Introduction
Oral squamous cell carcinoma (OSCC), one of the most
common malignant tumors in the head and neck, accounts for ~4% of
whole-body tumors and each year ~510,000 new OSCC cases are
diagnosed worldwide (1,2). Tongue squamous cell carcinoma (TSCC)
is one of the most common types of OSCC. Although tumor therapeutic
strategies have advanced in recent years, the prognosis of TSCC has
not substantially improvement. Metastasis is one of the main causes
of mortality in patients with TSCC (3); therefore, elucidating the mechanism by
which TSCC metastasis is regulated and developing novel
therapeutics to inhibit this metastasis is a potential way of
improving TSCC therapy (4).
The transcription factor, early growth response 1
(Egr-1), belongs to the early growth response protein family and is
ubiquitously expressed in various tissues, such as cardiovascular
(5), adipose (6), and muscle (7) tissues. Egr-1 is a zinc-finger protein
and regulates gene expression through the binding of the C2H2-type
zinc finger domain to GC-rich promoter regions of target genes.
Egr-1 participates in several physiological and pathological
processes, such as cell proliferation, cell apoptosis, cell
invasion, angiogenesis and the immune response (8–10).
Egr-1 is also associated with numerous diseases, including
interstitial inflammation (11),
diabetes (12), cardiovascular
disease (5), and gastric cancer
(13). Egr-1 primarily functions as
a tumor suppressor in a number of tumor types, such as in colon
cancer, mammary tumors and breast cancer (8,14,15),
while in certain other tumors, such as skin cancer, Egr-1 is
reported to be oncogenic (16).
Therefore, the role that Egr-1 may have in TSCC is not clear.
Dual-specificity protein phosphatase 1 (DUSP1), also
termed mitogen-activated protein kinase phosphatase 1 (17,18),
is involved in numerous physiological and pathological processes,
including endoplasmic reticulum stress (19), microglial polarization (20), homeostasis and inflammation
(21). Dysfunction of DUSP1 may
induce several diseases, including neuro-inflammation (22), renal fibrosis (23) and rheumatoid arthritis (24). DUSP1 is also associated with tumor
progression, and therefore, targeting DUSP1 has been suggested as a
potential neoadjuvant strategy for tumor therapy (25). By inhibiting zinc finger protein
SNAI1 (Snail), DUSP1 inhibits the migration and invasion of
prostate cancer cells (26).
Furthermore, micro RNA (miR)-133b inhibits bladder cancer cell
proliferation through targeting DUSP1 (27). DUSP1 can also restrict myeloid
cell-mediated OSCC progression (28). Long non-coding RNA (lncRNA)
LINC01111 upregulates DUSP1 expression through miR-3924 and
antagonizes pancreatic cancer aggressiveness (29). DUSP1 can also be regulated by
several factors. For instance, lncRNAs such as LINC00702, regulate
DUSP1 expression at the transcriptional level (30) and miR-152-3p downregulates DUSP1 and
participates in the progression of acute lung injury (31). Other factors such as upframeshift 1
and platelet-derived growth factor receptor also regulate DUSP1
expression (32,33). However, the regulation mechanisms of
DUSP1, especially at the transcriptional level, have not been fully
elucidated.
The present studyThe present study aimed to
demonstrate the role and mechanism of Egr-1 in regulating TSCC
migration and invasion, in order to determine whether Egr-1 could
be a potential therapeutic target for TSCC. To achieve this aim,
Egr-1 expression in TSCC was analyzed based on GEO datasets and the
effect of Egr-1 in TSCC tumor cell migration and invasion was
measured using a Transwell assay. By overexpressing DUSP1 in cells
with Egr-1 knockdown using Lentivirus infection, the role of DUSP1
in Egr-1-regulated TSCC cell migration and invasion was determined.
Furthermore, by using luciferase assay and ChIP assays, the
mechanism behind how DUSP1 was regulated by Egr-1 was detected.
Materials and methods
Cell lines
CAL-27 cells were purchased from the American Type
Culture Collection and cultured in DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere with 5%
CO2.
Analyzing the expression of Egr-1 and
DUSP1 in head and neck squamous cell carcinoma (HNSCC) based on the
University of Alabama at Birmingham Cancer Data Analysis Portal
(UALCAN) database
The UALCAN database (http://ualcan.path.uab.edu/) is a comprehensive web
resource for analyzing cancer omics data, which can be used for
performing gene expression analysis in various tumor types
(34). In the present study, the
expression profiles of Egr-1, Egr-2, Egr-3 and Egr-4 in HNSCC
samples were analyzed using the UALCAN database. The detail process
was as follows: The TCGA→TCGA genes links were chosen, and ‘Egr-1,
Egr-2, Egr-3, or Egr-4’ was entered in the ‘Enter gene symbol(s)’
dialog box. Next, in the ‘TCGA dataset’ drop-down list, ‘Head and
neck squamous cell carcinoma’ was chosen. Differences between the
normal and tumor groups were determined using unpaired Student's
t-test, and P<0.05 was considered to indicate a statistically
significant difference.
Analyzing Egr-1-associated genes based
on ULCAN database
DUSP1-associated genes were screened in ULCAN
database (http://ualcan.path.uab.edu/). The detailed process was
as follows: The TCGA→TCGA genes links were chosen and ‘Egr-1’ was
entered in the ‘Enter gene symbol(s)’ dialog box. Next, in the
‘TCGA dataset’ drop-down list, ‘Head and neck squamous cell
carcinoma’ was chosen. After the correlation dialog box was chosen,
the genes positively correlated with EGR1 in HNSC were provided
(http://ualcan.path.uab.edu/cgi-bin/TCGAExCorrel.pl?genenam=EGR1&cancer=HNSC).
The correlation was evaluated using Pearson's χ2
test.
Relative Egr-1 expression analyzed
using GEO dataset
The GSE31056 dataset was downloaded from GEO dataset
(https://www.ncbi.nlm.nih.gov/) (35). The expression of Egr-1 in OSCC and
normal tissue was analyzed using the R package DESeq2 based on the
data of the GEO dataset. The data were analyzed using a paired
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
CAL-27 cells were cultured in 6-well plates. To lyse
the cells, 1 ml TRIzol™ reagent (Thermo Fisher
Scientific, Inc.) was added to each well. The cell lysates were
then transferred to new microtubes and RNA was extracted by adding
500 µl chloroform to each tube. RNA was then separated by adding
200 µl isopropanol. The RNA concentration was determined using a
spectrophotometer (Implen GmbH). Total RNA was reverse transcribed
to cDNA using a Reverse Transcriptase Kit (cat no. D7160S; Beyotime
Institute of Biotechnology) following the manufacturer's
instructions. Egr-1 and DUSP1 mRNA expression levels were
determined by qPCR using a FastStart SYBR Green Master kit (cat.
no. D7260; Beyotime Institute of Biotechnology) and the following
thermocycling conditions: Initial denaturation at 95°C for 10 min;
40 cycles of 58°C for 15 sec and 72°C for 30 sec. The primers for
RT-qPCR are listed in Table SI.
The experiment was repeated three times, and the expression of
target genes was normalized to β-actin using the 2−ΔΔCq
method in accordance with a previous study (36).
Egr-1 knockdown
Egr-1 knockdown was conducted using the lentivirus
transduction technique. The lentiviral plasmid,
hU6-MCS-CBh-gcGFP-IRES-puromycin (Shanghai GeneChem Co., Ltd.), was
used to construct the plasmid for Egr-1 knockdown. The sequence for
knocking down Egr-1 was 5′-GCATCTGCATGCGCAACTTCA-3′. A
non-targeting sequence served as the negative control (shCon;
5′-CCGGTTCTCCGAACGTGTCACGTCTCGAGTTCTGGAAGTTCTCACGGCTTTTT-3′). Both
Lv-shEgr-1 and Lv-shCon were synthesized based on a 3rd lentivirus
generation system (Shanghai GeneChem Co., Ltd.) using 293T cells
(ATCC) as the interim cell line. For transfection, 1 µg lentivirus
plasmid for Egr-1 knockdown or negative control and 1 µl lentivirus
generation system were transfected into 293T cells at 37°C for 4 h.
After 48 h, the lentivirus particles were collected using a
Lentivirus concentration kit [Genomeditech; Jiman Biotechnology
(Shanghai) Co., Ltd.] according to the manufacture's instruction.
For lentivirus infection, CAL-27 cells were seeded in a 12-well
plate at 30,000 cells/well. After 24 h of culture, lentivirus (MOI:
10) was added to each well. After 4 h of transduction, the medium
containing lentivirus was replaced with lentivirus-free medium.
After a 48-h incubation, the cells transduced with lentivirus were
selected with 5 µg/ml puromycin (Sigma-Adrich; Merck KGaA). Each 3
weeks, the cells would be re-selected with 5 µg/ml puromycin. As
the lentivirus contained a green fluorescent protein gene, green
fluorescence was used to determine whether the cells had been
transduced. RT-qPCR was then used to confirm that Egr-1 had been
knocked down in transduced CAL-27 cells.
DUSP1 overexpression
DUSP1 overexpression was performed by lentivirus
transduction. A lentivirus for the overexpression of DUSP1
(Lv-oeDUSP1) and a negative control lentivirus (Lv-oeCon) were
synthesized based on a 3rd lentivirus generation system. The
Lv-oeDUSP1 was synthesized by inserting DUSP1 cDNA into the plasmid
backbone of pHBLV-CMV-MCS-IRES-Puro (Chemicalbook; Beijing Xilin
Buke Network Technology Co., Ltd.). To construct Lv-oeCon the
translation starting site codon (ATG) of Lv-oeDUSP1, was replaced
by AAG to abolish translation. The lentivirus transduction was
performed as aforementioned.
Construction of DUSP1 promoter
luciferase reporter vectors
The human DUSP1 promoter sequence was obtained from
GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The promoter
(−1,500 to +20 bp, where the transcription start site was defined
as +1) of human DUSP1 was amplified from the genomic DNA of CAL-27
cells using the following thermocycling conditions: Initial
denaturation at 98°C for 30 sec; 35 cycles of 98°C for 10 sec, 50°C
for 10 sec, 72°C for 40 sec; and a final extension step of 72°C for
2 min. The PCR primers used for amplifying the DUSP1 promoter are
shown in Table SII. After
amplification, the promoter fragments were cut by SacI and
MluI restriction enzymes. Then, the fragment was re-cloned
into the pGLuc-Dura plasmid (Beyotime Institute of Biotechnology)
between the SacI and MluI sites and the reconstructed
plasmid was termed pGLuc-DUSP1. The pGLuc-DUSP1 was confirmed by
DNA sequencing at Sangon Biotech Co., Ltd.
Identifying the putative binding sites
of Egr-1 in the DUSP1 promoter
The putative binding sites of Egr-1 in the DUSP1
promoter were predicated in JASPAR2022 software
(https://jaspar.genereg.net/analysis).
The explanation of the basis of the calculations and the mean of
the indexes in the results outputted by the software are described
in detail in a paper published by the manufacturer of the software
(37). Briefly, Egr-1 was submitted
to the search box and then human Egr-1 was added to the cart. The
sequence of the DUSP1 promoter was inserted into the scanning box
and the relative profile score threshold was set as 90%. The
putative binding site of Egr-1 in the DUSP1 promoter was then
presented.
Site-directed mutagenesis
Site-directed mutagenesis was performed according to
a previous study (38). Briefly,
Site A (−68/-81) and Site B (−1363/-1376) of pGLuc-DUSP1 were
mutated with a Q5® High-fidelity DNA Polymerase (New
England BioLabs, Inc.) and pGLuc-DUSP1 as the template, with the
following thermocycling conditions: Initial denaturation at 98°C
for 30 sec; 35 cycles of 98°C for 10 sec, 5°C for 10 sec, 72°C for
40 sec and a final extension at 72°C for 2 min. The PCR primers
used for mutating Site A (MTA) and Site B (MTB) are listed in
Table SIII. After the template was
digested with a DpnI restriction enzyme, the products were
transformed into TOP10 cells (Beyotime Institute of Biotechnology).
The mutant pGLuc-DUSP1 plasmids were isolated using a Plasmid Mini
Preparation Kit (Beyotime Institute of Biotechnology). The plasmid
was confirmed by Sanger sequencing technique performed by Sangon
Biotech, Co., Ltd.
Luciferase assay
The pGLuc-DUSP1 promoter reporter plasmid (1 µg) was
co-transfected with pGMLR-TK luciferase reporter (1 µg) into CAL-27
cells using Lipofectamine™ 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). At 24-h post transduction, the
cell culture supernatants were collected. The Gaussia
luciferase activity was measured by a Promega GloMax luminometer
(Promega Corporation) using a Gaussia-Dura luciferase kit following
the manufacturer's instructions (Thermo Fisher Scientific, Inc.).
Renilla luciferase activity was measured by the Promega
GloMax luminometer (Promega Corporation) using a
RenillaLumi™ Luciferase Assay Kit following the
manufacturer's instructions (Beyotime Institute of Biotechnology).
The experiment was repeated for three times. The promoter activity
was calculated by normalized Gaussia luciferase activity
with Renilla luciferase activity.
Transwell migration and invasion
assays
Transwell migration assay
Cells were seeded into the upper chambers of
Transwell plates at a density of 1×105 cells/well in
FBS-free DMEM, and DMEM with 10% FBS was added to the lower
chambers. The cells were incubated for 10 h in an incubator
containing 5% CO2 at 37°C. Subsequently, the cells on
the top surface of the semipermeable membrane were removed, while
the cells attached to the bottom surface of the membrane were
stained with crystal violet for 5 min at 20°C. Images of the
stained cells were captured under a light microscope (Olympus
Corporation). The experiment was repeated for three times and cells
of three randomly selected visual fields were counted manually and
averaged.
Transwell invasion assay
Transwell invasion assay shared same procedures as
Transwell migration assay, except that each upper chamber of the
Transwell plate was pre-coated with 20 µg Matrigel ECL
(MilliporeSigma; Merck KGaA) for 4 h at 37°C, and cells were seeded
at a density of 2×105 cells/well.
Western blotting
Cell lysates were extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology). The concentration of the
protein was determined using a BCA kit (Beyotime Institute of
Biotechnology). Then, 30 µg of protein in each group was loaded per
lane and separated on 10% gels using SDS-PAGE and transferred onto
a nitrocellulose (NC) membrane. After blocking using 5% fat-free
milk (Beyotime Institute of Biotechnology) for 30 min at 20°C, the
NC membrane was incubated with β-actin (1:1,000; cat. no. sc-8432;
Santa Cruz Biotechnology, Inc.) or Egr-1 (1:1,000; cat. no.
ab194357; Abcam) antibodies. The membranes were then incubated with
a Cy3-labelled goat anti-rabbit secondary antibody (1:10,000; cat.
no. A0516; Beyotime Institute of Biotechnology) or a Cy3-labelled
goat anti-mouse secondary antibody (1:10,000; cat. no. A0521;
Beyotime Institute of Biotechnology) for 1 h at 20°C. All
antibodies were diluted in TBST (containing 0.1% Tween 20).
Finally, the membrane was visualized in a Typhoon infrared scanning
imager (Cytiva). The experiment was repeated three times. The
densitometry was performed using ImageJ software (version 1.8.0;
National Institutes of Health).
Chromatin immunoprecipitation
(ChIP)
The ChIP assay was conducted using a ChIP assay kit
(Beyotime Institute of Biotechnology) following the manufacturer's
instructions. Briefly, CAL-27 cells were cultured in a 10-cm dish.
The chromatin in the cells was cross-linked by being incubated in
1% formaldehyde (Beyotime Institute of Biotechnology) at 20°C for
10 min and then in 1% glycine (Beyotime Institute of Biotechnology)
for 30 min. The cell were lysed in 0.1% SDS and then 500 µg cell
lysates were incubated with a Micrococcal nuclease (cat. no.
9013-53-0; Merck KGaA) for 15 min at 37°C to ensure the chromatin
in the cell lysate was digested into fragments of 200–1,000 bp. The
fragments were then incubated with 5 µg Egr-1 antibody (cat. no.
4154; Cell Signaling Technology, Inc.) at 4°C for 12 h and pulled
down using Protein A/G magnetic beads (cat. no. 26162; Thermo
Fisher Scientific, Inc.). The chromatin fragments were also
incubated with 5 µg anti-FLAG M2 antibody antibody (cat. no. 14793;
Cell Signaling Technology, Inc.), which served as the negative
control. After being washed in Immune Complex Wash Buffer (cat. no.
P2080S; Beyotime Institute of Biotechnology), the cross-linked
chromatin was released by incubation in 0.2 µM NaCl at 65°C for 6
h. The presence of DUSP1 promoter fragments containing Site A were
then determined by PCR amplification using a Taq DNA Polymerase
(cat. no. D7209; Beyotime Institute of Biotechnology) under the
following thermocycling conditions: Initial denaturation at 96°C
for 5 min; 35 cycles of 96°C for 15 sec, 55°C for 10 sec, 72°C for
30 sec; and a final extension step of 72°C for 5 min. The primers
for amplifying the fragments containing Site A were as follows:
Forward, 5′-ATTCAACGCAAAAAC-3′ and reverse, 5′-AACATAAACATTGCGC-3′.
The PCR products were analyzed using a 1.0% agarose gel
electrophoresis with 0.01% Gel-Green reagent (cat. no. D0143;
Beyotime Institute of Biotechnology) and then visualized using an
ultraviolet imager (Tanon Science and Technology Co., Ltd.).
Statistical analysis
Data analysis was performed using GraphPad Prism 5.0
(Dotmatics) for Windows. For The Cancer Genome Atlas (TCGA) data
comparing Egr-1 expression between normal tissues and HNSCC
tissues, data were compared between two groups using an unpaired
Student's t-test. For the Gene Expression Omnibus (GEO) dataset
comparing Egr-1 expression between normal tissues and TSCC tissues,
the data were analyzed using a paired Student's t-test. Data are
presented as the mean ± standard deviation. Differences between
groups with one variable were analyzed using the one-way ANOVA
followed by Tukey's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
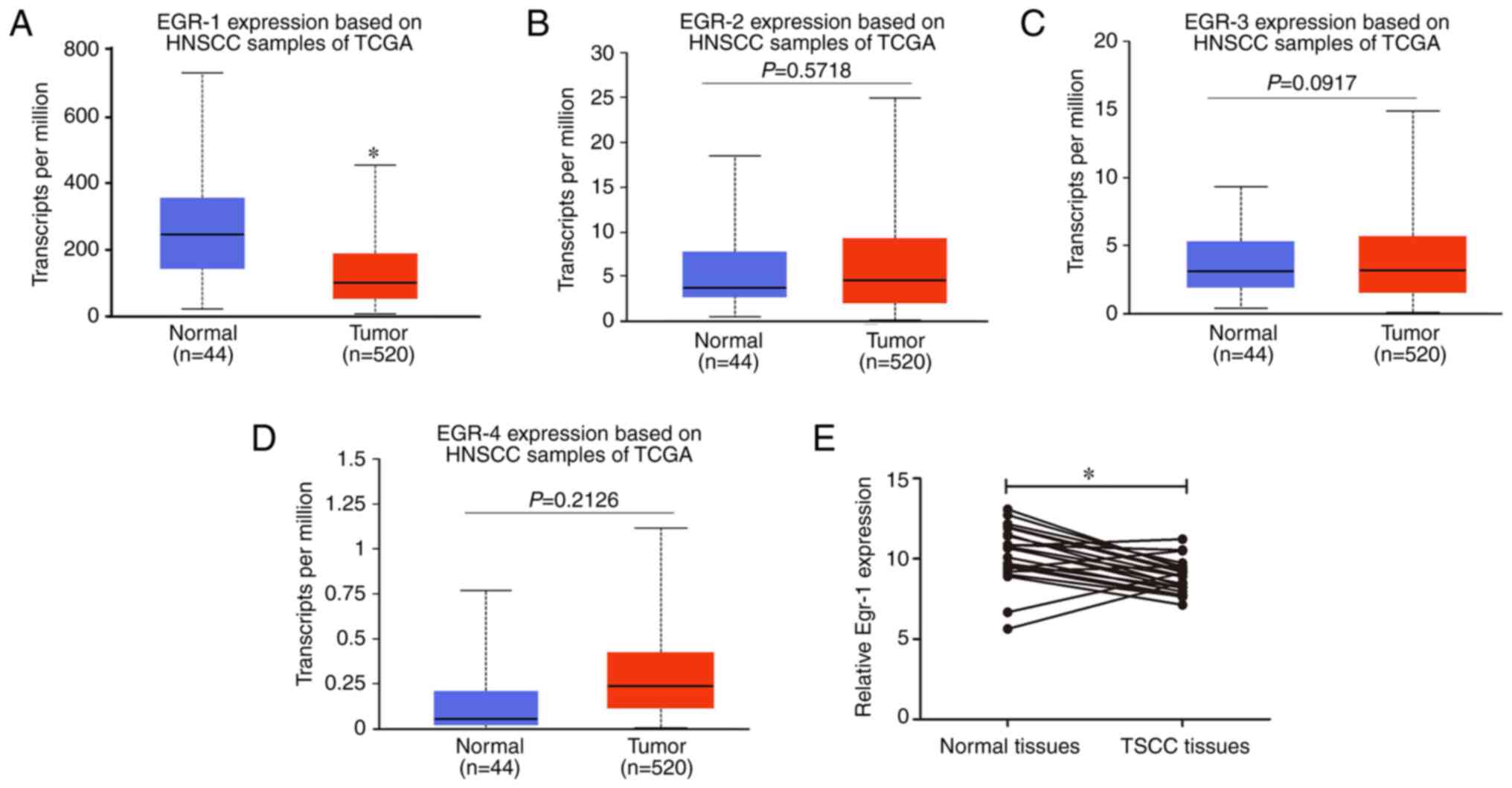
Egr-1 expression is decreased in TSCC
tumor tissues
The expression levels of members of the EGR family
in HNSCC were analyzed using the UALCAN database. The expression of
Egr-1 in HNSCC tissues was significantly lower compared with normal
tissues (Fig. 1A); however, there
was no statistically significant difference in Egr-2, Egr-3 and
Egr-4 mRNA expression between HNSCC tissues and normal tissues
(Fig. 1B-D). Furthermore, Egr-1
expression in TSCC tissues and adjacent normal tissues from a GEO
dataset (dataset no. GSE31056) (35) were compared, which demonstrated that
Egr-1 expression in TSCC tissues was significantly decreased
compared with normal tissues (Fig.
1E).
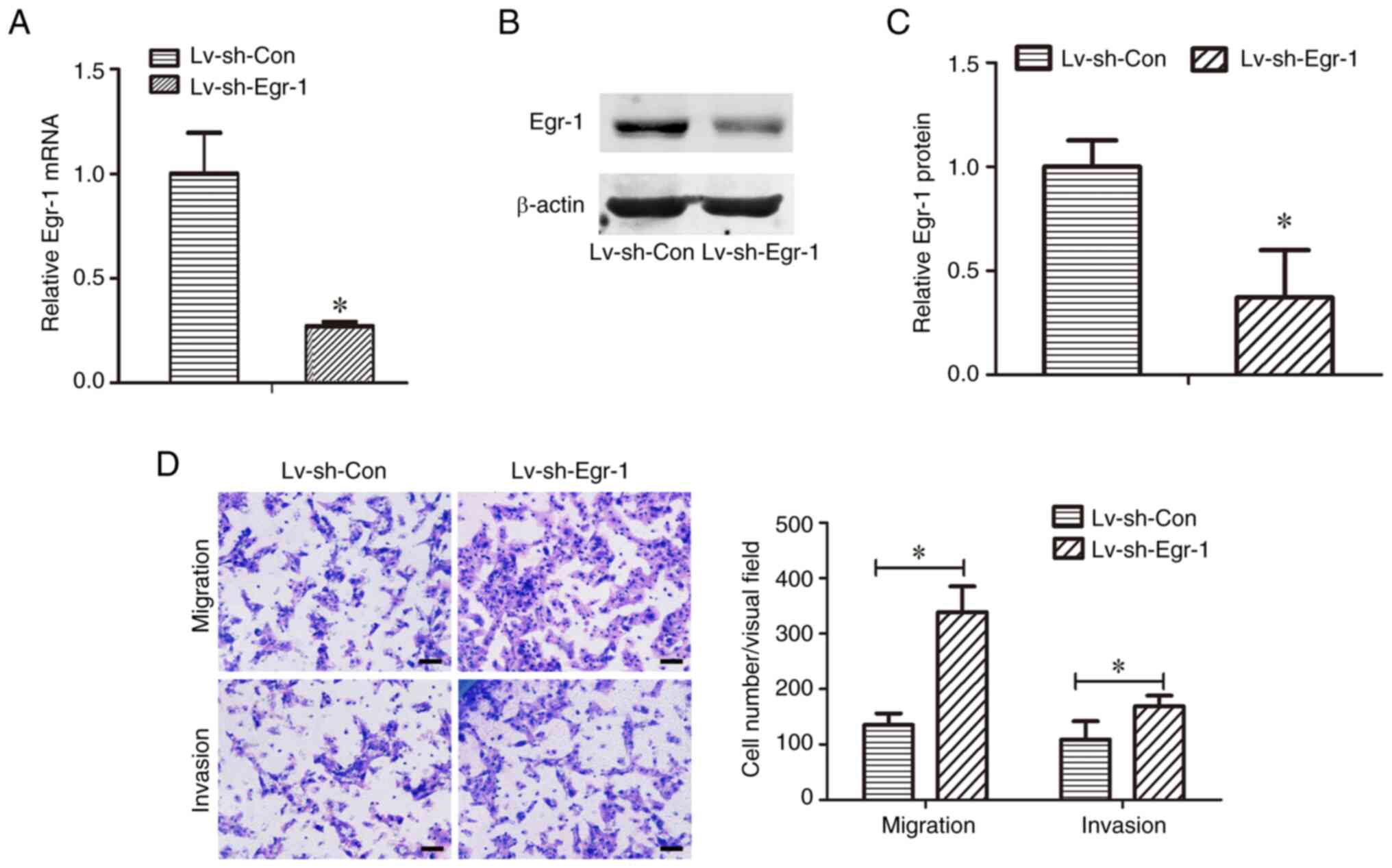
Knockdown of Egr-1 increases the
migration and invasion of CAL-27 cells
To determine the role of Egr-1 in TSCC metastasis,
the effects of Egr-1 knockdown on TSCC cell migration and invasion
were evaluated. First, it was demonstrated that Egr-1 mRNA levels
were significantly decreased in CAL-27 cells transduced with
Lv-shEgr-1 compared with Lv-shCon (Fig.
2A). The knockdown of Egr-1 was also confirmed using western
blotting, where the relative Erg-1 protein levels were
significantly decreased in Lv-shEgr-1 cells compared with the
Lv-shCon cells (Fig. 2B and C).
Transwell assays were used to evaluate the migration
and invasion capacities of the transduced cells. The Lv-shEgr-1
group demonstrated a significantly higher migration and invasion
capacity compared with the Lv-shCon group, indicating that the
knockdown of Egr-1 increased the migration and invasion capacities
of CAL-27 cells (Fig. 2D).
Egr-1 expression is correlated with
DUSP1 expression in TSCC
To elucidate how Egr-1 regulates TSCC cell migration
and invasion, Egr-1-associated genes were analyzed. According to
the UALCAN database, 242 genes were correlated with Egr-1 in HNSCC.
Among the correlated genes, FOS (Fig.
3A), DUSP1 (Fig. 3B), FOSB
(Fig. 3C) and ZFP36 (Fig. 3D) had the highest Pearson
correlation coefficients. Therefore, the expression of these genes
in cells with Egr-1 knockdown were further analyzed.
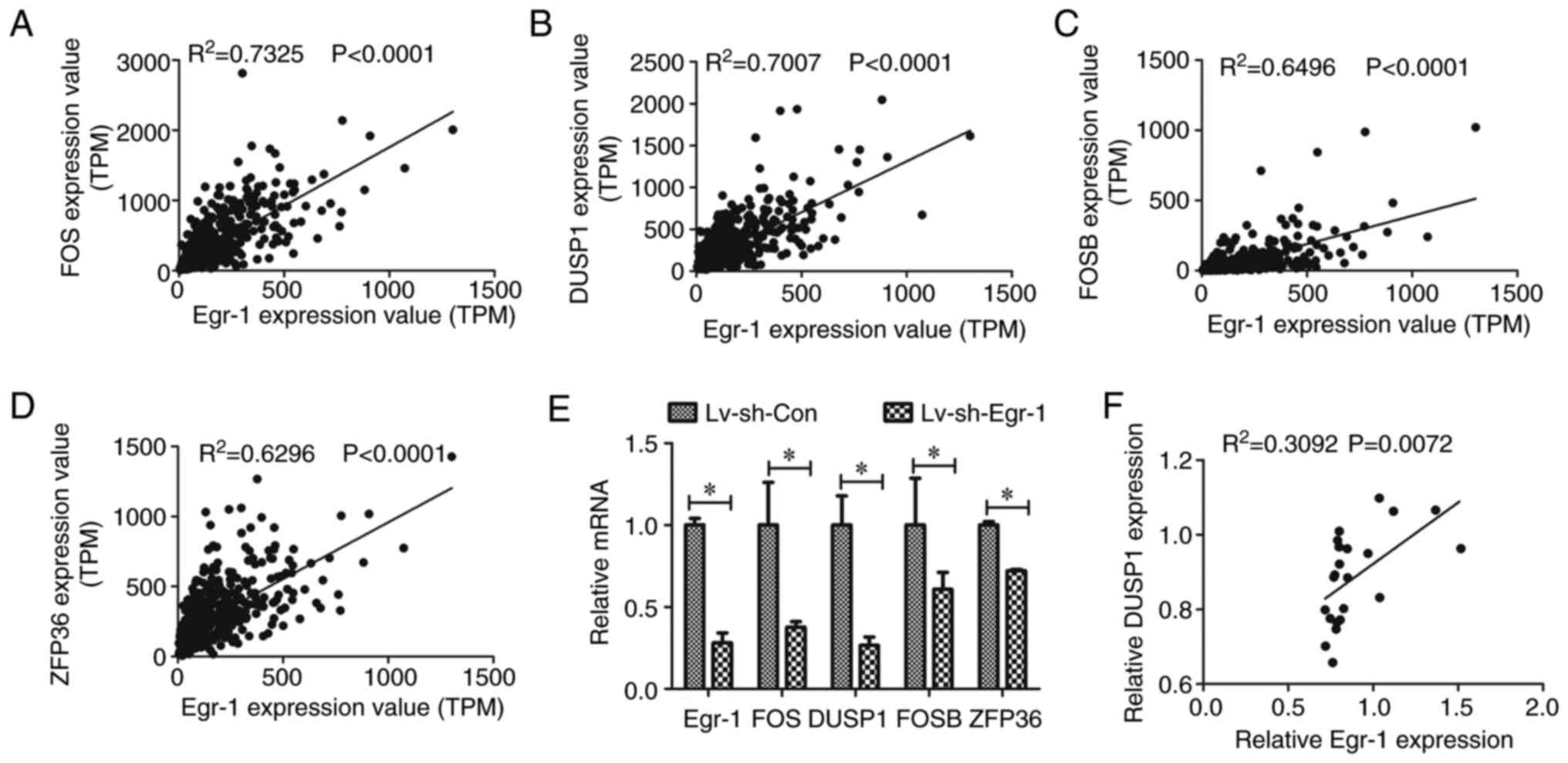 | Figure 3.Egr-1 expression is correlated with
DUSP1 expression in TSCC. Correlation analysis between Egr-1
expression and (A) FOS, (B) DUSP1, (C) FOSB and (D) ZFP36 based on
the University of Alabama at Birmingham Cancer Data Analysis Portal
database. (E) Egr-1, FOS, DUSP1, FOSB and ZFP36 mRNA expression in
CAL-27 cells with and without Egr-1 knockdown, *P<0.05. (F)
Correlation analysis between Egr-1 expression and DUSP1 based on
the Gene Expression Omnibus dataset, GSE31056. TPM, transcripts per
million; Lv, lentivirus; sh, short hairpin; DUSP1, dual-specificity
protein phosphatase 1; Egr-1, early growth response 1. |
The mRNA levels of FOS, DUSP1, FOSB and ZFP36 were
all significantly reduced in the sh-Egr-1 group compared with the
respective control, of which DUSP1 was the most reduced (Fig. 3E). Furthermore, the association
between Egr-1 and DUSP1 in TSCC was analyzed using the GEO dataset,
GSE31056, which demonstrated that Egr-1 was correlated with DUSP1
in TSCC tissues (Fig. 3F).
Egr-1-induced CAL-27 cell migration
and invasion are reversed by DUSP1 overexpression
A lentivirus, Lv-oeDUSP1, was constructed for
overexpressing DUSP1 in TSCC cells. Cells transduced with
Lv-oeDUSP1 demonstrated higher mRNA expression of DUSP1 compared
with cells infected with Lv-oeCon (Fig.
4A). In addition, Lv-oeDUSP1 CAL-27 cells underwent Egr-1
knockdown (Fig. 4B) and their
migration and invasion capacities were evaluated. Infection with
Lv-shEgr-1 significantly increased the migration and invasion cell
numbers of CAL-27 cells compared with the sh-Con; however, this
increase was reversed by infection with Lv-oeDUSP1, suggesting that
Egr-1 regulates CAL-27 cell migration and invasion through DUSP1
(Fig. 4C and D).
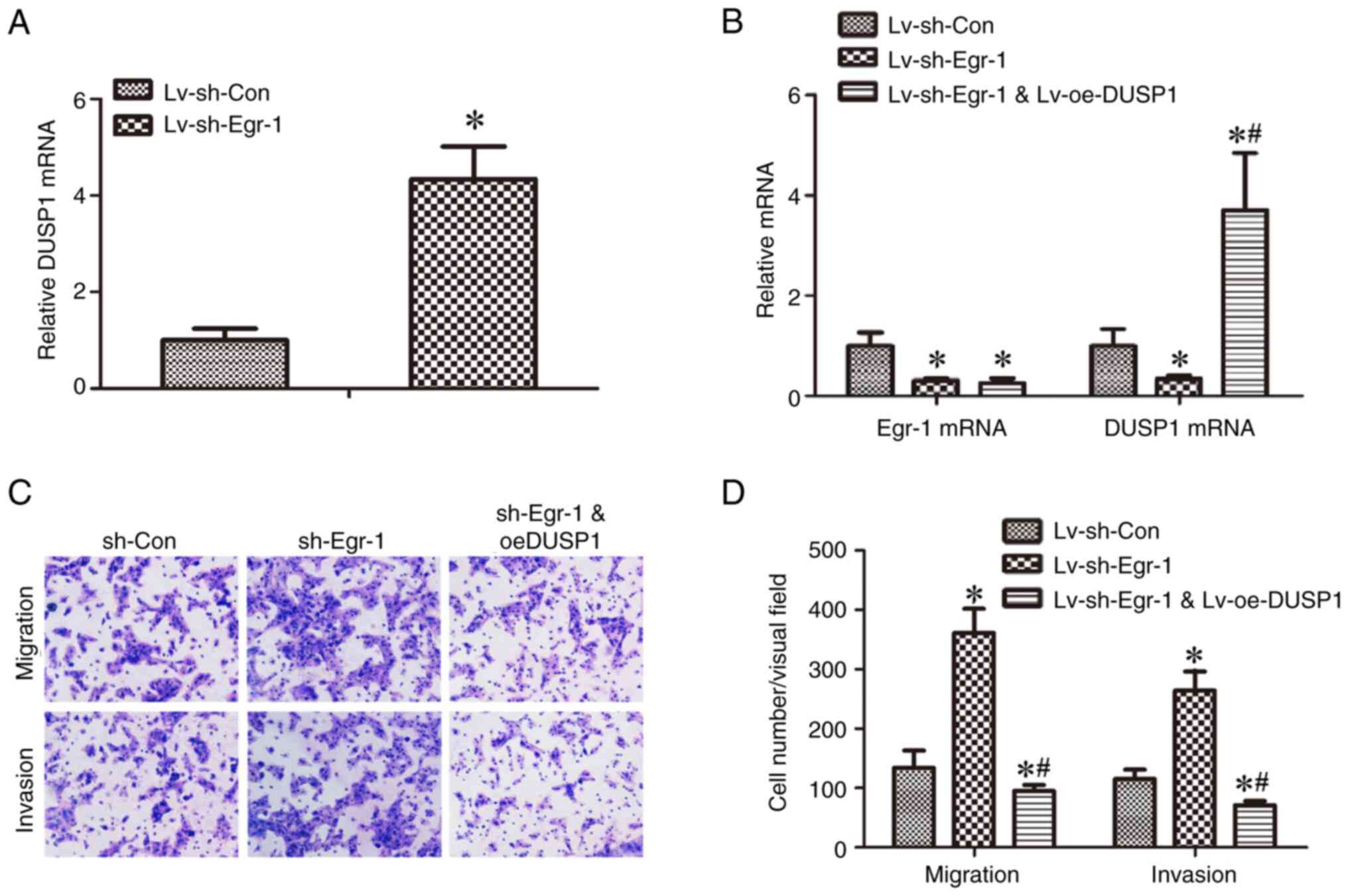 | Figure 4.Egr-1-induced decreased CAL-27 cell
migration and invasion is reversed by DUSP1 overexpression. (A)
DUSP1 mRNA expression in CAL-27 transduced with Lv-oeCon and
Lv-oeDUSP1 was detected by RT-qPCR, *P<0.05 vs. Lv-oeCon. (B)
Egr-1 and DUSP1 mRNA expression in CAL-27 cells with Lv-shCon,
Lv-shEgr-1 or Lv-shCon + Lv-oeDUSP1 was detected by RT-qPCR.
*P<0.05 vs. sh-Con, #P<0.05 vs. sh-Egr-1. (C)
CAL-27 cells transduced with Lv-shCon, Lv-shEgr-1 or Lv-shCon +
Lv-oeDUSP1 were assessed by Transwell migration and invasion
assays. (D) Cells in three randomly selected visual fields were
counted. *P<0.05 vs. sh-Con, #P<0.05 vs. sh-Egr-1.
RT-qPCR, reverse transcription quantitative-PCR; sh, short hairpin;
Lv, lentivirus; oe, overexpression; DUSP1, dual-specificity protein
phosphatase 1; Egr-1, early growth response 1. |
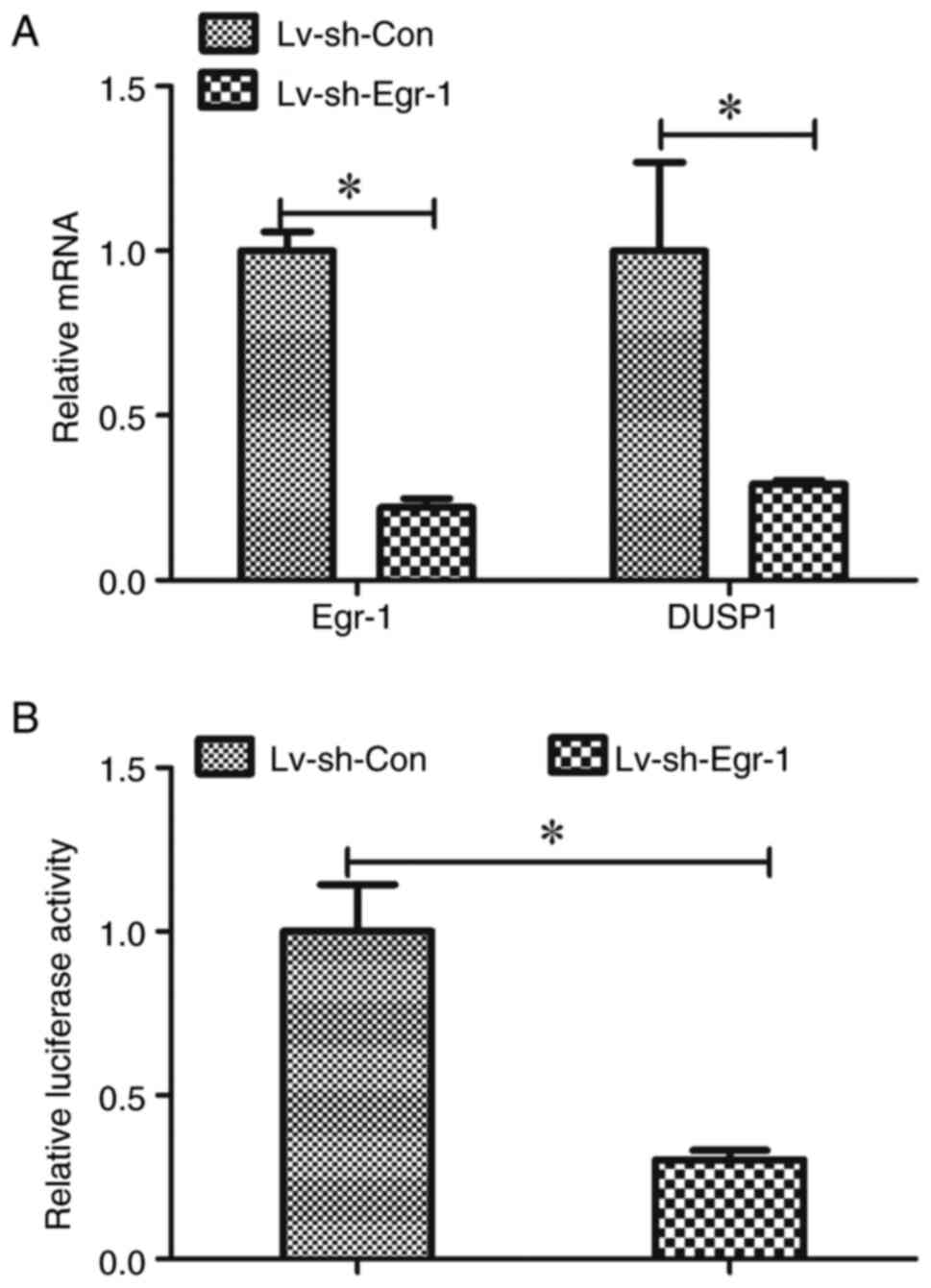
Knockdown of Egr-1 decreases DUSP1
expression at the transcriptional level
Since it was demonstrated that Egr-1 expression was
correlated with DUSP1 expression in TSCC, it was next determined
whether the expression of DUSP1 is regulated by Egr-1. DUSP1 mRNA
levels were measured in Egr-1 knockdown CAL-27 cells, and it was
demonstrated that these cells had significantly lower DUSP1 mRNA
expression compared with the control cells (Fig. 5A). Considering that Egr-1 is a
transcription factor, it was also assessed whether Egr-1 regulates
DUSP1 at the transcriptional level. A DUSP1 promoter-reporter
vector, pGLuc-DUSP1, was constructed and transduced into CAL-27
cells with or without Egr-1 knockdown and its activity was
measured. The luciferase activity of pGLuc-DUSP1 in the cells with
Egr-1 knockdown was significantly lower than the cells without
Egr-1 knockdown (Fig. 5B).
Identifying the site in the DUSP1
promoter through which Egr-1 regulates DUSP1
The Egr-1 binding site in the DUSP1 promoter was
predicted using JASPAR2022 software. A total of two
putative sites were predicted (Table
I), which were termed Site A and Site B. To determine which
site may be responsible for Egr-1 regulation of DUSP1
transcription, Site A and Site B in pGLuc-DUSP1 were mutated and
termed pGLuc-DUSP1-MTA and pGLuc-DUSP1-MTB, respectively (Fig. 6A). pGLuc-DUSP1, pGLuc-DUSP1-MTA and
PGLuc-DUSP1-MTB were transduced into CAL-27 cells with or without
Egr-1 knockdown and the promoter activity was measured.
pGLuc-DUSP1-MTA demonstrated significantly lower relative
luciferase activity compared with pGLuc-DUSP1 and pGLuc-DUSP1-MTB
in CAL-27 cells, while the activity of pGLuc-DUSP1-MTB was notably
increased compared with pGLuc-DUSP1 (Fig. 6B). In addition, knockdown of Egr-1
significantly decreased the relative luciferase activity of
pGLuc-DUSP1 (Fig. 5B) and
pGLuc-DUSP1-MTB (Fig. 6D) compared
with the relative controls; a slight decrease in the relative
luciferase activity was also demonstrated in pGLuc-DUSP1-MTA
(Fig. 6C), but this was not
statistically significant. These results indicate that Egr-1
regulates DUSP1 transcription through the putative binding site
A.
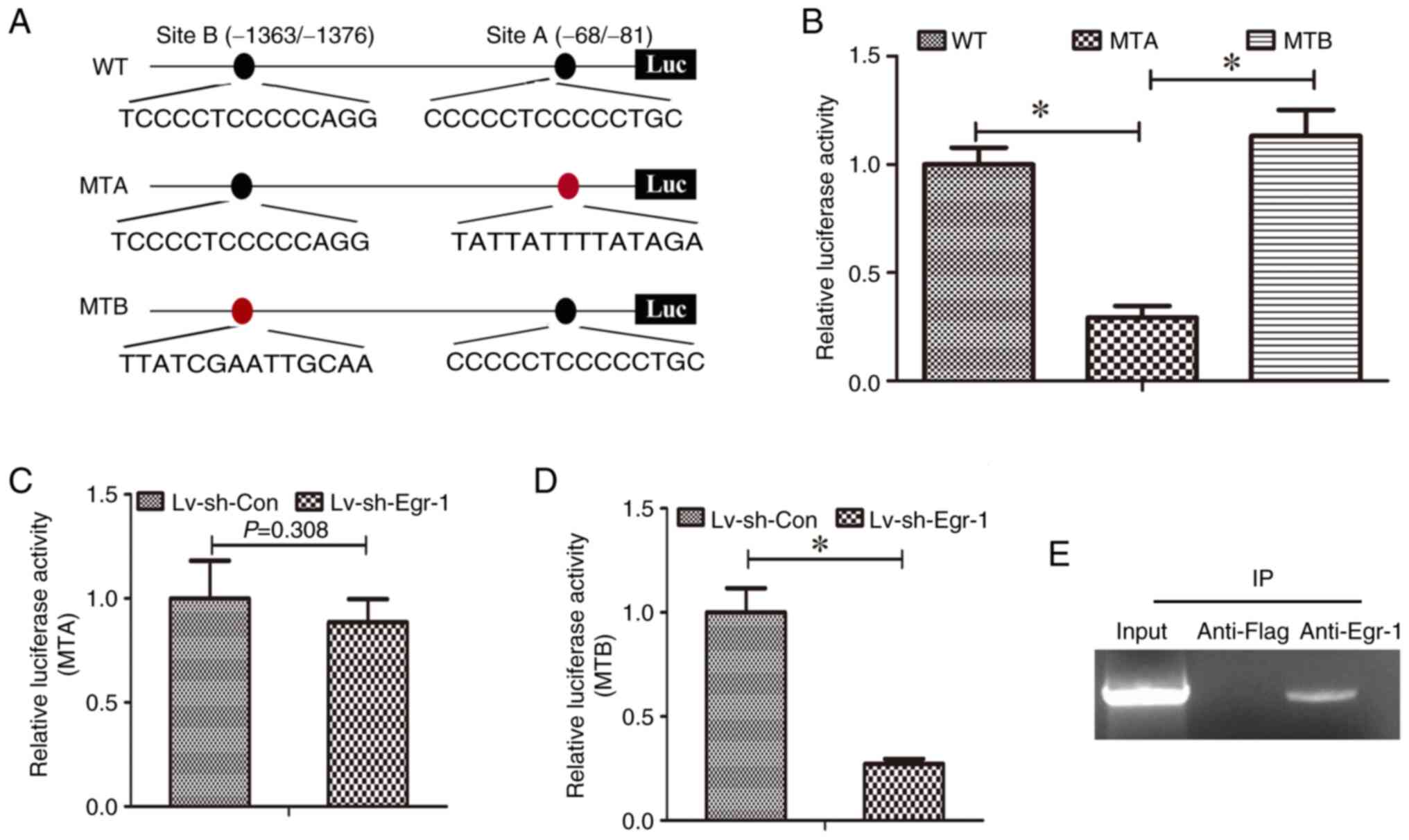 | Figure 6.Identifying the Erg-1 binding site on
the DUSP1 promoter. (A) The schematic of the DUSP1 promoter in
pGLuc-DUSP1, pGLuc-DUSP1-MTA and pGLuc-DUSP1-MTB. The black oval
represents the WT site and the red oval represents the mutant site.
(B) Relative luciferase activity of pGLuc-DUSP1, pGLuc-DUSP1-MTA
and pGLuc-DUSP1-MTB. MTA, pGLuc-DUSP1-MTA. MTB, pGLuc-DUSP1-MTB.
Relative luciferase activity of pGLuc-DUSP1-MTA (C) and
pGLuc-DUSP1-MTB (D) in CAL-27 cells with and without Egr-1
knockdown, *P<0.05. (E) Chromatin immunoprecipitation assay was
used to verify whether Egr-1 bound to the Site A in DUSP1 promoter.
WT, wild type; MTA, mutation Site A; MTB, mutation Site B; Lv,
lentivirus; sh, short hairpin; Luc, luciferase; Con, control;
DUSP1, dual-specificity protein phosphatase 1; Egr-1, early growth
response 1. |
 | Table I.Putative binding sites of Egr-1 in
DUSP1 promoter predicted using the JASPAR2022
software. |
Table I.
Putative binding sites of Egr-1 in
DUSP1 promoter predicted using the JASPAR2022
software.
| Matrix
IDa | Nameb | Scorec | Relative
scored | Starte | Endf |
Sequenceg |
|---|
| MA0162.2 | Egr-1 | 15.318353 |
0.9442267584698091 | −81 | −68 | CCCCCTCCCCCTGC |
| MA0162.2 | Egr-1 | 12.037096 |
0.9090872474628051 | −1,376 | −1,363 | TCCCCTCCCCCAGG |
Furthermore, the results of the ChIP assay
demonstrated that anti-Egr-1 antibodies immunoprecipitated
chromatin fragments containing Site A (Fig. 6E), which was not demonstrated for
the anti-Flag antibody control, suggesting that Egr-1 binds to Site
A. Therefore, it can be suggested that Egr-1 may regulate DUSP1 via
Site A.
Discussion
In the present study, the expression of Egr-1 in
TSCC was analyzed. It was demonstrated that Egr-1 served a role in
the downregulation of TSCC cell migration and invasion.
Furthermore, it was also demonstrated that Egr-1 affected TSCC cell
migration and invasion by regulating DUSP1 at the transcription
level.
TSCC is one of the most common types of HNSCC.
Metastasis is one of the main causes of the mortality of patients
with TSCC, and it is therefore necessary to elucidate the mechanism
of TSCC metastasis to identify possible therapeutic targets and
biomarkers (4). Egr-1 is a
ubiquitously expressed transcription factor and is dysregulated in
various tumor types, including colon cancer (39), esophageal squamous cell carcinoma
(ESCC) (40), breast cancer
(15) and pituitary tumors
(41); therefore, Egr-1 has been
suggested as a potential biomarker for tumor prognosis and a
candidate target for cancer therapy. In the present study it was
demonstrated that Egr-1 expression was downregulated in HNSCC and
TSCC and the knockdown of Egr-1 increased TSCC cell migration and
invasion, indicating a potential role of Egr-1 in TSCC
metastasis.
According to previous studies, Egr-1 has differing
effects in different tumor types. It has been reported that
knockdown of Egr-1 in ESCC significantly enhances cell migration
and invasion (40), whereas Egr-1
inhibits cell proliferation, migration and invasion in colon cancer
(8), suggesting that Egr-1 is a
tumor suppressor in ESCC and colon cancer, which supports the
results of the present study. However, in hepatocellular carcinoma
cells, Egr-1 enhances radioresistance and chemoresistance (42,43),
and Egr-1 is positively related to tumor size, lymph node
metastasis, tumor stage and poor survival in gastric cancer
(44), indicating that Egr-1 may
exert an oncogenic role in these tumor types. The different roles
of Egr-1 in tumor progression may be due to the histological
diversity, but this opinion needs further research.
Egr-1 participates in tumor progression by
transcriptionally regulating various target genes. Egr-1 regulates
LC3 expression through binding to the promoter of LC3, promotes
hypoxia-induced autophagy, and thus induces radioresistance in
hepatocellular carcinoma cells (42). In addition, Egr1 inhibits colon
cancer cell proliferation, migration and invasion by regulating
CDKL1 at the transcriptional level (8). In the present study, to assess the
mechanism by which Egr-1 regulates TSCC progression,
Egr-1-associated genes were analyzed using bioinformatics methods.
From this, four genes, DUSP1, FOS, FOSB and ZFP36, were selected
for further screening. It was then demonstrated that the mRNA
levels of these four genes were decreased following Egr-1
knockdown, of which DUSP1 was the most downregulated. It was
further demonstrated that overexpression of DUSP1 reversed
knockdown of Egr-1-induced TSCC migration and invasion, indicating
that Egr-1 regulates TSCC migration and invasion through DUSP1.
Furthermore, it was demonstrated that the DUSP1 binding site of
Egr-1 was demonstrated to be within Site A of the promotor.
Therefore, the present study demonstrated that DUSP1 may be a
target of Egr-1 and suggested the mechanism through which Egr-1 is
involved in TSCC metastasis. To regulate target gene expression in
general, Egr-1 interacts with numerous transcription and regulatory
factors, such as nuclear factor-κB, YAP-1 and Snail (45–48).
However, the roles of other factors in the regulation of DUSP1 by
Egr-1 requires further study.
In previous studies, DUSP1 has also been reported to
be regulated by other transcription factors. For instance,
LINC00702 was reported to promote DUSP1 expression by recruiting
JunD to the DUSP1 promoter (30).
In addition, AP-1 has been predicted to bind to the promoter region
of DUSP1 and regulate its expression (48). Furthermore, FOXM1 was reported to
directly activate the DUSP1 promoter in macrophages (49). Further studies are required to
assess whether DUSP1 is regulated by the aforementioned
transcription factors in TSCC, or whether these transcription
factors have synergistic effects with Egr-1 in regulating
DUSP1.
In the present study, other genes such as FOS, FOSB
and ZFP36 were demonstrated to be associated with Egr-1. The FOS
gene encodes a transcription factor that co-localise with members
of the Jun family, including c-Jun, JunB and JunD, and regulate
cell proliferation, tumor invasion, distant metastasis and
angiogenesis (50). FOSB is
upregulated in tumors and knockdown of FOSB decreases migration
ability (51). ZFP36 inhibits
prostate cancer progression by targeting CDK6 and oxidative stress
(52). In addition, ZFP36 interacts
with MCM3AP-AS1 to regulate the proliferation, apoptosis, migration
and invasion of breast cancer cells (53). Therefore, FOS, FOSB and ZFP36 are
also noteable regulators in tumor progression. However, whether
these genes are associated with the role of Egr-1 in TSCC
progression remains unknown, and this is a limitation of this
study. There are other limitations to the present study; for
example, the expression of Egr-1 in OSCC was analyzed based on data
from a public database, whereas it would be more convencing if the
expression of Egr-1 was detected in OSCC clinical tissues.
Furthermore, the function of Egr-1 in OSCC progression was analyzed
in vitro, whereas it will be better if there were also in
vivo experiments.
In conclusion, the results of the present study
demonstrated that Egr-1 was upregulated in TSCC and was associated
with tumor cell migration and invasion. Moreover, it was
demonstrated that DUSP1 may be a target gene of Egr-1 and DUSP1
participated in Egr-1-associated cell migration and invasion. The
findings of the present study suggested that Egr-1 and DUSP1 are
candidate biomarkers and therapeutic targets for the treatment of
TSCC metastasis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81602374), the Natural Science
Foundation of Shandong Provence (grant no. ZR2021MH176), China
Postdoctoral Science Foundation (grant no. 2021M701538) and the
Natural Science Foundation of Liaocheng People's Hospital (grant
no. LYQN201903).
Availability of data and materials
The data generated in the present study may be found
in the Figshare repository at the following URL: https://figshare.com/search?q=10.6084%2Fm9.figshare.21801127.
In addition, the data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZM and NG designed the study. LZ and JL performed
the cell culture, Transwell assay and bioinformatics analysis. YS
and ML performed the lentivirus infection, RT-qPCR and luciferase
assay. ZM drafted the manuscript. ZM and NG confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miller KD, Nogueira L, Devasia T, Mariotto
AB, Yabroff KR, Jemal A, Kramer J and Siegel RL: Cancer treatment
and survivorship statistics, 2022. CA Cancer J Clin. 72:409–436.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao M, Shi E, Wang H, Mao L, Wu Q, Li X,
Liang Y, Yang X, Wang Y and Li C: Personalized targeted therapeutic
strategies against oral squamous cell carcinoma. An evidence-based
review of literature. Int J Nanomedicine. 17:4293–4306. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khachigian LM: The MEK-ERK-Egr-1 axis and
its regulation in cardiovascular disease. Vascul Pharmacol.
153:1072322023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thomes PG and Donohue TM: Role of early
growth response-1 in the development of alcohol-induced steatosis.
Curr Mol Pharmacol. 10:179–185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Truong V, Jain A, Anand-Srivastava MB and
Srivastava AK: Angiotensin II-induced histone deacetylase 5
phosphorylation, nuclear export, and Egr-1 expression are mediated
by Akt pathway in A10 vascular smooth muscle cells. Am J Physiol
Heart Circ Physiol. 320:H1543–H1554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shao S, Ju M, Lei J, Lu X, Li H, Wang D
and Xia C: Egr-1 inhibits colon cancer cell proliferation,
migration and invasion via regulating CDKL1 at the transcriptional
level. Oncol Rep. 46:1692021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sheng J, Liu D, Kang X, Chen Y, Jiang K
and Zheng W: Egr-1 increases angiogenesis in cartilage via binding
Netrin-1 receptor DCC promoter. J Orthop Surg Res. 13:1252018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qi F, Wang X, Zhao S, Wang C, Sun R, Wang
H, Du P, Wang J, Wang X and Jiang G: miR-let-7c-3p targeting on
Egr-1 contributes to the committed differentiation of leukemia
cells into monocyte/macrophages. Oncol Lett. 24:2732022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen W, Zhao S, Xing J, Yu W, Rao T, Zhou
X, Ruan Y, Li S, Xia Y, Song T, et al: BMAL1 inhibits renal
fibrosis and renal interstitial inflammation by targeting the
ERK1/2/ELK-1/Egr-1 axis. Int Immunopharmacol. 125:1111402023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu J, Tao W, Bu D, Zhao Y, Zhang T, Chong
D, Xue B, Xing Z and Li C: Egr-1 transcriptionally activates
protein phosphatase PTP1B to facilitate hyperinsulinemia-induced
insulin resistance in the liver in type 2 diabetes. FEBS Lett.
593:3054–3063. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee SM, Park MS, Park SY, Choi YD, Chung
JO, Kim DH, Jung YD and Kim HS: Primary bile acid activates Egr-1
expression through the MAPK signaling pathway in gastric cancer.
Mol Med Rep. 25:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oh S, Kim H, Nam K and Shin I: Egr-1 is
required for neu/HER2-induced mammary tumors. Cell Signal.
45:102–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei LL, Wu XJ, Gong CC and Pei DS: Egr-1
suppresses breast cancer cells proliferation by arresting cell
cycle progression via down-regulating CyclinDs. Int J Clin Exp
Pathol. 10:10212–10222. 2017.PubMed/NCBI
|
|
16
|
Chuang KC, Chen FW, Tsai MH and Shieh JJ:
EGR-1 plays a protective role in AMPK inhibitor compound C-induced
apoptosis through ROS-induced ERK activation in skin cancer cells.
Oncol Lett. 21:3042021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Shepherd EG and Nelin LD: MAPK
phosphatases-regulating the immune response. Nat Rev Immunol.
7:202–212. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang TT, Wang Y, Zhang XW, Yang KY, Miao
XQ and Zhao GH: MiR-200c-3p regulates DUSP1/MAPK pathway in the
nonalcoholic fatty liver after laparoscopic sleeve gastrectomy.
Front Endocrinol (Lausanne). 13:7924392022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou X, Li L, Chen S, Ge C, Shen M and Fu
Z: MKP-1 overexpression reduces postischemic myocardial damage
through attenuation of ER stress and mitochondrial damage. Oxid Med
Cell Longev. 2021:89055782021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Jiang Y, Li J, Wang Y, Tian Y, Guo
Q and Cheng Z: DUSP1 promotes microglial polarization toward M2
phenotype in the medial prefrontal cortex of neuropathic pain rats
via inhibition of MAPK pathway. ACS Chem Neurosci. 12:966–978.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wilczek MP, Armstrong FJ, Geohegan RP,
Mayberry CL, DuShane JK, King BL and Maginnis MS: The MAPK/ERK
pathway and the role of DUSP1 in JCPyV infection of primary
astrocytes. Viruses. 13:18342021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang D and Jiang Y: MKP1 reduces
neuroinflammation via inhibiting endoplasmic reticulum stress and
mitochondrial dysfunction. J Cell Physiol. 235:4316–4325. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge Y, Wang J, Wu D, Zhou Y, Qiu S, Chen J,
Zhu X, Xiang X, Li H and Zhang D: lncRNA NR_038323 suppresses renal
fibrosis in diabetic nephropathy by targeting the miR-324-3p/DUSP1
axis. Mol Ther Nucleic Acids. 17:741–753. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ye Y, Bao C and Fan W: Overexpression of
miR-101 may target DUSP1 to promote the cartilage degradation in
rheumatoid arthritis. J Comput Biol. 26:1067–1079. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sanders BE, Yamamoto TM, McMellen A,
Woodruff ER, Berning A, Post MD and Bitler BG: Targeting DUSP
activity as a treatment for high-grade serous ovarian carcinoma.
Mol Cancer Ther. 21:1285–1295. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martinez-Martinez D, Toledo Lobo MV,
Baquero P, Ropero S, Angulo JC, Chiloeches A and Lasa M:
Downregulation of snail by DUSP1 impairs cell migration and
invasion through the inactivation of JNK and ERK and is useful as a
predictive factor in the prognosis of prostate cancer. Cancers
(Basel). 13:11582021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cai X, Qu L, Yang J, Xu J, Sun L, Wei X,
Qu X, Bai T, Guo Z and Zhu Y: Exosome-transmitted microRNA-133b
inhibited bladder cancer proliferation by upregulating
dual-specificity protein phosphatase 1. Cancer Med. 9:6009–6019.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Z, Zhang L, Liu FY, Li P, He J,
Kirkwood CL, Sohn J, Chan JM, Magner WJ and Kirkwood KL: MKP-1 is
required to limit myeloid-cell mediated oral squamous cell
carcinoma progression and regional extension. Oral Oncol.
120:1054012021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan S, Shen M, Zhou M, Shi X, He R, Yin T,
Wang M, Guo X and Qin R: Long noncoding RNA LINC01111 suppresses
pancreatic cancer aggressiveness by regulating DUSP1 expression via
microRNA-3924. Cell Death Dis. 10:8832019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan W, Han J, Wei N, Wu H, Wang Y and Sun
J: LINC00702-mediated DUSP1 transcription in the prevention of
bladder cancer progression: Implications in cancer cell
proliferation and tumor inflammatory microenvironment. Genomics.
114:1104282022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han J, Liu X and Wang L: Dexmedetomidine
protects against acute lung injury in mice via the DUSP1/MAPK/NF-κB
axis by inhibiting miR-152-3p. Pulm Pharmacol Ther.
1021312022.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee S, Hwang Y, Kim TH, Jeong J, Choi D
and Hwang J: UPF1 inhibits hepatocellular carcinoma growth through
DUSP1/p53 signal pathway. Biomedicines. 10:7932022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lane R, Cilibrasi C, Chen J, Shah K,
Messuti E, Mazarakis NK, Stebbing J, Critchley G, Song E, Simon T
and Giamas G: PDGF-R inhibition induces glioblastoma cell
differentiation via DUSP1/p38MAPK signalling. Oncogene.
41:2749–2763. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Reis PP, Waldron L, Perez-Ordonez B,
Pintilie M, Galloni NN, Xuan Y, Cervigne NK, Warner GC, Makitie AA,
Simpson C, et al: A gene signature in histologically normal
surgical margins is predictive of oral carcinoma recurrence. BMC
Cancer. 11:4372011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wasserman WW and Sandelin A: Applied
bioinformatics for the identification of regulatory elements. Nat
Rev Genet. 5:276–287. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu K, Meng Z, Xian XM, Deng MH, Meng QG,
Fang W, Zhang D and Long X: LncRNA PVT1 induces chondrocyte
apoptosis through upregulation of TNF-α in synoviocytes by sponging
miR-211-3p. Mol Cell Probes. 52:1015602020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang LF, Liu YS, Yang B, Li P, Cheng XS,
Xiao CX, Liu JJ, Li S, Ren JL and Guleng B: The extracellular
matrix protein mindin attenuates colon cancer progression by
blocking angiogenesis via Egr-1-mediated regulation. Oncogene.
37:601–615. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tseng YC, Shu CW, Chang HM, Lin YH, Tseng
YH, Hsu HS, Goan YG and Tseng CJ: Assessment of early growth
response 1 in tumor suppression of esophageal squamous cell
carcinoma. J Clin Med. 11:57922022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun SW, Fang XM, Li YF, Wang QB and Li YX:
Expression and clinical significance of EGR-1 and PTEN in the
pituitary tumors of elderly patients. Oncol Lett. 14:2165–2169.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peng WX, Xiong EM, Ge L, Wan YY, Zhang CL,
Du FY, Xu M, Bhat RA, Jin J and Gong AH: Egr-1 promotes
hypoxia-induced autophagy to enhance chemo-resistance of
hepatocellular carcinoma cells. Exp Cell Res. 340:62–70. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Peng WX, Wan YY, Gong AH, Ge L, Jin J, Xu
M and Wu CY: Egr-1 regulates irradiation-induced autophagy through
Atg4B to promote radioresistance in hepatocellular carcinoma cells.
Oncogenesis. 6:e2922017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Myung E, Park YL, Kim N, Chung CY, Park
HB, Park HC, Myung DS, Kim JS, Cho SB, Lee WS and Joo YE:
Expression of early growth response-1 in human gastric cancer and
its relationship with tumor cell behaviors and prognosis. Pathol
Res Pract. 209:692–699. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pang Z, Raudonis R, McCormick C and Cheng
Z: Early growth response 1 deficiency protects the host against
pseudomonas aeruginosa lung infection. Infect Immun. 88:e00678–19.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zagurovskaya M, Shareef MM, Das A, Reeves
A, Gupta S, Sudol M, Bedford MT, Prichard J, Mohiuddin M and Ahmed
MM: EGR-1 forms a complex with YAP-1 and upregulates Bax expression
in irradiated prostate carcinoma cells. Oncogene. 28:1121–1131.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu WS, You RI, Cheng CC, Lee MC, Lin TY
and Hu CT: Snail collaborates with EGR-1 and SP-1 to directly
activate transcription of MMP 9 and ZEB1. Sci Rep. 7:177532017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lei S, He X, Yang X, Gu X, He Y and Wang
J: A mechanism study of DUSP1 in inhibiting malignant progression
of endometrial carcinoma by regulating ERK/AP-1 axis and
dephosphorylation of EPHA2. J Cancer. 14:634–645. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Goda C, Balli D, Black M, Milewski D, Le
T, Ustiyan V, Ren X, Kalinichenko VV and Kalin TV: Loss of FOXM1 in
macrophages promotes pulmonary fibrosis by activating p38 MAPK
signaling pathway. PLoS Genet. 16:e10086922020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yin Y, Wang S, Sun Y, Matt Y, Colburn NH,
Shu Y and Han X: JNK/AP-1 pathway is involved in tumor necrosis
factor-alpha induced expression of vascular endothelial growth
factor in MCF7 cells. Biomed Pharmacother. 63:429–435. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qi M, Sun LA, Zheng LR, Zhang J, Han YL,
Wu F, Zhao J, Niu WH, Fei MX, Jiang XC and Zhou ML: Expression and
potential role of FOSB in glioma. Front Mol Neurosci.
15:9726152022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan D, Fang Y, Chen W, Jiang K, Zhu G,
Wang W, Zhang W, You G, Jia Z and Zhu J: ZFP36 inhibits tumor
progression of human prostate cancer by targeting CDK6 and
oxidative stress. Oxid Med Cell Longev. 2022:36115402022.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tang TP, Qin CX and Yu H: MCM3AP-AS1
regulates proliferation, apoptosis, migration, and invasion of
breast cancer cells via binding with ZFP36. Transl Cancer Res.
10:4478–4488. 2021. View Article : Google Scholar : PubMed/NCBI
|