Introduction
Cervical cancer is one of the most common
gynecological tumors and its incidence ranks fourth worldwide in
gynecological tumors (1). Although
its mortality rate has been declining in recent years, its
morbidity and the number of overall cancer-related deaths are on
the rise. There are still >500,000 new cases and >250,000
cancer-related deaths worldwide each year, and the 5-year survival
rate is only 64.2% (2). In
addition, given that certain women have exceeded the recommended
age for the human papillomavirus (HPV) vaccine, and not all cases
of cervical cancer are attributed to HPV infection, certain
patients still experience a progressive development into advanced
cancer (3). Therefore, it is
necessary to further explore the mechanism of cervical cancer and
identify novel therapeutic targets.
Chromobox homolog 7 (CBX7) belongs to the polycomb
repressive complex 1 (PRC1) family (4). The expression of CBX7 in tumors
varies, and it plays several functions in tumor occurrence and
development. The level of CBX7 is low in some tumors such as breast
cancer, bladder cancer, colon cancer and glioma, indicating that it
may be a tumor suppressor gene (5–8). In
vitro and in vivo experiments have revealed that
overexpression of CBX7 could inhibit the progression of glioma
(9,10). In pancreatic cancer, CBX7 was
revealed to negatively regulate the survival, drug resistance and
migration of pancreatic cancer cells (11). In lung cancer, CBX7 was demonstrated
to enhance the sensitivity of lung cancer cells to chemotherapeutic
drugs (12). However, in gastric
cancer, lymphoma and prostate, and ovarian cancer CBX7 is an
oncogene (13–16). For example, Ni et al
(13) found that CBX7
overexpression in gastric cancer cells promotes tumor progression.
However, the effect of CBX7 on cervical cancer remains unclear.
Gene ontology map of 44 genes revealed 13
physiologically important cellular pathways, such as integrin
(ITG), transforming growth factor β1 (TGFβ1),
phosphatidylinositol-3-kinase (PI3K), cadherin and other signaling
pathways (17). Rapisarda et
al revealed that downregulation of CBX7 expression promoted the
expression of ITG, which further activated TGFβ (18). TGFβ1 can reduce the expression of
E-cadherin (E-cad) and induce epithelial-mesenchymal transition
(EMT) (19,20). The integrin β3 (ITGβ3)/AKT signaling
pathway can promote the growth of platelet-induced
hemangioendothelioma (21). Ni
et al determined that the mechanism of CBX7 underlying its
inhibitory effect on cell growth may involve the AKT signaling
pathway (11). A previous study by
the authors also demonstrated that low CBX7 expression was
positively correlated with poor prognosis in patients with cervical
cancer (22). By overexpressing
CBX7, it was also revealed that CBX7 promoted apoptosis in cervical
cancer cells (23).
In the present study, the effects of CBX7 on the
metastasis and apoptosis of cervical cancer cells as well as the
underlying mechanisms were further studied.
Materials and methods
Subjects and human cervical cancer
samples
A total of 120 patients with cervical cancer
admitted to the First Affiliated Hospital of Xinjiang Medical
University (Urumqi, China) from January 2018 to Dec 2019 were
enrolled in the present study. The patients were aged from 28 to 72
years (mean, 56.88±0.92 years). All patients were diagnosed using
histolopathological examination of tissue samples by experts in
gynecological pathology. The tumor samples were obtained during
high resolution digital colposcopy, cervical colonization and
hysterectomy. The matching adjacent non-tumorous tissues were
collected and used as controls. Prior written and informed consent
was obtained from every patient and the study was approved
(approval no. 20120220-01) by the Ethics Review Board of Xinjiang
Medical University (Urumqi, China).
Immunohistochemistry
The expression levels of CBX7, ITGβ3, TGFβ1, PI3K,
AKT, phosphorylated (p)-AKT, vimentin (VIM) and E-cad in the tissue
were determined by immunohistochemistry, using a rabbit polymer
method detection system (cat. no. PV6001; OriGene Technologies,
Inc.). The tissue samples were fixed with 4% paraformaldehyde for
24 h at room temperature, dehydrated with graded alcohol series,
embedded in paraffin, and cut into 4-µm serial sections. Tissue
slides (4 µm) were then deparaffinized at 60°C, followed by
treatment with 100% xylene for 20 min and rehydration in graded
series of ethanol at room temperature. Followingly, the sections
were incubated in 3% H2O2 (cat. no. 1301;
Dezhou Dexinkang Disinfection Products Co., Ltd.) at room
temperature for 20 min. The antigen retrieval was performed with
0.01 M citrate buffer at 95°C for 10 min. After blocking with
normal sheep serum (1:100; cat. no. SAP-9100; OriGene Technologies,
Inc.) at room temperature for 30 min, the section was incubated
with anti-CBX7 (1:200; cat. no. ab21873; Abcam), anti-ITGβ3 (1:500;
cat. no. ab7166; Abcam), anti-TGFβ1 (1:200; cat. no. BA0290; Boster
Biological Technology; http://www.boster.com.cn/index/products/productslist?catid=all&keywords=BA0290),
anti-PI3K (1:100; cat. no. E-AB-22165; Elabscience Biotechnology,
Inc.), anti-AKT (1:1,000; cat. no. ab8805; Abcam), p-AKT (1:100;
cat. no. 4060; CST Biological Reagents Co., Ltd.), anti-E-cad
(1:500; cat. no. ab40772; Abcam) and anti-VIM (1:300; cat. no.
ab92547; Abcam) monoclonal antibodies, respectively, at 4°C
overnight. The section was then incubated with HRP-conjugated
anti-rabbit En Vision system (1:200; cat. no. PV-6001; OriGene
Technologies, Inc.) at 37°C for 20 min, followed by staining for
3–5 min with diaminobenzidine tetrahydrochloride (cat. no.
ZLI-9018; OriGene Technologies, Inc.). The sections were then
counterstained with hematoxylin for 20 s at room temperature.
Finally, the sections were observed under an IX71 inverted light
microscope (Magnificantion: 200×; Olympus, Tokyo, Japan).
Immunohistochemistry was assessed using a semiquantitative
approach, whereby the intensity of staining and the percentage of
positive cells were combined. The staining intensity was
categorized as absent (0 points), weak (1 point), moderate (2
points) or strong (3 points). The mean percentage of positively
stained cells was scored as follows: i) 0 points (<25%), ii) 1
point (25–50%), iii) 2 points (51–75%) and iv) 3 points (76–100%).
The total score was calculated by multiplying the staining
intensity and the percentage of positive cells, ranging from 0 to
9. The scores ≤1 were classified as negative expression, while
scores >1 were classified as positive expression.
Cell lines
HeLa (cat. no. ZQ0068) and SiHa (cat. no. ZQ0129)
cells were purchased from Shanghai Zhong Qiao Xin Zhou
Biotechnology Co., Ltd. These cell lines were incubated in DMEM
(cat. no. 11965-092; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (cat. no. 10091-148;
Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml of streptomycin (30-002-CI; Cellgro; Corning, Inc.). The
cells were maintained at 37°C and 95% humidity in a 5%
CO2 atmosphere. Once the cells reached 80–90%
confluency, they were subcultured.
Establishment of stable CBX7 knockdown
in cervical cancer cells
To stably knockdown CBX7 expression in cervical
cancer cells, four CBX7-specific siRNAs were designed and
synthesized by Beijing CorreGene Biotechnology Co., Ltd. Briefly,
the plasmids, including LentiCRTM-spKO-CBX7-sg1 (10 µg),
LentiCRTM-spKO-CBX7-sg2 (10 µg), LentiCRTM-spKO-CBX7-sg3 (10 µg),
LentiCRTM-spKO-CBX7-sg4 (10 µg), and LentiCRISPR E (10 µg), as well
as the auxiliary plasmids including psPAX2 (10 µg) and pVSVG (5 µg)
were packaged into lentiviruses and co-transfected into 293T cells
using LipofiterTM (Invitrogen; 11668-019). The culture
medium was changed after 18 h. The transfection was performed for
48 h. On days 3, 4 and 5, the lentiviruses were collected and
centrifuged at 4°C, 2,000 × g for 10 min. After removing cell
debris, the supernatant was collected and centrifuged at 4°C, 9,000
× g for 120 min. Next, low-passage SiHa and HeLa cells were seeded
in 10-cm culture dishes and infected with LentiCRTM-spKO-CBX7-sg1,
LentiCRTM-spKO-CBX7-sg2, LentiCRTM-spKO-CBX7-sg3,
LentiCRTM-spKO-CBX7-sg4 and LentiCRISPR E viruses for 24 h at 37°C
and 5% CO2 at a multiplicity of infection of 30. The
control cells were not infected. The cells were then incubated
overnight at 37°C and 5% CO2. At 24–48 h after
infection, cells were cultured in the medium containing 1 µg/ml of
puromycin. Every day, the DMEM containing 1 µg/ml puromycin was
replaced. After 7 days, the resistant cells were collected and
seeded into 25 cm2 culture flasks for further
cultivation. The transduction efficiency was confirmed using 24 h
after transduction. The method of constructing HeLa cell CBX7
gene-knockout cell lines is the same as that for SiHa cells. The
knockdown efficiency of CBX7-shRNA lentivirus was determined using
western blot analysis. LentiCRTM-spKO-CBX7-sg1
(GGAGCCAGAAGAGCACATCT) and LentiCRTM-spKO-CBX7-sg4
(gcTGCCGAGTGGGCACGTCA) were selected as the lentiviruses with the
optimal knockdown efficiency for HeLa and SiHa, respectively. HeLa
and SiHa cells were divided into 3 groups: the SiCBX7 group
(transfected with LentiCRTM-spKO-CBX7-sg), the SiCBX7-EVC group
(transfected with LentiCRISPR E), and the control (CTRL) group
(non-transfected cells).
Knockdown of TGFβ
TGFβ-specific shRNAs (TGFβ1-HOMO-875,
TGFβ1-HOMO-1158, and TGFβ1-HOMO-1408) and control shRNA were
designed and synthesized by Beijing Kerui Biotechnology Co., Ltd.
shRNA sequences used for the knockdown of TGFβ: TGFβ1-HOMO-875
forward, 5′-GCUACCGCUGCUGUGGCUATT-3′ and reverse,
5′-UAGCCACAGCAGCGGUAGCTT-3′; TGFβ1-HOMO-1158 forward,
5′-GAGGUCACCCGCGUGCUAATT' and reverse, 5′-UUAGCACGCGGGUGACCUCTT-3′
and TGFβ1-HOMO-1408 forward, 5′-GCGACUCGCCAGAGUGGUUTT' and reverse,
5′-AACCACUCUGGCGAGUCGCTT-3′. The sequence of control shRNA was
forward, 5′-UUCUCCGAACGUGUCACGUTT-3 and reverse,
5′ACGUGACACGUUCGGAGAATT-3. The transfection of control shRNA or
TGFβ shRNAs into SiHa and HeLa cells was carried out using
Lipofectamine 2000 transfection reagent (cat. no. 11668-019;
Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature.
Briefly, cells in the logarithmic phase of growth and in good
condition were used. The shRNAs (4 µg) and Lipofectamine 2000 (5
µl) were separately diluted in a 100 µl serum-free medium and
incubated at room temperature for 20 min. They were then mixed and
incubated with cells at 37°C and 5% CO2. After
incubation for 5 h, the culture medium was replaced with a complete
medium containing 10% fetal bovine serum, and the cells were
further cultured for 48 h. The RT-qPCR determined the TGFβ shRNA
with the optimal knockdown efficiency (24). The cells were divided into 6 groups:
siCBX7 + NC, siCBX7-EVC + NC, CTRL + NC group, siCBX7 + shRNATGFβ1,
siCBX7-EVC + shRNATGFβ1 and CTRL+ shRNATGFβ1 group.
MTT viability assay
HeLa and SiHa cells with knockdown of CBX7 or TGFβ1
(1.0×104 cells/well) were plated in 96 well plates. At
the following timepoints: 0, 12, 24, 48, 72 and 96 h, 20 µl of MTT
(cat. no. BS186; Biosharp Life Sciences) was added and incubated at
37°C for 4 h. The cells were then treated with 150 µl of DMSO (cat
no. D8370; Beijing Solarbio Science & Technology Co., Ltd.) for
10 min under agitation in a light-protected environment. The
absorbance was subsequently measured at 570 nm using a microplate
reader from Thermo Fisher Scientific, Inc.
Wound healing assay
HeLa and SiHa cells with stable knockdown of CBX7 or
TGFβ1 (5×105 cells/well) were seeded in six-well plates
and placed in a 37°C cell culture incubator. The cells were
cultured until cell confluence reached >90%. A wound was then
created vertically using a 1-ml pipette tip. After washing with
PBS, cells were incubated with fresh DMEM without FBS. The wound
was photographed at 0, 24 and 48 h of culture using an IX71
inverted light microscope (magnification, ×100×, Olympus
Corporation). The distance of the wound was measuredusing Image J
software (version 1.50; National Institutes of Health), and the
results were expressed as the percentage of the remaining wound
area. This assay was performed in triplicate.
Cell migration assay
Cell migration ability was analyzed using a
Transwell chamber (cat. no. 353097; Corning, Inc.). After
serum-starved culture overnight, a total of 1×104 HeLa
and SiHa cells with stable knockdown of CBX7 or TGFβ1 were plated
in the upper chambers of Transwell plates in 100 µl of serum-free
medium. The lower chamber was filled with a medium containing 10%
fetal bovine serum, which served as the chemoattractant. Following
incubation at 37°C for 24 h, cells that did not migrate through the
pores were carefully wiped out with cotton wool. The migratory
cells were fixed with pure methanol for 30 min, stained with 0.1%
crystal violet for 20 min at room temperature, and observed with an
IX71 inverted light microscope (Olympus Corporation).
Cell apoptotic analysis by flow
cytometry
Cell apoptotic analysis was performed using an
Annexin-V/PI assay kit (cat. no. KGA1026; Nanjing KeyGen Biotech
Co., Ltd.). Briefly, HeLa and SiHa cells (1×10*6/ml)
with stable knockdown of CBX7 or TGFβ1 were cultured for 24 h, the
cells were collected and washed. Next, the cells were stained with
Annexin-V-FITC/PI at room temperature in the dark for 10~15 min.
Finally, the samples were analyzed on a Cyto FLEX flow cytometer
(Beckman Coulter, Inc.). The data were analyzed using FlowJo v10
(FlowJo LLC).
Animals
Female BALB/c nude mice (age, 4–6 weeks old,
weighing 18±2g, n=36) were obtained from Beijing Vital River
Laboratory Animal Technology Co., Ltd. They were housed under
strict pathogen-free conditions with a temperature of 20–24°C, a
relative humidity of 50%, and a 12-h light-dark cycle. All mice
were provided food and water ad libitum. All animal experiment
procedures were approved (approval no. 20120220-01) by the Ethics
Committee of Xinjiang Medical University.
Xenograft tumor model
SiHa control cells and SiHa cells with stable
transfection of siCBX7-EVC and siCBX7 (5×106 cells in
100 µl PBS) were injected subcutaneously into the right flank of
each nude mouse (7 weeks old, n=12/group). The tumors were measured
every three days using a vernier caliper and the volumes of tumors
were calculated using the following formula: Volume
(mm3)=length × width2 × 0.5. After 30 days of
tumor formation, tumor measurements were conducted every three
days. On day 45 post-inoculation, the mice were euthanized by
intraperitoneal injection of 60 mg/kg pentobarbital, followed by
rapid cervical dislocation. To confirm death, the absence of
breathing and nerve reflexes were used as criteria. The tumors were
dissected and examined using a stereoscopic microscope.
Western blotting
Total protein was were extracted from HeLa and SiHa
cells (with stable knockdown of CBX7 or TGFβ1 knockdown) and tumor
tissues using RIPA lysis buffer (cat. no. 20188; Sigma-Aldrich;
Merck KGaA). The protein concentration was determined using a BCA
Protein Assay kit. Proteins (50 µg/lane) were then subjected to 10%
SDS-PAGE electrophoresis and PVDF membrane transfer. The membranes
were blocked with 5% skimmed milk and 0.1% Tween-20 in
Tris-buffered saline for 2 h at room temperature. After blocking,
the membrane was incubated with the corresponding primary
antibodies at 4°C overnight. The primary antibodies included
anti-TGFβ1 (1:300; BA0290; Boster Biological Technology), anti-CBX7
(1:500 dilution; cat. no. ab21837; Abcam), anti-vimentin (1:2,000;
cat. no. 10366-1-AP; Proteintech Group, Inc.), anti-p-AKT (1:2,000;
cat. no. 4060; CST Biological Reagents Co., Ltd.), anti-PI3K p85
(1:2,000; cat. no. bs-3332R; BIOSS), anti-AKT1 (1:1,000; cat. no.
ab8805; Abcam), anti-ITGβ3 (1:1,000; cat. no. ab7166; Abcam),
anti-E-cad (1:1,000; cat. no. ab40772; Abcam) and anti-β-actin
(1:500; cat. no. BM0627; Boster Biological Technology). Finally,
the membranes were incubated with horseradish peroxidase-conjugated
secondary antibody (1:10,000; cat. no. BA1054; Boster Biological
Technology) for 2 h at room temperature. The signal was visualized
using ECL western blotting detection reagent (cat. no. PE0010;
Beijing Solarbio Science & Technology Co., Ltd.). The blots
were analyzed with a gel imaging analysis system (Bio-Rad
Laboratories, Inc.). Relative protein expresion levels were
quantified using Quantity One 1-D software (version 4.62; Bio-Rad
Laboratories, Inc.).
RT-qPCR
Total RNA was extracted from cells and tumor tissues
using Trizol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and then reverse transcribed into cDNA using a
High Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.). The reaction was conducted at 42°C for 18 min
and 98°C for 5 min. The levels of mRNA were examined using the SYBR
Green Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.).
The primer sequences are presented in Table I. The thermocycling conditions were
as follows: Initial denaturation at 50°C for 2 min, 95°C for 10
min, 95°C for 30 sec and 60°C for 30 sec for 40 cycles. The
internal control was β-actin. The relative mRNA level was
calculated with the 2ΔΔCq method (24).
 | Table I.Primer sequences of reverse
transcription-quantitative PCR. |
Table I.
Primer sequences of reverse
transcription-quantitative PCR.
| Gene | Primer | Sequence | Size |
|---|
| Homo β-actin | Forward |
5′-AGCGAGCATCCCCCAAAGTT-3′ | 285 bp |
|
| Reverse |
5′-GGGCACGAAGGCTCATCATT-3′ |
|
| Homo TGFβ1 | Forward |
5′-CAGCAACAATTCCTGGCGATACCT-3′ | 140 bp |
|
| Reverse |
5′-CGCTAAGGCGAAAGCCCTCAAT-3′ |
|
| Homo ITGβ3 | Forward |
5′-TGGGGCTGATGACTGAGAAG-3′ | 206 bp |
|
| Reverse |
5′-ACGCACTTCCAGCTCTACTT-3′ |
|
| Homo AKT | Forward |
5′-ACACCAGGTATTTTGATGAGGA-3′ | 143 bp |
|
| Reverse |
5′-TCAGGCCGTGCCGCTGGCCGAGTAG-3′ |
|
| Homo VIM | Forward |
5′-TGAGTACCGGAGACAGGTGCAG-3′ | 119 bp |
|
| Reverse |
5′-TAGCAGCTTCAACGGCAAAGTTC-3′ |
|
| Homo CBX7 | Forward |
5′-CGGAAGAAGCGCGTGCGGAAGGGT-3′ | 372 bp |
|
| Reverse |
5′-GCGGAGCGGGAAGGGCAGGGTGGG-3′ |
|
| Homo PI3K | Forward |
5′-GACTCAAAAAGGTGTTCGG-3′ | 311 bp |
|
| Reverse |
5′-ACAAGTTATAGGGCTCGGC-3′ |
|
| Homo E-cad | Forward |
5′-ACGCATTGCCACATACACT-3′ | 149 bp |
|
| Reverse |
5′-CCATGACAGACCCCTTAAA-3′ |
|
Statistical analysis
All statistical analysis was performed using SPSS
21.0 software (IBM Corp.) and GraphPad software (v.7.0; GraphPad
Software; Dotmatics). Data are presented as the mean ± SD of three
independent experiments. The differences in the ITGβ3, TGFβ1, PI3K,
AKT, p-AKT, VIM and E-cad expression levels between the cervical
cancer tissues and adjacent non-tumorous cervical tissues were
analyzed by the χ2 test. The correlation of contingency
paired data of CBX7 with other indexes was analyzed by φ)
coefficient analysis. One-way ANOVA followed by Tukey's post hoc
test or χ2 method were used for multiple groups or
pairwise comparison of multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
CBX7 knockdown promotes the
proliferation and metastasis but inhibits the apoptosis of cervical
cancer cells
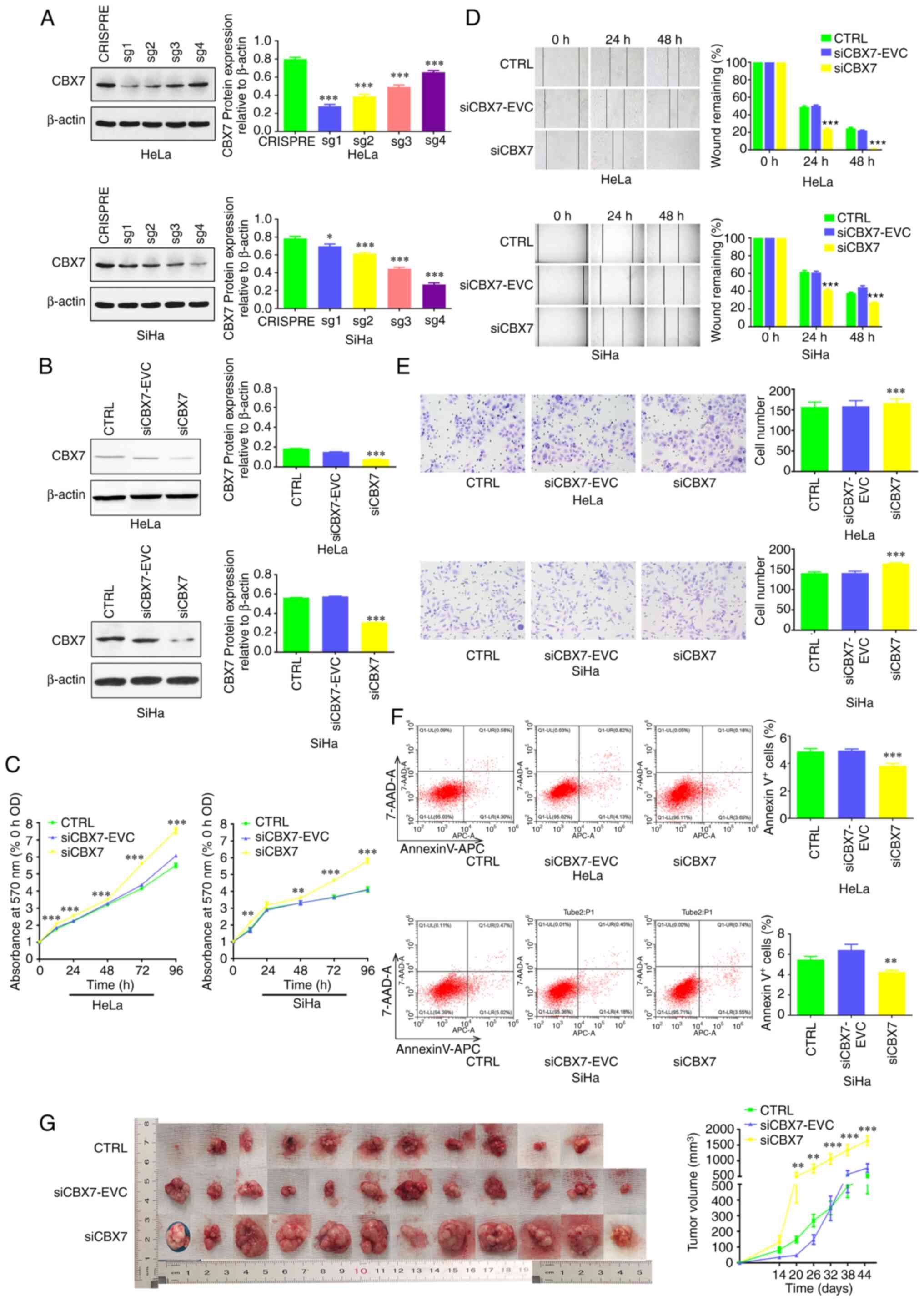
CBX7 was first knocked down in cervical cancer cell
lines. The results demonstrated that clones of HeLa-CBX7sg1 and
SiHa-CBX7sg4 had the lowest CBX7 expression (Fig. 1A). These clones were used in the
subsequent experiments. CBX7 expression in these clones was further
verified by western blot analysis, which revealed that CBX7 was
successfully knocked down (Fig.
1B). Subsequently, to determine the effects of CBX7 on cell
proliferation, an MTT assay was performed. The results showed that
the OD570 of the siCBX7 group significantly increased
compared with that of the siCBX7-EVC and CTRL groups, indicating
that CBX7 knockdown resulted in significantly increased
proliferation (P<0.05; Fig. 1C).
The wound healing assay revealed that the wound area of the siCBX7
group was significantly smaller than the other two groups at 48 h,
indicating that downregulation of CBX7 significantly promoted cell
migration (P<0.05; Fig. 1D).
Next, a Transwell assay was performed and revealed that cell
migration was also significantly increased by CBX7 knockdown. There
was a significant increase in the number of migrated cells in the
siCBX7 group (P<0.001; Fig. 1E).
Furthermore, flow cytometry revealed that the proportion of
apoptotic cells in the siCBX7 group was significantly lower than
that in the siCBX7-EVC and CTRL groups (P<0.05; Fig. 1F). Additionally, a xenograft tumor
model was subsequently established by injecting infected siCBX7
SiHa cells into nude mice to investigate whether knockdown of CBX7
exerts similar growth-promoting effects in vivo. As revealed
in Fig. 1G, one nude mouse in the
control group did not develop tumors, while the tumor incidence
rate for the other two groups was 100%. The largest tumor was
observed in the siCBX7 group, with a tumor volume of 1,812.54
mm3. Statistically, the tumor volume was increased in
the siCBX7 group compared with that in the siCBX7-EVC and CTRL
(P<0.05; Fig. 1G), demonstrating
that the downregulation of CBX7 may promote xenograft tumor growth
in nude mice.
Effect of CBX7 on the expression of
key genes in the ITGβ3/TGFβ1 signaling pathway and EMT markers
Immunohistochemistry was performed to detect the
protein expression of CBX7, ITGβ3, TGFβ1, PI3K, AKT, p-AKT, VIM and
E-cad in cancer tissues from humans and mice. As illustrated in
Fig. 2A and Table II, the expression of CBX7 and E-cad
in human cervical cancer tissues was significantly lower, whereas
the expression of ITGβ3, TGFβ1, PI3K, AKT, p-AKT and VIM was
significantly higher than those of adjacent tissues (P<0.05).
Correlation analysis demonstrated that CBX7 and E-cad expression
were positively correlated (φ=0.362; P<0.001); however, CBX7 was
negatively correlated with other proteins (all P<0.05; Table III). In xenograft tumor tissues of
mice, knockdown of CBX7 increased the level of VIM and decreased
expression of E-cad significantly (Fig.
2B; Table IV; P<0.01). In
addition, RT-qPCR revealed that CBX7 knockdown significantly
increased VIM, ITGβ3, TGFβ1, PI3K and AKT mRNA
levels, and reduced E-cad mRNA expression in xenograft tumor
tissues (P<0.05) (Fig. 2C).
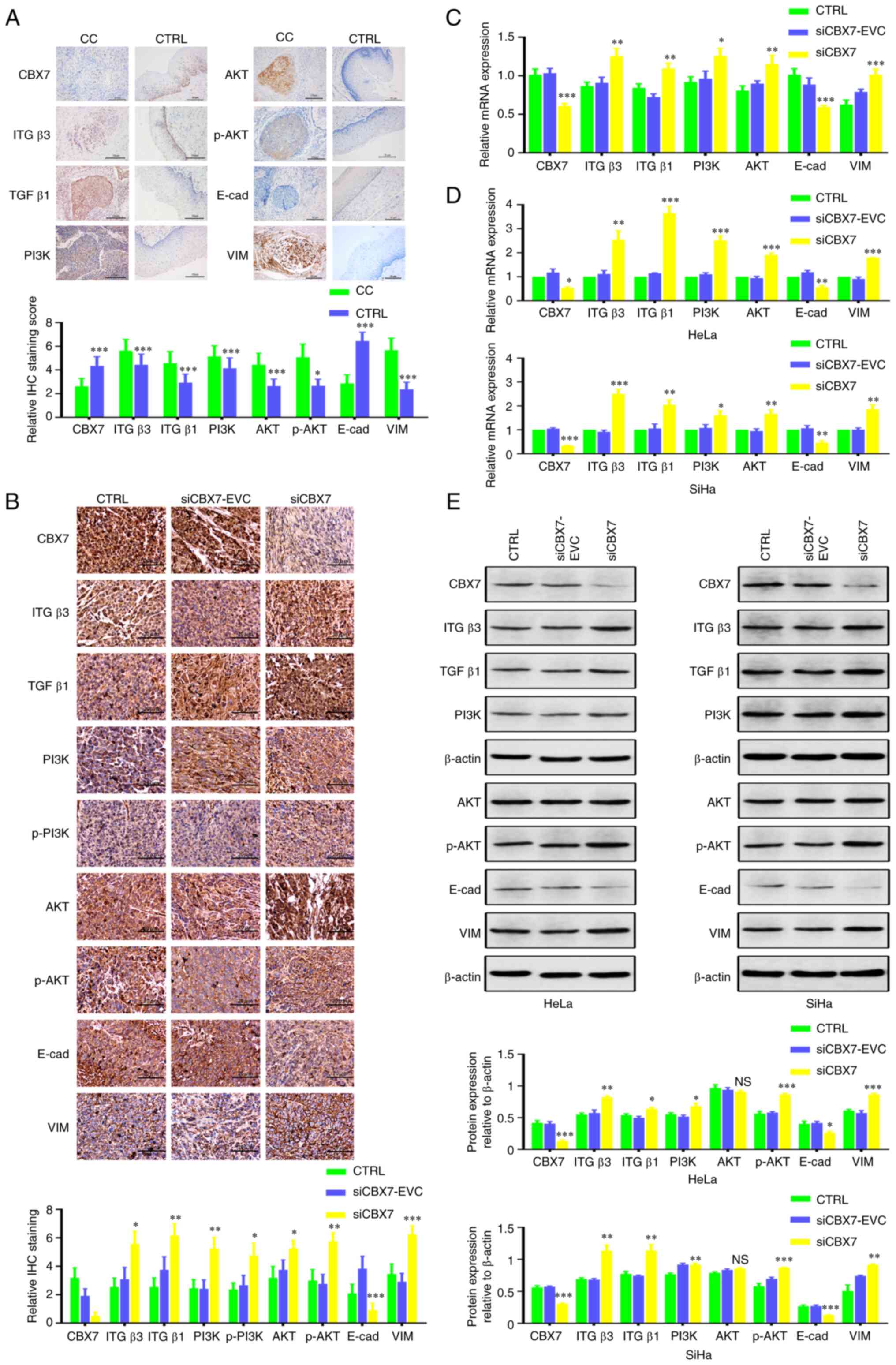 | Figure 2.Effect of CBX7 on the expression of
ITGβ3, TGFβ1, PI3K, AKT, p-AKT, VIM, and E-cad. (A) Representative
images and statistical analysis of IHC detection of CBX7, ITGβ3,
TGFβ1, PI3K, AKT, p-AKT, E-cad and VIM protein expression in CC and
CTRL tissues (magnificantion: 200×). (B) Representative images and
statistical analysis of IHC detection of CBX7, ITGβ3, TGFβ1, PI3K,
AKT, p-AKT, E-cad and VIM protein in transplanted tumor tissues
(Magnificantion: 200×). (C) The effect of CBX7 on CBX7, ITGβ3,
TGFβ1, PI3K, AKT, p-AKT, E-cad and VIM mRNA in nude
mouse-transplanted tumor tissues by reverse
transcription-quantitative PCR. (D) The effect of CBX7 on CBX7,
ITGβ3, TGFβ1, PI3K, AKT, p-AKT, E-cad and VIM mRNA in
HeLa and SiHa cells. (E) Expression of CBX7, ITGβ3, TGFβ1, PI3K,
AKT, p-AKT, E-cad and VIM protein in HeLa and SiHa cells. Error
bars in all panels represent the mean ± SD. *P<0.05, **P<0.01
and ***P<0.001 compared with the control and siCBX7-EVC. CBX7,
chromobox protein homolog 7; ITGβ3, integrin β3; TGFβ1,
transforming growth factor β1; PI3K, phosphatidylinositol-3-kinase;
p-, phosphorylated; VIM, vimentin; E-cad, E-cadherin; CC, cervical
cancer; CTRL, control; IHC, immunohistochemistry. |
 | Table II.Expression of CBX7, ITGβ3, TGFβ1,
PI3K, AKT, p-AKT, E-cad and VIM in cervical cancer and adjacent
non-tumorous cervical tissues (N, %). |
Table II.
Expression of CBX7, ITGβ3, TGFβ1,
PI3K, AKT, p-AKT, E-cad and VIM in cervical cancer and adjacent
non-tumorous cervical tissues (N, %).
|
| CBX7 | ITGβ3 | TGFβ1 | PI3K |
|---|
|
|
|
|
|
|
|---|
| Groups | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive |
|---|
| Cervical cancer
tissue | 71 (59.2) | 49 (40.8) | 51 (42.5) | 69 (57.5) | 18 (15.0) | 102 (85.0) | 29 (24.2) | 91 (75.8) |
| Adjacent
non-tumorous cervical tissues | 51 (42.5) | 69 (57.5) | 97 (80.8) | 23 (19.2) | 60 (50.0) | 60 (50.0) | 66 (55.0) | 54 (45.0) |
| χ2 | 6.018 |
| 37.297 |
| 33.504 |
| 23.852 |
|
| P-value | 0.014 |
| <0.001 |
| <0.001 |
| <0.001 |
|
|
|
| AKT | p-AKT | E-cad | VIM |
|
|
|
|
|
|
| Groups |
Negative |
Positive |
Negative |
Positive |
Negative |
Positive |
Negative |
Positive |
|
| CC | 59 (49.2) | 61 (50.8) | 56 (46.7) | 64 (53.3) | 89 (74.2) | 31 (25.8) | 27 (22.5) | 93 (77.5) |
| CTRL | 87 (72.5) | 33 (27.5) | 66 (60) | 44 (40) | 8 (6.2) | 122 (93.8) | 102 (85.0) | 18 (15.0) |
| χ2 | 13.71 |
| 4.096 |
| 118.710 |
| 101.950 |
|
| P-value | <0.001 |
| 0.043 |
| <0.001 |
| <0.001 |
|
 | Table III.Correlation of CBX7 with ITGβ3,
TGFβ1, PI3K, AKT, p-AKT, E-cad and VIM in cervical cancer tissues
(N, %). |
Table III.
Correlation of CBX7 with ITGβ3,
TGFβ1, PI3K, AKT, p-AKT, E-cad and VIM in cervical cancer tissues
(N, %).
|
| ITGβ3 frequency
(%) |
| TGFβ1 frequency
(%) |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| CBX7 | Negative | Positive | Total | Negative | Positive | Total |
|
|
|
|---|
| Negative | 21 (17.5) | 50 (41.7) | 71 | 5 (4.2) | 66 (55.0) | 71 |
|
|
|
| Positive | 30 (25.0) | 19 (15.8) | 49 | 13 (10.8) | 36 (30.0) | 49 |
|
|
|
| Total no. | 51 | 69 | 120 | 18 | 102 | 120 |
|
|
|
| Phi (φ) | −0.315 |
|
| −0.268 |
|
|
|
|
|
| P-value | 0.001 |
|
| 0.003 |
|
|
|
|
|
|
|
| PI3K frequency
(%) |
| AKT frequency
(%) |
| p-AKT frequency
(%) |
|
|
|
|
|
|
|
|
|
| CBX7 |
Negative |
Positive | Total |
Negative |
Positive | Total |
Negative |
Positive | Total |
|
| Negative | 6 (5.0) | 65 (54.2) | 71 | 26 (21.7) | 45 (37.5) | 71 | 13 (10.8) | 58 (48.3) | 71 |
| Positive | 23 (19.2) | 26 (21.7) | 49 | 33 (27.5) | 8 (13.3) | 49 | 43 (35.8) | 6 (5.0) | 49 |
| Total no. | 29 | 91 | 120 | 59 | 61 | 120 | 56 | 64 | 120 |
| Phi (φ | −0.442 |
|
| −0.302 |
|
|
| −0.684 |
|
| P-value | <0.001 |
|
| 0.001 |
|
|
| <0.001 |
|
|
|
| E-cad frequency
(%) |
| VIM frequency
(%) |
|
|
|
|
|
|
|
|
|
|
|
|
|
| CBX7 |
Negative |
Positive | Total |
Negative |
Positive | Total |
|
|
|
|
| Negative | 62 (51.7) | 9 (7.5) | 71 | 6 (5.0) | 65 (52.4) | 71 |
|
|
|
| Positive | 27 (22.5) | 22 (18.3) | 49 | 21 (17.5) | 28 (23.3) | 49 |
|
|
|
| Total no. | 89 | 31 | 120 | 27 | 93 | 120 |
|
|
|
| φ) | 0.362 |
|
| −0.405 |
|
|
|
|
|
| P-value | <0.001 |
|
| <0.001 |
|
|
|
|
|
 | Table IV.Comparison of positive expression
rates of key genes in the ITGβ3/TGFβ1 signaling pathway and EMT
markers in three groups of nude mice with xenograft tumors (N,
%). |
Table IV.
Comparison of positive expression
rates of key genes in the ITGβ3/TGFβ1 signaling pathway and EMT
markers in three groups of nude mice with xenograft tumors (N,
%).
|
|
| CBX7 | ITGβ3 | TGFβ1 | PI3K | p-PI3K |
|---|
|
|
|
|
|
|
|
|
|---|
| Groups | N | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive |
|---|
| CTRL | 11 | 4 (36.4) | 7 (73.6) | 4 (36.4) | 7 (73.6) | 3 (27.3) | 8 (72.7) | 4 (36.4) | 7 (73.6) | 3 (27.3) | 8 (72.7) |
| siCBX7-EVC | 12 | 6 (50.0) | 6 (50.0) | 5 (28.6) | 7 (71.4) | 5 (28.6) | 7 (71.4) | 5 (28.6) | 7 (71.4) | 5 (41.7) | 7 (58.3) |
| siCBX7 | 12 | 11 (91.7) | 1 (8.3) | 1 (8.3) | 11 (91.7) | 2 (16.7) | 10 (83.3) | 2 (16.7) | 10 (83.3) | 3 (25.0) | 9 (75.0) |
| χ2 |
| 4.216 |
| 4.274 |
| 1.874 |
| 2.038 |
| 0.886 |
|
| P-value |
| 0.040 |
| 0.118 |
| 0.392 |
| 0.383 |
| 0.642 |
|
|
|
|
| AKT | p-AKT | E-cad | VIM |
|
|
|
|
|
|
|
|
|
| Groups | N |
Negative |
Positive |
Negative |
Positive |
Negative |
Positive |
Negative |
|
Positive |
|
|
| CTRL | 11 | 3 (27.3) | 8 (72.7) | 4 (36.4) | 7 (73.6) | 6 (45.5) | 6 (54.5) | 3 (27.3) |
| 8 (72.7) |
|
| siCBX7-EVC | 12 | 3 (25.0) | 9 (75.0) | 5 (28.6) | 7 (71.4) | 4 (33.3) | 8 (66.7) | 4 (33.3) |
| 8 (66.7) |
|
| siCBX7 | 12 | 1 (8.3) | 11 (91.7) | 1 (8.3) | 11 (91.7) | 11 (91.7) | 1 (8.3) | 0 (0.0) |
| 12 (100.0) |
|
| χ2 |
| 1.757 |
| 4.274 |
| 10.485 |
| 6.861 |
|
|
|
| P-value |
| 0.415 |
| 0.118 |
| 0.005 |
| 0.032 |
|
|
|
Similar results were obtained in stable cells with
knockdown of CBX7 (Fig. 2D). In
HeLa and SiHa cells after CBX7 knockdown, RT-qPCR results showed
that knockdown of CBX7 reduced E-cad mRNA (57.0%) and
increased the mRNA levels of VIM (179.5%), ITGβ3
(253.3%), TGFβ1 (364.5%), PI3K (251.4%) and
AKT (191.2%) (Fig. 2D).
Similarly, the results of western blotting demonstrated that
knocking down CBX7 in HeLa and SiHa cells promoted the expression
of VIM, ITGβ3, TGFβ1, PI3K and AKT, and reduced the level of E-cad
(Fig. 2E and F).
The aforementioned results indicated that CBX7 may
regulate the ITGβ3/TGFβ1/AKT signaling pathway and may be involved
in the EMT process.
Knockdown of CBX7 promotes cervical
cancer progression through the ITGβ3/TGFβ1/AKT signaling
pathway
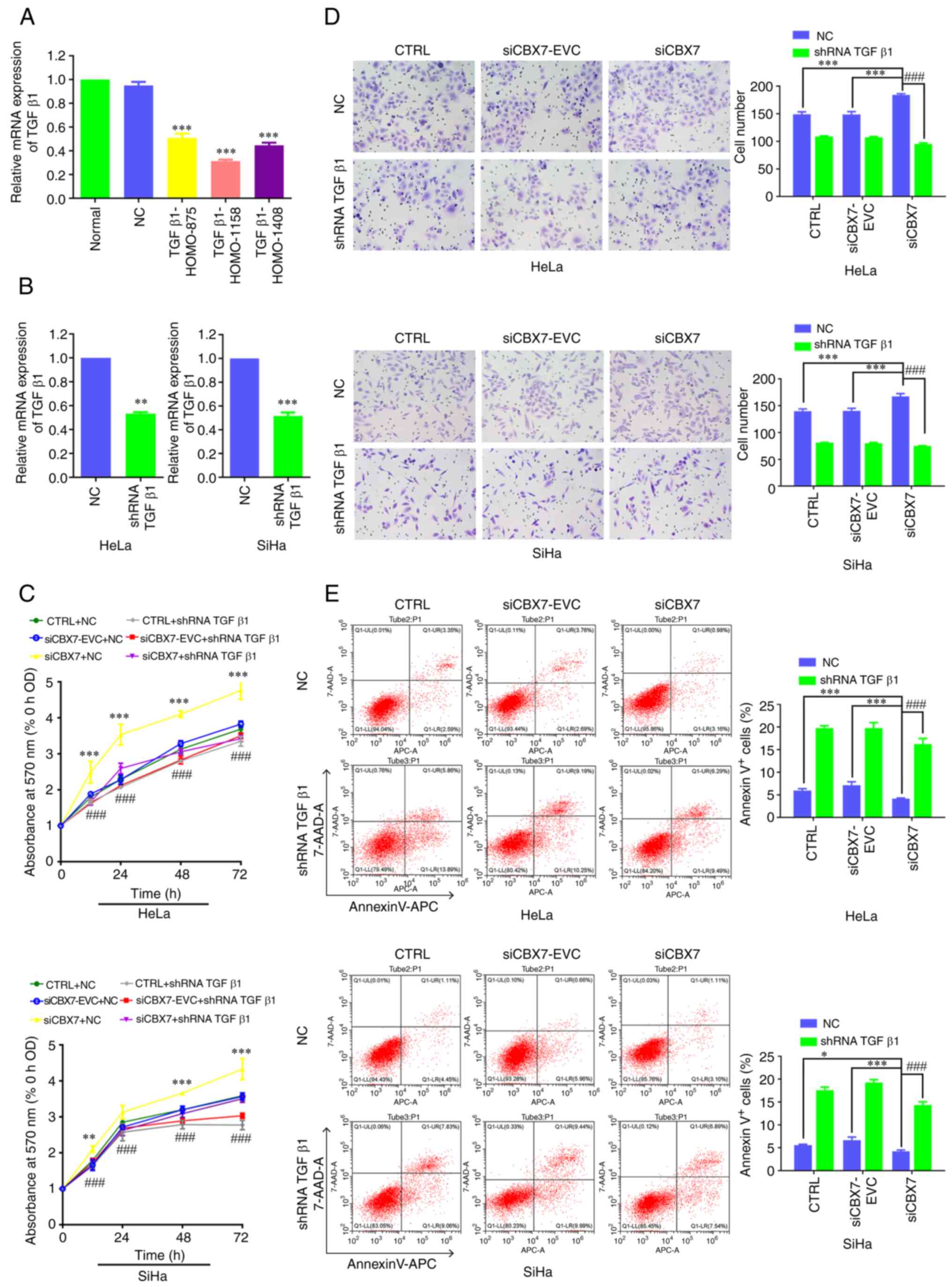
To verify the mechanism of CBX7 in cervical cancer,
TGFβ1 of the TGFβ1 pathway was further knocked down and its effect
on the proliferation, metastasis and apoptosis of cervical cancer
cells was explored. The expression of TGFβ1 mRNA after
transfection of TGFβ1-HOMO-875, TGFβ1-HOMO-1158 and TGFβ1-HOMO-1408
was 0.51, 0.31 and 0.47, respectively, significantly lower than
that in the control group (P<0.001) (Fig. 3A). The TGFβ1-HOMO-1158 fragment had
the highest knockdown efficiency and was therefore used in
subsequent experiments. TGFβ1 expression in control cells or cells
with stable knockdown of CBX7 was further verified by RT-qPCR. The
results revealed that TGFβ1 was successfully knocked down in each
group (Fig. 3B). The MTT assay and
Transwell assay demonstrated that the downregulation of CBX7
increased cell proliferation (Fig.
3C) and migration (Fig. 3D)
compared with siCBX7-EVC and CTRL groups. Furthermore, the
proliferation and migration ability of cells with knockdown of CBX7
and TGFβ1 decreased and were lower than the cells with CBX7
knockdown only at 12, 24, 48 and 72 h. Flow cytometry revealed that
the downregulation of CBX7 decreased apoptosis compared with
siCBX7-EVC and CTRL groups. Additionally, the apoptosis ability of
cells with knockdown of CBX7 and TGFβ1 increased and was higher
than the cells with CBX7 knockdown-only (Fig. 3E). Collectively, TGFβ1 silencing
reversed the increase in cell proliferation, and migration and
reversed the decrease in apoptosis. The aforementioned results
indicated that CBX7 may regulate the proliferation, metastasis and
apoptosis of cervical cancer cells through the TGFβ1 signaling
pathway. In addition, the expression of key genes of the
ITGβ3/TGFβ1 signaling pathway and the EMT process were also
analyzed. The RT-qPCR results revealed that the expression levels
of CBX7 and E-cad were significantly lower in the
siCBX7 group compared with the siCBX7-EVC and CTRL groups, whereas
ITGβ3, TGFβ1, PI3K, AKT and VIM exhibited higher
expression levels (P<0.05). No differences in CBX7 expression
were observed between the cells with silenced and unsilenced TGFβ1.
Silencing TGFβ1 in cells resulted in decreased mRNA levels of
ITGβ3, PI3K, AKT, and VIM, whereas E-cad mRNA
levels increased (Fig. 4A and B).
Western blot analysis demonstrated that silencing TGFβ1 resulted in
varying degrees of decrease in ITGβ3, p-PI3K, p-AKT and VIM, except
CBX7 and AKT, while E-cad expression increased (Fig. 4C and D). Thus, knockdown of TGFβ1
had no significant effect on CBX7 itself, but partially reversed
the activation of the ITGβ3/TGFβ1/AKT signaling pathway and
EMT-related genes caused by CBX7 knockdown. These results indicated
that knocking down CBX7 may regulate the EMT of cervical cancer
cells through the downregulated TGFβ1 signaling pathway.
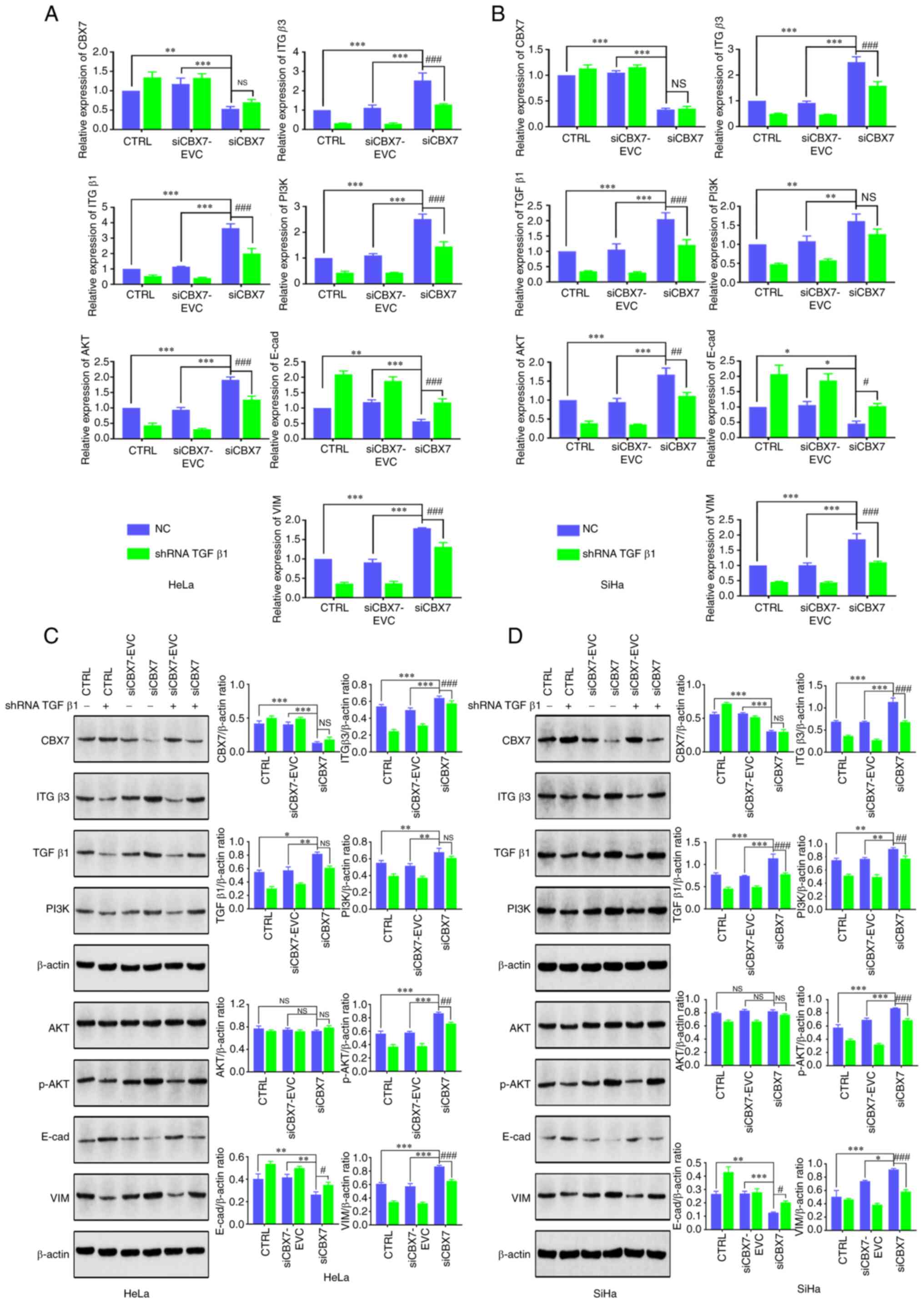 | Figure 4.Effect of TGFβ1 silencing and CBX7
knockdown on the expression of key genes in the PI3K/AKT and
ITGβ3/TGFβ1 signaling pathways. HeLa and SiHa cells with stable
expression of CBX7 knockdown were transfected with silencing
plasmids of TGFβ1. The mRNA expression of genes in the PI3K/AKT and
ITGβ3/TGFβ1 signaling pathways in (A) HeLa and (B) SiHa cells was
detected by reverse transcription-quantitative PCR. The protein
expression of genes in the PI3K/AKT and ITGβ3/TGFβ1 signaling
pathways in (C) HeLa and (D) SiHa cells was detected by western
blot analysis. Error bars in all panels represent the mean ± SD.
*P<0.05, **P<0.01 and ***P<0.001, siCBX7 compared with the
control and siCBX7-EVC; #P<0.05,
##P<0.01 and ###P<0.001, siCBX7 + shRNA
TGFβ1 compared with the siCBX7. TGFβ1, transforming growth factor
β1; CBX7, chromobox protein homolog 7; PI3K,
phosphatidylinositol-3-kinase; ITGβ3, integrin β3; shRNA, short
hairpin RNA; CTRL, control; NS, not significant; E-cad, E-cadherin;
VIM, vimentin; NC, negative control. |
Discussion
PRC1 is actively expressed in various cancers and
participates in tumor progression. As a member of PRC1, CBX7 is a
tumor suppressor involved in a variety of cancers (4). Previous studies have revealed that
inhibition of CBX7 promotes the migration of gliomas and tumor
invasion (10,25). In addition, in vitro
experiments have confirmed that inhibition of CBX7 promotes the
growth of pancreatic cancer (11)
and breast cancer (5). CBX7 has
also been confirmed to inhibit the growth of liver cancer cells
in vivo and in vitro (26). Recently, in CBX7 knockout mice
(9), CBX7 had an inhibitory effect
on tumors. CBX7 is involved in tumor growth (12), invasion (6) and migration (27). CBX7 overexpression in urinary
bladder cancer cells inhibits tumorigenicity, whereas CBX7
depletion promotes tumor development (28). Moreover, miR-19 affects the
migration and cell cycle of lung cancer cells by inhibiting CBX7
expression (29). Consistently, the
findings of the present study revealed that knockdown of CBX7
promoted the migration and invasion of cervical cancer cells in
vitro and inhibited cell apoptosis. Therefore, low expression
of CBX7 may promote tumor progression of cervical cancer.
By analyzing the expression of genes and proteins in
human cervical cancer tissues, transplanted tumors in nude mice and
cervical cancer cells, downregulation of CBX7 was found to increase
the expression of VIM, ITGβ3, TGFβ1, PI3K and AKT, and decrease the
expression of E-cad. Furthermore, correlation analysis demonstrated
that CBX7 was positively correlated with E-cad and negatively
correlated with VIM. Federico et al reported that CBX7
positively regulated E-cad expression by interacting with the
histone deacetylase 2 protein (30). E-cad and VIM are epithelial and
mesenchymal markers of EMT, respectively (31). E-cad can identify cell-cell
junctions and apicobasal polarity, limit the migratory potential of
epithelial cells and maintain the integrity of tissue structure and
function (32,33). The downregulation of E-cad can lead
to the destabilization of adherent junctions and promote the
transformation of epithelial cells to mesenchymal cells (34). A previous study has shown that the
reduction of epithelial phenotype and the increase of mesenchymal
phenotype may promote tumor progression (35). Additionally, it has been reported
that CBX7 upregulation enhanced E-cad expression (30). In the present study, consistently,
it was revealed that knockdown of CBX7 inhibited E-cad and promoted
VIM, suggesting that low expression of CBX may promote cervical
cancer progression through regulation of EMT. Furthermore, CBX7 can
upregulate E-cad (30) and plays a
key role in the occurrence of EMT and the progression of advanced
cancer (28). Previous research
conducted by the authors revealed that CBX7 was downregulated in
cervical cancer and its downregulation was associated with a poor
prognosis in patients with cervical cancer (22,23).
Moreover, it was revealed that CBX7 overexpression inhibited cell
proliferation and migration in cervical cancer cells (22,23).
Furthermore, the knockdown of CBX7 expression in the present study
significantly promoted the migration and invasion of cervical
cancer cells. These results strongly suggest that CBX7 may function
as a tumor suppressor in the development of cervical cancer.
Moreover, it was preliminarily inferred that the low expression of
CBX7 involved in the occurrence of cervical cancer EMT may be
related to ITGβ3, TGFβ1, PI3K, and AKT signaling pathways, which
all play an important role in tumor cell migration and invasion
(36,37).
Furthermore, TGFβ1 expression was silenced in cells
with stable knockdown of CBX7. TGFβ1 silencing was identified to
reduce the migration and invasion of stably transfected HeLa and
SiHa cells and increase cell apoptosis. Rapisarda et al
found that downregulation of CBX7 promoted the expression of ITGβ3
(18). IT5β3/AKT can promote the
progression of hemangioendothelioma. Furthermore, research has
revealed that TGFβ1 induces the upregulation of integrin alpha V,
leading to the promotion of EMT in ovarian cancer cells (38). Intercellular adhesion molecule 1 was
demonstrated to promote the migration of triple-negative breast
cancer cells through an integrin-mediated mechanism dependent on
TGF-β/EMT (39). There is an
interaction between TGFβ1 and ITGβ3 (40,41).
CBX7 has been revealed to activate PTEN, inhibit the downstream
PI3K/AKT pathway, and suppres the proliferation, metastasis and
invasion of pancreatic cancer cells (11). In addition, CBX7 was demonstrated to
enhance the sensitivity of bladder cancer cells to cisplatin
treatment through the inactivation of the PI3K/AKT signaling
pathway (42). The TGFβ1 and
PI3K/AKT signaling pathways are crucial in regulating diverse
cellular responses, such as proliferation, apoptosis and migration
(43). PI3K serves as a vital
intracellular kinase in organisms, while AKT acts as its most
significant downstream factor. The inhibition of TGFβ1 suppresses
the occurrence of EMT in cervical cancer cells, thereby restraining
cell migration and invasion (44).
In the present study, it was found that silencing of TGFβ1 reversed
the effects of CBX7 knockdown on the expression of key genes in the
ITGβ3/TGFβ1 signaling pathway and the EMT process. Therefore, CBX7
may regulate the progression of cervical cancer through
ITGβ3/TGFβ1/AKT signaling pathway (Fig.
5).
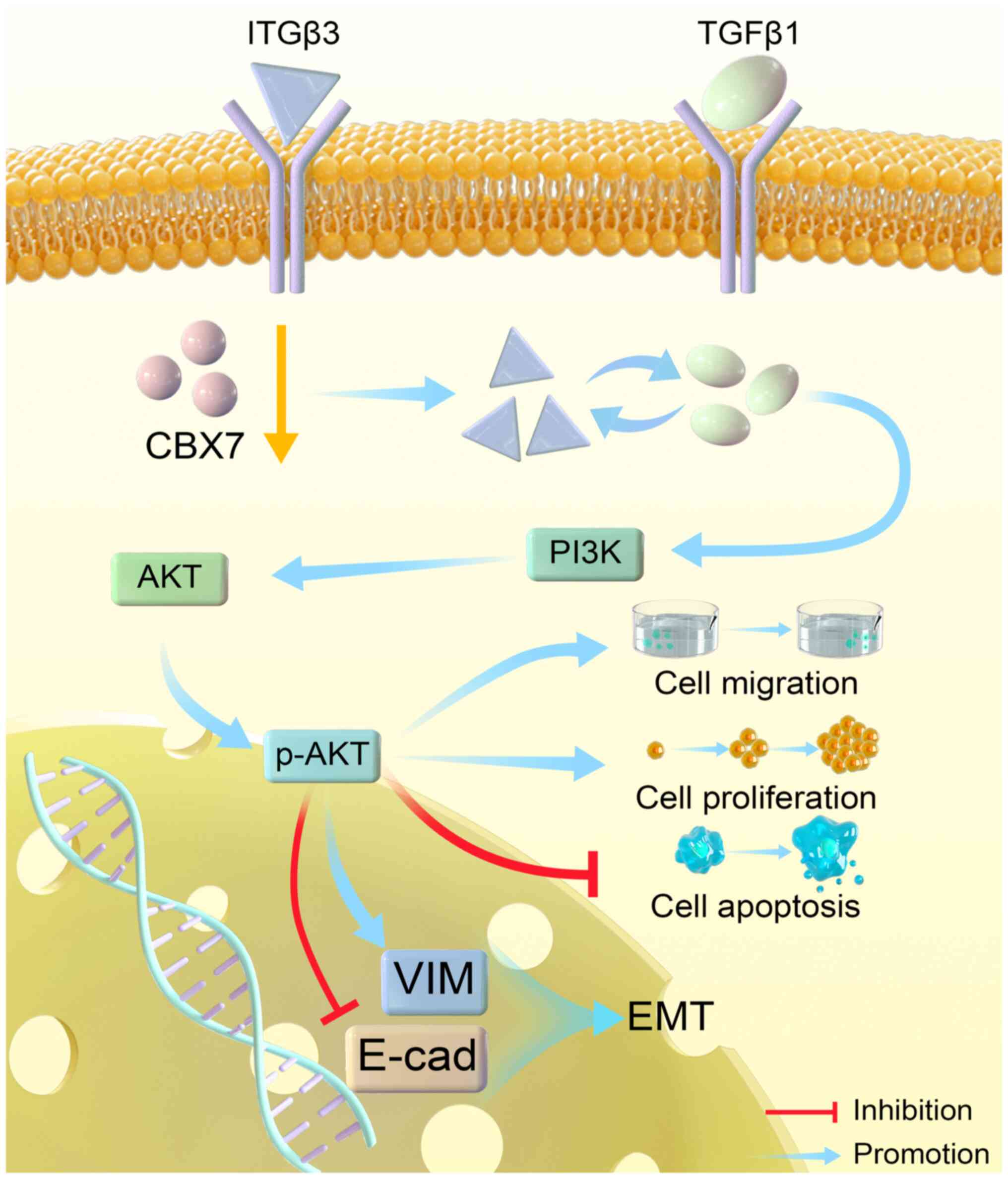 | Figure 5.Schematic diagram illustrating the
mechanism of CBX7 in cervical cancer. In cervical cancer,
downregulation of CBX7 may induce EMT, promote cell proliferation,
migration and inhibit cell apoptosis through the ITGβ3/TGFβ1/AKT
signaling pathway. CBX7, chromobox protein homolog 7; EMT,
epithelial-mesenchymal transition; ITGβ3, integrin β3; TGFβ1,
transforming growth factor β1; PI3K, phosphatidylinositol-3-kinase;
p-, phosphorylated; VIM, vimentin; E-cad, E-cadherin. |
The present study has some limitations. Firstly, a
subcutaneous xenograft model of cervical cancer cells in nude mice
was constructed. However, this model does not accurately replicate
the conditions within the nude mice. In the future, the orthotopic
transplantation tumor model, which involves transplantation to
anatomically relevant sites and exhibits stronger biological
relevance to cervical cancer, will be used. Secondly, the present
study validated the impact of CBX7 on proliferation, metastasis and
apoptosis. However, the factors related to proliferation,
metastasis and apoptosis were not examined. The inclusion of these
factors in the analysis can enhance the credibility of the
conclusions.
In summary, the present study show that low
expression of CBX7 is associated with high proliferation and
metastasis and low apoptosis of cervical cancer cells. In addition,
low expression of CBX7 may affect the PI3K/AKT signaling pathway by
upregulating ITGβ3/TGFβ1 and promoting the proliferation of
cervical cells and the occurrence of EMT. The findings of the
present study provide insights into the mechanisms of cervical
cancer and might help the development of potential therapeutic
targets for cervical cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the State Key Laboratory of
Pathogenesis, Prevention and Treatment of High Incidence Diseases
in Central Asia Fund (grant nos. SKL-HIDCA-2020-WF1 and
SKL-HIDCA-2022-GJ2), the Postdoctoral Science Foundation of China
(grant no. 2019M663963XB), the Xinjiang Autonomous Region
Collaborative Innovation Program (grant no. 2019E0282) and the
Xinjiang Medical University Student Innovation and Entrepreneurship
Training Program (grant no. CX2021046).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PT, JD and RL designed the study. PT and JD wrote
the manuscript. CM, AM, LD and GM performed experiments. QY, YaL,
HM, YuL, CZ and JR collected and analyzed data. All authors have
read and approved the final manuscript. PT and RL confirm the
authenticity of all raw data.
Ethics approval and consent to
participate
The present study was conducted following the
Declaration of Helsinki and approved (approval no. 20120220-01) by
the Institutional Review Board of Xinjiang Medical University
(Urumqi, China). Written informed consent was obtained from all
subjects involved in the study. The animal study protocol was
approved (approval no. 20120220-01) by the Ethics Committee of
Xinjiang Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CBX7
|
chromobox homolog 7
|
|
E-cad
|
E-cadherin
|
|
HPV
|
human papillomavirus
|
|
ITGβ3
|
integrin β3
|
|
PRC1
|
polycomb repressive complex 1
|
|
TGFβ1
|
transforming growth factor β1
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Razzaghi H, Saraiya M, Thompson TD, Henley
SJ, Viens L and Wilson R: Five-year relative survival for human
papillomavirus-associated cancer sites. Cancer. 124:203–211. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee BH: Commentary on: ‘Comprehensive
molecular characterization of papillary renal-cell carcinoma.’
Cancer Genome Atlas Research Network. N Engl J Med. 2016 Jan
14;374((2)): 135–45. Urol Oncol. 35:578–579. 2017.
|
|
4
|
Forzati F, Federico A, Pallante P, Abbate
A, Esposito F, Malapelle U, Sepe R, Palma G, Troncone G, Scarfò M,
et al: CBX7 is a tumor suppressor in mice and humans. J Clin
Invest. 122:612–623. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iqbal MA, Siddiqui S, Ur Rehman A,
Siddiqui FA, Singh P, Kumar B and Saluja D: Multiomics integrative
analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic
reprogramming of breast cancer. Mol Oncol. 15:1450–1465. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang Z, Yan Y, Zhu Z, Liu J, He X,
Dalangood S, Li M, Tan M, Cai J, Tang P, et al: CBX7 suppresses
urinary bladder cancer progression via modulating AKR1B10-ERK
signaling. Cell Death Dis. 12:5372021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forzati F, De Martino M, Esposito F, Sepe
R, Pellecchia S, Malapelle U, Pellino G, Arra C and Fusco A:
miR-155 is positively regulated by CBX7 in mouse embryonic
fibroblasts and colon carcinomas, and targets the KRAS oncogene.
BMC Cancer. 17:1702017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng ZQ, Yuan GQ, Kang NL, Nie QQ, Zhang
GG and Wang Z: Chromobox 7/8 serve as independent indicators for
glioblastoma via promoting proliferation and invasion of glioma
cells. Front Neurol. 13:9120392022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu T, Wu Y, Hu Q, Zhang J, Nie E, Wu W,
Wang X, Wang Y and Liu N: CBX7 is a glioma prognostic marker and
induces G1/S arrest via the silencing of CCNE1. Oncotarget.
8:26637–26647. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Xu Z, Zhou L and Hu K: Expression
profile and prognostic values of Chromobox family members in human
glioblastoma. Aging (Albany NY). 14:1910–1931. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ni S, Wang H, Zhu X, Wan C, Xu J, Lu C,
Xiao L, He J, Jiang C, Wang W and He Z: CBX7 suppresses cell
proliferation, migration, and invasion through the inhibition of
PTEN/Akt signaling in pancreatic cancer. Oncotarget. 8:8010–8021.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cacciola NA, Sepe R, Forzati F, Federico
A, Pellecchia S, Malapelle U, De Stefano A, Rocco D, Fusco A and
Pallante P: Restoration of CBX7 expression increases the
susceptibility of human lung carcinoma cells to irinotecan
treatment. Naunyn Schmiedebergs Arch Pharmacol. 388:1179–1186.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX,
Wan CH, Zhang JY, Zhang XW, Huang MZ, Gan L, et al: CBX7 regulates
stem cell-like properties of gastric cancer cells via p16 and
AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 11:172018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scott CL, Gil J, Hernando E,
Teruya-Feldstein J, Narita M, Martínez D, Visakorpi T, Mu D,
Cordon-Cardo C, Peters G, et al: Role of the chromobox protein CBX7
in lymphomagenesis. Proc Natl Acad Sci USA. 104:5389–5394. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gong L, Tang Y, Jiang L, Tang W and Luo S:
Regulation of circGOLPH3 and its binding protein CBX7 on the
proliferation and apoptosis of prostate cancer cells. Biosci Rep.
40:BSR202009362020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shinjo K, Yamashita Y, Yamamoto E,
Akatsuka S, Uno N, Kamiya A, Niimi K, Sakaguchi Y, Nagasaka T,
Takahashi T, et al: Expression of chromobox homolog 7 (CBX7) is
associated with poor prognosis in ovarian clear cell adenocarcinoma
via TRAIL-induced apoptotic pathway regulation. Int J Cancer.
135:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mosbah A, Barakat R, Nabiel Y and Barakat
G: High-risk and low-risk human papilloma virus in association to
spontaneous preterm labor: A case-control study in a tertiary
center, Egypt. J Matern Fetal Neonatal Med. 31:720–725. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rapisarda V, Borghesan M, Miguela V,
Encheva V, Snijders AP, Lujambio A and O'Loghlen A: Integrin Beta 3
regulates cellular senescence by activating the TGF-β pathway. Cell
Rep. 18:2480–2493. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao L, Zhu H, Shu J, Gong D, Zheng D and
Gao J: Overexpression of TGF-β1 and SDF-1 in cervical
cancer-associated fibroblasts promotes cell growth, invasion and
migration. Arch Gynecol Obstet. 305:179–192. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You X, Wang Y, Meng J, Han S, Liu L, Sun
Y, Zhang J, Sun S, Li X, Sun W, et al: Exosomal miR-663b exposed to
TGF-β1 promotes cervical cancer metastasis and
epithelial-mesenchymal transition by targeting MGAT3. Oncol Rep.
45:122021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu R, Sun X, Chi Y, Zhou Q, Xiang H, Bosco
DB, Lai X, Qin C, So KF, Ren Y and Chen XM: Integrin β3/Akt
signaling contributes to platelet-induced hemangioendothelioma
growth. Sci Rep. 7:64552017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian P, Zhang C, Ma C, Ding L, Tao N, Ning
L, Wang Y, Yong X, Yan Q, Lin X, et al: Decreased chromobox
homologue 7 expression is associated with epithelial-mesenchymal
transition and poor prognosis in cervical cancer. Open Med (Wars).
16:410–418. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li R, Yan Q, Tian P, Wang Y, Wang J, Tao
N, Ning L, Lin X, Ding L, Liu J and Ma C: CBX7 inhibits cell growth
and motility and induces apoptosis in cervical cancer cells. Mol
Ther Oncolytics. 15:108–116. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Larionov A, Krause A and Miller W: A
standard curve based method for relative real time PCR data
processing. BMC Bioinformatics. 6:622005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bao Z, Xu X, Liu Y, Chao H, Lin C, Li Z,
You Y, Liu N and Ji J: CBX7 negatively regulates migration and
invasion in glioma via Wnt/β-catenin pathway inactivation.
Oncotarget. 8:39048–39063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan C, Bei C, Zhu X, Zhang Y, Qin L and
Tan S: Single nucleotide polymorphisms of CBX4 and CBX7 decrease
the risk of hepatocellular carcinoma. Biomed Res Int.
2019:64368252019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pallante P, Forzati F, Federico A, Arra C
and Fusco A: Polycomb protein family member CBX7 plays a critical
role in cancer progression. Am J Cancer Res. 5:1594–1601.
2015.PubMed/NCBI
|
|
28
|
Huang Z, Liu J, Yang J, Yan Y, Yang C, He
X, Huang R, Tan M, Wu D, Yan J and Shen B: PDE4B induces
epithelial-to-mesenchymal transition in bladder cancer cells and is
transcriptionally suppressed by CBX7. Front Cell Dev Biol.
9:7830502021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peng X, Guan L and Gao B: miRNA-19
promotes non-small-cell lung cancer cell proliferation via
inhibiting CBX7 expression. Onco Targets Ther. 11:8865–8874. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Federico A, Sepe R, Cozzolino F, Piccolo
C, Iannone C, Iacobucci I, Pucci P, Monti M and Fusco A: The
complex CBX7-PRMT1 has a critical role in regulating E-cadherin
gene expression and cell migration. Biochim Biophys Acta Gene Regul
Mech. 1862:509–521. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cicchini C, Amicone L, Alonzi T, Marchetti
A, Mancone C and Tripodi M: Molecular mechanisms controlling the
phenotype and the EMT/MET dynamics of hepatocyte. Liver Int.
35:302–310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang C, Zhang J, Fok KL, Tsang LL, Ye M,
Liu J, Li F, Zhao AZ, Chan HC and Chen H: cd147 induces
epithelial-to-mesenchymal transition by disassembling cellular
apoptosis susceptibility Protein/E-Cadherin/β-Catenin complex in
human endometriosis. Am J Pathol. 188:1597–1607. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang N, Pan W, Li J, Cao T and Shen H:
Upregulated Circular RNA hsa_circ_0008433 regulates pathogenesis in
endometriosis via miRNA. Reprod Sci. 27:2002–2017. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vu T and Datta PK: Regulation of EMT in
colorectal cancer: A culprit in metastasis. Cancers (Basel).
9:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Loh CY, Chai JY, Tang TF, Wong WF, Sethi
G, Shanmugam MK, Chong PP and Looi CY: The E-Cadherin and
N-Cadherin switch in epithelial-to-mesenchymal transition:
signaling, therapeutic implications, and challenges. Cells.
8:11182019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luo R, Wei Y, Chen P, Zhang J, Wang L,
Wang W, Wang P and Tian W: Mesenchymal stem cells inhibit
epithelial-to-mesenchymal transition by modulating the IRE1α branch
of the endoplasmic reticulum stress response. Stem Cells Int.
2023:44837762023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
E M Eid E, S Alanazi A, Koosha S, A
Alrasheedy A, Azam F, M Taban I, Khalilullah H, Sadiq Al-Qubaisi M
and A Alshawsh M: Zerumbone induces apoptosis in breast cancer
cells by targeting αvβ3 integrin upon Co-administration with
TP5-iRGD peptide. Molecules. 24:25542019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dehghani-Ghobadi Z, Sheikh Hasani S,
Arefian E and Hossein G: Wnt5A and TGFβ1 Converges through YAP1
Activity and Integrin Alpha v Up-Regulation promoting epithelial to
mesenchymal transition in ovarian cancer cells and mesothelial cell
activation. Cells. 11:2372022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen M, Wu C, Fu Z and Liu S: ICAM1
promotes bone metastasis via integrin-mediated TGF-β/EMT signaling
in triple-negative breast cancer. Cancer Sci. 113:3751–3765. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shidal C, Singh NP, Nagarkatti P and
Nagarkatti M: MicroRNA-92 expression in CD133(+) melanoma stem
cells regulates immunosuppression in the tumor microenvironment via
integrin-dependent activation of TGFβ. Cancer Res. 79:3622–3635.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fullar A, Dudas J, Olah L, Hollósi P, Papp
Z, Sobel G, Karászi K, Paku S, Baghy K and Kovalszky I: Remodeling
of extracellular matrix by normal and tumor-associated fibroblasts
promotes cervical cancer progression. BMC Cancer. 15:2562015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ren J, Yu H, Li W, Jin X and Yan B:
Downregulation of CBX7 induced by EZH2 upregulates FGFR3 expression
to reduce sensitivity to cisplatin in bladder cancer. Br J Cancer.
128:232–244. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen LY, Cheng CS, Qu C, Wang P, Chen H,
Meng ZQ and Chen Z: CBX3 promotes proliferation and regulates
glycolysis via suppressing FBP1 in pancreatic cancer. Biochem
Biophys Res Commun. 500:691–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang C, Su T, Xue Y, Cheng C, Lay FD,
McKee RA, Li M, Vashisht A, Wohlschlegel J, Novitch BG, et al: Cbx3
maintains lineage specificity during neural differentiation. Genes
Dev. 31:241–246. 2017. View Article : Google Scholar : PubMed/NCBI
|