviation (SD) of UCN1 mRNA expression in groups A, B, and C was
24.86±1.01, 28.64±2.42, and 28.94±2.75, respectively. The product
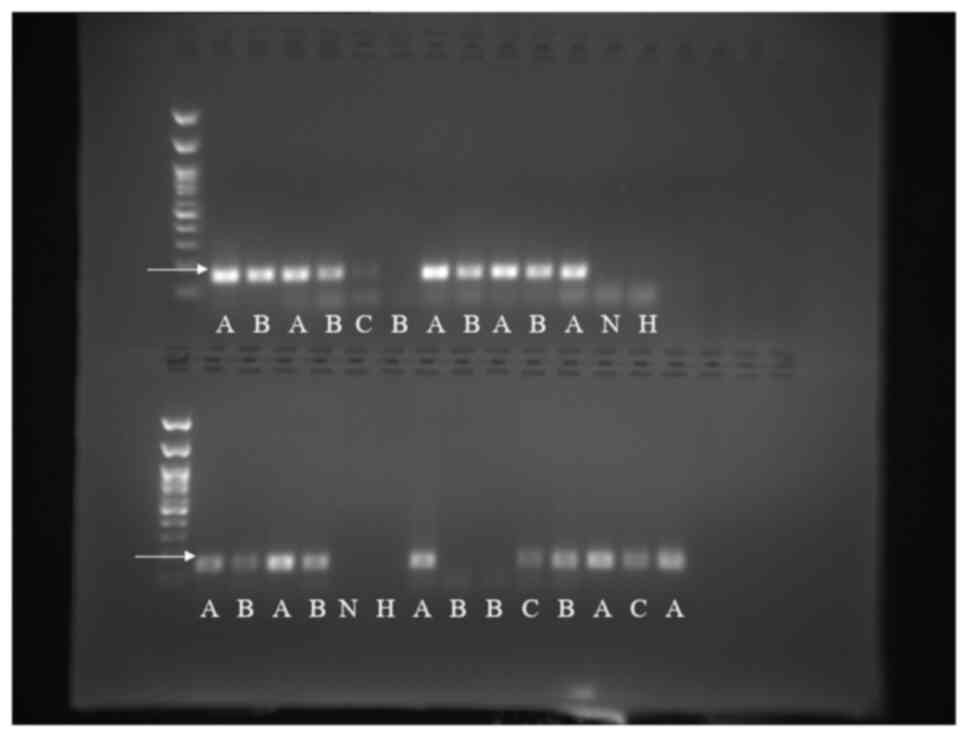
of qPCR, UCN1, was selectively confirmed by running agarose gel
(Fig. 1). Subsequently, statistical
analysis and comparison of relative UCN1 mRNA expression levels
between groups were performed using the Kruskal-Wallis test as the
levels did not follow a normal distribution (Kolmogorov-Smirnov
test, P<0.001). UCN1 mRNA expression levels were statistically
lower in group B compared to A (test statistics 11.717, mean rank
A=24.3, mean rank B=12.58, P=0.006) and statistically lower in
group C compared to A (test statistics 15.425, mean rank C=8.88,
P=0.001). Between groups B and C, there was no difference (P=1.000)
(Fig. 2A, B). The ANOVA test showed
no differences in the age of the patients between the groups
(P=0.112, f=2.38). Finally, in the Spearman's correlation test, the
relative UCN1 mRNA expression and the average raw Cq values did not
seem to relate to the age and sex of the patients, to the grade and
the T stage of tumour or to the risk of cancer progression (All
correlations showed P>0.05).
 | Table III.Main characteristics of the patients
per group. |
Table III.
Main characteristics of the patients
per group.
| A, Group A |
|---|
|
|---|
| Samples | Age (years) | Sex | T stage | Gradea | Riskb | Surgery |
|---|
| 1 | 65 |
Female | - | - | - | URS |
| 2 | 45 | Male | - | - | - | URS |
| 3 | 61 |
Female | - | - | - | URS |
| 4 | 79 | Male | - | - | - | TURP |
| 5 | 79 | Male | - | - | - | TURP |
| 6 | 73 | Male | - | - | - | URS |
| 7 | 80 | Male | - | - | - | TURP |
| 8 | 61 | Male | - | - | - | TURP |
| 9 | 70 | Male | - | - | - | URS |
| 10 | 65 | Male | - | - | - | URS |
| 11 | 41 | Male | - | - | - | URS |
|
| B, Group
B |
|
| Samples | Age
(years) | Sex | T stage |
Gradea |
Riskb | Surgery |
|
| 1 | 79 | Male | T1 | High | High | TURBT |
| 2 | 78 | Male | Ta | Low | Low | TURBT |
| 3 | 40 | Male | Ta | High | Low | TURBT |
| 4 | 56 | Male | T1 | High | Intermediate | TURBT |
| 5 | 67 | Male | Ta | High | Intermediate | TURBT |
| 6 | 74 | Male | Ta | Low | Low | TURBT |
| 7 | 74 | Male | T1 | High | High | TURBT |
| 8 | 64 | Male | T1 | High | Intermediate | TURBT |
| 9 | 87 | Male | T1 | High | High | TURBT |
| 10 | 51 | Male | Ta | High | Intermediate | TURBT |
| 11 | 70 | Male | Ta | High | Intermediate | TURBT |
| 12 | 77 | Male | Ta | Low | Low | TURBT |
| 13 | 72 | Male | Ta | High | Low | TURBT |
|
| C, Group
C |
|
| Samples | Age
(years) | Sex | T stage |
Gradea |
Riskb | Surgery |
|
| 1 | 79 | Male | Ta | High | Intermediate | TURBT |
| 2 | 77 | Male | Ta | High | Intermediate | TURBT |
| 3 | 65 | Male | Ta | High | Intermediate | TURBT |
| 4 | 84 | Male | T1 | High | High | TURBT |
| 5 | 81 |
Female | T1 | High | High | TURBT |
| 6 | 71 | Male | Ta | High | Intermediate | TURBT |
| 7 | 81 | Male | T1 | High | High | TURBT |
| 8 | 85 | Male | Ta | High | Intermediate | TURBT |
Regarding UCN3 mRNA expression, although the average
raw Cq values were higher in group A (26.29±1.83 in group A and
24.44±1.94 in group B), no statistically significant difference was
observed (Kruskal-Wallis test for relative UCN3 mRNA expression,
P=1.000). UCN3 mRNA expression was identified in four of eight
samples screened in group A, seven of twelve in group B and all
(n=8) from group C.
Regarding CRF, UCN2, CRFR1 and CRFR2 mRNAs, no
significant difference was observed between the groups
(Kruskal-Wallis test P-value for CRF and UCN2 0.429, and 0.514
respectively, ANOVA P-value for CRFR1, and CRFR2 0.523 and 0.640,
respectively). Overall, the expression of all CRF family genes was
identified in most samples (from 50 to 96.77%). The average raw Cq
values ± SD and the percentage of mRNA detection per group and
total are presented in Table
IV.
 | Table IV.Average means raw Cq values and the
percentage of gene detection in groups A, B, C and total. |
Table IV.
Average means raw Cq values and the
percentage of gene detection in groups A, B, C and total.
|
| Group A | Group B | Group C | Total
population |
|---|
|
|
|
|
|
|
|---|
| Gene | Average mean Cq
value ± SD value | Detection of the
gene (%) | Average mean Cq
value ± SD | Detection of the
gene (%) | Average mean Cq
value ± SD | Detection of the
gene (%) | Detection of the
gene (%) |
|---|
| CRF | 32.84±2.96 | 8/8 (100) | 30.56±4.01 | 7/9 (77.78) | 32.65±3.94 | 6/7 (85.7) | 21/24 (87.5) |
| UCN1 | 24.86±1.01 | 10/11 (90.9) | 28.64±2.42 | 12/13 (92.3) | 28.94±2.75 | 8/8 (100) | 30/32 (93.75) |
| UCN2 | 30.96±2.22 | 10/11 (90.9) | 29.72±2.2 | 12/12 (100) | 29.89±2.31 | 8/8 (100) | 30/31 (96.77) |
| UCN3 | 26.29±1.83 | 4/8 (50) | 24.44±1.94 | 7/12 (58.3) | 27.26±1.43 | 8/8 (100) | 19/28 (67.9) |
| CRFR1 | 35.08±4.68 | 2/7 (28.57) | 30.22±3.62 | 7/9 (77.78) | 32.82 | 1/2 (50) | 10/18 (55.56) |
| CRFR2 | 32.86±4.08 | 4/7 (57.14) | 31.47±4.36 | 3/9 (33.33) | 31.29±0.61 | 2/2 (100) | 9/18 (50) |
Discussion
Several studies have shown that the expression of
CRF family genes is involved in either the development or
progression of cancer (12).
Therefore, investigating their presence in different organs, such
as the bladder, is of great interest. The present study shows that
the mRNAs of the three UCNs, CRF and the two CRF receptors are
expressed in the normal human bladder. The second finding was that
the CFR family mRNAs are expressed in the samples received by the
bladder of cancer patients. Several cancer-related studies have
shown that UCN1 is expressed in the following human tissues:
gastric adenocarcinoma, pancreatic adenocarcinoma, liver carcinoma,
endometrial carcinoma, renal clear cell carcinoma and prostate
adenocarcinoma (22). It is also
expressed in the following cell lines: glioblastoma, pituitary
adenoma, malignant melanoma, insulinoma, pheochromocytoma, thyroid
carcinoma, adrenal carcinoma and breast cancer (22). However, to the extent of our
knowledge, this is the first study showing the expression of these
genes in urothelial cancer tissues.
We also found that UCN1 mRNA levels were
downregulated in tissues taken from bladder cancer compared to the
tissues received from the healthy group. Similar results were found
in the group of bladder cancer patients where samples were taken
from normal urothelium.
Downregulation of UCN1 mRNA expression in cancerous
and potentially cancerous tissues does not necessarily represent a
cause-effect relationship. It could be the effect rather than the
cause of carcinogenesis or a chance event. If the change in UCN1
expression results from urothelial cancer, it could be investigated
further as a biomarker. As a chance event, it could be presented
due to the non-study of possible confounding and interactive
factors related to the patient group. For example, patients'
exposure to certain chemicals could cause alterations in UCN1
expression in parallel and independently of carcinogenesis. Similar
results to our study, i.e. reduced expression of UCN1, have been
reported in endometrial cancer (23). The decreased expression of UCN1 in
endometrial carcinoma could occur through the activation of
estrogen receptor a (23).
Additional data showed that cell proliferation in endometrial
carcinoma was inhibited through the activation of the cAMP-protein
kinase A (PKA) pathway by CRF/UCN1 (24). In another study, UCN1 inhibited
differentiation in melanoma cell lines by regulating intracellular
Ca2+ homeostasis (25).
Furthermore, UCN1 controlled hepatocellular cell carcinoma
migration by decreasing the expression of the calcium-independent
phospholipase A2 enzyme (iPLA2) (23). Finally, UCN1 could inhibit
carcinogenesis by the cessation of angiogenesis (26). It is well known that UCN1 has a
strong affinity for both receptors (CRFR1 and CRFR2) (27), which could explain the conflicting
results of several cancer-related studies. Thus, activation of
CRFR1 by UCN1 could promote anticancer effects related to the PKA
pathway or intracellular Ca2+ signaling (24,25).
On the other hand, activation of CRFR2 may favor
cancer migration (23). Activating
CRFR2 by UCN1 could inhibit angiogenesis (26), one of the significant mechanisms in
oncogenesis. Also, the reduction in the expression of iPLA2, which
inhibits hepatocellular carcinoma migration, occurs via the
activation of CRFR2 (23).
All the mentioned mechanisms seem to be relevant to
bladder cancer. In particular, activation of the PKA pathway
appears to control cancer invasion by modulating
Microtubule-associated protein 4 (MAP4) (28). Goto and Miyamoto studied the
association of estrogen receptors in the pathogenesis of urothelial
carcinoma by analyzing their possible involvement. Thus, bladder
cancer could be associated with the endocrine system (29). Furthermore, there is evidence that
intracellular Ca2+ homeostasis plays a role in bladder
cancer (30). About iPLA2, the
observation of Cai et al was representative as its activity
was elevated in bladder cancer, while, as mentioned before, its
decrease controls cancer migration (31). Finally, angiogenesis seems to be
involved in the progression and recurrence of bladder cancer
(32).
The notable limitations of our work were the small
number of samples and the inability to use a second method, such as
western blot and immunohistochemistry, to confirm the specific
cells expressing UCN1. Unfortunately, several samples were either
inadequate or inappropriate due to the lack of expression of the
housekeeping gene. In addition, the exclusion criteria and the
invasive nature of our study made collecting large numbers of
tissues difficult. As for the second limitation, the experiments
were repeated three times to be as accurate as possible. Regarding
the cells expressing UCN1, we consider that the samples did not
include muscle layers as they were taken with cold biopsy forceps
(tissue thickness 2 to 3 mm). Also, the normal samples were taken
from the bladder dome. As regards the cancerous samples, the biopsy
was superficial from the tumour margin. As it is known in the
literature, only the lamina propria in the dome region can be thick
up to 3.1 mm (33). Also, the
muscularis mucosae layer is mainly located in the dome and
constitutes the firm boundary before the muscle layer (33).
Our study nature is mostly exploratory. Thus, our
results put the basis for future studies addressing the
pathophysiology of bladder cancer related to CRF family genes. In
our ongoing research, we could investigate whether the
administration of a UCN1 affects the progression and migration of
urothelial cancer cells in vitro. Also, a larger number of samples
would probably result in a statistically significant difference in
the expression of the other genes of the CRF family as UCN3, which
may be expressed to a greater extent in cancer samples. Finally,
whether altered gene expression would contribute to the prognosis
or diagnosis of urothelial cancer as a biomarker should be
examined.
In summary, our novel findings show that some of the
mRNAs of the CRF gene family are likely expressed in human bladder
cancer. Although no specific pathogenic mechanism was investigated,
UCN1 gene downregulation seems to be associated with urothelial
carcinoma. Further studies with a larger sample number could
investigate the relationship of CRF genes with cancer development,
invasion, progression and migration and their use as
biomarkers.
Acknowledgements
The authors would like to thank Mrs D. Pantartzi,
Scientific Secretary of the Department of Urology, University
General Hospital of Heraklion, Medical School, University of Crete
(Heraklion, Crete, Greece) for the administrative and technical
support.
Funding
This research project was supported by the Special Account for
Research Funds of University of Crete (SARF UoC grant no.
3550).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CMav, CMam, MV and GL conceptualized the project. MV
and ED designed the experiments. CMav, MD, ED and MV performed the
experiments. CMav, ED, MV, GL and CMam contributed to data
acquisition, analysis and interpretation. MV, GL and CMam provided
the resources. CMav and CMam performed the biopsies. CMav, ED, MD
and MV supplied the computer software used in the study. CMav wrote
the original draft. MV, MD, ED, GL, CMav and CMam reviewed, edited
and wrote the final draft. All authors read and approved the final
version of the manuscript. MV and ED confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
This study was approved by the University General
Hospital of Heraklion, Greece (approval no. 20/25-07-2018, 804), as
a research project. All patient candidates signed the informed
consent before any intervention under the Helsinki Declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vale W, Spiess J, Rivier C and Rivier J:
Characterization of a 41-residue ovine hypothalamic peptide that
stimulates secretion of corticotropin and beta-endorphin. Science.
213:1394–1397. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sakamoto R, Matsubara E, Nomura M, Wang L,
Kawahara Y, Yanase T, Nawata H and Takayanagi R: Roles for
corticotropin-releasing factor receptor type 1 in energy
homeostasis in mice. Metabolism. 62:1739–1748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grammatopoulos DK: Insights into
mechanisms of corticotropin-releasing hormone receptor signal
transduction. Br J Pharmacol. 166:85–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanna-Mitchell AT, Wolf-Johnston A,
Roppolo JR, Buffington TC and Birder LA: Corticotropin-releasing
factor family peptide signaling in feline bladder urothelial cells.
J Endocrinol. 222:113–121. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hillhouse EW and Grammatopoulos DK: The
molecular mechanisms underlying the regulation of the biological
activity of corticotropin-releasing hormone receptors: Implications
for physiology and pathophysiology. Endocr Rev. 27:260–286. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nozu T, Tsuchiya Y, Kumei S, Takakusaki K
and Okumura T: Peripheral corticotropin-releasing factor (CRF)
induces stimulation of gastric contractions in freely moving
conscious rats: Role of CRF receptor types 1 and 2.
Neurogastroenterol Motil. 25:190–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma S, Shen Q, Zhao LH, Mao C, Zhou XE,
Shen DD, de Waal PW, Bi P, Li C, Jiang Y, et al: Molecular basis
for hormone recognition and activation of corticotropin-releasing
factor receptors. Mol Cell. 77:669–680.e664. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fekete EM and Zorrilla EP: Physiology,
pharmacology, and therapeutic relevance of urocortins in mammals:
Ancient CRF paralogs. Front Neuroendocrinol. 28:1–27. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
LaBerge J, Malley SE, Zvarova K and
Vizzard MA: Expression of corticotropin-releasing factor and CRF
receptors in micturition pathways after cyclophosphamide-induced
cystitis. Am J Physiol Regul Integr Comp Physiol. 291:R692–R703.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saban MR, Nguyen NB, Hammond TG and Saban
R: Gene expression profiling of mouse bladder inflammatory
responses to LPS, substance P, and antigen-stimulation. Am J
Pathol. 160:2095–2110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jhang JF, Birder LA, Jiang YH, Hsu YH, Ho
HC and Kuo HC: Dysregulation of bladder corticotropin-releasing
hormone receptor in the pathogenesis of human interstitial
cystitis/bladder pain syndrome. Sci Rep. 9:191692019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaprara A, Pazaitou-Panayiotou K,
Kortsaris A and Chatzaki E: The corticotropin releasing factor
system in cancer: Expression and pathophysiological implications.
Cell Mol Life Sci. 67:1293–1306. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tezval H, Jurk S, Atschekzei F, Becker JU,
Jahn O, Serth J and Kuczyk MA: Urocortin and
corticotropin-releasing factor receptor 2 in human renal cell
carcinoma: Disruption of an endogenous inhibitor of angiogenesis
and proliferation. World J Urol. 27:825–830. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tezval H, Jurk S, Atschekzei F, Serth J,
Kuczyk MA and Merseburger AS: The involvement of altered
corticotropin releasing factor receptor 2 expression in prostate
cancer due to alteration of anti-angiogenic signaling pathways.
Prostate. 69:443–448. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin L, Zhang Q, Guo R, Wang L, Wang J, Wan
R, Zhang R, Xu Y and Li S: Different effects of
corticotropin-releasing factor and urocortin 2 on apoptosis of
prostate cancer cells in vitro. J Mol Endocrinol. 47:219–227. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dahm P and Gschwend JE: Malignant
non-urothelial neoplasms of the urinary bladder: A review. Eur
Urol. 44:672–681. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
World Medical Association: World Medical
Association Declaration of Helsinki: ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sylvester RJ, Rodríguez O, Hernández V,
Turturica D, Bauerová L, Bruins HM, Bründl J, van der Kwast TH,
Brisuda A, Rubio-Briones J, et al: European association of urology
(EAU) prognostic factor risk groups for non-muscle-invasive bladder
cancer (NMIBC) incorporating the WHO 2004/2016 and WHO 1973
classification systems for grade: An update from the EAU NMIBC
guidelines panel. Eur Urol. 79:480–488. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poulaki S, Rassouli O, Liapakis G,
Gravanis A and Venihaki M: Analgesic and anti-inflammatory effects
of the synthetic neurosteroid analogue BNN27 during CFA-induced
hyperalgesia. Biomedicines. 9:11852021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C, Wang YQ, Jin G, Wu S, Cui J and
Wang RF: Selection of reference genes for gene expression studies
in human bladder cancer using SYBR-Green quantitative polymerase
chain reaction. Oncol Lett. 14:6001–6011. 2017.PubMed/NCBI
|
|
21
|
De Spiegelaere W, Dern-Wieloch J, Weigel
R, Schumacher V, Schorle H, Nettersheim D, Bergmann M, Brehm R,
Kliesch S, Vandekerckhove L and Fink C: Reference gene validation
for RT-qPCR, a note on different available software packages. PLoS
One. 10:e01225152015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ikeda K, Akiyoshi K, Kamada M, Fujioka K,
Tojo K and Manome Y: Expression of urocortin I in normal tissues
and malignant tumors. Cancer Cell & Microenvironment. 1:45–50.
2014.PubMed/NCBI
|
|
23
|
Owens GL, Lawrence KM, Jackson TR, Crosbie
EJ, Sayan BS, Kitchener HC and Townsend PA: Urocortin suppresses
endometrial cancer cell migration via CRFR2 and its system
components are differentially modulated by estrogen. Cancer Med.
6:408–415. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Graziani G, Tentori L, Portarena I,
Barbarino M, Tringali G, Pozzoli G and Navarra P: CRH inhibits cell
growth of human endometrial adenocarcinoma cells via CRH-receptor
1-mediated activation of cAMP-PKA pathway. Endocrinology.
143:807–813. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carlson KW, Nawy SS, Wei ET, Sadée W,
Filov VA, Rezsova VV, Slominski A and Quillan JM: Inhibition of
mouse melanoma cell proliferation by corticotropin-releasing
hormone and its analogs. Anticancer Res. 21:1173–1179.
2001.PubMed/NCBI
|
|
26
|
Wang J, Xu Y, Xu Y, Zhu H, Zhang R, Zhang
G and Li S: Urocortin's inhibition of tumor growth and angiogenesis
in hepatocellular carcinoma via corticotrophin-releasing factor
receptor 2. Cancer Invest. 26:359–368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reyes TM, Lewis K, Perrin MH, Kunitake KS,
Vaughan J, Arias CA, Hogenesch JB, Gulyas J, Rivier J, Vale WW and
Sawchenko PE: Urocortin II: A member of the corticotropin-releasing
factor (CRF) neuropeptide family that is selectively bound by type
2 CRF receptors. Proc Natl Acad Sci USA. 98:2843–2848. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ou Y, Zheng X, Gao Y, Shu M, Leng T, Li Y,
Yin W, Zhu W, Huang Y, Zhou Y, et al: Activation of cyclic AMP/PKA
pathway inhibits bladder cancer cell invasion by targeting
MAP4-dependent microtubule dynamics. Urol Oncol. 32:47.e21–48.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goto T and Miyamoto H: The role of
estrogen receptors in urothelial cancer. Front Endocrinol
(Lausanne). 12:6438702021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stewart TA, Yapa KT and Monteith GR:
Altered calcium signaling in cancer cells. Biochim Biophys Acta.
1848:2502–2511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cai H, Chiorean EG, Chiorean MV, Rex DK,
Robb BW, Hahn NM, Liu Z, Loehrer PJ, Harrison ML and Xu Y: Elevated
phospholipase A2 activities in plasma samples from multiple
cancers. PLoS One. 8:e570812013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kopparapu PK, Boorjian SA, Robinson BD,
Downes M, Gudas LJ, Mongan NP and Persson JL: Expression of VEGF
and its receptors VEGFR1/VEGFR2 is associated with invasiveness of
bladder cancer. Anticancer Res. 33:2381–2390. 2013.PubMed/NCBI
|
|
33
|
Paner GP, Ro JY, Wojcik EM, Venkataraman
G, Datta MW and Amin MB: Further characterization of the muscle
layers and lamina propria of the urinary bladder by systematic
histologic mapping: Implications for pathologic staging of invasive
urothelial carcinoma. Am J Surg Pathol. 31:1420–1429. 2007.
View Article : Google Scholar : PubMed/NCBI
|