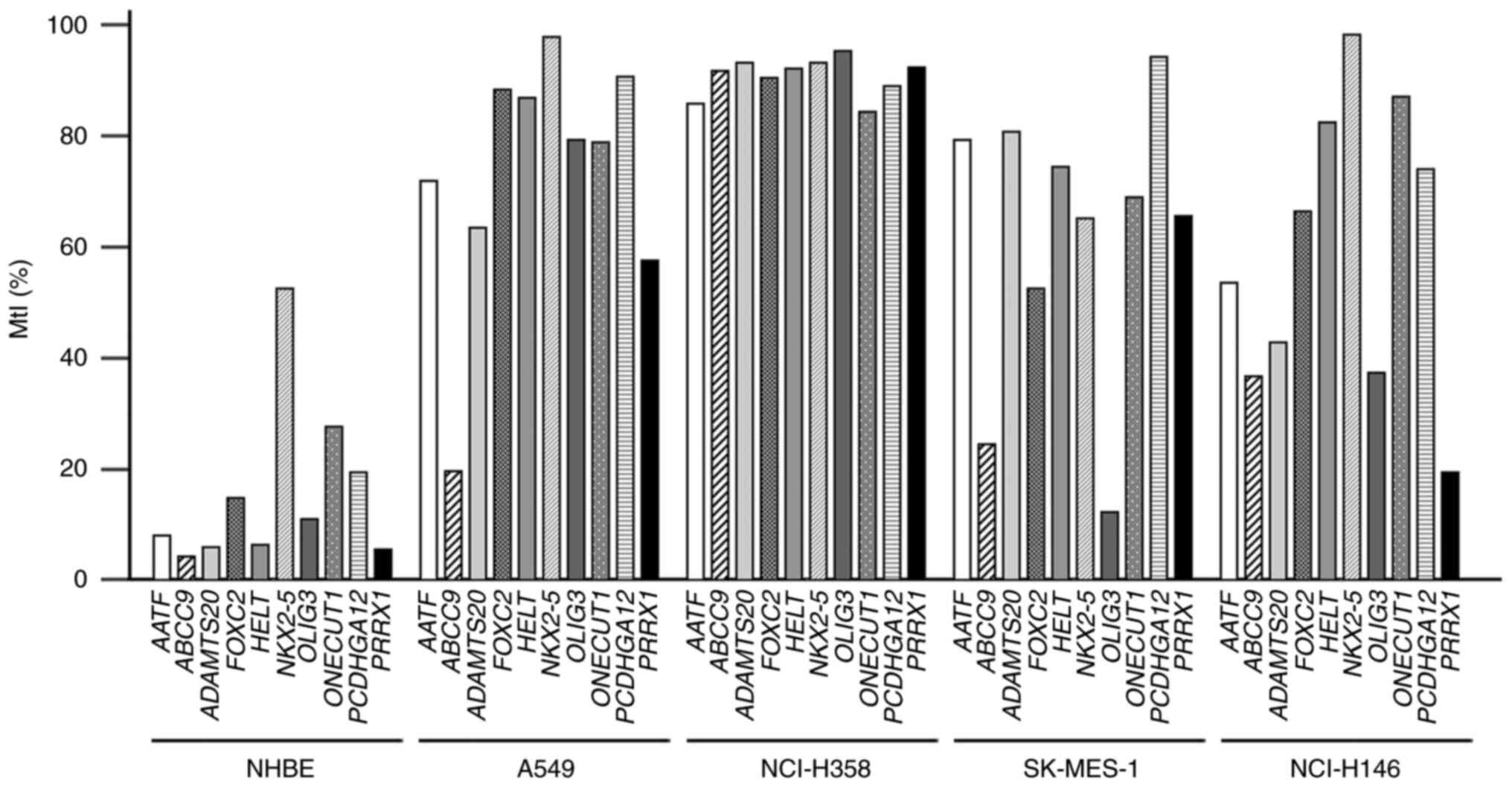
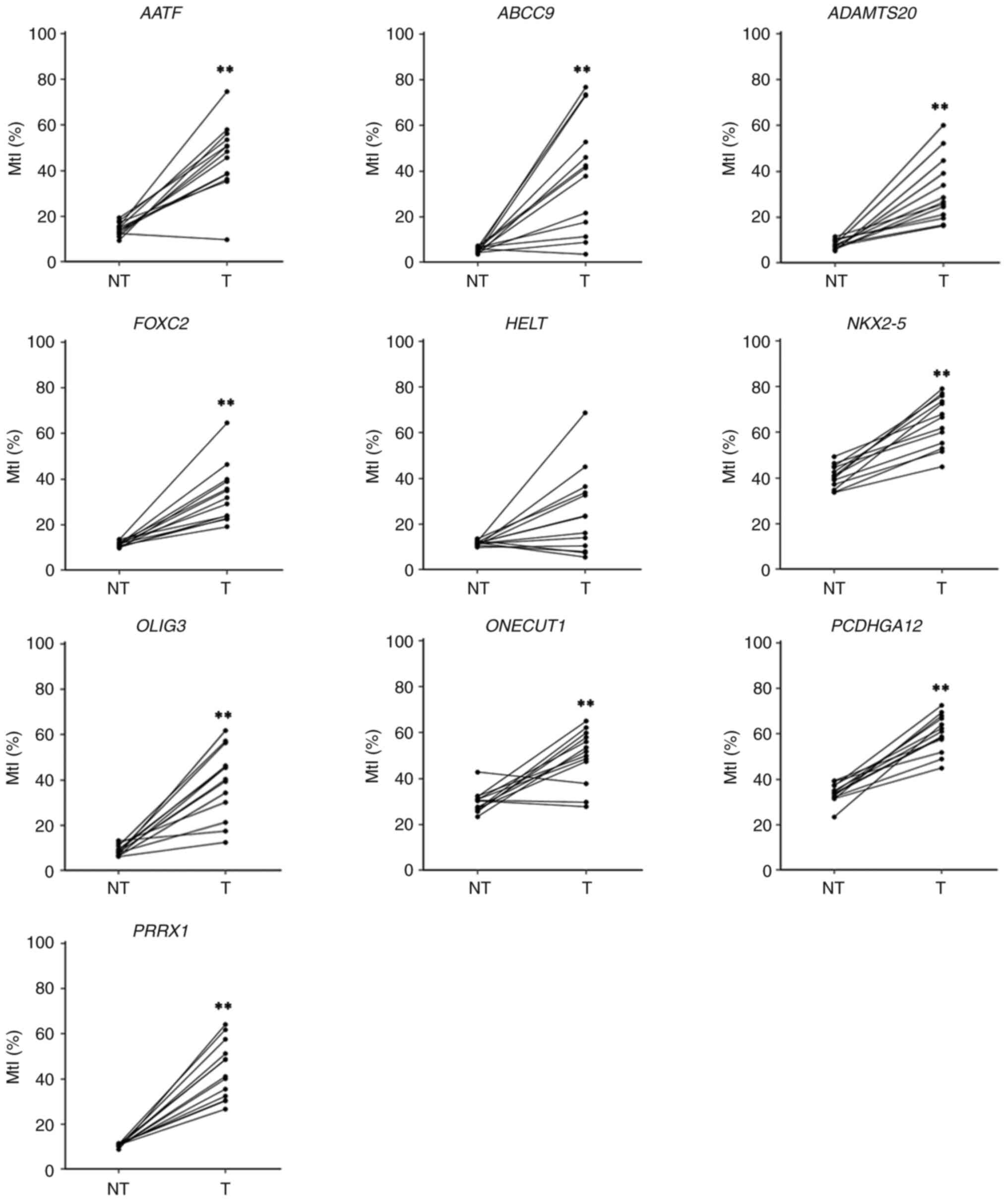
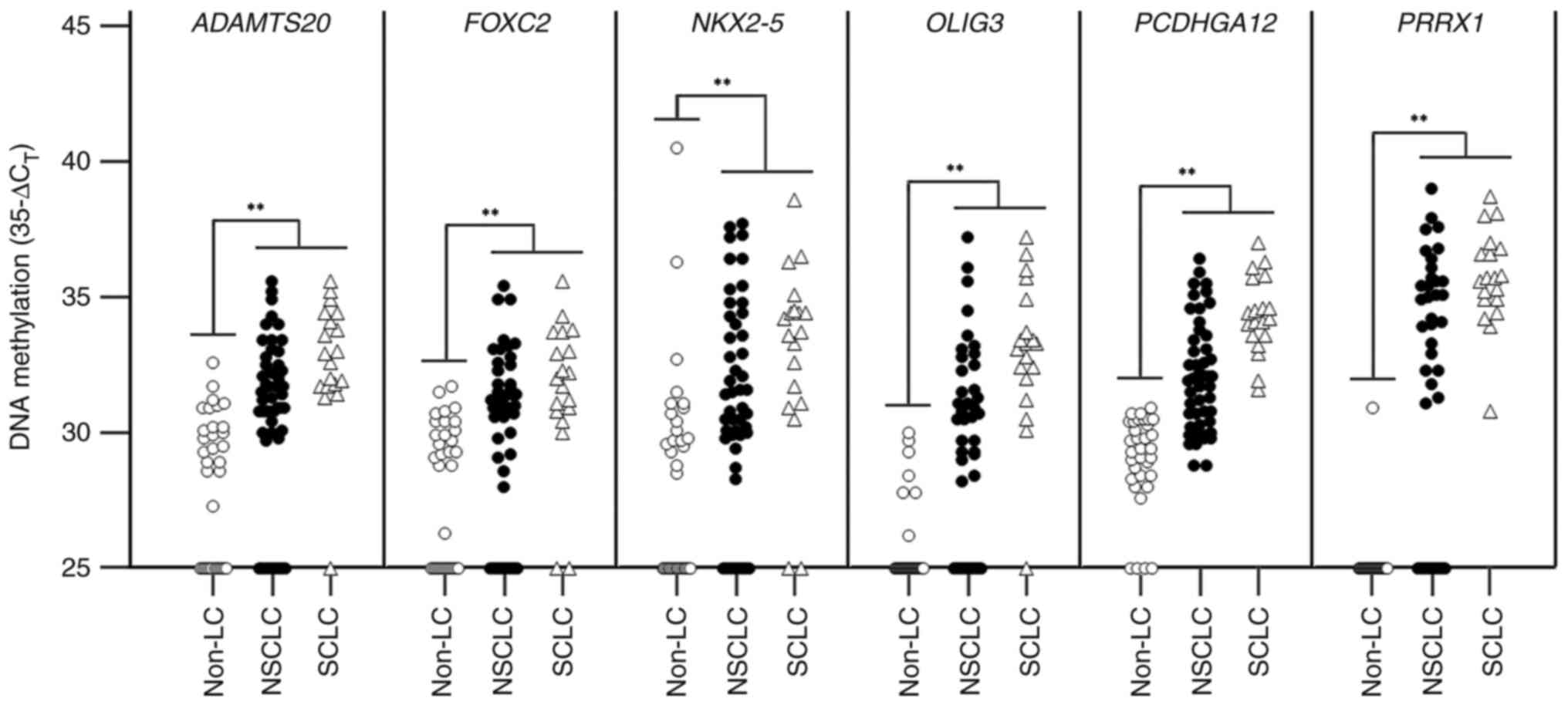
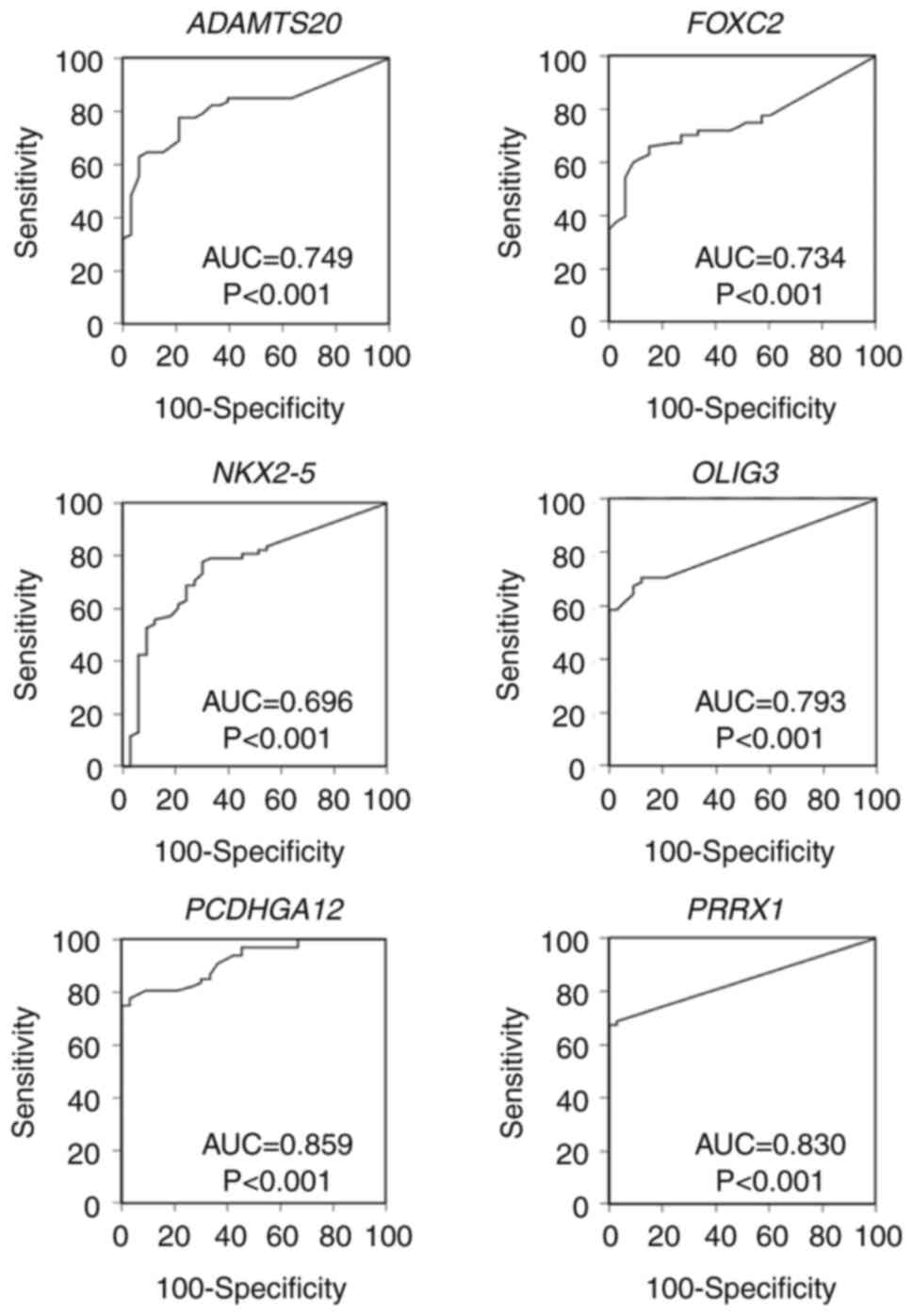
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soeromataram I, Jemal A and Bray F: Global caner statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tomasik B, Skrzypski M, Bieńkowski M,
Dziadziuszko R and Jassem J: Current and future applications of
liquid biopsy in non-small-cell lung cancer-a narrative review.
Trans Lung Cancer Res. 12:594–614. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7:1700702017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsou JA, Galler JS, Siegmund KD, Laird PW,
Turla S, Cozen W, Hagen JA, Koss MN and Laird-Offringa IA:
Identification of a panel of sensitive and specific DNA methylation
markers for lung adenocarcinoma. Mol Cancer. 6:702007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oken MM, Hocking WG, Kvale PA, Andriole
GL, Buys SS, Church TR, Crawford ED, Fouad MN, Isaacs C, Reding DJ,
et al: Screening by chest radiograph and lung cancer mortality: The
prostate, lung, colorectal, and ovarian (PLCO) randomized trial.
JAMA. 306:1865–1873. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
The National Lung Screening Trial Research
Team, . Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD,
Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks JD:
Reduced lung-cancer mortality with low-dose computed tomographic
screening. New Eng J Med. 365:395–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang C, Yu W, Wang L, Zhao M, Guo Q, Lv
S, Hu X and Lou J: DNA methylation analysis of the SHOX2 and
RASFF1A panel in bronchoavelolar lavage fluid for lung cancer
diagnosis. J Cancer. 8:3585–3591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu S, Tao J, Peng M, Ye Z, Chen Z, Chen H,
Yu H, Wang B, Fan J and Ni B: Accurate detection of early-stage
lung cancer using a panel of circulating cell-free DNA methylation
biomarkers. Biomarker Res. 11:452023. View Article : Google Scholar
|
|
9
|
Tsou JA, Hagen JA, Carpenter CL and
Laird-Offringa IA: DNA methylation analysis: A powerful new tool
for lung cancer diagnosis. Oncogene. 21:5450–5461. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmidt B, Liebenberg V, Dietrich D,
Schlegel T, Kneip C, Seegebarth A, Flemming N, Seemann S, Distler
J, Lewin J, et al: SHOX2 DNA methylation is a biomarker for
the diagnosis of lung cancer based on bronchial aspirates. BMC
Cancer. 10:6002010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dietrich D, Kneip C, Raji O, Liloglou T,
Seegebarth A, Schlegel T, Flemming N, Rausch S, Distler J,
Fleischhacker M, et al: Performance evaluation of the DNA
methylation biomarker SHOX2 for the aid in diagnosis of lung
cancer based on the analysis of bronchial aspirates. Int J Oncol.
40:825–832. 2012.PubMed/NCBI
|
|
12
|
Wiener RS, Wiener DC and Gould MK: Risks
of transthoracic needle biopsy: How high? Clin Plum Med. 20:29–35.
2013. View Article : Google Scholar
|
|
13
|
Li P, Liu S, Du L, Mohseni G, Zhang Y and
Wang C: Liquid biopsies based on DNA methylation as biomarkers for
the detection and prognosis of lung cancer. Clin Epigenetics.
14:1182022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma S, Yu X, Jin X, Qiu F, Chen X, Wang R
and Cao C: The usefulness of liquid-based cytology of
bronchoalveolar lavage fluids combined with bronchial brush
specimens in lung cancer diagnosis. J Int Med Res.
50:30006052211327082022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chrabańska M, Środa M, Kiczmer P and
Droddzowska B: Lung cancer cytology: Can any of the cytological
methods replace histopathology? J Cytol. 37:117–1121. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen SWC, Wen J, Hansen TF and Jakobsen A:
Cell free methylated tumor DNA in bronchial lavage as an additional
tool for diagnosing lung cancer-a systematic review. Cancers
(Basel). 14:22542022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang V, Li X, Li W, Zhu X and Li C: DNA
methylation in lung cancer patients: Opening a ‘window of life’
under precision medicine. Biomed Pharmacother. 144:1122022021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Esteller M: Epigenetics in cancer. N Eng J
Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ibrahim J, Peeters M, Van Camp G and Op de
Beeck K: Methylation biomarkers for early cancer detection and
diagnosis: Current and future perspectives. Eur J Cancer.
178:91–113. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Anglim PP, Alonzo TA and Laird-Offringa
IA: DNA methylation-based biomarkers for early detection of
non-small cell lung cancer: An update. Mol Cancer. 7:812008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Xie K, Zhu G, Ma C, Cheng C, Li Y,
Xiao X, Li C, Tang J, Wang H, et al: Early detection and
stratification of lung cancer aided by a cost-effective assay
targeting circulating tumor DNA (ctDNA) methylation. Respir Res.
24:1632023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rauch T, Wang Z, Zhang X, Zhong X, Wu X,
Lau SK, Kernstine KH, Riggs AD and Pfeifer GP: Homeobox gene
methylation in lung cancer studied by genome-wide analysis with a
microarray-based methylated CpG island recovery assay. Proc Natl
Acad Sci USA. 104:5527–5532. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hong Y and Kim WJ: DNA methylation markers
in lung cancer. Curr Genomics. 22:79–87. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dejeux E, Audard V, Cavard C, Gut IG,
Terris B and Tost J: Rapid identification of promoter
hypermethylation in hepatocellular carcinoma by pyrosequencing of
etiologically homogeneous sample pools. J Mol Diag. 9:510–520.
2007. View Article : Google Scholar
|
|
25
|
Chung W, Bondaruk J, Jelinek J, Lotan Y,
Liang S, Czerniak B and Issa JJ: Detection of bladder cancer using
novel DNA methylation biomarkers in urine sediments. Cancer
Epidemiol Biomarkers Prev. 20:1483–1491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grote HJ, Schiemann V, Gedder H, Rohr UP,
Kappes R, Gabbert HE and Böcking A: Aberrant promoter methylation
of p16(INKA4a), RARB2 and SEMA3B in bronchial aspirates from
patients with suspected lung cancer. Int J Cancer. 116:720–725.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ni S, Ye M and Huang T: Short stature
homeobox 2 methylation as a potential noninvasive biomarker in
bronchial aspirates for lung cancer diagnosis. Oncotarget.
8:61253–61263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roncarati R, Lupini L, Miotto E, Saccenti
E, Mascetti S, Morandi L, Bassi C, Rasio D, Callegari E, Conti V,
et al: Molecular testing on bronchial washings for the diagnosis
and predictive assessment of lung cancer. Mol Oncol. 14:2163–2175.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Silvestri GA, Vachani A, Whitney D,
Elashoff M, Porta Smith K, Ferguson JS, Parsons E, Mitra N, Brody
J, Lenburg ME, et al: A bronchial genomic classifier for the
diagnostic evaluation of lung cancer. N Eng J Med. 373:243–251.
2015. View Article : Google Scholar
|
|
30
|
Vachani A, Whitney DH, Parsons EC, Lenburg
M, Ferguson JS, Silvestri GA and Spira A: Clinical utility of a
bronchial genomic classifier in patients with suspected lung
cancer. Chest. 150:210–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeong IB, Yoon YS, Park SY, Cha EJ, Na MJ,
Kwon SJ, Kim JH, Oh TJ, An S, Park CR, et al: PCDHGA12
methylation biomarkers in bronchial washing specimens as an
adjunctive diagnostic tool to bronchoscopy in lung cancer. Oncol
Lett. 16:1039–1045. 2018.PubMed/NCBI
|
|
32
|
Ahrendt SA, Chow JT, Xu L, Yang SC,
Eisenberger CF, Esteller M, Herman JG, Wu L, Decker PA, Jen J and
Sidransky D: Molecular detection of tumor cells in bronchoalveolar
lavage fluid from patients with early stage lung cancer. J Natl
Cancer Inst. 91:332–339. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liam CK, Lee P, Yu CJ, Bai C and Yasufuku
K: The diagnosis of lung cancer in the era of interventional
pulmonology. Int J Tuberc Lung Dis. 25:6–15. 2021. View Article : Google Scholar : PubMed/NCBI
|