Breast cancer is the most common malignant tumor in
women worldwide, and the incidence of breast cancer is increasing
in all countries. In China, the incidence and mortality rates of
breast cancer are the highest of all tumors affecting women
(1–3). Annually, ~1 million cases of
triple-negative breast cancer (TNBC) are considered to be diagnosed
worldwide (4). TNBC is
characterized by decreased estrogen receptor (ER), progesterone
receptor (PR) and human epidermal receptor (HER2) expression.
According to previousa previous study, most patients with TNBC
harbor a chromosomal 5q deletion (5). Breast cancer can be categorized into
four subtypes (luminal A, luminal B, HER2+ and TNBC)
based on the expression of the ER, PR and HER2, and the different
subtypes can be treated accordingly (6,7). There
are four main signaling pathways related to TNBC, namely, the
JAK/STAT, PI3K/AKT/mTOR, TGF-β and INF-γ pathways (8).
MicroRNAs (miRNAs) can regulate parts of the
phosphatidylinositol 3-kinase (PI3K) signaling pathway (9). Complex interactions between the
PI3K/AKT/mammalian target of rapamycin (mTOR) pathway and several
interacting cellular signaling cascades can promote breast cancer
progression (10). PI3K is an
important protein in the AKT signaling pathway, which is involved
in cell survival, differentiation, growth, glucose transport and
glucose utilization (11).
Insulin-like growth factor (IGF-1) is a self-phosphorylating ligand
that stimulates downstream responses, transforming growth factor β
(TGF-β) alters cell growth and metabolism by entering the nucleus
through cross-linking complexes, and the kinase transcription
factor is an autophosphorylated ligand that stimulates downstream
responses. The pregnancy-associated plasma protein-A/IGF binding
protein/IGF axis plays an important role in TNBC motility and
epithelial-mesenchymal transition (EMT). This pathway influences
TNBC cell migration and motility (12). Janus kinase (JAK)/STAT, is
inextricably linked to TNBC progression, and has a role in breast
cell genesis and pathology. miRNAs are circulating non-coding RNA
molecules, 17–27 nucleotides in length, that regulate the
post-transcriptional expression of genes (such as oncogenes) in
oncogenic pathways (13). Depending
on the progression- or inhibition-promoting function of miRNAs,
they can be used as oncogenes or tumor suppressors, respectively
(14). Different signaling pathways
promote different cell growth modes, and miRNAs, as common
components, play a role in promoting or inhibiting cell growth.
In the present review, literature from the past 5
years was collected for examination In contrast to the previous
single study of a signaling pathway, the present review examines
four different signaling pathways and compares the microRNAs that
play a role in the signaling pathways, and describes the
co-expression of miR-21. Therefore, the aim of the present review
was to examine the specific role of miRNAs in four key signaling
pathways in breast cancer, and to understand the crosstalk and
influence of the signaling pathways with miRNA.
miRNAs are short-stranded non-coding RNAs consisting
of 17–27 nucleotides that are found in animals, plants and viruses,
and which can negatively regulate the gene expression of mRNA
(15,16). Gene expression relies on an effector
ribonucleoprotein complex termed the RNA-induced silencing complex
(RISC), for which mRNAs act as target recognizers. The Ago family
of proteins and their array of auxiliary components work together
to assemble RISCs (17). The
complex has an intrinsic ability to bind to typical 3′ untranslated
regions (3′UTRs) that are specific to its cytoplasmic mRNA targets.
The binding to the mRNA is based on the complementarity between the
base pairing at the 5′ end of the mature miRNA or open reading
frame and the cytoplasmic mRNA molecule. The binding site, termed
the seed region, is 6–8 bp from the 5′ end of the miRNA. The short
length of the binding site allows the miRNA to target and bind to a
large number of different mRNAs (18–20).
The combination of RISC and other genes is in turn the combination
of miRNA and other genes, and therefore both play a role in the
degradation, extension or activation of mRNA. If the composition of
RISC is modified, or the composition of Ago protein is changed, the
role of RISC will be changed. As such, the mechanism of specific
miRNAs in the regulation of genes becomes controllable, which can
be utilized to understand the role of miRNA in the expression of
different genes. The role of miRNAs in breast cancer is partly via
signaling pathways. There are four major signaling pathways in
breast cancer, and miRNAs are differentially expressed in different
signaling pathways and thus play different roles. Ultimately, all
miRNAs have an excitatory or inhibitory effect on tumor
development. Although different miRNAs play different roles in
different signaling pathways, the final effects are the same or
similar.
Breast cancer has different onset and progression
processes according to the different subtypes, which can be linked
to different signaling pathways. JAK/STAT is a signaling pathway
that has an impact on both breast cancer development and
metastasis. It has been demonstrated that the expression and active
form of STAT3 is detected in >50% of breast cancer cases, and
JAK/STAT signaling has become an emerging therapeutic target in a
wide range of malignant tumors (21–23).
STAT family members not only transduce signals used for
transcription, but also regulate mitochondrial synthesis and
metabolism, and have a role in compartmentalization of the nucleus
and genome integrity (24).
Numerous reports suggest that the acquisition of mutations that
alter the function of STAT proteins underlies tumor cell genesis
(25–28). STAT proteins do not accomplish tasks
independently and function alongside cytokines, inducible factor
expression or phosphorylation modifications (29). When JAK/STAT ligand and receptor
binding is activated, JAK binds non-covalently to the receptor
structure, after which the kinase activity of JAK is activated and
phosphorylates the tyrosine residues in the receptor region. Next,
the phosphorylated receptor residues recruit STAT through the SH2
structure, and the two combine to form a heterodimer, which
translocates to the nucleus to carry out a variety of biological
responses in the pathway and the occurrence of disease (30).
TGF-β is known to reprogram the tumor
microenvironment in TNBC; it suppresses the immune system by
increasing collagen production in cancer-associated fibroblasts
(31). The TGF-β pathway has
different roles in different stages of tumors. In the early stage
of tumors, the TGF-β pathway induces apoptosis and never curbs the
development of tumor cells, but in the late stage of the disease,
it promotes cell secretion and infiltration, and accelerates
disease progression (32). TGF-β
binds to two receptors to exert cellular effects, namely, TGFβ-type
RI (TGFβRI) and TGF-β type RII (TGFβRII). Initially, TGF-β binds to
TGFβRII, and then TGFβRII phosphorylates and activates TGFβRI. The
three form a heterodimeric complex to bind an intracellular
effector molecule termed SMAD, after which the
receptor-regulated-SMAD-common mediator-SMAD complex enters the
nucleus and recruits additional co-transcriptional activators,
repressors and/or cofactors (32–35).
IGF-1R is a cell-surface transmembrane tyrosine
kinase receptor consisting of two α and two β subunits, the β
subunits of which contain a tyrosine catalytic structural domain.
IGF-1R promotes a cascade reaction through ligand binding, the
basis of which is that IGF-1R autophosphorylates to activate a
variety of downstream signaling pathways, including the PI3K and
AKT signaling pathways (36). It is
possible that this pathway influences breast cancer development by
acting as an upstream signal, activated by autophosphorylation, and
then activating downstream signaling pathways for cell
proliferation, division and differentiation. This approach may
intersect with other signaling pathways in a one-to-many signaling
activation process.
PI3Ks are a class of lipid kinases that are
classified as classes I, II or III in mammals (37). The present review focuses on class
I, which has been shown to be strongly associated with the
development of cancer (38). The
target of PI3K downstream signaling is AKT, a serine/threonine
kinase with three isoforms, and this kinase acts in the biological
oxidation of glucose and in cell growth, reproduction and survival
(39). The next target is also a
serine/threonine kinase, mTOR, which consists of two complex
proteins that are important in the regulation of cell cycle
progression and growth factors (40). In total, 30–40% of patients with
breast cancer have mutations in the PI3K gene, which leads to
hyperactivation of PI3K isoforms (41), and this pathway is common to
multiple downstream target cells. The cascade reaction resulting
from hyperactivation of PI3K may underlie cellular carcinogenesis.
PI3Ks promote phosphatidylinositol (3,4,5)-trisphosphate
(PIP3) production through phosphorylation of phosphatidylinositol,
and docking of AKT to the N region of PIP3 facilitates
translocation to the cell membrane and promotes AKT activation,
resulting in phosphorylation of serine or threonine residues of AKT
(41). The pathways of various
signaling pathways may be different, but most pathogenesis caused
by signaling pathways is in the form of one-to-many, in which a
cascade reaction leads to ‘explosive’ growth. Therefore, in the
cases of mutations in breast cancer genes caused by alterations in
signaling pathways, it may be more optimal to treat the downstream
signaling targets and upstream trigger points, to curb the
subsequent cascade of changes (42,43).
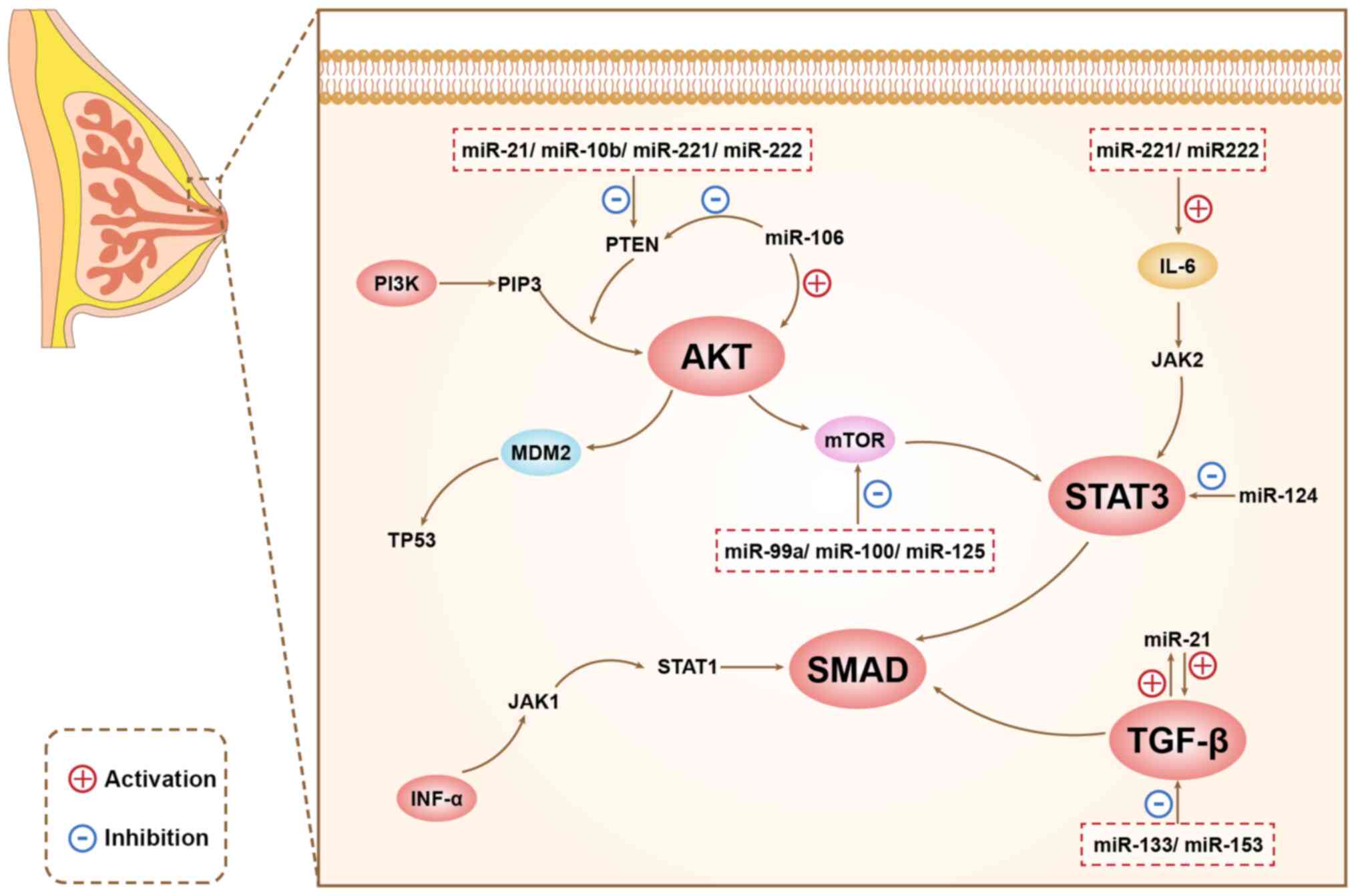
miRNAs play different roles in different signaling pathways, and
there are connections between different signaling pathways, as
shown in Figure 1.
Different miRNAs are associated with different
signaling pathways. In this section, the miRNAs in the signaling
pathways of TNBC are summarized. TNBC is currently the most
invasive and the most poorly treated type of breast cancer due to a
lack of hormone receptors to target. However, the emergence of
miRNAs as new biomarkers has brought new light to the treatment of
TNBC. The roles of different miRNAs in different signaling pathways
are summarized in Table I.
The PI3K/AKT/mTOR signaling pathway plays a key role
in causing cancer and promoting tumor growth and survival, and it
is typically activated in TNBC (44). Previous studies have shown that
miRNA expression is involved in carcinogenesis and is also a
notable regulator of PI3K/AKT (45,46).
PI3K can be activated by kinases, cells, receptors and other
factors to complete autophosphorylation to PIP3, after which PIP3
binds to and activates AKT. PTEN can also dephosphorylate PIP3 to
form PIP2, which inactivates downstream AKT by preventing it from
covalently binding to PIP3 (46).
miRNAs can also regulate the JAK/STAT pathway. JAK
is a tyrosine kinase, and the JAK/STAT signaling pathway is
activated when JAK binds to and interacts with receptors such as
cytokines (60). These transduction
and transcriptional activators ultimately enter the nucleus to
participate in cellular replication and transcription, apoptosis
and metastasis, and miRNAs are involved in these regulatory
processes. miR-221/222 can activate interleukin
(IL)-6-dependent-NF-κB and STAT3 via lipocalin receptor 1,
activation of STAT3 is important for breast cancer survival and
tumorigenesis, with important implications (61,62).
In addition to its oncogenic and tumor-suppressive effects, miR-205
has also been shown to have a notable role in the maintenance of
TNBC stem cell characteristics by SUM159PT (preventing downstream
cell renewal by blocking STAT downstream signaling), which has a
high probability of inhibiting breast cancer through blocking STAT
signaling, thus restricting cancer cell division and migration
(63). High expression of miR-31 in
mammary stem cells promotes mammary epithelial cell proliferation
and stem cell expansion, which also reduces tumor growth through
various signaling pathways such as the STAT pathway (64). miR-124, as an inhibitory factor,
directly targets STAT3 in breast cancer, an interaction which can
inhibit the growth and invasion of breast cancer cells. When
miR-124 is upregulated, it will reduce the level of STAT3 and
related proteins in breast cancer cells, and reduce the expansion
of breast cancer (65). In addition
to positive regulation, miRNAs also negatively regulate STAT.
miR-1207-5P and miR-505/148a can inhibit STAT expression to
activate the corresponding EMT/Wnt signaling molecule to inhibit
cancer metastasis (66,67).
A total of 33 members of the human TGF-β family bind
both TGFβRI and TGFβRII receptors. These receptors are
serine/threonine kinases that can be considered as intracellular
signaling pathways that initiate SMAD intracellular effectors in
the form of heterodimers. Compared with that in normal breast
tissues, the transcriptional expression of TGFβRI was higher in
breast cancer tissues, the expression of miR-133b was negatively
correlated with TGFβRI and miR-133b expression was lower in tumor
tissues (68). Similarly, TGFβRII
expression was higher in breast cancer tissues than in normal
breast tissues, but the expression of TGFβRII was negatively
correlated with the expression of miR-153, which was decreased in
breast cancer tissues (69).
Furthermore, overexpression of miR-133b significantly inhibited the
function of TGFβRI transduction in the TGF-β signaling pathway and
also inhibited TGFβ-induced endometrial stromal transformation and
breast cancer cell invasion (68).
It was found that miRNAs can not only affect the function of TGF-β,
but can also be affected by TGF-β signaling. Compared with its role
in normal breast tissues, TGFβI upregulated the expression of
miR-21 and downregulated the expression of miR-196a-3p, both of
which were significantly associated with breast cancer progression
(70,71). In TNBC, miR-21 is upregulated
through TGF-β1 and then through interaction with oncogenes.
Therefore, in TNBC, miR-21 upregulation or downregulation provides
a further explanation for tumor promotion or inhibition.
In the relationship between TGF-β and miRNAs, it is
difficult to say which influences the other, but it is more akin to
a mutually constraining relationship or fundamentally a crosstalk
between short-stranded non-coding RNAs and the signaling pathway as
a whole. Unlike other signaling pathways, this signaling pathway is
more of an initiator, as TGF-β can influence the transduction of
mTOR and EGFR, and in short, it is an alteration in TGF-β that
first leads to the subsequent signaling crosstalk (72).
In the 46% of patients with TNBC and IGF-1R
alterations, there is an association with poor survival (73). IGF-1R interacts with other pathways
to upregulate tumor stem cells in TNBC (74). As such, when the level of IGF-1 is
reduced, the number of tumor stem cells is also reduced. It has
also been shown that there is a significant relationship between
IGF-1 expression and drug resistance in TNBC (75). IGFs are categorized into two types,
IGF1 and IGF2, which are both classes of polypeptides with effects
on growth (76). IGF1 is a major
target gene of growth hormone, its post-transcriptional product has
a major role in growth and development, and high circulating levels
of IGF1 are present in human cancer (77). Moreover, IGF-1R is not mutated in
most cancer types, therefore expression levels are assessed to
predict the cancer outcome (77).
IGF1 promotes breast cancer progression by forming an insulin-like
substrate complex that preferentially activates the PI3K/AKT
pathway thereby altering survival genes (78). miR-148a-3p targets the mRNA of DNA
transmethylase 1, thereby enhancing target gene IGF1 expression
(79). In addition, miR-148a-3p
attenuates the expression of p53, which is a negative
transcriptional regulator of IGFR, and thus elevated levels of
miR-148a-3p are positively associated with breast cancer (80,81).
However, miR-21-5p is strongly associated with PI3K/AKT. miR-21-5p
inhibits PTEN expression thereby enhancing PI3K/AKT/mTOR signaling
(82).
In addition to crosstalk within and between
signaling pathways, miRNAs also influence cell chemotaxis and are
enriched in the tumor microenvironment (83). During tumorigenesis, tumor cells
produce non-coding RNAs that regulate monocyte recruitment and
differentiation to the tumor site by affecting the expression of
inflammatory factors or macrophages (84). miR-149 directly targets colony
stimulating factor 1 mRNA in breast cancer cells, resulting in a
significant reduction in macrophage levels in primary tumors
(85). In addition, miR-19 targets
PTEN to enhance the polarization of IL-17-producing T cells, and
miR-15/16 is targeted to induce the expression of FOXP3, thus
affecting the expression of CD4+ T cells (86–88).
In the tumor microenvironment, miRNAs not only affect increase or
decrease the number of tumor cells, but also have other mechanisms
that effect tumor cells. For instance, the production of
miR-146a-5p by breast cancer cells forms a crosstalk with tumor
cells and macrophages, thus altering the whole tumor
microenvironment. Specifically, miR-146a-5p inhibits macrophages,
which leads to the escape of tumor cells from the microenvironment
(89). Finally, in the tumor
microenvironment, the number of infiltrating lymphocytes indicates
the antitumor immune efficiency. miR-155 is able to activate the
immune system to infiltrate tumor cells, thus an increase in
miR-155 levels is beneficial to increase the fight against tumors
(90). Therefore, miRNAs play a
role in signaling pathways, as well as in the tumor
microenvironment where the signaling pathways are located.
The high incidence and mortality rates of breast
cancer, and its late detection, have indicated that early and
accurate screening of breast cancer is particularly important.
Diagnostic imaging has been widely used for breast cancer detection
and staging, but a high workload and the clarity of the images
greatly mislead clinicians when making accurate judgments (91). Image-based AI technology optimizes
the role of computer-aided diagnosis (CAD) in the clinic, improving
accuracy (92). Deep-learning
algorithms used for image recognition analysis mainly include
convolutional neural networks (CNNs), deep CNNs, fully
convolutional networks, recurrent neural networks and generative
adversarial networks (93). The
final target is decomposed into a number of visible layers and the
data input is used for multiple image extraction in these visible
layers. Next, a representative feature image is output from the
subsequent analysis of the extracted layers. The main part of
breast cancer AI examination includes image segmentation and
identification of benign and malignant tumors (94,95).
Mammography is the mainstay of early breast cancer screening
(96). Obtaining high-resolution
images allows the images to be preserved and further used
regardless of age and size. Full-field mammography has the
capability of inputting raw images and outputting processed images,
where AI analyzes the images and analyzes the breast mass, mass
segmentation, tissue density and risk assessment. The breast mass
is the most common manifestation of breast cancer assessment and
therefore CAD becomes one of the most important steps (97). Another method to assess the risk of
breast cancer is calcification. Calcification foci in X-rays are
classified as microcalcifications or macrocalcifications, and while
it is difficult to identify microcalcifications via manual
visualization, the CAD system is able to detect these foci
(98). This allows for timely
detection and intervention for patients with early stage breast
cancer. In addition, the use of AI can automate the process of
differentiating breast lumps from normal tissues in mammograms,
which was theorized to greatly improve the prognosis of patients
with early stage disease (99).
However, the reality is not as optimistic. CAD became part of
mammography screening 20 years ago, but the data so far have shown
that it does not necessarily have an advantage over a single
reading by a radiologist. The combination of the two methods has,
however, been reported to have an improved success rate in
determining the early stage of breast cancer in patients (100). AI for the detection of breast
cancer still has a greater outlook and development space, as the
performance of the algorithm, the identification of different
breast tissues and algorithm judgment still need to be updated. We
consider that, if the algorithms can be updated for differentiating
benign and malignant pathology, this method will be a more
convenient and efficient way for diagnosing breast cancer compared
with puncture or intraoperative pathology.
It is known that miR-21 is expressed at elevated
levels in patients with breast cancer, and miR-21 inhibitors have
been tested for their ability to reduce cell metastasis, suggesting
that inhibition of miR-21 could lead to a reduction in metastasis
and enhanced breast cancer treatment. This means that miR-21
inhibitor treatment brings the hope of new therapeutic approaches
for patients (104). In addition,
miR-21 also has a significant role in drug resistance.
Specifically, miR-21 regulates the resistance of breast cancer
cells to azithromycin by targeting phosphatases and PTEN (105). It has been reported that miR-21
also targets TGF-β, which can increase the rapid cell growth and
transformation of cancer cells. Therefore, knockdown of this
specific miRNA could be targeted to treat breast cancer (105,106). miR-155 is also a notable marker.
miR-155 is involved in the regulation of breast cancer growth and
is an oncogenic miRNA. It has been demonstrated that miR-155 leads
to telomere destabilization and that this mechanism is mediated by
telomeric repeat binding factor 1 (TRF1) expression (107,108). In addition, miR-125b has a
targeting effect on human invasive breast cancer. miR-125b
downregulated in breast cancer reduces the expression of mucin 1
(MUC1), an oncoprotein, and the silencing of MUC1 leads to DNA
damage-induced apoptosis in cancer cells (109). Jang et al (75) investigated the effect of miR-125
deregulation on the formation of metastasis and found that miR-125b
can cause metastasis by targeting MCF1, which is a TRF1. miR-125b
also induces metastasis by targeting StAR-related lipid transfer
domain containing 13 mRNA in MCF-7 and MDA-MB-231 breast cancer
cells (110). miR-17, as a
potential regulator, can directly negatively regulate c-Jun
activation domain binding protein-1 (JAB1) in vitro. In
tissue samples from patients with TNBC, both the JAB1 gene and
protein were highly expressed, while miR-21 expression was low. It
has been suggested that miR-17 inhibits JAB1 cell division through
E2F amplification, thus suppressing tumors (111). miR-21 is known to be expressed at
elevated levels in patients with breast cancer, and miR-21
inhibitors have been tested for their ability to reduce cellular
migration, suggesting that inhibition of miR-21 could lead to a
reduction in metastasis and cell division in breast cancer.
Therefore, miR-21 inhibitor therapy offers patients the hope of new
treatments (104,112). In addition, miR-21 also has a
major role in drug resistance. Specifically, miR-21 regulates
azithromycin resistance in breast cancer cells, which is mediated
by targeting phosphatases and PTEN. It has been reported that
miR-21 can also target TGF-β, which can increase the exponential
growth and transformation of cancer cells. Therefore, the knockdown
of this specific miRNA may be a targeted therapy for breast cancer
(105,106,113). miR-155 is also an important marker
that is involved in the growth regulation of breast cancer and is
an oncogenic miRNA that has been shown to cause telomere
destabilization through the expression of TRF1 (107,108). The therapeutic approach of miRNA
functions more like a source, paving the way for new strategies in
breast cancer drug therapy. Building upon the earlier discussion,
investigating the positive and negative regulatory mechanisms of
miR-21 and miR-155, and researching corresponding targeted drugs,
can either stimulate or inhibit the concentration of a specific
biomarker, thereby achieving treatment for breast cancer.
It has been shown that inflammatory factors and
other substances in the body are controlled by high levels of
miR-21, which in turn activates the expression of STAT3 (114). When miR-21 expression is knocked
down, the expression of STAT3 is suppressed in vivo
(115). Moreover, STAT3 in turn
affects the expression of miR-21 (116). When the expression level of STAT3
is changed, the oncogenic signaling pathway of breast cancer will
be altered, meaning that miR-21 influences STAT3 and thus affects
the development of breast cancer. Similarly, miR-21 is not only
involved in the expression of STAT3 pathway in breast cancer, but
also in the expression of other signaling pathways. miR-21 acts as
an oncogenic marker by targeting PTEN, which is an inhibitor of the
PI3K/AKT pathway. Therefore, miR-21 enhances the action of the
PI3K/AKT pathway (117–119). A study has shown that the levels
of miR-21 and TGF-β increase with breast cancer stage (120). With in-depth research, it was
found that miR-21 upregulation in breast cancer tissues was closely
related to TGF-β and mediated the effects of TGF-β on cell invasion
and chemotherapy through direct downregulation of PTEN and
phosphatase (70). In other words,
miR-21 indirectly affected the expression of TGF-β and thus the
treatment of breast cancer. Finally, miR-21 downregulation also
affects the expression of IGF-1 (121).
As a simple and detectable marker, miRNAs are
notably expressed in the early stages of tumors, which provides a
convenient method for subsequent analysis. Moreover, different
miRNAs have different roles among the four signaling pathways and
cannot be replaced. By decreasing or increasing the expression of
genes, miRNA expression leads to the alteration of gene expression
direction, which leads to the development of cancer. Moreover, the
miRNAs that play a role are different based on the type of breast
cancer and signaling pathway, raising the question of whether the
same miRNAs can also form a crosstalk between different signaling
pathways. Aberrant expression of miRNAs is closely related to the
development and progression of breast cancer and can be used as a
potential therapeutic target. Combining AI with miRNA therapy may
lead to more comprehensive and precise breast cancer treatment
programs. Furthermore, understanding the role of miRNAs in breast
cancer can be improved through big data analysis via AI, which can
optimize treatment strategies for individualized and efficient
treatment. However, numerous challenges remain to be faced in both
fields, including issues such as validation in clinical practice
and regulatory approval. With the deepening of research and the
continuous development of technology, more breakthroughs are
expected in the future.
The innovation of the present review lies in the
fact that miRNA, as a new biomarker for breast cancer diagnosis,
treatment and prognosis, has a profound future research prospect.
In addition, miRNA also targets downstream target genes and
proteins, which is a new research direction for breast cancer
treatment. miRNAs also link a variety of different signaling
pathways, which can be used to form a non-coding RNA network for
the treatment of breast cancer. It is also evident that miRNA
levels are significantly changed in the blood of patients with
breast cancer and are relatively easy indicators to extract and
obtain.
The roles of miRNAs, IGF-1 and IGF-1R have been
established in breast cancer, related receptors and related
signaling pathways. miRNA has been found to participate in a number
of signaling pathways, including PI3K/AKT and other pathways. The
present review mainly discussed the association and role of miR-21
and other related factors in four key signaling pathways, as miR-21
is expressed in all four of these pathways. miR-21 plays a similar
role in inhibiting tumor development in different breast cancer
signaling pathways; therefore, the present review also linked the
crosstalk between the four signaling pathways in breast cancer. The
present review supplements the previous shortcomings of the
influence of a single upstream target gene or signal on breast
cancer, and provides a new direction for future breast cancer
research. In addition, miR-21 is a key regulator of various
oncogenic pathways and can interact with circular RNA and long
non-coding RNA to regulate tumorigenesis. The present review
focused on the relationship between miRNA and signaling pathways,
which plays an important role in tumor formation and development,
and in diagnostic markers.
Not applicable.
This review was supported by The Natural Science Foundation of
Inner Mongolia Autonomous Region (grant no. 2022MS08010) and The
Graduate Student Excellence Program (grant no. YKDD2023ZY001).
Not applicable.
SY was responsible for drafting the manuscript,
building the logical framework, literature collection, retrieval,
reading, and embellishment of the manuscript's description and
typographical layout. DL was responsible for manuscript review and
revision, and communication with reviewers. Both authors have read
and approved the final version of the manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T,
Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, et
al: Global, regional, and national cancer incidence, mortality,
years of life lost, years lived with disability, and
disability-adjusted life-years for 29 cancer groups, 1990 to 2016:
A systematic analysis for the global burden of disease study. JAMA
Oncol. 4:1553–1568. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elizabeth MS, Cristina SBJ and Christian
CG: Immunotherapy in combination with chemotherapy for
triple-negative breast cancer. Mini Rev Med Chem. 24:431–439. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herschkowitz JI, Simin K, Weigman VJ,
Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S,
Chandrasekharan S, et al: Identification of conserved gene
expression features between murine mammary carcinoma models and
human breast tumors. Genome Biol. 8:R762007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pernas S and Tolaney SM: HER2-positive
breast cancer: New therapeutic frontiers and overcoming resistance.
Ther Adv Med Oncol. 11:1758835919833519. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ferrari P, Scatena C, Ghilli M, Bargagna
I, Lorenzini G and Nicolini A: Molecular mechanisms, biomarkers and
emerging therapies for chemotherapy resistant TNBC. Int J Mol Sci.
23:16652022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo XQ and Hua YM: Circular RNAs: novel
regulators of resistance to systemic treatments in breast cancer.
Neoplasma. 69:1019–1028. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Majidinia M and Yousefi B: DNA damage
response regulation by microRNAs as a therapeutic target in cancer.
DNA Repair (Amst). 47:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abu-Alghayth MH, Khan FR, Belali TM,
Abalkhail A, Alshaghdali K, Nassar SA, Almoammar NE, Almasoudi HH,
Hessien KBG, Aldossari MS and Binshaya AS: The emerging role of
noncoding RNAs in the PI3K/AKT/mTOR signalling pathway in breast
cancer. Pathol Res Pract. 255:1551802024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Elfaki I, Mir R, Abu-Duhier FM, Khan R and
Sakran M: Phosphatidylinositol 3-kinase Glu545Lys and His1047Tyr
Mutations are not Associated with T2D. Curr Diabetes Rev.
16:881–888. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Poddar A, Ahmady F, Rao SR, Sharma R,
Kannourakis G, Prithviraj P and Jayachandran A: The role of
pregnancy associated plasma protein-A in triple negative breast
cancer: A promising target for achieving clinical benefits. J
Biomed Sci. 31:232024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paszek S, Gabło N, Barnaś E, Szybka M,
Morawiec J, Kołacińska A and Zawlik I: Dysregulation of microRNAs
in triple-negative breast cancer. Ginekol Pol. 88:530–536. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Abdelfattah AM, Park C and Choi MY: Update
on non-canonical microRNAs. Biomol Concepts. 5:275–287. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawamata T and Tomari Y: Making RISC.
Trends Biochem Sci. 35:368–376. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J
and Jin Y: miR-24 regulates apoptosis by targeting the open reading
frame (ORF) region of FAF1 in cancer cells. PLoS One. 5:e94292010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Banerjee K and Resat H: Constitutive
activation of STAT3 in breast cancer cells: A review. Int J Cancer.
138:2570–2578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chung SS, Giehl N, Wu Y and Vadgama JV:
STAT3 activation in HER2-overexpressing breast cancer promotes
epithelial-mesenchymal transition and cancer stem cell traits. Int
J Oncol. 44:403–411. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Küçük C, Jiang B, Hu X, Zhang W, Chan JK,
Xiao W, Lack N, Alkan C, Williams JC, Avery KN, et al: Activating
mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK
cells. Nat Commun. 6:60252015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Heppler LN and Frank DA: Rare mutations
provide unique insight into oncogenic potential of STAT
transcription factors. J Clin Invest. 128:113–115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rajala HL, Eldfors S, Kuusanmäki H, van
Adrichem AJ, Olson T, Lagström S, Andersson EI, Jerez A, Clemente
MJ, Yan Y, et al: Discovery of somatic STAT5b mutations in large
granular lymphocytic leukemia. Blood. 121:4541–4550. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Araujo ED, Keserű GM, Gunning PT and
Moriggl R: Targeting STAT3 and STAT5 in Cancer. Cancers (Basel).
12:20022020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Owen KL, Brockwell NK and Parker BS:
JAK-STAT Signaling: A double-edged sword of immune regulation and
cancer progression. Cancers (Basel). 11:20022019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao L, Pang A and Li Y: Function of GCN5
in the TGF-β1-induced epithelial-to-mesenchymal transition in
breast cancer. Oncol Lett. 16:3955–3963. 2018.PubMed/NCBI
|
|
29
|
López-Mejía JA, Mantilla-Ollarves JC and
Rocha-Zavaleta L: Modulation of JAK-STAT Signaling by LNK: A
forgotten oncogenic pathway in hormone receptor-positive breast
cancer. Int J Mol Sci. 24:147772023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Budi EH, Duan D and Derynck R:
Transforming Growth Factor-β Receptors and Smads: Regulatory
complexity and functional versatility. Trends Cell Biol.
27:658–672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Said SS and Ibrahim WN: Breaking Barriers:
The promise and challenges of immune checkpoint inhibitors in
triple-negative breast cancer. Biomedicines. 12:3692024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heldin CH and Moustakas A: Signaling
Receptors for TGF-β Family Members. Cold Spring Harb Perspect Biol.
8:a0220532016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wrana JL, Attisano L, Wieser R, Ventura F
and Massagué J: Mechanism of activation of the TGF-beta receptor.
Nature. 370:341–347. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moustakas A, Souchelnytskyi S and Heldin
CH: Smad regulation in TGF-beta signal transduction. J Cell Sci.
114((Pt 24)): 4359–4369. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Christodoulou C, Oikonomopoulos G, Koliou
GA, Kostopoulos I, Kotoula V, Bobos M, Pentheroudakis G, Lazaridis
G, Skondra M, Chrisafi S, et al: Evaluation of the insulin-like
growth factor receptor pathway in patients with advanced breast
cancer treated with trastuzumab. Cancer Genomics Proteomics.
15:461–471. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JS, Tocheny CE and Shaw LM: The
insulin-like growth factor signaling pathway in breast cancer: An
elusive therapeutic target. Life (Basel). 12:19922022.PubMed/NCBI
|
|
37
|
Bilanges B, Posor Y and Vanhaesebroeck B:
PI3K isoforms in cell signalling and vesicle trafficking. Nat Rev
Mol Cell Biol. 20:515–534. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vanhaesebroeck B, Perry MWD, Brown JR,
André F and Okkenhaug K: PI3K inhibitors are finally coming of age.
Nat Rev Drug Discov. 20:741–769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Engelman JA: Targeting PI3K signalling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
9:550–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mayer IA and Arteaga CL: The PI3K/AKT
pathway as a target for cancer treatment. Annu Rev Med. 67:11–28.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tariq K and Luikart BW: Striking a
balance: PIP(2) and PIP(3) signaling in neuronal health and
disease. Explor Neuroprotective Ther. 1:86–100. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu M, Zhu S, Xiong S, Xue X and Zhou X:
MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review).
Oncol Rep. 41:1439–1454. 2019.PubMed/NCBI
|
|
43
|
Li YJ, Li XF, Yang EH and Shi M: Reaserch
Advances on the Role of PI3K/AKT Signaling Pathway and MiRNA in
Acute T-Cell Lymphocytic Leukemia-Review. Zhongguo Shi Yan Xue Ye
Xue Za Zhi. 27:1344–1347. 2019.(In Chinese). PubMed/NCBI
|
|
44
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang J, Wang X, Wen G and Ren Y:
miRNA-205-5p functions as a tumor suppressor by negatively
regulating VEGFA and PI3K/Akt/mTOR signaling in renal carcinoma
cells. Oncol Rep. 42:1677–1688. 2019.PubMed/NCBI
|
|
46
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Walter BA, Gómez-Macias G, Valera VA,
Sobel M and Merino MJ: miR-21 expression in pregnancy-associated
breast cancer: A possible marker of poor prognosis. J Cancer.
2:67–75. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gimm O, Perren A, Weng LP, Marsh DJ, Yeh
JJ, Ziebold U, Gil E, Hinze R, Delbridge L, Lees JA, et al:
Differential nuclear and cytoplasmic expression of PTEN in normal
thyroid tissue, and benign and malignant epithelial thyroid tumors.
Am J Pathol. 156:1693–1700. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li B, Lu Y, Wang H, Han X, Mao J, Li J, Yu
L, Wang B, Fan S, Yu X and Song B: RETRACTED: miR-221/222 enhance
the tumorigenicity of human breast cancer stem cells via modulation
of PTEN/Akt pathway. Biomed Pharmacother. 79:93–101. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li B, Lu Y, Wang H, Han X, Mao J, Li J, Yu
L, Wang B, Fan S, Yu X and Song B: miR-221/222 enhance the
tumorigenicity of human breast cancer stem cells via modulation of
PTEN/Akt pathway. Biomed Pharmacother. 79:93–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bahena-Ocampo I, Espinosa M,
Ceballos-Cancino G, Lizarraga F, Campos-Arroyo D, Schwarz A,
Maldonado V, Melendez-Zajgla J and Garcia-Lopez P: miR-10b
expression in breast cancer stem cells supports self-renewal
through negative PTEN regulation and sustained AKT activation. EMBO
Rep. 17:648–658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sarbassov DD, Guertin DA, Ali SM and
Sabatini DM: Phosphorylation and regulation of Akt/PKB by the
rictor-mTOR complex. Science. 307:1098–1101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang Z, Han Y, Cheng K, Zhang G and Wang
X: miR-99a directly targets the mTOR signalling pathway in breast
cancer side population cells. Cell Prolif. 47:587–595. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Imam JS, Plyler JR, Bansal H, Prajapati S,
Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, et
al: Genomic loss of tumor suppressor miRNA-204 promotes cancer cell
migration and invasion by activating AKT/mTOR/Rac1 signaling and
actin reorganization. PLoS One. 7:e523972012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang B, Zhao R, He Y, Fu X, Fu L, Zhu Z,
Fu L and Dong JT: MicroRNA 100 sensitizes luminal A breast cancer
cells to paclitaxel treatment in part by targeting mTOR.
Oncotarget. 7:5702–5714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Pakravan K, Babashah S, Sadeghizadeh M,
Mowla SJ, Mossahebi-Mohammadi M, Ataei F, Dana N and Javan M:
MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes
suppresses in vitro angiogenesis through modulating the
mTOR/HIF-1α/VEGF signaling axis in breast cancer cells. Cell Oncol
(Dordr). 40:457–470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Janaki Ramaiah M, Lavanya A, Honarpisheh
M, Zarea M, Bhadra U and Bhadra MP: MiR-15/16 complex targets p70S6
kinase 1 and controls cell proliferation in MDA-MB-231 breast
cancer cells. Gene. 552:255–264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hu Y, Zhu Q and Tang L: MiR-99a antitumor
activity in human breast cancer cells through targeting of mTOR
expression. PLoS One. 9:e920992014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang ZW, Guo RW, Lv JL, Wang XM, Ye JS,
Lu NH, Liang X and Yang LX: MicroRNA-99a inhibits insulin-induced
proliferation, migration, dedifferentiation, and rapamycin
resistance of vascular smooth muscle cells by inhibiting
insulin-like growth factor-1 receptor and mammalian target of
rapamycin. Biochem Biophys Res Commun. 486:414–422. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Banerjee S, Biehl A, Gadina M, Hasni S and
Schwartz DM: JAK-STAT signaling as a target for inflammatory and
autoimmune diseases: Current and Future Prospects. Drugs.
77:521–546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liang YK, Lin HY, Dou XW, Chen M, Wei XL,
Zhang YQ, Wu Y, Chen CF, Bai JW, Xiao YS, et al: MiR-221/222
promote epithelial-mesenchymal transition by targeting Notch3 in
breast cancer cell lines. NPJ Breast Cancer. 4:202018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Han M, Wang Y, Guo G, Li L, Dou D, Ge X,
Lv P, Wang F and Gu Y: microRNA-30d mediated breast cancer
invasion, migration, and EMT by targeting KLF11 and activating
STAT3 pathway. J Cell Biochem. 119:8138–8145. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mayoral-Varo V, Calcabrini A,
Sánchez-Bailón MP and Martín-Pérez J: miR205 inhibits stem cell
renewal in SUM159PT breast cancer cells. PLoS One. 12:e01886372017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng
X, Song Y, Meng Q, Yuan S, Luan L, et al: MiR-31 promotes mammary
stem cell expansion and breast tumorigenesis by suppressing Wnt
signaling antagonists. Nat Commun. 8:10362017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shi P, Chen C, Li X, Wei Z, Liu Z and Liu
Y: MicroRNA-124 suppresses cell proliferation and invasion of
triple negative breast cancer cells by targeting STAT3. Mol Med
Rep. 19:3667–3675. 2019.PubMed/NCBI
|
|
66
|
Qin Z, He W, Tang J, Ye Q, Dang W, Lu Y,
Wang J, Li G, Yan Q and Ma J: MicroRNAs Provide Feedback Regulation
of Epithelial-Mesenchymal Transition Induced by Growth Factors. J
Cell Physiol. 231:120–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tang Y, Wu B, Huang S, Peng X, Li X, Huang
X, Zhou W, Xie P and He P: Downregulation of miR-505-3p predicts
poor bone metastasis-free survival in prostate cancer. Oncol Rep.
41:57–66. 2019.PubMed/NCBI
|
|
68
|
Wang S, Huang M, Wang Z, Wang W, Zhang Z,
Qu S and Liu C: MicroRNA-133b targets TGFβ receptor I to inhibit
TGF-β-induced epithelial-to-mesenchymal transition and metastasis
by suppressing the TGF-β/SMAD pathway in breast cancer. Int J
Oncol. 55:1097–1109. 2019.PubMed/NCBI
|
|
69
|
Wang J, Liang S and Duan X: Molecular
mechanism of miR-153 inhibiting migration, invasion and
epithelial-mesenchymal transition of breast cancer by regulating
transforming growth factor beta (TGF-β) signaling pathway. J Cell
Biochem. 120:9539–9546. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Dai X, Fang M, Li S, Yan Y, Zhong Y and Du
B: miR-21 is involved in transforming growth factor β1-induced
chemoresistance and invasion by targeting PTEN in breast cancer.
Oncol Lett. 14:6929–6936. 2017.PubMed/NCBI
|
|
71
|
Chen Y, Huang S, Wu B, Fang J, Zhu M, Sun
L, Zhang L, Zhang Y, Sun M, Guo L and Wang S: Transforming growth
factor-β1 promotes breast cancer metastasis by downregulating
miR-196a-3p expression. Oncotarget. 8:49110–49122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yang Y, Hong M, Lian WW and Chen Z: Review
of the pharmacological effects of astragaloside IV and its
autophagic mechanism in association with inflammation. World J Clin
Cases. 10:10004–10016. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang GN, Zhang YK, Wang YJ, Gupta P,
Ashby CR Jr, Alqahtani S, Deng T, Bates SE, Kaddoumi A, Wurpel JND,
et al: Epidermal growth factor receptor (EGFR) inhibitor PD153035
reverses ABCG2-mediated multidrug resistance in non-small cell lung
cancer: In vitro and in vivo. Cancer Lett. 424:19–29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Farabaugh SM, Boone DN and Lee AV: Role of
IGF1R in breast cancer subtypes, stemness, and lineage
differentiation. Front Endocrinol (Lausanne). 6:592015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jang GB, Hong IS, Kim RJ, Lee SY, Park SJ,
Lee ES, Park JH, Yun CH, Chung JU, Lee KJ, et al: Wnt/β-Catenin
Small-Molecule Inhibitor CWP232228 preferentially inhibits the
growth of breast cancer stem-like cells. Cancer Res. 75:1691–1702.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Clemmons DR: Modifying IGF1 activity: An
approach to treat endocrine disorders, atherosclerosis and cancer.
Nat Rev Drug Discov. 6:821–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cao J and Yee D: Disrupting Insulin and
IGF receptor function in cancer. Int J Mol Sci. 22:5552021.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bowers LW, Cavazos DA, Maximo IX, Brenner
AJ, Hursting SD and deGraffenried LA: Obesity enhances nongenomic
estrogen receptor crosstalk with the PI3K/Akt and MAPK pathways to
promote in vitro measures of breast cancer progression. Breast
Cancer Res. 15:R592013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xu Y, Chao L, Wang J and Sun Y: miRNA-148a
regulates the expression of the estrogen receptor through
DNMT1-mediated DNA methylation in breast cancer cells. Oncol Lett.
14:4736–4740. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Melnik BC: Milk disrupts p53 and DNMT1,
the guardians of the genome: Implications for acne vulgaris and
prostate cancer. Nutr Metab (Lond). 14:552017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li X, Tang X, Li K and Lu L: Evaluation of
Serum MicroRNAs (miR-9-5p, miR-17-5p, and miR-148a-3p) as potential
biomarkers of breast cancer. Biomed Res Int.
2022:99614122022.PubMed/NCBI
|
|
82
|
Chawra HS, Agarwal M, Mishra A, Chandel
SS, Singh RP, Dubey G, Kukreti N and Singh M: MicroRNA-21′s role in
PTEN suppression and PI3K/AKT activation: Implications for cancer
biology. Pathol Res Pract. 254:1550912024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ruskovska T, Budić-Leto I, Corral-Jara KF,
Ajdžanović V, Arola-Arnal A, Bravo FI, Deligiannidou GE, Havlik J,
Janeva M, Kistanova E, et al: Systematic analysis of nutrigenomic
effects of polyphenols related to cardiometabolic health in
humans-Evidence from untargeted mRNA and miRNA studies. Ageing Res
Rev. 79:1016492022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Curtale G, Rubino M and Locati M:
MicroRNAs as molecular switches in macrophage activation. Front
Immunol. 10:7992019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sánchez-González I, Bobien A, Molnar C,
Schmid S, Strotbek M, Boerries M, Busch H and Olayioye MA: miR-149
suppresses breast cancer metastasis by blocking paracrine
interactions with macrophages. Cancer Res. 80:1330–1341. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zou X, Xia T, Li M, Wang T, Liu P, Zhou X,
Huang Z and Zhu W: MicroRNA profiling in serum: Potential
signatures for breast cancer diagnosis. Cancer Biomark. 30:41–53.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Warth SC, Hoefig KP, Hiekel A,
Schallenberg S, Jovanovic K, Klein L, Kretschmer K, Ansel KM and
Heissmeyer V: Induced miR-99a expression represses Mtor
cooperatively with miR-150 to promote regulatory T-cell
differentiation. EMBO J. 34:1195–1213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Singh Y, Garden OA, Lang F and Cobb BS:
MicroRNA-15b/16 enhances the induction of regulatory T cells by
regulating the expression of rictor and mTOR. J Immunol.
195:5667–5677. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Simanovich E, Brod V, Rahat MM and Rahat
MA: Function of miR-146a-5p in tumor cells as a regulatory switch
between cell death and angiogenesis: Macrophage therapy revisited.
Front Immunol. 8:19312018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zarogoulidis P, Petanidis S, Domvri K,
Kioseoglou E, Anestakis D, Freitag L, Zarogoulidis K,
Hohenforst-Schmidt W and Eberhardt W: Autophagy inhibition
upregulates CD4(+) tumor infiltrating lymphocyte expression via
miR-155 regulation and TRAIL activation. Mol Oncol. 10:1516–1531.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Giger ML: Update on the potential of
computer-aided diagnosis for breast cancer. Future Oncol. 6:1–4.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bilska-Wolak AO, Floyd CE Jr, Lo JY and
Baker JA: Computer aid for decision to biopsy breast masses on
mammography: Validation on new cases. Acad Radiol. 12:671–680.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Nassif AB, Talib MA, Nasir Q, Afadar Y and
Elgendy O: Breast cancer detection using artificial intelligence
techniques: A systematic literature review. Artif Intell Med.
127:1022762022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yanagawa M, Niioka H, Hata A, Kikuchi N,
Honda O, Kurakami H, Morii E, Noguchi M, Watanabe Y, Miyake J and
Tomiyama N: Application of deep learning (3-dimensional
convolutional neural network) for the prediction of pathological
invasiveness in lung adenocarcinoma: A preliminary study. Medicine
(Baltimore). 98:e161192019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tran WT, Sadeghi-Naini A, Lu FI, Gandhi S,
Meti N, Brackstone M, Rakovitch E and Curpen B: Computational
radiology in breast cancer screening and diagnosis using artificial
intelligence. Can Assoc Radiol J. 72:98–108. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Welch HG, Prorok PC, O'Malley AJ and
Kramer BS: Breast-Cancer tumor size, overdiagnosis, and mammography
screening effectiveness. N Engl J Med. 375:1438–1447. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
S P, N KV and S S: Breast cancer detection
using crow search optimization based intuitionistic fuzzy
clustering with neighborhood attraction. Asian Pac J Cancer Prev.
20:157–165. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cruz-Bernal A, Flores-Barranco MM,
Almanza-Ojeda DL, Ledesma S and Ibarra-Manzano MA: Analysis of the
Cluster Prominence Feature for Detecting Calcifications in
Mammograms. J Healthc Eng. 2018:28495672018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hmida M, Hamrouni K, Solaiman B and
Boussetta S: Mammographic mass segmentation using fuzzy contours.
Comput Methods Programs Biomed. 164:131–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lei YM, Yin M, Yu MH, Yu J, Zeng SE, Lv
WZ, Li J, Ye HR, Cui XW and Dietrich CF: Artificial intelligence in
medical imaging of the breast. Front Oncol. 11:6005572021.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Herranz H and Cohen SM: MicroRNAs and gene
regulatory networks: Managing the impact of noise in biological
systems. Genes Dev. 24:1339–1344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Nassar FJ, Nasr R and Talhouk R: MicroRNAs
as biomarkers for early breast cancer diagnosis, prognosis and
therapy prediction. Pharmacol Ther. 172:34–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Itani MM, Nassar FJ, Tfayli AH, Talhouk
RS, Chamandi GK, Itani ARS, Makoukji J, Boustany RN, Hou L, Zgheib
NK and Nasr RR: A signature of four circulating microRNAs as
potential biomarkers for diagnosing early-stage breast cancer. Int
J Mol Sci. 22:61212021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C,
Wang X, Luo Z, Wang J, Liu S, et al: microRNA-21 promotes breast
cancer proliferation and metastasis by targeting LZTFL1. BMC
Cancer. 19:7382019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Najjary S, Mohammadzadeh R, Mokhtarzadeh
A, Mohammadi A, Kojabad AB and Baradaran B: Role of miR-21 as an
authentic oncogene in mediating drug resistance in breast cancer.
Gene. 738:1444532020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Shi Y, Ye P and Long X: Differential
expression profiles of the transcriptome in breast cancer cell
lines revealed by next generation sequencing. Cell Physiol Biochem.
44:804–816. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Dinami R, Ercolani C, Petti E, Piazza S,
Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R, et
al: miR-155 drives telomere fragility in human breast cancer by
targeting TRF1. Cancer Res. 74:4145–4156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W,
Yu M, Lin J and Cui Q: MicroRNAs involved in carcinogenesis,
prognosis, therapeutic resistance and applications in human
triple-negative breast cancer. Cells. 8:14922019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rajabi H, Jin C, Ahmad R, McClary C, Joshi
MD and Kufe D: MUCIN 1 ONCOPROTEIN EXPRESSION IS SUPPRESSED BY THE
miR-125b ONCOMIR. Genes Cancer. 1:62–68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Tang F, Zhang R, He Y, Zou M, Guo L and Xi
T: MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7
and MDA-MB-231 breast cancer cells. PLoS One. 7:e354352012.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wang S, Oh DY, Leventaki V, Drakos E,
Zhang R, Sahin AA, Resetkova E, Edgerton ME, Wu W and Claret FX:
MicroRNA-17 acts as a tumor chemosensitizer by targeting JAB1/CSN5
in triple-negative breast cancer. Cancer Lett. 465:12–23. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Teichgraeber DC, Guirguis MS and Whitman
GJ: Breast cancer staging: Updates in the AJCC cancer staging
manual, 8th edition, and current challenges for radiologists, from
the AJR special series on cancer staging. AJR Am J Roentgenol.
217:278–290. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wang ZX, Lu BB, Wang H, Cheng ZX and Yin
YM: MicroRNA-21 modulates chemosensitivity of breast cancer cells
to doxorubicin by targeting PTEN. Arch Med Res. 42:281–290. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Filková M, Jüngel A, Gay RE and Gay S:
MicroRNAs in rheumatoid arthritis: Potential role in diagnosis and
therapy. BioDrugs. 26:131–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhou Q, Haupt S, Kreuzer JT, Hammitzsch A,
Proft F, Neumann C, Leipe J, Witt M, Schulze-Koops H and Skapenko
A: Decreased expression of miR-146a and miR-155 contributes to an
abnormal Treg phenotype in patients with rheumatoid arthritis. Ann
Rheum Dis. 74:1265–1274. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sun SS, Zhou X, Huang YY, Kong LP, Mei M,
Guo WY, Zhao MH, Ren Y, Shen Q and Zhang L: Targeting STAT3/miR-21
axis inhibits epithelial-mesenchymal transition via regulating CDK5
in head and neck squamous cell carcinoma. Mol Cancer. 14:2132015.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Carbognin L, Miglietta F, Paris I and
Dieci MV: Prognostic and predictive implications of PTEN in breast
cancer: Unfulfilled promises but intriguing perspectives. Cancers
(Basel). 11:14012019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yu X, Li R, Shi W, Jiang T, Wang Y, Li C
and Qu X: Silencing of MicroRNA-21 confers the sensitivity to
tamoxifen and fulvestrant by enhancing autophagic cell death
through inhibition of the PI3K-AKT-mTOR pathway in breast cancer
cells. Biomed Pharmacother. 77:37–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ,
Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, et al: Knockdown of miR-21 in
human breast cancer cell lines inhibits proliferation, in vitro
migration and in vivo tumor growth. Breast Cancer Res. 13:R22011.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wu X: Expressions of miR-21 and miR-210 in
breast cancer and their predictive values for prognosis. Iran J
Public Health. 49:21–29. 2020.PubMed/NCBI
|
|
121
|
Nivetha R, Arvindh S, Baba AB, Gade DR,
Gopal G, K C, Reddy KP, Reddy GB and Nagini S: Nimbolide, a neem
limonoid, inhibits angiogenesis in breast cancer by abrogating
aldose reductase mediated IGF-1/PI3K/Akt signalling. Anticancer
Agents Med Chem. 22:2619–2636. 2022. View Article : Google Scholar : PubMed/NCBI
|