Introduction
Currently, non-small cell lung cancer (NSCLC) is the
leading cause of lung cancer-associated mortality, which accounts
for up to 85% of all lung cancer cases (1). In spite of the improvement of the
existing methods of diagnosis and treatment, as well as targeted
therapy development, the recurrence and mortality rates remain high
and the rate of 5-year survival is <20% (2–4).
Therefore, it is crucial to deeply investigate the occurrence and
mechanisms of NSCLC in order to explore novel therapeutic drug
targets for early diagnosis and treatment, and to improve patient
prognosis.
MicroRNAs (miRNAs) have a length of 20–25
nucleotides and regulate gene expression at the
post-transcriptional stage by preventing transcription (5). In the past 10 years, it has become
clear that miRNAs have critical roles in the regulation of tumor
cell proliferation, as well as tumor invasion and metastasis
(6,7). Numerous miRNAs have been determined to
be ectopically expressed in NSCLC tissues and to promote the
development or progression of the disease (6–11). For
instance, miR-92a was found to be upregulated in NSCLC, and miR-92a
overexpression led to the promotion of NSCLC tumor growth in
vivo (12). miRNA-605 promotes
cell proliferation, migration and invasion in NSCLC by directly
targeting large tumor suppressor 2 (LATS2) (13). Furthermore, recent studies have
indicated that miR-373 has important roles in human cancers, but
the effect is controversial (14,15). A
previous study reported that miR-373 may be associated with the
progression of NSCLC (16).
However, the biological role of miR-373 during the development of
NSCLC requires further investigation.
The aim of the present study was to investigate the
role of miR-373 in the regulation of NSCLC cell proliferation and
invasion in vitro and the underlying molecular mechanism of
this process. The current findings provided a new therapeutic
strategy for clinical patients.
Materials and methods
Clinical specimens
From March 2019 to June 2020, 60 pairs of NSCLC
tissues and adjacent non-tumor tissues were collected from 60 NSCLC
patients who underwent lobectomy at the Department of Thoracic
Surgery at Huadong Hospital (Shanghai, China). Patients were
included when: i) They had primary NSCLC, excluding recurrence
cases; ii) they did not receive radiotherapy, chemotherapy or other
neoadjuvant treatment before surgery; iii) their diagnoses were
confirmed by professional pathologists according to the NSCLC
histopathological diagnostic criteria; and iv) had no history of
other malignant tumors. All of the patients provided written
informed consent prior to tissue collection. The
clinicopathological data are presented in Table I. The present study was approved by
the ethics committee of Huadong Hospital (Shanghai, China; approval
no. 2019-0037). The adjacent tissue was 5 cm away from the edge of
the tumour; all of the tissue samples were snap-frozen using liquid
nitrogen, and then stored at −80°C until further analysis.
 | Table I.Association between miR-373 and
clinicopathological features of patients with non-small cell lung
cancer. |
Table I.
Association between miR-373 and
clinicopathological features of patients with non-small cell lung
cancer.
|
|
| MiR-373
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Total (n=60) | High (n=35) | Low (n=25) | P-value |
|---|
| Gender |
|
|
| 0.4744 |
|
Male | 38 | 23 | 15 |
|
|
Female | 22 | 22 | 10 |
|
| Age, years |
|
|
| 0.8953 |
|
≥60 | 33 | 19 | 14 |
|
|
<60 | 27 | 16 | 11 |
|
| Tumor size, cm |
|
|
| 0.0455 |
| ≥5 | 42 | 21 | 21 |
|
|
<5 | 18 | 14 | 4 |
|
| Smoking |
|
|
| 0.5930 |
| No | 24 | 13 | 11 |
|
|
Yes | 36 | 22 | 14 |
|
| Differentiation
degree |
|
|
| 0.0395 |
|
Moderate | 21 | 16 | 5 |
|
|
Poor | 39 | 19 | 20 |
|
| Clinical stage |
|
|
| 0.0428 |
|
I–II | 26 | 19 | 7 |
|
|
III–IV | 34 | 16 | 18 |
|
| Distant
metastasis |
|
|
| 0.1736 |
| No | 15 | 11 | 4 |
|
|
Yes | 45 | 24 | 21 |
|
| Lymph node
involvement |
|
|
| 0.0804 |
| No | 11 | 9 | 2 |
|
|
Yes | 49 | 26 | 23 |
|
MiRNA expression profile data from
Gene Expression Omnibus (GEO)
The expression levels of miRNAs were assessed in
NSCLC tissues and normal tissue samples from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29248).
Differentially expressed miRNAs were identified using the R ‘limma’
package (version 4.2), which is a widely used tool that may be used
to analyze data from any GEO series and significance analysis of
microarray (SAM), to determine the differential expression of
miRNAs among groups. miRNAs were considered to be differentially
expressed according to the P<0.05 threshold from the limma
analysis and median false discovery rate <0.05 from SAM. Data
were visualized as heat maps using the online tool Morpheus (a
web-based tool; http://software.broadinstitute.org/morpheus/).
Reverse transcription-quantitative
(RT-q)PCR analyses
Total RNA from tumor tissues and transfected cells
was isolated using the miRNeasy Mini kit (Qiagen GmbH). RNA
concentrations were determined using the NanoDrop 2000 (Thermo
Fisher Scientific, Inc.). To obtain complementary DNA (cDNA), 1.0
µg total RNA was added for reaction using the OneScript Plus
Reverse Transcription Kit (GeneCopoeia) and the PrimeScript RT
reagent Kit (Takara Biotechnology, Inc.) in a 20-µl total volume
reaction according to the manufacturer's instructions. qPCR was
performed using PowerTrack SYBR Green kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) on a Bio-Rad CFX96 real-time PCR
detection system (Bio-Rad Laboratories, Inc.) according to the
manufacturer's instructions. PCR was conducted at 95°C for 10 min
followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec.
Primers for cDNA amplification were as follows: miR-21 forward,
5′-ATGCGCAACACCAGTCGATGG-3′ and universal reverse,
5′-GCAGGGTCCGAGGTATTC-3′; miR-224 forward,
5′-CGCAGAAAATGGTGCCCTAGT-3′; miR-96 forward,
5′-CGCTAATCATGTGCAGTGCC-3′; miR-30a forward,
5′-GCAGCGCTTTCAGTCGGATGTT-3′; miR-126, forward,
5′-GCACGTCGTACCGTGAGTAAT-3′; miR-7 forward,
5′-CAGTGGAAGACTAGTGATT-3′; miR-373 forward,
5′-CGAGAAGTGCTTCGATTTTG-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; Grb-associated binding protein 2
(GAB2) forward, 5′-AGAAGTTGAGGCGCTATGCC-3′ and reverse,
5′-AAGGTGCGTTCACTGGTCTT-3′; GAPDH forward,
5′-TCAACGACCCCTTCATTGACC-3′ and reverse,
5′-CTTCCCGTTGATGACAAGCTTC-3′. miRNA was normalized to U6 and GAB2
was normalized to GAPDH. Data were analyzed by using the
2−ΔΔCq method (17).
Cell lines and cell culture
The NSCLC cell lines (A549, H358 and H2170) were
purchased from the American Type Culture Collection (ATCC) and the
normal human bronchial epithelial cell line 16HBE was purchased
from the Cell Bank of the Chinese Academy of Sciences. All cells
were maintained in DMEM (Invitrogen; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich;
Merck KGaA) and specific antibiotics (100 U/ml penicillin and 0.1
mg/ml streptomycin) at 37°C in 95% air and 5% CO2.
Cell transfection
miR-373 mimics, mimics negative control (NC),
inhibitor and inhibitor NC were synthesized by Genepharma
(Shanghai, China). The sequences are as follows: MiR-373 mimics,
5′-GAAGUGCUUCGAUUUUGGGGUGU-3′; mimics NC,
5′-GCUUUAUUAGAGUGCUAUUGCUU-3′; miR-373 inhibitor,
5′-ACACCCCAAAAUCGAAGCACUUC-3′; and inhibitor NC,
5′-AUUCUCAUCACCAAUCACGAAAUA-3′. A549 and H358 cells were
transfected using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's
instructions with a GAB2-overexpression pcDNA3.1 plasmid (1 µg;
Invitrogen; Thermo Fisher Scientific, Inc.). As an NC, a pcDNA3.1
vector that was empty was utilized. After 48 h of transfection, the
cells were harvested for the following experiments. The efficiency
of overexpression or inhibition was verified by RT-qPCR and western
blot.
Cell proliferation
A Cell Counting Kit (CCK)-8 assay (Dojindo Molecular
Technologies, Inc.) was carried out in order to measure cell
proliferation according to the manufacturer's instructions. In
brief, A549 and H358 cells (2×104 cells) were seeded
into 96-well plates and transfected with miR-373 mimics, mimics-NC
or pcDNA-GAB2. The cells were then incubated for 1, 2 and 3 days at
37°C and 5% CO2 in an incubator with 95% air and
saturated humidity. Subsequently, 10 µl CCK-8 solution was added
into each well and incubated for a further 2 h at 37°C, and the
absorbance at 450 nm was measured using a microplate reader
(Bio-Rad Laboratories, Inc.).
Cell apoptosis
The apoptotic rate was analyzed using an Annexin
V-FITC/PI Apoptosis Detection kit (BD Biosciences) according to the
manufacturer's protocol. In brief, following miR-373 mimics,
mimics-NC or pcDNA-GAB2 transfection for 48 h, cells were digested
with trypsin, centrifuged at 600 × g for 5 min at 4°C and
resuspended in 400 µl binding buffer. Subsequently, 5 µl Annexin
V-fluorescein isothiocyanate (FITC) and 5 µl propidium iodide (PI)
solution (cat. no. 556547; BD Pharmingen; BD Biosciences) was added
and incubated in a dark room at room temperature for 20 min. The
apoptotic cell ratio was then detected on a BD FACSCalibur flow
cytometer (BD Biosciences) using FlowJo software (version 7.6.1;
FlowJo LLC). The results showed healthy viable cells in the lower
left quadrant on the scatter plot as (FITC-/PI-). The lower right
quadrant (Q3) represented the early-stage apoptotic cells as
(FITC+/PI-). The upper right quadrant (Q2) represented late-stage
apoptotic cells as (FITC+/PI+). The calculation was made as
follows: Apoptotic rate=percentage of early-stage apoptotic cells
(Q3) + percentage of late-stage apoptotic cells (Q2).
Caspase 3 activity assay
After treatment, total protein was extracted using
RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology)
and the protein concentration was evaluated using the bicinchoninic
acid assay (cat. no. P0010S; Beyotime Institute of Biotechnology)
according to the manufacturer's protocols. The caspase-3 activity
assay was performed using a Caspase-3 colorimetric assay kit (cat.
no. C1115; Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. The results were evaluated using a
microplate reader (Bio-Rad Laboratories, Inc.) at 405 nm.
Western blot analysis
Total cellular proteins were lysed in RIPA lysis
buffer (Beyotime Institute of Biotechnology) containing 2% protease
inhibitor PMSF and the protein concentration of each sample was
evaluated with a bicinchoninic acid protein assay kit (Beyotime
Institute of Biotechnology). Following separation by 10–12%
SDS-PAGE, proteins were transferred onto a PVDF membrane (GE
Healthcare; Cytiva). The membranes were blocked with a 5% skimmed
milk solution in PBS with 0.05% Tween-20 overnight at 4°C, and then
incubated with specific primary antibodies for 2 h at room
temperature, including GAB2 (1:500 dilution; cat. no. ab32365), Bax
(1:2,000; cat. no. ab32503), Bcl-2 (1:2,000; cat. no. ab32124),
cleaved caspase 3 (1:2,000; cat. no. ab32042), E-cadherin (1:2,000;
cat. no. ab40772), N-cadherin (1:1,500; cat. no. ab76011), Vimentin
(1:1,000; cat. no. ab92547), Fibronectin (1:1,000; cat. no.
ab2413), PI3K(p85) (1:1,000; cat. no. ab191606), mTOR (1:1,000;
cat. no. ab2732), phosphorylated (p)-mTOR (1:1,000; cat. no.
ab109268), Akt (1:1,000; cat. no. ab8805), p-Akt (1:2,000; cat. no.
ab81283) and β-actin (1:1,000; cat. no. ab8227; all from Abcam).
Subsequently, the membranes were incubated with the mouse
anti-rabbit IgG-HRP antibody (1:1,000 dilution; cat. no. sc2537;
Santa Cruz Biotechnology, Inc.) used as a secondary antibody for 60
min at room temperature. The proteins were visualized with an ECL
kit (cat. no. 34580; Thermo Fisher Scientific, Inc.).
Semi-quantification was performed using ImageJ software (version
1.46; National Institutes of Health).
Transwell assay
At 24 h after transfection, 1.0×105
cells/200 µl were added to the upper chambers of Transwell inserts
with 8-µm pores coated with Matrigel (Corning, Inc.). Furthermore,
600 µl 10% FBS in DMEM was added to matched bottom chambers. After
48 h at 37°C, cells on the bottom filter surfaces were fixed for 15
min at room temperature with 4% paraformaldehyde (Beyotime
Institute of Biotechnology) and stained for 10 min at room
temperature with 0.1% crystal violet. Under an IX81 microscope
(Olympus Corp.), images were acquired at a magnification of ×100
and the number of invaded cells was calculated by analyzing five
random fields per well.
Wound-healing assay
When cell confluence reached ~80%, a scratch wound
was generated using the tip of 10-µl pipette by making a straight
line and the culture medium was replaced by DMEM supplemented with
1% FBS. Digital photographs were obtained at 0 and 48 h after
scratching under a microscope at ×100 magnification (Olympus IX81;
Olympus Corp.) and the scratch area was measured using ImageJ
software (version 1.46; National Institutes of Health).
Luciferase assays
The biological targets of miRNA targets were
predicted using the algorithms TargetScan (https://www.targetscan.org/vert_80/) and microRNA.org (http://www.microrna.org/). The 3′-untranslated region
(UTR) of human GAB2 was amplified as previously described (18) and cloned into pmirGLO (E1330;
Promega Corp.) luciferase vector, named pGAB2-wild-type (WT). The
human GAB2 mRNA was extracted from adjacent non-tumor lung tissues
and reverse-transcribed to cDNA using a Reverse Transcription Kit
with gDNA eraser kit (Takara Bio, Inc.). The mutant form of the
3′-UTR of GAB2 was also cloned into the pmirGLO vector to construct
pGAB2-mutant (Mut) using the QuikChange Site-Directed Mutagenesis
kit (Thermo Fisher Scientific, Inc.). To examine whether miR-373
directly target GAB2 mRNA, the reporter plasmids, WT-GAB2-PGL3 and
Mut-GAB2-PGL3, were co-transfected with miR-373 mimics/inhibitor
into 293T cells (7×104; ATCC) using Lipofectamine® 2000
according to the manufacturer's instructions. The relative firefly
luciferase activity normalized to Renilla luciferase was
measured 48 h after transfection by using the Dual-Light
luminescent reporter gene assay (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 16.0; SPSS Inc.). Values are expressed as the
mean ± standard deviation. An unpaired Student's t-test was used to
perform comparisons of parameters between the two groups, while
one-way analysis of variance followed by Tukey's post-hoc test was
performed for comparing multiple groups. Differences in miR-373
expression and GAB2 gene expression between adjacent noncancerous
tissues and cancer tissues were examined using a paired t-test. The
correlation between miR-373 and clinicopathological features of
patients with NSCLC was analyzed using the chi-square test;
however, the variable ‘lymph node involvement’ was analyzed with
Fisher's exact test. Survival rates were calculated using the
Kaplan-Meier method and comparisons were performed using the
log-rank test. The correlation between the expression of miRNA and
GAB2 was analyzed using Pearson's correlation analysis. P<0.05
was considered to indicate statistical significance.
Results
miR-373 is downregulated in NSCLC
tissues and cell lines
Using the GSE29248 microarray dataset, the
differentially expressed miRNAs in NSCLC tissues and adjacent
noncancerous tissues were first examined. As presented in Fig. 1A, 35 miRNAs were elevated and 25
miRNAs were downregulated in NSCLC. In addition, some of these
miRNAs were confirmed by RT-qPCR in NSCLC tissues and adjacent
noncancerous tissues in order to evaluate the validity of the in
silico findings. According to the experimental results of the
present study, miR-21, miR-224 and miR-96 were elevated, whereas
miR-30a, miR-126 and miR-7 were downregulated (Fig. 1B), which was consistent with
previous studies (19–24). Consistent with the array data, the
results showed that miR-373 had the lowest expression and the most
significant difference. Considerable evidence suggests that miR-373
is downregulated in multiple tumor tissues and acts as a tumor
suppressor (14,15). Therefore, the subsequent research in
the present study focused on this miRNA.
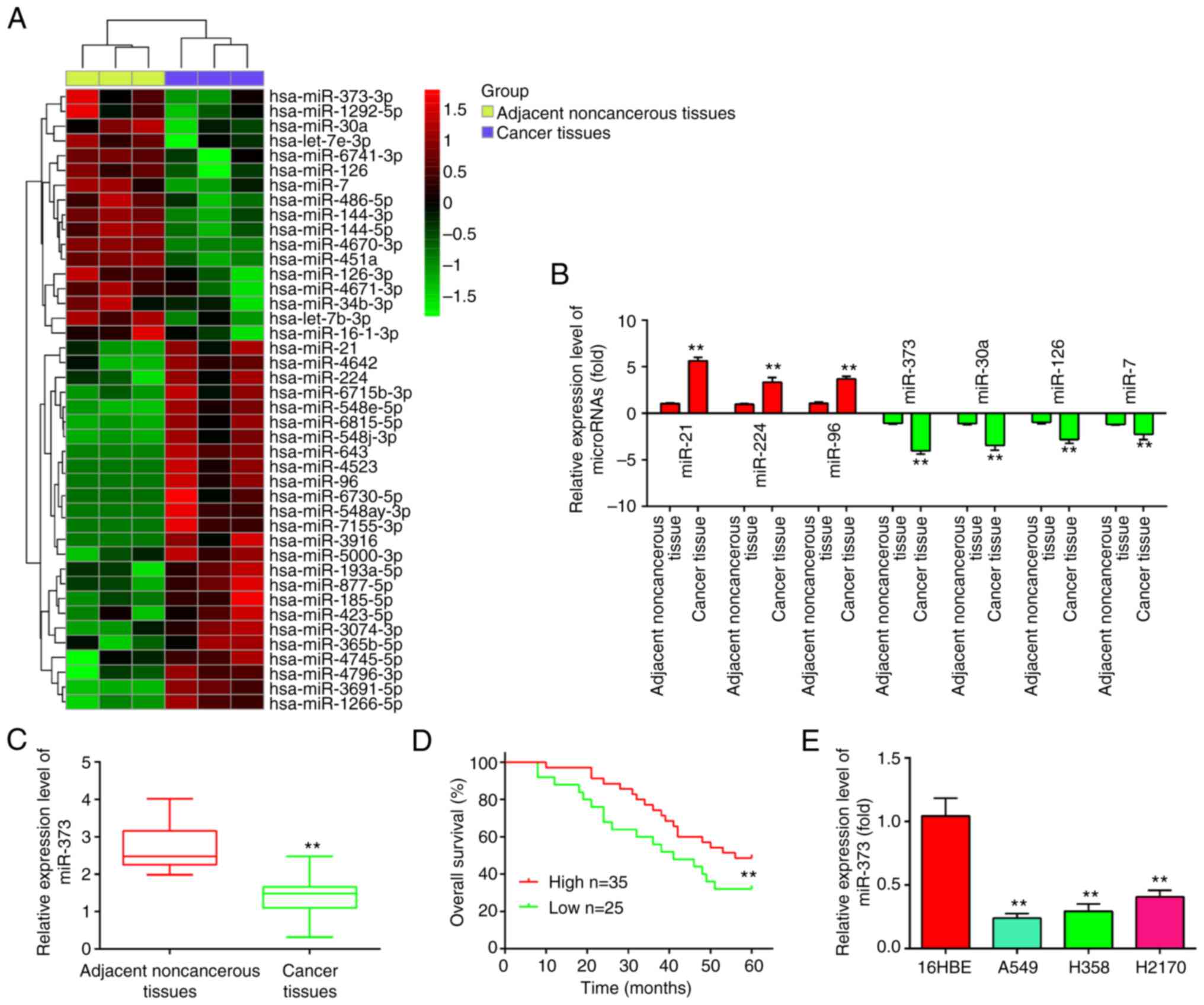 | Figure 1.miR-373 is downregulated in NSCLC
tissues and cell lines. (A) Differentially expressed miRNAs were
analyzed between NSCLC cancer tissue and the adjacent normal
tissue. The dataset was retrieved from Gene Expression Omnibus,
with the accession number GSE29248. The color code in the heat map
is linear and the expression levels of miRNAs that were upregulated
are shown in green to red, whereas the miRNAs that were
downregulated are shown in red to green. (B) The expression levels
of miR-373, miR-21, miR-224 and miR-96, miR-30a, miR-126 and miR-7
were analyzed by RT-qPCR in NSCLC tissues and noncancerous tissues.
(C) Relative expression of miR-373 was further analyzed by RT-qPCR
in 60 pairs of NSCLC tissue and noncancerous tissues. (D)
Kaplan-Meier overall survival curve for patients with NSCLC with
miR-373-high and miR-373-low character. (E) Relative expression of
miR-373 in three NSCLC cell lines (A549, H1299 and H358) and the
normal human bronchial epithelial cell line 16HBE. Values are
expressed as the mean ± standard deviation. **P<0.01 vs. paired
group/noncancerous tissues or 16HBE. miR-373, microRNA-373-3p;
RT-qPCR, reverse transcription-quantitative PCR; NSCLC, non-small
cell lung cancer. |
To confirm the downregulation of miR-373, its
expression was then examined in 60 pairs of NSCLC and adjacent
tissues by RT-qPCR. Compared to adjacent noncancerous tissues,
miR-373 was found to be downregulated in NSCLC tissues (Fig. 1C). The association between miR-373
expression and survival outcomes among patients with NSCLC was then
evaluated. According to the relative expression levels of
miR-373-3p in 60 paired tumor and normal tissues, the patients were
divided into two groups: High miR-373-3p group (miR-373-3p
expression above the median value; n=35) and a low miR-373-3p group
(miR-373-3p expression below the median value; n=25). The results
indicated that miR-373 low expression was closely associated with
the degree of differentiation, clinical stage and tumor size
(Table I). Patients with NSCLC with
low miR-373 expression had poorer overall survival than patients
with high miR-373 expression (Fig.
1D). Subsequently, RT-qPCR was used to determine the expression
of miR-373 in three NSCLC cell lines (A549, H358 and H2170) and a
healthy human bronchial epithelial cell line (16HBE) in order to
further determine whether miR-373 is downregulated in NSCLC cells.
As indicated in Fig. 1E, the three
NSCLC cell lines expressed miR-373 at a lower level than the 16HBE
cells, indicating that miR-373 may have a role in the occurrence of
NSCLC.
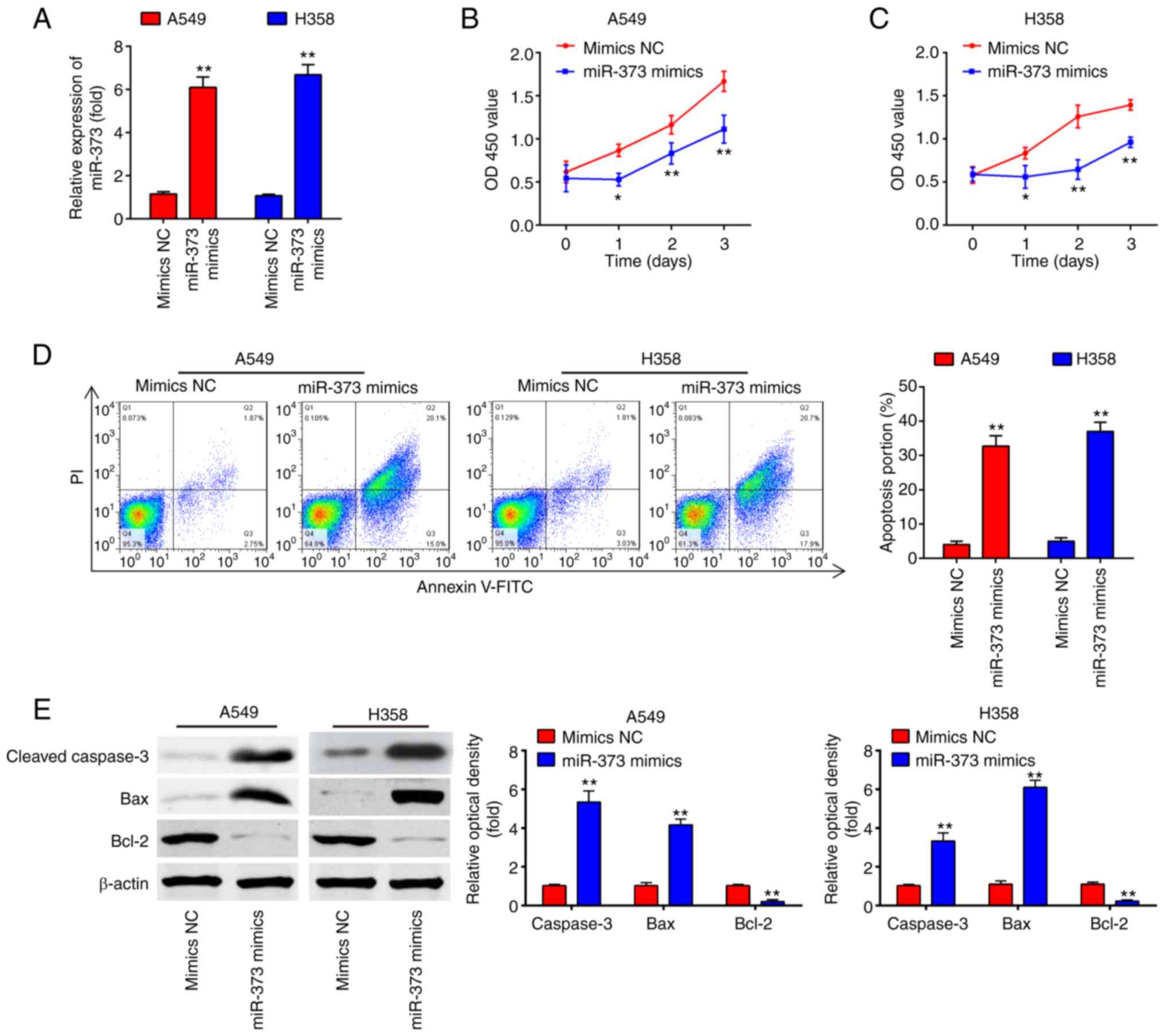
Overexpression of miR-373 suppresses
cell proliferation and promotes apoptosis
To investigate the role of miR-373 in NSCLC, a
gain-of-function analysis was performed by transfecting A549 and
H358 cells with chemically synthesized miR-373 mimics. As presented
in Fig. 2A, miR-373 levels were
effectively enhanced after miR-373 mimics transfection in A549 and
H358 cells. The results of the CCK-8 assay indicated that miR-373
mimics weakened the capacity of proliferation compared with that in
the mimics NC group (Fig. 2B and
C). Furthermore, the cell apoptosis rate was evidently
increased in the miR-373 mimics compared with that in the mimics NC
group (Fig. 2D). Consistently,
miR-373 overexpression markedly increased the expression of
cleaved-caspase-3 and Bax, and decreased the expression of Bcl2 in
A549 and H358 cells compared with the levels in the mimics NC group
(Fig. 2E). Collectively, these data
indicate that overexpression of miR-373 suppressed cell
proliferation and induced apoptosis in NSCLC cells.
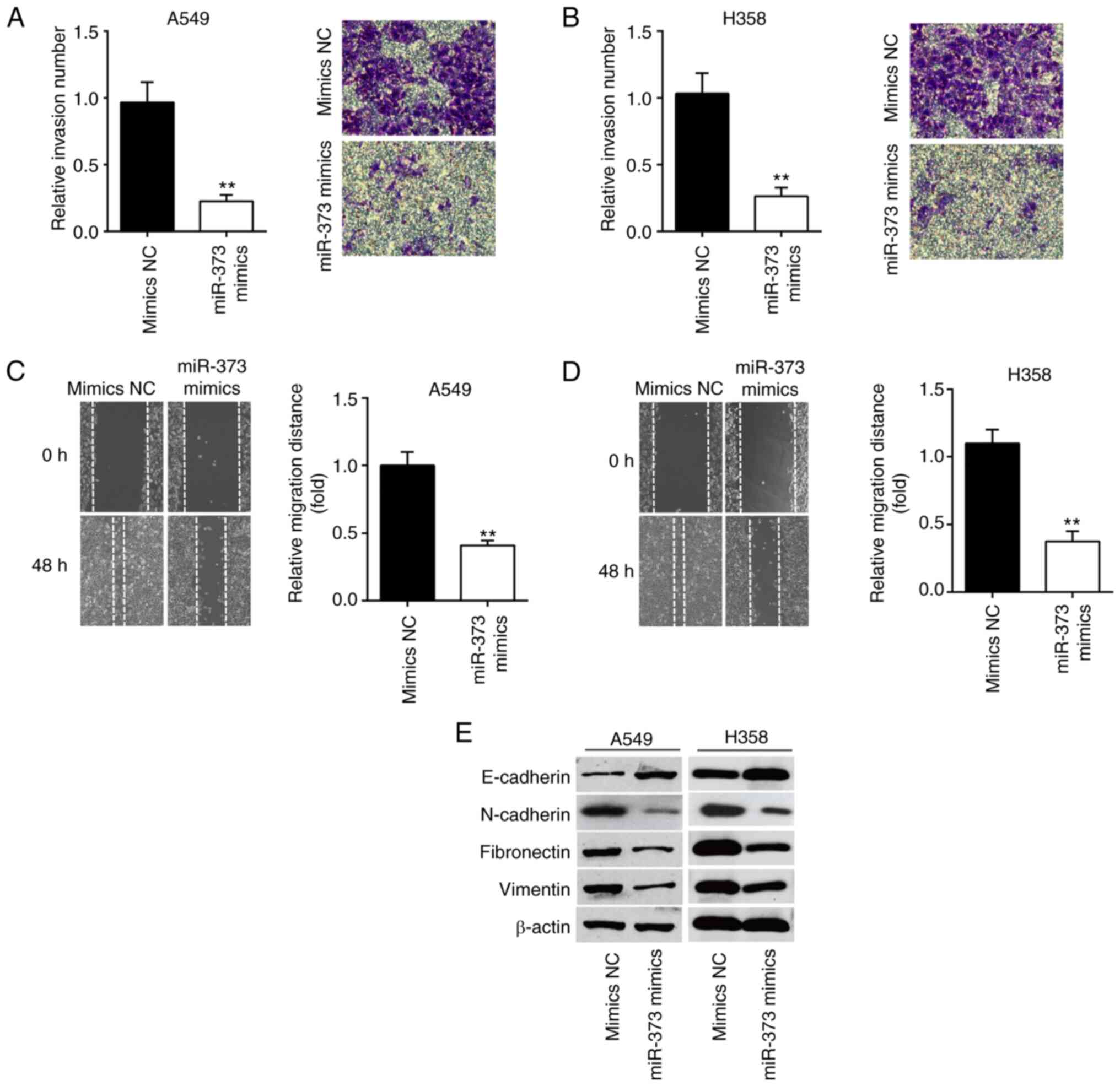
Overexpression of miR-373 suppresses
cell invasion and migration
Next, the migration and invasion ability of NSCLC
cells were assessed by Transwell and wound-healing assays. The
results of the Transwell assay indicated that miR-373 mimics led to
a significant reduction in cell invasion compared to the mimics NC
group (Fig. 3A and B). In the
wound-healing assay, it was shown that the cells' ability to
metastasize was significantly weakened by miR-373 mimics compared
with the mimics NC group (Fig. 3C and
D). The influence of miR-373 on the protein markers of
epithelial-mesenchymal transition (EMT) of A549 and H358 cells was
then determined. The western blot results showed that the
expression of E-cadherin, an epithelial marker, was markedly
increased, while N-cadherin, Fibronectin and Vimentin, three
mesenchymal markers, were significantly decreased in A549 and H358
cells after miR-373 mimics transfection (Fig. 3E). These results suggest that
miR-373 regulates NSCLC cell invasion and migration via inhibiting
the EMT process.
GAB2 is a direct target of
miR-373
TargetScan and miRanda, two publicly accessible
databases, were used to identify GAB2 as a novel target of miR-373
(Fig. 4A). Previous research has
shown that GAB2 is involved in a variety of carcinogenic
activities, such as cellular growth, survival, proliferation and
migration, through controlling the AKT pathway (25–27).
To verify whether GAB2 is a direct target of miR-373, a luciferase
reporter assay was performed. The results obtained with the
dual-luciferase reporter assay system indicated that miR-373 mimics
significantly suppressed the luciferase activity of the vector
carrying the WT-GAB2 3′UTR, but not of that with the mutant 3′UTR,
whereas knockdown of miR-373 led to the opposite result (Fig. 4B), indicating that miR-373 was able
to bind to the GAB2 3′-UTR.
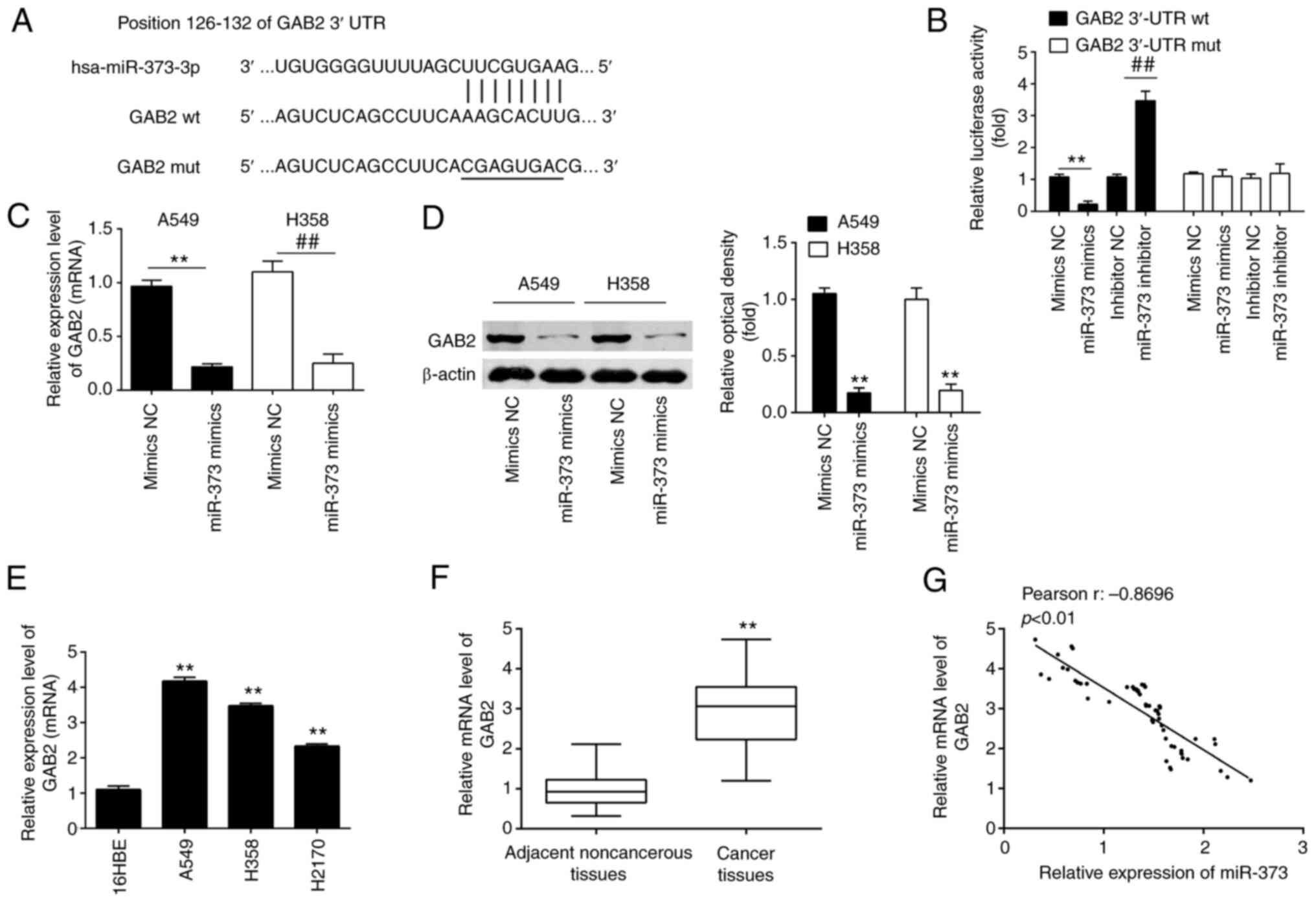 | Figure 4.GAB2 is a direct target of miR-373 in
NSCLC cells. (A) The predicted complementary sequences for miR-373
in the 3′-UTR of GAB2 and the mutations are shown in the seed
region of miR-373. (B) miR-373 mimics and luciferase plasmids,
which include WT GAB2 3′-UTR or Mut GAB2-3′-UTR, were
co-transfected into 293T cells. The relative luciferase activity
was determined. (C and D) GAB2 mRNA and protein expression levels
were measured in (C) A549 and (D) H358 cells transfected with
miR-373 mimics or inhibitor. Values are expressed as the mean ±
standard deviation. **P<0.01 vs. mimics-NC;
##P<0.01 vs. inhibitor-NC. (E and F) Relative
expression of GAB2 was further analyzed by RT-qPCR in (E) three
NSCLC cell lines (A549, H1299 and H358) and (F) in 60 pairs of
NSCLC tissue and noncancerous tissues. **P<0.01, compared with
16HBE cells or noncancerous tissues. (G) Pearson analysis for the
correlation of GAB2 and miR-373 expression levels in patients with
NSCLC (r=−0.8696; P<0.01). MiR, microRNA; NC, negative control;
GAB2, Grb-associated binding protein 2; RT-qPCR, reverse
transcription-quantitative PCR; NSCLC, non-small cell lung cancer;
WT, wild-type; Mut, mutant. |
Subsequent experiments revealed that overexpression
of miR-373 inhibited GAB2 expression at the mRNA and protein levels
in A549 and H358 cells (Fig. 4C and
D). Furthermore, RT-qPCR demonstrated that GAB2 expression
levels were markedly increased in NSCLC cell lines and NSCLC
tissues as compared with 16HBE and adjacent noncancerous tissues,
respectively (Fig. 4E and F). An
obvious inverse correlation between GAB2 and miR-373 expression
levels in NSCLC tissues was also observed (Fig. 4G). These results demonstrated that
miR-373 was able to suppress the expression of GAB2 in NSCLC cells
by directly targeting the GAB2 3′-UTR.
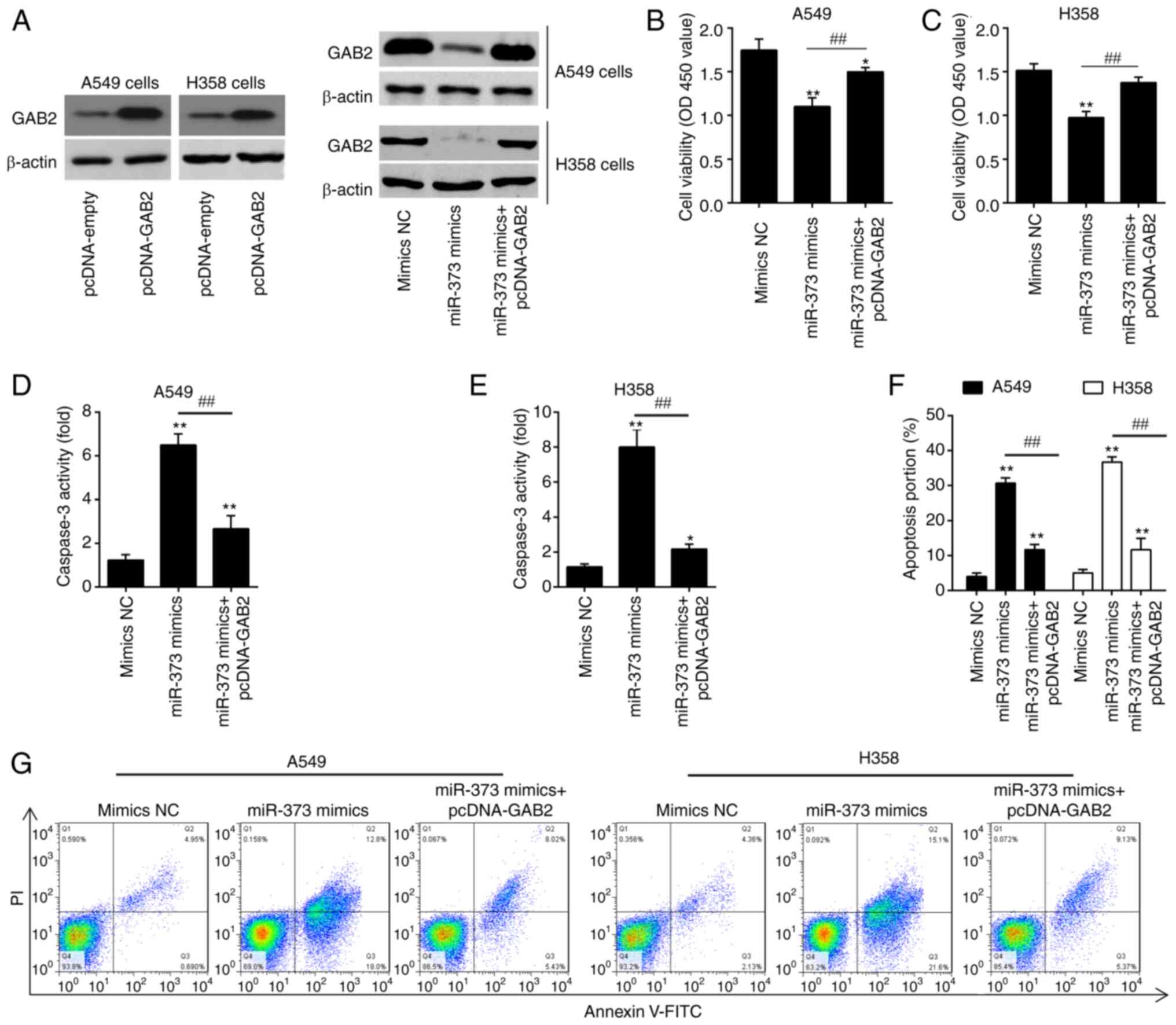
Overexpression of GAB2 attenuates the
effects of miR-373 on cell proliferation and apoptosis
In order to determine whether miR-373 controls cell
proliferation and apoptosis by targeting GAB2, pcDNA-GAB2 and
miR-373 mimics were transfected into A549 and H358 cells. Following
pcDNA-GAB2 transfection, GAB2 protein expression was markedly
enhanced in A549 and H358 cells, as indicated in Fig. 5A. After transfection with miR-373
mimics, the expression level of GAB2 was significantly inhibited.
However, this inhibitory effect was reversed by simultaneous GAB2
overexpression. A549 and H358 cells transfected with miR-373 mimics
had reduced cell proliferation compared with that in the mimics NC
group, according to the results of a CCK-8 assay, whereas the
inhibitory effect was efficiently attenuated by simultaneous
overexpression of GAB2 (Fig. 5B and
C). Furthermore, overexpression of GAB2 substantially reversed
the stimulatory effects of miR-373 on caspase 3 activity and cell
apoptosis in A549 and H358 cells (Fig.
5D-G). Together, these findings demonstrate that GAB2
contributes to the function of miR-373 in the proliferation and
apoptosis of NSCLC cells.
Overexpression of GAB2 attenuates the
inhibitory effects of miR-373 on the invasion and migration of
NSCLC cells
Next, it was investigated whether miR-373 regulates
cell invasion and migration by targeting GAB2. As anticipated,
miR-373 mimics-transfected A549 and H358 cell lines displayed
decreased invasion activity when compared to the mimics NC group,
while overexpression of GAB2 effectively reduced the inhibitory
effect of miR-373 mimics (Fig.
6A-C). Similarly, overexpression of GAB2 abolished the effects
of miR-373 mimics on migration (Fig.
6D-F). According to the above results, miR-373 mimics
significantly raised E-cadherin expression, while significantly
decreasing N-cadherin, Fibronectin and Vimentin levels, and
simultaneous GAB2 overexpression significantly reversed these
effects (Fig. 6G). These findings
demonstrated that miR-373 inhibited cell invasion and migration by
specifically targeting GAB2.
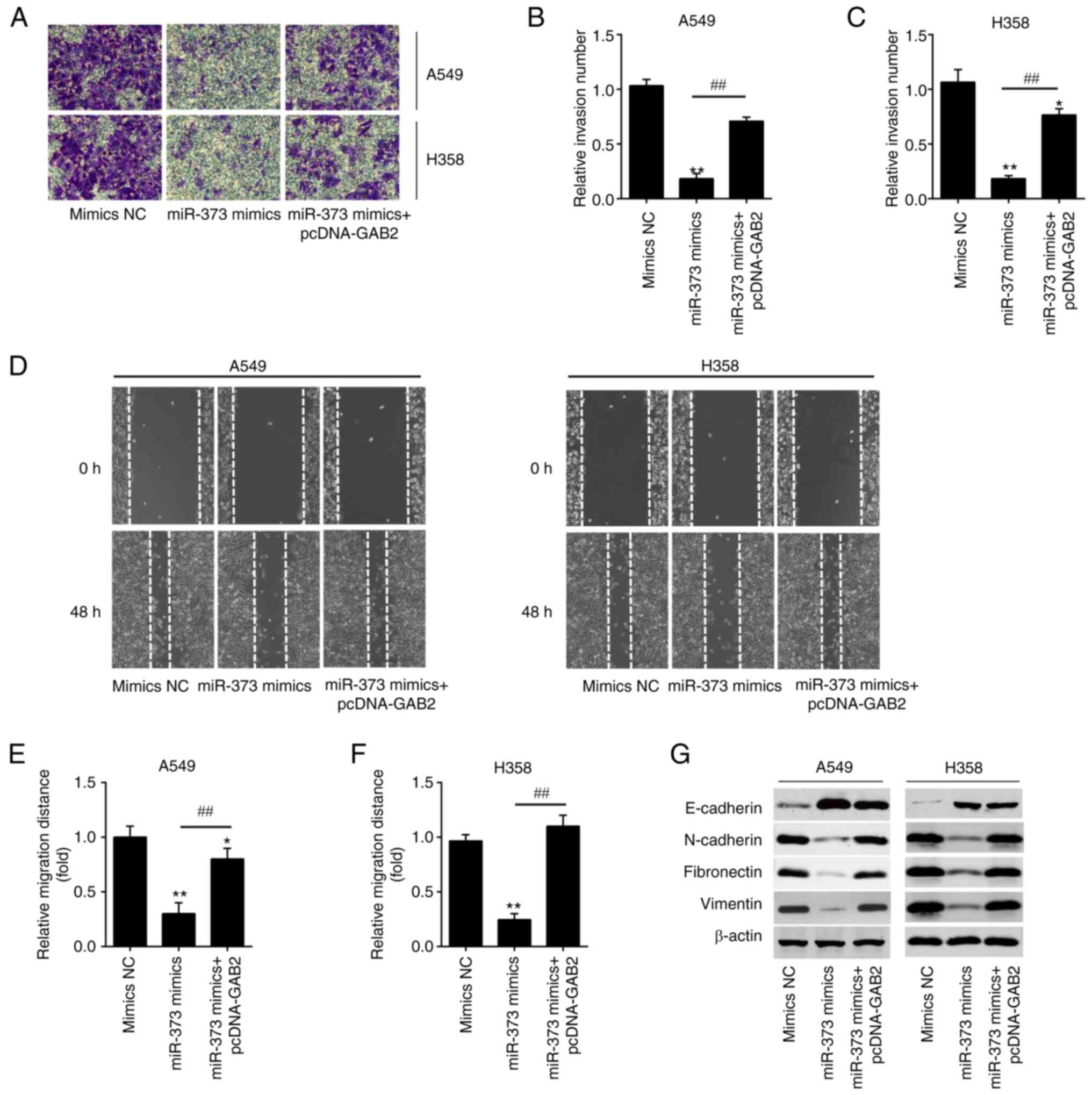 | Figure 6.Overexpression of GAB2 is required
for the miR-373-mediated suppression of migration and invasion in
non-small cell lung cancer cells. pcDNA-GAB2 was transfected into
A549 and H358 cells, along with miR-373 mimics, and the cells were
then harvested for subsequent experiments. (A-C) The invasion of
A549 and H358 cells was measured by Transwell assay after
transfection. (A) Representative images and quantified results for
(B) A549 and (C) H358 cells (magnification, ×200). (D-F) The
migration of A549 and H358 cells was measured by wound-healing
assay after transfection. (D) Representative images and quantified
results for (E) A549 and (F) H358 cells (magnification, ×200). (G)
The expression levels of E-cadherin, N-cadherin, Fibronectin and
Vimentin in A549 and H358 cells were detected by western blot after
transfection. Values are expressed as the mean ± standard
deviation. *P<0.05, **P<0.01 vs. mimics NC,
##P<0.01 vs. miR-373 mimics. MiR, microRNA; NC,
negative control; GAB2, Grb-associated binding protein 2. |
miR-373 suppresses the AKT/mTOR
signaling pathway via downregulation of GAB2
GAB2 is an important oncogenic protein that affects
the AKT/mTOR signaling pathway and regulates tumor cell
proliferation, apoptosis and migration in various types of cancer,
such as NSCLC and renal cell carcinoma (27–29).
Thus, the following experiment was designed to study whether the
miR-373/GAB2 regulatory axis affects the PI3K/AKT signaling pathway
in NSCLC. The effects of miR-373 on proteins relevant to the
AKT/mTOR signaling pathway were determined. It was shown that A549
and H358 cells with high expression of miR-373 had lower levels of
GAB2, PI3K(p85), p-AKT and p-mTOR protein than those in the NC
group, indicating that overexpression of miR-373 blocked AKT/mTOR
signaling pathway activation. However, the inhibitory effect was
reversed by overexpression of GAB2 (Fig. 7A-C). Collectively, these data
suggest that miR-373 suppressed the AKT/mTOR signaling pathway via
downregulation of GAB2.
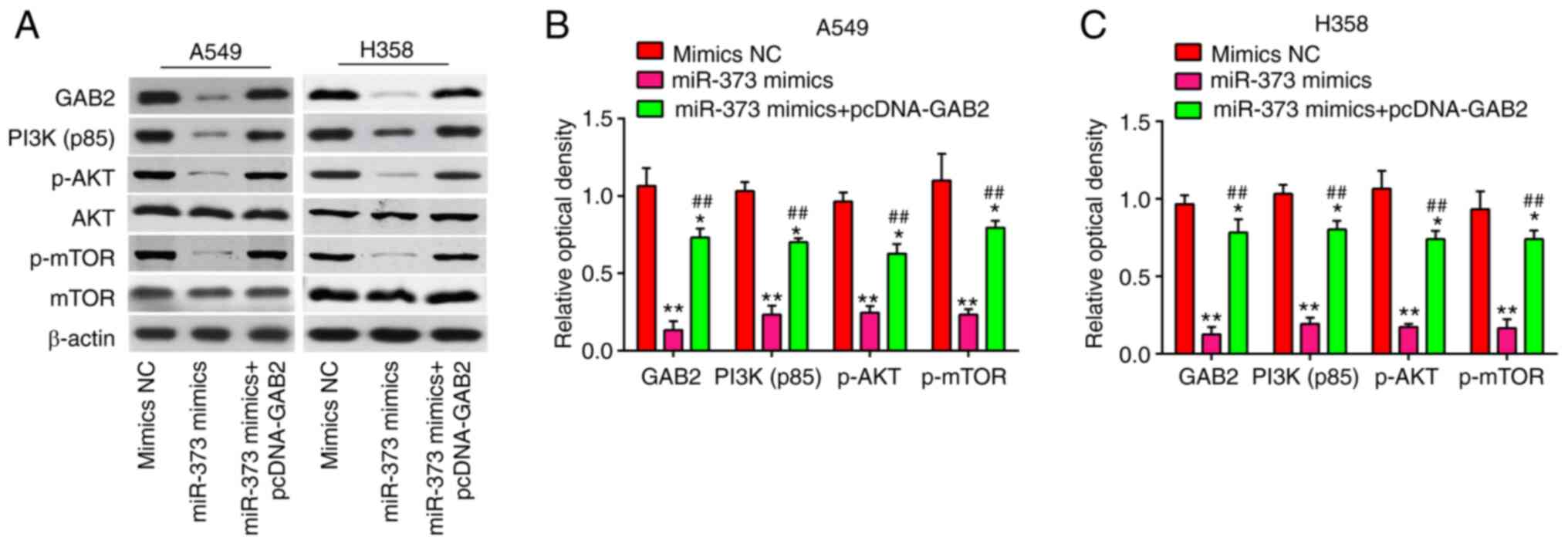 | Figure 7.MiR-373 regulates the GAB2-mediated
PI3K/AKT signaling pathway. pcDNA-GAB2 was transfected into A549
and H358 cells, along with miR-373 mimics, and the cells were then
harvested for western blot. (A) Protein levels of GAB2, PI3K(p85),
p-AKT, AKT, p-mTOR and mTOR were detected by western blot analysis.
Bands for (B) A549 and (C) H358 cells were semi-quantitatively
analyzed by using ImageJ software (version 1.46), normalized to
β-actin density. Values are expressed as the mean ± standard
deviation. *P<0.05, **P<0.01 vs. mimics NC,
##P<0.01 vs. miR-373 mimics. MiR, microRNA; NC,
negative control; GAB2, Grb-associated binding protein 2; p-AKT,
phosphorylated AKT. |
Discussion
In the present study, it was found that miR-373 was
downregulated in NSCLC tissues and cell lines, and closely
associated with the clinicopathological features and prognosis of
patients with NSCLC. Furthermore, upregulation of miR-373 had a
tumor suppressor effect in NSCLC cells by targeting GAB2/PI3K/AKT
signaling. These findings implicate miR-373 as a potential
therapeutic target for NSCLC.
Recent studies have indicated that miRNAs have a
crucial role in tumor cell proliferation, apoptosis, invasion and
migration (30,31). It has been reported that certain
miRNAs, including miR-383 (32),
miR-205 (33), miR-134 (34) and miR-132 (35), have important roles in regulating
the proliferation and function of NSCLC cells. miR-383
overexpression inhibited proliferation, invasion and migration of
A549 and H596 cells, and low tumorous miR-383 expression was
significantly associated with poor prognosis of patients with NSCLC
(32). It was previously reported
that in lung cancer, miR-205 targeted Smad4 to promote cell growth
in vitro and in vivo (33). Furthermore, miR-134 was observed to
suppress NSCLC cell migration and invasion via regulating integrin
beta 1 (34). To date, large-scale
miRNA expression profile analysis has been carried out (36,37).
In the present study, miRNA microarray profiling analysis was
performed in the public dataset GSE29248, which was searched and
downloaded from the GEO database. A signature of miRNAs that was
dysregulated in NSCLC tissues compared with that in normal samples
of the GSE29248 dataset was identified, including those that have
already been recognized by other studies: MiR-21, miR-224, miR-96,
miR-30a, miR-126 and miR-7 (14–19).
However, among these miRNAs, miR-373 showed the highest fold change
in NSCLC samples compared to adjacent noncancerous tissues.
miR-373, which has been shown to be aberrantly expressed in a
variety of human cancers (14,15,38).
However, the mechanism and function of miR-373 in NSCLC remain
unclear. Therefore, miR-373 was selected for further study. In the
present analysis, it was observed that miR-373 was significantly
downregulated in NSCLC tissues and cell lines. Furthermore, miR-373
had a close relationship with the degree of differentiation,
clinical stage, tumor size and prognosis of patients with NSCLC,
and it was indicated that miR-373 may be involved in the
development of NSCLC.
To date, miR-373 has raised considerable interest in
cancer research for its dual function as an oncogene and tumor
suppressor (39,40). For instance, overexpression of
miRNA-373-3p inhibited prostate cancer progression by targeting
AKT1 (41). miR-373 overexpression
decreased cell proliferation and invasion by targeting Ras-related
protein Rab22a, a well-known oncoprotein in liver cancer cells
(42). In cervical cancer,
miR-373-3p targeted AKT1 to suppress cell growth in vitro
and in vivo (43). Another
previous study showed that the expression of miR-373 was decreased
in NSCLC tissues and served as a tumor suppressor by targeting
TFIIB-related factor 2 (44). As an
oncogene, miR-373 promoted urinary bladder cancer cell
proliferation, migration and invasion through upregulating
epidermal growth factor receptor (45). The present results confirmed the
downregulation of miR-373 expression in NSCLC and revealed that
upregulation of miR-373 was able to suppress cell proliferation,
promote apoptosis and inhibit migration and invasion in NSCLC
cells. These findings suggested that miR-31-3p may act as a tumor
suppressor of NSCLC. However, the underlying mechanism remained
elusive.
After determining the biological functions of
miR-373 in NSCLC, a functional analysis was subsequently performed
to predict the molecular mechanisms underlying miR-373. According
to a Bioinformatics analysis, GAB2 is directly targeted by miR-373.
GAB2, a scaffolding protein, mediates interactions with various
signaling pathways and is involved in the regulation of tumor cell
proliferation, apoptosis and migration (46,47).
For instance, GAB2 promoted tumor cell migration and invasion, and
enhanced tumor growth and metastasis in vivo in melanoma
(48). GAB2 was overexpressed in
breast cancer cell lines and primary tumors, and its overexpression
increased the proliferative capacity of mammary epithelial cells
(49). Regarding NSCLC, GAB2
disruption impaired the migration of NSCLC cell lines H1975 and
H1299 (50). In the present study,
it was indicated that GAB2 was highly expressed in NSCLC tissues
and cell lines, and negatively correlated with the level of miR-373
in NSCLC tissues. These observations demonstrate that GAB2 can
promote migration and invasion, which counteract the role of
miR-373. To further explore whether the anti-tumor effect of
miR-373 is mediated by GAB2, rescue experiments were performed by
introducing GAB2. It was found that manipulation of GAB2 could
abrogate the regulative role of miR-373 in NSCLC cells.
Multiple cellular signaling pathways were reported
to be dysregulated in NSCLC, such as the Ras/mTOR pathway and
PI3K-AKT pathway. In the present study, GAB2 was identified as a
functional target of miR-373 in the regulation of NSCLC cell
proliferation, invasion and migration. It is known that GAB2 has
three tyrosine residues, one of which specifically binds to the P85
subunit in PI3K, and this interaction is important for the
activation of PI3K (51,52). The PI3K/Akt pathway has significant
regulatory roles in cell proliferation, apoptosis and invasion
(53). Furthermore, the PI3K/Akt
pathway is aberrantly activated in malignant tumors including
NSCLC, leading to upregulated growth (54). Therefore, PI3K/Akt pathway was
selected for further study. The present results demonstrated that
the phosphorylation status of AKT and mTOR and the expression of
PI3K(p85) were significantly decreased when miR-373 was
overexpressed, but it was reactivated by GAB2 upregulation.
Of note, the present study had certain limitations.
First, miR-373 is not the sole anti-tumor element in NSCLC.
Similarly, GAB2 is the only target gene of miR-373; there are other
genes as well. The results of the present study suggested that the
GAB2/PI3K/AKT signaling pathway may be one of the main mechanisms
through which miR-373 exerts its tumor suppressive role in NSCLC.
However, there may be other mechanisms by which miR-373 exerts its
tumor suppressive role in NSCLC and a study reported that miR-373
targeted the TGF-β-R2/SMAD pathway to suppress cell proliferation
and migration in prostate cancer (55). Another study also demonstrated that
miR-373 targeted LATS2 and oxidation resistance 1 to regulate the
Hippo and the p53 signaling pathway to promote the development of
esophageal squamous cell carcinoma (56); thus, further investigation is
required. In addition, more samples and further studies are needed
to unravel the relationship between miR-373-3p and the
clinicopathological features of patients with NSCLC.
In conclusion, the present study demonstrated that
miR-373 is a crucial regulator in the tumorigenesis of NSCLC, which
is at least partially associated with GAB2-mediated PI3K/Akt
pathway activation. In the future, they will be verified on a large
number of clinical samples to evaluate the utility of miR-373-3p as
a diagnostic marker or therapeutic target.
Acknowledgements
Not applicable.
Funding
This study was supported by the Clinical Science and Technology
Innovation Project of Shanghai Hospital Development Center (grant
no. SHDC22021218).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XXZ, XYC and LXZ performed the experiments,
contributed to data analysis and wrote the manuscript. XXZ, XYC and
LTZ analyzed the data. XYS conceptualized and designed the study,
and contributed to data analysis and experimental materials. XXZ
and XYS confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of Huadong Hospital (Shanghai, China). All patients
provided written informed consent to participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen P, Li Y, Liu R, Xie Y, Jin Y, Wang M,
Yu Z, Wang W and Luo X: Non-small cell lung cancer-derived exosomes
promote proliferation, phagocytosis, and secretion of microglia via
exosomal microRNA in the metastatic microenvironment. Transl Oncol.
27:1015942023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mercier O, Fadel E, de Perrot M, Mussot S,
Stella F, Chapelier A and Dartevelle P: Surgical treatment of
solitary adrenal metastasis from non-small cell lung cancer. J
Thorac Cardiovasc Surg. 130:136–140. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giovannetti E, Toffalorio F, De Pas T and
Peters GJ: Pharmacogenetics of conventional chemotherapy in
non-small-cell lung cancer: A changing landscape? Pharmacogenomics.
13:1073–1086. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi L, Zhu W, Huang Y, Zhuo L, Wang S,
Chen S, Zhang B and Ke B: Cancer-associated fibroblast-derived
exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to
promote the progression and chemoresistance of non-small cell lung
cancer. Clin Transl Med. 12:e9892022. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han B, Molins L, He Y, Viñolas N,
Sánchez-Lorente D, Boada M, Guirao A, Díaz T, Martinez D, Ramirez
J, et al: Characterization of the MicroRNA Cargo of Extracellular
Vesicles Isolated from a Pulmonary Tumor-Draining Vein Identifies
miR-203a-3p as a relapse biomarker for resected non-small cell lung
cancer. Int J Mol Sci. 23:71382022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yan X, Wang T and Wang J: Circ_0016760
Acts as a Sponge of MicroRNA-4295 to Enhance E2F transcription
factor 3 expression and facilitates cell proliferation and
glycolysis in non-small cell lung cancer. Cancer Biother
Radiopharm. 37:147–158. 2022.PubMed/NCBI
|
|
8
|
Gong J, Shen Y, Jiang F, Wang Y, Chu L,
Sun J, Shen P and Chen M: MicroRNA-20a promotes non-small cell lung
cancer proliferation by upregulating PD-L1 by targeting PTEN. Oncol
Lett. 23:1482022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
MicroRNA-211 promotes non-small-cell lung
cancer proliferation and invasion by targeting MxA [Retraction].
Onco Targets Ther. 15:1387–1388. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei J, Meng G, Wu J, Wang Y, Zhang Q, Dong
T, Bao J, Wang C and Zhang J: MicroRNA-326 impairs chemotherapy
resistance in non small cell lung cancer by suppressing histone
deacetylase SIRT1-mediated HIF1α and elevating VEGFA.
Bioengineered. 13:5685–5699. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang X, Shi H and Sun F: The
microRNA-520a-3p inhibits invasion and metastasis by targeting
NF-kappaB signaling pathway in non-small cell lung cancer. Clin
Transl Oncol. 24:1569–1579. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu C, Shan Z, Hong J and Yang L:
MicroRNA-92a promotes epithelial-mesenchymal transition through
activation of PTEN/PI3K/AKT signaling pathway in non-small cell
lung cancer metastasis. Int J Oncol. 51:235–244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye Y, Zhuang J, Wang G, He S, Ni J, Xia W
and Wang J: microRNA-605 promotes cell proliferation, migration and
invasion in non-small cell lung cancer by directly targeting LATS2.
Exp Ther Med. 24:4882022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eyking A, Reis H, Frank M, Gerken G,
Schmid KW and Cario E: MiR-205 and MiR-373 are associated with
aggressive human mucinous colorectal cancer. PLoS One.
11:e01568712016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seol HS, Akiyama Y, Shimada S, Lee HJ, Kim
TI, Chun SM, Singh SR and Jang SJ: Epigenetic silencing of
microRNA-373 to epithelial-mesenchymal transition in non-small cell
lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 353:232–241.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Xu J, Yan X, Jin K, Li W and Zhang
R: MicroRNA-485 plays tumour-suppressive roles in colorectal cancer
by directly targeting GAB2. Oncol Rep. 40:554–564. 2022.PubMed/NCBI
|
|
19
|
Wei J, Gao W, Zhu CJ, Liu YQ, Mei Z, Cheng
T and Shu YQ: Identification of plasma microRNA-21 as a biomarker
for early detection and chemosensitivity of non-small cell lung
cancer. Chin J Cancer. 30:407–414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan
M, Jeon YJ, Li B, Vicentini C, Peng Y, et al: MicroRNA-224 promotes
tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci
USA. 112:E4288–E4297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng
F, Liu X, Zhang Y and Yu J: An integrated analysis of miRNA and
mRNA expressions in non-small cell lung cancers. PLoS One.
6:e265022011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumarswamy R, Mudduluru G, Ceppi P,
Muppala S, Kozlowski M, Niklinski J, Papotti M and Allgayer H:
MicroRNA-30a inhibits epithelial-to-mesenchymal transition by
targeting Snai1 and is downregulated in non-small cell lung cancer.
Int J Cancer. 130:2044–2053. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Crawford M, Brawner E, Batte K, Yu L,
Hunter MG, Otterson GA, Nuovo G, Marsh CB and Nana-Sinkam SP:
MicroRNA-126 inhibits invasion in non-small cell lung carcinoma
cell lines. Biochem Biophys Res Commun. 373:607–612. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X
and Chu Y: MicroRNA-7 inhibits the growth of human non-small cell
lung cancer A549 cells through targeting BCL-2. Int J Biol Sci.
7:805–814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adams SJ, Aydin IT and Celebi JT: GAB2-a
scaffolding protein in cancer. Mol Cancer Res. 10:1265–1270. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding CB, Yu WN, Feng JH and Luo JM:
Structure and function of Gab2 and its role in cancer (Review). Mol
Med Rep. 12:4007–4014. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu DH, Mao JH, Pan XD, Zhu H, Chen X,
Zheng B and Shan Y: microRNA-302c-3p inhibits renal cell carcinoma
cell proliferation by targeting Grb2-associated binding 2 (Gab2).
Oncotarget. 8:26334–26343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mu L, Guan B, Tian J, Li X, Long Q, Wang
M, Wang W, She J, Li X, Wu D and Du Y: MicroRNA218 inhibits tumor
angiogenesis of human renal cell carcinoma by targeting GAB2. Oncol
Rep. 44:1961–1970. 2020.PubMed/NCBI
|
|
29
|
Yu S, Geng S and Hu Y: miR-486-5p inhibits
cell proliferation and invasion through repressing GAB2 in
non-small cell lung cancer. Oncol Lett. 16:3525–3530.
2018.PubMed/NCBI
|
|
30
|
Hu H, Tou FF, Mao WM, Xu YL, Jin H, Kuang
YK, Han CB and Guo CY: microRNA-1321 and microRNA-7515 contribute
to the progression of non-small cell lung cancer by targeting
CDC20. Kaohsiung J Med Sci. 38:425–436. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye J, Luo W, Luo L, Zhai L and Huang P:
MicroRNA-671-5p inhibits cell proliferation, migration and invasion
in non-small cell lung cancer by targeting MFAP3L. Mol Med Rep.
25:302022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shang Y, Zang A, Li J, Jia Y, Li X, Zhang
L, Huo R, Yang J, Feng J, Ge K, et al: MicroRNA-383 is a tumor
suppressor and potential prognostic biomarker in human non-small
cell lung caner. Biomed Pharmacother. 83:1175–1181. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W,
Liu Z and Huang JA: MicroRNA-205 targets SMAD4 in non-small cell
lung cancer and promotes lung cancer cell growth in vitro and in
vivo. Oncotarget. 8:30817–30829. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qin Q, Wei F, Zhang J and Li B: miR-134
suppresses the migration and invasion of nonsmall cell lung cancer
by targeting ITGB1. Oncol Rep. 37:823–830. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang JX, Zhai JF, Yang XT and Wang J:
MicroRNA-132 inhibits migration, invasion and
epithelial-mesenchymal transition by regulating TGFβ1/Smad2 in
human non-small cell lung cancer. Eur Rev Med Pharmacol Sci.
20:3793–3801. 2016.PubMed/NCBI
|
|
36
|
Jiang W, He Y, Shi Y, Guo Z, Yang S, Wei
K, Pan C, Xia Y and Chen Y: MicroRNA-1204 promotes cell
proliferation by regulating PITX1 in non-small-cell lung cancer.
Cell Biol Int. 43:253–264. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu HM, Yi WW, Ma YS, Wu W, Yu F, Fan HW,
Lv ZW, Yang HQ, Chang ZY, Zhang C, et al: Prognostic implications
of decreased microRNA-101-3p expression in patients with non-small
cell lung cancer. Oncol Lett. 16:7048–7056. 2018.PubMed/NCBI
|
|
38
|
Pang J, Dai L, Zhang C and Zhang Q:
MiR-373 Inhibits the epithelial-mesenchymal transition of prostatic
cancer via targeting runt-related transcription factor 2. J Healthc
Eng. 2021:69742252021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu N, Liu X, Xu X, Fan X, Liu M, Li X,
Zhong Q and Tang H: MicroRNA-373, a new regulator of protein
phosphatase 6, functions as an oncogene in hepatocellular
carcinoma. FEBS J. 278:2044–2054. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
et al: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Adv Exp Med Biol.
604:17–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qu HW, Jin Y, Cui ZL and Jin XB:
MicroRNA-373-3p inhibits prostate cancer progression by targeting
AKT1. Eur Rev Med Pharmacol Sci. 22:6252–6259. 2018.PubMed/NCBI
|
|
42
|
Ye Y, Zhang L, Song Y, Zhuang J, Wang G,
Ni J, Zhang S and Xia W: MicroRNA373 exerts antitumor functions in
human liver cancer by targeting Rab22a. Mol Med Rep. 20:3874–3882.
2019.PubMed/NCBI
|
|
43
|
Yu MM, Wang GJ, Wu KH, Xue SL, Ju LL, Li
QR, Xiong AW and Yin GP: MicroRNA-373-3p inhibits the growth of
cervical cancer by targeting AKT1 both in vitro and in vivo. Acta
Biochim Pol. 68:611–617. 2021.PubMed/NCBI
|
|
44
|
Wang L, Qu J, Zhou L, Liao F and Wang J:
MicroRNA-373 inhibits cell proliferation and invasion via targeting
BRF2 in human non-small cell lung cancer A549 cell line. Cancer Res
Treat. 50:936–949. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y, Xu Z and Wang X: miRNA-373
promotes urinary bladder cancer cell proliferation, migration and
invasion through upregulating epidermal growth factor receptor. Exp
Ther Med. 17:1190–1195. 2019.PubMed/NCBI
|
|
46
|
Mu L, Guan B, Tian J, Li X, Long Q, Wang
M, Wang W, She J, Li X, Wu D and Du Y: [Corrigendum] MicroRNA-218
inhibits tumor angiogenesis of human renal cell carcinoma by
targeting GAB2. Oncol Rep. 48:1912022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhuo Y, Li S, Hu W, Zhang Y, Shi Y, Zhang
F, Zhang J, Wang J, Liao M, Chen J, et al: Targeting SNORA38B
attenuates tumorigenesis and sensitizes immune checkpoint blockade
in non-small cell lung cancer by remodeling the tumor
microenvironment via regulation of GAB2/AKT/mTOR signaling pathway.
J Immunother Cancer. 10:e0041132022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Horst B, Gruvberger-Saal SK, Hopkins BD,
Bordone L, Yang Y, Chernoff KA, Uzoma I, Schwipper V, Liebau J,
Nowak NJ, et al: Gab2-mediated signaling promotes melanoma
metastasis. Am J Pathol. 174:1524–1533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bentires-Alj M, Gil SG, Chan R, Wang ZC,
Wang Y, Imanaka N, Harris LN, Richardson A, Neel BG and Gu H: A
role for the scaffolding adapter GAB2 in breast cancer. Nat Med.
12:114–121. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xu LJ, Wang YC, Lan HW, Li J and Xia T:
Grb2-associated binder-2 gene promotes migration of non-small cell
lung cancer cells via Akt signaling pathway. Am J Transl Res.
8:1208–1217. 2016.PubMed/NCBI
|
|
51
|
Zhang YM, Zhang ZQ, Liu YY, Zhou X, Shi
XH, Jiang Q, Fan DL and Cao C: Requirement of Galphai1/3-Gab1
signaling complex for keratinocyte growth factor-induced
PI3K-AKT-mTORC1 activation. J Invest Dermatol. 135:181–191. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shioyama W, Nakaoka Y, Higuchi K, Minami
T, Taniyama Y, Nishida K, Kidoya H, Sonobe T, Naito H, Arita Y, et
al: Docking protein Gab1 is an essential component of postnatal
angiogenesis after ischemia via HGF/c-met signaling. Circ Res.
108:664–675. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jiang N, Dai Q, Su X, Fu J, Feng X and
Peng J: Role of PI3K/AKT pathway in cancer: The framework of
malignant behavior. Mol Biol Rep. 47:4587–4629. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Papadimitrakopoulou V: Development of
PI3K/AKT/mTOR pathway inhibitors and their application in
personalized therapy for non-small-cell lung cancer. J Thorac
Oncol. 7:1315–1326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Weng W, Liu C, Li G, Ruan Q, Li H, Lin N
and Chen G: Long non-coding RNA SNHG16 functions as a tumor
activator by sponging miR-3733p to regulate the TGF-β-R2/SMAD
pathway in prostate cancer. Mol Med Rep. 24:8432021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang L and Wang L, Chang W, Li Y and Wang
L: MicroRNA-373 promotes the development of esophageal squamous
cell carcinoma by targeting LATS2 and OXR1. Int J Biol Markers.
34:148–155. 2019. View Article : Google Scholar : PubMed/NCBI
|