Introduction
The AT-rich interacting domain-containing protein 1A
(ARID1A) is a crucial subunit of the switch/sucrose non-fermentable
(SWI/SNF) chromatin remodeling complex (1). It contains an AT-rich interaction
domain (ARID) responsible for binding to DNA (2) and features multiple LXXLL motifs in
the C-terminal region, known to facilitate interactions with
nuclear hormone receptors (3).
ARID1A helps direct SWI/SNF complexes to specific chromatin targets
by interacting with other transcriptional regulatory factors and
enhancing the affinity for binding to these sites, thereby altering
chromatin accessibility for several nuclear factors (4). ARID1A serves essential roles in
chromatin remodeling, chromosome organization, transcriptional and
epigenetic regulation (5,6).
ARID1A is a tumor suppressor gene that has been
reported to undergo mutations in several types of cancer (7–9). These
mutations in ARID1A include missense or truncating mutations, as
well as in-frame insertions or deletions (indels), which contribute
to the loss of protein function (8,9).
Previous studies have demonstrated the participation of ARID1A in
several biological processes involved in carcinogenesis and tumor
progression, such as cell cycle control, modulation of the immune
response and regulation of several signaling pathways (10,11).
ARID1A mutations are found in 3.6–66.7% of
colorectal cancer (CRC) cases, and they are thought to be caused by
mismatch defects (1,12). Previous studies have reported the
prognostic and tumor suppressive roles of ARID1A in CRC.
Immunohistochemical analyses have revealed a relatively high
incidence of ARID1A protein loss (25.8%) in primary CRC tumors.
Loss of ARID1A protein expression is associated with
clinicopathological characteristics, including advanced
tumor-node-metastasis stage, distant metastasis, poor pathological
differentiation and worse overall survival in patients with CRC
(13,14). Additionally, ARID1A expression is
associated with immune cell infiltration and immunotherapeutic
response in patients with CRC (15). ARID1A knockdown enhances the
proliferation, migration, invasion and chemoresistance of CRC cells
(16,17). Furthermore, ARID1A has been reported
to promote the epithelial-mesenchymal transition (EMT) process by
regulating the expression of vimentin and E-cadherin (18,19).
However, the precise molecular mechanisms through which ARID1A
regulates CRC carcinogenesis and progression are not yet fully
understood.
Proteomics is a powerful tool that enables the
comprehensive analysis of global protein changes in biological
samples. It has been widely used in cancer research to unravel the
molecular mechanisms of diseases and facilitate the discovery of
biomarkers and therapeutic targets (20,21).
In the present study, a comparative proteomic analysis of
ARID1A-overexpressing colon cancer cells and empty vector control
cells was performed. Subsequently, bioinformatics analysis was
performed to gain novel insights into the molecular mechanisms
through which ARID1A is involved in CRC carcinogenesis and
progression. Moreover, the potential role of ARID1A in the
regulation of the Wnt signaling pathway was assessed using western
blotting.
Materials and methods
Establishment of ARID1A-overexpressing
SW48 cells
The pLenti-puro (cat. no. 39481; Addgene, Inc.) and
pLenti-puro-ARID1A (cat. no. 39478; Addgene, Inc.) were donated by
Professor Ie-Ming Shih from John Hopkins University (22). The plasmid vectors were cloned into
One Shot™ Stbl3™ Chemically Competent E.coli (cat. no. C737303;
Invitrogen™; Thermo Fisher Scientific, Inc.) and extracted using
the QIAprep Spin Miniprep Kit (Qiagen GmbH). Vector sizes were
confirmed using 1% agarose gel electrophoresis and visualized using
RedSafe™ (Intron Biotechnology, Inc.). SW48 and 293T cells were
purchased from the American Type Culture Collection. Both cells
were cultured in DMEM supplemented with 10% FBS and
penicillin/streptomycin (100 mg/ml; Invitrogen™; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2 in a humidified
chamber. Using a second-generation lentivirus system, 293T cell
transfection was performed using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 16 h at 37°C. For
each transfection, 1.5 µg of either the pLenti-puro-ARID1A or
pLenti-puro plasmid, 1 µg of packaging plasmid (psPAX2; cat. no.
12260; Addgene, Inc.), and 1 µg of envelope plasmid (pCMV–VSV-G;
cat. no. 8454; Addgene, Inc.) (23)
were used. The viral supernatants were harvested 48 h
post-transfection, centrifuged at 2,000 × g for 5 min, and filtered
through 0.45-µm pore size membrane filters. Lentivirus at a
multiplicity of infection value of 5 was used to transduce SW48
cells with an 8-mg/ml polybrene reagent (Sigma-Aldrich; Merck
KGaA). After 24 h of transduction, the medium was replaced with a
selection medium (culture medium supplemented with puromycin at a
final concentration of 500 ng/ml; Invitrogen; Thermo Fisher
Scientific, Inc.) to establish stable ARID1A-overexpressing cells.
The stable cells were maintained in the selection medium (500 ng/ml
puromycin).
Western blot analysis
The cells were lysed using RIPA buffer (Abcam) and
protein concentration was determined using a Bradford assay
(Bio-Rad Laboratories, Inc.). Equal amounts of protein from each
sample (20 µg) were separated by 12% SDS-PAGE and transferred onto
a nitrocellulose membrane. The membrane was incubated with 5%
skimmed milk in PBS at 25°C for 1 h. Subsequently, it was incubated
with rabbit polyclonal anti-ARID1A (1:1,000; cat. no. HPA005456;
Sigma-Aldrich; Merck KGaA), rabbit monoclonal anti-c-Myc (1:1,000;
cat. no. 5605; Cell Signaling Technology, Inc.), rabbit monoclonal
anti-T cell factor (TCF)1/7 (1:1,000; cat. no. 2203; Cell Signaling
Technology, Inc.), rabbit monoclonal anti-cyclin D1 (1:1,000; cat.
no. 2978; Cell Signaling Technology, Inc.), rabbit polyclonal
anti-zinc and ring finger 3 (ZNRF3; 1:1,000; cat. no. DF14289;
Affinity Biosciences), or mouse monoclonal anti-GAPDH (1:5,000;
cat. no. ab8245; Abcam) antibodies at 4°C overnight. Following
three washes with PBS containing 0.05% Tween 20 (PBS-T), the
membrane was further incubated with corresponding horseradish
peroxidase-conjugated secondary anti-rabbit IgG (1:2,000; cat. no.
65-6120; dilution, Invitrogen; Thermo Fisher Scientific, Inc.) or
anti-mouse IgG (1:5,000; cat. no. ab205719; Abcam) antibodies at
25°C for 1 h. Following three additional washes with PBS-T,
immunoreactive bands were visualized using Clarity™ Western ECL
Substrate (Bio-Rad Laboratories, Inc.) and imaged using the
ChemiDoc™ Touch Imaging System (Bio-Rad Laboratories, Inc.). The
intensity of immunoreactive bands was assessed using Image Lab
software (version 5.1; Bio-Rad Laboratories, Inc.). GAPDH was used
as a loading control. The band intensity of the target protein was
normalized to the corresponding loading control, and the relative
value was subjected to statistical analysis.
Sample preparation for shotgun
proteomics
The cells were placed in a 6-well plate at a density
of 200,000 cells per well and cultured in a complete growth medium
(DMEM supplemented with 10% FBS and 0.5 µg/ml puromycin) for 48 h
at 37°C. Following two washes with PBS, the cells were lysed with
0.5% SDS for effective extraction of both membrane-bound and
cytoplasmic proteins. Subsequent precipitation with acetone was
performed to remove contaminants and concentrate the proteins. The
lysate was mixed with 2 volumes of cold acetone (−20°C) and left to
incubate at −20°C for 12 h. Centrifugation at 10,000 × g for 15 min
at 4°C was used to collect protein precipitates. The resulting
pellets were air-dried and stored at −20°C until they were used for
subsequent proteomic analysis. The protein concentration was
determined using the Lowry assay with BSA as a standard protein
(24). Protein samples (5 µg) were
subjected to in-solution digestion. The samples were dissolved in
10 mM ammonium bicarbonate, disulfide bonds were reduced using 5 mM
dithiothreitol in 10 mM ammonium bicarbonate at 60°C for 1 h, and
sulfhydryl groups were alkylated using 15 mM iodoacetamide in 10 mM
ammonium bicarbonate at room temperature for 45 min in the dark.
Sequencing grade porcine trypsin (at a 1:20 ratio) was used to
digest the protein sample for 16 h at 37°C. The resulting tryptic
peptides were dried using a speed vacuum concentrator and
reconstituted in 0.1% formic acid for nano-liquid
chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
LC/MS-MS
The LC/MS-MS method was selected to achieve high
sensitivity and resolution for peptide separation and
identification. Label-free quantitative proteomic analysis was
chosen due to its ability to provide a comprehensive and
reproducible analysis of protein expression levels in complex
biological samples without the need for expensive labeling reagents
and intricate sample preparation (25,26).
The tryptic peptide samples were prepared for injection into the
UltiMate™ 3000 Nano/Capillary LC System (Thermo Fisher Scientific,
Inc.) coupled to a ZenoTOF 7600 mass spectrometer (SCIEX). Briefly,
1 µl peptide digests underwent enrichment on a µ-Precolumn (300 µm
i.d. × 5 mm) containing Acclaim™ PepMap™ 100 C18 HPLC column (5 µm;
100 A; Thermo Fisher Scientific, Inc.). Subsequently, separation
occurred on a 75 µm I.D. × 15 cm Acclaim PepMap RSLC C18 column (2
µm, 100Å; nanoViper; Thermo Fisher Scientific, Inc.). The C18
column was maintained in a thermostatted column oven set to 60°C.
Solvents A and B containing 0.1% formic acid in water and 0.1%
formic acid in 80% acetonitrile, respectively, were supplied to the
analytical column. A gradient of 5–55% solvent B was used to elute
the peptides at a constant flow rate of 0.30 µl/min over a period
of 30 min. The ZenoTOF 7600 system consistently used specific
source and gas settings throughout all acquisitions. These settings
included maintaining ion source gas 1 at 8 psi, curtain gas at 35
psi, CAD gas at 7 psi, a source temperature of 200°C, positive
polarity and a spray voltage set to 3,300 V. The top 50 precursor
ions with the highest abundance from the survey MS1 were chosen.
MS2 spectra were collected in the 100-1,800 m/z range with a 50
msec accumulation time, used the Zeno trap. LC-MS analysis of each
sample was performed in triplicate.
MS data analysis
Protein in individual samples was quantified using
MaxQuant 2.2.0.0 (27), using the
andromeda search engine to match MS/MS spectra with the UniProt
Homo sapiens database (https://www.uniprot.org/proteomes/UP000005640)
(27). Label-free quantitation was
performed using standard settings, including parameters such as
maximum of two miss cleavages, a mass tolerance of 0.6 daltons for
main search, trypsin as the digesting enzyme, carbamidomethylation
of cysteine as a fixed modification, and the oxidation of
methionine and acetylation of the protein N-terminus as variable
modifications. Protein identification required peptides with ≥7
amino acids and one unique peptide. Identified proteins with ≥2
peptides, including one unique peptide, were considered for further
data analysis. Protein false discovery rate was maintained at 1%,
determined using reversed search sequences. The maximal number of
modifications per peptide was set to 5. The Homo sapiens
proteome from UniProt served as the search FASTA file, whilst
potential contaminants listed in the contaminants.fasta file
provided by MaxQuant were automatically included to the search
space. The resulting MaxQuantProteinGroups.txt file was imported
into Perseus version 1.6.6.0 (28),
and contaminants unrelated to any UPS1 protein were eliminated from
the dataset. The three maximum intensities for each identified
protein were used for statistical comparisons using an unpaired
t-test with P<0.05 considered to indicate a statistically
significant difference. The log2 ratios of the
ARID1A-overexpressing sample and the control sample were
calculated. Any intensity value of 0 was set to 0.001 to prevent
division by 0. Differentially altered proteins between the
ARID1A-overexpressing sample and the control sample were identified
using a threshold of P<0.05 and a log2 ratio of ≥ 2
or ≤-2.
Bioinformatics analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.ncifcrf.gov/summary.jsp) online tool
(version v2023q4) (29,30) was used to perform Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analyses on differentially altered proteins. P<0.05
was considered to indicate a statistically significant enrichment.
Bubble plot visualization of the enrichment results was performed
using the GraphBio online tool (http://www.graphbio1.com/en/) (31). Interaction network analysis of
altered proteins enriched in the Wnt signaling pathway was
performed using GeneMANIA version 3.6.0 (https://genemania.org/) (32) with parameters set as follows:
Organism ‘Homo sapiens (human)’ and network weighting
‘Biological process based’. Survival analysis and expression
correlation of ARID1A with ZNRF3 were analyzed using The Cancer
Genome Atlas-colon adenocarcinoma (TCGA-COAD) dataset (33) using the Gene Expression Profiling
Interactive Analysis 2 database (http://gepia2.cancer-pku.cn/) with default parameters
(34).
Statistical analysis
GraphPad Prism version 8.0.1 (Dotmatics) was used
for statistical analysis. An unpaired t-test was used to compare
two datasets. P<0.05 was considered to indicate a statistically
significant difference.
Results
Establishment of ARID1A-overexpressing
SW48 cells
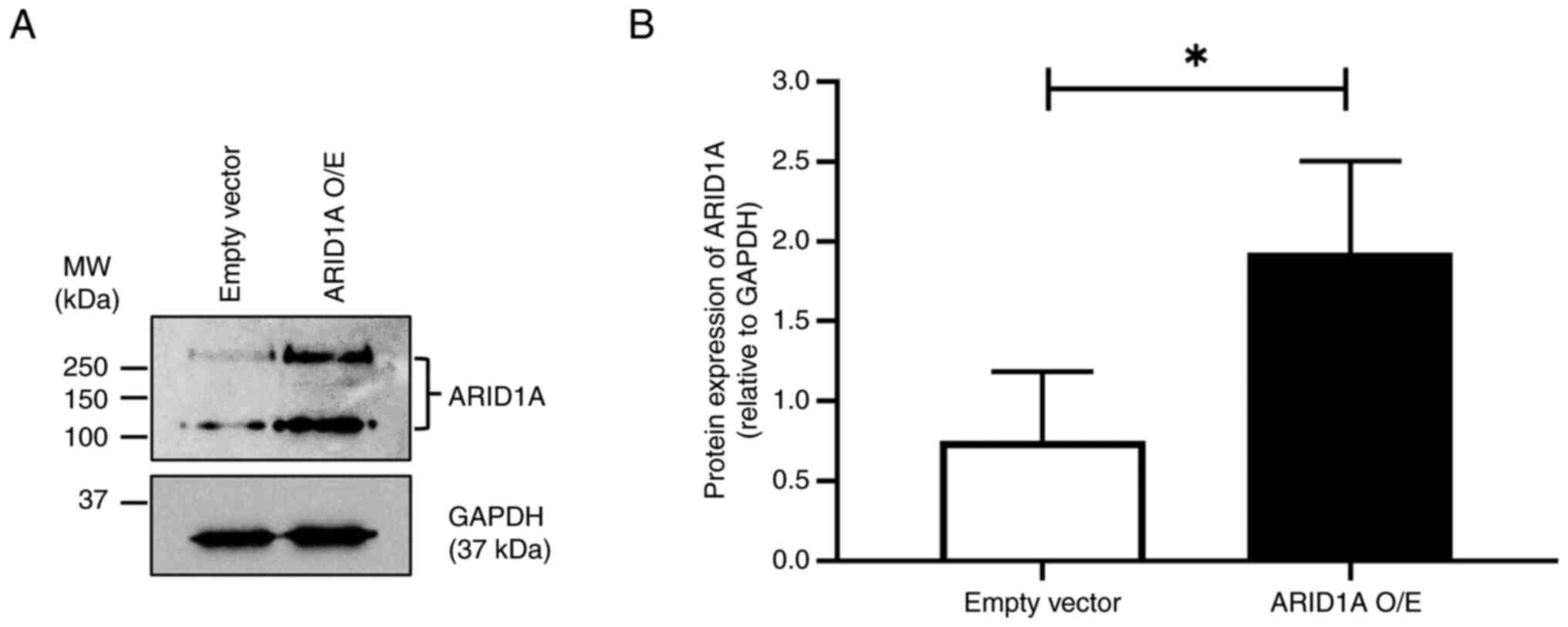
Following selection with puromycin, the level of
ARID1A protein in the empty vector control and
ARID1A-overexpressing cells was determined by western blotting. The
results demonstrated that the ARID1A protein level was
significantly higher in ARID1A-overexpressing cells compared with
the empty vector control, confirming the successful overexpression
of ARID1A in the cells (Fig.
1).
Proteomic analysis of altered proteins
in ARID1A- overexpressing cells
Proteomic analysis was performed to identify a set
of proteins differentially expressed following ARID1A
overexpression. Protein samples derived from ARID1A-overexpressing
and empty vector control cells were subjected to label-free
quantitative analysis by LC-MS/MS. The proteomic analysis
identified a total of 705 differentially altered proteins,
including 310 significantly increased proteins (P<0.05;
log2 ratio of ≥2) and 395 significantly decreased
proteins (P<0.05; log2 ratio of ≤-2) in the
ARID1A-overexpressing cells compared with the empty vector control
cells. Detailed information on differentially altered proteins is
presented in Table SI.
Functional enrichment analysis of
differentially altered proteins
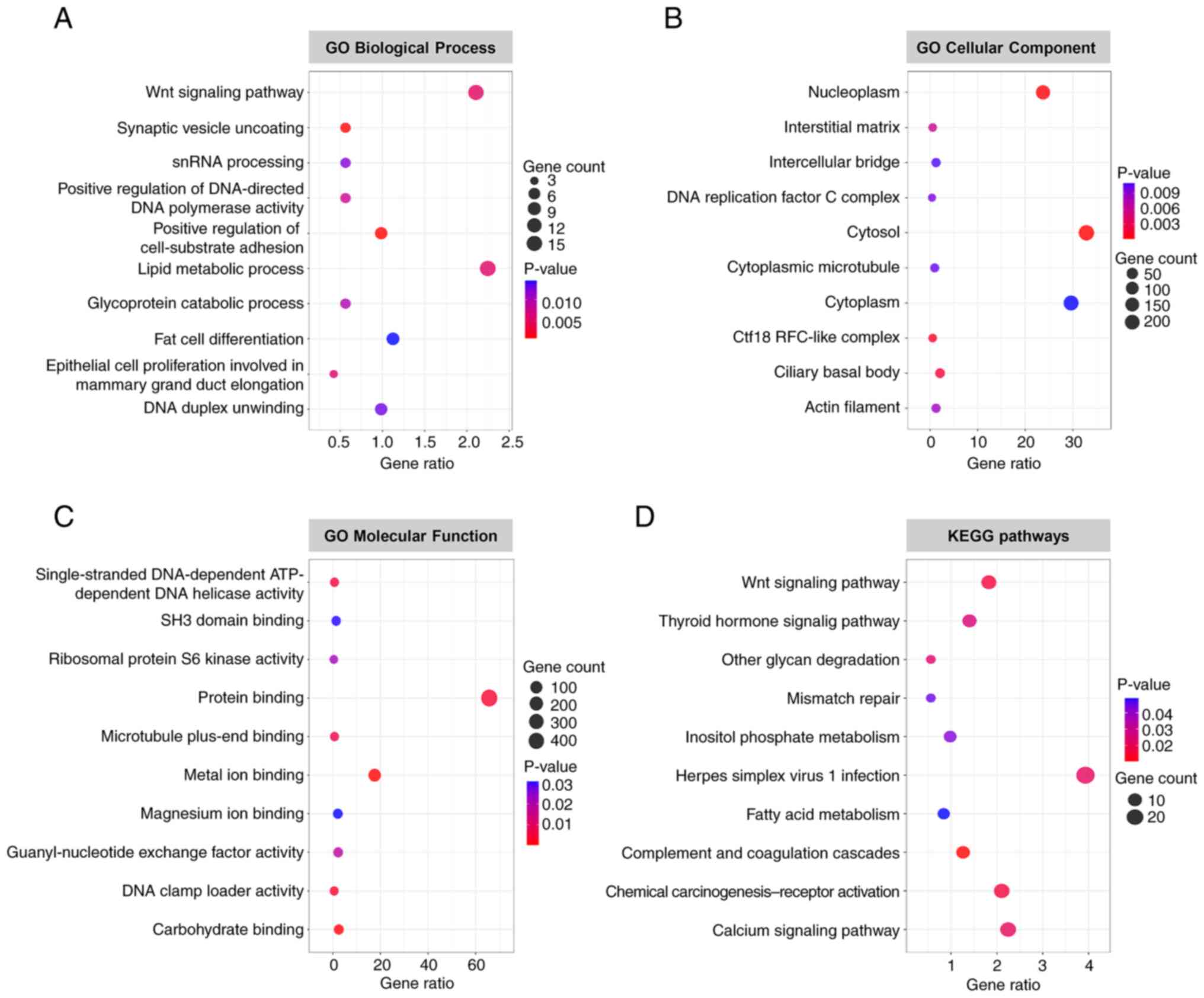
To assess the potential functions of ARID1A in colon
cancer cells, GO and KEGG pathway enrichment analyses were
performed on the differentially altered proteins. The results from
DAVID revealed that differentially altered proteins in
ARID1A-overexpressing cells were significantly enriched in several
biological processes, including the ‘Wnt signaling pathway’, ‘lipid
metabolic process’, and ‘fat cell differentiation’ (Fig. 2A). The majority of these altered
proteins were located in the ‘cytosol’, ‘cytoplasm’ and
‘nucleoplasm’ (Fig. 2B). This
analysis also identified several significantly enriched GO terms
for molecular function, such as ‘protein binding’, ‘metal ion
binding’ and ‘carbohydrate binding’ (Fig. 2C). KEGG pathway analysis indicated
that these altered proteins were enriched in several pathways,
including ‘herpes simplex virus 1 infection’, ‘calcium signaling
pathway’, ‘chemical carcinogenesis-receptor activation’ and ‘Wnt
signaling pathway’ (Fig. 2D).
Detailed information on significantly enriched GO terms and KEGG
pathways is summarized in Table
SII.
Involvement of ARID1A and altered
proteins in the Wnt signaling pathway
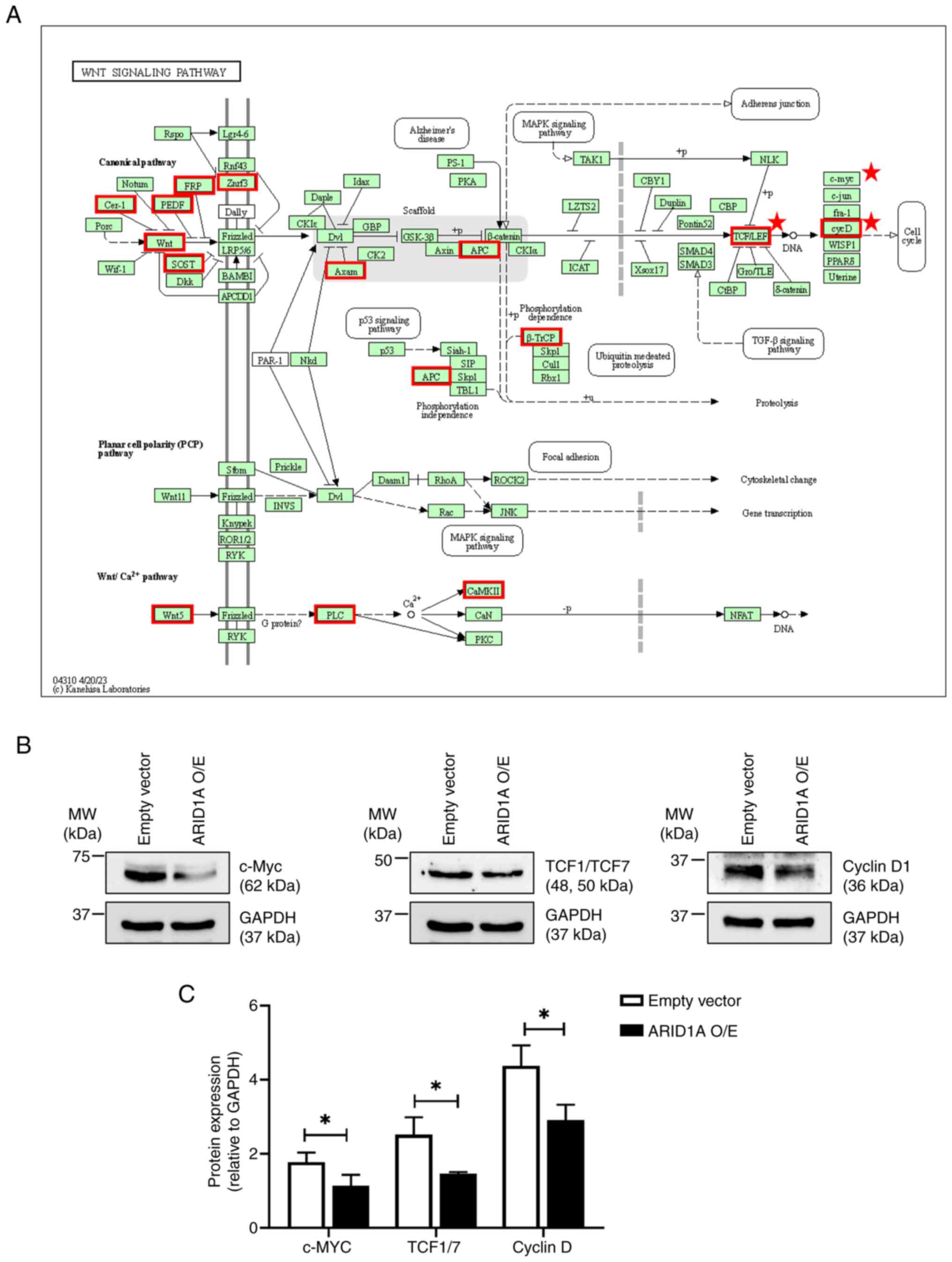
Functional enrichment analysis revealed the
potential involvement of ARID1A in the Wnt signaling pathway
(Fig. 3A). To assess this finding,
the protein levels of Wnt-target genes, including c-Myc, TCF1/TCF7
and cyclin D1 were compared between the ARID1A-overexpressing cells
and the empty vector control cells. The western blotting results
demonstrated that ARID1A overexpression resulted in significant
decreases of these Wnt-target genes compared with the empty vector
control cells, suggesting a potential regulatory role of ARID1A in
the Wnt signaling pathway (Fig. 3B and
C).
Relationship between ARID1A and
altered proteins in the Wnt signaling pathway
To gain a more comprehensive understanding of the
roles of ARID1A in the aforementioned pathway, an interaction
network was constructed using GeneMANIA to assess the relationships
between ARID1A and the altered proteins involved in the Wnt
signaling pathway. The results demonstrated that these proteins
interact with each other mainly through ‘Physical interactions’
(77.64%), ‘Co-expression’ (8.01%) and ‘Predicted’ interactions
(5.37%). Notably, ARID1A was demonstrated to directly interact with
transcription factor 7 like 1 (TCF7L1) and ZNRF3 through ‘Pathway’
and ‘Co-expression’, respectively (Fig.
4A). Further analyses using the TCGA-COAD public dataset
revealed that only ZNRF3, a negative regulator of the Wnt signaling
pathway (35), had prognostic
significance (Fig. 4B). Moreover,
patients with COAD with a low ZNRF3 expression demonstrated
significantly worse overall survival compared with those with a
high expression (Fig. 4C). ZNRF3
expression was also significantly correlated with ARID1A expression
in the database (Fig. 4D). These
findings highlight the potential relationship between ARID1A and
ZNRF3 in CRC carcinogenesis and progression.
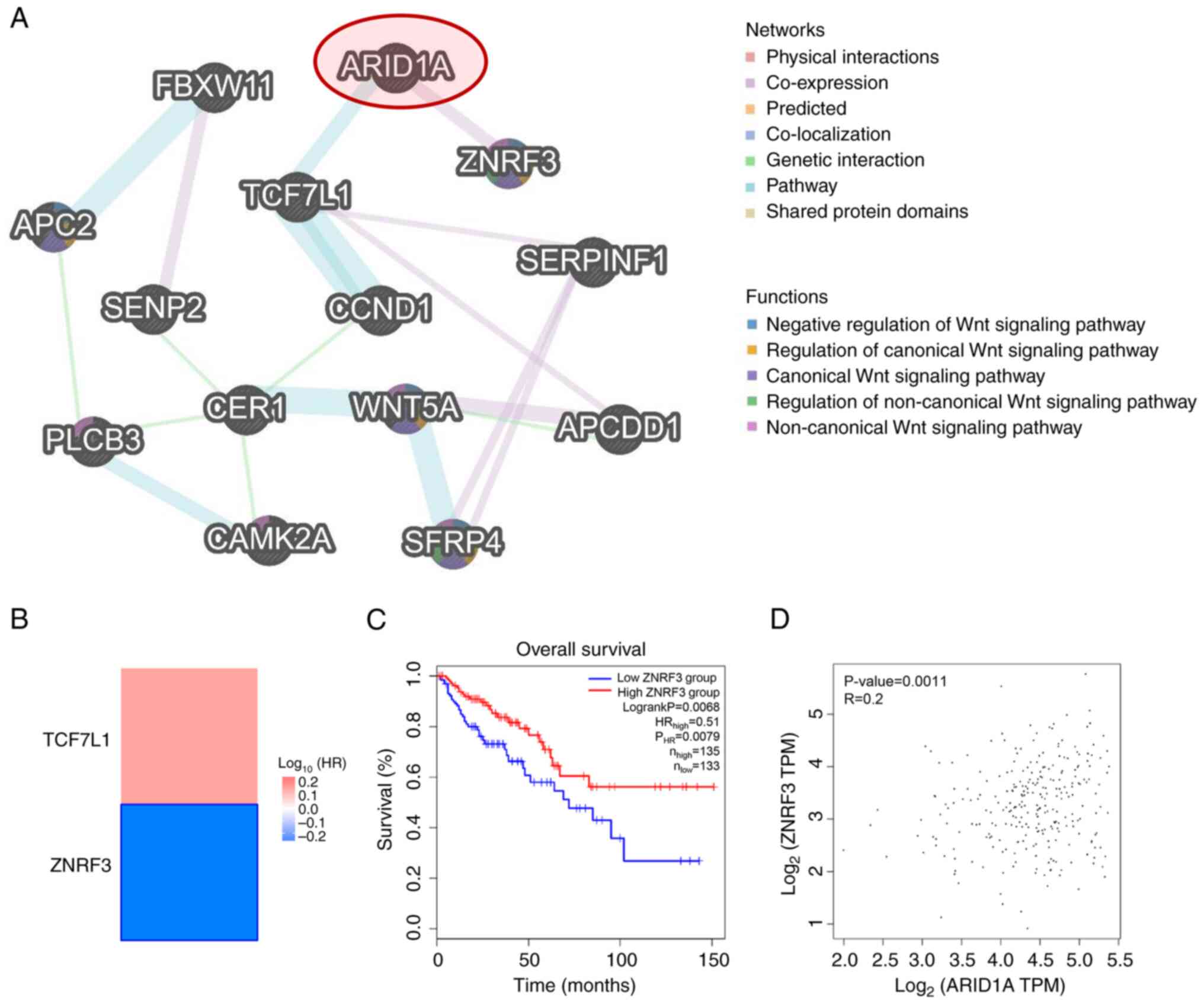 | Figure 4.Association between ARID1A and
altered proteins involved in the Wnt signaling pathway. (A)
Interaction network of ARID1A and altered proteins involved in the
Wnt signaling pathway constructed using GeneMANIA. (B) Heatmap of
the HR of TCF7L1 and ZNRF3 for the overall survival of patients
with COAD, created using GEPIA2. The framed block indicates a
significant prognostic value. (C) Kaplan-Meier curve for overall
survival of patients with COAD with different levels of ZNRF3
expression, plotted using GEPIA2. (D) Correlation between the
expression of ARID1A and ZNRF3 in COAD, analyzed using GEPIA2.
ARID1A, AT-rich interacting domain-containing protein 1A; TCF7L1,
transcription factor 7 like 1; ZNRF3, zinc and ring finger 3; COAD,
colon adenocarcinoma; GEPIA2, Gene Expression Profiling Interactive
Analysis 2; HR, hazard ratio; TPM, transcripts per million. |
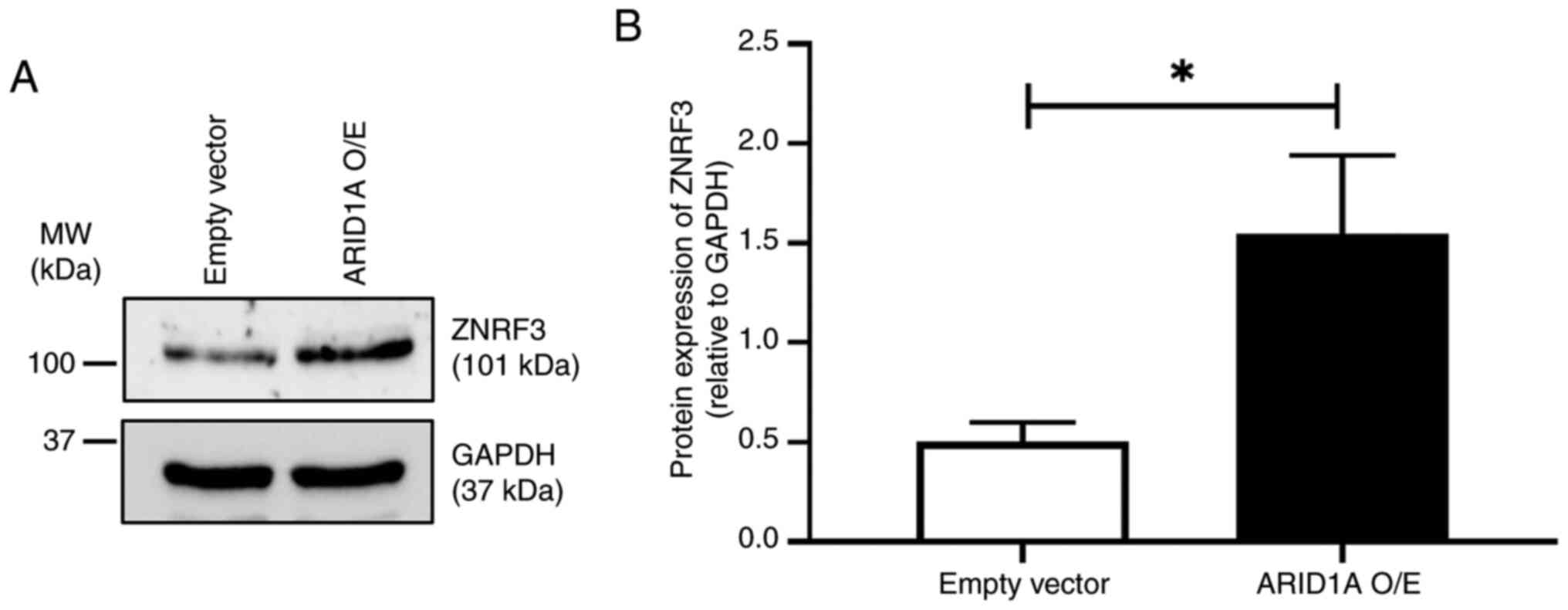
Expression of ZNRF3 protein in
ARID1A-overexpressing cells
Finally, the relationship between ARID1A and ZNRF3
was evaluated in the cells using western blotting. The results
revealed that ARID1A-overexpressing cells had a significantly
higher level of ZNRF3 protein expression compared with the empty
vector control, suggesting a positive regulatory axis between these
proteins (Fig. 5).
Discussion
ARID1A is an essential component of the SWI/SNF
chromatin remodeling complex, which serves a crucial role in
modulating the structure and accessibility of DNA (18). The diverse functions of ARID1A have
been extensively documented in previous studies. These functions
include chromatin remodeling, transcriptional regulation (5), oncogenic and tumor suppressor activity
(36), microsatellite instability
and immune checkpoint blockade (37), cell proliferation and
differentiation (38), and
epigenetic regulation (6). However,
the specific functions of ARID1A can vary depending on the cell
type and the cellular context (5,6,36–38).
Thus, an understanding of its precise function in a specific cancer
type is necessary.
In the present study, ARID1A-overexpressing SW48
cells were successfully established using lentivirus transduction.
Furthermore, western blotting demonstrated two immunoreactive bands
of ARID1A. ARID1A has a theoretical molecular weight of ~242 kDa
(UniProt accession no. O14497), corresponding to the upper band
detected at 250 kDa in the present study; however, several studies
have reported multiple bands of ARID1A using western blotting
(11,39,40).
Therefore, it appears that the multiple bands detected represent
ARID1A and possibly its degraded forms.
Previous studies have reported that ARID1A knockdown
promotes carcinogenic behaviors (16–18),
whilst its overexpression reverses such effects in CRC cells
(19). These findings underscored
the tumor suppressive role of ARID1A in CRC cells. Consequently,
the present study primarily focused on elucidating its underlying
mechanisms. Through proteomic analysis, 705 proteins that were
differentially altered in ARID1A-overexpressing cells compared with
empty vector control cells were identified. Functional enrichment
analysis revealed a significant enrichment of these altered
proteins in the Wnt signaling pathway, suggesting a potential
functional association between ARID1A and this pathway.
Label-free quantitative proteomic analysis was
chosen for the present study due to its cost-effectiveness and
ability to provide comprehensive protein expression profiles
without intricate sample preparation; however, it has limitations
such as susceptibility to technical variability and lower
sensitivity compared to labeled methods (25,26),
potentially missing low-abundance proteins. To address these issues
in future studies, rigorous sample preparation, use of internal
standards, and validation with complementary techniques such as
targeted proteomics or western blotting are recommended. Advances
in instrumentation and software are expected to further enhance the
reliability and accuracy of label-free proteomics (25,26).
The Wnt signaling pathway has been extensively
studied and is known to serve a crucial role in several cellular
processes associated with cancer, including cell proliferation,
apoptosis and EMT (41). A previous
study reported that the loss of ARID1A inhibited canonical
WNT/β-catenin activity in human gastric cancer organoid lines
(42). Downregulation of ARID1A and
the Notch/Wnt signaling pathway-related genes have also been
reported in gynecological tumors (43). In addition, ARID1A deletion in the
intestines of mice in a previous study led to a decrease in the
expression of target genes associated with the Wnt signaling
pathway, such as achaete-scute family bHLH transcription factor 2,
SRY-box transcription factor 9, axin 2, transcription factor 4, and
hes family bHLH transcription factor 1 (44). The data of the present study
demonstrated that ARID1A overexpression resulted in a decreased
expression of the Wnt-target genes in the cells. These findings
suggest that ARID1A may exert its tumor suppressive activity by
regulating the Wnt signaling pathway in CRC cells. However, the
expression of WNT/β-catenin signaling components, such as β-catenin
and its phosphorylated form, adenomatous polyposis coli, or
serine/threonine kinase GSK-3 and its phosphorylated form, were not
assessed in the present study. Clarifying the involvement of these
key markers in future studies would be essential to validate the
proposed relationship between ARID1A and the Wnt signaling pathway
in CRC.
Through proteomic analysis, a significant increase
of cyclin D1 in ARID1A-overexpressing cells was identified in the
present study; however, its decreased level was detected by western
blotting. Whilst this specific discrepancy in cyclin D1 levels
between LC-MS/MS and western blotting has not been reported before,
such discrepancies between these techniques are common in proteomic
studies (45–48). For instance, in a study by Pino
et al (47), discrepancies
between proteomic and western blot results were observed for
several proteins, which the authors attributed to differences in
sample preparation and detection techniques. Similarly, Zhang et
al (48) reported discrepancies
between 2D-difference gel electrophoresis and western blotting for
acyl-coA thioesterase 2. The study attributed these differences to
the low expression levels and issues with antibody specificity.
LC-MS/MS and western blotting differ in sample preparation,
detection methods and quantification approaches. LC-MS/MS
quantifies protein levels using retention time, mass-to-charge
ratio and ion intensities, whereas western blotting uses antibodies
to detect specific proteins (25,26).
The most likely explanation for this discrepancy is that cyclin D1
expression is influenced by post-translational modifications (PTMs)
and isoforms. Cyclin D1 is a key regulator in cell cycle control
and exists in two major isoforms (cyclin D1a and b) along with
other truncating variants and PTMs (49,50).
Aberrant expressions of cyclin D1 isoforms and variants are
involved in the pathogenesis of cancer (49,50).
Therefore, it is plausible that the observed increase in protein
abundance by LC-MS/MS may include specific isoforms or PTMs that
were not semi-quantified by western blotting. The observed
discrepancy in the present study may indicate a potential
alteration of cyclin D1 isoforms and PTMs induced by ARID1A
overexpression.
Among the altered proteins associated with the Wnt
signaling pathway, ARID1A was revealed to directly interact with
TCF7L1 and ZNRF3 in the present study; however, only the expression
of ZNRF3 had a significant prognostic impact on the overall
survival of patients with COAD. ZNRF3 is a transmembrane E3
ubiquitin ligase that acts as a negative regulator of the Wnt
signaling pathway (35). A previous
study reported an association between ZNRF3 expression and survival
outcomes in patients with CRC, with ZNRF3 overexpression promoting
the apoptosis and inhibiting the proliferation of CRC cells
(51). In the present study,
proteomic analysis identified a significant increase in ZNRF3
levels in ARID1A-overexpressing cells. Interaction network analysis
revealed a potential interaction with ARID1A through co-expression.
The TCGA-COAD dataset demonstrated a positive correlation between
ARID1A and ZNRF3 expression, further confirmed by western blotting.
Hence, these findings suggest that the interaction between ARID1A
with ZNRF3 may contribute to the carcinogenesis and progression of
CRC. However, the present study did not confirm the physical
interaction between these two proteins by co-immunoprecipitation or
double immunofluorescence staining experiments. Further studies are
needed to validate these interactions.
To the best of our knowledge, no previous study has
reported a direct relationship between ARID1A and ZNRF3. A proposed
mechanism for how ARID1A overexpression may result in increased
ZNRF3 expression is through transcriptional regulation. ARID1A
functions as a transcriptional regulator, modulating chromatin
accessibility and transcription factor binding (4). It is possible that ARID1A could
directly bind or interact with transcriptional co-factors or
chromatin remodelers at the ZNRF3 promoter, leading to enhanced
transcription and an increase in ZNRF3 mRNA levels. Alternatively,
ARID1A may interact with other proteins that stabilize ZNRF3 mRNA
or prevent its degradation, leading to increased ZNRF3 protein
levels. Further experimental studies are needed to elucidate the
precise molecular mechanisms underlying this interaction.
Whilst the present study provides insights into the
role of ARID1A in colon cancer and its potential interactions with
the Wnt signaling pathway, several limitations should be
acknowledged. First, the study primarily focused on ARID1A
overexpression and its effects on protein expression profiles in
one colon cancer cell line (SW48). Further studies in CRC cells
with different molecular characteristics are warranted to validate
the present findings across a broader spectrum. Additionally, the
lack of physiological assays, such as proliferation, migration,
invasion, apoptosis and cell cycle progression, is a limitation.
These assays are crucial to comprehensively understand the
functional consequences of ARID1A overexpression and its
relationship with the Wnt signaling pathway in CRC carcinogenesis
and metastasis. Experiments involving genetic manipulation or
inhibitor treatment specific to Wnt signaling molecules are also
necessary to obtain a deeper understanding. Furthermore, the
observed discrepancy between proteomic and western blot analyses
regarding cyclin D1 expression needs to be validated by different
experimental approaches. Finally, further investigations are
required to elucidate the underlying mechanisms of the regulatory
axis between ARID1A and ZNRF3 in CRC.
In conclusion, a set of 705 differentially altered
proteins were identified in ARID1A-overexpressing cells compared
with control cells. These altered proteins were mainly involved in
the Wnt signaling pathway. Bioinformatics analyses also highlighted
a potential functional interaction between ARID1A and ZNRF3 in CRC
carcinogenesis and progression. A better understanding of the role
of ARID1A in CRC would help in the development of targeted
therapies and diagnostic approaches for CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research received funding support from the National
Science, Research and Innovation Fund via the Program Management
Unit for Human Resources & Institutional Development, Research
and Innovation (grant no. B05F640168).
Availability of data and materials
The raw mass-spectrometric data generated in the
present study may be found in the ProteomeXchange Consortium via
the jPOST repository (https://repository.jpostdb.org) (52) under accession numbers PXD052413 in
the ProteomeXchange and JPST003117 in the jPOST repository, or at
the following URLs: https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD052413
and https://repository.jpostdb.org/entry/JPST003117,
respectively. The other data generated in the current study may be
requested from the corresponding author.
Authors' contributions
SA and KS contributed to the conception and design
of the study, performed experiments, data analysis and
visualization, and were major contributors to the writing of the
manuscript. SW, AM and AR performed experiments and data analysis.
SR performed the proteomic analysis. NS contributed to the
conception and design of the study, data analysis, supervision,
funding acquisition and manuscript editing. SA and NS confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhao S, Wu W, Jiang Z, Tang F, Ding L, Xu
W and Ruan L: Roles of ARID1A variations in colorectal cancer: A
collaborative review. Mol Med. 28:422022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilsker D, Probst L, Wain HM, Maltais L,
Tucker PW and Moran E: Nomenclature of the ARID family of
DNA-binding proteins. Genomics. 86:242–251. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang X, Nagl NG, Wilsker D, Van Scoy M,
Pacchione S, Yaciuk P, Dallas PB and Moran E: Two related ARID
family proteins are alternative subunits of human SWI/SNF
complexes. Biochem J. 383:319–325. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu RC, Wang TL and Shih IeM: The emerging
roles of ARID1A in tumor suppression. Cancer Biol Ther. 15:655–664.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Li Z, Wang Z, Liu F, Zhang L, Ke J,
Xu X, Zhang Y, Yuan Y, Wei T, et al: Chromatin remodeling induced
by ARID1A loss in lung cancer promotes glycolysis and confers JQ1
vulnerability. Cancer Res. 82:791–804. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilson MR, Reske JJ, Holladay J, Neupane
S, Ngo J, Cuthrell N, Wegener M, Rhodes M, Adams M, Sheridan R, et
al: ARID1A mutations promote P300-dependent endometrial invasion
through super-enhancer hyperacetylation. Cell Rep. 33:1083662020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang T, Chen X, Su C, Ren S and Zhou C:
Pan-cancer analysis of ARID1A alterations as biomarkers for
immunotherapy outcomes. J Cancer. 11:776–780. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fontana B, Gallerani G, Salamon I, Pace I,
Roncarati R and Ferracin M: ARID1A in cancer: Friend or foe? Front
Oncol. 13:11362482023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guan B, Gao M, Wu CH, Wang TL and Shih
IeM: Functional analysis of in-frame indel ARID1A mutations reveals
new regulatory mechanisms of its tumor suppressor functions.
Neoplasia. 14:986–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mullen J, Kato S, Sicklick JK and Kurzrock
R: Targeting ARID1A mutations in cancer. Cancer Treat Rev.
100:1022872021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Somsuan K, Peerapen P, Boonmark W,
Plumworasawat S, Samol R, Sakulsak N and Thongboonkerd V:
ARID1A knockdown triggers epithelial-mesenchymal transition
and carcinogenesis features of renal cells: Role in renal cell
carcinoma. FASEB J. 33:12226–12239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S and Tang C: The role of ARID1A
in tumors: Tumor initiation or tumor suppression? Front Oncol.
11:7451872021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL,
Zeng ZL, Wang RY, Huang YX, Jin Y, Wang F, et al: Clinicopathologic
and prognostic relevance of ARID1A protein loss in colorectal
cancer. World J Gastroenterol. 20:18404–18412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye J, Zhou Y, Weiser MR, Gönen M, Zhang L,
Samdani T, Bacares R, DeLair D, Ivelja S, Vakiani E, et al:
Immunohistochemical detection of ARID1A in colorectal carcinoma:
Loss of staining is associated with sporadic microsatellite
unstable tumors with medullary histology and high TNM stage. Hum
Pathol. 45:2430–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guan X, Cui L, Ruan Y, Fang L, Dang T,
Zhang Y and Liu C: Heterogeneous expression of ARID1A in colorectal
cancer indicates distinguish immune landscape and efficacy of
immunotherapy. Discov Oncol. 15:922024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie C, Fu L, Han Y, Li Q and Wang E:
Decreased ARID1A expression facilitates cell proliferation and
inhibits 5-fluorouracil-induced apoptosis in colorectal carcinoma.
Tumour Biol. 35:7921–7927. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peerapen P, Sueksakit K, Boonmark W,
Yoodee S and Thongboonkerd V: ARID1A knockdown enhances
carcinogenesis features and aggressiveness of Caco-2 colon cancer
cells: An in vitro cellular mechanism study. J Cancer. 13:373–384.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baldi S, Zhang Q, Zhang Z, Safi M, Khamgan
H, Wu H, Zhang M, Qian Y, Gao Y, Shopit A, et al: ARID1A
downregulation promotes cell proliferation and migration of colon
cancer via VIM activation and CDH1 suppression. J Cell Mol Med.
26:5984–5997. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Erfani M, Zamani M, Hosseini SY,
Mostafavi-Pour Z, Shafiee SM, Saeidnia M and Mokarram P: ARID1A
regulates E-cadherin expression in colorectal cancer cells: A
promising candidate therapeutic target. Mol Biol Rep. 48:6749–6756.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwon YW, Jo HS, Bae S, Seo Y, Song P, Song
M and Yoon JH: Application of proteomics in cancer: Recent trends
and approaches for biomarkers discovery. Front Med (Lausanne).
8:7473332021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aluksanasuwan S, Somsuan K, Ngoenkam J,
Chiangjong W, Rongjumnong A, Morchang A, Chutipongtanate S and
Pongcharoen S: Knockdown of heat shock protein family D member 1
(HSPD1) in lung cancer cell altered secretome profile and
cancer-associated fibroblast induction. Biochimica Biophys Acta Mol
Cell Res. 1871:1197362024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guan B, Wang TL and Shih IeM: ARID1A, a
factor that promotes formation of SWI/SNF-mediated chromatin
remodeling, is a tumor suppressor in gynecologic cancers. Cancer
Res. 71:6718–6727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stewart SA, Dykxhoorn DM, Palliser D,
Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et
al: Lentivirus-delivered stable gene silencing by RNAi in primary
cells. RNA. 9:493–501. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lowry OH, Rosebrough NJ, Farr AL and
Randall RJ: Protein measurement with the Folin phenol reagent. J
Biol Chem. 193:265–275. 1951. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu W, Smith JW and Huang CM: Mass
spectrometry-based label-free quantitative proteomics. J Biomed
Biotechnol. 2010:8405182010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Neilson KA, Ali NA, Muralidharan S,
Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC and
Haynes PA: Less label, more free: Approaches in label-free
quantitative mass spectrometry. Proteomics. 11:535–553. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tyanova S, Temu T and Cox J: The MaxQuant
computational platform for mass spectrometry-based shotgun
proteomics. Nat Protoc. 11:2301–2319. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tyanova S, Temu T, Sinitcyn P, Carlson A,
Hein MY, Geiger T, Mann M and Cox J: The perseus computational
platform for comprehensive analysis of (prote)omics data. Nat
Methods. 13:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao T and Wang Z: GraphBio: A shiny web
app to easily perform popular visualization analysis for omics
data. Front Genet. 13:9573172022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res(Web Server issue). 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 10:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koo BK, Spit M, Jordens I, Low TY, Stange
DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM
and Clevers H: Tumour suppressor RNF43 is a stem-cell E3 ligase
that induces endocytosis of Wnt receptors. Nature. 488:665–669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L,
Gopal P, Zhu M, Nassour I, Chuang JC, et al: Arid1a has
context-dependent oncogenic and tumor suppressor functions in liver
cancer. Cancer Cell. 32:574–589.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge
Z, Nagel ZD, Zou J, Wang C, Kapoor P, et al: ARID1A deficiency
promotes mutability and potentiates therapeutic antitumor immunity
unleashed by immune checkpoint blockade. Nat Med. 24:556–562. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu X, Dai SK, Liu PP and Liu CM: Arid1a
regulates neural stem/progenitor cell proliferation and
differentiation during cortical development. Cell Prolif.
54:e131242021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoodee S, Peerapen P, Plumworasawat S and
Thongboonkerd V: ARID1A knockdown in human endothelial cells
directly induces angiogenesis by regulating angiopoietin-2
secretion and endothelial cell activity. Int J Biol Macromol.
180:1–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoodee S, Peerapen P, Plumworasawat S,
Malaitad T and Thongboonkerd V: Identification and characterization
of ARID1A-interacting proteins in renal tubular cells and their
molecular regulation of angiogenesis. J Transl Med. 21:8622023.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao H, Ming T, Tang S, Ren S, Yang H, Liu
M, Tao Q and Xu H: Wnt signaling in colorectal cancer: Pathogenic
role and therapeutic target. Mol Cancer. 21:1442022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lo YH, Kolahi KS, Du Y, Chang CY,
Krokhotin A, Nair A, Sobba WD, Karlsson K, Jones SJ, Longacre TA,
et al: A CRISPR/Cas9-engineered ARID1A-deficient human
gastric cancer organoid model reveals essential and nonessential
modes of oncogenic transformation. Cancer Discov. 11:1562–1581.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vaicekauskaitė I, Dabkevičienė D, Šimienė
J, Žilovič D, Čiurlienė R, Jarmalaitė S and Sabaliauskaitė R:
ARID1A, NOTCH and WNT signature in gynaecological tumours.
Int J Mol Sci. 24:58542023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hiramatsu Y, Fukuda A, Ogawa S, Goto N,
Ikuta K, Tsuda M, Matsumoto Y, Kimura Y, Yoshioka T, Takada Y, et
al: Arid1a is essential for intestinal stem cells through Sox9
regulation. Proc Natl Acad Sci USA. 116:1704–1713. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Heunis T, Lamoliatte F, Marín-Rubio JL,
Dannoura A and Trost M: Technical report: Targeted proteomic
analysis reveals enrichment of atypical ubiquitin chains in
contractile murine tissues. J Proteomics. 229:1039632020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hong X, Wang T, Du J, Hong Y, Yang CP,
Xiao W, Li Y, Wang M, Sun H and Deng ZH: ITRAQ-based quantitative
proteomic analysis reveals that VPS35 promotes the expression of
MCM2-7 genes in HeLa cells. Sci Rep. 12:97002022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pino JMV, Nishiduka ES, da Luz MHM, Silva
VF, Antunes HKM, Tashima AK, Guedes PLR, de Souza AAL and Lee KS:
Iron-deficient diet induces distinct protein profile related to
energy metabolism in the striatum and hippocampus of adult rats.
Nutr Neurosci. 25:207–218. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Morikawa K, Mori Y, Zong C, Zhang
L, Garner E, Huang C, Wu W, Chang J, Nagashima D, et al: Proteomic
analysis of liver proteins of mice exposed to 1,2-dichloropropane.
Arch Toxicol. 94:2691–2705. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Van Dross R, Browning PJ and Pelling JC:
Do truncated cyclins contribute to aberrant cyclin expression in
cancer? Cell Cycle. 5:472–477. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang J, Su W, Zhang T, Zhang S, Lei H, Ma
F, Shi M, Shi W, Xie X and Di C: Aberrant cyclin D1 splicing in
cancer: From molecular mechanism to therapeutic modulation. Cell
Death Dis. 14:2442023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu N, Zhu H, Tao Y, Huang Y, Song X, Zhou
Y, Li Y, Pei Q, Tan Q and Pei H: Association between prognostic
survival of human colorectal carcinoma and ZNRF3 expression. Onco
Targets Ther. 9:6679–6687. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Okuda S, Watanabe Y, Moriya Y, Kawano S,
Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa
AC, et al: jPOSTrepo: An international standard data repository for
proteomes. Nucleic Acids Res. 45:D1107–D1111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|