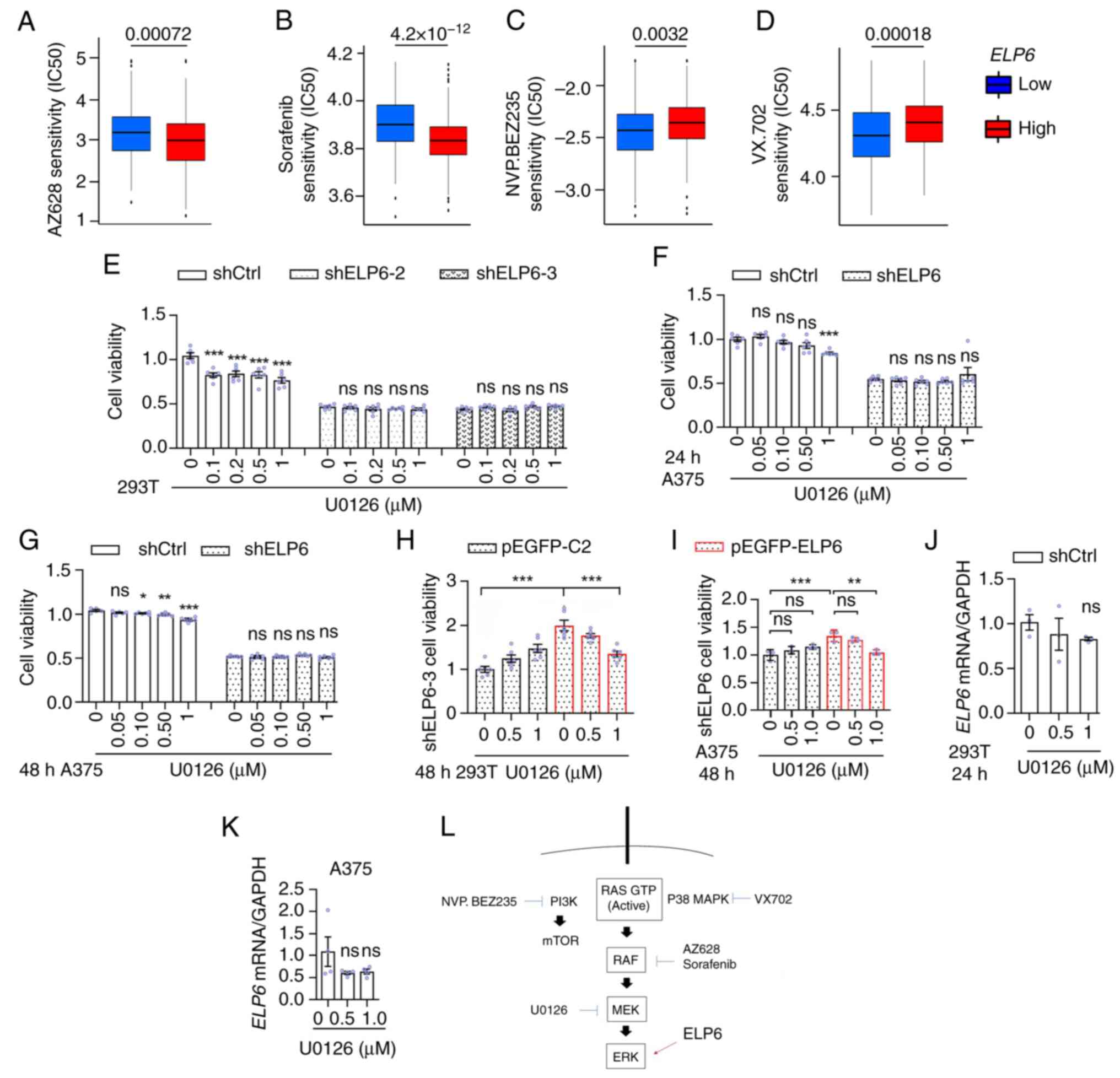
|
1
|
Garbe C, Keim U, Gandini S, Amaral T,
Katalinic A, Hollezcek B, Martus P, Flatz L, Leiter U and Whiteman
D: Epidemiology of cutaneous melanoma and keratinocyte cancer in
white populations 1943–2036. Eur J Cancer. 152:18–25. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dulskas A, Cerkauskaite D, Vincerževskiene
I and Urbonas V: Trends in incidence and mortality of skin melanoma
in Lithuania 1991–2015. Int J Environ Res Public Health.
18:41652021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Radomir MS, Tae-Kang K, Zorica J, Anna AB,
Ewa P, Katie MD, Rebecca SM, Robert CT, Rahul S, David KC, et al:
Malignant melanoma: An overview, new perspectives, and vitamin D
signaling. Cancers (Basel). 16:22622024. View Article : Google Scholar
|
|
4
|
Siegel R, Miller K and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alexandrov L, Nik-Zainal S, Wedge D,
Aparicio S, Behjati S, Biankin A, Bignell G, Bolli N, Borg A,
Børresen-Dale A, et al: Signatures of mutational processes in human
cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hodis E, Watson I, Kryukov G, Arold S,
Imielinski M, Theurillat J, Nickerson E, Auclair D, Li L, Place C,
et al: A landscape of driver mutations in melanoma. Cell.
150:251–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Radomir MS, Jake YC, Chander R and Andrzej
TS: Photo-neuro-immuno-endocrinology: How the ultraviolet radiation
regulates the body, brain, and immune system. Proc Natl Acad Sci
USA. 121:e23083741212024. View Article : Google Scholar
|
|
8
|
Álvarez-Prado ÁF, Maas RR, Soukup K, Klemm
F, Kornete M, Krebs FS, Zoete V, Berezowska S, Brouland JP,
Hottinger AF, et al: Immunogenomic analysis of human brain
metastases reveals diverse immune landscapes across genetically
distinct tumors. Cell Rep Med. 4:1009002023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanrahan AJ and Solit DB: BRAF mutations:
The discovery of Allele- and Lineage-specific differences. Cancer
Res. 82:12–14. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schadendorf D, van Akkooi ACJ, Berking C,
Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A and Ugurel
S: Melanoma. Lancet. 392:971–984. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flaherty K, Puzanov I, Kim K, Ribas A,
McArthur G, Sosman J, O'Dwyer P, Lee R, Grippo J, Nolop K and
Chapman PB: Inhibition of mutated, activated BRAF in metastatic
melanoma. N Engl J Med. 363:809–819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Otero G, Fellows J, Li Y, de Bizemont T,
Dirac A, Gustafsson C, Erdjument-Bromage H, Tempst P and Svejstrup
J: Elongator, a multisubunit component of a novel RNA polymerase II
holoenzyme for transcriptional elongation. Mol Cell. 3:109–118.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wittschieben B, Otero G, de Bizemont T,
Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C, Tempst P and
Svejstrup J: A novel histone acetyltransferase is an integral
subunit of elongating RNA polymerase II holoenzyme. Mol Cell.
4:123–128. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Winkler G, Kristjuhan A, Erdjument-Bromage
H, Tempst P and Svejstrup J: Elongator is a histone H3 and H4
acetyltransferase important for normal histone acetylation levels
in vivo. Proc Natl Acad Sci USA. 99:3517–3522. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Glatt S, Létoquart J, Faux C, Taylor N,
Séraphin B and Müller C: The Elongator subcomplex Elp456 is a
hexameric RecA-like ATPase. Nat Struct Mol Biol. 19:314–320. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rahl P, Chen C and Collins R: Elp1p, the
yeast homolog of the FD disease syndrome protein, negatively
regulates exocytosis independently of transcriptional elongation.
Mol Cell. 17:841–853. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu S, Jiang C, Lin R, Wang X, Hu X, Chen
W, Chen X and Chen T: Epigenetic activation of the elongator
complex sensitizes gallbladder cancer to gemcitabine therapy. J Exp
Clin Cancer Res. 40:3732021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu S, Zhan M, Jiang C, He M, Yang L, Shen
H, Huang S, Huang X, Lin R, Shi Y, et al: Genome-wide CRISPR screen
identifies ELP5 as a determinant of gemcitabine sensitivity in
gallbladder cancer. Nat Commun. 10:54922019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Waszak S, Robinson G, Gudenas B, Smith K,
Forget A, Kojic M, Garcia-Lopez J, Hadley J, Hamilton K, Indersie
E, et al: Germline elongator mutations in sonic hedgehog
medulloblastoma. Nature. 580:396–401. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feng X, Zhang H, Meng L, Song H, Zhou Q,
Qu C, Zhao P, Li Q, Zou C, Liu X and Zhang Z: Hypoxia-induced
acetylation of PAK1 enhances autophagy and promotes brain
tumori-genesis via phosphorylating ATG5. Autophagy. 17:723–742.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cruz-Gordillo P, Honeywell M, Harper N,
Leete T and Lee M: MCL1ELP-dependent expression of promotes
resistance to EGFR inhibition in triple-negative breast cancer
cells. Sci Signal. 13:eabb98202020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Y, Tang D, Yang S, Liu H, Luo S,
Stinchcombe T, Glass C, Su L, Shen S, Christiani DC and Wei Q:
ELP2novel variants of and in the interferon gamma signaling pathway
are associated with non-small cell lung cancer survival. Cancer
Epidemiol Biomarkers Prev. 29:1679–1688. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Close P, Gillard M, Ladang A, Jiang Z,
Papuga J, Hawkes N, Nguyen L, Chapelle J, Bouillenne F, Svejstrup
J, et al: DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity
and migration of melanoma cells as subunits of Elongator. J Biol
Chem. 287:32535–32545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Geeleher P, Cox N and Huang R: pRRophetic:
An R package for prediction of clinical chemotherapeutic response
from tumor gene expression levels. PLoS One. 9:e1074682014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davide C, Paola C, Marzia C, Mirella B,
Elisa S, Luca M, Jessica G, Elisabetta G, Veronica DG, Marcello M
and Presta M: The Pro-oncogenic Sphingolipid-metabolizing enzyme
β-galactosylceramidase modulates the proteomic landscape in
BRAF(V600E)-Mutated human melanoma cells. Int J Mol Sci.
24:105552023. View Article : Google Scholar
|
|
29
|
Whiteaker JR, Sharma K, Hoffman MA, Kuhn
E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C,
et al: Targeted mass spectrometry-based assays enable multiplex
quantification of receptor tyrosine kinase, MAP Kinase, and AKT
signaling. Cell Rep Methods. 1:1000152021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using Real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vecchiato A, Rossi CR, Montesco MC,
Frizzera E, Seno A, Piccoli A, Martello T, Ninfo V and Lise M:
Proliferating cell nuclear antigen (PCNA) and recurrence in
patients with cutaneous melanoma. Melanoma Res. 4:207–211. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roels S, Tilmant K and Ducatelle R: PCNA
and Ki67 proliferation markers as criteria for prediction of
clinical behaviour of melanocytic tumours in cats and dogs. J Comp
Pathol. 121:13–24. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hulleman E, Bijvelt JJ, Verkleij AJ,
Verrips CT and Boonstra J: Nuclear translocation of
mitogen-activated protein kinase p42MAPK during the ongoing cell
cycle. J Cell Physiol. 180:325–333. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nedialkova D and Leidel S: Optimization of
codon translation rates via tRNA modifications maintains proteome
integrity. Cell. 161:1606–1618. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo F, Cen S, Niu M, Javanbakht H and
Kleiman L: Specific inhibition of the synthesis of human lysyl-tRNA
synthetase results in decreases in tRNA(Lys) incorporation,
tRNA(3)(Lys) annealing to viral RNA, and viral infectivity in human
immunodeficiency virus type 1. J Virol. 77:9817–9822. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chatzilakou E, Hu Y, Jiang N and Yetisen
A: Biosensors for melanoma skin cancer diagnostics. Biosens
Bioelectron. 250:1160452024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mao Y, Yan R, Li A, Zhang Y, Li J, Du H,
Chen B, Wei W, Zhang Y, Sumners C, et al: Lentiviral vectors
mediate Long-term and high efficiency transgene expression in HEK
293T cells. Int J Med Sci. 12:407–415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stepanenko AA and Dmitrenko VV: HEK293 in
cell biology and cancer research: phenotype, karyotype,
tumorigenicity, and stress-induced genome-phenotype evolution.
Gene. 569:182–190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ascierto PA, Kirkwood JM, Grob JJ, Simeone
E, Grimaldi AM, Maio M, Palmieri G, Testori A, Marincola FM and
Mozzillo N: The role of BRAF V600 mutation in melanoma. J Transl
Med. 10:852012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Morris EJ, Jha S, Restaino CR, Dayananth
P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, et al:
Discovery of a novel ERK inhibitor with activity in models of
acquired resistance to BRAF and MEK inhibitors. Cancer Discov.
3:742–750. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Brożyna AA, Jóźwicki W, Carlson JA and
Slominski AT: Melanogenesis affects overall and disease-free
survival in patients with stage III and IV melanoma. Hum Pathol.
44:2071–2074. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Radomir MS, Tadeusz S, Przemysław MP,
Chander R, Anna AB and Andrzej TS: Melanoma, melanin, and
melanogenesis: The yin and yang relationship. Front Oncol.
12:8424962022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Slominski A, Kim TK, Brożyna AA,
Janjetovic Z, Brooks DL, Schwab LP, Skobowiat C, Jóźwicki W and
Seagroves TN: The role of melanogenesis in regulation of melanoma
behavior: Melanogenesis leads to stimulation of HIF-1α expression
and HIF-dependent attendant pathways. Arch Biochem Biophys.
563:79–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Radomir MS, Chander R, Jake YC and Andrzej
TS: How cancer hijacks the body's homeostasis through the
neuroendocrine system. Trends Neurosci. 46:263–275. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vitiello M, Tuccoli A, D'Aurizio R, Sarti
S, Giannecchini L, Lubrano S, Marranci A, Evangelista M, Peppicelli
S, Ippolito C, et al: Context-dependent miR-204 and miR-211 affect
the biological properties of amelanotic and melanotic melanoma
cells. Oncotarget. 8:25395–25417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Militaru IV, Rus AA, Munteanu CVA, Manica
G and Petrescu SM: New panel of biomarkers to discriminate between
amelanotic and melanotic metastatic melanoma. Front Oncol.
12:10618322023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kaczmarek-Szczepańska B, Kleszczyński K,
Zasada L, Chmielniak D, Hollerung MB, Dembińska K, Pałubicka K,
Steinbrink K, Swiontek Brzezinska M and Grabska-Zielińska S:
Hyaluronic Acid/ellagic acid as materials for potential medical
application. Int J Mol Sci. 25:58912024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Magina S, Vieira-Coelho MA, Serrão MP,
Kosmus C, Moura E and Moura D: Ultraviolet B radiation
differentially modifies catechol-O-methyltransferase activity in
keratinocytes and melanoma cells. Photodermatol Photoimmunol
Photomed. 28:137–141. 2012. View Article : Google Scholar : PubMed/NCBI
|