Introduction
Pathological complete response (pCR) refers to the
complete absence of cancer cells from both primary and metastatic
tumors after neoadjuvant chemotherapy (NAC). pCR is the optimal
outcome for patients with breast cancer (BC). However, numerous
patients with BC show resistance to NAC, with only a minority of
the patients (20–25%) attaining pCR (1,2).
Identifying prognostic indicators of drug resistance is essential
for predicting tumor response and clinical outcomes (3). Despite reports of drug resistance
mechanisms, the mechanisms underlying NAC resistance remain to be
fully elucidated (4–6). Additionally, reliable biomarkers for
predicting pCR are lacking, which complicates the effective
assessment of treatment outcomes.
Numerous mechanisms regarding drug resistance have
been proposed, including alterations in drug transport, metabolism,
DNA repair, and cell signaling pathways (7,8).
However, the mechanisms underlying NAC resistance remain unclear
and are likely to be multifactorial. Recently, several microRNAs
(miRNAs) were discovered to be significantly upregulated in
drug-resistant cancer cells, emerging as potential biomarkers for
evaluating tumor responses to treatment (9,10).
MiRNAs regulate gene expression by targeting multiple mRNAs for
degradation or translational inhibition (11). They are the most abundant biomarker
sources found in exosomes, for example, as cell-to-cell signal
transporters; they play a critical role in the pathogenesis of BC
by regulating target gene networks involved in key processes, such
as cell proliferation, apoptosis, and metastasis (12). Dysregulated exosomal miRNAs that
reach adjacent and distant cancer cells and regulate drug
resistance-related genes can potentially serve as predictive
indicators of tumor responses to treatment.
Numerous studies have demonstrated the involvement
of miRNAs in the development and progression of drug resistance in
cancer (13,14). However, a number of unidentified
miRNAs, target genes and molecular mechanisms remain associated
with drug resistance. Therefore, the construction of a miRNA-mRNA
network that uncovers the mechanisms underlying drug resistance
could help identify potential novel biomarkers for improving the
prediction accuracy of tumor response to treatment. In the present
study, tumor-derived exosomes (TDEs) were profiled in the plasma
using liquid biopsy to identify the specific molecules or
signatures responsible for transferring drug resistance. The
rationale of the authors of the present study was based on the
understanding that exosomes transport diverse molecules, including
proteins and nucleic acids, which influence the behavior of
recipient cells (15). Given the
well-documented cooperativity of exosomal miRNAs that regulate
tumorigenesis and patient survival in numerous cancers (16), miRNAs and their target gene networks
could potentially serve as predictive markers of metastasis,
relapse, and drug resistance. Previous studies by the authors
identified specific miRNA signatures in exosomes that are
significantly elevated in BC, thereby not only confirming the
presence of cancer cells but also allowing the evaluation of
molecular subtypes and metastatic potential in patients with BC
(17,18). Therefore, the role of exosomal
miRNAs was investigated, as a clinical tool to elucidate
diagnostic, prognostic, and treatment response assessments by
performing an integrated analysis. To ensure a more accurate and
precise analysis, the authors focused specifically on investigating
the changes in miRNA expression within TDEs because their clinical
value is relatively unexplored.
Investigating the role of exosomal miRNAs in BC drug
resistance can elucidate molecular mechanisms underlying BC drug
resistance and aid in the development of novel diagnostic and
therapeutic strategies. Furthermore, investigating epigenetic
alterations in drug-resistant tumors offers a deeper understanding
of how exosomal miRNAs affect the molecular landscape and
therapeutic responses of tumors. Analysis of such epigenetic
changes regulated in drug-resistant tumors can provide crucial
information regarding the complex interactions between exosomal
miRNAs and tumor-associated processes. In the present study, five
drug resistance-related miRNA combinations were identified,
particularly in TDEs and their predicted target genes, via network
analysis using public datasets. The findings of the present study
may aid in the development of targeted therapies that modulate
miRNA expression and regulate associated target gene expression.
Consequently, this may lead to improved treatment outcomes in
patients with BC.
Materials and methods
Patient selection
Informed consent for the use of plasma samples for
research purposes was obtained from all participants. Clinical
samples were obtained from subjects who visited Severance Hospital
(Seoul, South Korea), according to the guidelines of the
independent Ethics Committee of Yonsei University College of
Medicine (IRB approval no. 4-2020-1292; approval date January 4,
2021; Seoul, South Korea). A total of 35 patients with BC (20 with
BC exhibiting no pCR after NAC and 15 patients showing pCR after
NAC) who visited Severance Hospital between May 2015 and August
2020 were retrospectively registered in the present study.
Preoperative plasma samples were collected from the participants
before anesthesia. The criteria for inclusion in the analysis were
as follows: i) NAC prior to plasma specimen acquisition; ii)
confirmed pathological diagnosis of BC; and iii) hemolysis assessed
before the isolation of exosomes to evaluate plasma sample quality.
Moreover, the study excluded participants with breast cancer types
other than invasive ductal carcinoma and those with a history of
other malignancies or existing medical conditions. The clinical
characteristics of participants enrolled in the present study are
summarized in Table SI and the
research design is illustrated in Fig.
1.
Cell lines and chemo-drugs
Doxorubicin hydrochloride (brand name Adriamycin;
cat. no. D4000) and docetaxel (brand name Taxotere; cat. no. D1000)
were purchased from LC Laboratories. Cyclophosphamide (brand name
Cytoxan; cat. no. S1217) was purchased from Selleck Chemicals.
Adriamycin (5 mg/ml) and Cytoxan (20 mg/ml) were dissolved in
saline (JW Pharmaceutical Co., Ltd.). Taxotere (5 mg/ml) was
dissolved in dimethyl sulfoxide (Duchefa Biochemie; cat. no.
D1370). Aliquots were stored at −20°C for a maximum of 6 months and
thawed immediately before use. Human BC cell lines MDA-MB-231
(MM231; HTB-26), MDA-MB-468 (MM468; HTB-132), and HCC1395
(CRL-2324) were purchased from the American Type Culture
Collection. All cell lines were grown in Roswell Park Memorial
Institute-1640 medium (RPMI-1640; cat. no. 22400-089) supplemented
with 10% fetal bovine serum (FBS; cat. no. 12483-020) and 1%
penicillin-streptomycin (cat. no. 15140-122) (all from Gibco;
Thermo Fisher Scientific, Inc.). All cells were grown as monolayer
cultures and maintained in a humidified atmosphere of 5%
CO2 at 37°C.
Generating drug-resistant BC cell
lines
Drug-resistant BC cells were established by
consistently increasing the concentrations of clinically used
regimens, including Adriamycin, Cytoxan, and Taxotere, in the
culture medium (19). Adriamycin
and Cytoxan (1:10 weight ratio) followed by Taxotere were
alternately added to the cells at specific concentrations [1/120,
1/90, 1/60, 1/30, 1/10, and 1 time of the half-maximal inhibitory
concentration (IC50) values of each drug] when they
reached 60–70% confluency as per the protocol reported in the
previous study by the authors (20). After continuous exposure to
chemo-drugs for 6 months, the parental BC cells (herein referred to
as ‘MM231wild-type’, ‘MM468wild-type’ and
‘HCC1395wild-type’) were allowed to grow in fresh medium
for an additional month until the surviving cells recovered
favorably. After 7 months, the stabilized drug-resistant BC cells
(herein referred to as ‘MM231resistant-type’,
‘MM468resistant-type’ and
‘HCC1395resistant-type’) were generated and stored in a
deep freezer (−70 to −80°C) for further investigation. The drug
resistance characteristics were confirmed by comparison of the
IC50 values between the cells (Fig. S1 and Table SII).
Microarray analysis
Small RNA was isolated from cultured cells using a
mirVana™ miRNA Isolation kit (Thermo Fisher Scientific, Inc.; cat.
no. AM1560) according to the manufacturer's instructions, using
TRIzol reagent (Thermo Fisher Scientific, Inc.; cat. no. 15596026).
The extracted small RNA was quantified using UV absorption at a
wavelength of 260 and 280 nm with a spectrophotometer (NanoDrop
3000; Thermo Fisher Scientific, Inc.) and then stored at −80°C
until further analysis. Affymetrix GeneChip microarray (Thermo
Fisher Scientific, Inc.) runs were performed on the RNA eluates.
Intensity values of the CEL files were normalized to remove bias
between the arrays using the Robust Multiarray Average and
Detection Above BackGround algorithms implemented using the
Affymetrix Expression Console software (version 1.4.1; Thermo
Fisher Scientific, Inc.). Overall signal distributions of each
array were plotted and compared using tools available from the
Bioconductor Project (https://www.bioconductor.org/) to verify for
normalization. After confirming that the data were properly
normalized, differentially expressed miRNAs (DEMs) that exhibited
>2-fold difference between the average signal values of the
control and treatment groups were selected manually. In addition,
the normalized data of selected DEMs were imported into R software
(version 4.1.2; R Core Team) for t-test. Genes with a P<0.05
were extracted as significant DEMs for further study.
Characterization of tumor-derived
exosomes
Microbeads attached to TDEs were fixed for 24 h in
Karnovsky's fixative consisting of 2% glutaraldehyde (Merck KGaA;
cat. no. 354400) and 2% paraformaldehyde (Merck KGaA; cat. no.
818715) dissolved in 0.1 M phosphate buffer (pH 7.4). Next, the
fixed samples were washed twice for 30 min with 0.1 M phosphate
buffer (Merck KGaA; cat. nos. S7907 and S9638). The beads were
post-fixed with 1% OsO4 for 2 h and dehydrated using a gradually
ascending ethanol series (50–100%) with a Critical Point Dryer
(Leica Microsystems; cat. no. CPD300). They were then coated with
platinum using an ion sputter (Leica Microsystems; cat. no. ACE600)
and observed under a field-emission scanning electron microscope at
×20,000 magnification (SEM; MERLIN; Carl Zeiss AG). Confocal
microscopy measurements were obtained to confirm the presence of
TDEs using 3-µm microbeads. The captured exosomes were detected
using a primary PE-Cy7-labeled antibody (BD Biosciences; cat. no.
561982) against the general exosome marker CD63. Fluorescence
images were obtained using a Zeiss LSM 780 confocal microscope
(Carl Zeiss AG). All data were obtained from at least three
independent experimental replicates. The concentrations and size
distributions of the TDEs resuspended in phosphate-buffered saline
(PBS) were measured using a Nanoparticle Tracking Analyzer (NTA;
NanoSight NS300 system; Malvern Panalytical Ltd.) Analysis was
performed using NTA 3.1 software (Malvern Panalytical Ltd.) with
default settings according to the manufacturer's software manual.
The camera focus was adjusted to distinctly visualize TDEs that did
not exceed a particle signal.
Tumor-derived exosomal miRNA
analysis
To validate the miRNA profile of exosomes, TDEs were
isolated following a previously reported method (18,21).
miRNAs were extracted from three pairs of TDEs originating from
wild-type and resistant-type BC cell lines using a mirVana™ miRNA
Isolation kit (Thermo Fisher Scientific, Inc.). RNA concentration
was measured using a Qubit™ microRNA Assay kit (Thermo Fisher
Scientific, Inc.; cat. no. Q32880) with a Qubit® 2.0
Fluorometer (Thermo Fisher Scientific, Inc.; cat. no. Q32866). The
extracted RNA was reverse transcribed using the TaqMan microRNA
Reverse Transcription Kit (Thermo Fisher Scientific, Inc.; cat. no.
4366597). Candidate miRNAs were selected from the cellular
microarray data (Table SIII),
public datasets (GSE71142; Fig.
S2) (22), and other reference
studies on drug-resistant exosomal miRNAs (23–27).
Differential expression levels of 12 miRNAs (miR-21-5p, miR-122-5p,
miR-125b, miR-146a-5p, miR-148-5p, miR-155, miR-210-3p, miR-222-5p,
miR-484, miR-501-5p, miR-1246-5p and miR-1260b) were measured via
cDNA amplification reactions with a TaqMan Universal PCR Master
Mix, No AmpErase UNG (Thermo Fisher Scientific, Inc.; cat. no.
4324018) and TaqMan microRNA Assay kit (Thermo Fisher Scientific,
Inc.; cat. no. 4440887) in a CFX96 Real-time PCR system (Bio-Rad
Laboratories, Inc.; cat. no. 3600037). Individual miRNAs were
reverse-transcribed with the following conditions: 30 min at 16°C
to anneal primers, 30 min at 42°C for the extension, and 5 min at
85°C to stop the reaction. qPCR was run using cDNA with the
following conditions: 10 min at 95°C for enzyme activation,
followed by 40 cycles consisting of denaturing at 95°C for 15 sec,
and annealing and elongation at 60°C for 10 min. miRNA expression
levels were normalized using miR-16 as a normalization control for
exosomal miRNAs. All experiments were performed according to the
manufacturer's protocol and repeated in duplicate. The
2−ΔΔCq method was used to determine the relative
expression of exosomal miRNAs (28).
Exosome education
To compare miRNA signature profiles of wild-type,
educated-type (BC cells treated with drug-resistant exosomes), and
resistant-type BC cells using quantitative PCR in BC cells,
2×105 wild-type BC cells were seeded in 6-well plates
and incubated at 37°C for 24 h, followed by treatment with
1×1010 of drug-resistant exosomes re-suspended in a
complete culture medium and incubated at 37°C for 24 h.
Subsequently, the exosome-treated cells were harvested for
real-time PCR to evaluate miRNA expression levels. To verify the
drug tolerance of wild-type, educated-type, and resistant-type BC
cells, 5×103 of both wild-type and resistant-type BC
cells were seeded in 96-well plates. After a 24-h exosome education
using 1×109 of drug-resistant exosomes, the cells were
incubated with chemo-drugs for 48 h and their cell viabilities were
measured using MTT assay. The purple formazan was then dissolved in
methanol at room temperature for 30 min on an orbital shaker and
the absorbance was recorded at 570 nm with a correction wavelength
of 690 nm with a spectrophotometer (NanoDrop 3000; Thermo Fisher
Scientific, Inc.).
Receiver operating characteristic
(ROC) analysis
ROC analysis of drug-resistant exosomal miRNA
markers was performed on data from plasma samples of 35 patients
with BC using MedCalc (version 20.014; MedCalc Software Ltd.).
Univariate ROC analysis was utilized for each miRNA target to
obtain the ROC curve, area under the curve (AUC), AUC standard
error (SE), and 95% confidence interval (CI) for evaluating the
diagnostic power of the drug-resistant miRNA marker combinations.
After performing a univariate ROC analysis on each combination of
drug-resistant miRNA targets, the ‘best’ combination with the
highest AUC was selected and also the lowest SE of AUC.
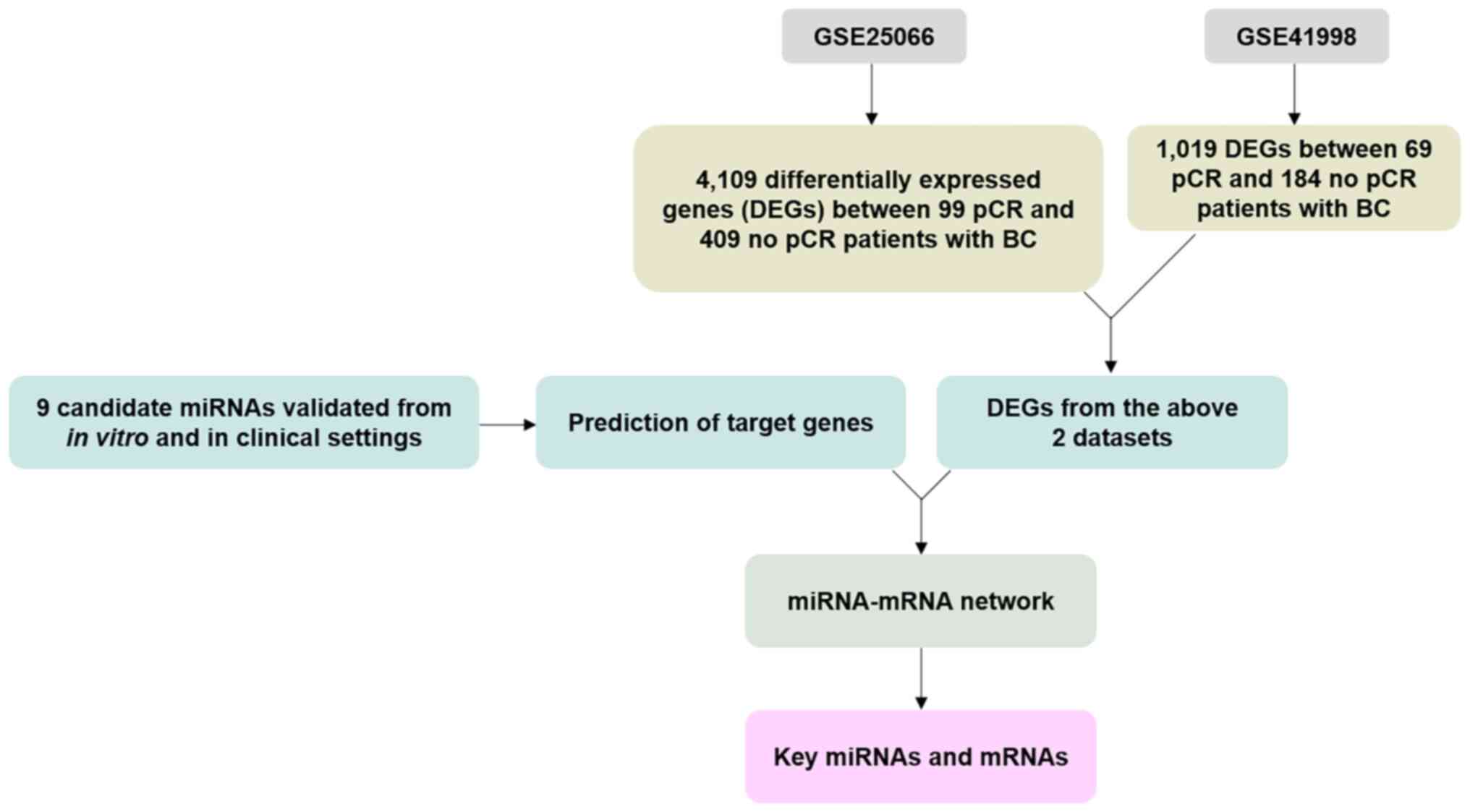
Identification of DEGs from the
dataset and network analysis
The present study included the gene expression
profiles corresponding to the GSE25066 (29) and GSE41998 (30) datasets from the Gene Expression
Omnibus (GEO) database. GSE25066 (508 samples) included 409 and 99
patients with BC exhibiting no response and complete response after
NAC, respectively. GSE41998 (253 samples) included 184 and 69
patients with BC showing no response and complete response after
NAC, respectively. DEGs were identified using DEGSeq (version
1.48.0) (31) with P<0.05,
whereas log2FC ≥1 and log2FC ≤-1 cutoffs were used to denote
upregulated and downregulated DEGs, respectively. Volcano plots
were generated using ggplot2 (32)
(version 3.3.5). Gene ontology (GO) was analyzed using
ClusterProfiler (33).
Representations of GO were generated by the DAVID tool (http://www.geneontology.org/) for functionally
annotated molecular functions, biological processes, and biological
pathways. To predict the miRNA-mRNA targets, TargetScan software
(version 6.0) (https://www.targetscan.org/vert_60/) was used, an open
platform for the prediction of miRNA targets. Gene function was
annotated based on the biological process of GO gene set (34) (C5; MSigDB collections, BROAD
Institute) and gene reference into function (GeneRIF; http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene)
database.
cBioportal for survival analysis
The cBioPortal for Cancer genomics is an open-access
resource (http://www.cbioportal.org/),
providing survival analysis for >10,000 tumor samples from 23
breast cancer studies in TCGA pipeline. This database was applied
to predict the patient's survival using miRNA target gene sets
(BAK1, NOVA1, PTGER4, RTKN2, AGO1, CAP1 and ETS1)
that are highly related to drug resistance (miRNAhigh R)
and miRNA target gene sets (E2F2, ITGA3, SKP2, RIPK2 and
STAT3) that are moderately related to drug resistance
(miRNAmoderate R) as queries. The generated results were
displayed as Kaplan-Meier curves with P-values from the log-rank
test. The search parameters included alterations (missense
mutations, splice mutations, truncating mutation, structural
variant, deep deletion and copy number alterations) from whole
genome/exome sequencing and targeted sequencing data with the
default setting. OS and DFS were calculated on the basis of
cBioPortal's online instruction.
Statistical analysis
Each experiment was repeated at least three times
independently. Data are presented as the mean ± standard deviation.
All statistical analyses were performed with either unpaired
Student's t-test, or multiple comparison tests following one-way
analysis of variance (ANOVA), using R and R studio (version 4.1.2;
R Core Team) and GraphPad Prism software (version 9.0.0;
Dotmatics). P<0.05 was considered to indicate a statistically
significant difference.
Results
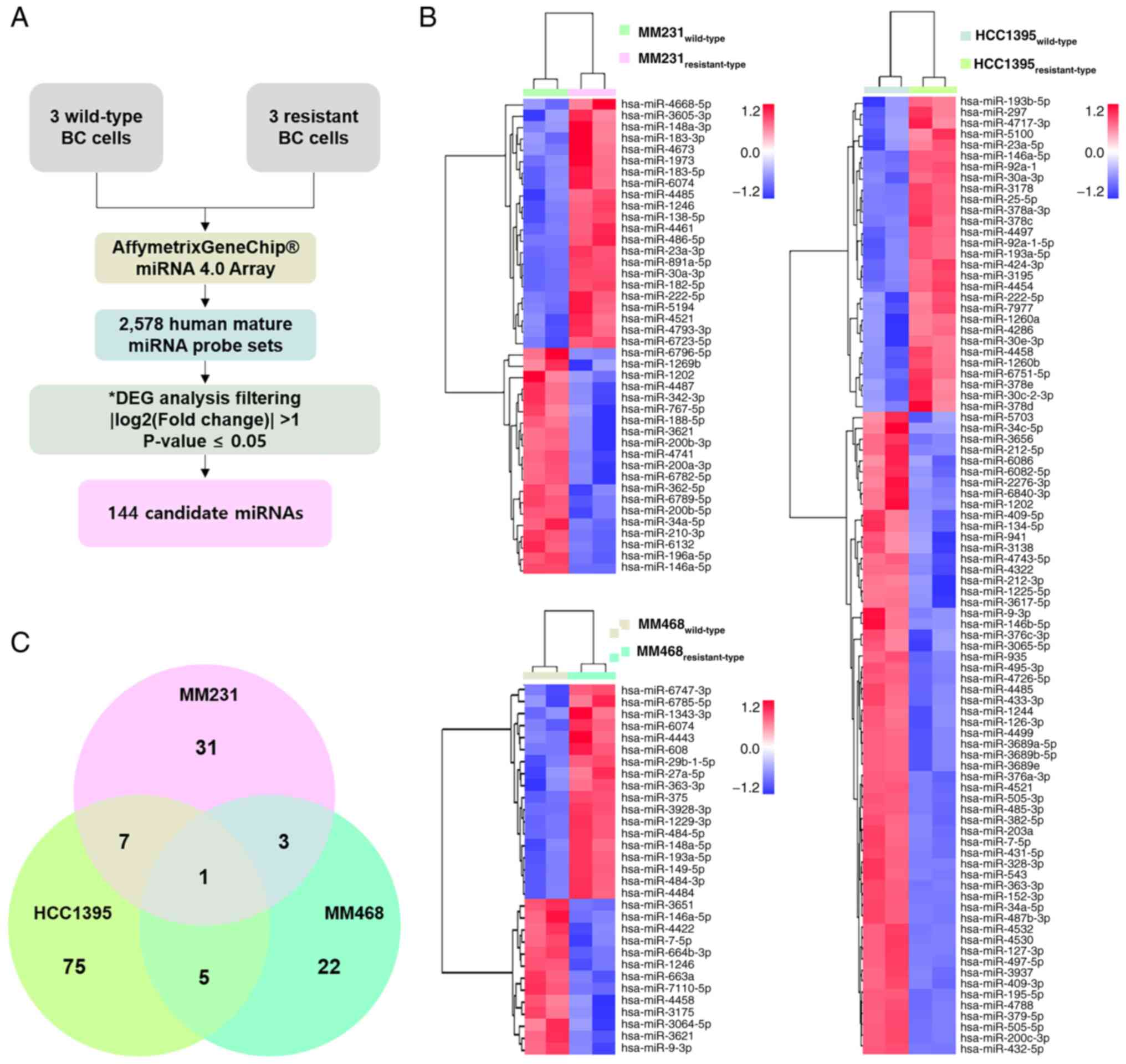
Identification of drug
resistance-associated miRNAs in BC
Microarrays were used to identify DEMs with drug
resistance-associated properties. MiRNA expression levels were
compared with BC cells that were untreated and continuously treated
with drugs. After evaluating miRNA expression using 2,578 human
mature miRNA probe sets, 144 DEMs were identified in resistant-type
BC (compared with wild-type controls) using the following cut-off
criteria: P<0.05; fold change ≥2 (Fig. 2A). To visualize the expression
patterns of the identified DEMs, hierarchical clustering was
performed, and heatmaps were generated for each cell line. The
results showed that MM231, MM468, and HCC1395 cells contained 42,
31 and 88 DEMs, respectively (Fig.
2B). Among the 144 DEMs identified, only 64 miRNAs were
significantly upregulated, indicating that these were
high-confidence candidates involved in drug resistance. When the
common DEMs were assessed, 16 were common between at least two cell
lines, and 1 was found to be common across all three cell lines
(Fig. 2C). These observations
highlighted the heterogeneity of miRNA expression patterns across
different BC cell lines. The upregulated miRNAs could be further
investigated as potential targets or biomarkers for drug
resistance, considering both the common and unique regulatory
mechanisms in BC.
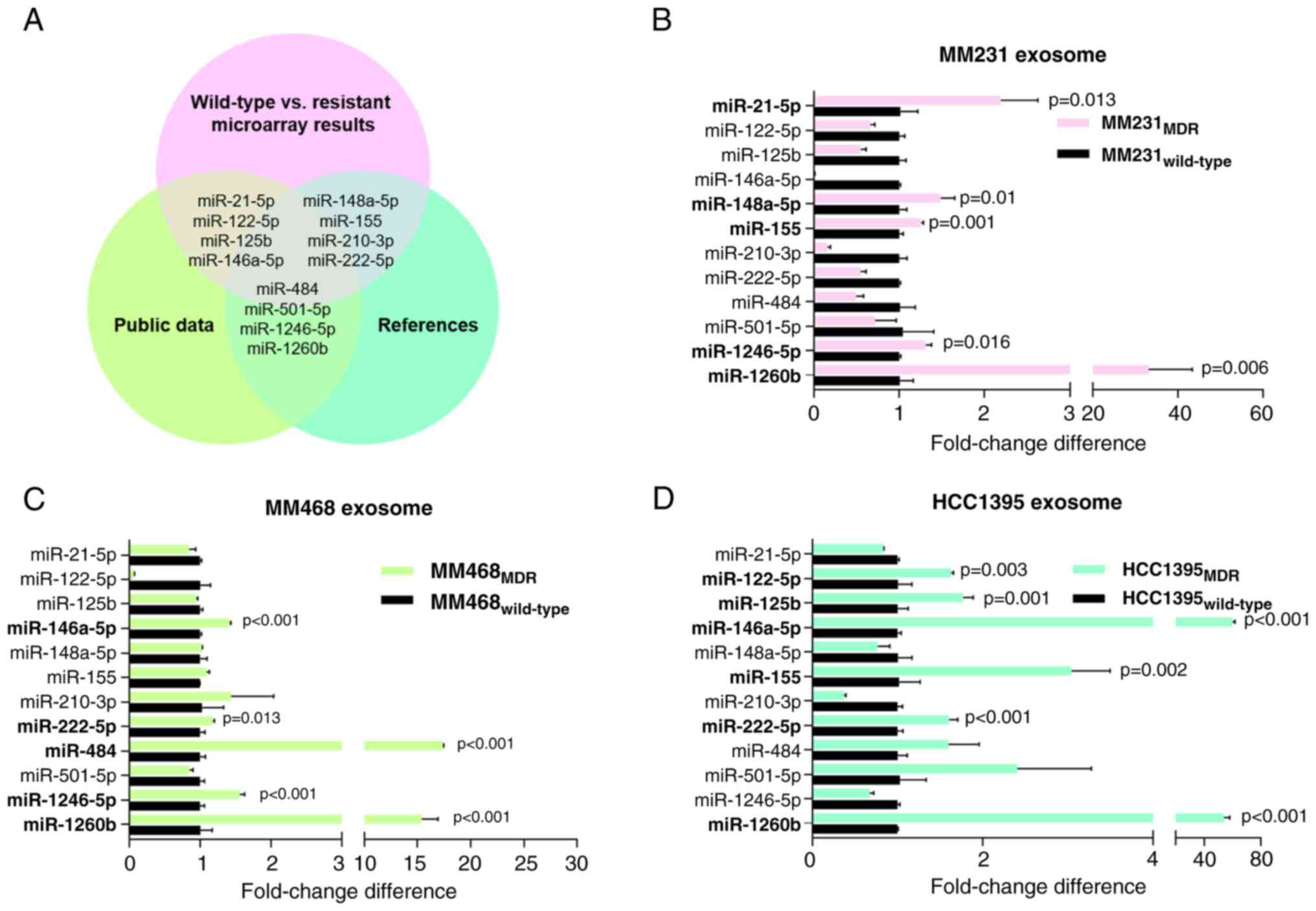
Evaluation of exosomal miRNAs for
predicting drug resistance
A panel of 12 candidate exosomal miRNAs was selected
for analysis. These miRNAs (miR-21-5p, miR-122-5p, miR-125b,
miR-146a-5p, miR-148a-5p, miR-155, miR-210-3p, miR-222-5p, miR-484,
miR-501-5p, miR-1246-5p, and miR-1260b) were selected based on a
comprehensive assessment that incorporated microarray data, public
datasets, and relevant studies on drug-resistant exosomal miRNAs
(Fig. 3A). This crosschecking
approach provided a basis for selecting specific miRNAs as
potential candidates for drug-resistant exosomal miRNAs and
strengthened the reliability of the selection process. These miRNAs
were further analyzed in exosomes isolated from the three BC cell
lines (MM231, MM468, and HCC1395) and their expression patterns
between the wild-type and resistant-type (Fig. 3B-D) were compared. Although the
overall miRNA expression patterns varied among exosomes derived
from BC cells, a significant finding emerged. Notably, miR-21-5p,
miR-122-5p, miR-125b, miR-146a-5p, miR-148a-5p, miR-155, miR-484,
miR-1246-5p and miR-1260b exhibited a marked increase in
drug-resistant exosomes compared with the wild-type counterparts.
This intriguing result suggested that these confirmed miRNAs
exhibited consistent expression patterns in both exosomes and
parental cells and were potentially involved in the drug resistance
mechanisms of BC through exosomal communication.
Exosome education for acquisition of
drug resistance
To investigate the impact of drug-resistant exosomal
miRNAs on drug tolerance, wild-type BC cells were exposed to
resistant-type exosomes and the relationship between miRNA
expression levels and cell viability was analyzed. The radar plots
revealed significant differences in miRNA expression patterns
between wild-type and resistant-type BC cells (Fig. 4A). Notably, the miRNA expression
patterns of wild-type BC cells became nearly identical to those of
resistant-type BC cells after exposure to resistant-type exosomes.
Furthermore, examination of the cell viability of wild-type BC
cells following treatment with exosomes derived from resistant-type
BC cells revealed a substantial increase in chemoresistance to both
Adriamycin and Taxotere (Fig. 4B).
Specifically, after Adriamycin treatment at a drug concentration
equivalent to the IC50 value of wild-type BC cells, a
survival increase of 36.9, 10.1 and 9.6% was noted for
MM231wild-type, MM468wild-type, and
HCC1395wild-type cells, respectively. Under the same
conditions, treatment with Taxotere resulted in a survival increase
of 27.9, 28.3 and 21.8% for BC cells. These findings indicated that
exosomal miRNAs from resistant-type BC cells have the capacity to
reshape miRNA expression patterns in wild-type BC cells and can
confer the wild-type BC cells with an increased resistance to
chemotherapy drugs, causing their behavior to more closely resemble
that of drug-resistant BC cells.
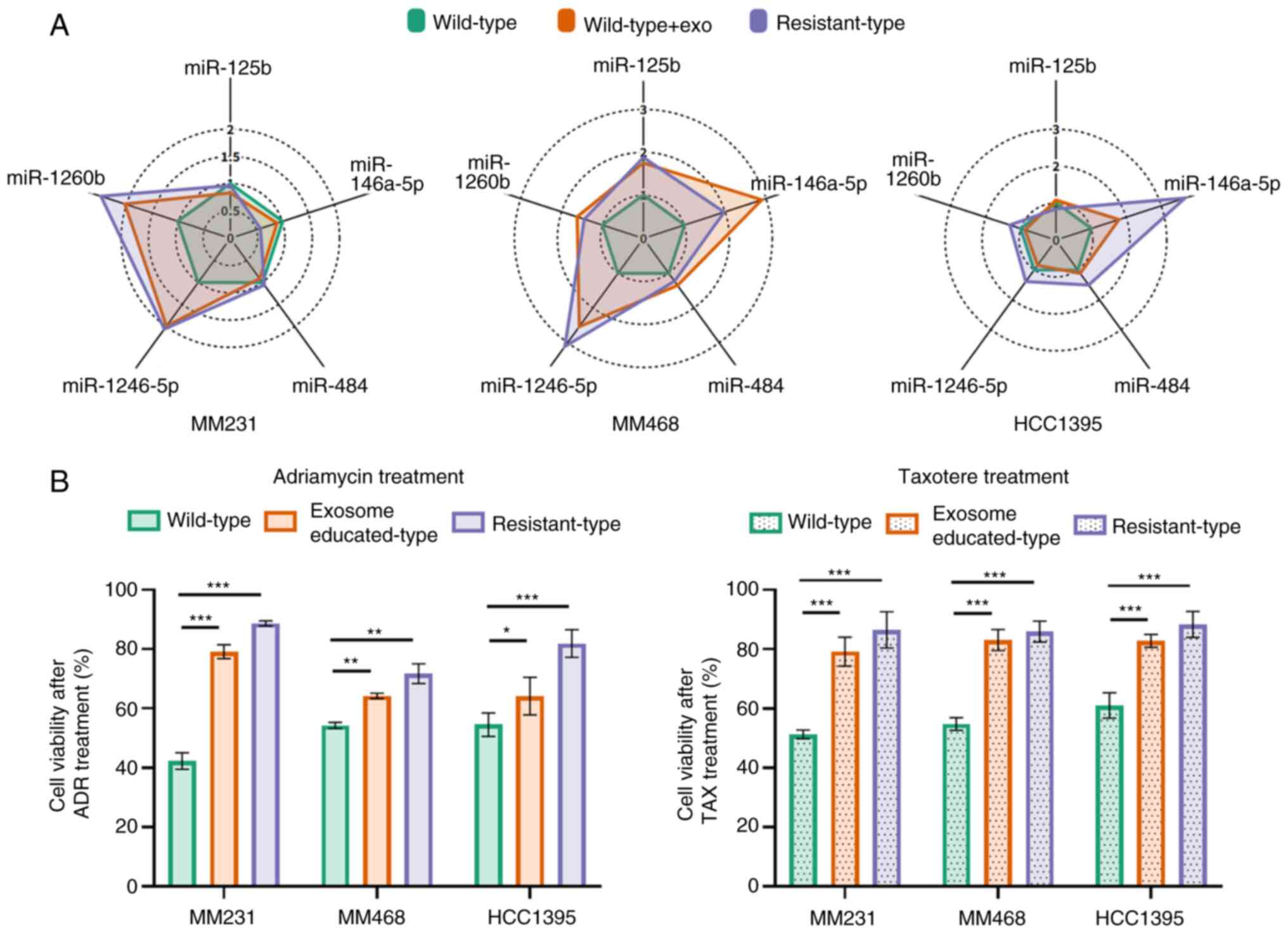 | Figure 4.(A) Radar plots of miRNA signature
profiles from quantitative PCR in BC cells, including MDA-MB-231
(MM231), MDA-MB-468 (MM468), and HCC1395. The relative expression
levels of miRNAs in wild-type BC cells, drug-resistant exosome 24
h-treated (educated-type) BC cells, and resistant-type BC cells
were compared. The line labels represent the drug-resistant miRNAs,
and the ring labels represent the fold change calculated from each
group/wild-type difference. (B) The cell viabilities of wild-type,
educated-type, and resistant-type BC cells were compared after 48 h
of incubation with Adriamycin and Taxotere. miRNA or miR, microRNA;
BC, breast cancer; ADR, Adriamycin; TAX, Taxotere. |
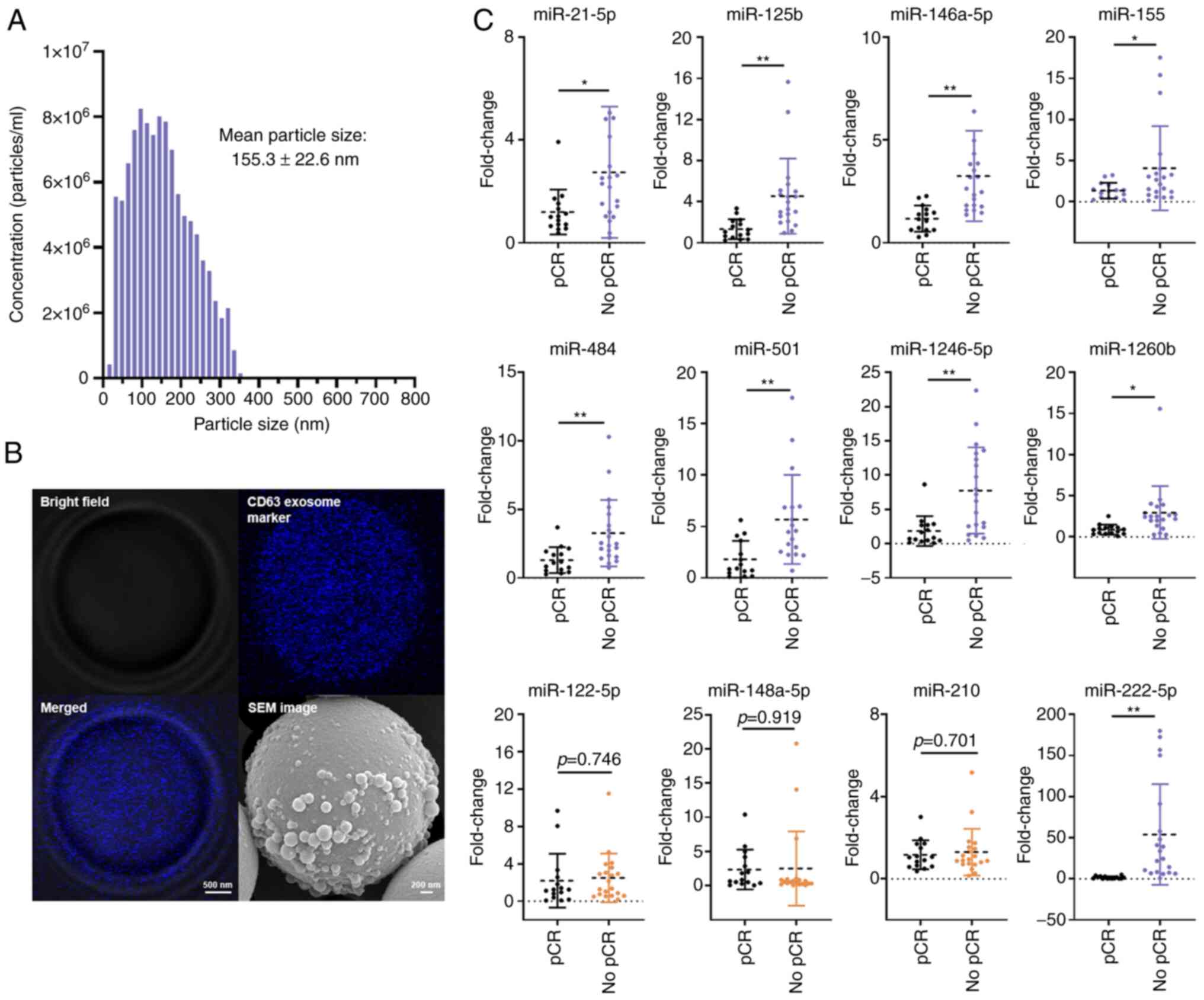
Validation of selected exosomal miRNAs
in patient samples
Exosomes were isolated from the plasma of patients
with BC. These plasma exosomes exhibited a particle size
distribution of 30–350 nm in diameter, with an average of 155.3 nm
(Fig. 5A). TDEs were specifically
targeted using the immunoaffinity method, and then a specific
population of exosomes of interest released from the tumor cells
was isolated. This specificity allowed for focused analysis of
exosomes relevant to research on drug-resistant miRNAs.
Furthermore, SEM was employed to confirm whether the isolated TDEs
possessed the characteristic features of exosomes. Microscopy
revealed that the isolated TDEs expressed CD63, a positive
tetraspanin marker associated with exosomes (35), and possessed the expected
morphological and structural characteristics of exosomes (Fig. 5B). The expression of selected 12
drug-resistant miRNAs in 35 patients with BC were analyzed,
including 20 and 15 patients showing no response and complete
response following NAC, respectively, using real-time PCR. The raw
Ct values of the 12 miRNAs are listed in Table SIV. The results showed that nine
miRNAs (miR-21-5p, miR-125b, miR-146a-5p, miR-155, miR-222-5p,
miR-484, miR-501, miR-1246-5p, and miR-1260b) were significantly
upregulated in patients with BC who showed no response following
NAC (Fig. 5C). These findings
suggested that the upregulated miRNAs may be strongly associated
with drug resistance in BC. No significant differences were
observed in the other three miRNAs (miR-122-5p, miR-148a-5p and
miR-210) between the groups in the patient cohort.
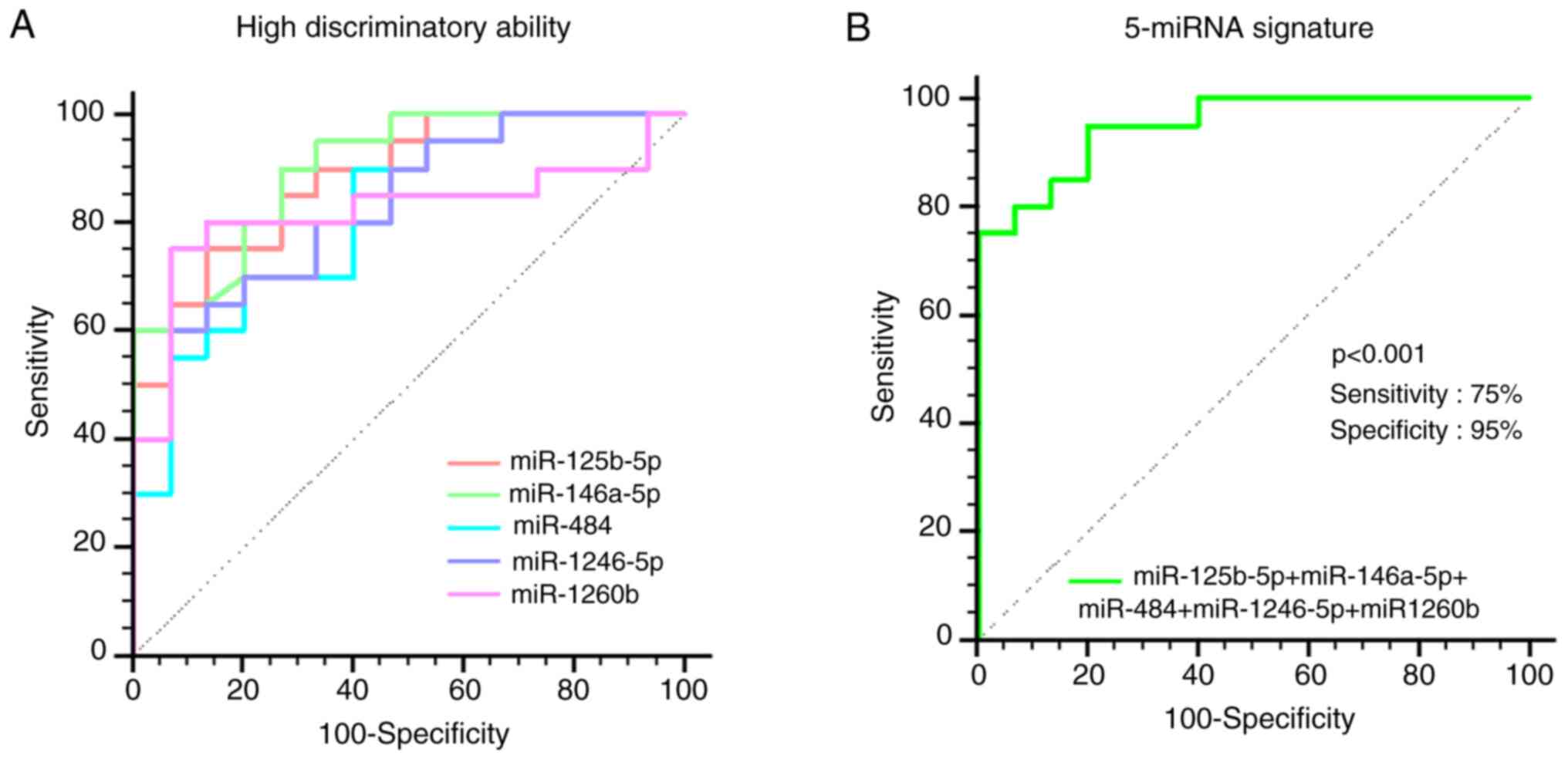
ROC curve analysis of validated
exosomal miRNAs as prognosis markers
Considering the elevated levels of several miRNAs in
patients with BC who exhibited no response to NAC, their roles as
prognostic markers were determined using ROC curve analysis. A
total of five miRNAs (miR-125b-5p, miR-146a-5p, miR-484,
miR-1246-5p and miR-1260b) demonstrated relatively high
discriminatory abilities, as indicated by their AUC values ranging
from 0.817 to 0.898 (Fig. 6A).
These miRNAs had the potential to serve as prognostic markers for
identifying patients with BC likely to develop drug resistance
following NAC. Conversely, miR-21-5p, miR-155-5p, miR-222-5p, and
miR-501-5p showed moderate outcomes, with AUC values <0.8 and
miR-122, miR-148a-5p, and miR-210 exhibited poor outcomes, with AUC
values <0.6 (Table SV). The
integrated model consisting of the five miRNAs showed even greater
discriminatory ability, with an AUC of 0.950 (95% CI: 0.819–0.995;
P<0.001), 75% sensitivity and 95% specificity (Fig. 6B). These results suggested that the
combined 5-miRNA signature, among various combinations, could
provide enhanced predictive outcomes for identifying patients with
BC showing drug resistance (Fig.
S3; Tables SVI and SVII). Therefore, compared with standalone
miRNA markers, more effective combinations are proprosed, to
maximize AUC and achieve improved diagnostic/prognostic accuracy
(Table SVIII).
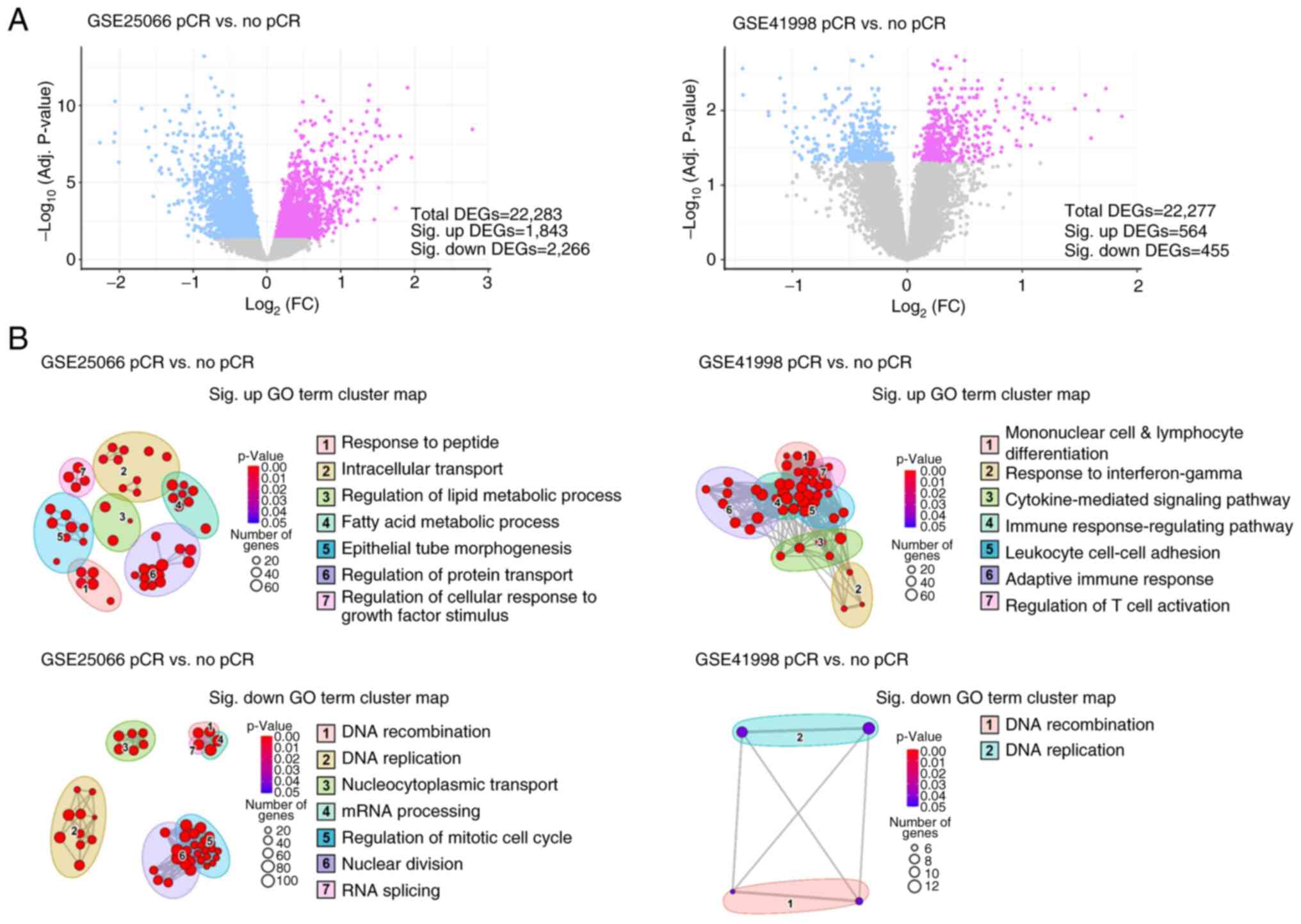
Meta-analysis of drug-resistant gene
expression patterns in BC with NAC case studies
To reinforce the limited scale of clinical
validation, a meta-analysis of datasets containing gene expression
profiles in patients who received sequential NAC, such as
doxorubicin, cyclophosphamide and paclitaxel, after histologically
confirming primary invasive breast adenocarcinoma was performed to
identify DEGs between the response and no-response groups. Analysis
of GSE25066 identified 2,266 downregulated and 1,843 upregulated
DEGs. Analysis of GSE41988 identified 455 downregulated and 564
upregulated DEGs (Fig. 7A).
Subsequently, cutoffs of adjusted P-values <0.05 and
|log2FC|>1 were used for each analysis. Moreover, GO analysis
was performed for each significantly dysregulated DEG to identify
GO terms representing the biological function of the genes.
Clustering of GO terms based on their semantic similarity suggested
two and seven GO clusters for the upregulated and downregulated
genes, respectively (Fig. 7B).
Notably, the analysis demonstrated that the upregulated GO clusters
were enriched in the ‘regulation of cellular response to growth
factor stimulus’, ‘intracellular transport’ and ‘cytokine-mediated
signaling pathway’, strongly suggesting their involvement in drug
resistance mechanisms. Conversely, the downregulated GO clusters
were associated with the regulation of ‘DNA recombination’, ‘DNA
replication’, ‘RNA splicing’, ‘mRNA processing’, and ‘regulation of
mitotic cell cycle’, suggesting a potential impairment of the G2/M
checkpoint and DNA repair pathways. These findings aligned with the
documented effects of drug resistance mechanisms in solid tumors,
as elucidated by several pivotal studies (36–38).
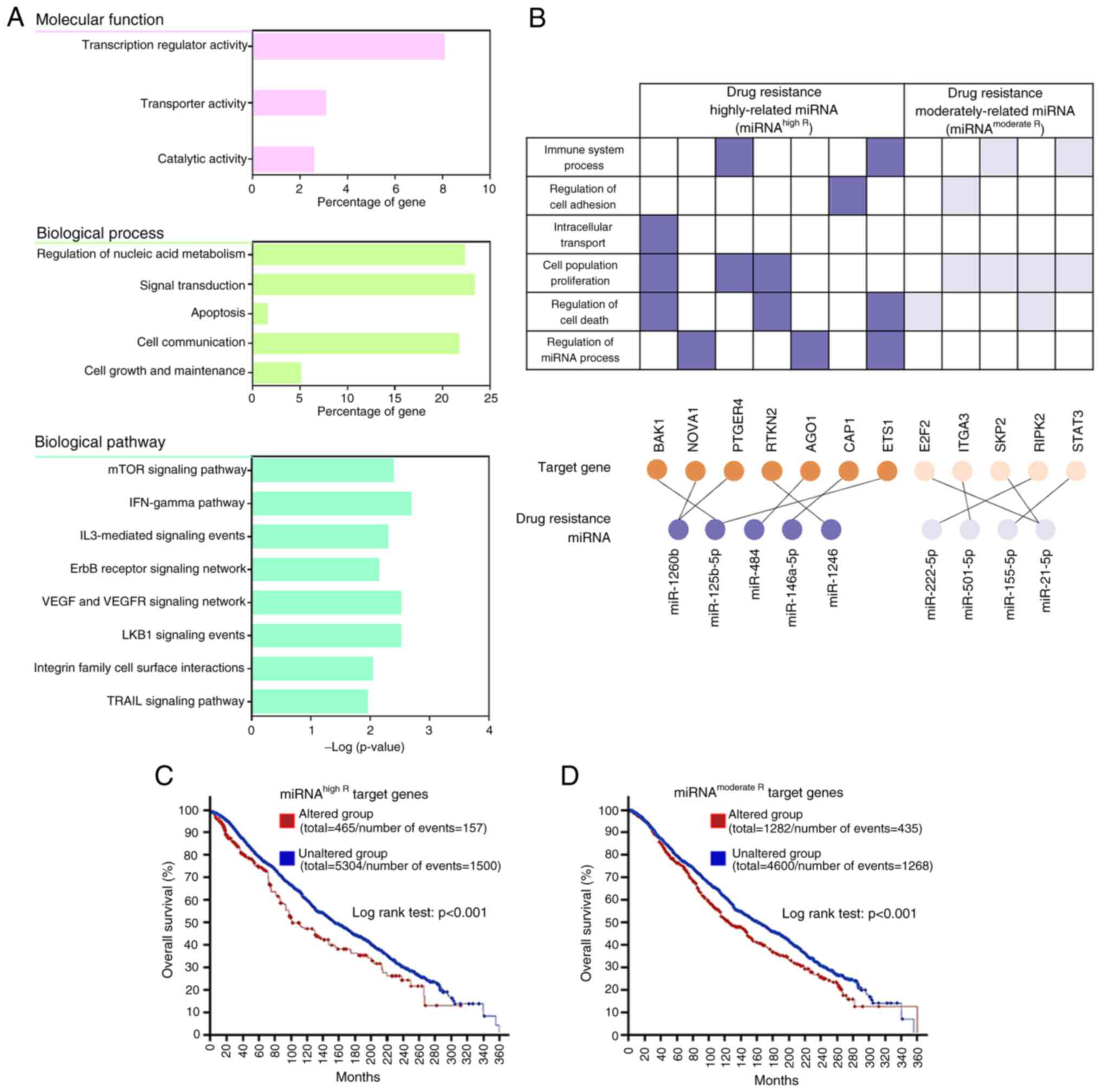
Network analysis of drug-resistant
genes
By integrating miRNAs, their respective target
genes, and GO data, the authors successfully confirmed a drug
resistance regulatory network. TargetScan predictions were utilized
for identifying the target genes of the nine candidate miRNAs. To
ensure reliability, the authors focused on high-confidence
interactions with binding scores ≥90 (Table SIX). Subsequently, the miRNA-target
gene interactions were subjected to GO analysis, with specific
emphasis on the GO terms showing an association with drug-resistant
genes. This narrowed the analysis to the molecular functions,
biological processes, and biological pathways most relevant to drug
resistance in BC (Fig. 8A). The
significant drug-related categories of molecular function were
‘transcription regulator activity’, ‘transporter activity’ and
‘catalytic activity’. GO biological processes included ‘regulation
of nucleic acid metabolism’, ‘signal transduction’, ‘apoptosis’,
‘cell communication’, and ‘cell growth and maintenance’. Moreover,
the GO terms for target mRNAs of the nine candidate miRNAs were
associated with multiple biological pathways participating in ‘mTOR
signaling pathway’, ‘IFN-gamma pathway’, ‘IL3-mediated signaling
events’, ‘ErbB receptor signaling network’, ‘VEGF and VEGFR
signaling network’, ‘LKB1 signaling events’, ‘integrin family cell
surface interactions’ and ‘TRAIL signaling pathway’. Notably, these
results were similar to the findings in Fig. 7, presenting the analysis of the
metadata for drug resistance genes, implying that the nine miRNAs
are related to drug resistance and potentially regulate target
genes to influence tumor response to chemotherapy. Furthermore,
miRNAhigh R target gene sets (BAK1, NOVA1, PTGER4,
RTKN2, AGO1, CAP1 and ETS1) and miRNAmoderate
R target gene sets (E2F2, ITGA3, SKP2, RIPK2 and
STAT3) that are related to multiple biological activities
related to drug resistance were revealed (Fig. 8B). Notably, NOVA1, ITGA3,
SKP2 and RIPK2 were common after cross-validation of
metadata analysis, DEMs and target DEGs, suggesting that these four
genes are more importantly involved in drug resistance.
Survival analyses of drug-resistant
genes
To evaluate the clinical significance of the
identified drug-resistant miRNAs and their target genes, survival
analyses was performed using data from a comprehensive collection
of cancer studies, accessed through cBioPortal, encompassing
thousands of patient samples filtered by BC classification.
Kaplan-Meier curves and log-rank tests were performed for
individual genes identified in the present study, followed by the
analysis of combined gene-altered signatures involving
miRNAhigh R or miRNAmoderate R, which showed
significant results (Fig. S4). The
gene-altered group exhibited a decreased lifespan than the
unaltered group, indicating a high correlation with patient
prognosis. For example, the median overall survival in the
miRNAhigh R target gene signature-altered and unaltered
groups was determined to be 110.77 and 154.50 months, respectively
(Fig. 8C). Similarly, the median
overall survival rates in the miRNAmoderate R target
gene signature-altered and unaltered groups were 124.20 and 161.13
months, respectively (Fig. 8D). In
addition, both gene signatures showed significant differences in
progression-free survival and relapse-free survival (Fig. S5). These findings highlighted the
prognostic significance of the drug resistance target genes,
including BAK1, NOVA1, PTGER4, RTKN2, AGO1, CAP1, ETS1, E2F2,
ITGA3, SKP2, RIPK2 and STAT3, in patients with BC. Taken
together, these results provided compelling evidence for a strong
association between the altered expression of drug-resistant genes
induced by the regulation of drug-resistant miRNAs and poor
prognosis among patients with BC.
Discussion
In the present study, it was identified that
exosomal miR-125b-5p, miR-146a-5p, miR-484, miR-1246-5p and
miR-1260b were highly enriched in TDEs from patients with BC who
displayed tolerance to chemotherapy. To the best of the authors'
knowledge, only a few studies have reported the potential roles of
these miRNAs in tumorigenesis and cancer treatment (39). Furthermore, research on the
functional aspects of exosomal miRNAs remains relatively rare. One
of the key challenges in exosome research is the uncertainty
regarding whether exosomal miRNAs accurately reflect the miRNA
expression patterns in the originating tissue. This ambiguity can
sometimes lead to conflicting findings, and it is partly attributed
to the inherent heterogeneity of both tumors and exosomes.
Additionally, the isolation and analysis of tumor-derived exosomes
have not yet reached an optimized standard. The present study aimed
to mitigate these challenges by isolating a comprehensive set of
tumor-derived exosomes, focusing on those associated with drug
resistance. The comparative analysis in the present study, linking
these candidate exosomal miRNAs with their target genes, elucidated
a clear and significant involvement of these five aforementioned
exosomal miRNAs in the response of the tumor to chemotherapy and,
subsequently, in the clinical outcomes of the patients.
When investigating the individual functions of each
miRNA in drug resistance, the previous research of the authors
(40) on the function of miRNAs in
tumor tissue, provided valuable insights. It was reported in that
study that high miR-1260b expression was markedly associated with
bulky tumor size, advanced stage, lymph node invasion, and a
shorter period of overall survival. In addition to the oncogenic
function of miR-1260b, it was identified that its target is
CASP8, a key gene in the p53 tumor suppressor pathway,
implicating its involvement in drug resistance (40). In the present study, a significant
association between exosomal miR-1260b and regulation of the key
drug-resistance genes, NOVA1 and PTGER4, was
revealed. As an RNA-binding protein, NOVA1 is known to exert
influence over miRNA activity and the regulation of RNA splicing
(41,42). Its versatile role in
post-transcriptional gene regulation renders it a highly promising
candidate in the context of drug resistance. In addition,
PTGER4 is a critical target in the transduction pathways
essential for cancer cell survival and tumor progression, including
the AKT and ERK pathways implicated in numerous other cancers
(43). Further investigations are
required to elucidate the precise role of exosomal miR-1260b in BC
drug resistance.
Zheng et al (44) reported that increased miR-125b-5p
expression in tumor tissue was associated with a lack of pCR after
anthracycline-taxane-based chemotherapy in BC. Similarly, Zhou
et al (45) revealed that
the downregulation of the pro-apoptotic gene BAK1, which is
a direct target of miR-125-5p, could suppress Taxol-induced
apoptosis and result in increased resistance to Taxol. In the
present study it was revealed that ETS1 is another possible
target of exosomal miR-125-5p. ETS1 has shown ambiguous
functions as an oncogene and a tumor suppressor gene in numerous
types of cancers, but its function as a tumor suppressor has been
clearly reported recently in BC (46). Another drug-resistant miRNA
candidate, miR-146a-5p, has been reported to be overexpressed in
cisplatin-resistant BC cells, affecting the cell levels of the
tumor suppressor BRCA1, HOXD10 homeobox family, tumor
suppressor CDKN1B, and ESR1 gene (47). Moreover, Dai et al (48) demonstrated that miR-1246 affected
cell migration, invasion and doxorubicin resistance in BC by
targeting the transcription factor NFE2L3. According to the
analysis in the present study, CAP1 and RTKN2 are
potent targets of miR-146a-5p and miR-1246, respectively. These
genes are known to be involved in tumor invasion, apoptosis, and
immune response in lung cancer, but the molecular mechanisms
underlying drug resistance in BC remain unclear (49–51).
Notably, the previously reported roles of miR-484 in regulating
drug resistance in BC are in contrast with the findings of the
present study. For example, Ye et al (52) demonstrated that the upregulation of
miR-484 reduced cell proliferation and reversed chemo-resistance to
gemcitabine in BC. Jia et al (53) argued that miR-484 is typically
described as a tumor suppressor; however, this claim could be
simplistic and one-sided. Existing evidence primarily addresses
miR-484 expression in tumor tissues rather than exosomal miR-484
expression. Considering the potential discrepancies among
circulating cell-free, tumor tissues, and exosomal miRNAs, the
function of miR-484 in TDEs requires careful investigation.
Contrary to the initial expectations, limited
consistency was observed between the target genes identified using
TargetScan and those suppressed in drug-resistant tumors. This
suggested that the regulatory relationship between them is not
straightforward, indicating the presence of additional regulatory
mechanisms or complexities in the context of drug resistance. The
regulatory roles of miRNAs are complex and involve various
mechanisms that mediate gene regulation. To fully understand the
regulatory networks of the exosomal miRNAs involved in drug
resistance, it is crucial to understand the specifics of these
mechanisms. Drug-resistant miRNAs present in the blood of patients
undergoing treatment may not exclusively target or suppress tumor
suppressor genes or oncogenes. The involvement of miRNA-mediated
feedback and feed-forward loops, which are regulatory circuits
involving reciprocal regulation between miRNAs and their target
genes, should also be considered. These loops add an additional
layer of complexity to the control of gene expression and could
contribute to the observed inconsistencies (54). Moreover, although the expression of
miRNAs and their targets is often highly correlated, it is possible
that additional factors, such as epigenetic modifications,
transcriptional factors, or protein-protein interactions, influence
the observed inconsistencies. A recent study has emphasized the
significance of these factors in miRNA-mediated gene regulation
(55). Therefore, it is essential
to explore these alternative mechanisms and conduct further
experimental validation to gain a more comprehensive understanding
of the regulatory networks of exosomal miRNAs involved in drug
resistance. By investigating the interplay between miRNAs, their
targets, and other regulatory elements, the complexities underlying
miRNA-mediated gene regulation and mechanisms that contribute to
drug resistance can be better understood.
In summary, the present study provided valuable
insights regarding the molecular mechanisms underlying drug
resistance in BC. By constructing a miRNA-mRNA network, key
exosomal miRNAs were identified (miR-125b-5p, miR-146a-5p, miR-484,
miR-1246-5p and miR-1260b) and their target genes (BAK1, NOVA1,
PTGER4, RTKN2, AGO1, CAP1 and ETS1) that may be involved
in the development of drug resistance in patients with BC. These
miRNAs and their target genes are promising candidate biomarkers
for predicting tumor response to treatment and may serve as
potential therapeutic targets for patients with BC. An interesting
finding, in the present study, is based on how the response of
cells sensitive to chemotherapy drugs can change due to
transference of drug-resistant exosomes (a process called exosome
education). Overall, the present study highlights the importance of
early diagnosis with liquid biopsy because it provides valuable
information for predicting patient prognosis by extracting and
analyzing drug-resistant exosomes, which can affect tumor response
to treatment. Given the heterogeneity of exosomes and their dynamic
nature during treatment, it is crucial to explore changes in
exosomal miRNA profiles to enhance the understanding of treatment
responses for improving patient outcomes. Further investigations
are warranted for establishing exosomal miRNA signatures derived
from liquid biopsies as reliable indicators of tumor response in
BC.
However, there are still a few limitations to
acknowledge. First, circulating biomarkers, including exosomes,
ctDNAs, and miRNAs, possess other contaminants that exist in the
blood, and their origin is difficult to define. Despite the efforts
of the authors to analyze tumor-derived exosomes using the
immuno-affinity isolation method, it is not considered that all the
exosomes isolated in the present study reflect tumor information.
An innovative exosome isolation technology is essential for a more
accurate analysis. Second, the results obtained in the present
study were based on a relatively small sample size, which may limit
the generalizability of the findings, thus further large-scale
validation is required. Finally, experimental validation through
gain/loss-of-function approaches has not been carried out in the
present study. It is expected that future investigations will
address the impact of these miRNAs and target genes in cancer
treatment and provide directions for overcoming drug
resistance.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Severance Hospital
Research Fund for Clinical Excellence (grant no. C-2023-0006) and
the National Research Foundation of Korea Grants (grant nos.
2021R1I1A1A01051594 and 2022R1F1A1074605).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The microarray data generated in the present study are
available in the GEO database under accession code GSE237873.
Authors' contributions
MWK, JeYK and SIK conceived the study. SL, SM, YK,
JoYK and HL were responsible for the collection, curation and
analysis of the experimental data. YK, JoYK and HL prepared the
methodology and software. MWK and SM validated the analysis, and
confirm the authenticity of all the raw data. SM wrote the original
draft and MWK revised the manuscript. SIK and JeYK were responsible
for study supervision. All authors contributed to writing of the
manuscript and have read and approved the final manuscript.
Ethics approval and consent to
participate
Clinical samples were obtained from subjects who
visited Severance Hospital in South Korea, according to the
guidelines of the independent Ethics Committee of Yonsei University
College of Medicine (IRB approval no. 4-2020-1292, approved on
January 4, 2021; Seoul, South Korea). The study was performed
following the principles of the Declaration of Helsinki. Informed
consent for the use of plasma samples for research purposes was
obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bianchini G, De Angelis C, Licata L and
Gianni L: Treatment landscape of triple-negative breast
cancer-expanded options, evolving needs. Nat Rev Clin Oncol.
19:91–113. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spring LM, Fell G, Arfe A, Sharma C,
Greenup R, Reynolds KL, Smith BL, Alexander B, Moy B, Isakoff SJ,
et al: Pathologic complete response after neoadjuvant chemotherapy
and impact on breast cancer recurrence and survival: A
comprehensive meta-analysis. Clin Cancer Res. 26:2838–2848. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Echeverria GV, Ge Z, Seth S, Seth S, Zhang
X, Jeter-Jones S, Zhou X, Cai S, Tu Y, McCoy A, et al: Resistance
to neoadjuvant chemotherapy in triple-negative breast cancer
mediated by a reversible drug-tolerant state. Sci Transl Med.
11:eaav09362019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoogstraat M, Lips EH, Mayayo-Peralta I,
Mulder L, Kristel P, van der Heijden I, Annunziato S, van Seijen M,
Nederlof PM, Sonke GS, et al: Comprehensive characterization of
pre- and post-treatment samples of breast cancer reveal potential
mechanisms of chemotherapy resistance. NPJ Breast Cancer. 8:602022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lips EH, Michaut M, Hoogstraat M, Mulder
L, Besselink NJ, Koudijs MJ, Cuppen E, Voest EE, Bernards R,
Nederlof PM, et al: Next generation sequencing of triple negative
breast cancer to find predictors for chemotherapy response. Breast
Cancer Res. 17:1342015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Y, Schaafsma E and Cheng C: Gene
signature-based prediction of triple-negative breast cancer patient
response to Neoadjuvant chemotherapy. Cancer Med. 9:6281–6295.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nedeljkovic M and Damjanovic A: Mechanisms
of chemotherapy resistance in triple-negative breast cancer-how we
can rise to the challenge. Cells. 8:9572019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Robey RW, Pluchino KM, Hall MD, Fojo AT,
Bates SE and Gottesman MM: Revisiting the role of ABC transporters
in multidrug-resistant cancer. Nat Rev Cancer. 18:452–464. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fu Y, Yang Q, Yang H and Zhang X: New
progress in the role of microRNAs in the diagnosis and prognosis of
triple negative breast cancer. Front Mol Biosci. 10:11624632023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sueta A, Fujiki Y, Goto-Yamaguchi L,
Tomiguchi M, Yamamoto-Ibusuki M, Iwase H and Yamamoto Y: Exosomal
miRNA profiles of triple-negative breast cancer in neoadjuvant
treatment. Oncol Lett. 22:8192021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O'Brien K, Breyne K, Ughetto S, Laurent LC
and Breakefield XO: RNA delivery by extracellular vesicles in
mammalian cells and its applications. Nat Rev Mol Cell Biol.
21:585–606. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11:252019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
An X, Sarmiento C, Tan T and Zhu H:
Regulation of multidrug resistance by microRNAs in anti-cancer
therapy. Acta Pharm Sin B. 7:38–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Niel G, Carter DRF, Clayton A, Lambert
DW, Raposo G and Vader P: Challenges and directions in studying
cell-cell communication by extracellular vesicles. Nat Rev Mol Cell
Biol. 23:369–382. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mitra R, Adams CM, Jiang W, Greenawalt E
and Eischen CM: Pan-cancer analysis reveals cooperativity of both
strands of microRNA that regulate tumorigenesis and patient
survival. Nat Commun. 11:9682020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gwak H, Park S, Kim J, Lee JD, Kim IS, Kim
SI, Hyun KA and Jung HI: Microfluidic chip for rapid and selective
isolation of tumor-derived extracellular vesicles for early
diagnosis and metastatic risk evaluation of breast cancer. Biosens
Bioelectron. 192:1134952021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim MW, Park S, Lee H, Gwak H, Hyun KA,
Kim JY, Jung HI and Il Kim S: Multi-miRNA panel of tumor-derived
extracellular vesicles as promising diagnostic biomarkers of
early-stage breast cancer. Cancer Sci. 112:5078–5087. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gradishar WJ, Anderson BO, Abraham J, Aft
R, Agnese D, Allison KH, Blair SL, Burstein HJ, Dang C, Elias AD,
et al: Breast Cancer, Version 3.2020, NCCN Clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 18:452–478. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim MW, Niidome T and Lee R: Glycol
Chitosan-Docosahexaenoic acid liposomes for drug delivery:
Synergistic effect of doxorubicin-rapamycin in drug-resistant
breast cancer. Mar Drugs. 17:5812019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim MW, Koh H, Kim JY, Lee S, Lee H, Kim
Y, Hwang HK and Kim SI: Tumor-specific miRNA signatures in
combination with CA19-9 for liquid biopsy-based detection of PDAC.
Int J Mol Sci. 22:136212021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi
Y, Xiong W, Li G, Lu J, Fodstad O, et al: MicroRNA-125b confers the
resistance of breast cancer cells to paclitaxel through Suppression
of Pro-apoptotic Bcl-2 Antagonist Killer 1 (Bak1) Expression. J
Biol Chem. 285:21496–21507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tao L, Wu YQ and Zhang SP: MiR-21-5p
enhances the progression and paclitaxel resistance in
drug-resistant breast cancer cell lines by targeting PDCD4.
Neoplasma. 66:746–755. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu DD, Lv MM, Chen WX, Zhong SL, Zhang XH,
Chen L, Ma TF, Tang JH and Zhao JH: Role of miR-155 in drug
resistance of breast cancer. Tumour Biol. 36:1395–1401. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Lu P, Wang DD, Yang SJ, Wu Y, Shen
HY, Zhong SL, Zhao JH and Tang JH: The role of miRNAs in drug
resistance and prognosis of breast cancer formalin-fixed
paraffin-embedded tissues. Gene. 595:221–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Si Z, Zhong Y, Lao S, Wu Y, Zhong G and
Zeng W: The Role of miRNAs in the resistance of anthracyclines in
breast cancer: A systematic review. Front Oncol. 12:8991452022.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(−Delta Delta C) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hatzis C, Pusztai L, Valero V, Booser DJ,
Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, et
al: A genomic predictor of response and survival following
taxane-anthracycline chemotherapy for invasive breast cancer. JAMA.
305:1873–1881. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Horak CE, Pusztai L, Xing G, Trifan OC,
Saura C, Tseng LM, Chan S, Welcher R and Liu D: Biomarker analysis
of neoadjuvant doxorubicin/cyclophosphamide followed by ixabepilone
or paclitaxel in early-stage breast cancer. Clin Cancer Res.
19:1587–1595. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang LK, Feng ZX, Wang X, Wang XW and
Zhang XG: DEGseq: An R package for identifying differentially
expressed genes from RNA-seq data. Bioinformatics. 26:136–138.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ito K and Murphy D: Application of ggplot2
to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol.
2:e792013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu GC, Wang LG, Han YY and He QY:
clusterProfiler: An R Package for comparing biological themes among
gene clusters. Omics. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liberzon A, Subramanian A, Pinchback R,
Thorvaldsdóttir H, Tamayo P and Mesirov JP: Molecular signatures
database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Escola JM, Kleijmeer MJ, Stoorvogel W,
Griffith JM, Yoshie O and Geuze HJ: Selective enrichment of
tetraspan proteins on the internal vesicles of multivesicular
endosomes and on exosomes secreted by human B-lymphocytes. J Biol
Chem. 273:20121–20127. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong X, Bai X, Ni J, Zhang H, Duan W,
Graham P and Li Y: Exosomes and breast cancer drug resistance. Cell
Death Dis. 11:9872020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Marra A, Trapani D, Viale G, Criscitiello
C and Curigliano G: Practical classification of triple-negative
breast cancer: Intratumoral heterogeneity, mechanisms of drug
resistance, and novel therapies. NPJ Breast Cancer. 6:542020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang M, Yu F, Ding H, Wang Y, Li P and
Wang K: Emerging function and clinical values of exosomal MicroRNAs
in cancer. Mol Ther Nucleic Acids. 16:791–804. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park S, Kim J, Cho Y, Ahn S, Kim G, Hwang
D, Chang Y, Ha S, Choi Y, Lee MH, et al: Promotion of tumorigenesis
by miR-1260b-targeting CASP8: Potential diagnostic and prognostic
marker for breast cancer. Cancer Sci. 113:2097–2108. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Storchel PH, Thummler J, Siegel G,
Aksoy-Aksel A, Zampa F, Sumer S and Schratt G: A large-scale
functional screen identifies Nova1 and Ncoa3 as regulators of
neuronal miRNA function. Embo J. 34:2237–2254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang YA, Liu HN, Zhu JM, Zhang DY, Shen
XZ and Liu TT: RNA binding protein Nova1 promotes tumor growth in
vivo and its potential mechanism as an oncogene may due to its
interaction with GABAA Receptor-γ2. J Biomed Sci.
23:712016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Catalano RD, Wilson MR, Boddy SC, McKinlay
AT, Sales KJ and Jabbour HN: Hypoxia and prostaglandin E receptor 4
signalling pathways synergise to promote endometrial adenocarcinoma
cell proliferation and tumour growth. PLoS One. 6:e192092011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng Y, Li S, Boohaker RJ, Liu X, Zhu Y,
Zhai L, Li H, Gu F, Fan Y, Lang R, et al: A MicroRNA expression
signature In Taxane-anthracycline-Based neoadjuvant chemotherapy
response. J Cancer. 6:671–677. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi
Y, Xiong W, Li G, Lu J, Fodstad O, et al: MicroRNA-125b confers the
resistance of breast cancer cells to paclitaxel through suppression
of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J
Biol Chem. 285:21496–21507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim GC, Lee CG, Verma R, Rudra D, Kim T,
Kang K, Nam JH, Kim Y, Im SH and Kwon HK: ETS1 suppresses
tumorigenesis of human breast cancer via trans-activation of
canonical tumor suppressor genes. Front Oncol. 10:6422020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dai YC, Pan Y, Quan MM, Chen Q, Pan Y,
Ruan YY and Sun JG: MicroRNA-1246 mediates drug resistance and
metastasis in breast cancer by targeting NFE2L3. Front Oncol.
11:6771682021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li LP, Huang YC, Zhang Y, Peng A, Qin J,
Lu S and Huang Y: RTKN2 is associated with unfavorable prognosis
and promotes progression in non-small-cell lung cancer. Onco
Targets Ther. 13:10729–10738. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen K, Ye C, Gao Z, Hu J, Chen C, Xiao R,
Lu F and Wei K: Immune infiltration patterns and identification of
new diagnostic biomarkers GDF10, NCKAP5, and RTKN2 in non-small
cell lung cancer. Transl Oncol. 29:1016182023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zeng J, Li X, Liang L, Duan H, Xie S and
Wang C: Phosphorylation of CAP1 regulates lung cancer
proliferation, migration, and invasion. J Cancer Res Clin Oncol.
148:137–153. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ye FG, Song CG, Cao ZG, Xia C, Chen DN,
Chen L, Li S, Qiao F, Ling H, Yao L, et al: Cytidine deaminase axis
modulated by miR-484 differentially regulates cell proliferation
and chemoresistance in breast cancer. Cancer Res. 75:1504–1515.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jia YZ, Liu J, Wang GQ and Song ZF:
miR-484: A Potential Biomarker in Health and Disease. Front Oncol.
12:8304202022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tsang J, Zhu J and van Oudenaarden A:
MicroRNA-mediated feedback and feedforward loops are recurrent
network motifs in mammals. Mol Cell. 26:753–767. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nersisyan S, Galatenko A, Galatenko V,
Shkurnikov M and Tonevitsky A: miRGTF-net: Integrative
miRNA-gene-TF network analysis reveals key drivers of breast cancer
recurrence. PLoS One. 16:2021. View Article : Google Scholar : PubMed/NCBI
|