Introduction
Lung cancer is the second most common malignant
tumor worldwide (1). Lung cancer is
primarily divided into non-small cell lung cancer (NSCLC) and small
cell lung cancer based on histopathological features (2). Lung adenocarcinoma (LUAD) accounts for
>40% of NSCLC (3) and is a
common type of lung cancer tissue (4). LUAD patients are typically older when
diagnosed, at a later disease stage, possess a worse prognosis, and
have a 5-year survival rate <20% (5). Surgery, radiotherapy and chemotherapy
are the primary treatment modalities for lung cancer. However,
their efficacy is often limited in patients with metastatic lung
cancer. Therefore, an in-depth exploration of the molecular
mechanisms related to the pathogenesis of LUAD could support the
search for optimized approaches to early diagnosis and treatment
targeting specific genes.
The role of the immune system in the occurrence and
development of tumors is complex. It can eliminate tumor cells in
specific tissues, establish an inflammatory environment that
prevents tumorigenesis by removing pathogens and inflammation, and
promote tumor growth through immunoediting amongst other methods
(6). Moreover, tumor cells can
adopt various methods to evade immune system attack, including
changing surface antigens and inhibiting immune cell activity. The
primary components of tumor immunity are an immunosuppressive tumor
microenvironment (TME) and dysfunctional anti-tumor T cells
(7). The complex surroundings
around the tumor, including components such as molecules, blood
vessels, and other non-tumor cells, are known as the TME and
influence both the anti-tumor immune response and immunotherapy
effectiveness (8). Development in
immunotherapy have been remarkably successful in treating certain
cancers. In LUAD, patients were administered immune checkpoint
inhibitors (ICIs) to target programmed cell death 1, programmed
cell death ligand 1 and cytotoxic T lymphocyte antigen 4 which
improved survival (9). However,
there are some issues with immunotherapy, such as acquired
resistance and severe side effects (10). A previous study reported that tumor
cell and TME interaction in spatiotemporal dynamics was essential
for tumor progression (11). Thus,
tumor growth promotes the development of immunological tolerance in
the body, weakening the therapeutic effect of ICIs (12). Not every lung adenocarcinoma patient
responds to ICI treatment due to the effects of immune tolerance.
Moreover, the abnormal expression of tumor immune-related genes in
tumor escape has become a novel direction of focus in tumor
research (13). However, to the
best of our knowledge, only a few reports exist on how abnormal
immune-related genes affect lung adenocarcinoma prognosis and
immune cell infiltration. Therefore, elucidating the immune genes
associated with lung adenocarcinoma prognosis and constructing a
LUAD prognosis model based on immune cell infiltration in such
patients, could have clinical value.
In the present study, LUAD-related data were
retrieved from The Cancer Genome Atlas (TCGA) database and
bioinformatics were used to analyze the immune-related genes
significantly linked with the prognosis of LUAD patients. An
immune-related genes prognostic model was constructed and its
independent prediction power was assessed. The study also used
reverse transcription-quantitative (RT-qPCR) to analyze
immune-related prognostic gene expression while evaluating immune
cell infiltration and function levels in LUAD. Thus, an
immune-related prognostic risk model was developed for LUAD based
on disease transcriptomics, offering novel targets for therapeutic
immunotherapy.
Materials and methods
Data sources
The RNA sequencing and clinical data of 535 LUAD
samples and 59 normal tissues were retrieved from the TCGA database
(http://www.cancer.gov/). The gene expression data
were corrected in batches and those with missing information
regarding clinical features were eliminated. Finally, there were
522 LUAD samples and normal tissues. A total of 2,483
immune-related genes were downloaded from the Immunology Database
and Analysis Portal (ImmPort) database (http://www.immport.org/). LM22 marker genes were
downloaded from the CIBERSORT website (https://cibersortx.stanford.edu). The Human Protein
Atlas (https://www.proteinatlas.org)
database was used to retrieve prognostic immune-related gene
protein expression data. The TIDE algorithm was used to predict
responses to ICIs therapy (14).
Screening of immune-related
differential genes and enrichment analysis
Differential analysis was performed across all LUAD
patient genes using the limma package (https://bioinf.wehi.edu.au/limma/), with
fdrFilter=0.05 and logFCfilter=2, followed by construction of the
differential gene volcano diagram with graphics (https://www.R-project.org/). The differentially
expressed immune-related genes were obtained by assessing the
intersection of the differentially expressed genes (DEGs) with 2483
immune-related genes with the Venn diagram depicting the
intersection results by venn package (https://CRAN.R-project.org/package=venn). Gene Set
Enrichment Analysis (GSEA) (15),
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) (https://ggplot2.tidyverse.org)
enrichment analysis were performed on the differentially expressed
immune-related genes previously identified. Univariate Cox
regression analysis was used to identify the immune-related genes
associated with prognosis and DEGs with P<0.05 were included in
the multifactor Cox proportional hazards model analysis to obtain
the immune-related genes which were subsequently used to construct
the prognostic model.
Construction and analysis of
prognostic risk models
The expression levels of the prognostic
immune-related genes, identified by the aforementioned multivariate
Cox regression analyses, were multiplied using the coefficients
obtained from their respective multivariate Cox regression analyses
to determine the risk score of each patient. A prognostic risk
model was then constructed using multifactor Cox proportional
hazards model analysis. Patients were divided into high and
low-risk groups with the median risk score as the cutoff value,
followed by the plotting of the Kaplan-Meier survival curve
(https://CRAN.R-project.org/package=survival), receiver
operating characteristic (ROC) curve (16) and nomogram (https://CRAN.R-project.org/package=rms). Univariate
and multivariate Cox regression analyses were also performed for
the hazard ratio (HR) of the risk score and clinical
characteristics including age, gender, stage and, tumor (T), node
(N) and metastasis (M) scores.
Immune infiltration analysis in
LUAD
The LUAD immune cells of LM22 marker genes were
analyzed using the CIBERSORT algorithm, with P<0.05 being
considered to indicate a statistically significantly difference.
The Wilcox.test was used to assess the differences in immune cell
expression in LUAD, and a differential heatmap of lung
adenocarcinoma immune cells was drawn. Student's t-test was used to
evaluate differences in immune cells and the relationship between
prognostic genes and immune cells in high and low-risk groups.
Single sample GSEA was used to compare Tumor Immune Dysfunction and
Exclusion (TIDE) scores between high and low-risk groups and to
determine immune-related functional activity.
Cell culture
Human normal lung epithelial MRC5 cells and human
NSCLC A549 and H1975 cells were purchased from The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences. The
cells were cultured in DMEM high glucose (H) medium containing 10%
fetal bovine serum and incubated at 37°C with 5% CO2,
with the medium replaced every 2–3 days.
RNA extraction and RT-qPCR
Total RNA was extracted from MRC5, A549 and H1975
cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
RT-PCR was performed after removing genomic DNA, according to the
instructions of the PrimeScript™ RT reagent kit with gDNA Eraser
(Takara Biotechnology Co., Ltd.). RT-qPCR primers were designed and
synthesized by Sangon Biotech Co., Ltd. (Table I). The RT-qPCR solution comprised
12.5 µl TB Green Premix Ex TaqII (Takara Biotechnology Co., Ltd.),
1.0 µl PCR forward primer (10 µΜ), 1.0 µl PCR reverse primer (10
µΜ), 2 µl RT reaction solution (complimentary DNA solution), and
8.5 µl sterilized water, for a total volume of 25 µl. The RT-qPCR
reaction conditions were as follows: 95°C for 30 sec, then 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The relative gene
expression was determined using the 2−∆∆Cq method
(17), with GAPDH used as an
internal control. Each experiment was performed in triplicate.
 | Table I.Sequences of primers used for reverse
transcriptionquantitative PCR. |
Table I.
Sequences of primers used for reverse
transcriptionquantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| S100A16 | F:
CAGCCTGGTCAAGAACAAGAT |
|
|
ATGATTGGCATCCAGGTTC |
| FURIN | F:
AACACTGTGCCCTGGTGGA |
|
| R:
CAGATGGCTGGGTACCAGGA |
| FGF-2 | F:
ACCGTTACCTGGCTATGAAG |
|
| R:
CCAGTTCGTTTCAGTGCC |
| LGR4 | F:
ACTCAAAGTTCTAACGCTCCAG |
|
| R:
CAGCCACAGATGCCGTAAC |
|
TNFRSF11A | F:
CCATCATCTTTGGCGTTTG |
|
| R:
CTCCTCCAGAGTCAGCAGTAAG |
| VIPR1 | F:
CATTTTGAGGATTATGGGTGC |
|
| R:
GCAGTTTCTGAAGCAGGATT |
| GAPDH | F:
TGGAAATCCCATCACCATCT |
|
| R:
GTCTTCTGGGTGGCAGTGAT |
Statistical analysis
All statistical analysis and presentations were
performed using the R 4.3.0 software package (https://mirrors.tuna.tsinghua.edu.cn/CRAN/). The
Wilcox.test was used to compare the differential expression of
immune-related genes in LUAD and normal tissues. Immune-related
genes linked with poor prognosis in LUAD were identified using
multivariate Cox regression analysis. The correlation of prognostic
genes and transcription factors was assessed using t-test. The
comparison of the expression levels of prognostic genes in MRC5,
H1975 and A549 cells was performed using one-way ANOVA with
Dunnett's multiple comparisons test as the post-hoc test.
Kaplan-Meier curves were used for survival analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
Differential immune-related gene
expression and enrichment analysis in LUAD
The clinicopathological characteristics of the 522
LUAD samples from the TCGA database were assessed (Table II). The intersection of DEGs and
immune related genes in LUAD identified 505 differentially
immune-related genes using logFCfilter=1 and fdrFilter=0.05 for
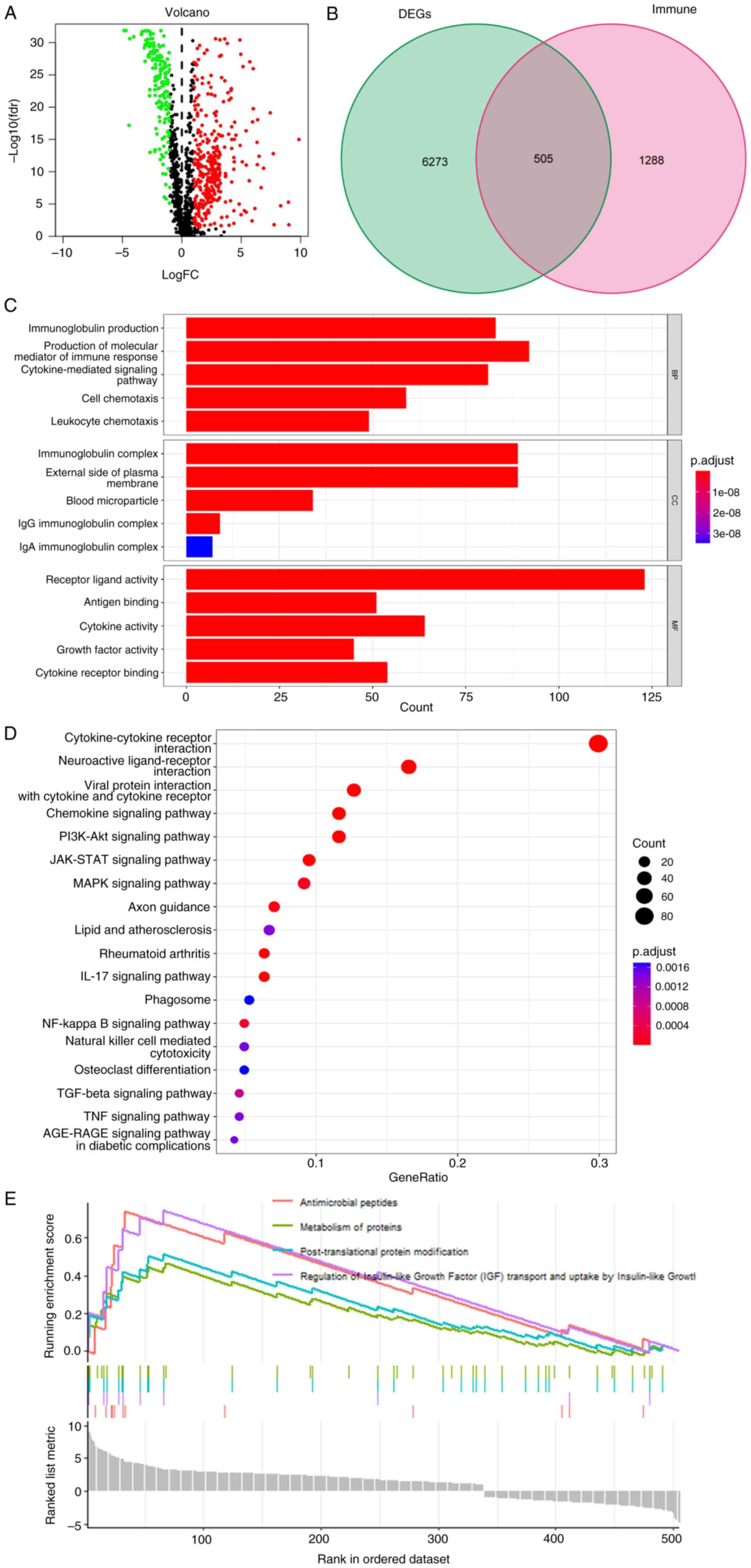
these genes in LUAD (Fig. 1B).
Among these, 338 genes were up-regulated and 167 were
down-regulated (Fig. 1A and B). The
volcano diagram was drawn using the DEGs. GO enrichment analysis of
these differentially immune-related genes indicated that they were
primarily enriched in biological processes such as ‘immunoglobulin
production’ and ‘production of molecular mediator of immune
response’, cellular components such as ‘immunoglobulin complex’ and
‘external side of plasma membrane’, and molecular functions such as
‘antigen binding’ and ‘receptor ligand activity’ (Fig. 1C). KEGG analysis indicated the
differentially immune-related genes were mainly enriched in
‘cytokine-cytokine receptor interaction’, ‘neuroactive
receptor-ligand interaction’, ‘viral protein ineraction with
cytokine and cytokine receptor’ and ‘chemokine signaling pathway’
pathways (Fig. 1D). GSEA analysis
demonstrated primary enrichment in ‘antimicrobial peptides’,
‘regulation of insulin-like growth factor (IGF) transport and
uptake by IGF’, ‘post-translational protein modification’ and
‘metabolism of proteins’ (Fig.
1E).
 | Table II.Clinical characteristics of patients
involved in the study. |
Table II.
Clinical characteristics of patients
involved in the study.
|
Characteristics | n (%) |
|---|
| Age, years |
|
|
<65 | 223 (42.7) |
|
≥65 | 280 (53.7) |
|
Unknown | 19 (3.6) |
| Gender |
|
|
Female | 280 (53.6) |
|
Male | 242 (46.4) |
| T
classification |
|
| T1 | 172 (33.0) |
| T2 | 281 (53.8) |
| T3 | 47 (9.0) |
| T4 | 19 (3.6) |
| TX | 3 (0.6) |
| N
classification |
|
| N0 | 335 (64.2) |
| N1 | 98 (18.8) |
| N2 | 75 (14.4) |
| N3 | 2 (0.3) |
| NX | 11 (2.1) |
|
Unknown | 1 (0.2) |
| M
classification |
|
| M0 | 353 (67.6) |
| M1 | 25 (4.8) |
| MX | 140 (26.8) |
|
Unknown | 4 (0.8) |
| Tumor stage |
|
| I | 279 (53.4) |
| II | 124 (23.8) |
|
III | 85 (16.3) |
| IV | 26 (5.0) |
|
Unknown | 8 (1.5) |
| Vital status |
|
|
Alive | 167 (32.0) |
|
Dead | 355 (68.0) |
Construction of a prognostic model
using immune-related genes
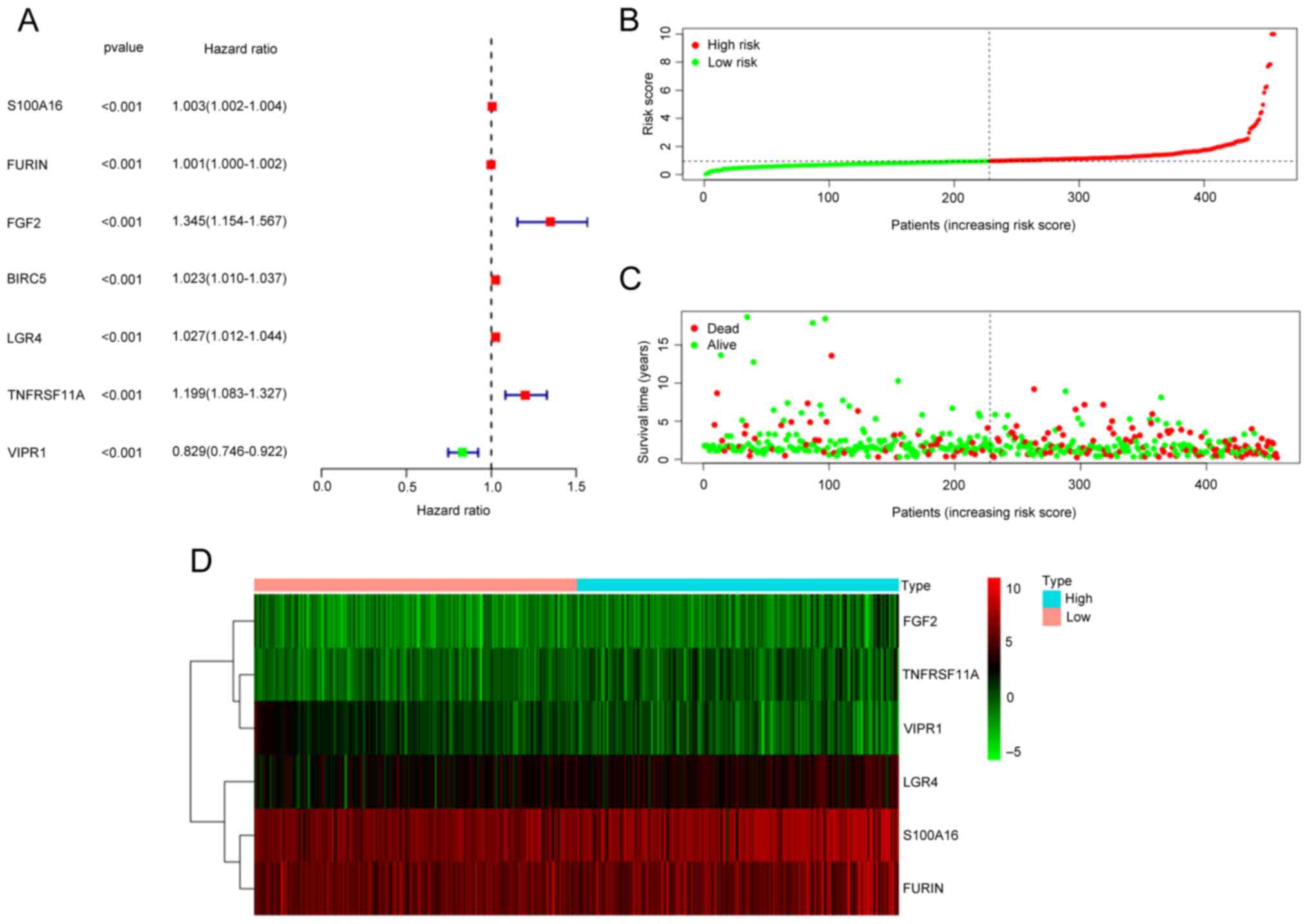
Seven differential immune-related genes associated
with prognosis were obtained using univariate Cox regression
analysis (Fig. 2A). Multifactor Cox
proportional hazards model analysis was performed on these seven
genes, from which six prognostic genes (S100A16, FURIN, FGF2,
LGR4, TNFRSF11A and VIPR1) were significantly associated
with the occurrence and development of LUAD (Table III). The multivariate regression
coefficients of the six prognostic immune-related genes were
multiplied with their respective expression levels in each sample.
Thus, the risk score of each sample was calculated as follows: risk
score=S100A16 expression level × 0.002303 + FURIN
expression level × 0.00073 + FGF2 expression level × 0.24979
+ LGR4 expression level × 0.015309 + TNFRSF11A
expression level × 0.176773 - VIPR1 expression level ×
0.14724. Depending on the median risk score, the samples were
differentiated into high and low-risk groups. The risk curve
indicated the mortality of most patients in the high-risk group;
with an increase in the patient's risk value, more patients died
(Fig. 2B and C). The survival heat
map indicated that the expression levels of S100A16, FURIN,
FGF2, LGR4 and TNFRSF11A gradually increased with the
risk score. In contrast, VIPR1 gene expression gradually
decreased (Fig. 2D).
 | Table III.Multifactorial Cox proportional
hazard model analysis. |
Table III.
Multifactorial Cox proportional
hazard model analysis.
| Gene | coef | HR | HR.95L | HR.95H | P-value |
|---|
| S100A16 | 0.002303 | 1.002306 | 1.000988 | 1.003626 | 0.000604 |
| FURIN | 0.00073 | 1.00073 | 1.000044 | 1.001416 | 0.036931 |
| FGF2 | 0.24979 | 1.283756 | 1.108018 | 1.487367 | 0.000882 |
| LGR4 | 0.015309 | 1.015427 | 0.998317 | 1.032829 | 0.077442 |
|
TNFRSF11A | 0.176773 | 1.19336 | 1.075284 | 1.324403 | 0.000883 |
| VIPR1 | −0.14724 | 0.863084 | 0.777428 | 0.958178 | 0.005761 |
Independent prognostic analysis
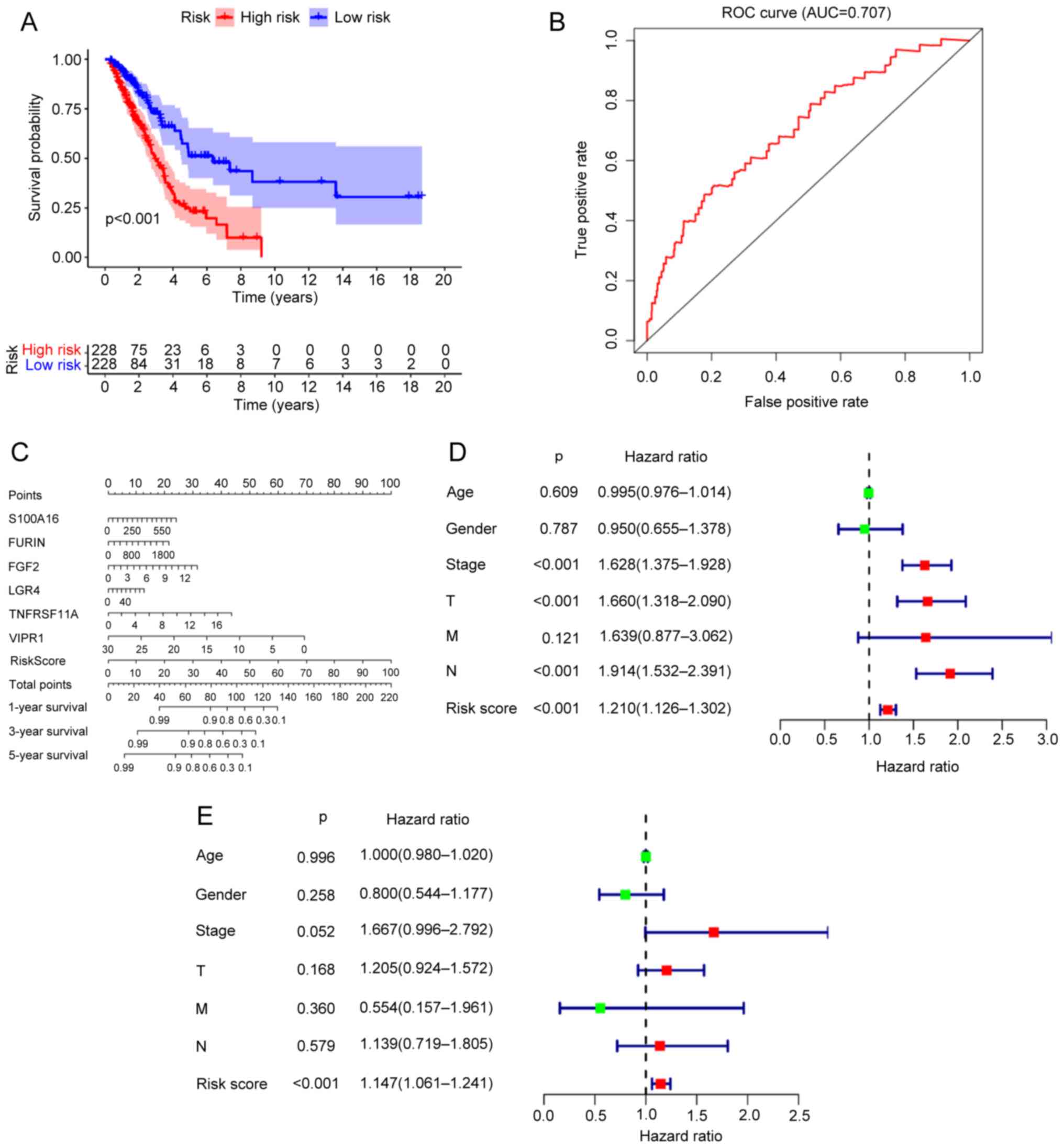
The survival analysis of the prognostic risk model
indicated that the survival time of patients with LUAD in the
high-risk score group was significantly lower than those in the
low-risk score group. The 5-year overall survival rate of patients
in the high-risk score group was ~23%, while that of those in the
low-risk score group was ~51% (Fig.
3A). The ROC curve indicated an area under the curve of 0.707
(Fig. 3B), which suggested that the
prognostic model possessed strong predictive ability. A nomogram
was built as a quantitative method for the six immune-related genes
to predict LUAD prognosis. The nomogram point scale made it
possible to assign a value to a single variable by constructing a
straight line between the predicted curve and the total point axis.
Moreover, the nomogram could be used to estimate the 1, 3 and
5-year overall survival rates of the patients with LUAD (Fig. 3C) through the predictive power of
the prognostic risk model. The univariate Cox regression analysis
of the risk score of the prognostic model and the clinical
characteristics of LUAD patients (age, sex, stage and TNM scores)
revealed that the stage, T, N and risk score all independently
predicted the prognosis of patients LUAD (Fig. 3D). Multivariate Cox regression
analysis indicated that the risk score independently predicted the
prognosis of patients with LUAD (Fig.
3E).
Prognostic immune-related gene
expression and survival analysis
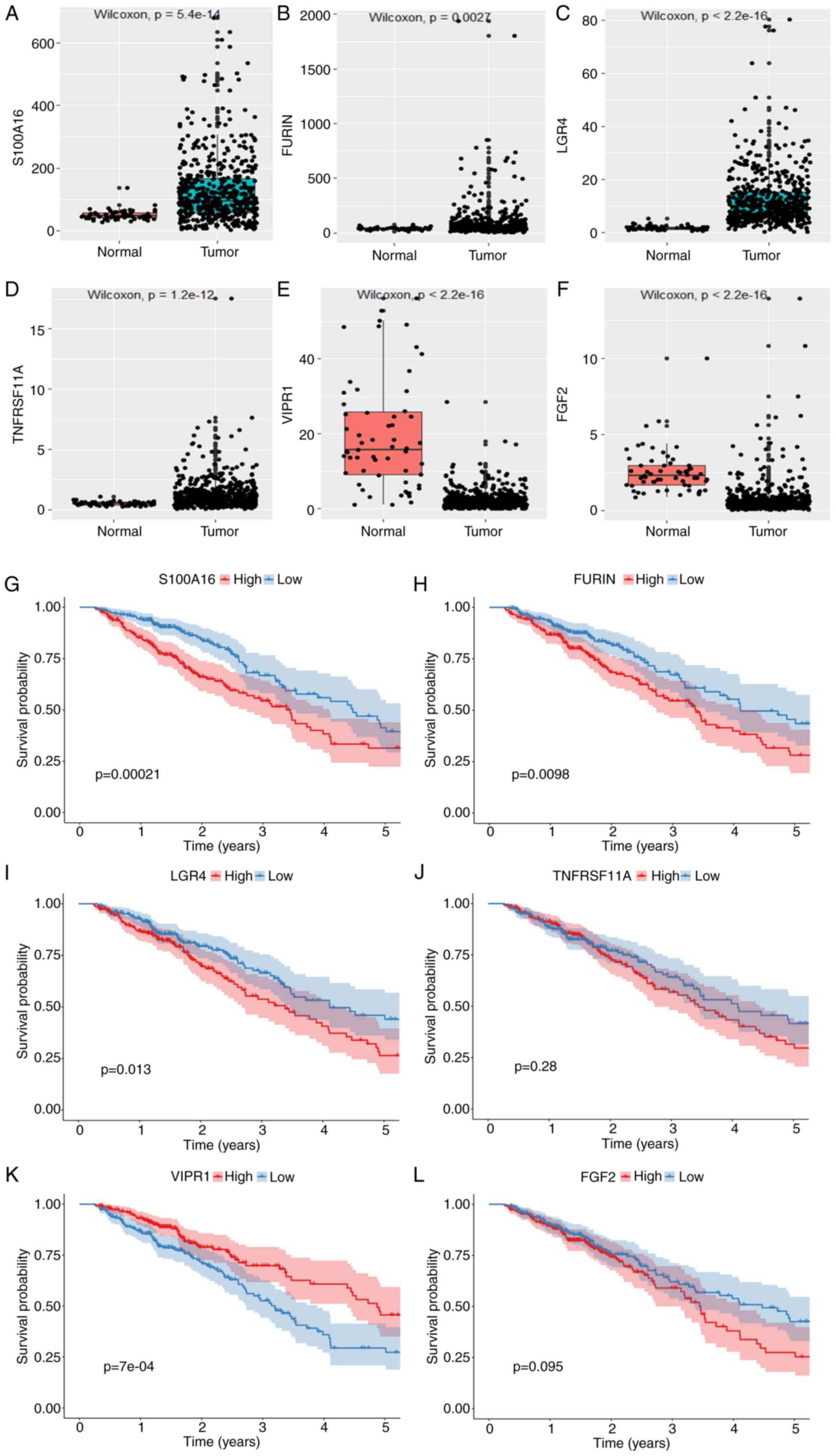
The expression levels of S100A16, FURIN, LGR4
and TNFRSF11A were significantly higher in tumor tissues
compared with normal tissues and the expression levels of
VIPR1 and FGF2 were significantly lower in tumor
tissues compared with normal tissues (Fig. 4A-F). Patients with high S100A16,
FURIN and LGR4 gene expression had a significantly worse
prognosis compared with the low expression group, while patients
with low VIPR1 gene expression had a significantly worse
prognosis compared with the high expression group. However,
FGF2 and TNFRSF11A did not demonstrate a significant
impact on the survival time (Fig.
4G-L).
Prognostic gene validation
S100A16, FURIN, LGR4, TNFRSF11A and
VIPR1 mRNA expression levels were significantly higher in
human lung cancer H1975 and A549 cells compared with normal human
embryonic lung MRC5 cells. FURIN mRNA expression levels were
significantly higher in human lung cancer H1975 compared with
normal human embryonic lung MRC5 cells. However, FGF2
expression was significantly lower in the human lung cancer H1975
and A549 cells compared with normal human embryonic lung MRC5 cells
(Fig. 5A). Due to the low copy
numbers of S100A16, FURIN and FGF2 in all three cell lines,
the significance of gene expression differences between the cell
lines should be treated with caution. S100A16, FURIN,
TNFRSF11A and VIPR1 were not stained for in normal
alveolar tissue cells, with weak/moderate expression in human lung
tumor tissue cells. Additionally, FGF2 staining was weak in
normal alveolar and lung tumor tissue cells (Fig. 5B-D). These results indicated the
inconsistency of the VIPR1 expression between LUAD and lung
cancer and differences in FGF2 expression in cell lines and
tissues.
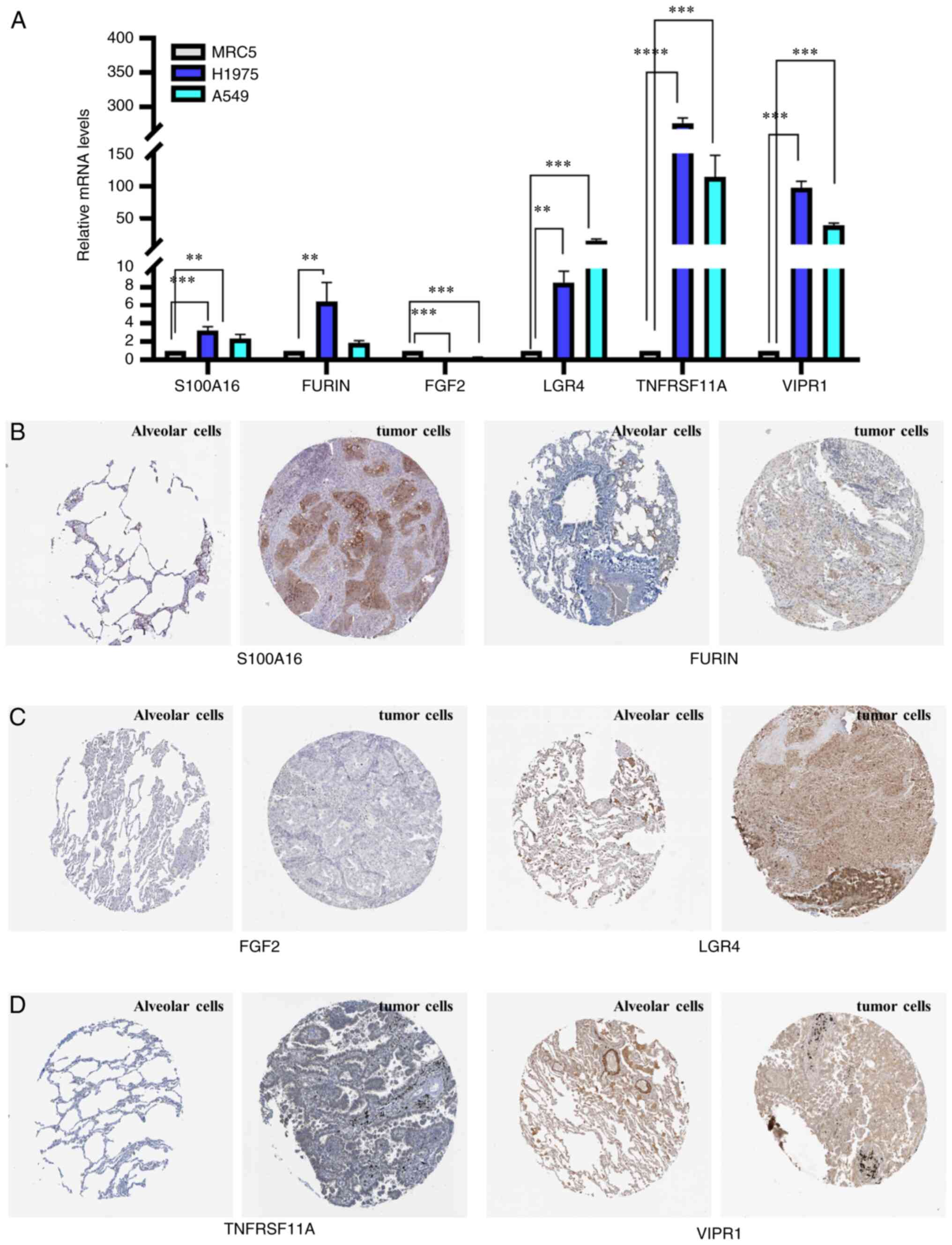 | Figure 5.Reverse transcription-quantitative
PCR and staining analysis. (A) S100A16, FURIN, FGF2, LGR4,
TNFRSF11A and VIPR1 mRNA expression levels in MRC5, H1975 and A549
cells. (B) S100A16, FURIN, (C) FGF2, LGR4, (D) TNFRSF11A, and VIPR1
protein expression in normal alveolar and tumor cells. The
immunohistochemistry images were downloaded from the Human Protein
Atlas database (https://www.proteinatlas.org/ENSG00000188643-S100A16/pathology/lung+cancer;
https://www.proteinatlas.org/ENSG00000140564-FURIN/pathology/lung+cancer;
https://www.proteinatlas.org/ENSG00000205213-LGR4/pathology/lung+cancer;
https://www.proteinatl-as.org/ENSG00000141655-TNFRSF11A/pathology/lung+cancer;
and http://www.proteina-tlas.org/ENSG00000114812-VIPR1/pathology/lung+cancer).
**P<0.01, ***P<0.001 and ****P<0.0001. |
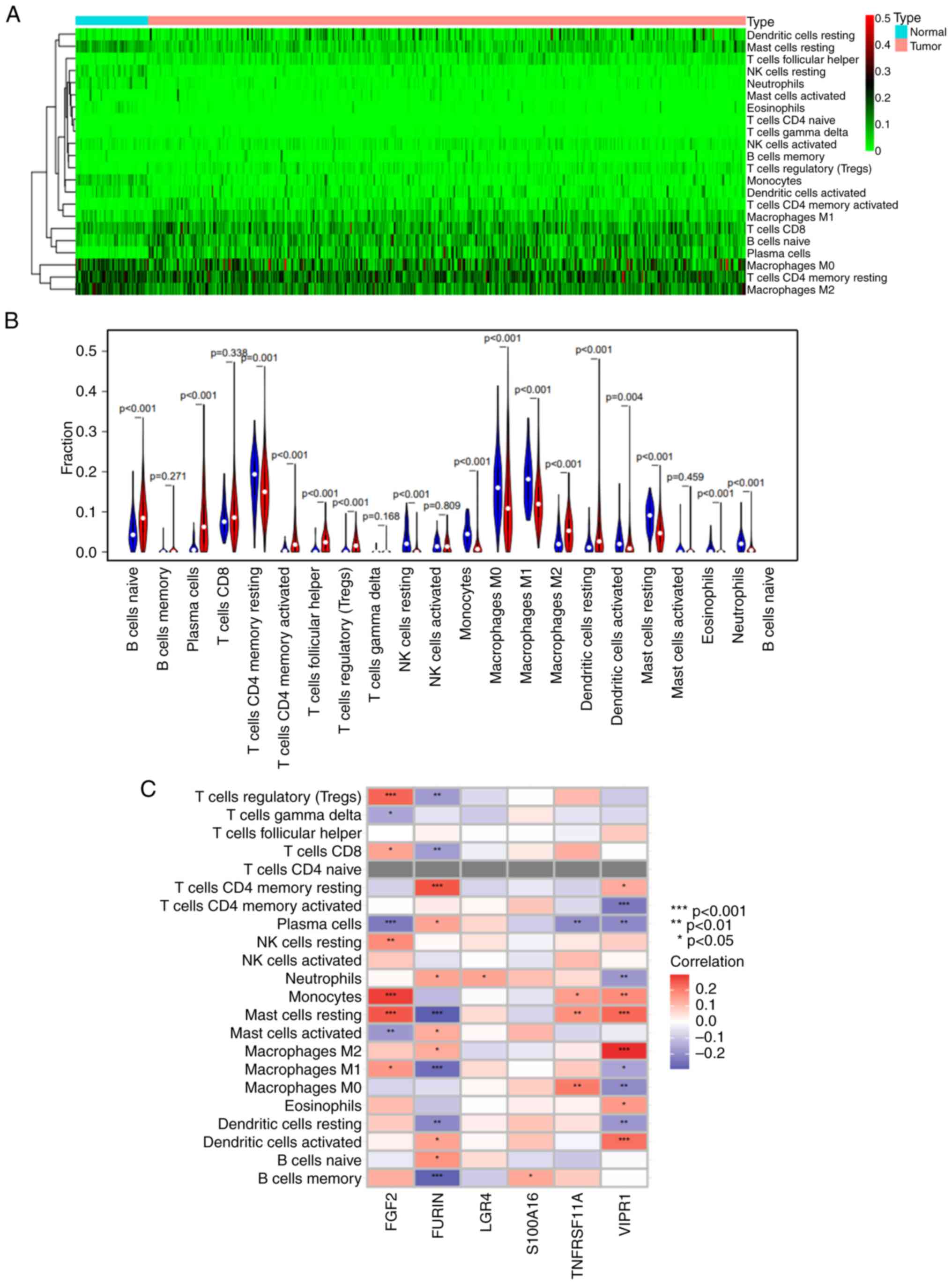
Analysis of immune cell infiltration
in LUAD
The heat map of immune cell infiltration indicated
higher levels of macrophages M0, T cells CD4 memory resting and
macrophages M2 cells within LUAD tissue (Fig. 6A). Immune cell component analysis
indicated that most immune cells were differentially distributed
between the lung adenocarcinoma samples and adjacent tissues
(Fig. 6B). The degree of
infiltration of B cells naive, plasma cells, T cell CD4 memory
activated, T cell follicular helper, T cells regulatory (Tregs),
macrophages M2 and dendritic cells resting were significantly
higher in lung adenocarcinoma tissue than normal tissues. However
degree of infiltration of T cells CD4 memory resting, NK cells
resting, macrophages M0, macrophages M1, mast cells resting,
eosinophils and neutrophils were significantly lower in lung
adenocarcinoma tissue than normal tissues. The correlation between
risk genes and immune cells indicated that FURIN was
significantly positively associated with T cells CD4 memory
resting, and significantly negatively associated with T cells
regulatory (Tregs), T cells CD8, B cells memory, macrophages M1,
dendritic cells resting and mast cells resting. FGF2 was
significantly positively associated with mast cells resting,
monocytes, NK cells resting, Tregs and significantly negatively
associated with plasma cells and mast cells activated.
TNFRSF11A was significantly positively associated with
macrophages M0 and mast cells resting, and was significantly
negatively associated with plasma cells. VIPR1 was
positively associated with dendritic cells activated, macrophages
M2, mast cells resting, monocytes, and significantly negatively
associated with T cells CD4 memory activated, dendritic cells
resting, neutrophils, plasma cells and macrophages M0 (Fig. 6C).
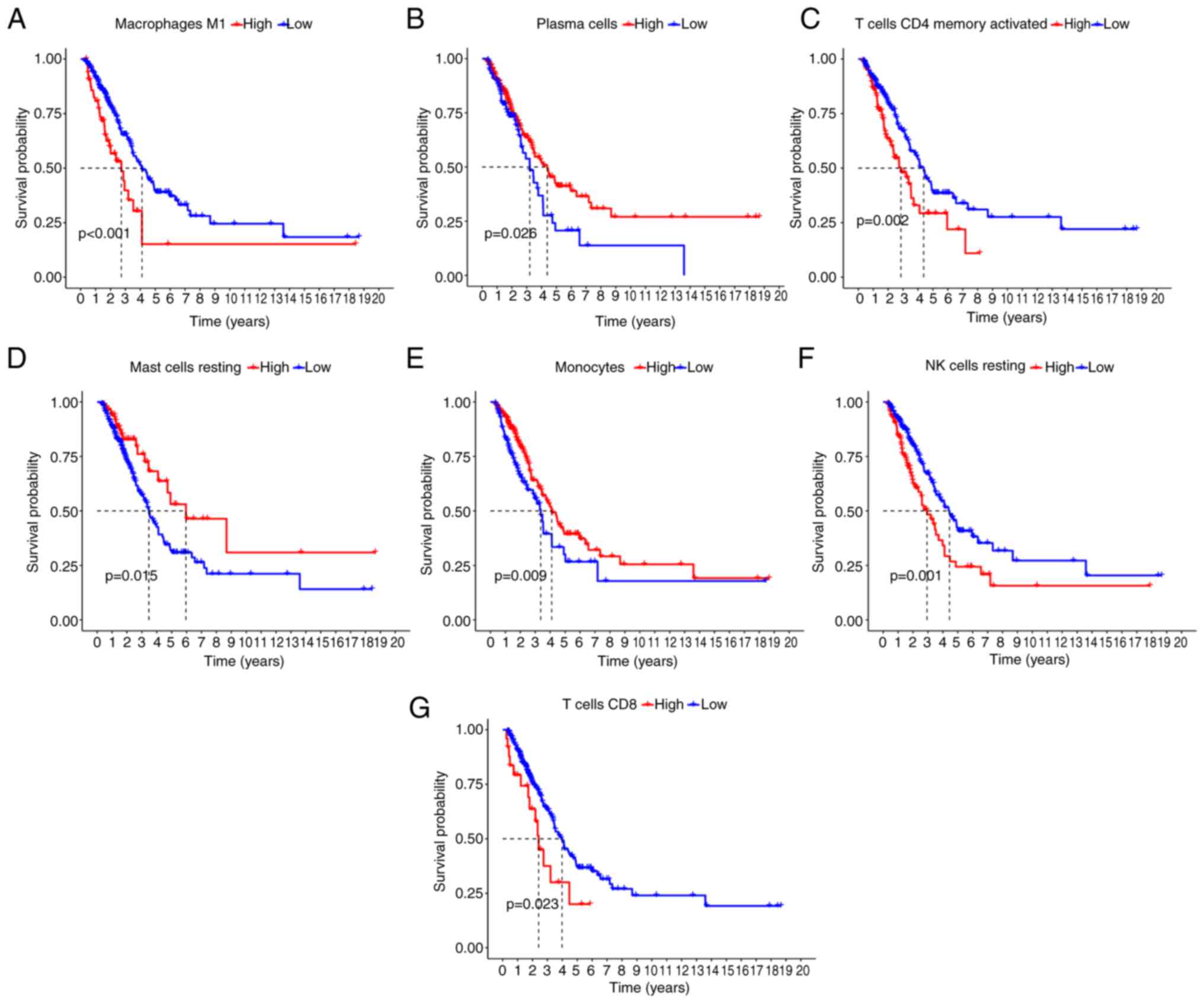
Immune-infiltrating cell survival
analysis
The prognosis of patients with LUAD with high
expression of macrophages M1, T cells CD4 memory activated, NK
cells resting and T cells CD8 activated was significantly worse
compared with the low expression group (Fig. 7A-D). However, the low expression of
plasma cells, mast cells resting and monocytes indicated a
significantly worse prognosis compared with the high expression
group (Fig. 7E-G).
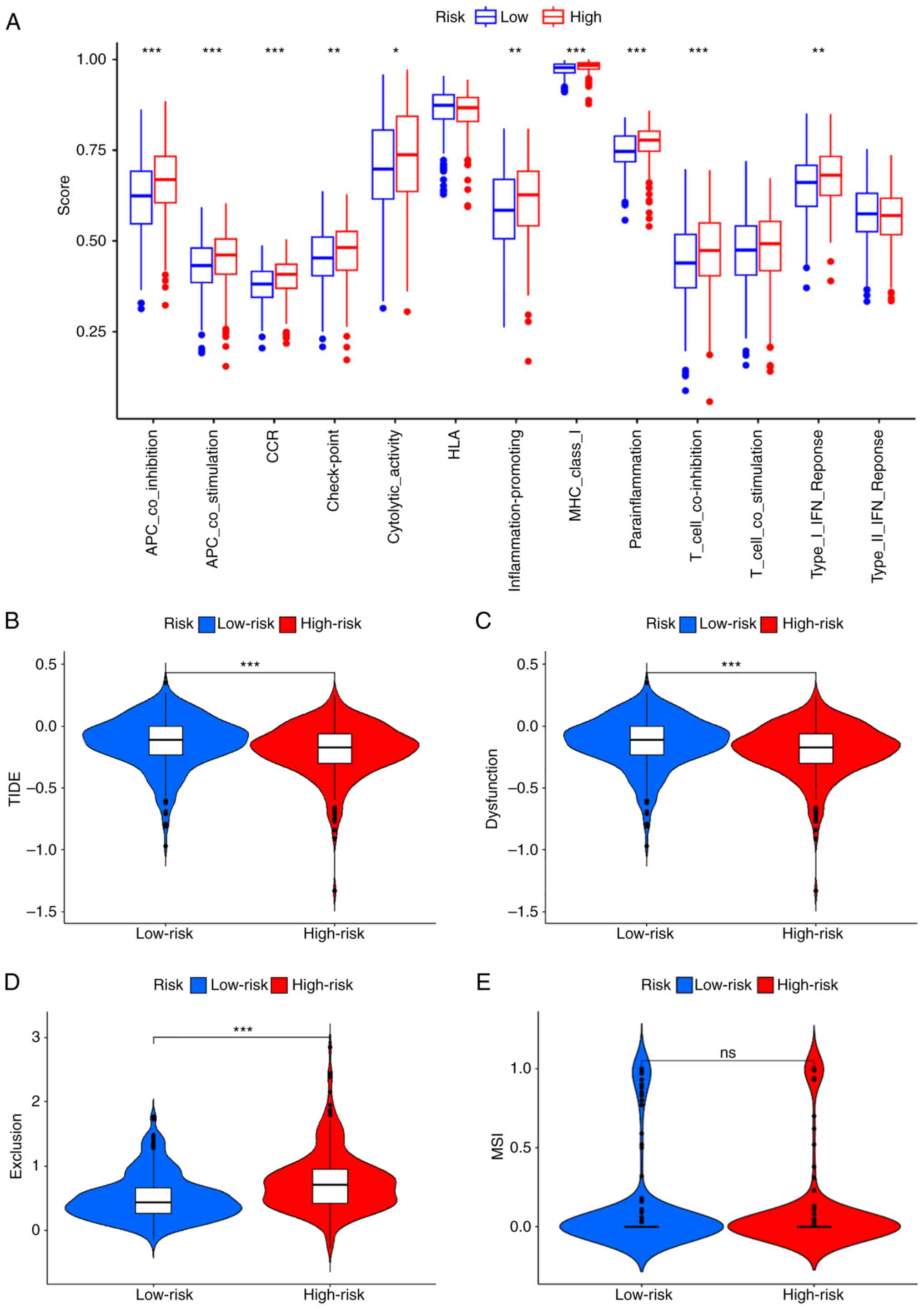
Immune-related functional
analysis
Antigen presenting cell (APC) co-stimulation, APC
co-inhibition, chemokine receptor (CCR), check-point,
cytolytic-activity, inflammation-promoting, major
histocompatibility complex (MHC)-class-I, parainflammation,
T-cell-co-inhibition and Type-I–IFN-Response immune function were
expressed at a significantly higher level within the high-risk
group (Fig. 8A). Most tumor
proliferation-associated function was improved in the high-risk
group, which suggested that tumor proliferation was more active
(Fig. 8A). The risk model tumor
immune escape analysis demonstrated significantly lower TIDE and
dysfunction scores within the high-risk group compared with the
low-risk group, however the exclusion score was significantly
higher in the high-risk group compared with the low-risk group
(Fig. 8B-E).
Discussion
Non-small cell lung cancer is the leading cause of
lung cancer related deaths worldwide (18). LUAD is the primary lung cancer
subtype with specific epidemiological, molecular and clinical
features (19). Currently, lung
cancer treatment comprises surgery, radiotherapy, chemotherapy
(20), targeted drug therapy and
immunotherapy (2). For patients
with metastatic lung cancer, conventional radiotherapy and
chemotherapy may have limited efficacy (21). Tumor immunotherapy is a promising
treatment for use following surgery, radiotherapy, chemotherapy or
targeted therapy. Previous clinical studies have reported that many
tumors are insensitive to immunotherapy due to the immune tumor
microenvironment, with immune genes serving a vital role in this
insensitivity. Previous studies have reported that CD3, IL-12 and
IL-17 expression in the tumor stroma is significantly linked with
postoperative recurrence of early LUAD (22–24).
Furthermore, many immune factors and cells (such as, neutrophils,
macrophages and lymphocytes) are associated with angiogenesis, cell
proliferation and invasion in LUAD (25). Therefore, the elucidation of
immune-related genes with biological significance and assessment of
their prognostic value could aid the diagnosis and treatment of
patients with LUAD.
Previous studies have demonstrated that abnormally
expressed genes can be utilized as prognostic LUAD markers
(26–28). However, only a few previous studies
have reported the abnormally expressed immune genes and their
immune infiltration role (29–31).
The present study first identified six immune-related genes closely
related to the prognosis of LUAD patients, including S100A16,
FURIN, FGF2, LGR4, TNFRSF11A and VIPR1. These genes
serve an important biological role in the occurrence and
development of tumors. Among these genes, S100A16 is highly
conserved in mammals (32), and its
high expression is linked with tumor cell growth and EMT (33). S100A16 is a prognostic marker
for various tumors, including LUAD and colorectal cancer (34,35).
The present study showed that S100A16 was highly expressed
in NSCLC cells and that its high expression was associated with a
worse prognosis in LUAD. FURIN is a protein conversion
enzyme expressed in breast cancer, squamous cell carcinoma and
striated muscle tumors, which indicates that it may hold an
important role. FURIN expression is elevated in NSCLC, and
upregulated RURIN activity correlates with accelerated tumor
progression (36,37). The present study found high
FURIN expression was associated with a worse prognosis in
patients with LUAD and with an increased number of infiltrated
immune cells. FGF2 is a fibroblast growth factor family
member and exerts mitogenic activity by binding to fibroblast
factor receptors (38). It is a
tumor angiogenesis factor, and neutrophils are reported to enhance
the growth of liver metastases through FGF2-dependent angiogenesis,
converting tumor-associated macrophages into pro-tumorigenic M2
macrophages (39). The present
study demonstrated that the low FGF2 expression in LUAD
cells was closely linked with increased numbers of infiltrated
immune cells. LGR4 is a receptor of the G protein-coupled
receptors superfamily, which regulate signaling pathways in normal
and pathological processes. LGR4 is commonly activated by
R-spondins, Norrin and receptor activators of NF-κβ ligand.
LGR4 activation leads to signaling in the Wnt/β-catenin and
G protein-associated pathways (40). LGR4 is highly expressed in
several cancers, enhances tumorigenesis and metastasis, regulates
cancer stem cells and is linked with poor patient prognosis
(41–43). The present study showed a high level
of LGR4 expression in NSCLC cells, with a worse prognosis
for patients with LUAD. TNFRSF11A is a crucial regulator of
cell differentiation, proliferation and survival, an inducer for
the activation of dendritic cells, and a critical factor in
maintaining immune tolerance (44).
TNFRSF11A promotes cervical cancer cell proliferation and
inhibits cell apoptosis (45). The
present study showed high TNFRSF11A expression in LUAD cells
which was related to increased infiltrated immune cells. VIP
is a vital neuropeptide controlling lung physiology and mainly
functions through two receptor subtypes, VAPC1 and
VAPC2 (46). VIP or
VPAC1 receptor antagonist strengthens the ability of
chemotherapy to kill breast cancer cells (47) while improving anti-viral immunity
(48). VIPR1 inhibits the
growth of certain cancers including prostate cancer and
lymphoblastoma, and VIPR1 overexpression inhibits the
proliferation of LUAD H1299 cells (49). The present study found a low
VIPR1 expression in LUAD cells, which was associated with
infiltrated immune cells, and patients with low expression of VIPR1
had a worse prognosis.
The prognostic risk model generated in the present
study depicted significant prognostic differences between the high
and low-risk groups. Immune cell infiltration, immune cell-related
functions and TIDE were compared in high and low-risk patient
groups to elucidate the potential reasons behind the prognostic
differences. We jypothesize that the differences in the degree of
tumor immune cell infiltration, immune-related functions and low
TIDE score may be potential mechanisms affecting the prognosis of
patients with LUAD. The present study indicated that immune cells,
including T cells CD4 memory resting, T cells CD8, macrophages M0,
macrophages M1, plasma cells and B cells naive were dominant in
both LUAD and normal tissue. Thus, monocyte macrophages, NK cells
resting and T cells follicular helper were also expressed in LUAD.
Macrophages M1 and M2 are crucial immune cells polarized by
circulating monocytes (50).
Macrophages M1 generate reactive oxygen/nitrogen species and
pro-inflammatory factors critical for host defense and tumor cell
death. The increase in the levels of macrophages M1 has been
reported to promote the tumor-antagonistic effect of M1-polarized
macrophages and enhances the survival trend of colorectal cancer
patients (51). However,
macrophages M2 promote angiogenesis and matrix remodeling, which
enhance tumor progression and metastasis, and suppress immune
surveillance of tumor cells (52).
Although NK cells are essential for tumor immune surveillance, a
previous study reported that their ability to kill target cells and
produce IFN-γ may be decreased in NSCLC tumors (53). The current study demonstrated that
levels of NK cells activated were lower in LUAD; thus, infiltrating
NK cells is inhibited. NK cells resting in normal tissues express
more without releasing their toxic particles before non-specific
target cell recognition. In recent years, increasing evidence has
demonstrated effective interaction between NK cells and B cells
(54). Although B cells can release
cytokines with cytotoxic T-cell activity and function as effective
antigen-presenting cells, their anti-tumor role in the TME remains
debatable. Nevertheless, previous studies have described that B
cells can recruit myeloid-derived suppressor cells, improve
vascular survival by producing cytokines (55) and potentially induce tumors by
blocking cytotoxic T lymphocyte function (56).
Immune function scores, including APC-co-inhibition,
APC co-stimulation, CCR, MHC-class-I, parainflammation and
T-cell-co-inhibition were significantly elevated in the high-risk
group. Thus, the density of immune dysfunction was higher, with a
worse prognosis in the high-risk group. The present study also
assessed the difference in TIDE scores between high and low-risk
groups, and the result showed the high-risk group with a lower TIDE
score. TIDE analysis focuses on the function and status of T cells
without reflecting the complexity of the immune cell infiltration
in the TME which is associated with immunotherapy responses
(57). However, TIDE can predict
patient response to immunotherapy (58). The TIDE score calculates the
efficacy of ICIs for treating tumors. A high TIDE score indicates
poor ICI efficacy with a short survival period post-ICI treatment.
The current study demonstrated a low TIDE score within the
prognostic model for the high-risk group. Thus, LUAD tumors of the
high-risk group are less likely to escape immune killing, enhancing
their treatment using better immunotherapy. Higher TIDE scores are
linked with a greater likelihood of immune evasion and reduced
survival in patients receiving ICIs, rendering immunotherapy less
effective (14). The low-risk group
had higher immune evasion potential. This indicated that the
high-risk group could derive greater benefit from immunotherapy
with an improved prognosis.
The current study has certain limitations. First,
some of the roles and mechanisms of genes in the LUAD model remain
unclear. Therefore, further experiments are required to assess
their functions and mechanisms. Second, the study data came from
public databases and most patients did not receive immunotherapy.
Deficiencies also exist in predicting the prognosis of lung cancer
patients who underwent immunotherapy, highlighting the need for
additional clinical samples. Third, these results were primarily
obtained using bioinformatic analysis and lack clinical, in
vitro and in vivo studies to support the results.
In conclusion, the present study identified six
immune-related genes linked with patient prognosis, and a
prognostic risk model was constructed for LUAD based on
bioinformatic analysis. The survival times of patients with LUAD
could be independently predicted using the prognostic risk model.
It is anticipated that future treatments for LUAD molecular targets
will require targeting of these genes to guide the diagnosis and
treatment of LUAD patients.
Acknowledgments
The authors would like to thank Ms. Mao Yang, Miss
Wenjun Wang and Miss Xianyu Zhu (Institute for Cancer Medicine,
School of Basic Medicine Sciences, Southwest Medical University,
Luzhou, Sichuan, China) for their support in performing
experiments.
Funding
The present study was supported by the Sichuan Science and
Technology Program (grant no. 2022YFS0623), the Sichuan Science and
Technology Program Joint Innovation Grant (grant no.
2022YFS0623-B3), Southwest Medical University Grant (grant no.
2021ZKZD005) and Sichuan Science and Technology Grant (grant no.
2020YFH0121).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JY and WY designed the study, drafted the manuscript
and confirm the authenticity of all the raw data. JY performed TCGA
data analysis and drew the figures. CT performed the cellular
experiments and qPCR, and analyzed the data. CL helped perform the
cellular experiments and analyzed the data. XL helped design the
study, provided the cells and critically revised the drafted work
for content. All the authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lahiri A, Maji A, Potdar PD, Singh N,
Parikh P, Bisht B, Mukherjee A and Paul MK: Lung cancer
immunotherapy: progress, pitfalls, and promises. Mol Cancer.
22:402023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi J, Hua X, Zhu B, Ravichandran S, Wang
M, Nguyen C, Brodie SA, Palleschi A, Alloisio M, Pariscenti G, et
al: Somatic genomics and clinical features of lung adenocarcinoma:
A retrospective study. PLoS Med. 13:e10021622016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ortega MA, Pekarek L, Navarro F,
Fraile-Martínez O, García-Montero C, Álvarez-Mon MÁ, Diez-Pedrero
R, Boyano-Adánez MDC, Guijarro LG, Barrena-Blázquez S, et al:
Updated views in targeted therapy in the patient with non-small
cell lung cancer. J Pers Med. 13:1672023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Byun J, Schwartz AG, Lusk C, Wenzlaff AS,
de Andrade M, Mandal D, Gaba C, Yang P, You M, Kupert EY, et al:
Genome-wide association study of familial lung cancer.
Carcinogenesis. 39:1135–1140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schreiber RD, Old LJ and Smyth MJ: Cancer
immunoediting: Integrating immunity's roles in cancer suppression
and promotion. Science. 331:1565–1570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Joyce JA and Fearon DT: T cell exclusion,
immune privilege, and the tumor microenvironment. Science.
348:74–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karasaki T, Nagayama K, Kuwano H, Nitadori
JI, Sato M, Anraku M, Hosoi A, Matsushita H, Morishita Y,
Kashiwabara K, et al: An immunogram for the cancer-immunity cycle:
Towards personalized immunotherapy of lung cancer. J Thorac Oncol.
12:791–803. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liede A, Hernandez RK, Wade SW, Bo R,
Nussbaum NC, Ahern E, Dougall WC and Smyth MJ: An observational
study of concomitant immunotherapies and denosumab in patients with
advanced melanoma or lung cancer. Oncoimmunology. 7:e14803012018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mahalingam P and Newsom-Davis T: Cancer
immunotherapy and the management of side effects. Clin Med (Lond).
23:56–60. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo W: Nasopharyngeal carcinoma ecology
theory: Cancer as multidimensional spatiotemporal ‘unity of ecology
and evolution’ pathological ecosystem. Theranostics. 13:1607–1631.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Y, Yu Y and Lu S: Effectiveness of
PD-1/PD-L1 inhibitors in the treatment of lung cancer: Brightness
and challenge. Sci China Life Sci. 63:1499–1514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang S, Zeng Z, Liu Y, Huang J, Long J,
Wang Y, Peng X, Hu Z and Ouyang Y: Prognostic landscape of
tumor-infiltrating immune cells and immune-related genes in the
tumor microenvironment of gastric cancer. Aging (Albany NY).
12:17958–17975. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li S, Zhu R, Li D, Li N and Zhu X:
Prognostic factors of oligometastatic non-small cell lung cancer: A
meta-analysis. J Thorac Dis. 10:3701–3713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ortega MA, Navarro F, Pekarek L,
Fraile-Martínez O, García-Montero C, Saez MA, Arroyo M, Monserrat J
and Alvarez-Mon M: Exploring histopathological and serum biomarkers
in lung adenocarcinoma: Clinical applications and translational
opportunities (review). Int J Oncol. 61:1542022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leduc C, Antoni D, Charloux A, Falcoz PE
and Quoix E: Comorbidities in the management of patients with lung
cancer. Eur Respir J. 49:16017212017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan T, Lu J, Niu D, Zhang Y, Wang B, Zhang
B, Zhang Z, He X, Peng N, Li B, et al: Immune and non-immune cell
subtypes identify novel targets for prognostic and therapeutic
strategy: A study based on intratumoral heterogenicity analysis of
multicenter scRNA-seq datasets in lung adenocarcinoma. Front
Immunol. 13:10461212022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Airoldi I, Di Carlo E, Cocco C, Caci E,
Cilli M, Sorrentino C, Sozzi G, Ferrini S, Rosini S, Bertolini G,
et al: IL-12 can target human lung adenocarcinoma cells and normal
bronchial epithelial cells surrounding tumor lesions. PLoS One.
4:e61192009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Q, Duan L, Qian X, Fan J, Lv Z,
Zhang X, Han J, Wu F, Guo M, Hu G, et al: IL-17 promotes angiogenic
factors IL-6, IL-8, and Vegf production via Stat1 in lung
adenocarcinoma. Sci Rep. 6:365512016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Suzuki K, Kadota K, Sima CS, Nitadori J,
Rusch VW, Travis WD, Sadelain M and Adusumilli PS: Clinical impact
of immune microenvironment in stage I lung adenocarcinoma: Tumor
interleukin-12 receptor β2 (IL-12Rβ2), IL-7R, and stromal FoxP3/CD3
ratio are independent predictors of recurrence. J Clin Oncol.
31:490–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Domagala-Kulawik J, Kwiecien I, Pankowski
J, Pasieka-Lis M, Wolosz D and Zielinski M: Elevated Foxp3/CD8
ratio in lung adenocarcinoma metastatic lymph nodes resected by
transcervical extended mediastinal lymphadenectomy. Biomed Res Int.
2017:51850342017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma C, Li F, Wang Z and Luo H: A novel
immune-related gene signature predicts prognosis of lung
adenocarcinoma. Biomed Res Int. 2022:49958742022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu X, Zhu J, Liu W, Jin M, Xiong M and Hu
K: A novel prognostic and predictive signature for lung
adenocarcinoma derived from combined hypoxia and infiltrating
immune cell-related genes in TCGA patients. Int J Gen Med.
14:10467–10481. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang D, Ma X and Song P: A prognostic
model of non small cell lung cancer based on TCGA and ImmPort
databases. Sci Rep. 12:4372022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun L, Zhang Z, Yao Y, Li WY and Gu J:
Analysis of expression differences of immune genes in non-small
cell lung cancer based on TCGA and ImmPort data sets and the
application of a prognostic model. Ann Transl Med. 8:5502020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun S, Guo W, Wang Z, Wang X, Zhang G,
Zhang H, Li R, Gao Y, Qiu B, Tan F, et al: Development and
validation of an immune-related prognostic signature in lung
adenocarcinoma. Cancer Med. 9:5960–5975. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qi C, Ma J, Sun J, Wu X and Ding J: The
role of molecular subtypes and immune infiltration characteristics
based on disulfidptosis-associated genes in lung adenocarcinoma.
Aging (Albany NY). 15:5075–5095. 2023.PubMed/NCBI
|
|
32
|
Babini E, Bertini I, Borsi V, Calderone V,
Hu X, Luchinat C and Parigi G: Structural characterization of human
S100A16, a low-affinity calcium binder. J Biol Inorg Chem.
16:243–256. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Basnet S, Vallenari EM, Maharjan U, Sharma
S, Schreurs O and Sapkota D: An update on S100A16 in human cancer.
Biomolecules. 13:10702023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen D, Luo L and Liang C: Aberrant
S100A16 expression might be an independent prognostic indicator of
unfavorable survival in non-small cell lung adenocarcinoma. PLoS
One. 13:e01974022018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun X, Wang T, Zhang C, Ning K, Guan ZR,
Chen SX, Hong TT and Hua D: S100A16 is a prognostic marker for
colorectal cancer. J Surg Oncol. 117:275–283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bassi DE, Mahloogi H, Al-Saleem L, Lopez
De Cicco R, Ridge JA and Klein-Szanto AJ: Elevated furin expression
in aggressive human head and neck tumors and tumor cell lines. Mol
Carcinog. 31:224–232. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Braun E and Sauter D: Furin-mediated
protein processing in infectious diseases and cancer. Clin Transl
Immunology. 8:e10732019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Babina IS and Turner NC: Advances and
challenges in targeting FGFR signalling in cancer. Nat Rev Cancer.
17:318–332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Im JH, Buzzelli JN, Jones K, Franchini F,
Gordon-Weeks A, Markelc B, Chen J, Kim J, Cao Y and Muschel RJ:
FGF2 alters macrophage polarization, tumour immunity and growth and
can be targeted during radiotherapy. Nat Commun. 11:40642020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gong X, Yi J, Carmon KS, Crumbley CA,
Xiong W, Thomas A, Fan X, Guo S, An Z, Chang JT and Liu QJ:
Aberrant RSPO3-LGR4 signaling in Keap1-deficient lung
adenocarcinomas promotes tumor aggressiveness. Oncogene.
34:4692–4701. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G,
Huang J, Dai W, Li C, Zheng C, et al: LGR4 is a receptor for RANKL
and negatively regulates osteoclast differentiation and bone
resorption. Nat Med. 22:539–546. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Glinka A, Dolde C, Kirsch N, Huang YL,
Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM and Niehrs C:
LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and
Wnt/PCP signalling. EMBO Rep. 12:1055–1061. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ruffner H, Sprunger J, Charlat O,
Leighton-Davies J, Grosshans B, Salathe A, Zietzling S, Beck V,
Therier M, Isken A, et al: R-Spondin potentiates Wnt/β-catenin
signaling through orphan receptors LGR4 and LGR5. PLoS One.
7:e409762012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ono T, Hayashi M, Sasaki F and Nakashima
T: RANKL biology: Bone metabolism, the immune system, and beyond.
Inflamm Regen. 40:22020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shang WQ, Li H, Liu LB, Chang KK and Yu
JJ, Xie F, Li MQ and Yu JJ: RANKL/RANK interaction promotes the
growth of cervical cancer cells by strengthening the dialogue
between cervical cancer cells and regulation of IL-8 secretion.
Oncol Rep. 34:3007–3016. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Szilasi M, Buglyo A, Treszl A, Kiss L,
Schally AV and Halmos G: Gene expression of vasoactive intestinal
peptide receptors in human lung cancer. Int J Oncol. 39:1019–1024.
2011.PubMed/NCBI
|
|
47
|
Moody TW, Leyton J, Chan D, Brenneman DC,
Fridkin M, Gelber E, Levy A and Gozes I: VIP receptor antagonists
and chemotherapeutic drugs inhibit the growth of breast cancer
cells. Breast Cancer Res Treat. 68:55–64. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yassin MA, Datiko DG, Tulloch O, Markos P,
Aschalew M, Shargie EB, Dangisso MH, Komatsu R, Sahu S, Blok L, et
al: Innovative community-based approaches doubled tuberculosis case
notification and improve treatment outcome in Southern Ethiopia.
PLoS One. 8:e631742013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao L, Yu Z and Zhao B: Mechanism of
VIPR1 gene regulating human lung adenocarcinoma H1299 cells. Med
Oncol. 36:912019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tong Z, Wang X, Liu H, Ding J, Chu Y and
Zhou X: The relationship between tumor infiltrating immune cells
and the prognosis of patients with lung adenocarcinoma. J Thorac
Dis. 15:600–610. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Aras S and Zaidi MR: TAMeless traitors:
Macrophages in cancer progression and metastasis. Br J Cancer.
117:1583–1591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wolf D, Sopper S, Pircher A, Gastl G and
Wolf AM: Treg(s) in cancer: Friends or foe? J Cell Physiol.
230:2598–2605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Platonova S, Cherfils-Vicini J, Damotte D,
Crozet L, Vieillard V, Validire P, André P, Dieu-Nosjean MC,
Alifano M, Régnard JF, et al: Profound coordinated alterations of
intratumoral NK cell phenotype and function in lung carcinoma.
Cancer Res. 71:5412–5422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yuan D, Koh CY and Wilder JA: Interactions
between B lymphocytes and NK cells. FASEB J. 8:1012–1018. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Garaud S, Buisseret L, Solinas C,
Gu-Trantien C, de Wind A, Van den Eynden G, Naveaux C, Lodewyckx
JN, Boisson A, Duvillier H, et al: Tumor infiltrating B-cells
signal functional humoral immune responses in breast cancer. JCI
Insight. 5:e1296412019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tsou P, Katayama H, Ostrin EJ and Hanash
SM: The emerging role of B cells in tumor immunity. Cancer Res.
76:5597–5601. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tian J, Fu C, Peng Q, Yang J, Fan X, Zeng
X, Qing W and Wu Y: Construction of immune cell infiltration score
model to assess prognostic ability of tumor immune environment in
lung adenocarcinoma. Am J Transl Res. 15:1730–1743. 2023.PubMed/NCBI
|
|
58
|
Chen C, Huang H and Wu CH: Protein
bioinformatics databases and resources. Methods Mol Biol. 694:3–24.
2011. View Article : Google Scholar : PubMed/NCBI
|