Introduction
Neuroblastoma is a tumor originating from the
sympathetic nervous system and is a common extra-cranial pediatric
tumor, accounting for >8% of childhood malignancies and ~15% of
cancer-associated mortality in children worldwide (1,2).
Neuroblastoma is a disease with broad biological and clinical
heterogeneity, occurring predominantly in children <5 years old,
which is characterized by a poor prognosis and high recurrence rate
(3–6). Numerous factors, including age,
chromosomal abnormalities, tumor grade, MYCN status, DNA
ploidy and diagnostic category, influence the prognosis of
neuroblastoma (7,8).
Cancers are genetically unstable and have numerous
aberrations, including genetic mutations, gene translocations,
genomic copy number aberrations (CNAs), gene expression profiles
and epigenetic changes (9–12). Previous studies have reported that
DNA CNAs commonly occur in neuroblastoma, including amplification
of the MYCN oncogene and anaplastic lymphoma kinase
(ALK) oncogene, loss of heterozygosity (LOH) of chromosomes
1p, 3p, 4p, 11q and 14q, and gains of chromosomes 1q, 2p and 17q
(13–18). These chromosomal aberrations are
essential to predict the outcome in neuroblastoma, and several have
been incorporated into treatment stratification (19).
CNAs can be utilized to distinguish between
different genetic subgroups of neuroblastoma that have certain
prognostic characteristics. Whole chromosome abnormalities often
accumulate in localized tumors and in children <1 year of age,
and are associated with a good prognosis (20). Segmental chromosome aberrations
often occur in advanced stages or older children, and are
associated with a high risk of relapse (20). In addition, MYCN
amplification has been identified in ~20% of all neuroblastoma
cases, and is associated with stage 4 neuroblastoma and a poor
prognosis (21). ALK
amplification occurs in ~4% of high-risk neuroblastoma cases and
ALK is almost entirely co-amplified with MYCN
(14,15). However, there is a notable issue in
classifying individuals into distinct therapy groups based on their
anticipated risk. Since pediatric neuroblastoma has an extensive
spectrum of clinical outcomes, novel markers that could be used to
build reliable prognostic classifications need to be
identified.
Chemotherapy-related adverse drug reactions (ADRs)
are a major factor in cancer treatment-associated mortality and are
a pressing clinical problem that has to be addressed. Drug efficacy
and toxicity are associated with specific single nucleotide
polymorphisms (SNPs), and the analysis of SNPs could reveal novel
markers to improve tailored therapy (22). Numerous studies have reported that
multiple SNPs within the dihydropyrimidine dehydrogenase
(DPYD) gene are linked to the toxicity risk and
metabolism/pharmacokinetics in patients treated with fluorouracil,
and the Clinical Pharmacogenetics Implementation Consortium and
Dutch Pharmacogenetics Working Group guidelines provide information
for the interpretation of the clinical DPYD genotype
(23–26). The relationship between toxicity of
commonly prescribed chemotherapy medications, such as cisplatin,
carboplatin, cyclophosphamide, doxorubicin, methotrexate and
docetaxel, and different SNPs provides a reference for clinical
pharmacology in adult cancer (27–30).
Nevertheless, there are still numerous gaps in drug-gene
interaction research on long-term clinical interventions for
pediatric patients. According to the PHarmGKB database, only 633 of
the 5,112 clinical annotations are related to pediatrics, and only
2,572 of the 27,452 variant annotations are applicable to
pediatrics (https://www.pharmgkb.org/pediatric/dashboard).
Pediatric medications are different from those for adults because
of their unique characteristics of physiological development and
pharmacokinetic behaviors (31,32).
Children of different ages have their own specific reactions to
drugs because of the continuous maturation process (33). Therefore, the treatment of pediatric
neuroblastoma is different from that of adult tumors, and screening
for genetic markers that predict toxicity can reduce the use of
high-risk medicines in infants and children.
At present, there are few reports on the association
between whole chromosome or segmental chromosomal aberrations and
clinical characteristics in pediatric neuroblastoma, with a small
number of studies on SNPs associated with drug-induced toxicity
(34–38). In addition, the genetic
characteristics of pediatric neuroblastoma are still unclear. The
present study investigated the CNA profile of a cohort of 45
patients with neuroblastoma from Wuhan Children's Hospital (Wuhan,
China), with the intention of assessing CNA patterns in this tumor
type and validating putative CNA regions linked to clinical
characteristics. Furthermore, in a small clinical cohort of
neuroblastoma cases, the present study assessed the association
between certain SNPs and chemotherapy-associated ADRs, which may
offer novel insights into the genetic origins of ADRs, as well as
some warning signs that might be utilized in clinical settings.
Materials and methods
Studied subjects
A total of 45 continuous pediatric patients with
newly diagnosed neuroblastoma at Wuhan Children's Hospital were
enrolled in the present study between January 2020 and January
2023, including 20 patients with ganglioneuroblastoma (GNB) and 25
patients with neuroblastoma whose formalin-fixed paraffin-embedded
(FFPE) surgical specimens or bone marrow specimens were collected.
All experiments and bioinformatics services were entrusted with
Shanghai Cinopath Medical Laboratory Co., Ltd. The protocol of the
present study was approved by the Wuhan Children's Hospital
Institutional Ethics Committee. The legal guardians of all patients
provided written informed consent.
Therapy and toxicity evaluation
The International Neuroblastoma Staging System
(INSS) and International Neuroblastoma Risk Group (INRG) criteria
were used to establish the neuroblastoma stage and risk
categorization. Homogeneous treatment cohorts were defined
according to the INRG based on the Image-Defined Risk Factors
classification system, and patients were treated according to
pediatric neuroblastoma diagnosis and treatment expert consensus in
China (39). Tumors with low-risk
biological characteristics were treated with carboplatin (2,800
mg/m2), etoposide (1,440 mg/m2), doxorubicin
(120 mg/m2) and cyclophosphamide (6,000
mg/m2) for eight rounds of chemotherapy. Patients with
intermediate-risk neuroblastoma received eight cycles of
chemotherapy with cisplatin (720 mg/m2), vincristine (12
mg/m2), etoposide (640 mg/m2), doxorubicin
(120 mg/m2) and cyclophosphamide (9,600
mg/m2). Patients with high-risk biological
characteristics were treated with cisplatin (600 mg/m2),
vincristine (4 mg/m2), topotecan (18 mg/m2),
etoposide (1,800 mg/m2), doxorubicin (150
mg/m2) and cyclophosphamide (7,600 mg/m2).
Drug toxicity standards referred to the Common Terminology Criteria
for Adverse Events v5.0 and the toxicity was divided into five
categories (grades 1–5) (40). ADR
grades were assessed by independent clinicians before any
genotyping was performed. The highest-grade adverse reactions
observed in each patient during the induction therapy were
recorded.
Sample preparation and SNP array
analysis
DNA was extracted from FFPE tissues or bone marrow
using QIAamp DNA FFPE Advanced Kit (cat. no. 56604; Qiagen, Inc.).
The DNA quality was verified based on optical density (OD)260/OD280
and OD260/OD230 ratios measured by NanoDrop One (Thermo Fisher
Scientific, Inc.). Eligible samples were confirmed to have an
OD260/OD280 ratio between 1.7 and 2.0 and an OD260/OD230 ratio
>1.6. The integrity of the processed samples was verified by
Qsep400 (BiOptic. Inc.), with DNA quality number >3 as the
qualification standard. DNA concentration was determined using
Qubit 4 (Thermo Fisher Scientific, Inc.). A minimum of 200 ng
genomic DNA was aliquoted into 96-well plates and genotyped using
the Illumina BeadStation (Illumina, Inc.). Briefly, genome-wide
amplification of DNA was performed, followed by fragmentation,
hybridization, fluorescence tagging and scanning according to
standard Infinium protocols (41).
Sample analyses were performed using the Infinium CytoSNP-850K v1.2
BeadChip Kit (cat. no. 20103480; Illumina, Inc.), which contained
~850,000 SNP probes distributed throughout the whole genome.
Samples with call rates <90% were excluded from the
analysis.
Copy number analysis
In the present study, copy number variations were
visually inferred from SNP arrays by GenomeStudio v2.0 (Illumina,
Inc.) and normalized using MoChA v2021-05-14 (https://software.broadinstitute.org/software/mocha/)
to eliminate the artifactual CNAs due to GC content across the
genome. CNAs, including gain, loss, amplification and copy neutral
LOH, were detected by measuring log R ratios (as assessed by
aberrations in probe intensities) along with B-allele frequency (as
assessed by the shift in genotype frequencies of the SNP probes)
(42).
The copy number of mosaic changes was associated
with the percentage of aberrant cells. The mosaicism detection
limit in the present study was set at 10%. For gain, the copy
number was required to be more than diploidy; for loss, the copy
number was required to be less than diploidy. Furthermore,
chromosome ploidy is defined as hypodiploid with 35–45 chromosomes,
diploid with 46 chromosomes, hyperdiploid with 47–57 chromosomes
and near-triploid with 58–80 chromosomes. MYCN and
ALK amplifications were defined as regions with >10
copies. Focal changes refer to regions <5 Mb. The present study
focused on CNAs with regions >5 Mb (p or q), and the frequency
of CNAs was compared between neuroblastoma and GNB. Subsequently,
the present study also assessed the association between CNAs and
clinical features in neuroblastoma.
Correlation analysis could not be carried out in
this study due to sample limitations, so frequency index was chosen
as an alternative analysis. The frequency index of two variables
was defined as the incidence of patients with variable A in
patients with variable B; i.e., frequency index=patients with both
variable A and variable B/variable B.
SNP genotyping analysis
A total of 567 SNPs potentially associated with
chemotherapy drugs in neuroblastoma were analyzed through a review
of the relevant literature and comparison with information from the
PharmGKB database (www.pharmgkb.org). Selected SNPs were genotyped using
GenomeStudio software v2.0 (Illumina, Inc.), and those with a
GenCall Score <0.15 were excluded from the analysis. Therefore,
79 SNPs (Table SI) remained and
were used to analyze the impact of drug-related SNPs on the
toxicity of neuroblastoma treatment. The reference alleles of SNPs
in in the forest plots were derived from aggregate allele frequency
in the East Asian Group from the Allele Frequency Aggregator
project (https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/).
Statistical analysis
In the present study, odds ratios (ORs) and 95% CIs
were used to evaluate the risk of developing various manifestations
of toxicity after therapy in neuroblastoma with specific genotypes
using univariate analysis with Fisher's exact test and binary
logistic regression model. To estimate the influence of the
specific SNP on different manifestations of toxicity, two series of
analyses were performed: The first explored SNPs associated with
adverse reaction manifestations in the whole cohort and the second
aimed to further evaluate whether polymorphisms were associated
with toxicities during high-risk induction chemotherapy. All
statistical analyses were carried out using IBM SPSS Statistics V27
(IBM Corp.). Statistical comparisons of categorical variables were
performed using Fisher's exact test. For all statistical analyses,
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical samples
Among the 45 cases, 27 were male, indicating no
apparent sex bias. In all but one case, patients were diagnosed for
the first time. The median age at primary diagnosis was 43 months
(2–132 months), and eight cases were <18 months old. Metastasis
occurred in 29 patients. Tumors were classified as low-risk in 16
cases, intermediate-risk in 11 cases and high-risk in 18 cases.
Most of the patients (91.1%) were in stage 2, 3 and 4, and the
retroperitoneum, adrenal gland and mediastinum were the most common
primary sites of neuroblastoma (88.9%). After a median follow-up
time of 20 months (range, 4–42 months), 35 patients had stable
disease, five patients suffered from tumor relapse or progression,
three patients were not followed up and the remaining two patients
died. The clinical characteristics of the 45 patients with
neuroblastoma are shown in Table
I.
 | Table I.Clinical characteristics of the study
patients. |
Table I.
Clinical characteristics of the study
patients.
| Characteristic | n (%) |
|---|
| Sex |
|
|
Male | 27 (60.0) |
|
Female | 18 (40.0) |
| Age, months |
|
|
<18 | 8 (17.8) |
|
18-60 | 24 (53.3) |
|
≥60 | 13 (28.9) |
| INSS stage |
|
| Stage
1 | 3 (6.7) |
| Stage
2 | 18 (40.0) |
| Stage
3 | 11 (24.4) |
| Stage
4 | 12 (26.7) |
| Stage
4s | 1 (2.2) |
| Risk
stratification |
|
|
Low | 16 (35.6) |
|
Intermediate | 11 (24.4) |
|
High | 18 (40.0) |
| Primary
diagnosis |
|
|
Yes | 44 (97.8) |
| No | 1 (2.2) |
| Metastasis |
|
|
Yes | 29 (64.4) |
| No | 15 (33.3) |
| Data
not available | 1 (2.2) |
| Primary tumor
site |
|
|
Retroperitoneum | 17 (37.8) |
| Adrenal
gland | 11 (24.4) |
|
Mediastinum | 12 (26.7) |
|
Other | 5 (11.1) |
| State at last
follow-up |
|
|
Alive | 40 (88.9) |
|
Dead | 2 (4.4) |
|
Other | 3 (6.7) |
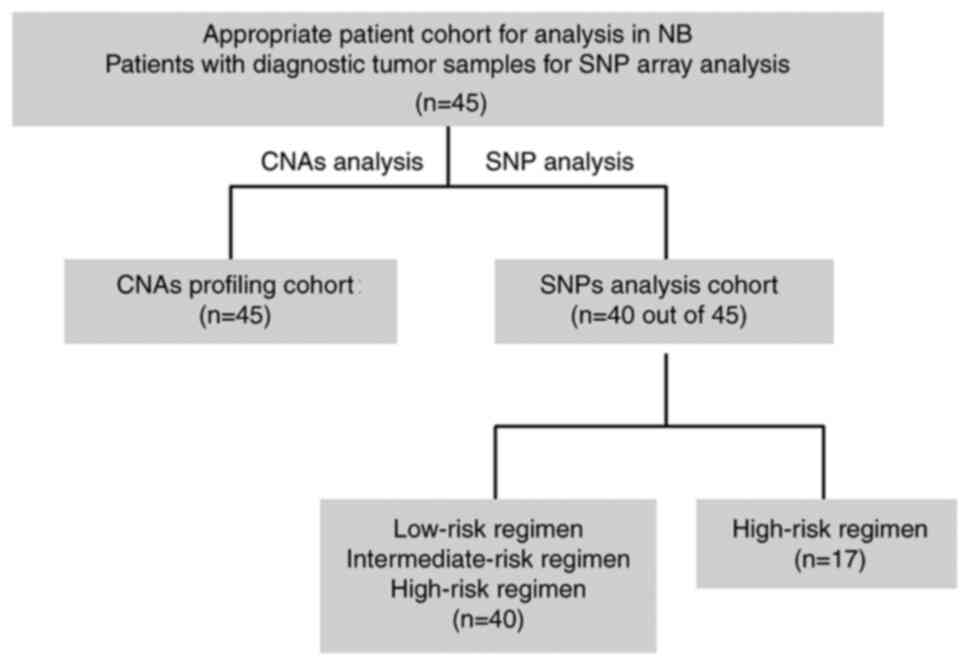
Fig. 1 shows the
number of cases included throughout the study, indicating the
subsets of cases that were selected for the different analyses
reported. Among the 45 newly diagnosed cases that were available in
the retrospective collection, five did not have available therapy
information and 40 remained for the analysis of genotypes. There
were 45 patients in the entire study cohort, and the CNAs and SNP
studies were performed on subgroups of the cohort. A total of 40 of
the 45 patients completed the entire induction treatment at the
same hospital; thus, analysis of chemotherapy toxicity associated
with SNPs could be performed. Among the 45 patients, only 17 were
treated with the high-risk regimen, and the association between
toxicity and SNPs was analyzed.
Biological characteristics of GNB and
neuroblastoma
A high number of CNAs was observed in most
neuroblastoma cases (92%; 23/25); however, the incidence rate of
CNAs was relatively low in GNB (15%; 3/20). The present study
compared CNAs between neuroblastoma and GNB, and significant
differences in CNAs were found, including differences in 1p LOH, 2
gain, 7 gain, 12 gain, 17 gain, 17q gain and Y LOH (P<0.05;
Table SII). Furthermore, there
were significant differences in age (P=0.047), INSS stage
(P<0.001), risk stratification (P<0.001) and tumor size
(P<0.001) between neuroblastoma and GNB. The clinical
characteristics of GNB included a relatively small tumor size, low
occurrence of CNAs, low INSS stage and low risk at diagnosis,
suggesting slow proliferation, low invasiveness and good genomic
stability as biological features of GNB. These results are
summarized in Table SIII.
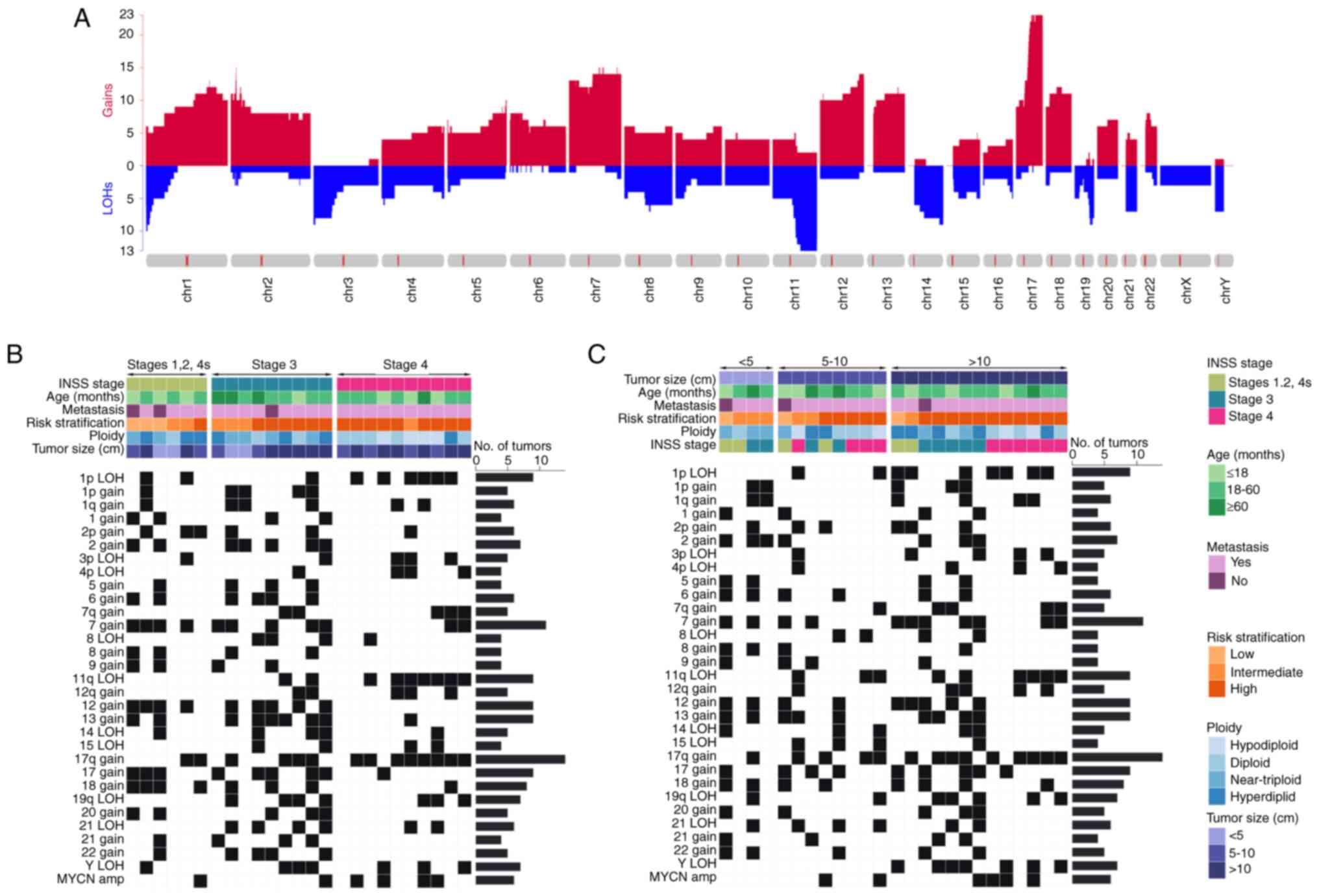
CNA landscape in neuroblastoma
The median total number of chromosomal aberrations
per patient with neuroblastoma was 16 (range, 0–33), with a median
of 8 gains (range, 0–25), four LOHs (range, 0–16) and 0
amplifications (range, 0–6). As shown in Fig. 2 and Table SII, >16% of gains included 1p,
1q, 1, 2p, 2, 5, 6, 7q, 7, 8, 9, 12q, 12, 13, 17q, 17, 18, 20, 21
and 22, and >16% of LOHs were 1p, 3p, 4p, 8, 11q, 14, 15, 19q,
21q and Y, and high-frequency amplification of MYCN was
observed. The overwhelming majority of these gains and LOHs were
clonal, indicating that genomic instability is a genetic feature of
neuroblastoma.
To assess CNAs associated with clinical
characteristics such as INSS stage and tumor size, the gathered
data were examined. The percentage of 1p LOH, 11q LOH and
MYCN amplification in stage 4 was markedly higher, whereas
4p LOH, 7q gain, 8 LOH, 15 LOH, 12q gain, 19q LOH and 21 LOH were
almost absent in stages 1, 2 and 4s. Gain in chromosome regions 1,
2, 2p, 5, 6, 7, 8, 9, 12, 13, 17, 18, 20, 21 and 22 was common in
stages 1, 2 and 4s, and stage 3. Similarly, the hyperdiploid and
near-triploid karyotypes were common in stages 1, 2 and 4s, and
stage 3, while the diploid and hypodiploid karyotypes were more
common in stage 4, which was consistent with the CNA distribution.
Furthermore, the largest proportion of 1p LOH, 3p LOH, 7q gain and
12q gain was observed in neuroblastoma cases with tumor sizes
>10 cm, and 8 gain, 9 gain and 15 LOH were common in patients
with a tumor size <10 cm. Notably, loss of the Y chromosome
(LoY) occurred only in boys with tumor sizes >10 cm. Fig. 2 provides a summary of these
findings. There was >3-fold difference in these CNAs between
stages 1, 2 and 4s, and stage 3 and 4 and between tumor sizes
>10 cm and <10 cm, indicating that these CNAs were associated
with tumor invasion and proliferation, even though the small sample
size meant that the results were not significant.
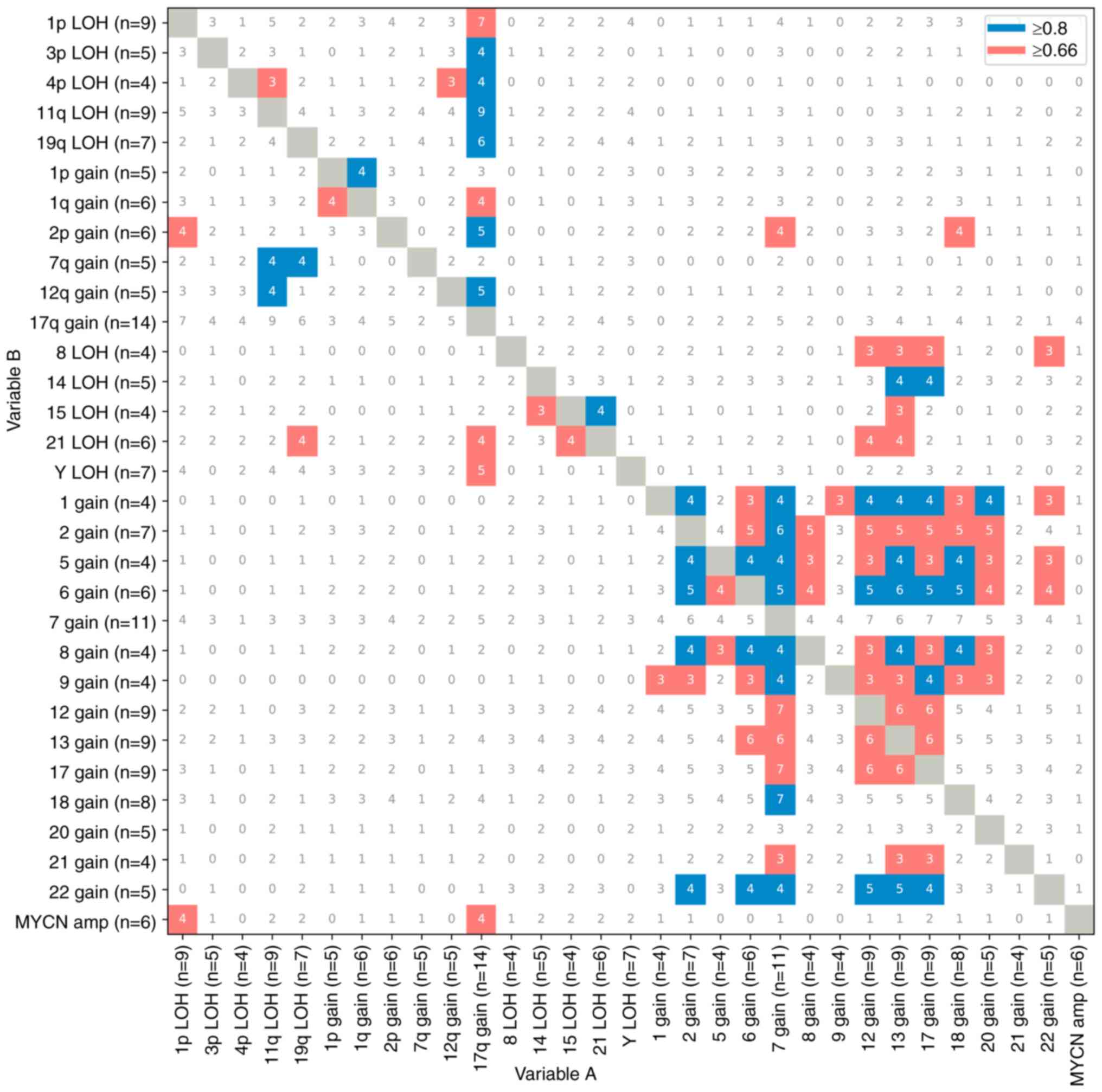
Exploratory analysis of co-occurrence
patterns of CNAs in neuroblastoma
Among the 25 neuroblastoma cases, 1p LOH and 17q
gain accounted for 67% of MYCN amplifications. Previous
reports have indicated positive associations between MYCN
amplification and 1p LOH and 17q gain, and a negative association
between MYCN amplification and 11q aberration (43–45).
In accordance with this finding, if the frequency index of variable
A and variable B was greater than 67%, they were considered to be
positively associated (Fig. 3).
Conversely, if it was less than 67%, the association between them
was considered weak. Nevertheless, there was a lack of negative
relationship evidence available and further investigation was not
performed. The frequency index plot of CNAs in 25 neuroblastoma
cases showed that MYCN amplification (67%), 1p LOH (78%), 3p
LOH (80%), 4p LOH (100%), 11q LOH (100%), 19q LOH (86%), 1q gain
(67%), 2p gain (83%), 12q gain (100%), 21 LOH (67%) and LoY (71%)
were positively associated with 17q gain (Fig. 3). In addition, 4p LOH and 11q LOH
(75%) or 12q gain (75%), 1p gain and 1q gain (80%), 2p gain and 1p
LOH (67%), 7q gain and 11q LOH (80%) or 19q LOH (80%), and 12q gain
and 11q LOH (80%) showed positive associations. In this regard,
segmental chromosomal abnormalities frequently occurred together.
Furthermore, segmental chromosomal abnormalities occurred in
conjunction with whole chromosome abnormalities in 68% of
cases.
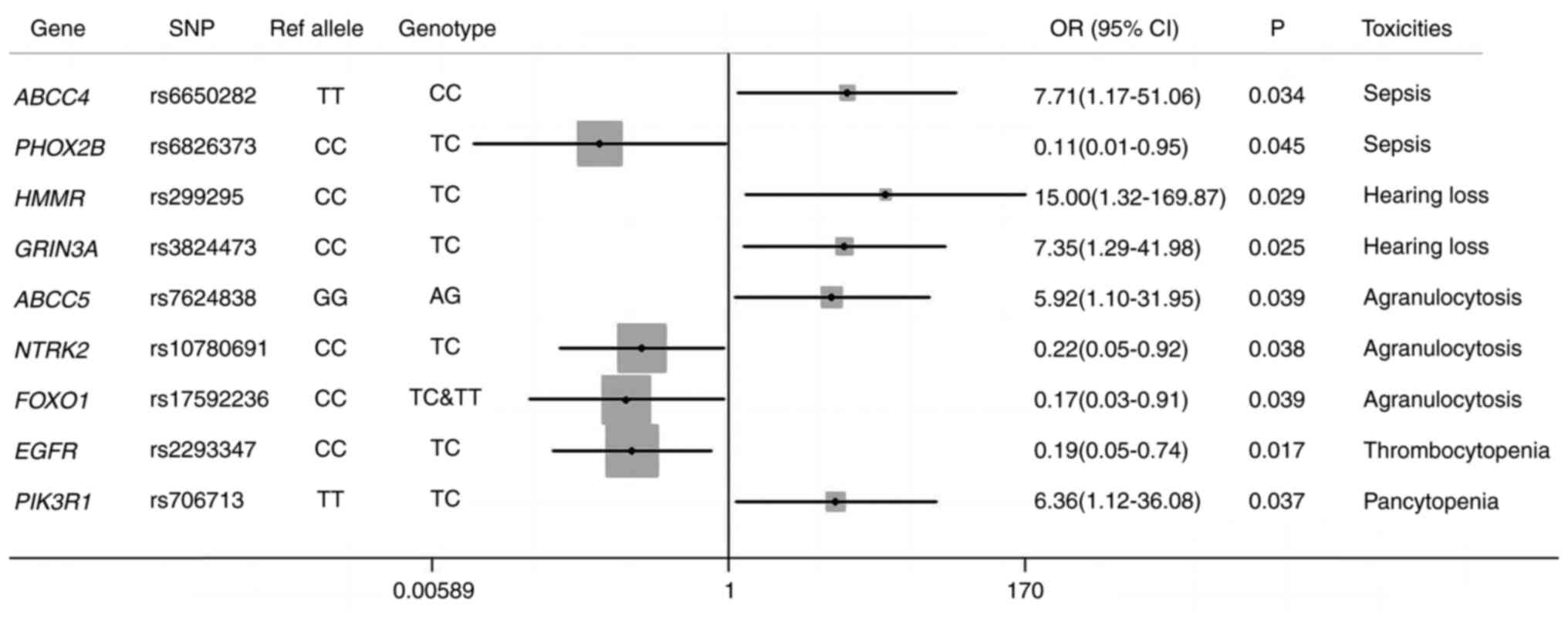
SNPs related to toxicity during
induction treatment
A total of 40 patients had clinical data regarding
toxicities during induction therapy. All cases of toxicity in
response to the three treatment regimens were counted and were
analyzed together. The prevalence of developing toxicities (n=40)
in all patients was as follows: Sepsis (n=11; 27.5%), moderate or
severe hearing loss (n=9; 22.5%), pancytopenia (n=9; 22.5%),
agranulocytosis (n=27; 67.5%) and thrombocytopenia (n=24; 60%).
Univariate analysis was performed to assess the manifestations of
toxicity of the induction chemotherapy in neuroblastoma to identify
meaningful associations between SNPs and chemotherapy toxicity.
Fig. 4 shows the risk assessment of
various toxicity manifestations for the SNPs under investigation
based on the binary logistic regression model.
A total of 11 SNPs were chosen by the binary
logistic regression model based on toxicity results. As shown in
Fig. 4, patients with rs6650282 CC
in ATP binding cassette subfamily C member (ABCC)4
had an increased risk of developing sepsis (P=0.034; OR, 7.71; 95%
CI, 1.17–51.06) compared with patients with TT/TC. A total of three
of the selected SNPs, rs6826373 TC in paired-like homeobox 2b
(P=0.045; OR, 0.11; 95% CI, 0.01–0.95), rs3814057 CC in nuclear
receptor subfamily 1 group I member 2 (NR1I2; P=0.024) and
rs7624838 AA in ABCC5 (P=0.024), were associated with a
significant risk reduction of sepsis among patients. The risk of
moderate or severe hearing loss was increased in patients with
rs299295 TC in hyaluronan-mediated motility receptor (P=0.029; OR,
15.00; 95% CI, 1.32–169.87) and rs3824473 TC in glutamate
ionotropic receptor NMDA type subunit 3A (GRIN3A; P=0.025;
OR, 7.35; 95% CI, 1.29–41.98). Conversely, rs3745551 TC in insulin
receptor was significantly associated with reduced occurrence of
moderate or severe hearing loss (P=0.037). Similarly, rs7624838 AG
in ABCC5 (P=0.039; OR, 5.92; 95% CI, 1.10–31.95) predicted
severe agranulocytosis, and rs10780691 TC in neurotrophic receptor
tyrosine kinase 2 (P=0.038; OR, 0.22; 95% CI, 0.05–0.92) and
rs17592236 TC+TT in forkhead box O1 (P=0.039; OR, 0.17; 95% CI,
0.03–0.91) were significantly associated with reduced risk of
agranulocytosis. Thrombocytopenia was reduced among patients
carrying rs2293347 TC in EGFR (P=0.017; OR, 0.19; 95% CI,
0.05–0.74). Furthermore, pancytopenia was significantly more
frequent in patients with rs706713 TC in PIK3R1 compared
with those with the TT/CC genotype, and the increased risk was
statistically significant (P=0.037; OR, 6.36; 95% CI, 1.12–36.08).
Overall, a set of SNPs associated with ADRs was identified.
SNPs related to toxicity in the
high-risk treatment group
The most homogeneously treated patients were chosen
for the high-risk regimen, and all were treated with the same
induction therapy at Wuhan Children's Hospital. These requirements
were met by 17 patients. An analysis was performed to evaluate the
associations between SNPs and chemotherapy toxicity after
collecting the data for the high-risk treatment group. Fig. 5 shows the SNPs associated with
chemotherapy toxicity according to the logistic regression
model.
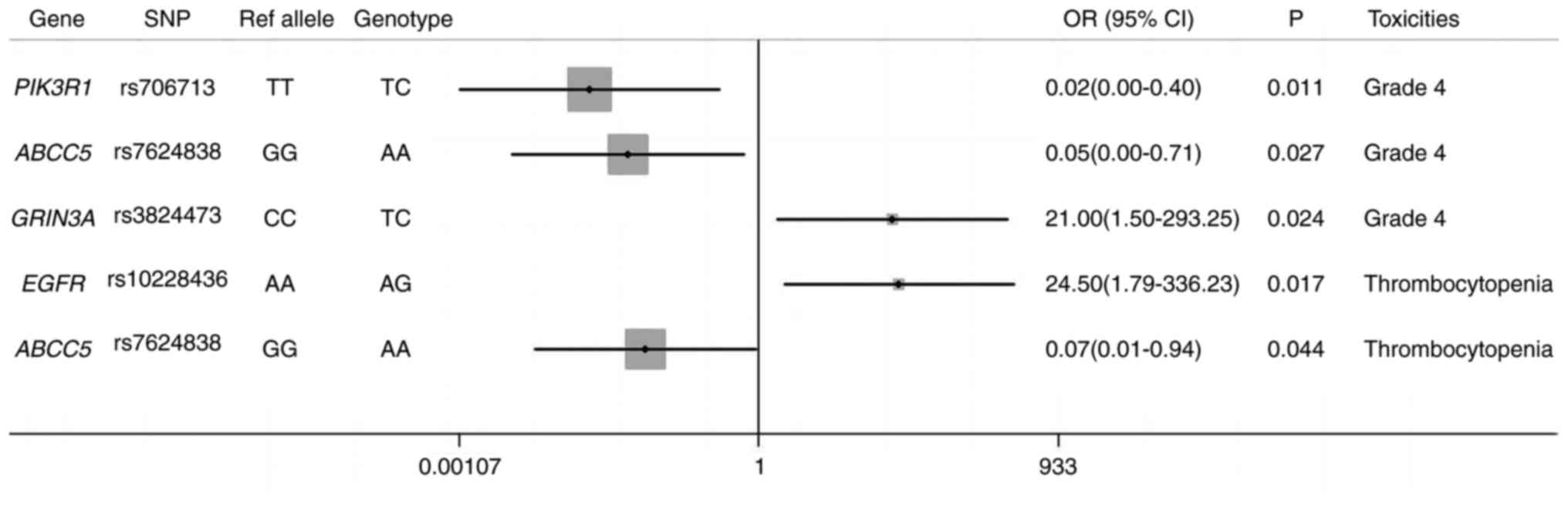 | Figure 5.SNPs related to the toxicity in
high-risk treatment. Reference allele, aggregate allele frequency
in the East Asian Group from the Allele Frequency Aggregator
project. Patient cohort, n=17. Gray box, the midpoint of the box
symbolizes the point estimate of the OR, and its size (area) is
proportionate to the weight of the study. Vertical line, the solid
vertical line corresponds to ‘no effect’-an OR of 1.0. Horizontal
line, if the horizontal line falls to the left of the vertical
line, it can be concluded that the studied factor favors the
occurrence of the outcome; if the horizontal line falls to the
right of the vertical line, it can be concluded that the studied
factor is detrimental to the occurrence of the outcome. OR, odds
ratio; SNP, single nucleotide polymorphism. |
Two SNPs significantly reduced the risk of grade 4
adverse reactions; rs706713 TC in PIK3R1 (P=0.011; OR, 0.02;
95% CI, 0.00–0.40) and rs7624838 AA in ABCC5 (P=0.027; OR,
0.05; 95% CI, 0.00–0.71). Conversely, rs3824473 TC in GRIN3A
(P=0.024; OR, 21.00; 95% CI, 1.50–293.25) predicted a significantly
increased risk of grade 4 adverse reactions. The statistical model
selected two SNPs, rs10228436 AG in EGFR and rs7624838 AA in
ABCC5, which predicted opposite toxicity information.
rs10228436 AG in EGFR (P=0.017; OR, 24.50; 95% CI,
1.79–336.23) was more likely to cause thrombocytopenia, while
rs7624838 AA in ABCC5 (P=0.044; OR, 0.07; 95% CI, 0.01–0.94)
had the opposite effect. Additionally, rs7624838 AA in ABCC5
(P=0.037) and rs2809244 AC in TSC complex subunit 1 (P=0.017)
significantly reduced the risk of sepsis and agranulocytosis.
Discussion
Neuroblastoma causes notable mortality worldwide.
Effective molecular indicators are still lacking in some cases,
despite the fact that the prognosis of neuroblastoma can presently
be predicted using risk markers such as INSS, MYCN, 11q and
DNA ploidy. The potential biomarkers obtained from CNA and SNP
testing could improve precision oncology therapy, predicting
patient prognosis, and predicting the response to treatments and
their potential toxicities. A SNP microarray is one of the methods
used to detect ADR markers, which may provide a comprehensive assay
for simultaneous detection of SNP markers and tumor-associated
CNAs. In the present study, a comprehensive genetic investigation
relying on a SNP microarray for 45 neuroblastoma cases demonstrated
the global features of the CNAs and validated the variety of CNAs
in neuroblastomas. In neuroblastoma, a high incidence of CNA
variation (92%) was associated with genomic instability. This
suggests that the accumulation of CNAs and other genomic variations
may be the primary cause of the bulk of these cancers. The therapy
schedule was not always followed by all patients due to
heterogeneity, and ADRs were frequent. Based on the present
hypothesis, research on possible genetic markers of ADRs might help
predict treatment-related toxicities. As aforementioned, SNPs may
be linked to a variety of treatment-related ADRs.
Chromosome alterations are a major feature of cancer
cells (46). Numerous chromosome
regions of gain and LOH have been revealed in neuroblastoma;
however, CNAs specific to GNB have been infrequently studied
(47,48). Using a SNP array, the present study
compared the CNAs in neuroblastoma and GNB in order to examine
chromosomal changes. GNB exhibited a low occurrence of CNAs,
smaller tumor size, lower INSS stage and lower risk at diagnosis,
which indicated that GNB was characterized by slow proliferation,
low invasiveness and good genomic stability, and may have a
different mechanism of cancer cell evolution compared with
neuroblastoma. The small number of samples that were available
prevented accurate assessment of every anomaly in the present
investigation.
In neuroblastoma, segmental chromosome alterations
and MYCN amplification were almost absent in stages 1, 2 and
4s, including LOH of 1p, 11q, 4p and 19q, and gain of 7q and 12q.
Additionally, 1p LOH, 3p LOH, 7q gain and 12q gain were observed in
neuroblastoma cases with tumor sizes >10 cm, indicating that
segmental alterations promoted invasion and proliferation of tumor
cells. The majority of the entire chromosome modifications occurred
in stages 1, 2, 4s and 3, even in the presence of segmental
chromosomal abnormalities. Thus, chromosomal aneuploidy may be a
primary feature of localized NB. Whole chromosome aberrations are
associated with an improved prognosis in patients with
neuroblastoma (20,38). LOH of 8, 15, 21 and Y was mainly
observed in patients with stage 4 and 3, suggesting that LOH of the
whole chromosome may have similar effects on invasion as segmental
alterations in neuroblastoma. LOH of the whole chromosome may be a
subset of whole chromosome alterations that could help identify a
novel biological subtype and may be associated with prognosis.
Examining these chromosomal abnormalities might aid in clarifying
the processes involved in neuroblastoma tumor growth. There was
limited long-term follow-up information to identify independent
associations between these copy number variations and
prognosis.
In previous reports, LoY was frequently observed in
elderly men; however, LoY mutations were increased in tumor tissues
and associated with an overall worse prognosis and sensitization to
programmed cell death protein 1-targeted immunotherapy (49,50).
In the present study, LoY was only found in individuals whose
tumors measured >10 cm and was markedly increased in
neuroblastoma compared with in GNB. LoY was mainly present in
patients diagnosed at stage 4 and 3, and was associated with larger
tumor size (>10 cm), implying an association between LoY and the
invasion and proliferation of tumor cells. LoY may represent a
novel prognosis and treatment marker for male patients. Due to the
limited sample cohort used in the current investigation, more
large-scale cohort verification is still required.
According to the present findings, entire chromosome
modifications exhibited a comparable likelihood for co-occurrence
as segmental abnormalities. As aforementioned, segmental
chromosomal abnormalities occurred in conjunction with whole
chromosome abnormalities in 68% of cases. Previous studies have
reported that segmental chromosome alterations may derive from an
intermediate stage characterized by whole chromosome alterations
(49,51,52).
Co-occurrence exacerbates genomic instability during the evolution
of cancer cells and promotes clonal dominance of tumor cells.
Standardized chemotherapy was administered to 40
patients in the present cohort who had neuroblastoma. Distinct
toxicity symptoms were noted in equal measure across the various
treatment plans. Therefore, multiple toxicity manifestations were
collected for SNP analyses. The model selected 11 SNPs related to
sepsis, hearing loss, agranulocytosis, thrombocytopenia and
pancytopenia, including rs3814057 in the NR1I2 gene, which
has been associated with major adverse cardiovascular events in
clopidogrel-treated patients and hepatotoxicity risk in
anti-tuberculosis drugs-treated patients in the literature
(53,54); and the rs2293347 in the EGFR
gene, which has been reported to be associated with skin toxicity
in patients with colorectal cancer receiving an antibody against
EGFR (55). For high-risk
regimen-treated patients, the model selected five SNPs, including
EGFR rs10228436 which has been reported to be associated
with the incidence of skin rash for imatinib in gastrointestinal
stromal tumors and hepatotoxicity risk in gefitinib-treated
patients with lung cancer (56,57).
ADRs of the remaining polymorphisms have rarely been reported by
the scientific community, suggesting that there is a need to
further confirm the effects of these SNPs. In the present study,
using the candidate SNPs approach, associations between multiple
SNPs and chemotherapy-induced ADRs were identified. Although the
mechanism to induce ADRs should be further validated in different
cohorts or by molecular analysis, the present study has provided
novel evidence for ADR prediction systems, which may lead to an
improved outcome and quality of life for patients. If these results
could be validated in different cohorts, the set of SNPs could
become a predictive signature for identifying patients at risk of
developing these toxicities.
The present study revealed that rs7624838 in the
ABCC5 gene, which was significantly associated with a
reduced incidence of grade 4 high-risk regimen-induced ADRs,
exhibited similar trends for both sepsis and thrombocytopenia,
indicating that rs7624838 AA may prevent the development of severe
ADRs. rs7624838 might act as an important marker associated with a
lower incidence of ADRs in response to high-risk regimens. Previous
studies have shown that the ABCC5 gene can influence the
disposition of endogenous metabolites, toxins and drugs in human
cancer (58–60). rs7624838 is located in intron 2 of
the ABCC5 gene. It may be hypothesized that the impairment
of rs7624838 in ABCC5 could lead to a sufficient drug
clearance and subsequent decrease of the drug concentration in the
body. However, this hypothesis should be validated using larger
samples as well as by a functional analysis of rs7624838.
The present study has certain limitations. First,
selection bias was likely present, since a retrospective,
single-center study was performed with a relatively small sample
size. Second, the present study was not validated in conjunction
with a public database. Third, there was a relatively short
observation period. Further prospective, large, multi-center
studies with longer follow-up periods are needed to validate the
results of the present study.
According to the present study, individuals with
neuroblastoma exhibited genomic instability that was noticeably
higher compared with patients with GNB, and segmental changes aided
the invasion and proliferation of tumor cells. LOH of whole
chromosomes and segmental alterations may both be associated with
invasion. In addition, LOH of the whole chromosome was associated
with increased aggressiveness compared with whole chromosome gain.
Furthermore, a group of SNPs linked to toxic symptoms was
identified; patients with these SNPs should be assessed during the
initial phase of therapy. Treatment decisions for patients with
neuroblastoma should consider multiple molecular changes as
predictive elements to make treatment adjustments for patients with
varying probabilities of treatment success; however, the results
should be validated in different comparable cohorts.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The microarray data generated in the present study
may be found in the NCBI Gene Expression Omnibus under accession
number GSE288908 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE288908.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
KLC, YL, QX, HPL and YJL initiated the conception
and design of the study, participated in the interpretation of the
data and wrote the article. HCL, JQL, LC and YX performed the
experiments and participated in the analysis of the data. YLT and
YFY performed bioinformatics analysis including data processing,
formal analysis, statistics and visualization. YLT and HPL confirm
the authenticity of all the raw data. All authors read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the local Ethics
Committee of Wuhan Children's Hospital (approval no. 2024R089-E01).
Written informed consent was obtained from their legal
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gurney JG, Davis S, Severson RK, Fang JY,
Ross JA and Robison L: Trends in cancer incidence among children in
the US. Cancer. 78:532–541. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakagawara A, Li Y, Izumi H, Muramori K,
Inada H and Nishi M: Neuroblastoma. Jpn J Clin Oncol. 48:214–241.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maris J, Hogarty MD, Bagatell R and Cohn
SL: Neuroblastoma. Lancet. 369:2106–2120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park JR, Kreissman SG, London WB, Naranjo
A, Cohn SL, Hogarty MD, Tenney SC, Haas-Kogan D, Shaw PJ, Kraveka
JM, et al: Effect of tandem autologous stem cell transplant vs
single transplant on event-free survival in patients with high-risk
neuroblastoma: A randomized clinical trial. JAMA. 322:746–755.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park JA and Cheung NKV: Targets and
antibody formats for immunotherapy of neuroblastoma. J Clin Oncol.
38:1836–1848. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zafar A, Wang W, Liu G, Wang X, Xian W,
McKeon F, Foster J, Zhou J and Zhang R: Molecular targeting
therapies for neuroblastoma: Progress and challenges. Med Res Rev.
41:961–1021. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cohn SL, Pearson AD, London WB, Monclair
T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et
al: The international neuroblastoma risk group (INRG)
classification system: An INRG task force report. J Clin Oncol.
27:289–297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sokol E, Desai AV, Applebaum MA,
Valteau-Couanet D, Park JR, Pearson ADJ, Schleiermacher G, Irwin
MS, Hogarty M, Naranjo A, et al: Age, diagnostic category, tumor
grade, and mitosis-karyorrhexis index are independently prognostic
in neuroblastoma: An INRG project. J Clin Oncol. 38:1906–1918.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Asgharzadeh S, Pique-Regi R, Sposto R,
Wang H, Yang Y, Shimada H, Matthay K, Buckley J, Ortega A and
Seeger RC: Prognostic significance of gene expression profiles of
metastatic neuroblastomas lacking MYCN gene amplification. J Natl
Cancer Inst. 98:1193–1203. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi Y, Yuan J, Rraklli V, Maxymovitz E,
Cipullo M, Liu M, Li S, Westerlund I, Bedoya-Reina OC, Bullova P,
et al: Aberrant splicing in neuroblastoma generates RNA-fusion
transcripts and provides vulnerability to spliceosome inhibitors.
Nucleic Acids Res. 49:2509–2521. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hiwatari M, Seki M, Matsuno R, Yoshida K,
Nagasawa T, Sato-Otsubo A, Yamamoto S, Kato M, Watanabe K,
Sekiguchi M, et al: Novel TENM3-ALK fusion is an alternate
mechanism for ALK activation in neuroblastoma. Oncogene.
41:2789–2797. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fetahu IS and Taschner-Mandl S:
Neuroblastoma and the epigenome. Cancer Metastasis Rev. 40:173–189.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brodeur GM, Seeger RC, Schwab M, Varmus HE
and Bishop JM: Amplification of N-myc in untreated human
neuroblastomas correlates with advanced disease stage. Science.
224:1121–1124. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bresler SC, Weiser DA, Huwe PJ, Park JH,
Krytska K, Ryles H, Laudenslager M, Rappaport EF, Wood AC, McGrady
PW, et al: ALK mutations confer differential oncogenic activation
and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer
Cell. 26:682–694. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bellini A, Pötschger U, Bernard V,
Lapouble E, Baulande S, Ambros PF, Auger N, Beiske K, Bernkopf M,
Betts DR, et al: Frequency and prognostic impact of ALK
amplifications and mutations in the European neuroblastoma study
group (SIOPEN) high-risk neuroblastoma trial (HR-NBL1). J Clin
Oncol. 39:3377–3390. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Spitz R, Hero B, Ernestus K and Berthold
F: Deletions in chromosome arms 3p and 11q are new prognostic
markers in localized and 4s neuroblastoma. Clin Cancer Res.
9:52–58. 2003.PubMed/NCBI
|
|
17
|
Schleiermacher G, Mosseri V, London WB,
Maris JM, Brodeur GM, Attiyeh E, Haber M, Khan J, Nakagawara A,
Speleman F, et al: Segmental chromosomal alterations have
prognostic impact in neuroblastoma: A report from the INRG project.
Br J Cancer. 107:1418–1422. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fong C, White PS, Peterson K, Sapienza C,
Cavenee WK, Kern SE, Vogelstein B, Cantor AB, Look AT and Brodeur
GM: Loss of heterozygosity for chromosomes 1 or 14 defines subsets
of advanced neuroblastomas. Cancer Res. 52:1780–1785.
1992.PubMed/NCBI
|
|
19
|
Brady SW, Liu Y, Ma X, Gout AM, Hagiwara
K, Zhou X, Wang J, Macias M, Chen X, Easton J, et al:
Pan-neuroblastoma analysis reveals age-and signature-associated
driver alterations. Nat Commun. 11:51832020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Janoueix-Lerosey I, Schleiermacher G,
Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier
J, Peuchmaur M, Valent A, et al: Overall genomic pattern is a
predictor of outcome in neuroblastoma. J Clin Oncol. 27:1026–1033.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park JR, Eggert A and Caron H:
Neuroblastoma: Biology, prognosis, and treatment. Pediatr Clin N
Am. 55:97–120. 2008. View Article : Google Scholar
|
|
22
|
Olivera GG, Yáñez Y, Gargallo P, Sendra L,
Aliño SF, Segura V, Sanz MÁ, Cañete A, Castel V, Mora JF, et al:
MTHFR and VDR polymorphisms improve the prognostic
value of MYCN status on overall survival in neuroblastoma
patients. Int J Mol Sci. 21:27142020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Caudle KE, Thorn CF, Klein TE, Swen JJ,
McLeod HL, Diasio RB and Schwab M: Clinical pharmacogenetics
implementation consortium guidelines for dihydropyrimidine
dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol
Ther. 94:640–645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Amstutz U, Henricks LM, Offer SM,
Barbarino J, Schellens JHM, Swen JJ, Klein TE, McLeod HL, Caudle
KE, Diasio RB and Schwab M: Clinical Pharmacogenetics
implementation consortium (CPIC) guideline for dihydropyrimidine
dehydrogenase genotype and fluoropyrimidine dosing: 2017 update.
Clin Pharmacol Ther. 103:210–216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Swen JJ, Nijenhuis M, de Boer A, Grandia
L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH,
Schalekamp T, Touw DJ, et al: Pharmacogenetics: From bench to
byte-an update of guidelines. Clin Pharmacol Ther. 89:662–673.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lunenburg CATC, van der Wouden CH,
Nijenhuis M, Crommentuijn-van Rhenen MH, de Boer-Veger NJ, Buunk
AM, Houwink EJF, Mulder H, Rongen GA, van Schaik RHN, et al: Dutch
pharmacogenetics working group (DPWG) guideline for the gene-drug
interaction of DPYD and fluoropyrimidines. Eur J Hum Genet.
28:508–517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pu X, Hildebrandt MAT, Lu C, Lin J,
Stewart DJ, Ye Y, Gu J, Spitz MR and Wu X: PI3K/PTEN/AKT/mTOR
pathway genetic variation predicts toxicity and distant progression
in lung cancer patients receiving platinum-based chemotherapy. Lung
Cancer. 71:82–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jabeen S, Holmboe L, Alnæs GIG, Andersen
AM, Hall KS and Kristensen VN: Impact of genetic variants of RFC1,
DHFR and MTHFR in osteosarcoma patients treated with high-dose
methotrexate. Pharmacogenomics J. 15:385–390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sugishita M, Imai T, Kikumori T, Mitsuma
A, Shimokata T, Shibata T, Morita S, Inada-Inoue M, Sawaki M,
Hasegawa Y and Ando Y: Pharmacogenetic association between GSTP1
genetic polymorphism and febrile neutropenia in Japanese patients
with early breast cancer. Breast Cancer. 23:195–201. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bosó V, Herrero MJ, Santaballa A, Palomar
L, Megias JE, de la Cueva H, Rojas L, Marqués MR, Poveda JL,
Montalar J and Aliño SF: SNPs and taxane toxicity in breast cancer
patients. Pharmacogenomics. 15:1845–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Verscheijden LFM, Koenderink JB, Johnson
TN, de Wildt SN and Russel FGM: Physiologically-based
pharmacokinetic models for children: Starting to reach maturation?
Pharmacol Ther. 211:1075412020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Min S: Design of formulations for
pediatric drugs. Prog Pharm Sci. 43:655–666. 2019.
|
|
33
|
Fernandez E, Perez R, Hernandez A, Tejada
P, Arteta M and Ramos JT: Factors and mechanisms for
pharmacokinetic differences between pediatric population and
adults. Pharmaceutics. 3:53–72. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ognibene M, De Marco P, Amoroso L, Fragola
M, Zara F, Parodi S and Pezzolo A: Neuroblastoma patients' outcome
and chromosomal instability. Int J Mol Sci. 24:155142023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Körber V, Stainczyk SA, Kurilov R, Henrich
KO, Hero B, Brors B, Westermann F and Höfer T: Neuroblastoma arises
in early fetal development and its evolutionary duration predicts
outcome. Nat Genet. 55:619–630. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Olivera GG, Urtasun A, Sendra L, Aliño SF,
Yáñez Y, Segura V, Gargallo P, Berlanga P, Castel V, Cañete A and
Herrero MJ: Pharmacogenetics in neuroblastoma: What can already be
clinically implemented and what is coming next? Int J Mol Sci.
22:98152021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Urtasun A, Olivera GG, Sendra L, Aliño SF,
Berlanga P, Gargallo P, Hervás D, Balaguer J, Juan-Ribelles A,
Andrés MDM, et al: Personalized medicine in infant population with
cancer: Pharmacogenetic pilot study of polymorphisms related to
toxicity and response to chemotherapy. Cancers (Basel).
15:14242023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paolini L, Hussain S and Galardy PJ:
Chromosome instability in neuroblastoma: A pathway to aggressive
disease. Front Oncol. 12:9889722022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pediatric Oncology Committee, Chinese
Anti-Cancer Association, Oncology Group, Chinese Association of
Pediatric Surgeons, . Expert consensus on diagnosing and treating
of neuroblastoma in children. Chin J Pediatr Surg. 36:3–7.
2015.
|
|
40
|
Department of Health and Human Services, .
Common Terminology Criteria for Adverse Events (CTCAE). Version
5.0. 2017.
|
|
41
|
Gunderson KL, Steemers FJ, Lee G, Mendoza
LG and Chee MS: A genome-wide scalable SNP genotyping assay using
microarray technology. Nat Genet. 37:549–554. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Conlin LK, Thiel BD, Bonnemann CG, Medne
L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H and
Spinner NB: Mechanisms of mosaicism, chimerism and uniparental
disomy identified by single nucleotide polymorphism array analysis.
Hum Mol Genet. 19:1263–1275. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Attiyeh EF, London WB, Mossé YP, Wang Q,
Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et
al: Chromosome 1p and 11q deletions and outcome in neuroblastoma. N
Engl J Med. 353:2243–2253. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ambros PF, Ambros IM, Brodeur GM, Haber M,
Khan J, Nakagawara A, Schleiermacher G, Speleman F, Spitz R, London
WB, et al: International consensus for neuroblastoma molecular
diagnostics: Report from the international neuroblastoma risk group
(INRG) biology committee. Br J Cancer. 100:1471–1482. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Thompson D, Vo KT, London WB, Fischer M,
Ambros PF, Nakagawara A, Brodeur GM, Matthay KK and DuBois SG:
Identification of patient subgroups with markedly disparate rates
of MYCN amplification in neuroblastoma: A report from the
international neuroblastoma risk group project. Cancer.
122:935–945. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sansregret L and Swanton C: The role of
aneuploidy in cancer evolution. Cold Spring Harb Perspect Med.
7:a0283732017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bourdeaut F, Ribeiro A, Paris R, Pierron
G, Couturier J, Peuchmaur M and Delattre O: In neuroblastic
tumours, Schwann cells do not harbour the genetic alterations of
neuroblasts but may nevertheless share the same clonal origin.
Oncogene. 27:3066–3071. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Toraman AD, Keser I, Lüleci G, Tunali N
and Gelen T: Comparative genomic hybridization in
ganglioneuroblastomas. Cancer Genet Cytogen. 132:36–40. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Abdel-Hafiz HA, Schafer JM, Chen X, Xiao
T, Gauntner TD, Li Z and Theodorescu D: Y chromosome loss in cancer
drives growth by evasion of adaptive immunity. Nature. 619:624–631.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Müller P, Camacho OV, Yazbeck AM, Wölwer
C, Zhai W, Schumacher J, Heider D, Buettner R, Quaas A and Hillmer
AM: Why loss of Y? A pan-cancer genome analysis of tumors with loss
of Y chromosome. Comput Struct Biotechnol J. 21:1573–1583. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ognibene M, De Marco P, Parodi S, Meli M,
Di Cataldo A, Zara F and Pezzolo A: Genomic analysis made it
possible to identify gene-driver alterations covering the time
window between diagnosis of neuroblastoma 4s and the progression to
stage 4. Int J Mol Sci. 23:65132022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Janssen A, Van Der Burg M, Szuhai K, Kops
GJ and Medema RH: Chromosome segregation errors as a cause of DNA
damage and structural chromosome aberrations. Science.
333:1895–1898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wu Y, Yu H, Tang H, Su Y, Shi TL, Liu S,
Xia Q and Xu DJ: PXR polymorphisms have impact on the clinical
efficacy of clopidogrel in patients undergoing percutaneous
coronary intervention. Gene. 653:22–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Richardson M, Kirkham J, Dwan K, Sloan DJ,
Davies G and Jorgensen AL: Association of variants within the GST
and other genes with anti-tubercular agents related toxicity: A
systematic review and meta-analysis. BioRxiv. 9:5158172019.
|
|
55
|
Saito R, Suzuki H, Yamada T, Endo S,
Moriwaki T, Ueno T, Hirose M, Hirai S, Yamato K, Mizokami Y and
Hyodo I: Predicting skin toxicity according to EGFR polymorphisms
in patients with colorectal cancer receiving antibody against EGFR.
Anticancer Res. 33:4995–4998. 2013.PubMed/NCBI
|
|
56
|
Zhuang W, Qiu H, Wang X, Xin S, Huang M
and Zhou Z: Impact of pharmacogenomics on imatinib toxicity in
gastrointestinal stromal tumors. J Clin Oncol. 35:110432017.
View Article : Google Scholar
|
|
57
|
Wei F, Xi C and Shaoxing G: Relationship
between MAPK1 gene polymorphism and gefitinib hepatotoxicity in
NSCLC patients with activating EGFR mutations. Acta Pharm
Sin. 53:760–764. 2018.
|
|
58
|
Jansen RS, Mahakena S, de Haas M, Borst P
and van de Wetering K: ATP-binding cassette subfamily C member 5
(ABCC5) functions as an efflux transporter of glutamate conjugates
and analogs. J Biol Chem. 290:30429–30440. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen J, Wang Z, Gao S, Wu K, Bai F, Zhang
Q, Wang H, Ye Q, Xu F, Sun H, et al: Human drug efflux transporter
ABCC5 confers acquired resistance to pemetrexed in breast cancer.
Cancer Cell Int. 21:1–10. 2021.PubMed/NCBI
|
|
60
|
Belkahla S, Khan AUH, Gitenay D, Alexia C,
Gondeau C, Vo DN, Orecchioni S, Talarico G, Bertolini F, Cartron G,
et al: Changes in metabolism affect expression of ABC transporters
through ERK5 and depending on p53 status. Oncotarget. 9:1114–1129.
2018. View Article : Google Scholar : PubMed/NCBI
|