Introduction
Gastric cancer (GC) was the fourth most common
cancer globally in 2020, and 769,000 people died from GC that year
according to GLOBOCAN estimates (1). A high sodium intake explains many
cases of GC, and this association can be explained by two important
factors: i) Salt strongly irritates the stomach wall and promotes
chemical gastric carcinogenesis; and ii) excess salt induces
gastric colonization of Helicobacter pylori in the stomach,
which is a known risk factor for GC (2).
Genetic factors serve an important role in gastric
carcinogenesis due to aberrant gene expression, which leads to a
malignant phenotype (3). A total of
≥44 GC-related genes have been reported to date (4). Adducin (ADD) family members are
involved in oncogenic signal transduction pathways in several types
of cancer (5). The ADD1 gene
encodes adducin 1, which is a cytoskeletal protein that is
ubiquitously expressed and implicated in the formation of
actin-spectrin complexes, actin polymerization and cell signal
transduction (6,7). The rs4961 c.1378 G>T p.Gly460Trp
variant of the ADD1 gene located at chromosome 4p16.3 has
been associated with salt-sensitive hypertension, and it is
reported that ethnic differences and genetic background may be the
main causes of salt sensitivity (8–11).
This variant is also related to other diseases, such as hemorrhagic
stroke, atherosclerosis, myocardial infarction and renal disease
(5).
Genetic variation in several genes may account for
the increased salt sensitivity of certain individuals (12). Moreover, when regarding the
predisposition towards GC, individuals who are not sensitive to
salt may ingest larger amounts of salt than necessary or normal
when attempting to taste salt in food, thus increasing the
likelihood of developing GC. To the best of our knowledge, the
association between the rs4961 variant of the ADD1 gene and
the risk of developing GC has not yet been assessed. Therefore, the
aim of the present study was to evaluate the association between
the rs4961 variant and the development of preneoplastic gastric
lesions (PGLs) and GC in a population from western Mexico.
Materials and methods
The present study assessed 225 subjects recruited
from March 2018 to November 2022 from the gastroenterology services
of the National Medical Center of the West, Hospital 110, Hospital
14 and Ambulatory Care Medical Unit 52 of the Mexican Social
Security Institute (Guadalajara, Mexico).
The subjects underwent endoscopy as part of their
diagnosis, and a biopsy was taken for histopathological analysis
(GC, n=71; PGL, n=53; and controls, n=101). Briefly, 5 µm-thick
tissue sections were placed on electrocharged slides, and
hematoxylin and eosin staining was applied as follows, the
electrocharged slides were placed at 55°C for 15 min, placed in
xylol for 5 min and then in decreasing ethanol solutions (100, 90,
70 and 30%), and finally in distilled water. Subsequently, the
samples were placed in hematoxylin for 8 min at room temperature,
rinsed and placed in an acidic alcohol bath, and rinsed and placed
in lithium chloride for 1 min. After which, the samples were rinsed
and left for 30 sec in alcohol, and then they were placed in eosin
for 1 min at room temperature. The sections were rinsed and
dehydrated in solutions of increasing concentrations of ethanol,
and subsequently placed in xylol for 5 min. Finally, the sections
were covered with resin and coverslips, and allowed to dry for 8 h
before observation under an optical microscope. All of the subjects
in the GC group were adults, and the ages in the PGL and control
groups ranged from 2–89 years. Diagnoses was made using a
histopathological assessment of the biopsies by a pathologist, and
based on Lauren's classification (13), with GC including diffuse and
intestinal types and PGL involving atrophic gastritis and
intestinal metaplasia. The control group consisted of subjects with
non-atrophic gastritis (Fig. S1).
The inclusion criteria was as follows: i) Individuals who would
undergo an endoscopy study as part of their diagnosis; ii) those
who were not undergoing any treatment for chronic gastritis or
gastric cancer; and iii) those who agreed to participate in the
present study and signed a letter of informed consent. Individuals
whose gastric tissue or DNA samples did not have sufficient quality
or quantity for analysis were excluded from the present study.
DNA was extracted from peripheral blood leukocytes
using the salting out method, as described by Miller et al
(14). To identify the rs4961
G>T variant of the ADD1 gene, a 415 bp DNA fragment
including the region of interest was amplified using PCR. The
following primers were designed by Oligo software (version 6.0;
http://oligo.net/oligo_updates.htm):
Forward, 5′-GGGCTACAGAACTGGCTACC-3′ and reverse,
5′-GCCTCCGAAGCCCCAGCTACCCA-3′. The reaction was performed under the
following conditions: 100 ng of genomic DNA, 5 pM of each primer,
0.5 U of Taq polymerase (Invitrogen™; Thermo Fisher
Scientific, Inc.), 1X PCR buffer, 1.5 mM MgCl2 and 2 mM
deoxynucleotide triphosphate mix (dNTP Set; Vivantis Technologies
Sdn. Bhd.). The PCR conditions were as follows: 94°C for 5 min; 30
cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min; then
72°C for 5 min. This was performed by using a 2720 Thermal Cycler
(Applied Biosystems™; Thermo Fisher Scientific, Inc.).
The resulting fragment was purified using ExoSAP-IT™ PCR
Product Cleanup Reagent (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and Sanger capillary sequencing was performed
using the BigDye™ Terminator v3.1 Cycle Sequencing Kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with the
SeqStudio Genetic Analyser (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and analyzed using Chromas DNA Sequencing
Software (https://technelysium.com.au/wp/chromas/).
The allelic and genotypic frequencies were obtained
by direct counting, and comparisons between groups were analyzed
using Fisher's exact test using the PASW Statistic Base 18 software
(SPSS, Inc.). Other variables, such as sex and age, were analyzed
using the χ2 and unpaired Student's t-tests. P<0.05
was considered to indicate a statistically significant
difference.
In addition, a cohort from the Genomic Data Commons
(GDC) portal (https://portal.gdc.cancer.gov/; v.39.0) was obtained,
which included patients for whom the stomach was the primary cancer
site (n=824). The datasets (appears as ‘projects’ in the GDC
portal) included were EXCEPTIONAL_RESPONDERS-ER, FM-AD, HCMI-CMDC,
MATCH-S1, MATCH-Z1D, TARGET-NBL, TCGA-DLBC, TCGA-ESCA, TCGA-SARC
and TCGA-STAD. These datasets (projects) belong to the following
programs: EXCEPTIONAL_RESPONDERS, FM, HCMI, MATCH, TARGET and TCGA.
Mutations in the ADD1 gene and demographic data in the
cohort were searched for using ProteinPaint, Mutation Frequency and
OncoMatrix analysis tools, which are all freely available at the
GDC portal (https://portal.gdc.cancer.gov/).
Results
The demographic data of the three groups are
reported in Table I. The GC group
had a significantly greater proportion of males (63%) than females
(37%) compared with the control group (41% vs. 59%, respectively;
P=0.003); however, the PGL group had a similar proportion compared
with the control group (P=0.737) (Table
I). Furthermore, the odds ratio (OR) for male sex was
evaluated, and the results demonstrated an increased risk of GC in
males compared with that in controls, with an OR of 2.5 and a 95%
confidence interval (CI) of 1.35–4.73 (P=0.003; Table I). Conversely, mean age was
significantly higher in the GC group (59.8 years) than in the PGL
and control groups (58.9 and 46.5 years, respectively; P<0.05;
Table I).
 | Table I.Demographic data of the three groups
studied. |
Table I.
Demographic data of the three groups
studied.
| Variable | Gastric cancer
(n=71) | P-value | Preneoplastic
gastric lesions (n=53) | P-value | Controls
(n=101) | P-value |
|---|
| Sex |
|
0.00300a |
|
0.73700b |
|
0.02600c |
|
Female | 26 (36.6) |
| 30 (56.6) |
| 60 (59.4) |
|
|
Male | 45 (63.4) |
| 23 (43.4) |
| 41 (40.6) |
|
| Age, years | 59.8±14.8 |
0.00003a | 58.9±18.1 |
0.00090b | 46.5±23.0 |
0.38500c |
The frequency of the mutated homozygous genotype
(TT) of the rs4961 variant was <10% in the three evaluated
groups, and the frequency of the minor allele (T) was <21% in
the GC, PGL and control groups (Table
II). The allele and genotype frequencies of rs4961 polymorphism
were in the Hardy-Weinberg equilibrium (P>0.05; data not
shown).
 | Table II.Genotypic and allelic frequencies of
the ADD1 rs4961 variant observed in the studied groups. |
Table II.
Genotypic and allelic frequencies of
the ADD1 rs4961 variant observed in the studied groups.
| Frequency | Gastric cancer
(n=71) | P-value | Preneoplastic
gastric lesions (n=53) | P-value | Controls
(n=101) | P-value |
|---|
| Genotype |
| 0.301a |
| 0.408b |
| 0.705c |
|
Wildtype GG | 52 (73.2) |
| 35 (66.0) |
| 67 (66.3) |
|
|
Heterozygous GT | 15 (21.1) |
| 14 (26.4) |
| 31 (30.7) |
|
| Mutated
homozygous TT | 4 (5.6) |
| 4 (7.5) |
| 3 (3.0) |
|
| Allele |
| 0.732a |
| 0.603b |
| 0.406c |
| Allele
wildtype (G) | 119 (83.8) |
| 84 (79.2) |
| 166 (81.7) |
|
| Allele
mutated (T) | 23 (16.2) |
| 22 (20.8) |
| 36 (18.3) |
|
The distributions of allelic and genotypic
frequencies of rs4961 were compared between the three groups;
however, no differences were observed in genotypic or allelic
frequencies between GCs and controls or between PGLs and controls
(P>0.05; Table II). The
wild-type, heterozygous, and homozygous mutated genotypes observed
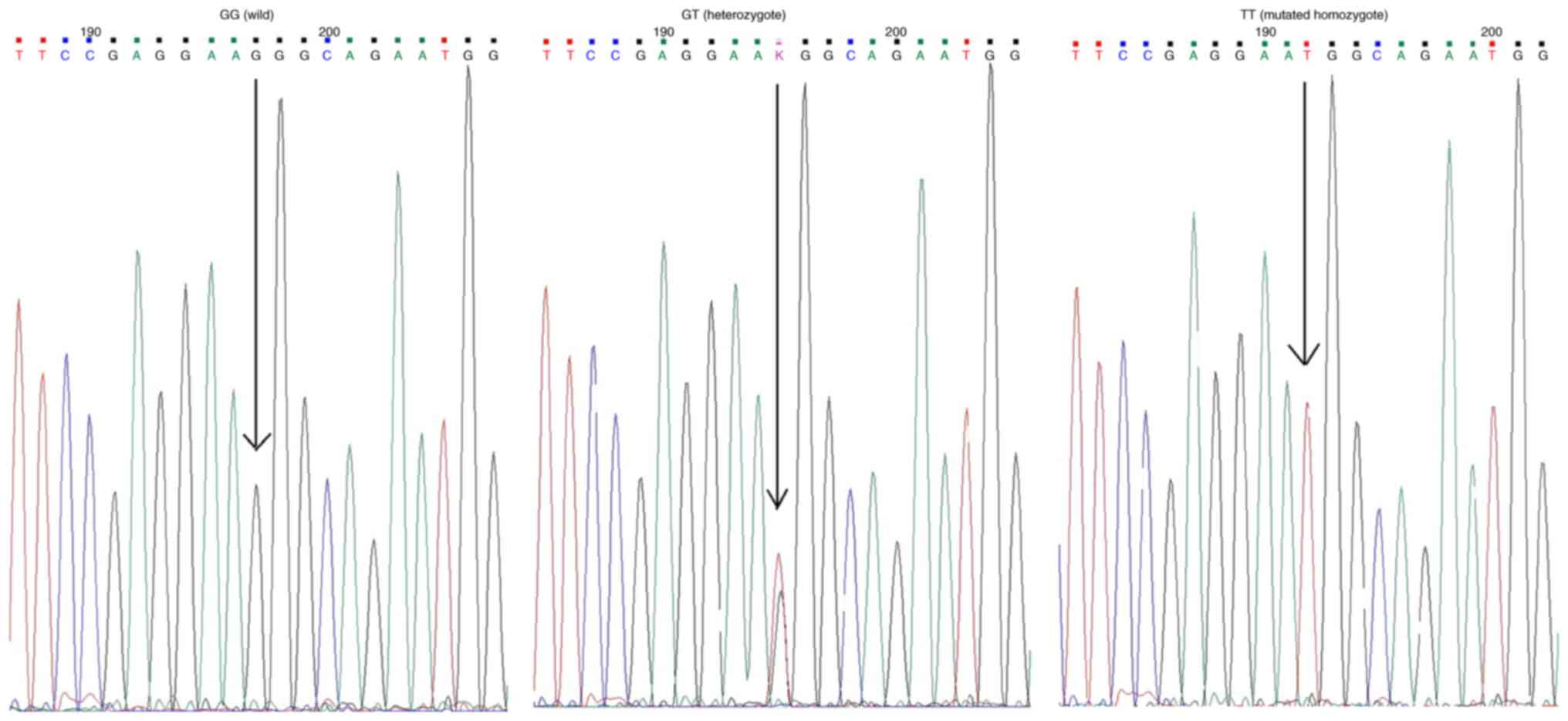
by Sanger sequencing are shown in Fig.
1.
Furthermore, data from the GDC portal revealed that
the ADD1 gene was altered in 4,103 patients with cancer
affected by 3,975 copy number variation (CNV) events (duplications
+ deletions) across 47 datasets (Fig.
2). In addition, 182 patients were affected by 189 other
non-CNV mutations in the ADD1 gene across 27 projects
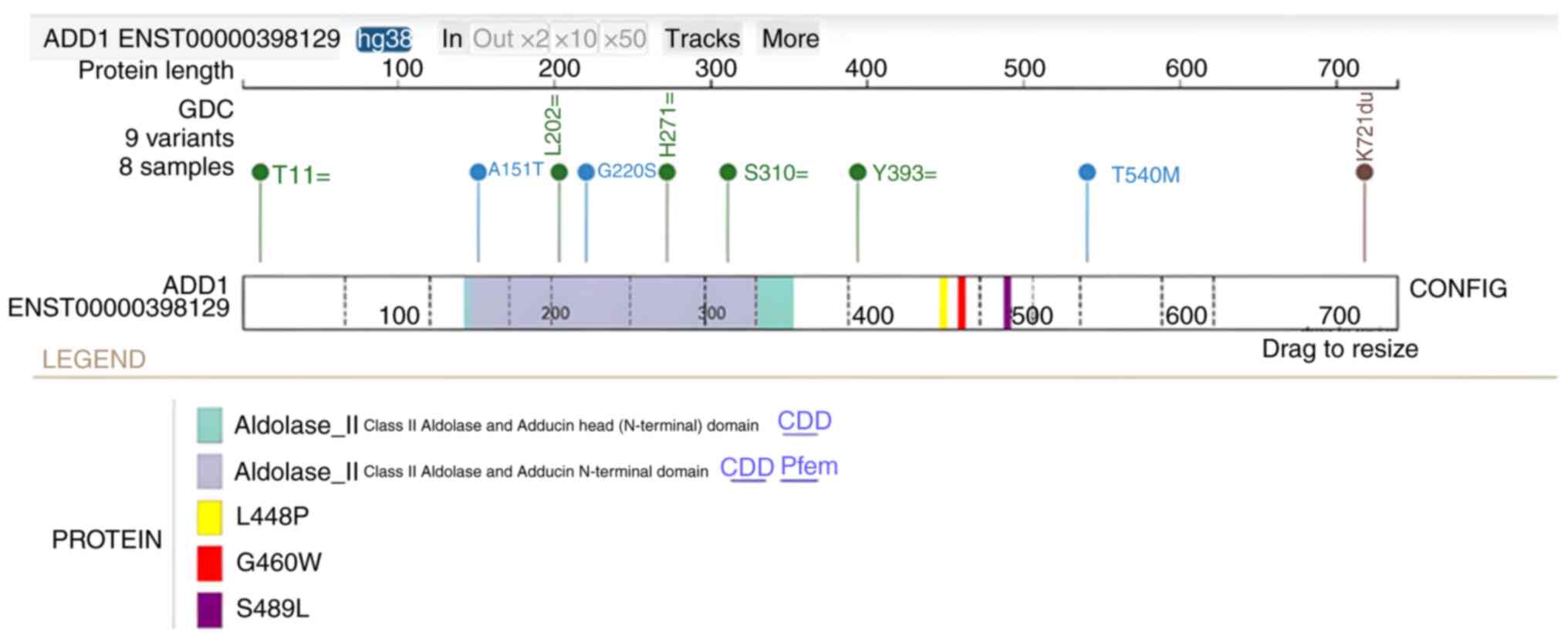
(Fig. 3). Subsequently, analysis of
the GC cohort (n=824) demonstrated that there were 8 patients with
5 silent, 3 missense and 1 stop-gain mutations in ADD1
(Fig. 4). Notably, L448P and S489L
are two likely damaging mutations (Polyphen score >0.99;
http://www.ensembl.org/info/genome/variation/prediction/protein_function.html)
and both mutations are in proximity to rs4961 (Fig. 4). The L448P mutation was found to be
present in two Caucasian male patients with skin cancer and the
S489L mutation was present in two Caucasian male patients with
colon and mouth cancer in the database. Within the same cohort, but
excluding CNVs (female, n=9 and male, n=28), the most commonly
mutated genes were Mucin 16, AT-rich interactive domain-containing
protein 1A and tumor protein 53 (Fig.
5). CNVs in the GC cohort are presented in Fig. 6.
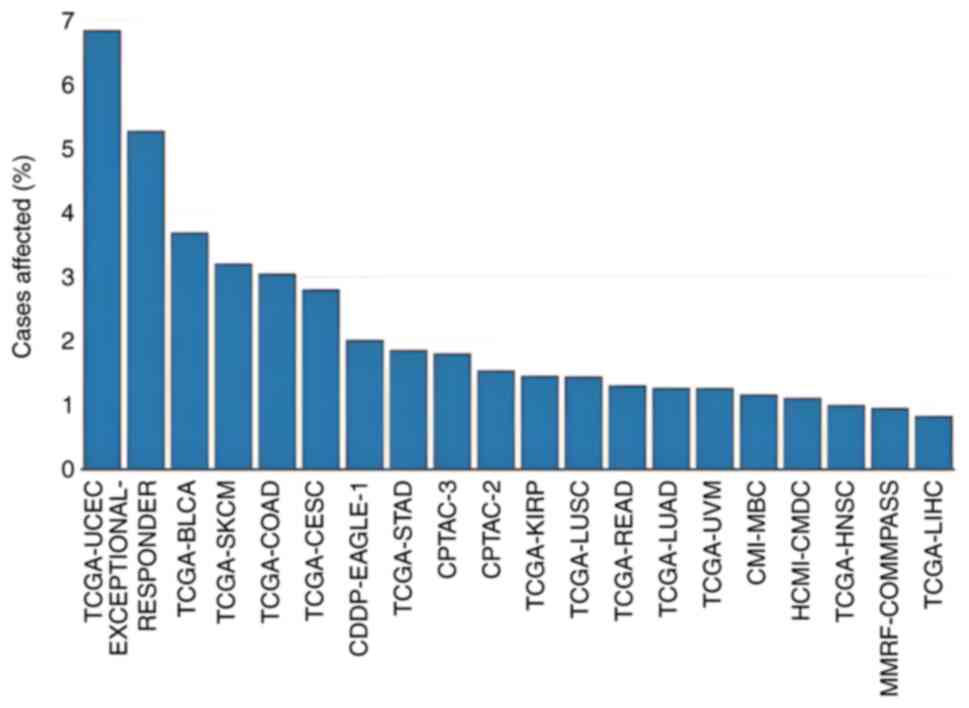 | Figure 2.Proportion of cancer cases in each
dataset in the Genomic Data Commons portal where there is a
mutation (except copy number variants) in the adducin 1 gene. BLCA,
bladder urothelial carcinoma; CDDP-EAGLE-1, CDDP Integrative
Analysis of Lung Adenocarcinoma (Phase 2); CESC, cervical squamous
cell carcinoma and endocervical adenocarcinoma; CMI-MBC, count Me
In (CMI): The Metastatic Breast Cancer (MBC) Project; COAD, colon
adenocarcinoma; CPTAC-2, CPTAC-Breast, Colon, Ovary; CPTAC-3,
CPTAC-Brain, Head and Neck, Kidney, Lung, Pancreas, uterus;
HCMI-CMDC, NCI Cancer Model Development for the Human Cancer Model
Initiative; HNSC, head and neck squamous cell carcinoma; KIRP,
kidney renal papillary cell carcinoma; LIHC, liver hepatocellular
carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell
carcinoma; MMRF-COMMPASS, Multiple Myeloma CoMMpass Study; READ,
rectum adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach
adenocarcinoma; TCGA, The Cancer Genome Atlas; UCEC, uterine corpus
endometrial carcinoma; UVM, uveal melanoma. |
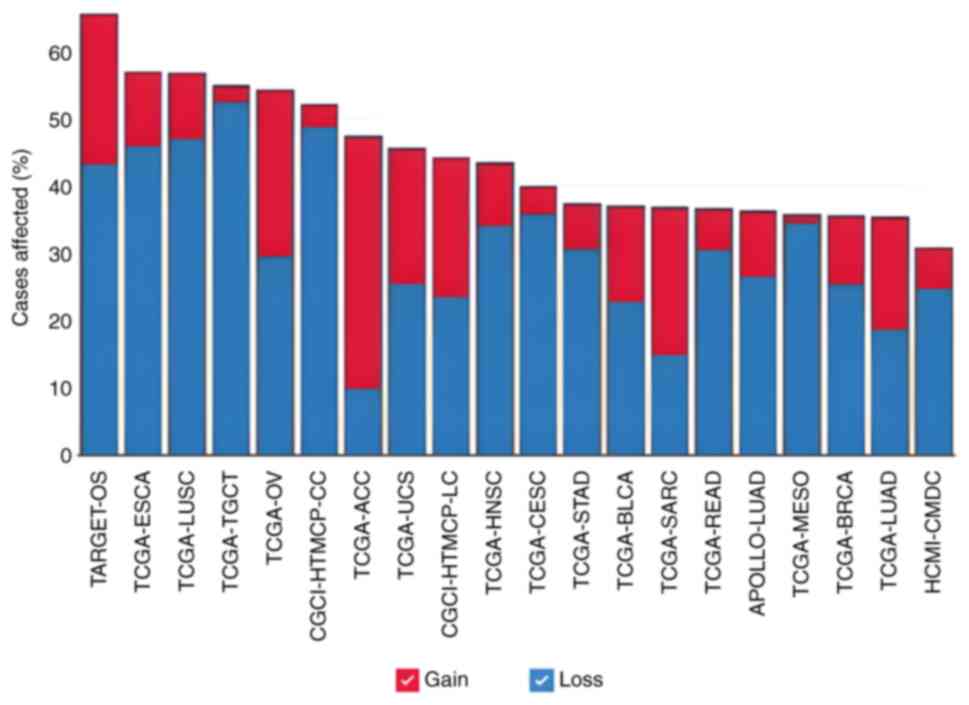 | Figure 3.Proportion of cancer cases in each
dataset in the Genomic Data Commons portal where there are copy
number variants mutations in adducin 1 gene. ACC, adrenocortical
carcinoma; APOLLO-LUAD, APOLLO1: Proteogenomic characterization of
lung adenocarcinoma; BLCA, bladder urothelial carcinoma; BRCA,
breast invasive carcinoma; CESC, cervical squamous cell carcinoma
and endocervical adenocarcinoma; CGCI-HTMCP-CC, HIV+ Tumor
Molecular Characterization Project-Cervical Cancer; CGCI-HTMCP-LC,
HIV+ Tumor Molecular Characterization Project-Lung Cancer; ESCA,
esophageal carcinoma; HCMI-CMDC, NCI Cancer Model Development for
the Human Cancer Model Initiative; HNSC, head and neck squamous
cell carcinoma; LUSC, lung squamous cell carcinoma; MESO,
mesotelioma; OV, ovarian serous cystadenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; STAD, stomach adenocarcinoma;
TARGET-OS, TARGET osteosarcoma; TCGA, The Cancer Genome Atlas;
TGCT, testicular germ cell tumors; UCS, uterine carcinosarcoma. |
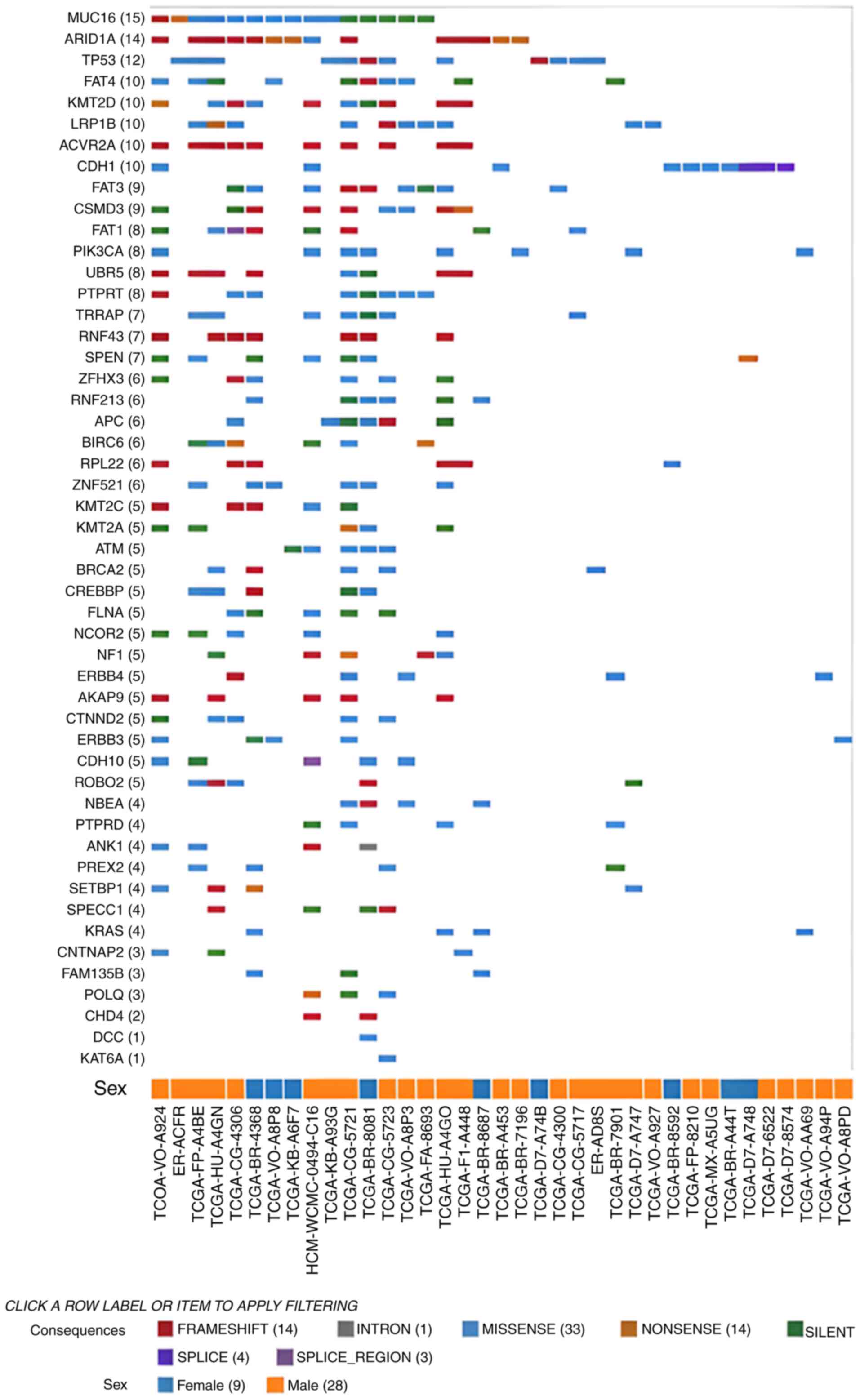 | Figure 5.Mutations (excluding copy number
variants) found in the gastric cancer cohort (n=824) of the Genomic
Data Commons portal. Each row is a gene with the highest mutation
frequencies from top to bottom. Each column refers to a patient:
Males (orange squares) and females (blue squares). The codes at the
bottom of the figure refers to the unique ID of each patient.
ACVR2A, activin A receptor type 2A; AKAP9, A-kinase
anchoring protein 9; ANK1, ankyrin 1; APC, APC
regulator of WNT signaling pathway; ARID1A, AT-rich
interaction domain 1A; ATM, ATM serine/threonine kinase;
BIRC6, baculoviral IAP repeat containing 6; BRCA2,
BRCA2 DNA repair associated; CDH1, cadherin 1; CDH10,
cadherin 10; CHD4, chromodomain helicase DNA binding protein
4; CNTNAP2, contactin associated protein 2; CREBBP,
CREB binding protein; CSMD3, CUB and Sushi multiple domains
3; CTNND2, catenin delta 2; DCC, DCC netrin 1
receptor; ERBB4, erb-b2 receptor tyrosine kinase 4;
FAM135B, family with sequence similarity 135 member B;
FAT1, FAT atypical cadherin 1; FAT3, FAT atypical
cadherin 4; FAT4, FAT atypical cadherin 4; FLNA,
filamin A; KAT6A, lysine acetyltransferase 6A; KMT2A,
lysine methyltransferase 2A; KMT2C, lysine methyltransferase
2C; KMT2D, lysine methyltransferase 2D; KRAS, KRAS
proto-oncogene, GTPase; LRP1B, LDL receptor related protein
1B; MUC16, mucin 16; NBEA, neurobeachin;
NCOR2, nuclear receptor corepressor 2; NF1,
neurofibromin 1; PIK3CA,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha; POLQ, DNA polymerase theta; PTPRD, protein
tyrosine phosphatase receptor type D; PTPRT, protein
tyrosine phosphatase receptor type T; PREX2,
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange
factor 2; ROBO2, roundabout guidance receptor 2;
RPL22, ribosomal protein L22; RNF43, ring finger
protein 43; RNF213, ring finger protein 213; SETBP1,
SET binding protein 1; SPECC1, sperm antigen with calponin
homology and coiled-coil domains; SPEN, spen family
transcriptional repressor; TP53, tumor protein p53;
TRRAP, transformation/transcription domain associated
protein; UBR5, ubiquitin protein ligase E3 component
n-recognin 5; ZFHX3, zinc finger homeobox 3; ZNF521,
zinc finger protein 521. |
 | Figure 6.Copy number variants found in the
gastric cancer cohort (n=824) of the Genomic Data Commons portal,
with copy number gains (pink) and losses (green). Each column
refers to each patient: Males (orange squares) and females (blue
square). The codes at the bottom of the figure refers to the unique
ID of each patient. ACVR2A, activin A receptor type 2A;
AKAP9, A-kinase anchoring protein 9; ANK1, ankyrin 1;
APC, APC regulator of WNT signaling pathway; ARID1A,
AT-rich interaction domain 1A; ATM, ATM serine/threonine
kinase; BIRC6, baculoviral IAP repeat containing 6;
BRCA2, BRCA2 DNA repair associated; CDH1, cadherin 1;
CDH10, cadherin 10; CHD4, chromodomain helicase DNA
binding protein 4; CNTNAP2, contactin associated protein 2;
CREBBP, CREB binding protein; CSMD3, CUB and Sushi
multiple domains 3; CTNND2, catenin delta 2; DCC, DCC
netrin 1 receptor; ERBB4, erb-b2 receptor tyrosine kinase 4;
FAM135B, family with sequence similarity 135 member B;
FAT1, FAT atypical cadherin 1; FAT3, FAT atypical
cadherin 4; FAT4, FAT atypical cadherin 4; FLNA,
filamin A; KAT6A, lysine acetyltransferase 6A; KMT2A,
lysine methyltransferase 2A; KMT2C, lysine methyltransferase
2C; KMT2D, lysine methyltransferase 2D; KRAS, KRAS
proto-oncogene, GTPase; LRP1B, LDL receptor related protein
1B; MUC16, mucin 16; NBEA, neurobeachin;
NCOR2, nuclear receptor corepressor 2; NF1,
neurofibromin 1; PIK3CA,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha; POLQ, DNA polymerase theta; PTPRD, protein
tyrosine phosphatase receptor type D; PTPRT, protein
tyrosine phosphatase receptor type T; PREX2,
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange
factor 2; ROBO2, roundabout guidance receptor 2;
RPL22, ribosomal protein L22; RNF43, ring finger
protein 43; RNF213, ring finger protein 213; SETBP1,
SET binding protein 1; SPECC1, sperm antigen with calponin
homology and coiled-coil domains; SPEN, spen family
transcriptional repressor; TP53, tumor protein p53;
TRRAP, transformation/transcription domain associated
protein; UBR5, ubiquitin protein ligase E3 component
n-recognin 5; ZFHX3, zinc finger homeobox 3; ZNF521,
zinc finger protein 521. |
Discussion
The rs4961 c.1378 G>T, p.Gly460Trp variant of the
ADD1 gene has been associated with salt-sensitive
hypertension and renal cell cancer risk and is a candidate for
breast cancer susceptibility (15,16).
To the best of our knowledge, the role of the rs4961 variant in GC
risk has not been previously assessed.
In the present study, the rs4961 variant of the
ADD1 gene was evaluated in relation to GC risk, and the
results excluded the rs4961 variant as being a risk factor for
gastric carcinogenesis in the Mexican population. However,
additional studies are needed to determine the role of this variant
in GC in this and other populations, as well as the relationship
between the ADD1 gene and cancer.
Notably, interethnic differences may explain the
observed differences in the effect of the rs4961 variant. For
instance, a previous study reported that in patients with
hypertension and coronary artery disease, Black patients and
ADD1 variant carriers (GT or TT) were at greater risk of a
primary outcome event than those with wild-type homozygotes [GG;
adjusted hazard ratio (HR), 2.62; 95% CI, 1.23–5.58; P=0.012), with
an 8-fold increase in the risk of death and a similar trend in
White (HR, 1.24; 95% CI, 0.90–1.71) and Hispanic patients (adjusted
HR, 1.43; 95% CI, 0.86–2.39); however, the difference was not-
significant and had a smaller magnitude of effect (17).
In a previous study, the ADD1 rs4961 variant
was reported to be related to cancer renal cell risk, with an HR of
1.24 (95% CI, 1.01–1.53) for the GT + TT vs. GG genotype, yet these
results were not statistically significant after adjustments for
multiple testing (15).
Additionally, the variant rs4961 has been reported to be a
candidate for breast cancer susceptibility (16). Although there is no direct evidence
of the role of ADD1 in breast cancer progression and
tumorigenesis, variants of this gene, such as rs4961 and rs4963,
have been reported to occur more frequently in patients with
higher-grade malignant breast tumors (III vs. II) (18). Hypertension is also a common
comorbidity in patients with breast cancer and is associated with a
poor prognosis. Li et al (18) reported that 77% of patients with
grade III breast cancer carries both rs4961 and rs4963 variants,
thus indicating an increased risk of developing hypertension
compared with 16% of patients with grade III breast cancer.
Furthermore, the present study demonstrated that
male sex confers a significantly increased risk of GC, with an OR
of 2.5 (95% CI, 1.35–4.73; P=0.003). Male sex is a well-known risk
factor for GC and other types of cancer (19).
Nonetheless, the present study had several
limitations, such as the small sample size of patients with GC who
were included in the analysis. It is therefore important to
consider matching samples by age and sex in future studies. A
further limitation of the present study is that the
clinicopathological characteristics of patients with GC were not
collected, such as tumor size, lymph nodes and tumor infiltration,
therefore, it is difficult to specify the relationship of the
rs4961 variant of the ADD1 gene with these variables, as
well as with the prognosis of the patients.
In conclusion, in the present population, the
ADD1 rs4961 variant was not associated with GC risk;
however, its role in other populations and in other types of cancer
is worthy of future research.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Bill Wysocki
(Center for Translational Science, The University of Chicago,
Chicago, USA) for providing training with the GDC portal tools.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MYAF and KRP performed the PCR and DNA sequencing
experiments, contributed to the acquisition, analysis and
interpretation of the data and were the major contributors to the
writing of the manuscript. SRDLTG contributed to the analysis and
interpretation of the data. FJPD, MTMT and EPMDO contributed to the
acquisition, analysis and interpretation of data, and manuscript
drafting or critical revisions of the intellectual content. JYSL
contributed to the conception and design of the study, as well as
analysis and interpretation of the data and approval of the final
manuscript version to be published. JYSL also agreed to be
accountable for all aspects of the work so that any questions
relating to research integrity or scientific accuracy in any part
of the study could be appropriately investigated and resolved. FJPD
and JYSL confirmed the authenticity of all of the raw data. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the local
Committee of Health Research and Ethics of the Western Biomedical
Research Center, Mexican Social Security Institute (Guadalajara,
Mexico; approval no. R-2023-1305-009). As the research involved
human subjects, written informed consent was obtained from all of
the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar
|
|
2
|
Wu X, Chen L, Cheng J, Qian J, Fang Z and
Wu J: Effect of dietary salt intake on risk of gastric cancer: A
systematic review and meta-analysis of case-control studies.
Nutrients. 14:42602022. View Article : Google Scholar
|
|
3
|
Fu DG: Epigenetic alterations in gastric
cancer (review). Mol Med Rep. 12:3223–3230. 2015. View Article : Google Scholar
|
|
4
|
Lee S, Yang HK, Lee HJ, Park DJ, Kong SH
and Park SK: Systematic review of gastric cancer-associated genetic
variants, gene-based meta-analysis, and gene-level functional
analysis to identify candidate genes for drug development. Front
Genet. 13:9287832022. View Article : Google Scholar
|
|
5
|
Kiang KM and Leung GK: A review on adducin
from functional to pathological mechanisms: Future direction in
cancer. Biomed Res Int. 2018:34659292018. View Article : Google Scholar
|
|
6
|
Hughes CA and Bennett V: Adducin: A
physical model with implications for function in assembly of
spectrin-actin complexes. J Biol Chem. 270:18990–18996. 1995.
View Article : Google Scholar
|
|
7
|
Matsuoka Y, Li X and Bennett V: Adducin:
Structure, function and regulation. Cell Mol Life Sci. 57:884–895.
2000. View Article : Google Scholar
|
|
8
|
Jin H, Huang Y and Yang G: Association
between α-adducin rs4961 polymorphism and hypertension: A
meta-analysis based on 40 432 subjects. J Cell Biochem.
120:4613–4619. 2019. View Article : Google Scholar
|
|
9
|
Ju Z, Zhang H, Sun K, Song Y, Lu H, Hui R
and Huang X: Alpha-adducin gene polymorphism is associated with
essential hypertension in Chinese: A case-control and family-based
study. J Hypertens. 21:1861–1868. 2003. View Article : Google Scholar
|
|
10
|
Barlassina C, Norton GR, Samani NJ,
Woodwiss AJ, Candy GC, Radevski I, Citterio L, Bianchi G and Cusi
D: Alpha-adducin polymorphism in hypertensives of South African
ancestry. Am J Hypertens. 13:719–723. 2000. View Article : Google Scholar
|
|
11
|
Tamaki S, Iwai N, Tsujita Y, Nakamura Y
and Kinoshita M: Polymorphism of alpha-adducin in Japanese patients
with essential hypertension. Hypertens Res. 21:29–32. 1998.
View Article : Google Scholar
|
|
12
|
Sousa AC, Palma Dos Reis R, Pereira A,
Borges S, Freitas AI, Guerra G, Góis T, Rodrigues M, Henriques E,
Freitas S, et al: Relationship between ADD1 Gly460Trp gene
polymorphism and essential hypertension in Madeira Island. Medicine
(Baltimore). 96:e78612017. View Article : Google Scholar
|
|
13
|
Lauren P: The two histological main types
of gastric carcinoma: Diffuse and so-called intestinal-type
carcinoma. An attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965. View Article : Google Scholar
|
|
14
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar
|
|
15
|
Deckers IA, van den Brandt PA, van
Engeland M, van Schooten FJ, Godschalk RW, Keszei AP, Hogervorst JG
and Schouten LJ: Potential role of gene-environment interactions in
ion transport mechanisms in the etiology of renal cell cancer. Sci
Rep. 6:342622016. View Article : Google Scholar
|
|
16
|
Savas S, Schmidt S, Jarjanazi H and
Ozcelik H: Functional nsSNPs from carcinogenesis-related genes
expressed in breast tissue: Potential breast cancer risk alleles
and their distribution across human populations. Hum Genomics.
2:287–296. 2006. View Article : Google Scholar
|
|
17
|
Gerhard T, Gong Y, Beitelshees AL, Mao X,
Lobmeyer MT, Cooper-DeHoff RM, Langaee TY, Schork NJ, Shriver MD,
Pepine CJ, et al: Alpha-adducin polymorphism associated with
increased risk of adverse cardiovascular outcomes: Results from
GENEtic Substudy of the INternational VErapamil SR-trandolapril
STudy (INVEST-GENES). Am Heart J. 156:397–404. 2008. View Article : Google Scholar
|
|
18
|
Li Y, Wang X, Vural S, Mishra NK, Cowan KH
and Guda C: Exome analysis reveals differentially mutated gene
signatures of stage, grade and subtype in breast cancers. PLoS One.
10:e01193832015. View Article : Google Scholar
|
|
19
|
Jackson SS, Marks MA, Katki HA, Cook MB,
Hyun N, Freedman ND, Kahle LL, Castle PE, Graubard BI and
Chaturvedi AK: Sex disparities in the incidence of 21 cancer types:
Quantification of the contribution of risk factors. Cancer.
128:3531–3540. 2022. View Article : Google Scholar
|