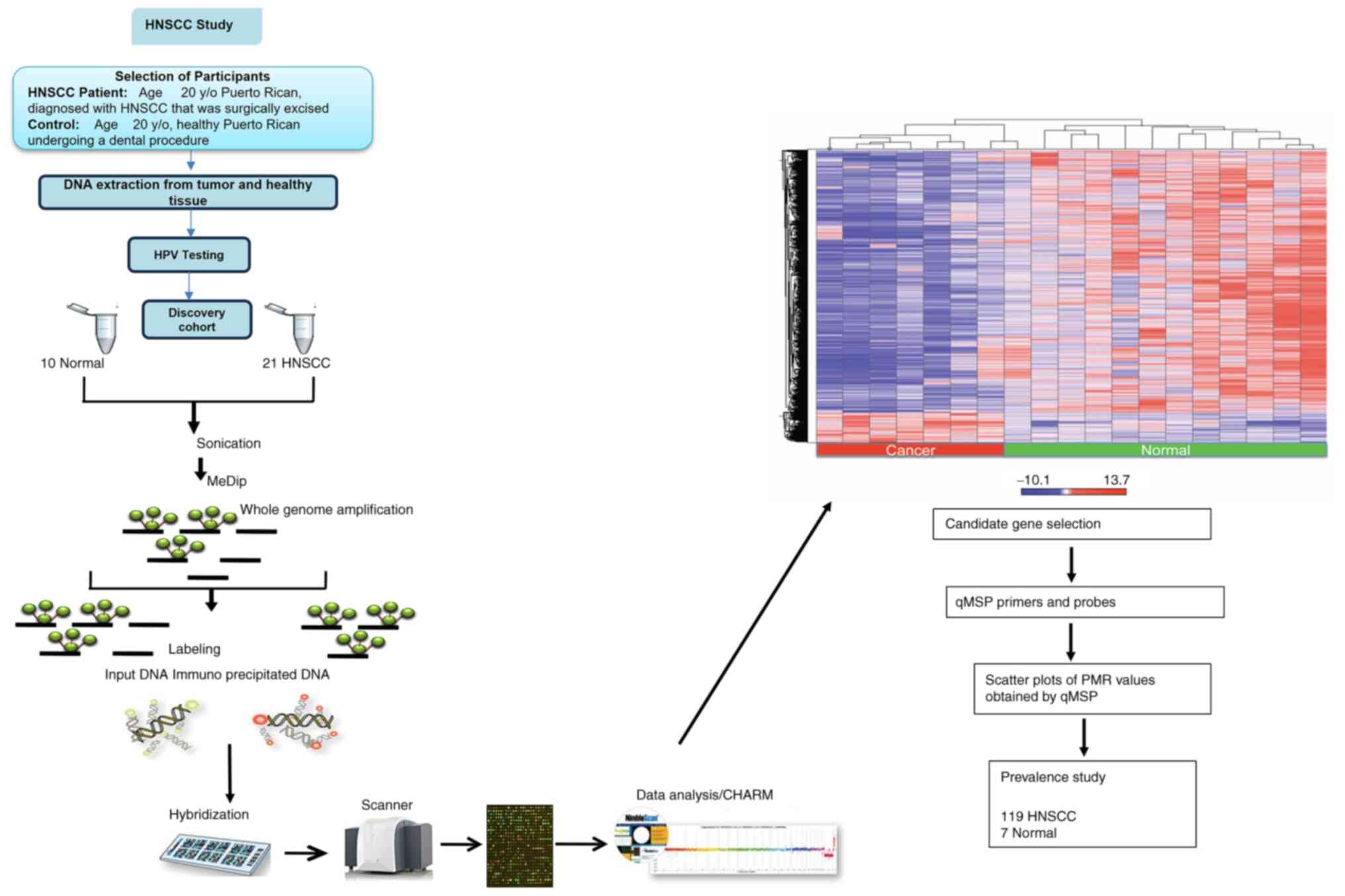
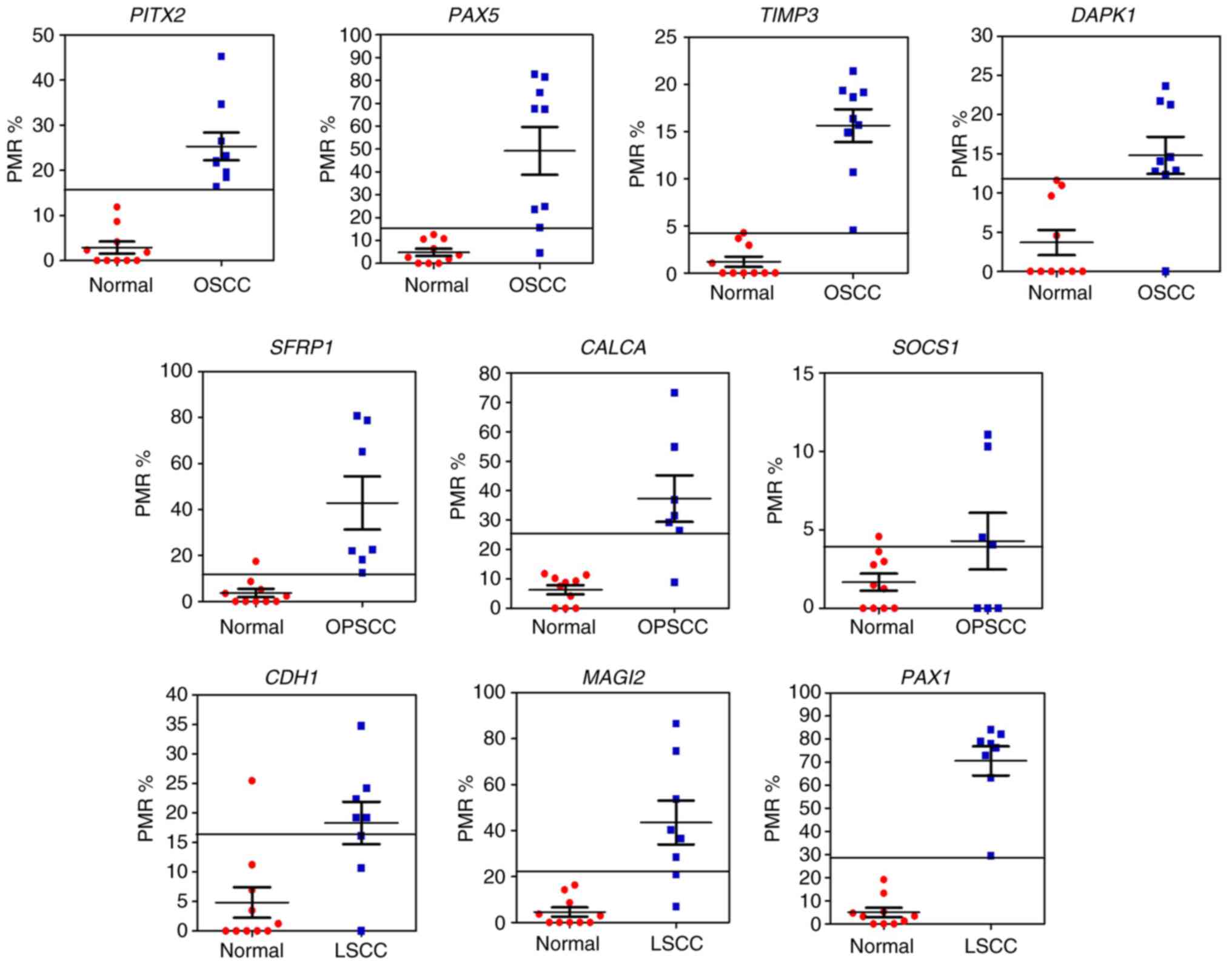
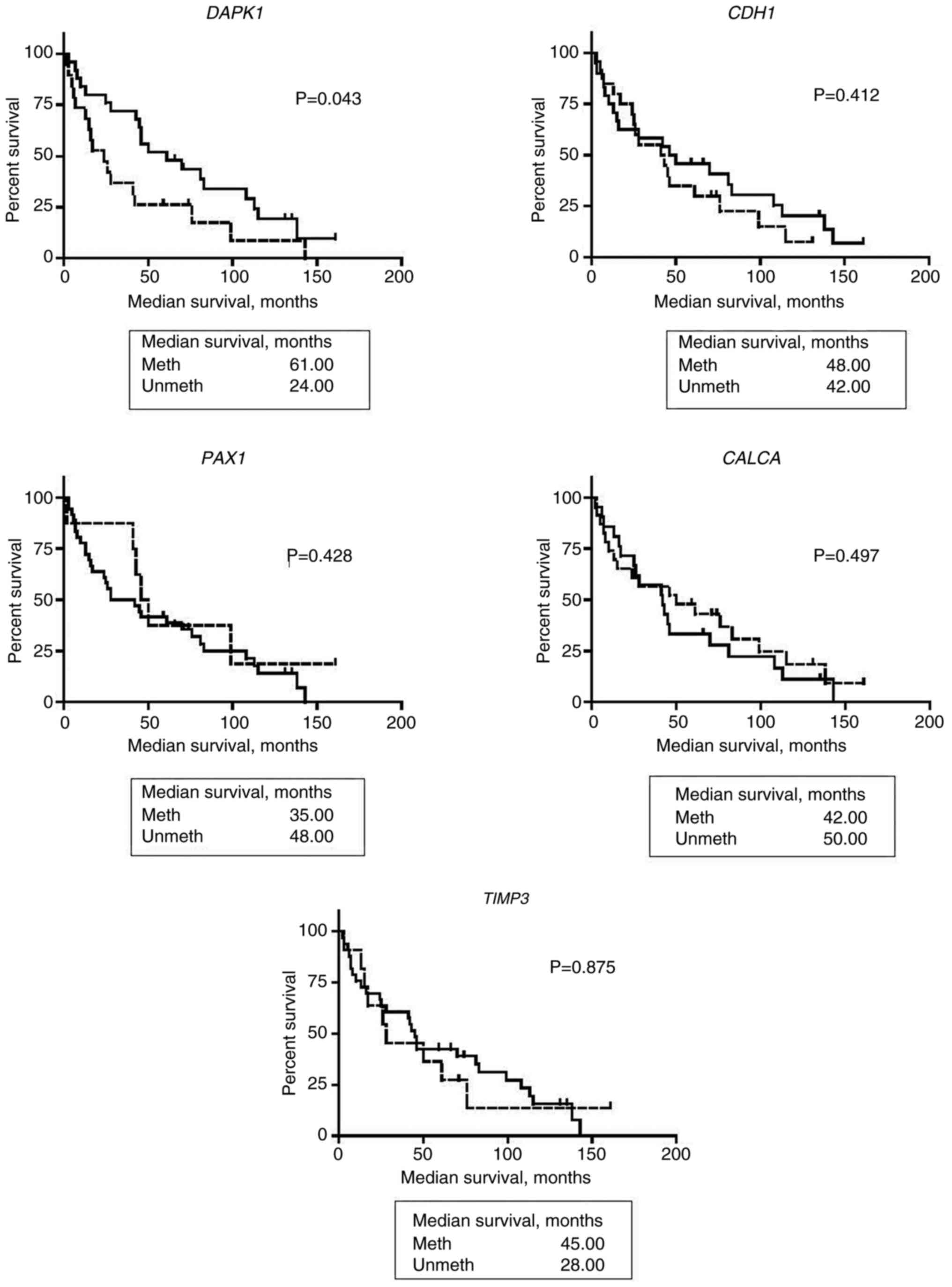
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
O'Rorke MA, Ellison MV, Murray LJ, Moran
M, James J and Anderson LA: Human papillomavirus related head and
neck cancer survival: A systematic review and meta-analysis. Oral
Oncol. 48:1191–1201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Báez A: Genetic and environmental factors
in head and neck cancer genesis. J Environ Sci Health C Environ
Carcinog Ecotoxicol Rev. 26:174–200. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saracci R: The interactions of tobacco
smoking and other agents in cancer etiology. Epidemiol Rev.
9:175–193. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blot WJ, McLaughlin JK, Winn DM, Austin
DF, Greenberg RS, Preston-Martin S, Bernstein L, Schoenberg JB,
Stemhagen A and Fraumeni JF Jr: Smoking and drinking in relation to
oral and pharyngeal cancer. Cancer Res. 48:3282–3287.
1988.PubMed/NCBI
|
|
6
|
Hashibe M, Brennan P, Benhamou S,
Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova
E, Fernandez L, et al: Alcohol drinking in never users of tobacco,
cigarette smoking in never drinkers, and the risk of head and neck
cancer: Pooled analysis in the International Head and Neck Cancer
Epidemiology Consortium. J Natl Cancer Inst. 99:777–789. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gillison ML, Koch WM, Capone RB, Spafford
M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et
al: Evidence for a causal association between human papillomavirus
and a subset of head and neck cancers. J Natl Cancer Inst.
92:709–720. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mork J, Lie AK, Glattre E, Hallmans G,
Jellum E, Koskela P, Møller B, Pukkala E, Schiller JT, Youngman L,
et al: Human papillomavirus infection as a risk factor for
squamous-cell carcinoma of the head and neck. N Engl J Med.
344:1125–1131. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Báez A, Almodóvar JI, Cantor A, Celestin
F, Cruz-Cruz L, Fonseca S, Trinidad-Pinedo J and Vega W: High
frequency of HPV16-associated head and neck squamous cell carcinoma
in the Puerto Rican population. Head Neck. 26:778–784. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rivera-Peña B, Ruíz-Fullana FJ,
Vélez-Reyes GL, Rodriguez-Benitez RJ, Marcos-Martínez MJ,
Trinidad-Pinedo J and Báez A: HPV-16 infection modifies overall
survival of Puerto Rican HNSCC patients. Infect Agent Cancer.
11:472004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park A, Alabaster A, Shen H, Mell LK and
Katzel JA: Undertreatment of women with locoregionally advanced
head and neck cancer. Cancer. 125:3033–3039. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Katzel JA, Merchant M, Chaturvedi AK and
Silverberg MJ: Contribution of demographic and behavioral factors
on the changing incidence rates of oropharyngeal and oral cavity
cancers in northern California. Cancer Epidemiol Biomarkers Prev.
24:978–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dayyani F, Etzel CJ, Liu M, Ho CH, Lippman
SM and Tsao AS: Meta-analysis of the impact of human papillomavirus
(HPV) on cancer risk and overall survival in head and neck squamous
cell carcinomas (HNSCC). Head Neck Oncol. 2:152010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shiboski CH, Schmidt BL and Jordan RC:
Tongue and tonsil carcinoma: Increasing trends in the U.S.
population ages 20–44 years. Cancer. 103:1843–1849. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
American Joint Committee on Cancer, . AJCC
Cancer Staging Manual. 8. New York; Springer: 2016
|
|
17
|
Baliga S, Yildiz VO, Bazan J, Palmer JD,
Jhawar SR, Konieczkowski DJ, Grecula J, Blakaj DM, Mitchell D,
Henson C, et al: Disparities in survival outcomes among
Racial/Ethnic minorities with head and neck squamous cell cancer in
the united states. Cancers (Basel). 15:17812023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suárez E, Calo WA, Hernández EY, Diaz EC,
Figueroa NR and Ortiz AP: Age-standardized incidence and mortality
rates of oral and pharyngeal cancer in Puerto Rico and among
Non-Hispanics Whites, Non-Hispanic Blacks, and Hispanics in the
USA. BMC Cancer. 9:1292009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fernandez AF, Assenov Y, Martin-Subero JI,
Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC,
Galm O, et al: A DNA methylation fingerprint of 1628 human samples.
Genome Res. 22:407–419. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taby R and Issa JP: Cancer epigenetics. CA
Cancer J Clin. 60:376–392. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ibáñez de Cáceres I and Cairns P:
Methylated DNA sequences for early cancer detection, molecular
classification and chemotherapy response prediction. Clin Transl
Oncol. 9:429–437. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calmon MF, Colombo J, Carvalho F, Souza
FP, Filho JF, Fukuyama EE, Camargo AA, Caballero OL, Tajara EH,
Cordeiro JA and Rahal P: Methylation profile of genes CDKN2A (p14
and p16), DAPK1, CDH1, and ADAM23 in head and neck cancer. Cancer
Genet Cytogenet. 173:31–37. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Demokan S and Dalay N: Role of DNA
methylation in head and neck cancer. Clin Epigenetics. 2:123–150.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ovchinnikov DA, Cooper MA, Pandit P, Coman
WB, Cooper-White JJ, Keith P, Wolvetang EJ, Slowey PD and
Punyadeera C: Tumor-suppressor gene promoter hypermethylation in
saliva of head and neck cancer patients. Transl Oncol. 5:321–326.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lleras RA, Smith RV, Adrien LR, Schlecht
NF, Burk RD, Harris TM, Childs G, Prystowsky MB and Belbin TJ:
Unique DNA methylation loci distinguish anatomic site and HPV
status in head and neck squamous cell carcinoma. Clin Cancer Res.
19:5444–5455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bernabe RD: INK4a/ARF/INK4b tumor
suppressor locus: Its potential role in head and neck cancer
tumorigenesis (Doctoral dissertation) (order no. 3221909). ProQuest
Dissertations and Theses Global. 2006.Available from. https://search.proquest.com/docview/304984294?accountid=44820
|
|
29
|
Rivera-Peña B and Báez A: Abstract 4793:
Aberrant methylation of CDH1 correlates with poor prognosis in
patients with head and neck squamous cell carcinoma (abstract). In:
Proceedings of the 102nd Annual Meeting of the American Association
for Cancer Research. Cancer Res. 71 (Suppl 8):4793. 2011.
View Article : Google Scholar
|
|
30
|
Irizarry RA, Ladd-Acosta C, Carvalho B, Wu
H, Brandenburg SA, Jeddeloh JA, Wen B and Feinberg AP:
Comprehensive high-throughput arrays for relative methylation
(CHARM). Genome Res. 18:780–790. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jaffe AE, Murakami P, Lee H, Leek JT,
Fallin MD, Feinberg AP and Irizarry RA: Bump hunting to identify
differentially methylated regions in epigenetic epidemiology
studies. Int J Epidemiol. 41:200–209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du P, Zhang X, Huang CC, Jafari N, Kibbe
WA, Hou L and Lin SM: Comparison of Beta-value and M-value methods
for quantifying methylation levels by microarray analysis. BMC
Bioinformatics. 11:5872010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
The Cancer Genome Atlas Network, .
Comprehensive genomic characterization of head and neck squamous
cell carcinomas. Nature. 517:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aryee MJ, Jaffe AE, Corrada-Bravo H,
Ladd-Acosta C, Feinberg AP, Hansen KD and Irizarry RA: Minfi: A
flexible and comprehensive Bioconductor package for the analysis of
Infinium DNA methylation microarrays. Bioinformatics. 30:1363–1369.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D, Danenberg PV and Laird PW: MethyLight: A
high-throughput assay to measure DNA methylation. Nucleic Acids
Res. 28:E322000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rettori MM, de Carvalho AC, Longo AL, de
Oliveira CZ, Kowalski LP, Carvalho AL and Vettore AL: TIMP3 and
CCNA1 hypermethylation in HNSCC is associated with an increased
incidence of second primary tumors. J Transl Med. 11:3162013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Valle B, Rodriguez-Torres S, Kuhn E,
Díaz-Montes T, Parrilla-Castellar E, Lawson F, Folawiyo O,
Ili-Gangas C, Brebi-Mieville P, Eshleman J, et al: HIST1H2BB and
MAGI2 methylation and somatic mutations as precision medicine
biomarkers for diagnosis and prognosis of high-grade serous ovarian
cancer. Cancer Prev Res (Phila). 13:783–794. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guerrero-Preston R, Valle BL, Jedlicka A,
Turaga N, Folawiyo O, Pirini F, Lawson F, Vergura A, Noordhuis M,
Dziedzic A, et al: Molecular triage of premalignant lesions in
Liquid-Based cervical cytology and circulating Cell-Free DNA from
urine, using a panel of methylated human papilloma virus and host
genes. Cancer Prev Res (Phila). 9:915–924. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guerrero-Preston R, Michailidi C,
Marchionni L, Pickering CR, Frederick MJ, Myers JN,
Yegnasubramanian S, Hadar T, Noordhuis MG, Zizkova V, et al: Key
tumor suppressor genes inactivated by ‘greater promoter’
methylation and somatic mutations in head and neck cancer.
Epigenetics. 9:1031–1046. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Anglim PP, Galler JS, Koss MN, Hagen JA,
Turla S, Campan M, Weisenberger DJ, Laird PW, Siegmund KD and
Laird-Offringa IA: Identification of a panel of sensitive and
specific DNA methylation markers for squamous cell lung cancer. Mol
Cancer. 7:622008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Müller HM, Widschwendter A, Fiegl H,
Ivarsson L, Goebel G, Perkmann E, Marth C and Widschwendter M: DNA
methylation in serum of breast cancer patients: An independent
prognostic marker. Cancer Res. 63:7641–7665. 2003.PubMed/NCBI
|
|
43
|
Eads CA, Lord RV, Wickramasinghe K, Long
TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester
TR, Skinner KA and Laird PW: Epigenetic patterns in the progression
of esophageal adenocarcinoma. Cancer Res. 61:3410–3418.
2001.PubMed/NCBI
|
|
44
|
van Asperen CJ, Brohet RM,
Meijers-Heijboer EJ, Hoogerbrugge N, Verhoef S, Vasen HF, Ausems
MG, Menko FH, Gomez Garcia EB, Klijn JG, et al: Cancer risks in
BRCA2 families: Estimates for sites other than breast and ovary. J
Med Genet. 42:711–719. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Seiwert TY, Zuo Z, Keck MK, Khattri A,
Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, et
al: Integrative and comparative genomic analysis of HPV-positive
and HPV-negative head and neck squamous cell carcinomas. Clin
Cancer Res. 21:632–641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Califano J, Van Der Riet P, Westra W,
Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B,
Koch W and Sidransky D: Genetic progression model for head and neck
cancer: Implications for field cancerization. Cancer Res.
56:2488–2492. 1996.PubMed/NCBI
|
|
47
|
Cai F, Xiao X, Niu X and Zhong Y:
Association between promoter methylation of DAPK gene and HNSCC: A
meta-analysis. PLoS One. 12:e01731942017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ngan HL, Liu Y, Fong AY, Poon PHY, Yeung
CK, Chan SSM, Lau A, Piao W, Li H, Tse J, et al: MAPK pathway
mutations in head and neck cancer affect immune microenvironments
and ErbB3 signaling. Life Sci Alliance. 3:e2019005452020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cilona M, Locatello LG, Novelli L and
Gallo O: The mismatch repair system (MMR) in head and neck
carcinogenesis and its role in modulating the response to
immunotherapy: A critical review. Cancers (Basel). 12:30062020.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Meng RW, Li YC, Chen X, Huang YX, Shi H,
Du DD, Niu X, Lu C and Lu MX: Aberrant Methylation of RASSF1A
closely associated with HNSCC, a Meta-Analysis. Sci Rep.
6:207562016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Moon SM, Kim SA, Yoon JH and Ahn SG: HOXC6
is deregulated in human head and neck squamous cell carcinoma and
modulates Bcl-2 expression. J Biol Chem. 287:35678–35688. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Carla C, Daris F, Cecilia B, Francesca B,
Francesca C and Paolo F: Angiogenesis in head and neck cancer: A
review of the literature. J Oncol. 2012:3584722012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mineta H, Miura K, Ogino T, Takebayashi S,
Misawa K, Ueda Y, Suzuki I, Dictor M, Borg A and Wennerberg J:
Prognostic value of vascular endothelial growth factor (VEGF) in
head and neck squamous cell carcinomas. Br J Cancer. 83:775–781.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Iwai S, Katagiri W, Kong C, Amekawa S,
Nakazawa M and Yura Y: Mutations of the APC, beta-catenin, and axin
1 genes and cytoplasmic accumulation of beta-catenin in oral
squamous cell carcinoma. J Cancer Res Clin Oncol. 131:773–782.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Leethanakul C, Patel V, Gillespie J,
Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M and
Gutkind JS: Distinct pattern of expression of differentiation and
growthrelated genes in squamous cell carcinomas of the head and
neck revealed by the use of laser capture microdissection and cDNA
arrays. Oncogene. 19:3220–3224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Paska AV and Hudler P: Aberrant
methylation patterns in cancer: A clinical view. Biochem Med
(Zagreb). 25:161–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hao X, Luo H, Krawczyk M, Wei W, Wang W,
Wang J, Flagg K, Hou J, Zhang H, Yi S, et al: DNA methylation
markers for diagnosis and prognosis of common cancers. Proc Natl
Acad Sci USA. 114:7414–7419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen D, Wang M, Guo Y, Wu W, Ji X, Dou X,
Tang H, Zong Z, Zhang X and Xiong D: An aberrant DNA methylation
signature for predicting the prognosis of head and neck squamous
cell carcinoma. Cancer Med. 10:5936–5947. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Stadler ME, Patel MR, Couch ME and Hayes
DN: Molecular biology of head and neck cancer: Risks and pathways.
Hematol Oncol Clin North Am. 22:1099–1124. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bialik S and Kimchi A: The
death-associated protein kinases: Structure, function, and beyond.
Annu Rev Biochem. 75:189–210. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gade P, Manjegowda SB, Nallar SC, Maachani
UB, Cross AS and Kalvakolanu DV: Regulation of the death-associated
protein kinase 1 expression and autophagy via ATF6 requires
apoptosis signal-regulating kinase 1. Mol Cell Biol. 34:4033–448.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li C, Wang L, Su J, Zhang R, Fu L and Zhou
Y: mRNA expression and hypermethylation of tumor suppressor genes
apoptosis protease activating factor-1 and death-associated protein
kinase in oral squamous cell carcinoma. Oncol Lett. 6:280–286.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jayaprakash C, Varghese VK, Bellampalli R,
Radhakrishnan R, Ray S, Kabekkodu SP and Satyamoorthy K:
Hypermethylation of Death-associated protein kinase (DAPK1) and its
association with oral carcinogenesis-An experimental and
meta-analysis study. Arch Oral Biol. 80:117–129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Melchers LJ, Clausen MJ, Mastik MF,
Slagter-Menkema L, van der Wal JE, Wisman GB, Roodenburg JL and
Schuuring E: Identification of methylation markers for the
prediction of nodal metastasis in oral and oropharyngeal squamous
cell carcinoma. Epigenetics. 10:850–860. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wei DM, Liu DY, Lei DP, Jin T, Wang J and
Pan XL: Aberrant methylation and expression of DAPk1 in human
hypopharyngeal squamous cell carcinoma. Acta Otolaryngol.
135:70–78. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
van Kempen PM, van Bockel L, Braunius WW,
Moelans CB, van Olst M, de Jong R, Stegeman I, van Diest PJ,
Grolman W and Willems SM: HPV-positive oropharyngeal squamous cell
carcinoma is associated with TIMP3 and CADM1 promoter
hypermethylation. Cancer Med. 3:1185–1196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Stephen JK, Chen KM, Shah V, Havard S,
Kapke A, Lu M, Benninger MS and Worsham MJ: DNA hypermethylation
markers of poor outcome in laryngeal cancer. Clin Epigenetics.
1:61–69. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Strzelczyk JK, Krakowczyk Ł and Owczarek
AJ: Aberrant DNA methylation of the p16, APC, MGMT, TIMP3 and CDH1
gene promoters in tumours and the surgical margins of patients with
oral cavity cancer. J Cancer. 9:1896–1904. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Schmezer P and Plass C: Epigenetic aspects
in carcinomas of the head and neck. HNO. 56:594–602. 2008.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Singh P, Ravanan P and Talwar P: Death
associated protein kinase 1 (DAPK1): A regulator of apoptosis and
autophagy. Front Mol Neurosci. 9:462016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Humphries MJ and Newham P: The structure
of cell-adhesion molecules. Trends Cell Biol. 8:78–83. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Riethmacher D, Brinkmann V and Birchmeier
C: A targeted mutation in the mouse E-cadherin gene results in
defective preimplantation development. Proc Natl Acad Sci USA.
92:855–859. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pećina-Slaus N: Tumor suppressor gene
E-cadherin and its role in normal and malignant cells. Cancer Cell
Int. 3:172003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Starska K, Forma E, Lewy-Trenda I, Papież
P, Woś J and Bryś M: Diagnostic impact of promoter methylation and
E-cadherin gene and protein expression levels in laryngeal
carcinoma. Contemp Oncol (Pozn). 17:263–271. 2013.PubMed/NCBI
|
|
75
|
Fan CC, Wang TY, Cheng YA, Jiang SS, Cheng
CW, Lee AY and Kao TY: Expression of E-cadherin, Twist, and p53 and
their prognostic value in patients with oral squamous cell
carcinoma. J Cancer Res Clin Oncol. 139:1735–1744. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fujii R, Imanishi Y, Shibata K, Sakai N,
Sakamoto K, Shigetomi S, Habu N, Otsuka K, Sato Y, Watanabe Y, et
al: Restoration of E-cadherin expression by selective Cox-2
inhibition and the clinical relevance of the
epithelial-to-mesenchymal transition in head and neck squamous cell
carcinoma. J Exp Clin Cancer Res. 33:402014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Shen Z, Zhou C, Li J, Deng H, Li Q and
Wang J: The association, clinicopathological significance, and
diagnostic value of CDH1 promoter methylation in head and neck
squamous cell carcinoma: A meta-analysis of 23 studies. Onco
Targets Ther. 9:6763–6773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
McGaughran JM, Oates A, Donnai D, Read AP
and Tassabehji M: Mutations in PAX1 may be associated with
Klippel-Feil syndrome. Eur J Hum Genet. 11:468–474. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wallin J, Eibel H, Neubüser A, Wilting J,
Koseki H and Balling R: Pax1 is expressed during development of the
thymus epithelium and is required for normal T-cell maturation.
Development. 122:23–30. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xu J, Xu L, Yang B, Wang L, Lin X and Tu
H: Assessing methylation status of PAX1 in cervical scrapings, as a
novel diagnostic and predictive biomarker, was closely related to
screen cervical cancer. Int J Clin Exp Pathol. 8:1674–1681.
2015.PubMed/NCBI
|
|
81
|
Dietrich S and Gruss P: Undulated
phenotypes suggest a role of Pax-1 for the development of vertebral
and extravertebral structures. Dev Biol. 167:529–548. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Su D, Ellis S, Napier A, Lee K and Manley
NR: Hoxa3 and pax1 regulate epithelial cell death and proliferation
during thymus and parathyroid organogenesis. Dev Biol. 236:316–329.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lorincz AT: Virtues and Weaknesses of DNA
methylation as a test for cervical cancer prevention. Acta Cytol.
60:501–512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Juodzbalys G, Kasradze D, Cicciù M,
Sudeikis A, Banys L, Galindo-Moreno P and Guobis Z: Modern
molecular biomarkers of head and neck cancer. Part I. Epigenetic
diagnostics and prognostics: Systematic review. Cancer Biomark.
17:487–502. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Huang YK, Peng BY, Wu CY, Su CT, Wang HC
and Lai HC: DNA methylation of PAX1 as a biomarker for oral
squamous cell carcinoma. Clin Oral Investig. 18:801–808. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sun R, Juan YC, Su YF, Zhang WB, Yu Y,
Yang HY, Yu GY and Peng X: Hypermethylated PAX1 and ZNF582 genes in
the tissue sample are associated with aggressive progression of
oral squamous cell carcinoma. J Oral Pathol Med. 49:751–760. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Cheng SJ, Chang CF, Ko HH, Lee JJ, Chen
HM, Wang HJ, Lin HS and Chiang CP: Hypermethylated ZNF582 and PAX1
genes in mouth rinse samples as biomarkers for oral dysplasia and
oral cancer detection. Head Neck. 40:355–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Morandi L, Gissi D, Tarsitano A, Asioli S,
Gabusi A, Marchetti C, Montebugnoli L and Foschini MP: CpG location
and methylation level are crucial factors for the early detection
of oral squamous cell carcinoma in brushing samples using bisulfite
sequencing of a 13-gene panel. Clin Epigenetics. 9:852017.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cheng SJ, Chang CF, Ko HH, Liu YC, Peng
HH, Wang HJ, Lin HS and Chiang CP: Hypermethylated ZNF582 and PAX1
genes in oral scrapings collected from cancer-adjacent normal oral
mucosal sites are associated with aggressive progression and poor
prognosis of oral cancer. Oral Oncol. 75:169–177. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hsu YW, Huang RL, Su PH, Chen YC, Wang HC,
Liao CC and Lai HC: Genotype-specific methylation of HPV in
cervical intraepithelial neoplasia. J Gynecol Oncol. 28:e562017.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Brain SD, Williams TJ, Tippins JR, Morris
HR and MacIntyre I: Calcitonin gene-related peptide is a potent
vasodilator. Nature. 313:54–56. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Vidal DO, Paixão VA, Brait M, Souto EX,
Caballero OL, Lopes LF and Vettore AL: Aberrant methylation in
pediatric myelodysplastic syndrome. Leuk Res. 31:175–181. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Martinelli CMDS, Lengert AVH, Cárcano FM,
Silva ECA, Brait M, Lopes LF and Vidal DO: MGMT and CALCA promoter
methylation are associated with poor prognosis in testicular germ
cell tumor patients. Oncotarget. 8:50608–50617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Brait M, Begum S, Carvalho AL, Dasgupta S,
Vettore AL, Czerniak B, Caballero OL, Westra WH, Sidransky D and
Hoque MO: Aberrant promoter methylation of multiple genes during
pathogenesis of bladder cancer. Cancer Epidemiol Biomarkers Prev.
17:2786–2794. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wang Y, Zhang D, Zheng W, Luo J, Bai Y and
Lu Z: Multiple gene methylation of nonsmall cell lung cancers
evaluated with 3-dimensional microarray. Cancer. 112:1325–1336.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhang B, Liu S, Zhang Z, Wei J, Qu Y, Wu
K, Yang Q, Hou P and Shi B: Analysis of BRAF(V600E) mutation and
DNA methylation improves the diagnostics of thyroid fine needle
aspiration biopsies. Diagn Pathol. 9:452014. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Loyo M, Brait M, Kim MS, Ostrow KL, Jie
CC, Chuang AY, Califano JA, Liégeois NJ, Begum S, Westra WH, et al:
Asurvey of methylated candidate tumor suppressor genes in
nasopharyngeal carcinoma. Int J Cancer. 128:1393–1403. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ismail EA, El-Mogy MI, Mohamed DS and
El-Farrash RA: Methylation pattern of calcitonin (CALCA) gene in
pediatric acute leukemia. J Pediatr Hematol Oncol. 33:534–542.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Paixão VA, Vidal DO, Caballero OL, Vettore
AL, Tone LG, Ribeiro KB and Lopes LF: Hypermethylation of CpG
island in the promoter region of CALCA in acute lymphoblastic
leukemia with central nervous system (CNS) infiltration correlates
with poorer prognosis. Leuk Res. 30:891–894. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ji M, Guan H, Gao C, Shi B and Hou P:
Highly frequent promoter methylation and PIK3CA amplification in
non-small cell lung cancer (NSCLC). BMC Cancer. 11:1472011.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Morán A, Fernández-Marcelo T, Carro J, De
Juan C, Pascua I, Head J, Gómez A, Hernando F, Torres AJ, Benito M
and Iniesta P: Methylation profiling in non-small cell lung cancer:
Clinical implications. Int J Oncol. 40:739–746. 2012.PubMed/NCBI
|
|
102
|
Guerrero-Preston R, Soudry E, Acero J,
Orera M, Moreno-López L, Macía-Colón G, Jaffe A, Berdasco M,
Ili-Gangas C, Brebi-Mieville P, et al: NID2 and HOXA9 promoter
hypermethylation as biomarkers for prevention and early detection
in oral cavity squamous cell carcinoma tissues and saliva. Cancer
Prev Res (Phila). 4:1061–1072. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Jithesh PV, Risk JM, Schache AG, Dhanda J,
Lane B, Liloglou T and Shaw RJ: The epigenetic landscape of oral
squamous cell carcinoma. Br J Cancer. 108:370–379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hypermethylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
105
|
Bachman KE, Herman JG, Corn PG, Merlo A,
Costello JF, Cavenee WK, Baylin SB and Graff JR:
Methylation-associated silencing of the tissue inhibitor of
metalloproteinase-3 gene suggest a suppressor role in kidney,
brain, and other human cancers. Cancer Res. 59:798–802.
1999.PubMed/NCBI
|
|
106
|
Darnton SJ, Hardie LJ, Muc RS, Wild CP and
Casson AG: Tissue inhibitor of metalloproteinase-3 (TIMP-3) gene is
methylated in the development of esophageal adenocarcinoma: Loss of
expression correlates with poor prognosis. Int J Cancer.
115:351–358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Smookler DS, Mohammed FF, Kassiri Z,
Duncan GS, Mak TW and Khokha R: Tissue inhibitor of
metalloproteinase 3 regulates TNF-dependent systemic inflammation.
J Immunol. 176:721–725. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Mohammed FF, Smookler DS, Taylor SE,
Fingleton B, Kassiri Z, Sanchez OH, English JL, Matrisian LM, Au B,
Yeh WC and Khokha R: Abnormal TNF activity in Timp3-/-mice leads to
chronic hepatic inflammation and failure of liver regeneration. Nat
Genet. 36:969–977. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhang L, Zhao L, Zhao D, Lin G, Guo B, Li
Y, Liang Z, Zhao XJ and Fang X: Inhibition of tumor growth and
induction of apoptosis in prostate cancer cell lines by
overexpression of tissue inhibitor of matrix metalloproteinase-3.
Cancer Gene Ther. 17:171–179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ahonen M, Ala-Aho R, Baker AH, George SJ,
Grénman R, Saarialho-Kere U and Kähäri VM: Antitumor activity and
bystander effect of adenovirally delivered tissue inhibitor of
metalloproteinases-3. Mol Ther. 5:705–715. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Spurbeck WW, Ng CY, Strom TS, Vanin EF and
Davidoff AM: Enforced expression of tissue inhibitor of matrix
metalloproteinase-3 affects functional capillary morphogenesis and
inhibits tumor growth in a murine tumor model. Blood.
100:3361–3368. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Bian J, Wang Y, Smith MR, Kim H, Jacobs C,
Jackman J, Kung HF, Colburn NH and Sun Y: Suppression of in vivo
tumor growth and induction of suspension cell death by tissue
inhibitor of metalloproteinases (TIMP)-3. Carcinogenesis.
17:1805–1811. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Arantes LM, de Carvalho AC, Melendez ME,
Centrone CC, Góis-Filho JF, Toporcov TN, Caly DN, Tajara EH,
Goloni-Bertollo EM and Carvalho AL; GENCAPO: Validation of
methylation markers for diagnosis of oral cavity cancer. Eur J
Cancer. 51:632–641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Righini CA, de Fraipont F, Timsit JF,
Faure C, Brambilla E, Reyt E and Favrot MC: Tumor-specific
methylation in saliva: A promising biomarker for early detection of
head and neck cancer recurrence. Clin Cancer Res. 13:1179–1185.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhang HZ, Shan CG, Huang AP and Wang JM:
Characterization of gene methylation in human papillomavirus
associated-head and neck squamous cell carcinoma. Genet Mol Res.
152016.doi: 10.4238/gmr.15038206.
|
|
116
|
Sartor MA, Dolinoy DC, Jones TR, Colacino
JA, Prince ME, Carey TE and Rozek LS: Genome-wide methylation and
expression differences in HPV(+) and HPV(−) squamous cell carcinoma
cell lines are consistent with divergent mechanisms of
carcinogenesis. Epigenetics. 6:777–787. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Nakagawa T, Matsusaka K, Misawa K, Ota S,
Takane K, Fukuyo M, Rahmutulla B, Shinohara KI, Kunii N, Sakurai D,
et al: Frequent promoter hypermethylation associated with human
papillomavirus infection in pharyngeal cancer. Cancer Lett.
407:21–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Nakagawa T, Kurokawa T, Mima M, Imamoto S,
Mizokami H, Kondo S, Okamoto Y, Misawa K, Hanazawa T and Kaneda A:
DNA Methylation and HPV-Associated Head and Neck Cancer.
Microorganisms. 9:8012021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Nakahara Y, Shintani S, Mihara M, Ueyama Y
and Matsumura T: High frequency of homozygous deletion and
methylation of p16(INK4A) gene in oral squamous cell carcinomas.
Cancer Lett. 163:221–228. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Richards KL, Zhang B, Baggerly KA, Colella
S, Lang JC, Schuller DE and Krahe R: Genome-wide hypomethylation in
head and neck cancer is more pronounced in HPV-negative tumors and
is associated with genomic instability. PLoS One. 4:e49412009.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
van Kempen PM, Noorlag R, Braunius WW,
Stegeman I, Willems SM and Grolman W: Differences in methylation
profiles between HPV-positive and HPV-negative oropharynx squamous
cell carcinoma: A systematic review. Epigenetics. 9:194–203. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Camuzi D, Buexm LA, Lourenço SQC, Esposti
DD, Cuenin C, Lopes MdSA, Manara F, Talukdar FR, Herceg Z, Ribeiro
Pinto LF, et al: HPV infection leaves a DNA methylation signature
in oropharyngeal cancer affecting both coding genes and
transposable elements. Cancers. 13:36212021. View Article : Google Scholar : PubMed/NCBI
|