Introduction
Acute leukemia constitutes the most common
malignancy in childhood, accounting for ~25% of new cancer cases in
the USA (1). There are ~0.6
deaths/100,000 per year attributed to the disease, with the highest
rate recorded in adolescents aged 15–19 years (1). Acute lymphoblastic leukemia (ALL)
accounts for ~85% of leukemia cases in childhood (2). Survival rates have increased over
previous decades and are >90% at present, an improvement which
can be attributed to the implementation of efficacious risk-adapted
treatment, high-dose chemotherapy, novel targeted therapies and
advances in supportive care (2).
Currently, risk stratification is based on demographic
characteristics (age and sex), and clinical (presence of
extramedullary disease) and laboratory [number of white blood cells
(WBCs) at diagnosis, karyotype and cytogenetic findings]
parameters. Regarding cytogenetic findings, it is noteworthy that
the presence of the translocation (12;21)(p13;q22), which gives
rise to the fusion gene erythroblast transformation specific (ETS)
variant transcription factor 6 (ETV6)-RUNX family transcription
factor 1 (RUNX1; previously known as
translocation-Ets-leukemia-acute myeloid leukemia 1 protein), which
occurs in ~25% of pediatric patients with ALL, is associated with a
favorable prognosis and is part of the risk stratification system
of chemotherapeutic protocols (3).
Response to treatment during induction is assessed based on the
number of blasts in the peripheral blood on day 8 of the
therapeutic protocol and the minimal residual disease (MRD) on day
15 and 33 of the therapeutic protocol (3). However, relapsed or refractory disease
still poses a therapeutic challenge, as it remains one of the most
common causes of mortality attributed to disease in childhood. The
event-free-survival and overall survival rates of relapsed patients
with B-ALL and T-ALL do not exceed 33% even with the use of the new
protocols (4). Furthermore, the
fact that patients with favorable prognosis according to the
established criteria do not exhibit the anticipated response to
treatment, while at the same time a subset of patients is
overtreated (in risk of treatment-associated toxicities),
underlines the need for additional prognostic factors that can be
used for risk stratification (5,6).
MicroRNAs (miRNAs/miRs) are small (19–22
nucleotides), endogenous, non-coding, single-stranded RNA molecules
that act as epigenetic regulators and are implicated in several
biological processes, including cell division, differentiation and
death, thus guarding cell homeostasis (7). Therefore, disruption of normal miRNA
expression can lead to oncogenesis and aberrant miRNA expression
has been implicated in a plethora of solid tumors, including
breast, cervical, ovarian, prostate, lung, colorectal and brain
cancer, as well as in hematological malignancies (7). Multiple miRNAs regulate hematopoiesis
lineage differentiation (8).
Particularly, in pediatric ALL, the miRNA expression profile has
been evaluated as a marker for diagnosis and classification, as
well as to determine prognosis (9–11).
Recently, miRNAs have also been proposed as therapeutic targets in
leukemia (12). However, the high
heterogeneity among studies hinders the recognition of a set of
miRNAs that can be effectively used for risk stratification and
assessment of response to treatment.
The aim of the present pilot study was to assess the
miRNA expression profile in bone marrow aspirates of pediatric
patients with newly diagnosed ALL at two crucial time points: i) At
diagnosis (day 0 of induction therapy); and ii) at day 33 [end of
induction therapy in the Acute Lymphoblastic Leukemia
Intercontinental Berlin-Frankfurt-Münster (ALL IC BFM) 2009
protocol (13)]. Additionally, the
present study aimed to evaluate the association of miRNA expression
levels with established prognostic factors, to identify miRNAs that
could be used as prognostic biomarkers or are involved in the
mechanisms underlying resistance to therapy and that could serve as
novel therapeutic targets. To the best of our knowledge, the
present study was the first to assess such a large number of
candidate miRNAs in the bone marrow not only at diagnosis, but also
at the end of induction, in a Greek population. The set of miRNAs
used in the present study comprised 84 miRNAs that have been
previously associated with tumorigenesis, but the majority of them
had not been studied in pediatric acute lymphoblastic leukemia.
Materials and methods
Clinical samples
Patients aged <18 years with newly diagnosed ALL
and no history of malignancy or autoimmune disease were enrolled
between October 2021 and October 2022. Patients were diagnosed and
treated in the pediatric hematology units of AHEPA University
Hospital (Thessaloniki, Greece) and Hippokration General Hospital
(Thessaloniki, Greece). A total of 10 patients who fulfilled the
inclusion criteria were included in the study. A total of 4
patients were excluded due to unavailable samples (bone marrow
aspirates) either at diagnosis or on day 33 of induction. All
patients were treated according to the ALL IC BFM 2009 protocol.
The biological samples included two bone marrow aspirates in
EDTA-containing tubes for each patient obtained at diagnosis (day 0
of induction therapy) and at day 33 (end of induction therapy).
Sample collection and storage complied with the General Data
Protection Regulation. The present study was approved by the
Committee for Bioethics and Ethics of the School of Medicine of the
Aristotle University of Thessaloniki (Thessaloniki, Greece;
approval no. 1.463; 19/10/2021). Written informed consent was
obtained from patients or legal caregivers of all patients, as well
as from patients aged >12 years.
Separation of bone marrow mononuclear
cells (BMNCs)
BMNCs were separated using
Histopaque®−1077 (MilliporeSigma; Merck KGaA), according
to the manufacturer's instructions. Briefly, 1.5 ml bone marrow
aspirate was diluted with PBS solution to a final volume of 3 ml
and layered on top of 3 ml Histopaque-1077. The gradient was
centrifuged at 400 × g for 30 min at room temperature, and the
BMNCs were then collected. Once collected, the BMNCs were washed
with PBS and stored at −80°C until use.
miRNA isolation
Total RNA was isolated from BMNCs using the miRNeasy
Mini Kit with QIAzol Lysis reagent (cat. no. 217004; Qiagen GmbH),
according to the manufacturer's protocol. As a quality control and
to determine the efficiency of RNA extraction, three synthetic RNAs
(spike-ins: UniSp2, UniSp4 and UniSp5) included in the RNA Spike-in
Kit, For RT (cat. no. 339390; Qiagen GmbH) were added as
recommended by the manufacturer. The quantity and quality (purity)
of the isolated RNA was assessed spectrophotometrically, with
measurements at wavelengths of 230, 260 and 280 nm.
Reverse transcription-quantitative
(RT-q)PCR
Purified RNA was used for RT using the miRCURY LNA
RT Kit (cat. no. 339340; Qiagen GmbH). During RT, a spike-in mix of
two synthetic RNAs (UniSp6 and cel-miR-39-3p) was used as a quality
control for complementary (c)DNA synthesis. The RT conditions were
60 min at 42°C, followed by 5 min at 95°C (inactivation step).
cDNA samples were then prepared using the miRCURY
LNA SYBR® Green PCR Kit (cat. no. 339345; Qiagen GmbH).
A Human Cancer Focus miRCURY LNA miRNA Focus PCR Panel (cat. no.
339325; Qiagen GmbH) was used to assess the RNA expression levels
of 84 cancer-relevant human miRNAs. Each plate included lyophilized
primer sets for 84 miRNAs and additionally contained primers for
interplate calibrators, candidate reference genes, RNA spike-in
controls and one water blank. qPCR was performed using a
StepOnePlus™ Real-Time PCR System (Applied Biosystems™; Thermo
Fisher Scientific, Inc.) with the following conditions: 95°C for 2
min, followed by 45 cycles at 95°C for 10 sec and 56°C for 1 min.
Melting curve analysis was performed at the end of the PCR cycles.
UniSp3 synthetic RNA was used as an interplate calibrator.
Data analysis
Raw Cq values from each PCR panel
(Table SI) were uploaded to the
GeneGlobe Data Analysis tool (Qiagen GmbH) for normalization, using
let-7a-5p as a reference gene, as this miRNA was one of the most
stably expressed candidates (Table
SI). The relative quantity of each miRNA was calculated using
the 2−ΔCq method (14)
using let-7a-5p as the endogenous control. The fold change (FC) in
miRNA expression between the two timepoints was calculated using
the 2−ΔΔCq method and treated as a dichotomous variable
(FC <1 corresponding to an increase in expression levels at the
end of induction and FC >1 corresponding to a decrease).
Statistical analysis
The Mann-Whitney U test was used to evaluate
differences in the expression levels of the miRNAs of interest at
diagnosis between the patient groups defined by the following
demographic and clinical characteristics: Age; sex; WBCs/µl;
immunophenotype; hyperploidy; presence of the t(12;21)(p13;q22)
translocation; prednisone response; risk group; and complete
response at the end of induction (day 33) according to the
definitions of the ALL IC BFM 2009 protocol. Spearman's rank
correlation coefficient was used to evaluate the correlation
between miRNA expression levels at diagnosis and MRD values on days
15 and 33. The Wilcoxon signed-rank test was used to assess
differences between expression levels at diagnosis and day 33.
Fisher's exact test was performed to assess possible associations
between FC and the risk group, and between FC and complete response
at the end of induction. SPSS software version 25 (IBM Corp.) was
used for statistical analysis. P<0.05 was considered to indicate
a statistically significant difference.
Bioinformatics analysis
The MicroRNA ENrichment TURned NETwork (15) web tool (release 3.4.4, March 2018)
was used to identify experimentally proven targets of miRNAs of
interest from miRTarBase release 9.0 beta (16), construct the interaction networks
between miRNAs of interest and mRNAs, and identify the associated
molecular pathways based on the Reactome database (17), version 85. The false discovery rate
was set at ≤0.05 in all cases.
Results
Demographic and clinical
characteristics of the patients
A total of 10 patients who fulfilled the inclusion
criteria were included in the present study. The majority were
female (6/10), aged 1–6 years (8/10) and without underlying
conditions (8/10). One patient was diagnosed with congenital
hearing impairment, neuropsychomotor developmental delay and
epilepsy, and one patient with a ventricular septal defect. Upon
diagnosis, 7/10 patients had <20,000 WBCs per µl [risk
stratification cut-off (13)] and
8/10 were diagnosed with B-cell ALL (B-ALL). No patients exhibited
extramedullary manifestations. The cytogenetic findings
demonstrated that, 4/10 patients had hyperploidy and 5/10 had the
translocation t(12;21)(p13;q22), which gives rise to the fusion
gene ETV6-RUNX1 (3). None of the
translocations t(1;19)(q23;p13), t(4;11)(q21;q23) and
t(9;22)(q34;q11) were detected. Regarding response to treatment,
good prednisone response with an absolute blast count of <1,000
in the peripheral blood on day 8 of the therapeutic protocol was
observed in 8/10 patients. The mean MRD on days 15 and 33 was
7.1±15.3% (range, 0–50%) and 0.9±2.6% (range, 0–8.35%),
respectively. A total of 2 patients were allocated to the
standard-risk group, 5 to the intermediate-risk group and 3 to the
high-risk group, as defined by the criteria of the ALL IC BFM 2009
protocol. A total of 8/10 patients achieved complete remission at
the end of induction, based on the criteria of the ALL IC BFM 2009
protocol. The demographic and clinical characteristics of the
patients are presented in Table
I.
 | Table I.Demographic and clinical
characteristics of the patients included in the study. |
Table I.
Demographic and clinical
characteristics of the patients included in the study.
|
|
| Complete remission
at day 33 |
|---|
|
|
|
|
|---|
|
Characteristics | Total (n=10) | Yes (n=8) | No (n=2) |
|---|
| Age, years |
|
|
|
|
1-6 | 8 | 7 | 1 |
| <1
or >6 | 2 | 1 | 1 |
| Sex |
|
|
|
|
Female | 6 | 5 | 1 |
|
Male | 4 | 3 | 1 |
| Syndrome or
underlying disease |
|
|
|
|
Yes | 2 | 2 | 0 |
| No | 8 | 6 | 2 |
| White blood cells
at diagnosis, /µl |
|
|
|
|
<20,000 | 7 | 6 | 1 |
|
≥20,000 | 3 | 2 | 1 |
|
Immunophenotype |
|
|
|
|
B-ALL | 8 | 8 | 0 |
|
T-ALL | 2 | 0 | 2 |
| Hyperploidy |
|
|
|
|
Yes | 4 | 3 | 1 |
| No | 6 | 5 | 1 |
|
t(12;21)(p13;q22) |
|
|
|
|
Yes | 5 | 5 | 0 |
| No | 5 | 3 | 2 |
| Prednisone
response |
|
|
|
| Good
(<1,000 blasts) | 8 | 7 | 1 |
| Poor
(≥1,000 blasts) | 2 | 1 | 1 |
| BFM risk group |
|
|
|
|
Standard/intermediate | 7 | 7 | 0 |
|
High | 3 | 1 | 2 |
Association of miRNA levels at
diagnosis with demographic and clinical characteristics of the
patients
Among the evaluated miRNAs, the following results
were demonstrated: The expression levels of nine miRNAs were
significantly upregulated in patients aged 1–6 years compared with
patients aged <1 year or >6 years; the expression levels of
26 miRNAs were significantly upregulated in male patients compared
with female patients; the expression levels of three miRNAs were
significantly upregulated in patients with <20,000/µl WBCs at
diagnosis; the expression levels of three miRNAs were significantly
upregulated in patients with B-ALL compared with T-cell ALL
(T-ALL); and the expression levels of 13 miRNAs were significantly
upregulated in patients with hyperploidy compared with those in
patients without hyperploidy. No statistically significant
differences between patients with and without t(12;21)(p13;q22)
were observed. The miRNAs with significantly different expression
levels between the aforementioned patient groups are presented in
Table SII.
Association of miRNA levels at
diagnosis with response to treatment
Patients with a good prednisone response exhibited
markedly higher expression levels of seven miRNAs at diagnosis,
namely let-7c-5p, miR-106b-5p, miR-26a-5p, miR-155-5p, miR-191-5p,
miR-30b-5p and miR-31-5p, compared with those with poor prednisone
response (Table II). The 10 most
significant protein-coding genes targeted by the seven miRNAs,
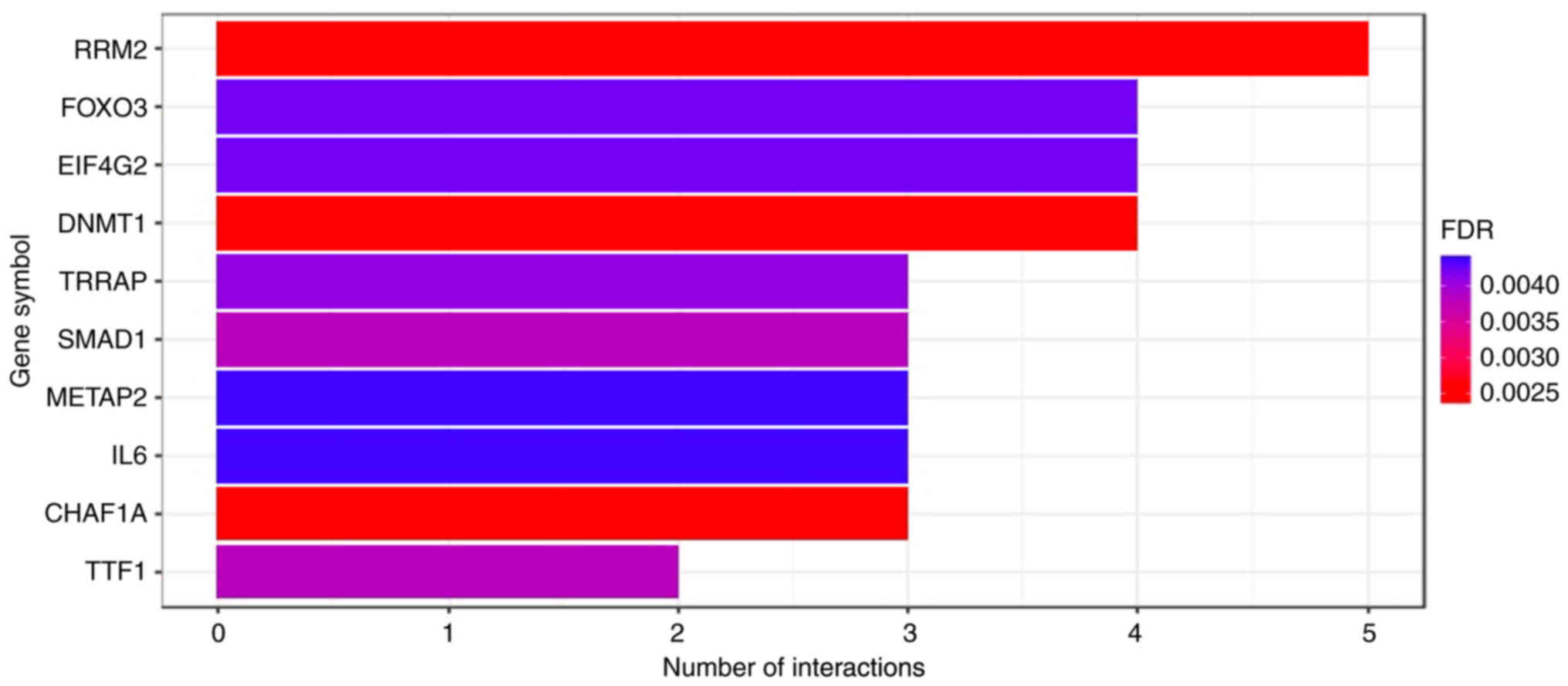
identified by miRTarBase, are presented in Fig. 1. The associated molecular pathways
were identified using the Reactome database and are presented in
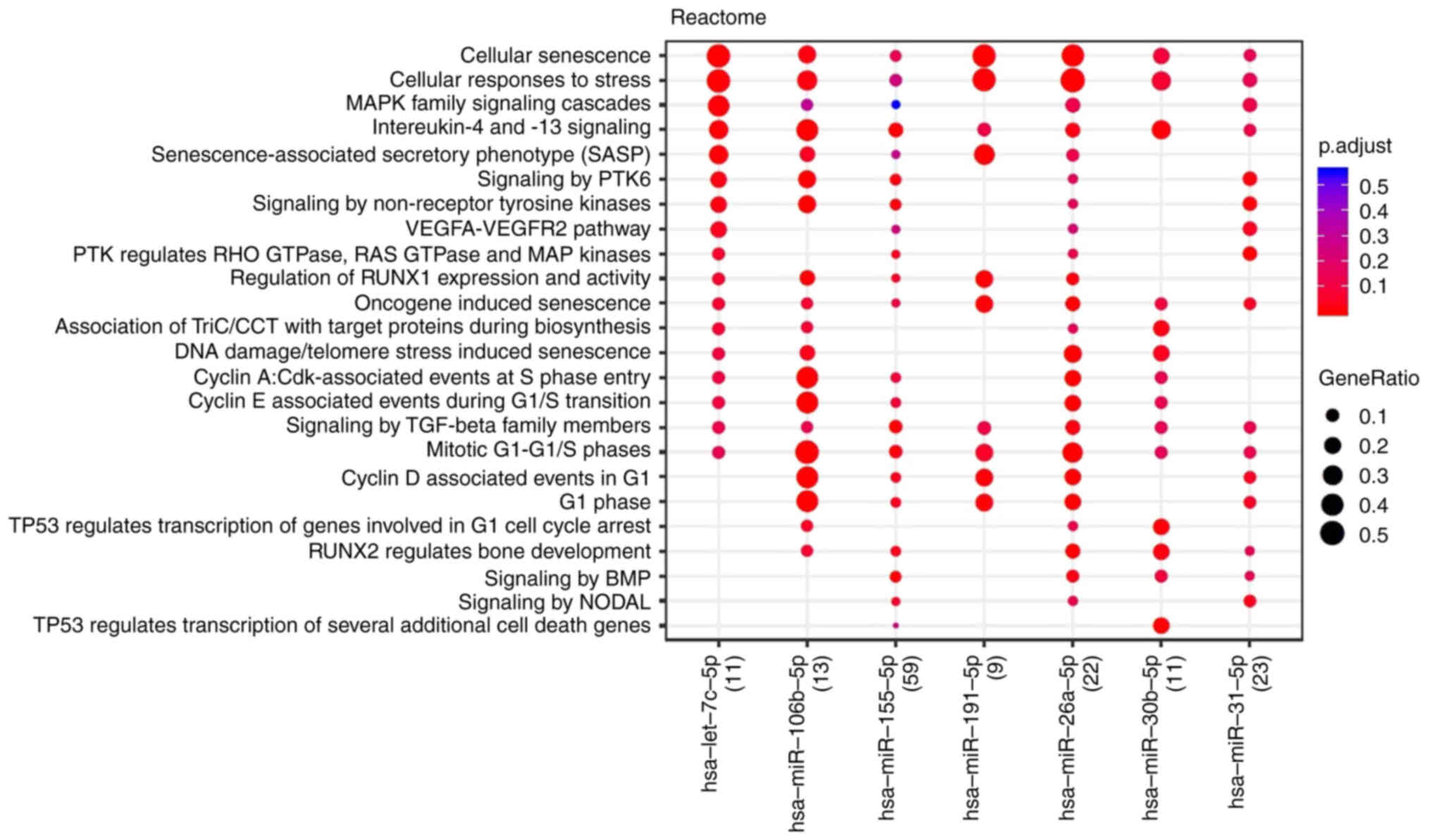
Fig. 2. Cellular senescence,
responses to stress and cell cycle regulation by cyclins appeared
to be the key pathways involved.
 | Table II.miRNAs with expression levels at
diagnosis that were significantly associated with at ≥2 parameters
(demographic/clinical characteristics, laboratory findings and
measures of response to treatment). |
Table II.
miRNAs with expression levels at
diagnosis that were significantly associated with at ≥2 parameters
(demographic/clinical characteristics, laboratory findings and
measures of response to treatment).
|
|
P-value |
|---|
|
|
|
|---|
| miRNA | Age (1–6
years) | Sex (Male) | WBCs
(<20,000/µl) | Immuno-phenotype
(B-ALL) | Karyotype
(Hyperploidy) | PR (Good) | MRD day 15
(Negative correlation) | MRD day 33
(Negative correlation) | BFM risk group
(SR/IR) | CR day 33
(Yes) |
|---|
| hsa-let-7c-5p | - | - | - | - | - | 0.044 | 0.005 | 0.037 | 0.017 | - |
| hsa-let-7g-5p | - | - | - | - | - | - | 0.035 | - | 0.017 | - |
| hsa-miR-100-5p | 0.044 | - | 0.033 | - | - | - | - | - | - | - |
| hsa-let-7f-5p | - | - | - | - | - | - | 0.020 | 0.034 | 0.017 | - |
|
hsa-miR-106b-5p | - | - | - | - | - | 0.044 | 0.001 | 0.014 | 0.017 | - |
|
hsa-miR-125b-5p | - | - | - | 0.044 | - | - | 0.012 | 0.002 | 0.017 | 0.044 |
| hsa-miR-126-3p | - | - | - | - | - | - | 0.018 | 0.004 | 0.017 | - |
| hsa-miR-132-3p | - | 0.010 | - | - | 0.038 | - | - | - | - | - |
| hsa-miR-10b-5p | 0.044 | - | 0.017 | - | - | - | - | - | - | - |
|
hsa-miR-133a-3p | 0.044 | - | 0.033 | - | - | - | - | - | - | - |
|
hsa-miR-146a-5p | - | - | - | - | - | - | 0.005 | 0.016 | 0.017 | - |
| hsa-miR-150-5p | - | - | - | 0.044 | - | - | - | - | 0.033 | 0.044 |
| hsa-miR-155-5p | - | - | - | - | - | 0.044 | 0.003 | - | - | - |
| hsa-miR-186-5p | - | - | - | - | - | - | 0.018 | 0.010 | 0.033 | - |
| hsa-miR-191-5p | - | - | - | - | - | 0.044 | 0.001 | 0.007 | 0.017 | - |
|
hsa-miR-148a-3p | 0.044 | - | - | - | 0.038 | - | - | - | - | - |
| hsa-miR-195-5p | - | - | - | - | - | - | 0.011 | 0.049 | 0.017 | - |
|
hsa-miR-200b-3p | - | 0.019 | - | - | 0.010 | - | - | - | - | - |
| hsa-miR-206 | - | - | - | 0.044 | 0.038 | - | - | - | - | - |
| hsa-miR-26b-5p | - | - | - | - | - | - | 0.030 | 0.034 | 0.017 | - |
| hsa-miR-29a-3p | 0.044 | - | - | - | 0.038 | - | - | - | 0.017 | - |
| hsa-miR-30b-5p | - | - | - | - | - | 0.044 | 0.009 | - | - | - |
| hsa-miR-31-5p | - | - | - | - | - | 0.044 | - | - | 0.017 | - |
| hsa-miR-7-5p | 0.044 | 0.038 | - | - | 0.010 | - | - | - | - | - |
| hsa-miR-99a-5p | - | - | - | 0.044 | - | - | 0.012 | 0.001 | 0.017 | 0.044 |
Regarding other measures of response to treatment, a
strong or moderate-to-strong negative correlation was observed
between the expression levels of 18 (Table SIII) and 12 (Table SIII) miRNAs at diagnosis and MRD on
days 15 and 33 of the therapeutic protocol, respectively.
Conversely, a strong positive correlation was observed between the
levels of miR-206 and MRD on day 33, based on the calculated
Spearman's correlation coefficient.
The expression levels of 14 miRNAs differed
significantly between the risk groups (patients in the standard- or
intermediate-risk groups vs. patients in the high-risk group;
Table SIV). Mann-Whitney U test
was performed in order to detect the miRNAs whose expression
differed significantly between patients with and without complete
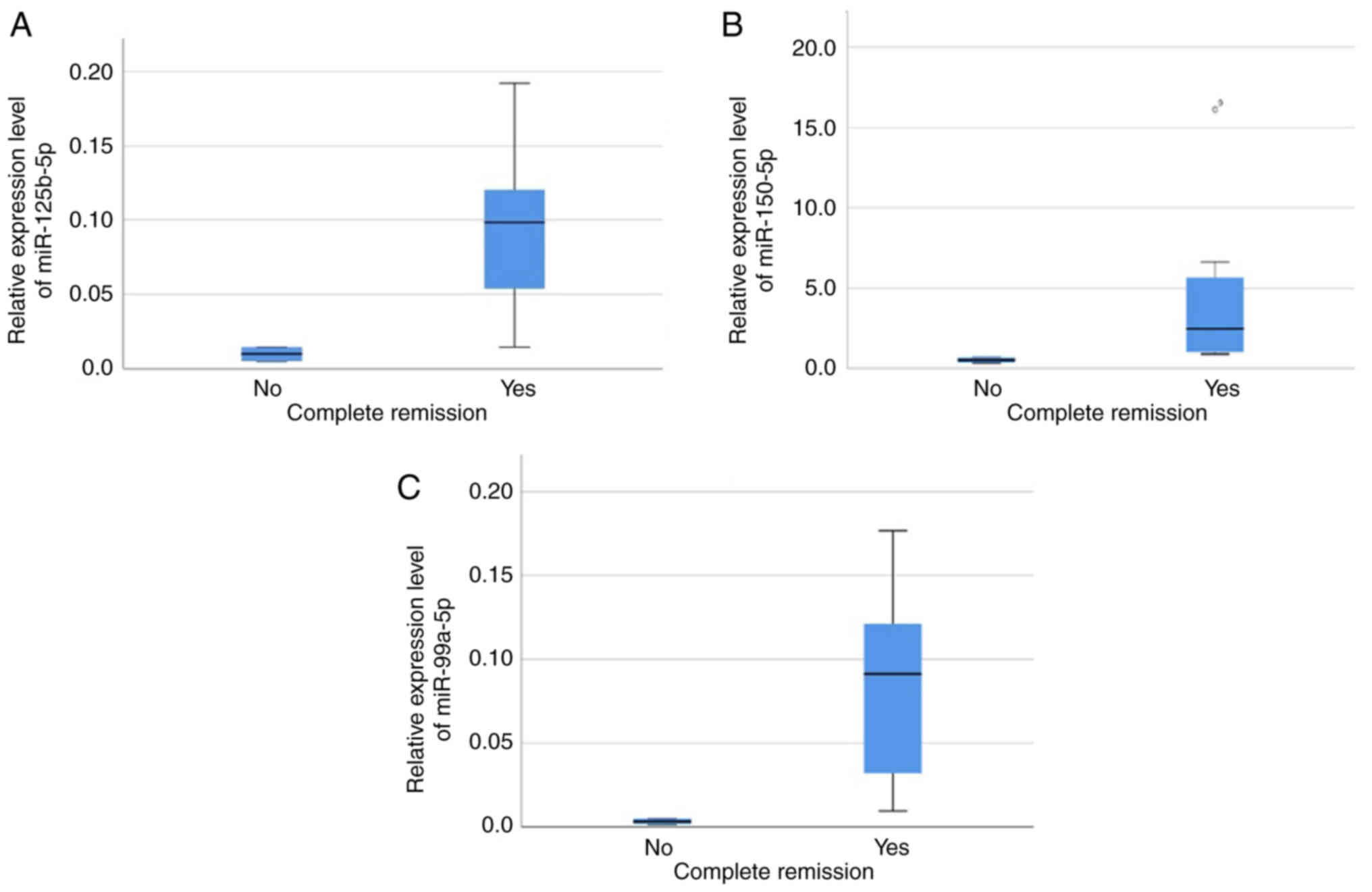
remission at the end of induction (day 33). Higher expression
levels of miR-125b-5p, miR-150-5p and miR-99a-5p were associated
with a complete response at the end of induction (Mann-Whitney U
P=0.044 for all 3 miRNAs; Table
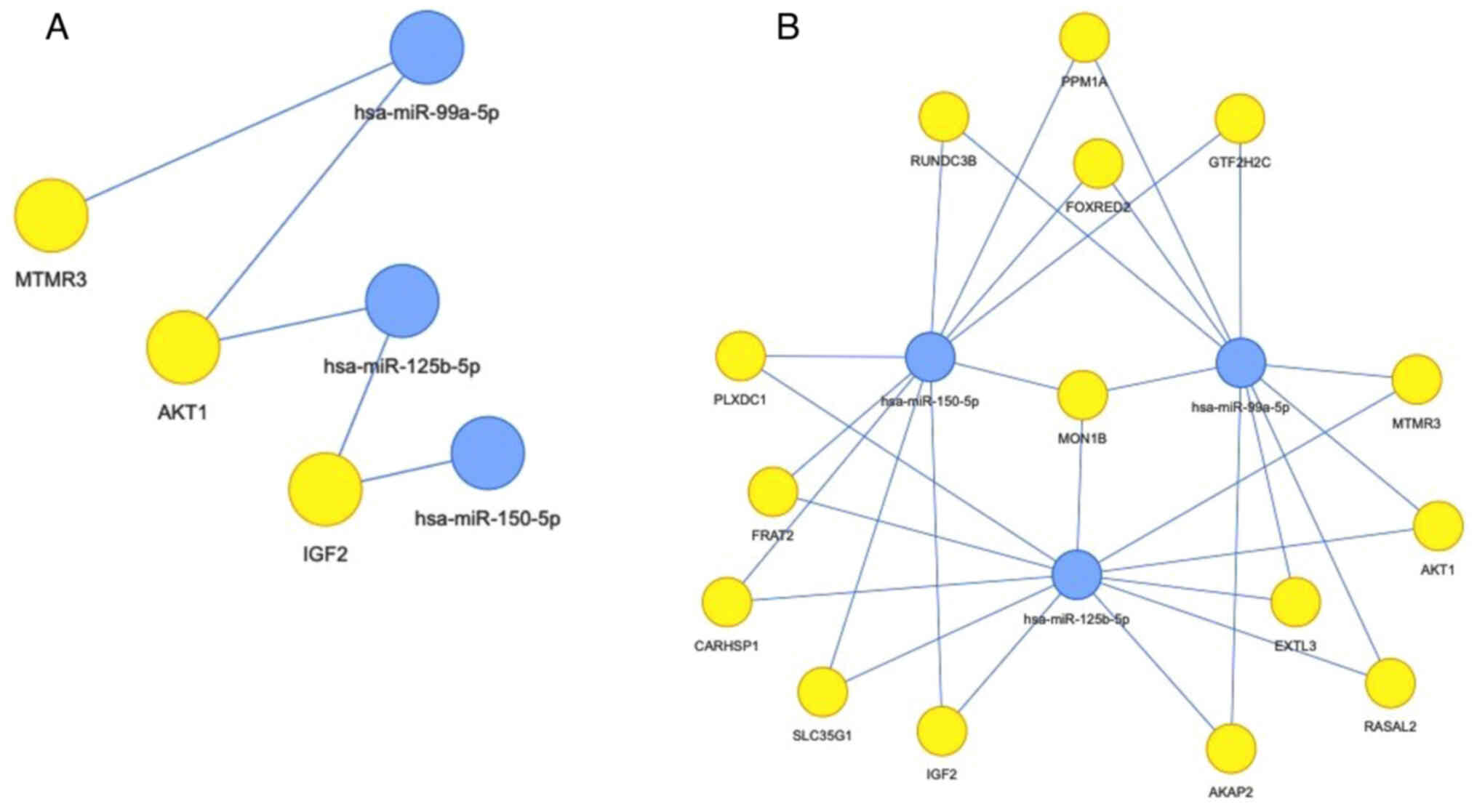
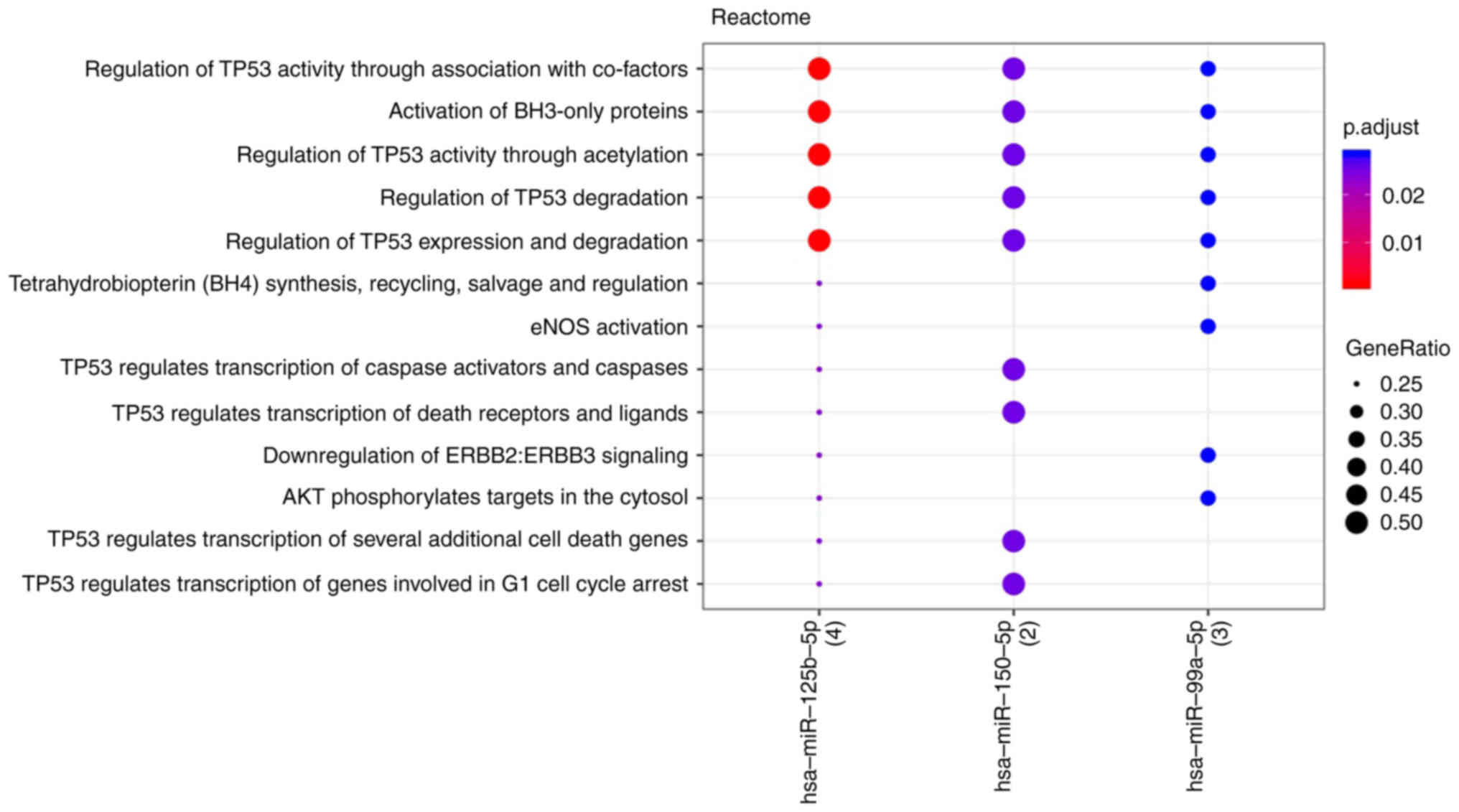
II), as shown in Fig. 3. The
interaction networks of these three miRNAs and their targets (based
on miRTarBase) are presented in Fig.
4 and the associated molecular pathways are shown in Fig. 5. TP53 and BH3 interacting domain
death agonist associated signaling appeared to serve a central role
in response to treatment.
miRNAs significantly associated with at least two
parameters (demographic and clinical characteristics, laboratory
findings and measures of response to treatment) and the respective
P-values are summarized in Table
II.
Effect of induction chemotherapy on
the miRNA expression profile
The expression levels of seven miRNAs (miR-100-5p,
miR-10b-5p, miR-143-3p, miR-145-5p, miR-182-5p, miR-10a-5p and
miR-22-3p) were markedly upregulated at diagnosis, whilst those of
eight miRNAs (miR-130a-3p, miR-146a-5p, miR-181a-5p, miR-181b-5p,
miR-195-5p, miR-20b-5p, miR-210-3p and miR-222-3p) were markedly
upregulated at the end of induction (day 33 of the therapeutic
protocol) (Table SV).
The FC between diagnosis and day 33 (as a
dichotomous variable, namely increase/decrease) differed
significantly between risk groups for three miRNAs (Table SVI). A significant increase in
expression levels of miR-206 was observed in all high-risk
patients, compared with a significant decrease in 6/7
standard/intermediate-risk patients (P=0.033). Conversely, an
increase in miR-210 levels was significantly associated with
standard/intermediate-risk disease, as it was observed in all
standard/intermediate-risk patients, compared with the significant
decrease in miR-210 levels observed in 2/3 high-risk patients
(P=0.047). An increase in miR-99a was also significantly associated
with standard/intermediate-risk disease, as all
standard/intermediate-risk patients had higher levels at the end of
induction compared with diagnosis, whilst its expression levels
were decreased in all high-risk patients (P=0.008).
Discussion
In the present study, differentially expressed
miRNAs were identified among pediatric ALL patient groups with
favorable and poor prognostic factors. The present study used a
commercially available cancer panel which included 84 miRNAs that
were tested in hematological malignancies or evaluated in solid
tumors. This allowed for the detection of novel associations that
have not been described previously, to the best of our knowledge.
Furthermore, certain associations that have been described
previously were further demonstrated in the present study.
Prednisone response was initially associated with
event-free survival by the Berlin-Frankfurt-Munster Group in 1983
(18) and its importance regarding
disease outcome has since been reported in a series of studies
(19–21). In the current pilot study, good
prednisone response was associated with increased expression levels
of let-7c-5p, miR-106b-5p, miR-26a-5p, miR-155-5p, miR-191-5p,
miR-30b-5p and miR-31-5p at diagnosis. Notably, 5/7 of this set of
miRNAs target the mRNA of the gene encoding the ribonucleotide
reductase regulatory subunit M2 (RRM2). RRM2 is an essential enzyme
for DNA synthesis and cell homeostasis as it catalyzes the
formation of deoxyribonucleotides from ribonucleotides, and
imbalanced deoxyribonucleoside triphosphate pools hinder proper DNA
replication and repair (22). RRM2
expression has been reported to be upregulated in cancer as may be
expected due to the increased DNA synthesis in the rapidly
proliferating neoplastic cells. It has also been used as a
biomarker associated with poor prognosis in solid tumors (22,23).
Thomas et al (24) first
reported that administration of dexamethasone reduced RRM2 levels
in multiple myeloma through glucocorticoid receptor binding within
100 kB of the transcription start site. Apart from its enzymatic
function, RRM2 has been implicated in the regulation of cell death
and its knockdown induces both autophagy and ferroptosis (25). Additionally, RRM2 is associated with
immune escape through macrophage polarization and the programmed
death protein 1/programmed death-ligand 1 signaling pathway
(22). However, to the best of our
knowledge, no studies have addressed the potential role of RRM2 in
ALL. The present study provided a basis for the use of RRM2 and/or
its pathway as a biomarker predictive of response, and as a
therapeutic target in case of high expression levels. Regarding the
other genes that serve as targets of more than two of the miRNAs
associated with prednisone response, forkhead box O3 has previously
been associated with poor prognosis and relapse in pediatric ALL
(26). DNA methyltransferase 1 and
transformation/transcription domain associated protein have been
implicated in epigenetic dysregulation, a defining feature of ALL
(27). Methionine aminopeptidase 2
is a metallopeptidase that catalyzes the removal of the N-terminal
methionine from newly synthesized proteins, an essential step for
their proper function, and has been implicated in the pathogenesis
of numerous solid tumors (28).
Chromatin assembly factor 1 subunit A (CHAF1A) mediates the
assembly of histone octamers onto replicating DNA during the
S-phase of the cell cycle and is required for normal hematopoiesis,
as CHAF1A deletion results in failure of stem and progenitor cells
to enter S-phase from G0/G1. Its
overexpression is associated with leukemogenesis (29).
The expression levels of three miRNAs (miR-125b-5p,
miR-99a-5p and miR-150-5p) were significantly upregulated at
diagnosis in standard/intermediate-risk patients (P=0.017, P=0.033
and P=0.017, respectively), as well as in those who achieved a
complete response on day 33 of induction (P=0.044 for all three
miRNAs).
The expression levels of miR-125b were higher in
patients with B-ALL, standard/intermediate-risk disease and a
complete response on day 33, and were negatively correlated with
MRD values on day 15 and 33. These findings are in-line with
previous studies, which reported that low levels of miR-125b may be
associated with a poor prognosis in pediatric ALL (30,31).
Low levels of miR-125b have not only been associated with older age
(>9 years), increased WBCs (>50.000/µl) and the high-risk
group, but have also been reported to be an independent prognostic
factor based on multivariate analysis (30). Increased miR-125b RNA expression
levels have also been associated with the presence of the
ETV6-RUNX1 fusion gene (10). In
the present study, higher expression levels of miR-125b were
observed in patients with the translocation t(12;21)(p13;q22)
compared with those without; however, the observation did not reach
statistical significance (P=0.22), possibly due to the small sample
size. Nevertheless, both El-Khazragy et al (30) and Piatopoulou et al (31) previously reported an association of
an increase in miR-125b levels post-chemotherapy with unfavorable
clinicopathological prognostic features, poor survival and relapse.
These findings are in accordance with the association of increased
miR-125b levels with resistance to vincristine and daunorubicin
(32), which are both included in
the ALL IC BFM 2009 protocol (13).
Regarding the underlying molecular mechanism, miR-125b serves an
essential role in normal B-cell development and protection from
carcinogenesis by targeting lin-28 homolog A (33). Furthermore, miR-125b regulates
apoptosis by targeting the antiapoptotic gene BCL2 (30,34).
miR-99a RNA expression levels were significantly
higher in patients with B-ALL, standard/intermediate-risk disease
and complete response on day 33 and were significantly negatively
correlated with MRD values on day 15 and 33. These findings are
also in-line with previous studies. Li et al (35) reported that miR-99a was
downregulated in high-risk patients and its expression levels were
associated with survival. In vitro restoration of miR-100
and miR-99a expression has been reported to result in suppression
of cell proliferation and to enhance the effect of dexamethasone
regarding induction of apoptosis by targeting two important
signaling pathways: i) FK506-binding protein 51, which along with
heat shock proteins regulates the translocation of the
glucocorticoid receptor from the cytoplasm to the nucleus; and ii)
insulin like growth factor 1 receptor/mTOR, and subsequently the
antiapoptotic MCL1 gene of the BCL2 family (35). Notably, in the present study, the
expression levels of miR-99a were increased in all
standard/intermediate-risk patients and decreased in all high-risk
patients. The present results supported the previously reported
observation that miR-99a acts as a tumor suppressor miRNA in
pediatric B-ALL, compared with its activity as an oncomiR in acute
myeloid leukemia (36).
Furthermore, similar to miR-125b, miR-99a is part of the molecular
signature of ETV6-RUNX1+ ALL and is also associated with
resistance to vincristine and daunorubicin (9,37,38).
In the present study, the levels of miR-99a were higher in the
patient group with the translocation t(12;21)(p13;q22); however,
this was not statistically significant (P=0.22), possibly due to
the small sample size (data not shown).
The expression levels of miR-150 were higher in
patients with B-ALL, standard/intermediate-risk disease and a
complete response on day 33 of induction chemotherapy. In
accordance with the present findings, previous studies have
reported downregulation of miR-150 at diagnosis to be associated
with unfavorable prognostic factors, namely age <1 year and
>6 years, increased WBCs (>20,000/µl) and T-ALL, as well as
with a greater risk of relapse (39,40).
Fang et al (41) reported
that miR-150 suppressed cell proliferation and induced apoptosis in
ALL cell lines and had a synergistic effect with cytarabine
administration, which is also an agent used in the ALL IC BFM 2009
protocol (13). miR-150 exerts its
antileukemic activity by regulating key cellular processes,
including transcription and cell metabolism, and its experimentally
identified targets include eukaryotic translation initiation factor
4B, forkhead box O4, protein kinase C a and Tet methylcytosine
dioxygenase 3 (41).
A total of three miRNAs, namely let-7c-5p,
miR-106b-5p and miR-191-5p, were associated with four prognostic
factors. let-7c-5p is known to be a part of the molecular signature
of ETV6-RUNX1+ ALL (10). Naderi et al (42) reported that miR-106b was implicated
in the pathogenesis of T-ALL by targeting cyclin D1, a key gene in
the Notch signaling pathway. Furthermore, miR-106b has been
reported to be upregulated in relapsed lysine methyltransferase
2A-rearranged acute myeloid leukemia in pediatric patients
(43). However, to the best of our
knowledge, there are no data regarding the role of miR-106b in
pediatric ALL at present. A total of two studies have associated
lower levels of miR-191-5p with relapse in ALL in the Chinese
pediatric population (39,44).
Bioinformatics analysis revealed that all three
miRNAs (miR-125b-5p, miR-99a-5p and miR-150-5p) that were
associated with complete response at the end of induction, targeted
the mRNA encoding MON1 homolog B (MON1B), which interferes with
NF-κB signaling. Knockdown of MON1B has been demonstrated to
increase the expression levels of of NF-κB inhibitor A and reduce
the expression levels of p65 (45).
Constitutively activated NF-κB complexes are found in the majority
of childhood ALL cases, regardless of subtype (46). Regarding the molecular pathways
associated with response to treatment, TP53 signaling seems to
serve a key role. Unlike in solid tumors, the role of TP53 in ALL
has only recently been reported. TP53 mutations are very rare in
pediatric ALL, accounting for 2–4% of B-ALL cases at diagnosis and
12% at relapse, and they are associated with a poor prognosis
(47). However, functional
deregulation of TP53 has been observed in B-ALL due to variations
in isoform expression (48).
Furthermore, Nakagawa et al (49) recently reported that TP53 was
implicated in the nucleolar stress response, through which
6-mercaptopurine, methotrexate, daunorubicin and cytarabine exert
their antileukemic activity. Further studies are required to
elucidate the role of miRNAs in TP53 deregulation in pediatric ALL
and possible associations with clinicopathological features and
prognosis.
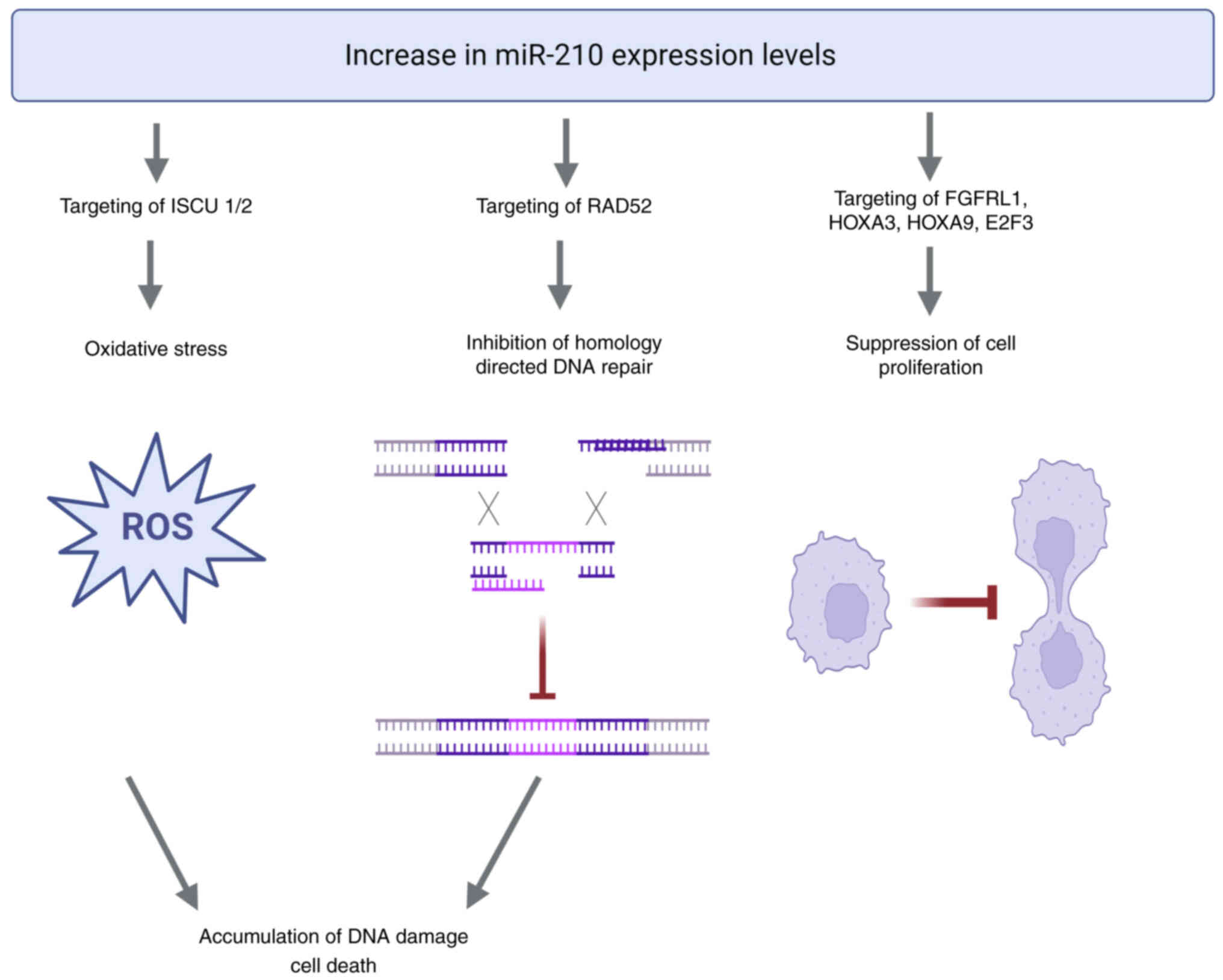
In the present study, miR-210 levels exhibited
changes pre- and post-therapy that markedly differed between risk
groups. miR-210 is a well-known hypoxamir, the expression of which
is induced by hypoxia inducible factor (HIF); it regulates key
cellular processes, including mitochondrial metabolism,
angiogenesis, DNA repair and cell survival (50,51).
Increased levels of miR-210 have been observed in several solid
tumors, serving at the same time as a biomarker associated with a
poor prognosis (50). However, its
role in ALL remains unclear with contradicting results in the few
studies with clinical samples (39,52,53).
In the present pilot study, there was no significant difference
between risk groups at diagnosis, but a significant increase in
expression levels was observed in all standard/intermediate-risk
patients compared with a decrease in 2/3 high-risk patients
(P=0.047) after chemotherapy administration. Contrary to its
protective effect in hypoxia, increased expression of miR-210 in
normoxia leads to the accumulation of reactive oxygen species and
oxidative stress mainly due to repression of the iron-sulfur
cluster scaffold proteins, ISCU1 and ISCU2. Furthermore, by
targeting DNA repair protein RAD52, miR-210 inhibits
homology-dependent DNA repair (50,51).
This causes accumulation of DNA damage and suppression of cell
proliferation via additional targeting of fibroblast growth factor
receptor like 1, homeobox A3, homeobox A9 and E2F transcription
factor 3 (50,51). As the extremely hypoxic state due to
rapid uncontrolled proliferation of leukemic cells in the bone
marrow at diagnosis is normalized with therapy administration, the
mechanism may provide a plausible explanation of the present
results. The proposed mechanism is presented in Fig. 6. Further studies are needed to
elucidate the complex mechanisms of the hypoxia-induced epigenetic
dysregulation in ALL and the effect of therapy administration, the
HIF-independent actions of miR-210 and ultimately of its possible
use as a biomarker for monitoring response to treatment in
pediatric ALL.
Furthermore, in the present study, the expression
levels of miR-206 were significantly increased in all high-risk
patients, compared with their decrease in 6/7
standard/intermediate-risk patients (P=0.033) after chemotherapy
administration. miR-206 was also significantly positively
correlated with MRD on day 33 (r=0.724; P=0.018). The role of
miR-206 has been extensively investigated in solid tumors, where it
acts as a tumor suppressor miRNA in the majority of cases (54); however, there is a lack of data
regarding its role in hematological malignancies. Existing evidence
suggests that miR-206 is upregulated in pediatric patients with
B-ALL compared with age-matched controls and that it inhibits cell
proliferation by targeting neuroepithelial cell-transforming 1
(55). Although the mechanism
behind the results of the present study is not yet clear, it can be
hypothesized that miR-206 participates in pathways associated with
cell proliferation. However, the potential for its use as a
prognostic biomarker needs to be addressed by larger studies.
In conclusion, the present study identified miRNAs
that were associated with established prognostic factors, including
age, sex, WBCs at diagnosis, immunophenotype and cytogenetic
findings. Furthermore, analysis revealed miRNA signatures
associated with the prednisone response, risk group and complete
response at the end of induction, as well as miRNAs, whose change
with chemotherapy administration could be monitored to assess the
response to treatment. These miRNAs could be added to the existing
measures of evaluating response to treatment, namely, prednisone
response and MRD, especially as RT-qPCR detection of molecular
markers is more sensitive than flow cytometry, which is routinely
used for MRD measurement (56).
Moreover, the present findings provide the basis for further
exploration of certain miRNA targets, whose role in pediatric ALL
has not yet been investigated. However, the short follow-up time
and the small sample size constitute the main limitations of the
present study, thus necessitating the validation of these findings
in larger patient cohorts to evaluate the extent to which these
miRNAs could serve as independent prognostic factors and whether
they could be successfully integrated into the current risk
stratification systems. Furthermore, evaluation of the target genes
at the mRNA and protein level could provide confirmation of the
role of the miRNAs and elucidate the molecular mechanisms of
treatment resistance.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by Pfizer Hellas S.A. (grant no.
75239833).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ET, EH, AT and EG were responsible for
conceptualization, EG and GT were responsible for the methodology,
ET and EG were responsible for formal analysis, ET, CA, ML, EP and
AGT performed the investigation, ML, EP, EH, AT, AGT and GT
provided resources, ET, CA and EG were responsible for data
curation, ET and CA prepared the original draft of the manuscript,
EH, AT, EG, GT, EP, and AGT reviewed and edited the manuscript, ET
was responsible for visualization of the study results, EH, AT, EG,
AGT and GT supervised the project, EH, AT, EG, GT and AGT were
responsible for administration and AT was responsible for funding
acquisition. ET and EG confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki and approved by the Committee for
Bioethics and Ethics of the School of Medicine of the Aristotle
University of Thessaloniki (Thessaloniki, Greece; approval no.
1.463; 15/10/2021).
Written informed consent was obtained from parents
or legal caregivers of all patients, as well as from patients aged
>12 years.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Howlader N, Noone AM, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS,
Feuer EJ and Cronin KA: SEER Cancer Statistics Review, 1975–2018.
National Cancer Institute; Bethesda, MD: 2011
|
|
2
|
Hunger SP, Lu X, Devidas M, Camitta BM,
Gaynon PS, Winick NJ, Reaman GH and Carroll WL: Improved survival
for children and adolescents with acute lymphoblastic leukemia
between 1990 and 2005: A report from the children's oncology group.
J Clin Oncol. 30:1663–1669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pui CH, Carroll WL, Meshinchi S and Arceci
RJ: Biology, risk stratification, and therapy of pediatric acute
leukemias: An update. J Clin Oncol. 29:551–565. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eckert C, Parker C, Moorman AV, Irving JA,
Kirschner-Schwabe R, Groeneveld-Krentz S, Révész T, Hoogerbrugge P,
Hancock J, Sutton R, et al: Risk factors and outcomes in children
with high-risk B-cell precursor and T-cell relapsed acute
lymphoblastic leukaemia: Combined analysis of ALLR3 and ALL-REZ BFM
2002 clinical trials. Eur J Cancer. 151:175–189. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pui CH, Mullighan CG, Evans WE and Relling
MV: Pediatric acute lymphoblastic leukemia: Where are we going and
how do we get there? Blood. 120:1165–1174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Woo JS, Alberti MO and Tirado CA:
Childhood B-acute lymphoblastic leukemia: A genetic update. Exp
Hematol Oncol. 3:162014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Montagner S, Dehó L and Monticelli S:
MicroRNAs in hematopoietic development. BMC Immunol. 15:142014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carvalho de Oliveira J, Molinari Roberto
G, Baroni M, Bezerra Salomão K, Alejandra Pezuk J and Sol Brassesco
M: MiRNA dysregulation in childhood hematological cancer. Int J Mol
Sci. 19:26882018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gutierrez-Camino A, Garcia-Obregon S,
Lopez-Lopez E, Astigarraga I and Garcia-Orad A: MiRNA deregulation
in childhood acute lymphoblastic leukemia: A systematic review.
Epigenomics. 12:69–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Szczepanek J: Role of microRNA
dysregulation in childhood acute leukemias: Diagnostics, monitoring
and therapeutics: A comprehensive review. World J Clin Oncol.
11:348–369. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Durmaz B, Bagca BG, Cogulu O, Susluer SY,
Alpay A, Aksoylar S and Gunduz C: Antileukemic Effects of
Anti-miR-146a, Anti-miR-155, Anti-miR-181a, and prednisolone on
childhood acute lymphoblastic leukemia. Biomed Res Int.
2021:32073282021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stary J, Zimmermann M, Campbell M,
Castillo L, Dibar E, Donska S, Gonzalez A, Izraeli S, Janic D,
Jazbec J, et al: Intensive chemotherapy for childhood acute
lymphoblastic leukemia: Results of the randomized intercontinental
trial ALL IC-BFM 2002. J Clin Oncol. 32:174–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Licursi V, Conte F, Fiscon G and Paci P:
MIENTURNET: An interactive web tool for microRNA-target enrichment
and network-based analysis. BMC Bioinformatics. 20:5452019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:(Database issue). D163–D169.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gillespie M, Jassal B, Stephan R, Milacic
M, Rothfels K, Senff-Ribeiro A, Griss J, Sevilla C, Matthews L,
Gong C, et al: The reactome pathway knowledgebase 2022. Nucleic
Acids Res. 50((D1)): D687–D692. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Riehm H, Reiter A, Schrappe M, Berthold F,
Dopfer R, Gerein V, Ludwig R, Ritter J, Stollmann B and Henze G:
Corticosteroid-dependent reduction of leukocyte count in blood as a
prognostic factor in acute lymphoblastic leukemia in childhood
(therapy study ALL-BFM 83). Klin Padiatr. 199:151–160. 1987.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dördelmann M, Reiter A, Borkhardt A,
Ludwig WD, Götz N, Viehmann S, Gadner H, Riehm H and Schrappe M:
Prednisone response is the strongest predictor of treatment outcome
in infant acute lymphoblastic leukemia. Blood. 94:1209–1217. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schrappe M, Möricke A, Reiter A, Henze G,
Welte K, Gadner H, Ludwig WD, Ritter J, Harbott J, Mann G, et al:
Key treatment questions in childhood acute lymphoblastic leukemia:
results in 5 consecutive trials performed by the ALL-BFM study
group from 1981 to 2000. Klin Padiatr. 225 (Suppl):S62–S72. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lauten M, Möricke A, Beier R, Zimmermann
M, Stanulla M, Meissner B, Odenwald E, Attarbaschi A, Niemeyer C,
Niggli F, et al: Prediction of outcome by early bone marrow
response in childhood acute lymphoblastic leukemia treated in the
ALL-BFM 95 trial: Differential effects in precursor B-cell and
T-cell leukemia. Haematologica. 97:1048–1056. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aye Y, Li M, Long MJ and Weiss RS:
Ribonucleotide reductase and cancer: Biological mechanisms and
targeted therapies. Oncogene. 34:2011–2021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Su YF, Wu TF, Ko JL, Tsai HT, Tee YT,
Chien MH, Chou CH, Lin WL, Low HY, Chou MY, et al: The expression
of ribonucleotide reductase M2 in the carcinogenesis of uterine
cervix and its relationship with clinicopathological
characteristics and prognosis of cancer patients. PLoS One.
9:e916442014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thomas AL, Coarfa C, Qian J, Wilkerson JJ,
Rajapakshe K, Krett NL, Gunaratne PH and Rosen ST: Identification
of potential glucocorticoid receptor therapeutic targets in
multiple myeloma. Nucl Recept Signal. 13:e0062015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zuo Z, Zhou Z, Chang Y, Liu Y, Shen Y, Li
Q and Zhang L: Ribonucleotide reductase M2 (RRM2): Regulation,
function and targeting strategy in human cancer. Genes Dis.
11:218–233. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han BW, Feng DD, Li ZG, Luo XQ, Zhang H,
Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, et al: A set of miRNAs
that involve in the pathways of drug resistance and leukemic
stem-cell differentiation is associated with the risk of relapse
and glucocorticoid response in childhood ALL. Hum Mol Genet.
20:4903–4915. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuan B, Zhang J, Wang H, Xiong L, Cai Q,
Wang T, Jacobsen S, Pradhan S and Wang Y: 6-Thioguanine reactivates
epigenetically silenced genes in acute lymphoblastic leukemia cells
by facilitating proteasome-mediated degradation of DNMT1. Cancer
Res. 71:1904–1911. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Selvakumar P, Lakshmikuttyamma A, Dimmock
JR and Sharma RK: Methionine aminopeptidase 2 and cancer. Biochim
Biophys Acta. 1765:148–154. 2006.PubMed/NCBI
|
|
29
|
Volk A, Liang K, Suraneni P, Li X, Zhao J,
Bulic M, Marshall S, Pulakanti K, Malinge S, Taub J, et al: A
CHAF1B-Dependent molecular switch in hematopoiesis and leukemia
pathogenesis. Cancer Cell. 34:707–723.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
El-Khazragy N, Elshimy AA, Hassan SS,
Matbouly S, Safwat G, Zannoun M and Riad RA: Dysregulation of
miR-125b predicts poor response to therapy in pediatric acute
lymphoblastic leukemia. J Cell Biochem. 120:7428–7438. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Piatopoulou D, Avgeris M, Marmarinos A,
Xagorari M, Baka M, Doganis D, Kossiva L, Scorilas A and Gourgiotis
D: MiR-125b predicts childhood acute lymphoblastic leukaemia poor
response to BFM chemotherapy treatment. Br J Cancer. 117:801–812.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schotte D, Chau JCK, Sylvester G, Liu G,
Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R and
den Boer ML: Identification of new microRNA genes and aberrant
microRNA profiles in childhood acute lymphoblastic leukemia.
Leukemia. 23:313–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chaudhuri AA, So AY, Mehta A, Minisandram
A, Sinha N, Jonsson VD, Rao DS, O'Connell RM and Baltimore D:
Oncomir miR-125b regulates hematopoiesis by targeting the gene
Lin28A. Proc Natl Acad Sci USA. 109:4233–4238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Willimott S and Wagner SD: miR-125b and
miR-155 contribute to BCL2 repression and proliferation in response
to CD40 ligand (CD154) in human leukemic B-cells. J Biol Chem.
287:2608–2617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li XJ, Luo XQ, Han BW, Duan FT, Wei PP and
Chen YQ: MicroRNA-100/99a, deregulated in acute lymphoblastic
leukaemia, suppress proliferation and promote apoptosis by
regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J
Cancer. 109:2189–2198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Li X, Ke Z, Huang L, Liang Y, Wu
J, Zhang X, Chen Y, Zhang H and Luo X: MiR-99a may serve as a
potential oncogene in pediatric myeloid leukemia. Cancer Cell Int.
13:1102013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schotte D, De Menezes RX, Akbari Moqadam
F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R and Den
Boer ML: MicroRNA characterize genetic diversity and drug
resistance in pediatric acute lymphoblastic leukemia.
Haematologica. 96:703–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Akbari Moqadam F, Lange-Turenhout EA,
Ariës IM, Pieters R and den Boer ML: MiR-125b, miR-100 and miR-99a
co-regulate vincristine resistance in childhood acute lymphoblastic
leukemia. Leuk Res. 37:1315–1321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Luo XQ, Zhang P, Huang LB, Zheng
YS, Wu J, Zhou H, Qu LH, Xu L and Chen YQ: MicroRNA patterns
associated with clinical prognostic parameters and CNS relapse
prediction in pediatric acute leukemia. PLoS One. 4:e78262009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Avigad S, Verly IR, Lebel A, Kordi O,
Shichrur K, Ohali A, Hameiri-Grossman M, Kaspers GJ, Cloos J,
Fronkova E, et al: miR expression profiling at diagnosis predicts
relapse in pediatric precursor B-cell acute lymphoblastic leukemia.
Genes Chromosomes Cancer. 55:328–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fang ZH, Wang SL, Zhao JT, Lin ZJ, Chen
LY, Su R, Xie ST, Carter BZ and Xu B: miR-150 exerts antileukemia
activity in vitro and in vivo through regulating genes in multiple
pathways. Cell Death Dis. 7:e23712016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Naderi T, Mohammadi-Yeganeh S,
Mohammadi-Hezaveh N, Hadavi R, Gharehbaghian A, Vazifeh-Shiran N,
Fallah Azad V and Paryan M: Investigating the inhibitory effect of
miR-34a, miR-449a, miR-1827, and miR-106b on target genes including
NOTCH1, c-Myc, and CCND1 in human T cell acute lymphoblastic
leukemia clinical samples and cell line. Iran J Basic Med Sci.
23:376–382. 2020.PubMed/NCBI
|
|
43
|
Verboon LJ, Obulkasim A, De Rooij JD,
Katsman-Kuipers JE, Sonneveld E, Baruchel A, Trka J, Reinhardt D,
Pieters R, Cloos J, et al: MicroRNA-106b ~ 25 cluster is
upregulated in relapsed MLL-rearranged pediatric acute myeloid
leukemia. Oncotarget. 7:48412–48422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu L, Liang YN, Luo XQ, Liu XD and Guo HX:
Association of miRNAs expression profiles with prognosis and
relapse in childhood acute lymphoblastic leukemia. Zhonghua Xue Ye
Xue Za Zhi. 32:178–181. 2011.(In Chinese). PubMed/NCBI
|
|
45
|
Jiang L, Qian J, Yang Y and Fan Y:
Knockdown of MON1B exerts anti-tumor effects in colon cancer in
vitro. Med Sci Monit. 24:7710–7718. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kordes U, Krappmann D, Heissmeyer V,
Ludwig WD and Scheidereit C: Transcription factor NF-κB is
constitutively activated in acute lymphoblastic leukemia cells.
Leukemia. 14:399–402. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ueno H, Yoshida K, Shiozawa Y, Nannya Y,
Iijima-Yamashita Y, Kiyokawa N, Shiraishi Y, Chiba K, Tanaka H,
Isobe T, et al: Landscape of driver mutations and their clinical
impacts in pediatric B-cell precursor acute lymphoblastic leukemia.
Blood Adv. 4:5165–5173. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Oh L, Hainaut P, Blanchet S and Ariffin H:
Expression of p53 N-terminal isoforms in B-cell precursor acute
lymphoblastic leukemia and its correlation with clinicopathological
profiles. BMC Cancer. 20:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nakagawa S, Kawahara K, Okamoto Y, Kodama
Y, Nishikawa T, Kawano Y and Furukawa T: Association between
dysfunction of the nucleolar stress response and multidrug
resistance in pediatric acute lymphoblastic leukemia. Cancers
(Basel). 14:51272022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Devlin C, Greco S, Martelli F and Ivan M:
miR-210: More than a silent player in hypoxia. IUBMB Life.
63:94–100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chan SY and Loscalzo J: MicroRNA-210: A
unique and pleiotropic hypoxamir. Cell Cycle. 9:1072–1083. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mei Y, Gao C, Wang K, Cui L, Li W, Zhao X,
Liu F, Wu M, Deng G, Ding W, et al: Effect of microRNA-210 on
prognosis and response to chemotherapeutic drugs in pediatric acute
lymphoblastic leukemia. Cancer Sci. 105:463–472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mei Y, Li Z, Zhang Y, Zhang W, Hu H, Zhang
P, Wu M and Huang D: Low miR-210 and CASP8AP2 expression is
associated with a poor outcome in pediatric acute lymphoblastic
leukemia. Oncol Lett. 14:8072–8077. 2017.PubMed/NCBI
|
|
54
|
Khalilian S, Hosseini Imani SZ and
Ghafouri-Fard S: Emerging roles and mechanisms of miR-206 in human
disorders: A comprehensive review. Cancer Cell Int. 22:4122022.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sun H, Zhang Z, Luo W, Liu J, Lou Y and
Xia S: NET1 enhances proliferation and chemoresistance in acute
lymphoblastic leukemia cells. Oncol Res. 27:935–944. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Contreras Yametti GP, Ostrow TH, Jasinski
S, Raetz EA, Carroll WL and Evensen NA: Minimal residual disease in
acute lymphoblastic leukemia: Current practice and future
directions. Cancers (Basel). 13:18472021. View Article : Google Scholar : PubMed/NCBI
|