Introduction
Liposarcoma (LPS) is one of the most common types of
soft tissue sarcoma (1–3). Among its four subtypes,
well-differentiated LPS (WDLPS) and dedifferentiated LPS (DDLPS)
share similar genomic changes, notably the amplification of the
12q13-15 chromosomal region, where mouse double minute 2 homolog
(MDM2) is the primary oncogene. WDLPS rarely metastasizes and can
remain stable for several years. By contrast, DDLPS is a highly
aggressive disease characterized by frequent local recurrence and
distant metastasis (4). Wide
surgical excision with curative intent is the preferred treatment
for localized disease. However, for advanced DDLPS, despite
recommended first-line chemotherapy with doxorubicin (5) and second-line options including
eribulin or trabectedin (6–8), treatment outcomes remain poor.
Ferroptosis is a form of cell death characterized by
the oxidative modification of phospholipid membranes, resulting in
the accumulation of lipid-based reactive oxygen species (ROS)
(9–11). Cysteine metabolism is crucial in the
regulation of ferroptosis (9–11). The
cystine-glutamate antiporter (xCT), composed of solute carrier
family 7 member 11 (SLC7A11) and SLC3A2, facilitates the uptake of
cystine from the extracellular environment. Once imported, cystine
is reduced to cysteine, a key component of the tripeptide
glutathione. Glutathione peroxidase 4 (GPX4) requires glutathione
as a cofactor to reduce lipid peroxidation and prevent ferroptosis
(12,13). Inactivation of GPX4, either through
cystine deprivation by the xCT inhibitor erastin or direct
inhibition by the GPX4 inhibitor Ras-selective lethal small
molecule 3 (RSL3), leads to the accumulation of lipid-based ROS and
ultimately ferroptotic cell death (13,14).
Few studies have explored the role of ferroptosis in
LPS, a tumor originating from adipocytic differentiation. Tumor
protein p53 (TP53), a well-known tumor suppressor mutated in nearly
half of all cancers (15), plays a
key role in regulating ferroptosis through multiple, sometimes
paradoxical, pathways. These include suppressing SLC7A11 expression
to promote ferroptosis (16);
upregulating 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase
(HMGCR), which produces molecules with anti-ferroptotic properties
(17); and inhibiting
dipeptidyl-peptidase-4 (DPP4)-dependent lipid peroxidation, thereby
reducing ferroptosis (18). MDM2,
the key oncogene in DDLPS (19),
and its homolog MDM4 (20)
negatively regulate TP53 (21–24),
potentially influencing ferroptotic death through both
TP53-dependent and TP53-independent mechanisms (25). These findings suggest that exploring
the ferroptosis pathway may lead to the discovery of novel
therapeutic strategies, with MDM2 playing a critical role in the
regulation of ferroptosis in DDLPS.
In the present study, an investigation of the
expression of genes involved in the ferroptosis pathway in DDLPS
was performed via bioinformatics, to identify differentially
expressed ferroptosis-related genes. Additionally, the sensitivity
of DDLPS cell lines to the ferroptosis-inducing agents erastin and
RSL3 was investigated. The effects of nutlin-3, an MDM2 inhibitor,
as a co- or pre-treatment with erastin or RSL3 were also
investigated, and the effect of TP53 knockdown (KD) was
explored.
Materials and methods
Exploration of ferroptosis-related
gene expression regulated by the MDM2-TP53 pathway through
bioinformatics analysis
Bioinformatics analysis was performed as previously
described (26). Microarray data
from the Affymetrix Human Genome U133 Plus 2.0 and Affymetrix Human
Genome U133A platforms were obtained from the National Center for
Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/gds/). The datasets
comprised DDLPS samples from GSE21050 (U133 Plus 2.0) (27) and GSE30929 (U133A) (28), WDLPS samples from GSE20559 (U133
Plus 2.0) (29) and GSE30929
(U133A) (28), and adipose tissue
samples from GSE41168 (U133 Plus 2.0) (30) and GSE35710 (U133A) (31). The gene expression data were
normalized using dChip (32,33).
Specific genes associated with ferroptosis were selected, and their
expression levels were expressed as Z-scores following Z-score
transformation.
Cell lines and reagents
DDLPS cell lines LPS853 and NDDLS-1, provided by Dr
Fletcher JA (Department of Pathology, Brigham and Women's Hospital,
Boston, MA 02115, USA) and Dr Ariizumi (Division of Orthopedic
Surgery, Niigata University Graduate School of Medical and Dental
Sciences, Niigata City, Niigata 951-8510, Japan), respectively,
were used as in vitro models to test the efficacy of various
agents. The cells were cultured in RPMI 1640 Medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with Fetal Bovine Serum
(Corning, Inc.) and Penicillin-Streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) and incubated at 37°C with 5% CO2. The
xCT inhibitor erastin (CAS no. 571203-78-6; Cayman Chemical
Company), GPX4 inhibitor RSL3 (CAS no. 1219810-16-8; Cayman
Chemical Company) and HMGCR inhibitors lovastatin (S2061; Selleck
Chemicals) and simvastatin (S1792; Selleck Chemicals) were
evaluated for their ferroptosis-inducing effects. Nutlin-3a (S8059;
Selleck Chemicals) was used as an MDM2 antagonist; this is the
active enantiomer of nutlin-3, and for simplicity is referred to as
nutlin-3 throughout the manuscript. Ferrostatin-1 (Fer-1; CAS no.
347174-05-4; Cayman Chemical Company) was used to inhibit
ferroptosis. The antibodies used for immunoblotting were as
follows: p53 (cat. no. 2527S; 1:1,000; Cell Signaling Technology,
Inc.), MDM2 (cat. no. 86934S; 1:1,000; Cell Signaling Technology,
Inc.), SLC7A11 (cat. no. A13685; 1:1,000; ABclonal Biotech Co.,
Ltd.), GPX4 (cat. no. A1933; 1:1,000; ABclonal Biotech Co., Ltd.),
SLC3A2/CD98hc (cat. no. A3658; 1:1,000; ABclonal Biotech Co.,
Ltd.), 4EBP1 (cat. no. 9644; 1:1,000; Cell Signaling Technology,
Inc.), phosphorylated (p)-4EBP1 (cat. no. 9459; 1:1,000; Cell
Signaling Technology, Inc.), p70S6 (cat. no. 9202; 1:1,000; Cell
Signaling Technology, Inc.), p-p70S6 (cat. no. 9205; 1:1,000; Cell
Signaling Technology, Inc.) and β-actin (cat. no. ab6276; 1:1,000;
Abcam).
Lipid peroxidation assay
The DDLPS cell lines (2.5×105 cells/well)
were seeded in six-well plates and incubated at 37°C with different
concentrations of test agents for specific durations. For single
agent treatment, cells were treated with erastin (12 µM for
NDDLS-1, 20 µM for LPS853), RSL3 (0.2 µM for both), lovastatin (18
µM for both), or simvastatin (18 µM for both) for 24 h. For
combination with Fer-1, cells were treated with erastin (8 µM for
LPS853, 20 µM for NDDLS-1), or RSL3 (0.05 µM for both), and
combined with Fer-1 (4, 6, or 8 µM), with co-treatment of erastin
and Fer-1 for 48 h and RSL3 and Fer-1 for 4 h. For combination with
nutlin-3, cells were treated with erastin (8 or 16 µM for LPS853,
12 or 20 µM for NDDLS-1), or RSL3 (0.2 or 0.4 µM for both), and
combined with nutlin-3 (10 µM for both). Co-treatment with erastin
and nutlin-3 or RSL3 and nutlin-3 was performed for 24 h, whereas
in the sequential treatment, cells were first exposed to nutlin-3
for 24 h, followed by treatment with either erastin or RSL3 for an
additional 24 h. After treatment, cells were incubated with 2 µM
BODIPY 581/591 C11 (Thermo Fisher Scientific, Inc.) for 30 min at
37°C. After this, the cells were collected, resuspended in 500 µl
phosphate-buffered saline (PBS), and analyzed using a BD FACSCanto™
II flow cytometry system (BD Biosciences) (34). Data were analyzed using FlowJo
software (version 7.6; FlowJo LLC). The experiment was performed in
triplicate.
Cell viability assay
Cell viability was assessed using the Cell Counting
Kit-8 (CCK-8) assay (Abbkine Scientific Co., Ltd.) according to the
manufacturer's instructions. In brief, cells were plated in 96-well
plates at a concentration of 5,000 cells in 100 µl/well. The
following day, different concentrations of the test agents were
added to individual wells and incubated at 37°C for the appropriate
duration. The treatment conditions were as follows: for single
agent treatment, Erastin (4, 8, 12, 16, 20 µM), RSL3 (1, 2, 3, 4, 5
µM), lovastatin (6, 12, 18, 24, 30 µM), or simvastatin (6, 12, 18,
24, 30 µM) was used for 24-h treatment; for combination with Fer-1,
Erastin (3, 6, 9, 12, 15 µM) or RSL3 (0.01, 0.02, 0.03, 0.04, 0.05
µM) were combined with Fer-1 (1, 2, 3, 4, 5 µM) for 48-h treatment;
for combination with nutlin-3, Erastin (3, 6, 9, 12, 15 µM) or RSL3
(0.01, 0.02, 0.03, 0.04, 0.05 µM) were combined with nutlin-3 (4,
8, 12, 16, 20 µM). Co-treatment with erastin and nutlin-3 or RSL3
and nutlin-3 was performed for 24 h, whereas in the sequential
treatment, cells were first exposed to nutlin-3 for 24 h, followed
by treatment with either erastin or RSL3 for an additional 24 h.
After incubation, 10 µl CCK-8 solution was added to each well,
followed by incubation for 1.5–2 h. The absorbance at 450 nm was
measured for each well using a microplate reader. The combination
index (CI) for two-drug combinations was calculated using CalcuSyn
software (Biosoft), where CI <1, CI=1 and CI >1 indicate
synergism, additivity and antagonism, respectively. All experiments
were performed in triplicate (35).
Apoptosis assessment
Apoptosis was assessed as previously described
(26). Specifically, for single
agent treatment, cells (1×105 cells/well) were treated
with erastin (2, 4, 6, 8 µM for LPS853, 3, 6, 9, 12 µM for NDDLS-1)
or RSL3 (0.1, 0.2, 0.3, 0.4 µM for both), with LPS853 cells exposed
to erastin for 24 h, NDDLS-1 cells exposed to erastin for 48 h, and
both cell lines treated with RSL3 for 24 h. For combination with
nutlin-3, cells (1×105 cells/well) were treated with
erastin (10 µM), or RSL3 (0.4 µM), and combined with nutlin-3 (10
µM). Co-treatment with erastin and nutlin-3 or RSL3 and nutlin-3
was performed for 24 h, whereas in the sequential treatment, cells
were first exposed to nutlin-3 for 24 h, followed by treatment with
either erastin or RSL3 for an additional 24 h. After treatment,
cells were washed with 1X PBS and resuspended in 100 µl staining
solution containing annexin V-fluorescein isothiocyanate and
propidium iodide in HEPES buffer (BD Pharmingen). The cells were
incubated at room temperature for 15 min, then diluted in 1X
annexin V-binding buffer (BD Pharmingen) and analyzed by flow
cytometry with a BD FACSCanto II system, BD FACSDiva Software
v8.0.2 operating software (all BD Biosciences) and FlowJo (version
7.6; FlowJo LLC) analysis software. The experiment was performed in
triplicate.
Immunoblotting
Immunoblotting was performed as previously described
(36). Cells were treated with
erastin (8 µM), RSL3 (0.2 µM), or nutlin-3 (12 µM), either as
single agent or various combinations, for 24 h. After treatment,
cultured monolayer cells were rinsed with PBS and lysed using RIPA
Lysis and Extraction Buffer (Thermo Fisher Scientific, Inc.). The
cell suspensions were then incubated at 4°C for 30 min, followed by
centrifugation at 15,974 × g for 30 min at 4°C. Total protein
concentrations were measured using the Pierce BCA Protein Assay Kit
(Thermo Fisher Scientific, Inc.). Proteins (50 µg per lane) were
separated by 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinylidene
difluoride (PVDF) membranes (PerkinElmer, Inc.). The membranes were
blocked with 5% bovine serum albumin (BSA; Bionovas Biotechnology
Co., Ltd.) at room temperature for 30 min. Primary antibodies were
incubated at 4°C overnight, while secondary antibodies were
incubated at room temperature for 45 min. Primary and secondary
antibodies were prepared in 5% bovine serum albumin (BSA). The
secondary antibodies, Anti-rabbit IgG (cat. no. 7074S) and
Anti-mouse IgG (cat. no. 7076S), were diluted at 1:4,500 and
purchased from Cell Signaling Technology, Inc. The immunoreactive
bands were visualized using Immobilon Western Chemiluminescent HRP
Substrate (MilliporeSigma) and a UVP ChemStudio PLUS Touch Western
Blot Imaging System (Analytik Jena AG). Densitometric analysis was
performed using ImageJ software (version 1.51, National Institutes
of Health).
Cystine uptake assay
Cystine uptake was measured using a Cystine Uptake
Assay Kit (Dojindo Laboratories, Inc.) according to the
manufacturer's instructions. Cells (1×104 cells/well)
were seeded in a black 96-well plate for 24 h. The medium was then
replaced and cells were treated with single agent erastin (40 µM)
or nutlin-3 (10 µM) or combination for the time period specified by
the manufacturer. After treatment, the culture medium was
aspirated, and the cells were washed three times with serum-free
RPMI 1640 prior to incubation in serum-free RPMI 1640 for 5 min at
37°C. Subsequently, the cells were incubated with Cystine Analog
Solution (selenocystine) from the kit in serum-free RPMI 1640 for
30 min at 37°C. Fluorescence was measured using a fluorescence
microplate reader with an excitation wavelength of 490 nm and an
emission wavelength of 535 nm.
Small interfering RNA (siRNA)-mediated
knockdown (KD) of TP53
The TP53 KD experiment was conducted using the
ON-TARGETplus system (Horizon Discovery, Ltd.). Cells
(1×106) were first treated with DharmaFECT 1
Transfection Reagent (cat. no. T-2001-03; GE Healthcare Dharmacon,
Inc.; ON-TARGETplus system) and subsequently transfected with
ON-TARGETplus Human TP53 (cat. no. 7157) siRNA-SMARTpool (cat. no.
L-003329-00-0005) target sequences: GAAAUUUGCGUGUGGAGUA,
GUGCAGCUGUGGGUUGAUU, GCAGUCAGAUCCUAGCGUC and GGAGAAUAUUUCACCCUUC;
20 µM) or the ON-TARGETplus Non-targeting Pool (cat. no.
D-001810-10-20; GE Healthcare Dharmacon, Inc.) target sequences:
UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, and
UGGUUUACAUGUUUUCCUA; 20 µM as a negative control. However, the
sense and antisense strand sequences were not provided for either
siRNA. Following transfection, cells were incubated at 37°C for 48
h.
The KD efficiency of TP53 was confirmed by reverse
transcription-quantitative PCR (RT-qPCR). RNA extraction was
performed using the LabPrep™ RNA Plus Mini Kit (LabPrep). RT was
carried out using the HiScript™ First Strand cDNA Synthesis Kit
(Bionovas Biotechnology Co., Ltd.) according to the manufacturer's
protocol. qPCR was performed using SYBR Green PCR Master Mix
(Thermo Fisher Scientific, Inc.) with the following primers: TP53
forward, 5′-GCCATCTACAAGCAGTCACAG-3′ and reverse,
5′-TCATCCAAATACTCCACACGC-3′; GAPDH forward,
5′-GCCAAGGTCATCCATGACAACT-3′ and reverse,
5′-GAGGGGCCATCCACAGTCTT-3′. The thermocycling conditions were as
follows: 95°C for 10 min, followed by 36 cycles of 95°C for 15 sec
and 55°C for 60 sec. Amplification and analysis were performed
using the QuantStudio3 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Gene copy numbers were calculated
according to the 2−ΔΔCq method (37).
Statistical analysis
Differences in the expression of genes among the
DDLPS, WDLPS and adipose tissues were analyzed using the
Kruskal-Wallis test, followed by Dunn's post hoc test for pairwise
comparisons. In the cell-based experiments, differences between two
groups were analyzed using unpaired Student's t-test and among
multiple groups were analyzed by one-way ANOVA followed by
Bonferroni's post hoc test. P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
conducted using SPSS Statistics for Windows, version 17.0 (SPSS
Inc.).
Results
Ferroptosis-related genes regulated by
the MDM2-TP53 pathway are differentially expressed in DDLPS
Publicly available data were analyzed to identify
potential differences in the expression of ferroptosis-related
genes regulated by the MDM2-TP53 pathway. The expression levels of
MDM2 were significantly upregulated in WDLPS and DDLPS compared
with those in adipose tissue (Fig.
1A). By contrast, the GPX4 expression levels in DDLPS were
significantly lower than those in both adipose tissue and WDLPS
(Fig. 1B), which may contribute to
a pro-ferroptosis phenotype. The expression levels of HMGCR
observed in DDLPS were higher compared with those in adipose tissue
and WDLPS (Fig. 1E), potentially
contributing to an anti-ferroptosis effect. The expression levels
of SLC7A11 and SLC3A2 were higher in both types of LPS compared
with those in adipose tissue. However, the differential expression
of DPP4 in adipose tissue, WDLPS and DDLPS was inconsistent between
the two platforms (Fig. 1C, D and
F).
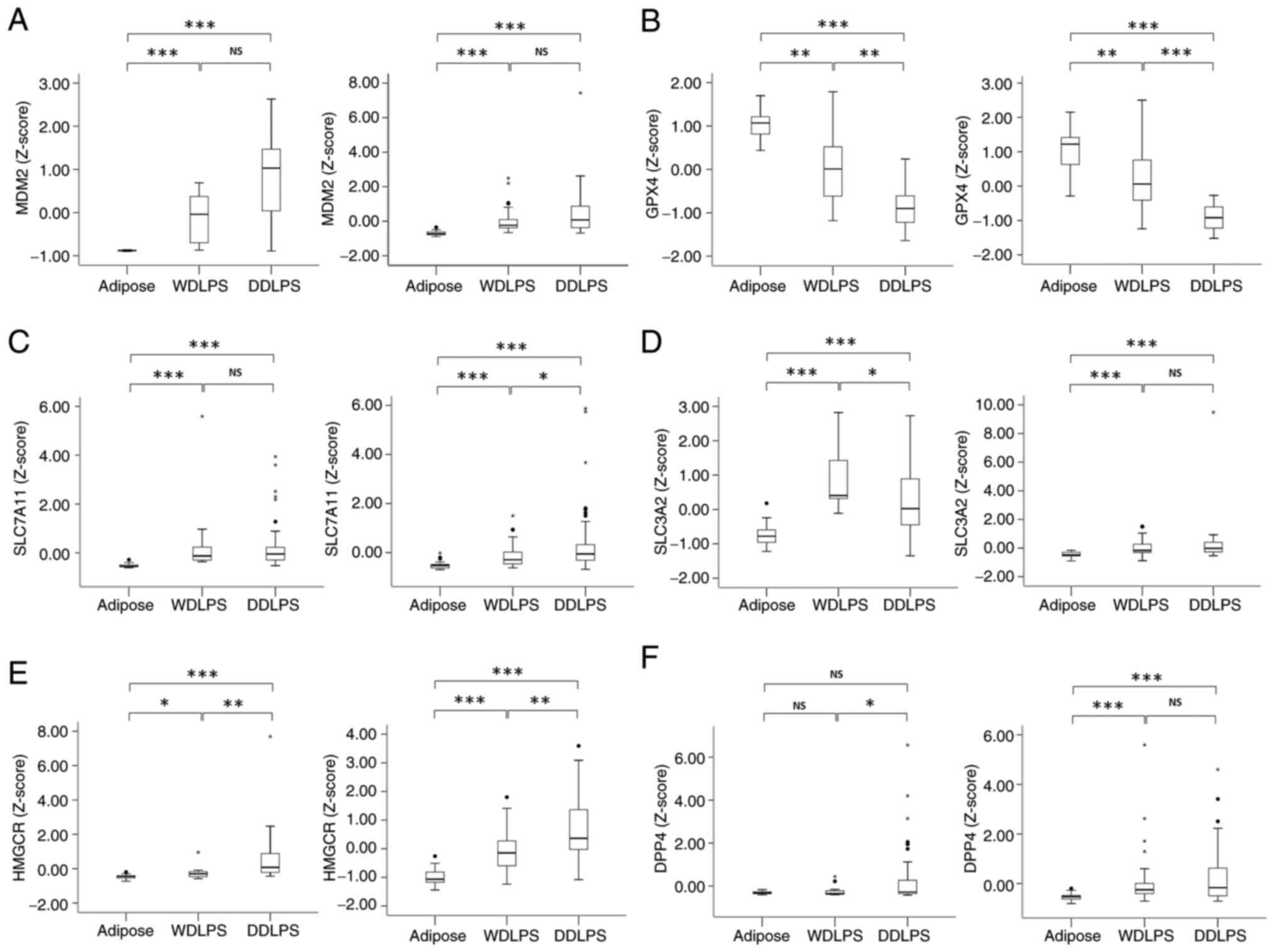 | Figure 1.Comparison of the expression levels
of six ferroptosis-related genes among adipose, WDLPS and DDLPS
tissues. Expression levels of (A) MDM2, (B) GPX4, (C) SLC7A11, (D)
SLC3A2, (E) HMGCR and (F) DPP4. In each panel, data in the left
graph are from GSE21050, GSE20559 and GSE41168 (U133 Plus 2.0) and
data in the right graph are from GSE30929 and GSE35710 (U133A).
*P<0.05, **P<0.01 and ***P<0.001 as indicated. Differences
in levels among the different tissue types were analyzed using
Kruskal-Wallis followed by Dunn's post hoc test. NS, not
significant; WDLPS, well-differentiated liposarcoma; DDLPS,
dedifferentiated liposarcoma; MDM2, mouse double minute 2 homolog;
GPX4, glutathione peroxidase 4; SLC7A11, solute carrier family 7
member 11; SLC3A2, solute carrier family 3 member 2; HMGCR,
3-hydroxy-3-methyl-glutaryl-coenzyme A reductase; DPP4, dipeptidyl
peptidase-4. |
DDLPS cell lines show variable
sensitivity to ferroptosis-inducing agents
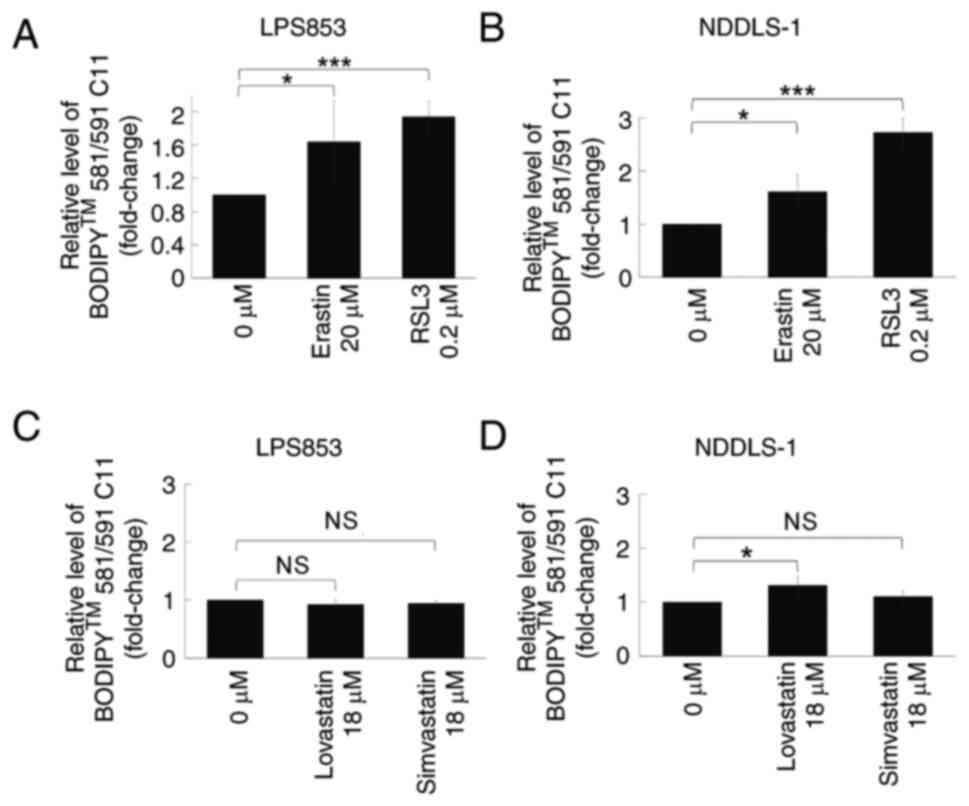
The potential lipid peroxidation effects of
ferroptosis-inducing agents on two DDLPS cell lines were evaluated.
Both erastin and RSL3 treatment significantly increased lipid
peroxidation levels in both cell lines compared with those in
untreated cells, as demonstrated by flow cytometry (Figs. 2 and S1). However, regarding the two statins
with HMGCR inhibitory activity, only lovastatin exhibited a
significant effect, inducing an increase in lipid peroxidation
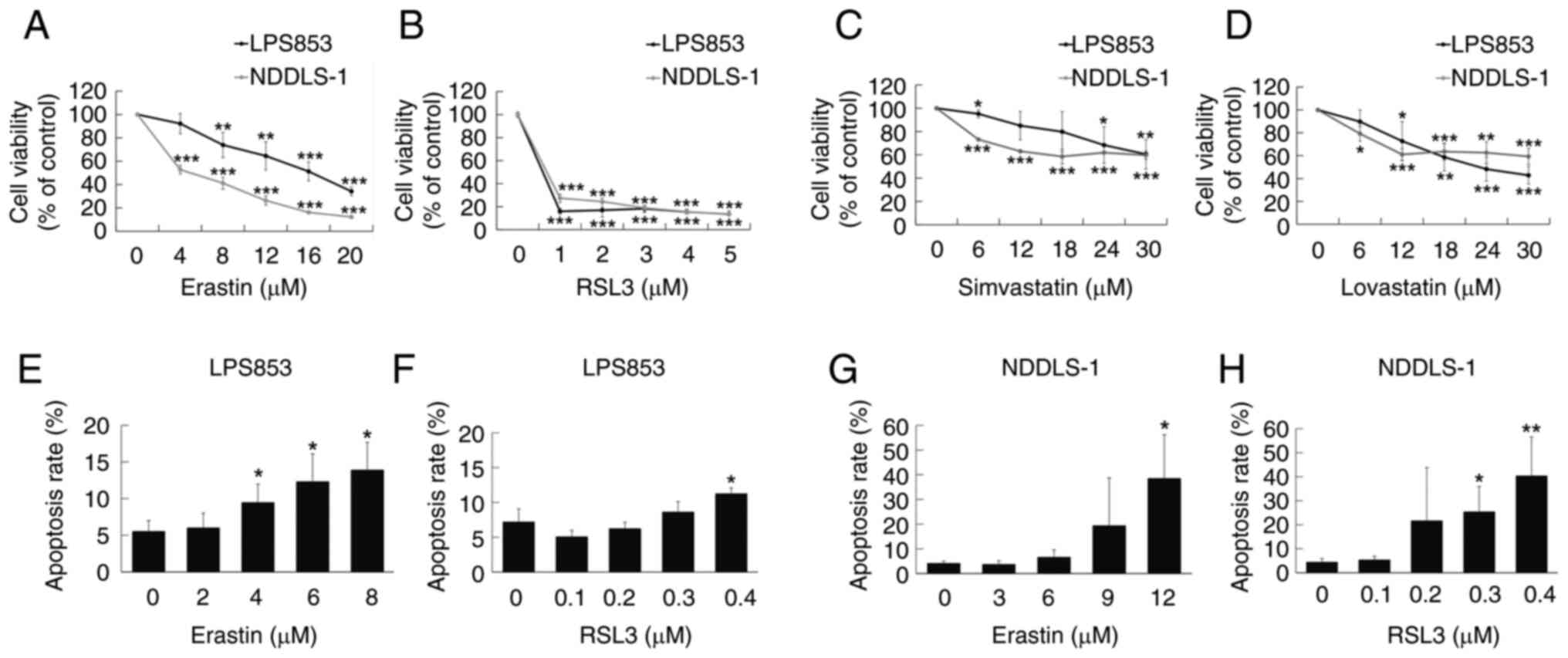
levels in NDDLS-1 cells only. The cytotoxic effects of these four
agents were then assessed in the two cell lines. Erastin and RSL3
exerted significant cytotoxic effects on both DDLPS cell lines at
relatively low doses (Fig. 3A and
B). Lovastatin and simvastatin also showed significant,
although less potent, efficacy against the two DDLPS cell lines
(Fig. 3C and D). In addition,
erastin (Figs. 3E and G, S2A and C) and RSL3 (Figs. 3F and H, S2B and D) induced apoptosis in both DDLPS
cell lines. On the basis of these results, only erastin and RSL3
were used in subsequent experiments.
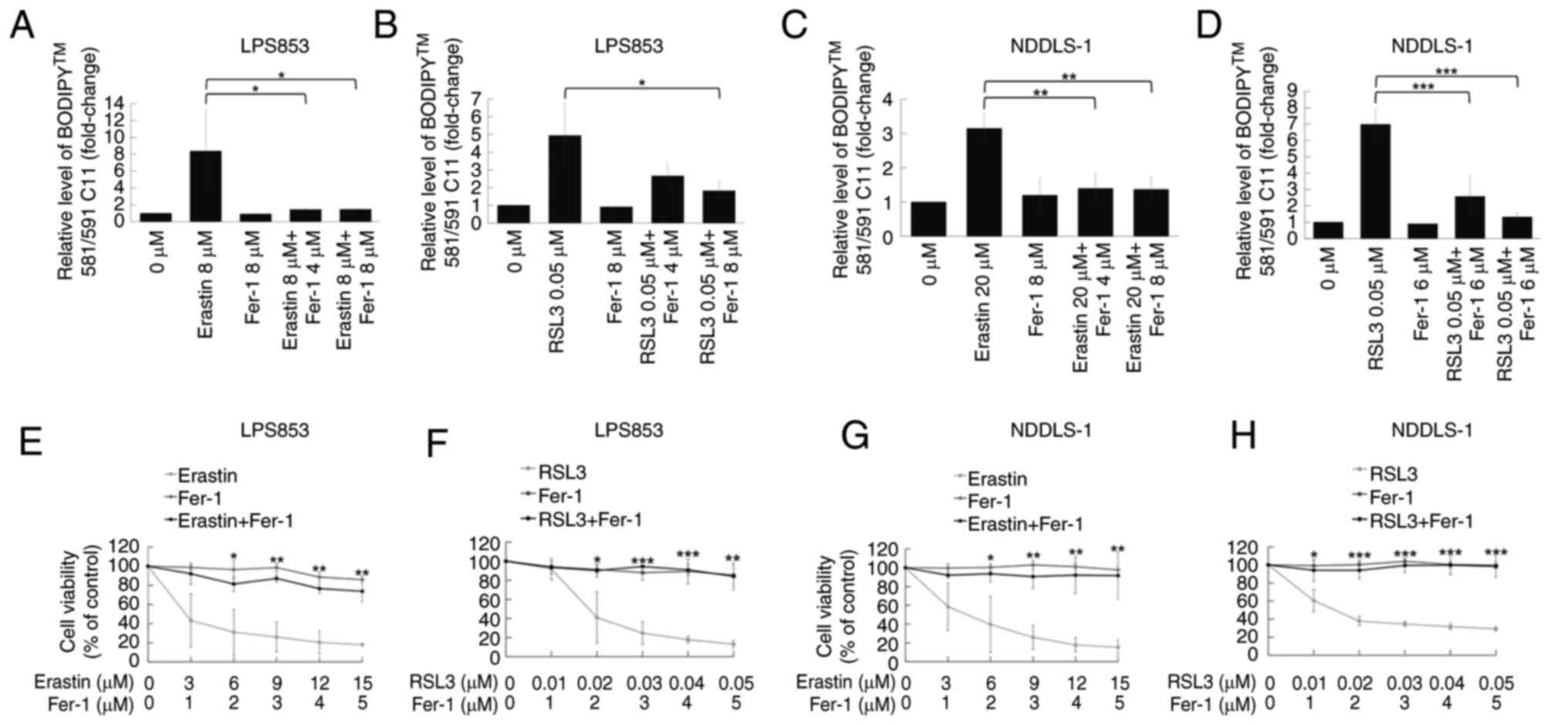
Erastin and RSL3 exert cytotoxic
effects primarily through ferroptosis
Further analysis was performed to determine whether
ferroptosis is the primary mechanism underlying the cytotoxic
effects of erastin and RSL3. The ferroptosis inhibitor Fer-1
partially diminished the lipid peroxidation induced by erastin or
RSL3 in the LPS853 and NDDLS-1 cell lines (Figs. 4A-D and S3). Furthermore, CCK-8 assay results
revealed that Fer-1 attenuated the cytotoxic effects of both
erastin and RSL3 in both DDLPS cell lines (Fig. 4E-H). These results indicate that
both erastin and RSL3 exert their cytotoxic effects at least in
part via the induction of ferroptosis.
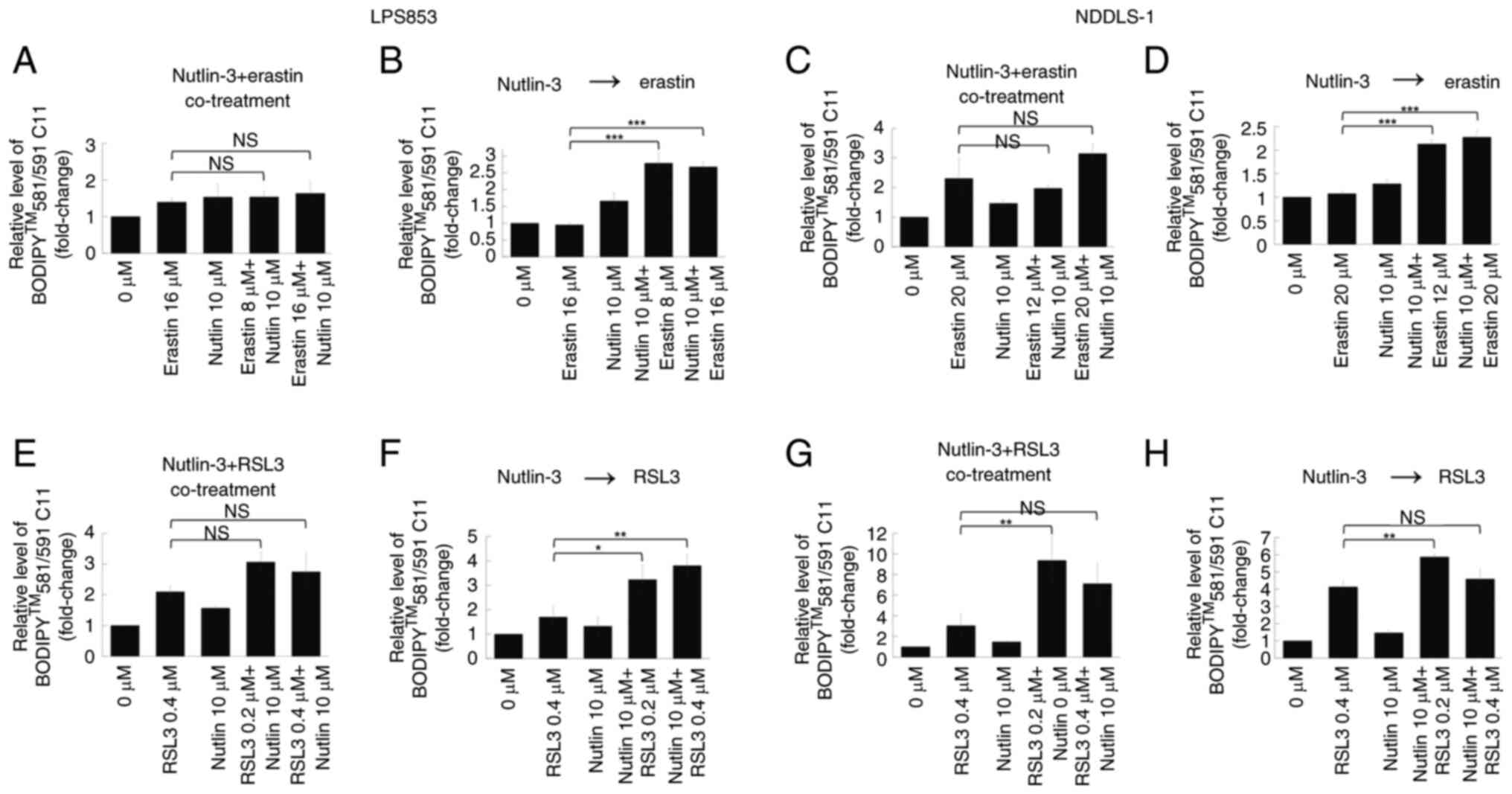
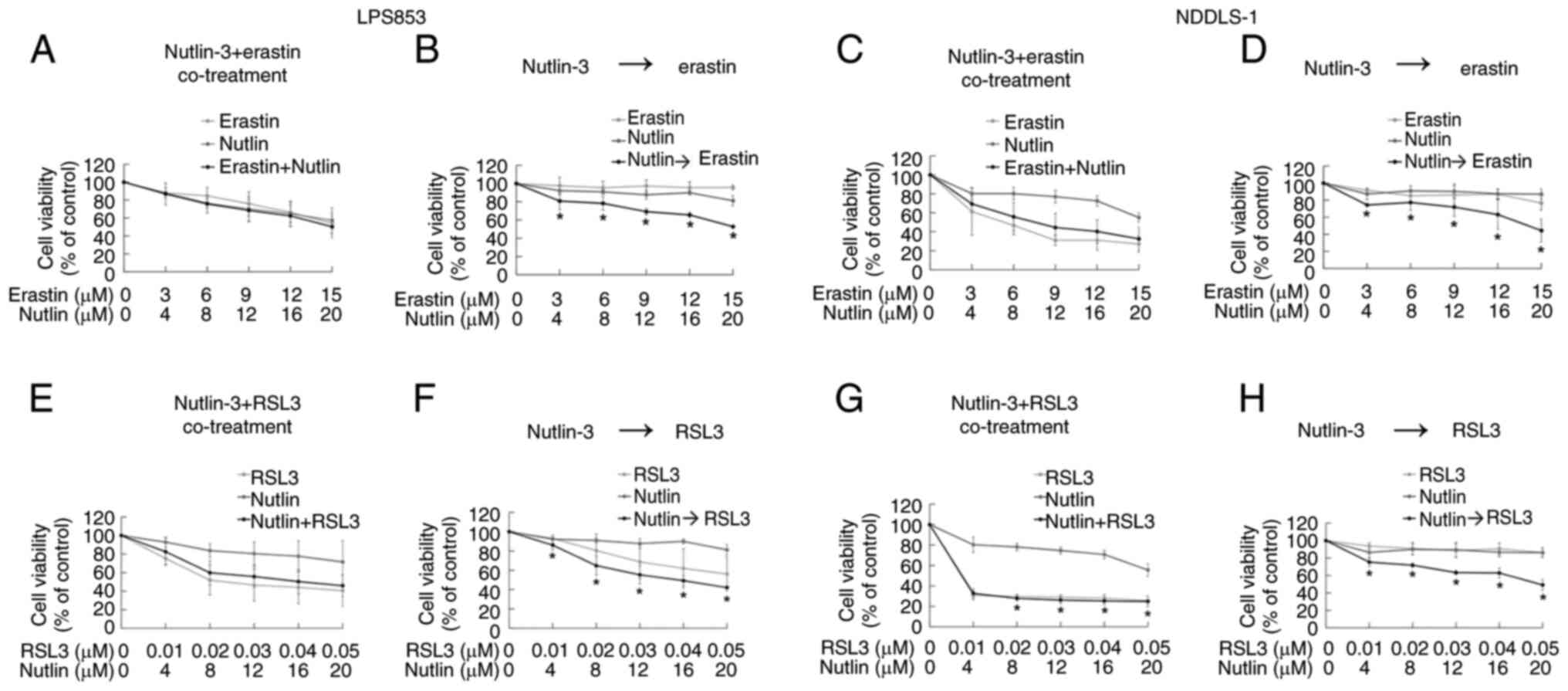
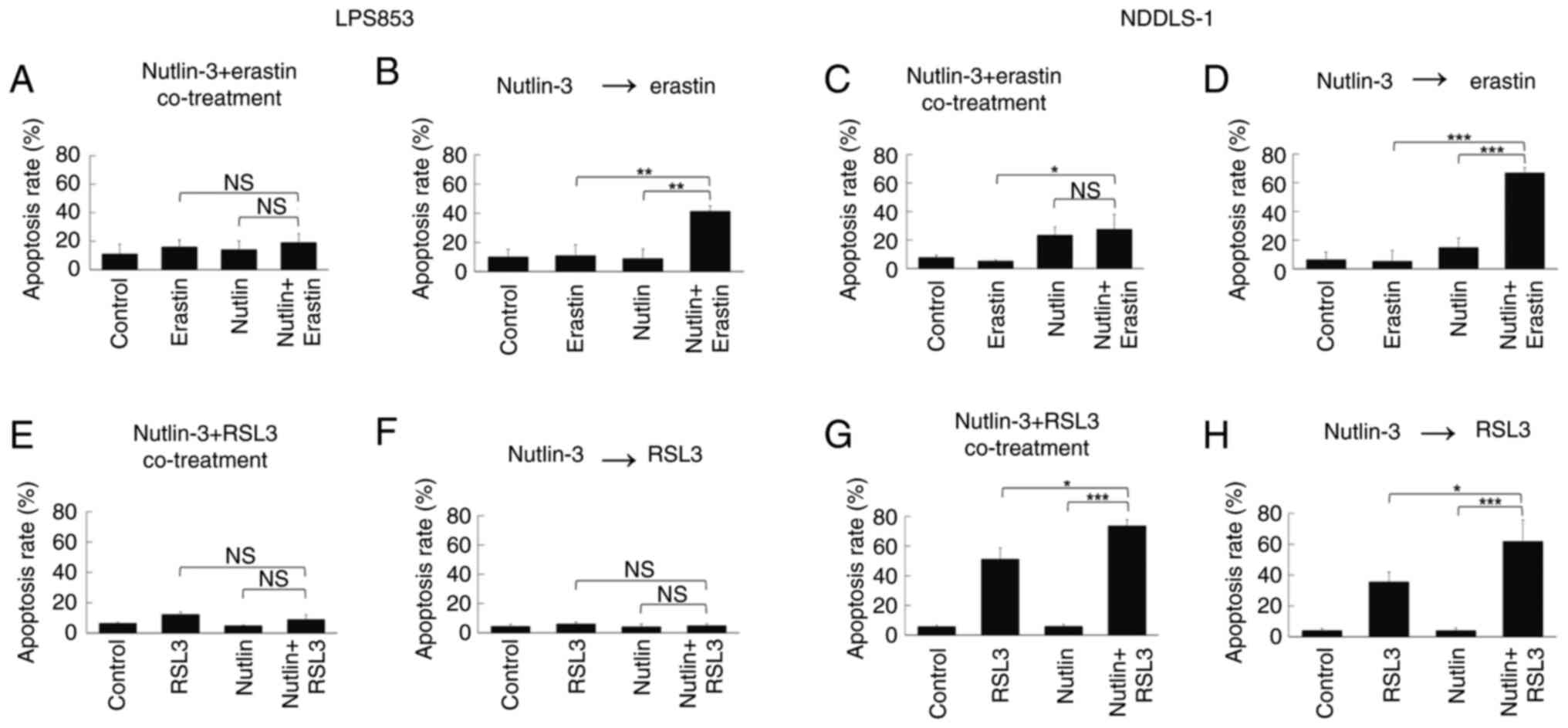
Treatment sequence is crucial for the
synergy of nutlin-3 with erastin but not RSL3 in ferroptosis
induction and cytotoxicity
The potential synergistic effects of combining
nutlin-3 with erastin or RSL3 were evaluated. First, their effects
on lipid peroxidation were tested. Co-treatment with nutlin-3 and
erastin (Figs. 5A and C, S4A and C) did not significantly alter the
extent of lipid peroxidation compared with that induced by erastin
alone in either DDLPS cell line. However, pre-treatment with
nutlin-3 for 24 h significantly increased the lipid
peroxidation-inducing effects of erastin in both DDLPS cell lines
(Figs. 5B and D, S4B and D). By contrast, whether nutlin-3
and RSL3 were co-administered or applied sequentially did not
clearly impact the lipid peroxidation-inducing effects in either
DDLPS cell line (Figs. 5E-H and
S4E-H).
The synergistic cytotoxic effects of nutlin-3
combined with erastin or RSL3 were evaluated using the CCK-8 assay.
Co-treatment with nutlin-3 and erastin did not exhibit a
significant synergy in cytotoxicity (Fig. 6A and C). However, a 24-h
pre-treatment with nutlin-3 followed by treatment with erastin
synergistically induced cytotoxicity in both DDLPS cell lines
(Fig. 6B and D). For nutlin-3
combined with RSL3, the treatment sequence affected the synergy of
the cytotoxicity in LPS853 cells (Fig.
6E and F) but not in NDDSL-1 cells (Fig. 6G and H). Similarly, in the apoptosis
assay, the apoptosis-inducing effects of nutlin-3 and erastin
varied according to the treatment sequence (Figs. 7A-D, S5A and B, S6A and B), whereas those of nutlin-3 and
RSL3 did not (Figs. 7E-H, S5C and D, S6C and D). These findings indicate that
nutlin-3 pre-treatment significantly enhances the
ferroptosis-inducing and cytotoxic effects of erastin but not those
of RSL3.
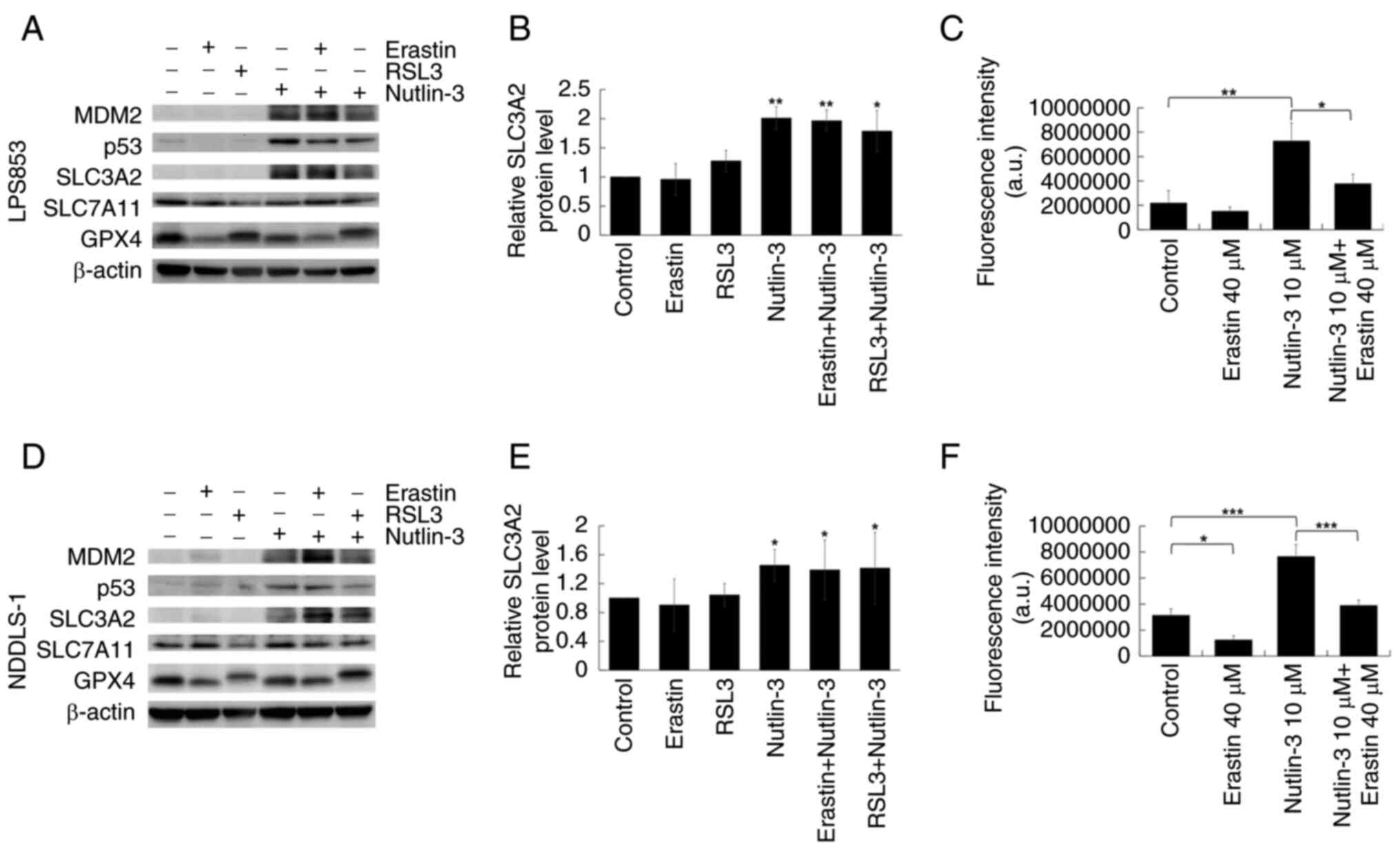
Nutlin-3-induced upregulation of
SLC3A2 and cystine uptake in DDLPS cell lines is suppressed by
erastin
As shown in Fig. 8A and
D, nutlin-3 treatment increased the expression of MDM2 and
TP53. This is consistent with previous studies which have shown
that nutlin-3 disrupts the interaction between MDM2 and TP53,
prevents their ubiquitination and leads to increased expression of
both proteins (38–40). Nutlin-3 treatment also increased the
expression of SLC3A2 (Fig. 8B and
E), while its effects on SLC7A11 and GPX4 expression were
inconsistent between the two cell lines. In the cystine uptake
assay, nutlin-3 treatment significantly increased the uptake of a
selenocystine fluorescent probe, and this effect was significantly
suppressed by erastin (Fig. 8C and
F). This suggests that nutlin-3 treatment increased the
expression of SLC3A2 in DDLPS, leading to enhanced cystine uptake
and greater sensitivity to erastin. This may explain why nutlin-3
pre-treatment synergistically promoted the cytotoxic effects of
erastin but not those of RSL3.
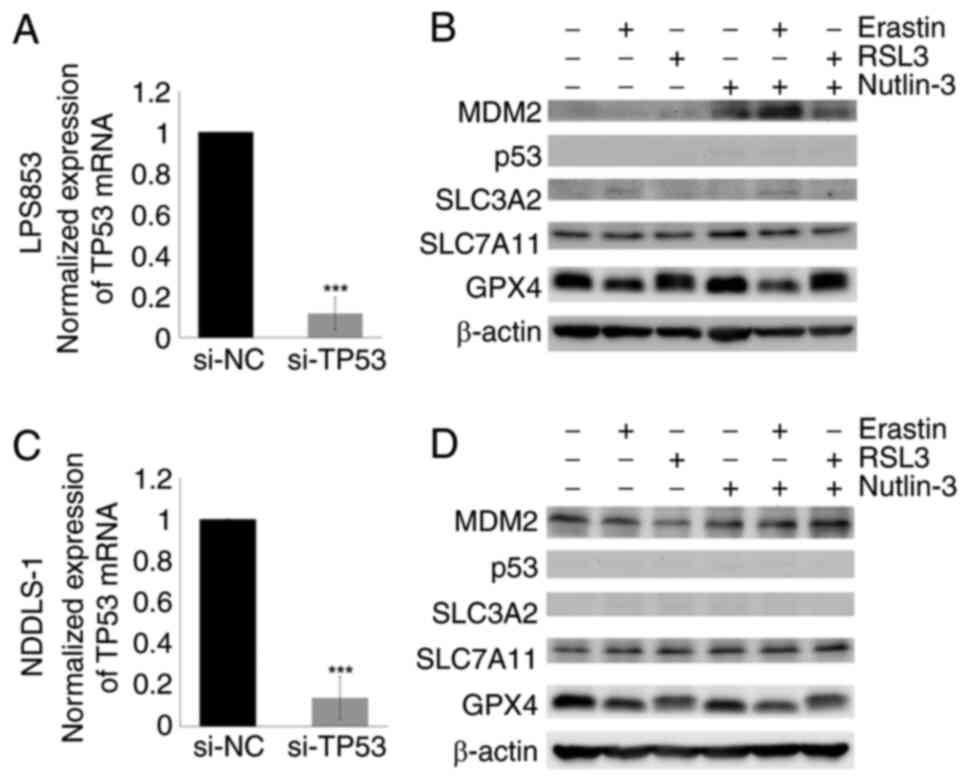
Nutlin-3-induced SLC3A2 upregulation
is abolished by TP53 KD
The mechanism by which nutlin-3 induces SLC3A2
upregulation was investigated. Given the role of TP53 in the
regulation of SLC3A2 expression, TP53 KD experiments were
performed. As shown in Fig. 9, the
nutlin-3-induced SLC3A2 upregulation previously observed in the
untransfected DDLPS cell lines was abolished by TP53 KD. This
result highlights the critical role of TP53 in nutlin-3-induced
SLC3A2 upregulation.
Combination of nutlin-3 with erastin
or RSL3 suppresses the mTOR pathway
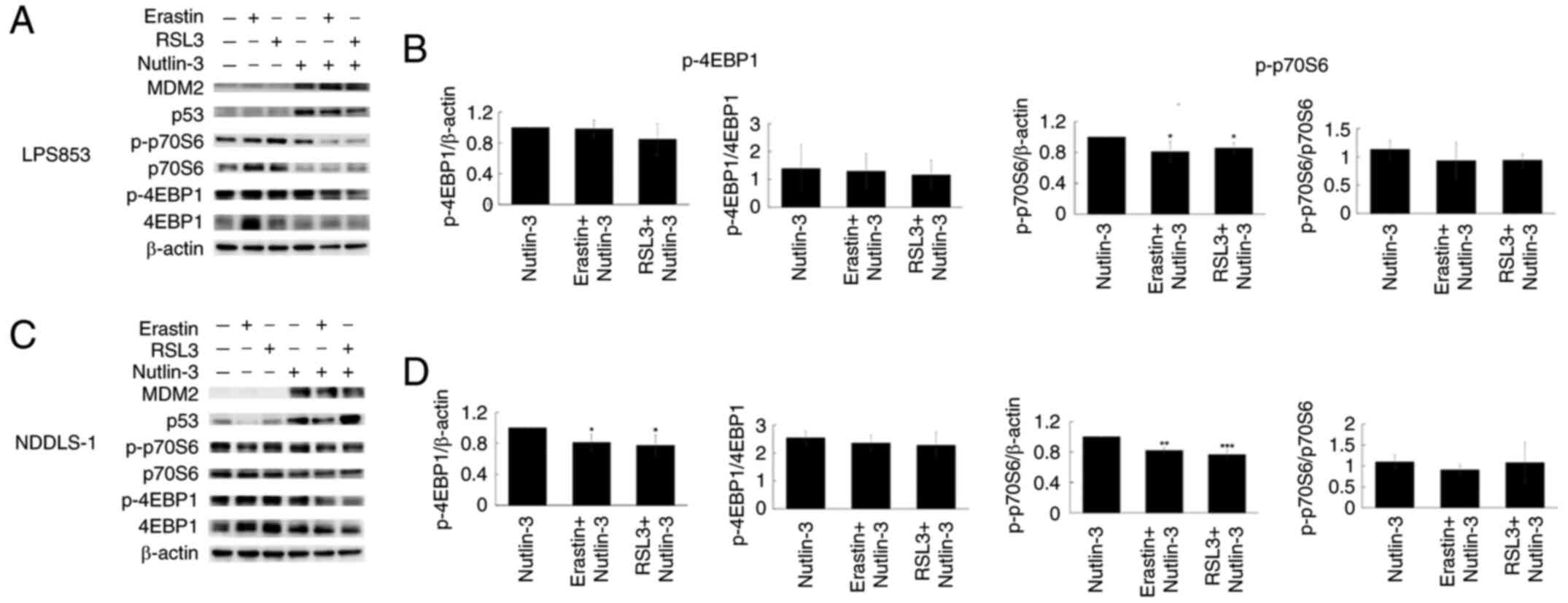
Notably, the present study found that the
combination of nutlin-3 with either erastin or RSL3 can increase
apoptosis. The mTOR pathway has been reported to play a role in the
regulation of ferroptosis (41–43).
In addition, inhibition of mTOR pathway is known to induce
apoptosis (44–46). Therefore, the involvement of this
pathway was assessed via the evaluation of the mTOR downstream
effectors eukaryotic translation initiation factor 4E binding
protein 1 (4EBP1) and p70 ribosomal protein S6 kinase (p70S6) in
DDLPS cells using western blotting. As shown in Fig. 10, the combination of nutlin-3 with
erastin or RSL3 suppressed the absolute p-4EBP1 levels in NDDLS-1
cells and p-p70S6 levels in both cell lines. However, these
treatment combinations had no significant effect on the
p-4EBP1/4EBP1 and p-p70S6/p70S6 ratios. This suppression of the
mTOR pathway may contribute to the apoptosis-inducing effects
observed with these combinations, but more studies are needed for
clarity.
Discussion
In the present study, bioinformatics analysis
identified that several ferroptosis-related genes are
differentially expressed in DDLPS. Additionally, in vitro
experiments demonstrated that two DDLPS cell lines were sensitive
to ferroptosis-inducing agents, particularly erastin and RSL3.
Furthermore, treatment of the cells with nutlin-3, an MDM2
inhibitor, followed by erastin, resulted in increased
ferroptosis-inducing and cytotoxic effects. Nutlin-3 also
upregulated the expression of SLC3A2 in the DDLPS cell lines, which
increased cystine uptake, and erastin attenuated these effects.
TP53 KD diminished the effect of nutlin-3 on SLC3A2, indicating
that TP53 contributes to SLC3A2 upregulation. Combining nutlin-3
with either of these ferroptosis-inducing agents reduced the
absolute p-4EBP1 levels in NDDLS-1 cells and p-p70S6 levels in both
cell lines, without significantly affecting the p-4EBP1/4EBP1 and
p-p70S6/p70S6 ratios. This suggests a potential role of the mTOR
pathway in the pro-apoptotic effect of these combinations, which
warrants further investigation.
MDM2 and CDK4 are oncogenes located in the 12q13-15
amplified chromosomal regions in both WDLPS and DDLPS (47). CDK4 phosphorylates Rb, promoting
cell cycle progression (48), while
MDM2 suppresses the function of TP53 by downregulating its
expression, exporting it from the nucleus to the cytoplasm, and
cooperating with MDM4 to induce TP53 polyubiquitination and
subsequent degradation (20–24).
CDK4/6 inhibitors have shown efficacy in hormone receptor-positive
breast cancer (49–51), and several MDM2 inhibitors have been
developed, with some advancing to clinical trials (52–58).
However, CDK4/6 inhibitors (59,60)
and MDM2 inhibitors (52–58) have demonstrated only limited
activity in WDLPS/DDLPS.
Ferroptosis, a form of necrotic cell death
characterized by the oxidative modification of phospholipid
membranes through an iron-dependent mechanism (9), has been identified as a potential
mechanism of action for various anticancer treatments across
multiple cancers (61–65), including sarcomas (66). MDM2, a key oncogene in DDLPS
(19), negatively regulates TP53
(20), which itself is a critical
regulator of ferroptosis (16–18).
Therefore, the investigation of ferroptosis regulation in DDLPS may
lead to the identification of new therapeutic strategies for this
deadly disease.
GPX4, SLC7A11 and SLC3A2 are three key regulators of
ferroptosis resistance. GPX4 dependency has been identified as a
unique characteristic of therapy-resistant cancer (67). In the present study, the
bioinformatics analysis of publicly available revealed that the
expression level of GPX4 in DDLPS is significantly lower compared
with that in adipose tissue and WDLPS. In vitro experiments
revealed that DDLPS cells are highly sensitive to RSL3 and erastin,
indicating a susceptibility to ferroptosis. Conversely, SLC7A11 and
SLC3A2, two components of xCT, were found to be more highly
expressed in LPS than in benign tissue. The upregulation of SLC7A11
may be partially due to the downregulation of TP53 by MDM2 in WDLPS
and DDLPS. The mechanism by which TP53 mediates SLC3A2 expression
is complex; previous studies have shown that SLC3A2 is upregulated
by mutant, but not wild-type, TP53 (68,69).
Therefore, further exploration of the regulatory mechanisms of
ferroptosis in DDLPS is warranted.
Given that MDM2 is a well-known oncogene in DDLPS,
the potential synergistic effect of nutlin-3, a MDM2 inhibitor,
with erastin and RSL3 was explored. The treatment sequence was
identified as a critical determinant of the efficacy of erastin.
Co-treatment with nutlin-3 did not markedly affect the
lipid-peroxidation-inducing and cytotoxic effects of erastin.
However, treatment with nutlin-3 for 24 h prior to treatment with
erastin significantly augmented the induction of lipid-peroxidation
and cytotoxicity of erastin in both DDLPS cell lines. This effect
of nutlin-3 was not observed with RSL3. These findings suggest that
nutlin-3 may sensitize DDLPS cell lines to erastin, potentially by
modulating the expression of ferroptosis-related genes underlying
its effects.
Immunoblotting revealed that nutlin-3 upregulated
SLC3A2 expression in both DDLPS cell lines. Subsequently, nutlin-3
was shown to increase cystine uptake in an erastin-suppressible
manner, which may be attributed to SLC3A2 upregulation.
Additionally, the nutlin-3-induced expression of SLC3A2 was
abolished by TP53 KD. These findings suggest that nutlin-3
treatment induces TP53-mediated SLC3A2 upregulation, leading to
increased cystine uptake and erastin sensitivity in DDLPS.
Notably, SLC7A11 expression levels remained largely
unchanged after nutlin-3 treatment. TP53, a key suppressor of
SLC7A11, is expected to decrease the levels of SLC7A11 when TP53
function is restored (16), as
nutlin-3 dissociates MDM2 from TP53, thereby preventing the
degradation of TP53 by ubiquitination (38–40).
However, as MDM2 is also released and is known to upregulate
SLC7A11 (70,71), the enhancing effect of MDM2 on
SLC7A11 expression may be counterbalanced by the suppressive effect
of restored TP53.
The present study found that treatment with nutlin-3
prior to treatment with erastin or RSL3 increased lipid
peroxidation, a hallmark of ferroptosis, and apoptosis. Nutlin-3
alone is known to induce apoptosis via cell cycle arrest or the
restoration of TP53 function (72–74).
mTOR is crucial in different types of programmed cell death
(75), including apoptosis
(44–46) and ferroptosis (41–43).
In the present study, combining nutlin-3 with either erastin or
RSL3 reduced the absolute levels of phosphorylated proteins in the
mTOR pathway but not the phosphorylated/total protein ratios,
suggesting the potential role of the mTOR pathway in the apoptosis
induction of these combinations. Further investigations are needed
to confirm this finding.
The present bioinformatics analysis revealed that
HMGCR expression is upregulated in LPS. However, lovastatin and
simvastatin, two inhibitors of HMGCR, showed minimal effects on
lipid peroxidation and only modest cytotoxicity against the two
DDLPS cell lines, suggesting that the HMGCR pathway may not play a
major role in the regulation of ferroptosis in DDLPS. DPP4
expression was also found to be upregulated in LPS, likely due to
TP53 downregulation in WDLPS and DDLPS (18). DPP4 is an exoprotease that is
expressed by different cell types and plays a role in plasma
membrane-associated lipid peroxidation (18). However, previous studies have shown
that its role in cancer is primarily restricted to the tumor
microenvironment (76). Therefore,
these two genes were not pursued further in the present study.
In summary, the present study showed that several
ferroptosis-related genes are differentially expressed in DDLPS.
Additionally, in vitro experiments demonstrated that DDLPS
cell lines are highly sensitive to the lipid peroxidation-inducing
and cytotoxic effects of erastin and RSL3. Furthermore,
pre-treatment with nutlin-3 significantly increased the efficacy of
erastin in DDLPS cell lines, potentially via the TP53-mediated
upregulation of SLC3A2, resulting in increased cystine uptake and
vulnerability to ferroptosis. Suppression of the mTOR pathway may
contribute to the apoptosis-inducing effects observed with the
nutlin-3-containing combinations. This mechanism may contribute to
resistance to MDM2 inhibitors in DDLPS. These findings provide
valuable insights into the potential development of novel
treatments for DDLPS. In future studies, it is planned to examine
the combined effect of MDM2 inhibitors and ferroptosis-inducing
agents in greater detail, and to explore the underlying mechanisms
in an in vivo model.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was jointly supported by grants from the National
Science and Technology Council (grant nos. MOST 110-2314-B-075-070
and MOST 111-2314-B-075-022), and Taipei Veterans General Hospital
(grant nos. V110D56-002-MY2-1, V110D56-002-MY2-2, V110C-208,
V112C-093 and V113C-116). This study was also supported by the
Taiwan Clinical Oncology Research Foundation, Melissa Lee Cancer
Foundation (grant no. MLCF_V114_A11403), and the Chong Hin Loon
Memorial Cancer and Biotherapy Research Center of National Yang
Ming Chiao Tung University.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CCY, PCHC, TCC and JAF were responsible for the
design and conception of the study. MHY, CHY and YCL were
responsible for data acquisition. SCC, WCW, PKW, CMC and JYW were
responsible for data interpretation. CCY was responsible for
writing the original draft of the manuscript. PCHC and JAF reviewed
and edited the manuscript. CHY and YCL edited the figures. TCC and
MHY confirm the authenticity of all the raw data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
This study was deemed as being exempt from ethical
review by the Institutional Review Board (IRB) of Taipei Veterans
General Hospital (IRB no. 2021-07-003AE).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence (AI) tools were used to improve the readability and
language of the manuscript, and subsequently, the authors revised
and edited the content produced by the AI tools as necessary,
taking full responsibility for the ultimate content of the
manuscript.
References
|
1
|
Hung GY, Yen CC, Horng JL, Liu CY, Chen
WM, Chen TH and Liu CL: Incidences of primary soft tissue sarcoma
diagnosed on extremities and trunk wall: A population-based study
in taiwan. Medicine (Baltimore). 94:e16962015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hung GY, Horng JL, Chen PC, Lin LY, Chen
JY, Chuang PH, Chao TC and Yen CC: Incidence of soft tissue sarcoma
in Taiwan: A nationwide population-based study (2007–2013). Cancer
Epidemiol. 60:185–192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ducimetiere F, Lurkin A, Ranchere-Vince D,
Decouvelaere AV, Peoc'h M, Istier L, Chalabreysse P, Muller C,
Alberti L, Bringuier PP, et al: Incidence of sarcoma histotypes and
molecular subtypes in a prospective epidemiological study with
central pathology review and molecular testing. PLoS One.
6:e202942011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee ATJ, Thway K, Huang PH and Jones RL:
Clinical and molecular spectrum of liposarcoma. J Clin Oncol.
36:151–159. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seddon B, Strauss SJ, Whelan J, Leahy M,
Woll PJ, Cowie F, Rothermundt C, Wood Z, Benson C, Ali N, et al:
Gemcitabine and docetaxel versus doxorubicin as first-line
treatment in previously untreated advanced unresectable or
metastatic soft-tissue sarcomas (GeDDiS): A randomised controlled
phase 3 trial. Lancet Oncol. 18:1397–1410. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schoffski P, Chawla S, Maki RG, Italiano
A, Gelderblom H, Choy E, Grignani G, Camargo V, Bauer S, Rha SY, et
al: Eribulin versus dacarbazine in previously treated patients with
advanced liposarcoma or leiomyosarcoma: A randomised, open-label,
multicentre, phase 3 trial. Lancet. 387:1629–1637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Demetri GD, Schoffski P, Grignani G, Blay
JY, Maki RG, Van Tine BA, Alcindor T, Jones RL, D'Adamo DR, Guo M
and Chawla S: Activity of eribulin in patients with advanced
liposarcoma demonstrated in a subgroup analysis from a randomized
phase III study of eribulin versus dacarbazine. J Clin Oncol.
35:3433–3439. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Demetri GD, von Mehren M, Jones RL,
Hensley ML, Schuetze SM, Staddon A, Milhem M, Elias A, Ganjoo K,
Tawbi H, et al: Efficacy and safety of trabectedin or dacarbazine
for metastatic liposarcoma or leiomyosarcoma after failure of
conventional chemotherapy: Results of a phase III randomized
multicenter clinical trial. J Clin Oncol. 34:786–793. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Conrad M, Angeli JP, Vandenabeele P and
Stockwell BR: Regulated necrosis: Disease relevance and therapeutic
opportunities. Nat Rev Drug Discov. 15:348–366. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie Y, Hou W, Song X, Yu Y, Huang J, Sun
X, Kang R and Tang D: Ferroptosis: Process and function. Cell Death
Differ. 23:369–379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ingold I, Berndt C, Schmitt S, Doll S,
Poschmann G, Buday K, Roveri A, Peng X, Porto Freitas F, Seibt T,
et al: Selenium utilization by GPX4 is required to prevent
hydroperoxide-induced ferroptosis. Cell. 172:409–422. e4212018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheok CF, Verma CS, Baselga J and Lane DP:
Translating p53 into the clinic. Nat Rev Clin Oncol. 8:25–37. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang L, Kon N, Li T, Wang SJ, Su T,
Hibshoosh H, Baer R and Gu W: Ferroptosis as a p53-mediated
activity during tumour suppression. Nature. 520:57–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moon SH, Huang CH, Houlihan SL, Regunath
K, Freed-Pastor WA, Morris JP, Tschaharganeh DF, Kastenhuber ER,
Barsotti AM, Culp-Hill R, et al: p53 represses the mevalonate
pathway to mediate tumor suppression. Cell. 176:564–580. e5192019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J,
Zhong M, Yuan H, Zhang L, Billiar TR, et al: The tumor suppressor
p53 limits ferroptosis by blocking DPP4 activity. Cell Rep.
20:1692–1704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dei Tos AP, Doglioni C, Piccinin S, Sciot
R, Furlanetto A, Boiocchi M, Dal CP, Maestro R, Fletcher CD and
Tallini G: Coordinated expression and amplification of the MDM2,
CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol.
190:531–536. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karni-Schmidt O, Lokshin M and Prives C:
The roles of MDM2 and MDMX in cancer. Annu Rev Pathol. 11:617–644.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leslie PL and Zhang Y: MDM2 oligomers:
Antagonizers of the guardian of the genome. Oncogene. 35:6157–6165.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hock AK and Vousden KH: The role of
ubiquitin modification in the regulation of p53. Biochim Biophys
Acta. 1843:137–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
do Patrocinio AB, Rodrigues V and Guidi
Magalhaes L: P53: Stability from the ubiquitin-proteasome system
and specific 26S proteasome inhibitors. ACS Omega. 7:3836–3843.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bang S, Kaur S and Kurokawa M: Regulation
of the p53 family proteins by the ubiquitin proteasomal pathway.
Int J Mol Sci. 21:2612019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Venkatesh D, O'Brien NA, Zandkarimi F,
Tong DR, Stokes ME, Dunn DE, Kengmana ES, Aron AT, Klein AM, Csuka
JM, et al: MDM2 and MDMX promote ferroptosis by PPARalpha-mediated
lipid remodeling. Genes Dev. 34:526–543. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yen CC, Chen LT, Li CF, Chen SC, Chua WY,
Lin YC, Yen CH, Chen YC, Yang MH, Chao Y and Fletcher JA:
Identification of phenothiazine as an ETV1targeting agent in
gastrointestinal stromal tumors using the connectivity map. Int J
Oncol. 55:536–546. 2019.PubMed/NCBI
|
|
27
|
Chibon F, Lagarde P, Salas S, Perot G,
Brouste V, Tirode F, Lucchesi C, de Reynies A, Kauffmann A, Bui B,
et al: Validated prediction of clinical outcome in sarcomas and
multiple types of cancer on the basis of a gene expression
signature related to genome complexity. Nat Med. 16:781–787. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gobble RM, Qin LX, Brill ER, Angeles CV,
Ugras S, O'Connor RB, Moraco NH, Decarolis PL, Antonescu C and
Singer S: Expression profiling of liposarcoma yields a multigene
predictor of patient outcome and identifies genes that contribute
to liposarcomagenesis. Cancer Res. 71:2697–2705. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doyle KR, Mitchell MA, Roberts CL, James
S, Johnson JE, Zhou Y, von Mehren M, Lev D, Kipling D and Broccoli
D: Validating a gene expression signature proposed to differentiate
liposarcomas that use different telomere maintenance mechanisms.
Oncogene. 31:265–266; author reply 267–268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshino J, Conte C, Fontana L,
Mittendorfer B, Imai S, Schechtman KB, Gu C, Kunz I, Rossi Fanelli
F, Patterson BW and Klein S: Resveratrol supplementation does not
improve metabolic function in nonobese women with normal glucose
tolerance. Cell Metab. 16:658–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nookaew I, Svensson PA, Jacobson P, Jernas
M, Taube M, Larsson I, Andersson-Assarsson JC, Sjostrom L, Froguel
P, Walley A, et al: Adipose tissue resting energy expenditure and
expression of genes involved in mitochondrial function are higher
in women than in men. J Clin Endocrinol Metab. 98:E370–E378. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li C and Wong WH: Model-based analysis of
oligonucleotide arrays: Model validation, design issues and
standard error application. Genome Biol. 2:RESEARCH00322001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li C and Wong WH: Model-based analysis of
oligonucleotide arrays: Expression index computation and outlier
detection. Proc Natl Acad Sci USA. 98:31–36. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yoshida Y, Shimakawa S, Itoh N and Niki E:
Action of DCFH and BODIPY as a probe for radical oxidation in
hydrophilic and lipophilic domain. Free Radic Res. 37:861–872.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chou TC and Talalay P: Quantitative
analysis of dose-effect relationships: The combined effects of
multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 22:27–55.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chou YS, Yen CC, Chen WM, Lin YC, Wen YS,
Ke WT, Wang JY, Liu CY, Yang MH, Chen TH and Liu CL: Cytotoxic
mechanism of PLK1 inhibitor GSK461364 against osteosarcoma: Mitotic
arrest, apoptosis, cellular senescence, and synergistic effect with
paclitaxel. Int J Oncol. 48:1187–1194. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vassilev LT, Vu BT, Graves B, Carvajal D,
Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et
al: In vivo activation of the p53 pathway by small-molecule
antagonists of MDM2. Science. 303:844–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tovar C, Rosinski J, Filipovic Z, Higgins
B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al:
Small-molecule MDM2 antagonists reveal aberrant p53 signaling in
cancer: Implications for therapy. Proc NatI Acad Sci USA.
103:1888–1893. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Henze J, Muhlenberg T, Simon S, Grabellus
F, Rubin B, Taeger G, Schuler M, Treckmann J, Debiec-Rychter M,
Taguchi T, et al: p53 modulation as a therapeutic strategy in
gastrointestinal stromal tumors. PLoS One. 7:e377762012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi Z, Naowarojna N, Pan Z and Zou Y:
Multifaceted mechanisms mediating cystine starvation-induced
ferroptosis. Nat Commun. 12:47922021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Z, Zong H, Liu W, Lin W, Sun A, Ding
Z, Chen X, Wan X, Liu Y, Hu Z, et al: Augmented ERO1alpha upon
mTORC1 activation induces ferroptosis resistance and tumor
progression via upregulation of SLC7A11. J Exp Clin Cancer Res.
43:1122024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yin J, Chen J, Hong JH, Huang Y, Xiao R,
Liu S, Deng P, Sun Y, Chai KXY, Zeng X, et al: 4EBP1-mediated
SLC7A11 protein synthesis restrains ferroptosis triggered by MEK
inhibitors in advanced ovarian cancer. JCI Insight. 9:e1778572024.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Han J, Wang L, Lv H, Liu J, Dong Y, Shi L
and Ji Q: EphA2 inhibits SRA01/04 cells apoptosis by suppressing
autophagy via activating PI3K/Akt/mTOR pathway. Arch Biochem
Biophys. 711:1090242021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang J, Pi C and Wang G: Inhibition of
PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy
in hepatocellular carcinoma cells. Biomed Pharmacother.
103:699–707. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li W, Li D, Ma Q, Chen Y, Hu Z, Bai Y and
Xie L: Targeted inhibition of mTOR by BML-275 induces
mitochondrial-mediated apoptosis and autophagy in prostate cancer.
Eur J Pharmacol. 957:1760352023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fletcher CD: The evolving classification
of soft tissue tumours-An update based on the new 2013 WHO
classification. Histopathology. 64:2–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sherr CJ, Beach D and Shapiro GI:
Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov.
6:353–367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Finn RS, Martin M, Rugo HS, Jones S, Im
SA, Gelmon K, Harbeck N, Lipatov ON, Walshe JM, Moulder S, et al:
Palbociclib and letrozole in advanced breast cancer. N Engl J Med.
375:1925–1936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hortobagyi GN, Stemmer SM, Burris HA, Yap
YS, Sonke GS, Paluch-Shimon S, Campone M, Petrakova K, Blackwell
KL, Winer EP, et al: Updated results from MONALEESA-2, a phase III
trial of first-line ribociclib plus letrozole versus placebo plus
letrozole in hormone receptor-positive, HER2-negative advanced
breast cancer. Ann Oncol. 29:1541–1547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Goetz MP, Toi M, Campone M, Sohn J,
Paluch-Shimon S, Huober J, Park IH, Tredan O, Chen SC, Manso L, et
al: MONARCH 3: Abemaciclib as initial therapy for advanced breast
cancer. J Clin Oncol. 35:3638–3646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang S, Zhao Y, Aguilar A, Bernard D and
Yang CY: Targeting the MDM2-p53 protein-protein interaction for new
cancer therapy: Progress and challenges. Cold Spring Harb Perspect
Med. 7:a0262452017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kocik J, Machula M, Wisniewska A, Surmiak
E, Holak TA and Skalniak L: Helping the released guardian: Drug
combinations for supporting the anticancer activity of HDM2 (MDM2)
antagonists. Cancers (Basel). 11:10142019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fang Y, Liao G and Yu B: Small-molecule
MDM2/X inhibitors and PROTAC degraders for cancer therapy: Advances
and perspectives. Acta Pharm Sin B. 10:1253–1278. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhao Y, Aguilar A, Bernard D and Wang S:
Small-molecule inhibitors of the MDM2-p53 protein-protein
interaction (MDM2 Inhibitors) in clinical trials for cancer
treatment. J Med Chem. 58:1038–1052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
LoRusso P, Yamamoto N, Patel MR, Laurie
SA, Bauer TM, Geng J, Davenport T, Teufel M, Li J, Lahmar M and
Gounder MM: The MDM2-p53 Antagonist brigimadlin (BI 907828) in
patients with advanced or metastatic solid tumors: Results of a
phase Ia, first-in-human, dose-escalation study. Cancer Discov.
13:1802–1813. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ray-Coquard I, Blay JY, Italiano A, Le
Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, et al:
Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients
with MDM2-amplified, well-differentiated or dedifferentiated
liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol.
13:1133–1140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gounder MM, Bauer TM, Schwartz GK, Weise
AM, LoRusso P, Kumar P, Tao B, Hong Y, Patel P, Lu Y, et al: A
first-in-human Phase I study of milademetan, an MDM2 inhibitor, in
patients with advanced liposarcoma, solid tumors, or lymphomas. J
Clin Oncol. 41:1714–1724. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Assi T, Kattan J, Rassy E, Nassereddine H,
Farhat F, Honore C, Le Cesne A, Adam J and Mir O: Targeting CDK4
(cyclin-dependent kinase) amplification in liposarcoma: A
comprehensive review. Crit Rev Oncol Hematol. 153:1030292020.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dickson MA, Schwartz GK, Keohan ML,
D'Angelo SP, Gounder MM, Chi P, Antonescu CR, Landa J, Qin LX,
Crago AM, et al: Progression-free survival among patients with
well-differentiated or dedifferentiated liposarcoma treated with
CDK4 inhibitor palbociclib: A phase 2 Clinical Trial. JAMA Oncol.
2:937–940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C,
Dai X, Li Z and Wu G: Ferroptosis: A novel anti-tumor action for
cisplatin. Cancer Res Treat. 50:445–460. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ma S, Henson ES, Chen Y and Gibson SB:
Ferroptosis is induced following siramesine and lapatinib treatment
of breast cancer cells. Cell Death Dis. 7:e23072016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Trujillo-Alonso V, Pratt EC, Zong H,
Lara-Martinez A, Kaittanis C, Rabie MO, Longo V, Becker MW, Roboz
GJ, Grimm J and Guzman ML: FDA-approved ferumoxytol displays
anti-leukaemia efficacy against cells with low ferroportin levels.
Nat Nanotechnol. 14:616–622. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yamaguchi Y, Kasukabe T and Kumakura S:
Piperlongumine rapidly induces the death of human pancreatic cancer
cells mainly through the induction of ferroptosis. Int J Oncol.
52:1011–1022. 2018.PubMed/NCBI
|
|
65
|
Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R
and Tang D: Activation of the p62-Keap1-NRF2 pathway protects
against ferroptosis in hepatocellular carcinoma cells. Hepatology.
63:173–184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Brashears CB, Prudner BC, Rathore R,
Caldwell KE, Dehner CA, Buchanan JL, Lange SES, Poulin N, Sehn JK,
Roszik J, et al: Malic enzyme 1 absence in synovial sarcoma shifts
antioxidant system dependence and increases sensitivity to
ferroptosis induction with ACXT-3102. Clin Cancer Res.
28:3573–3589. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Viswanathan VS, Ryan MJ, Dhruv HD, Gill S,
Eichhoff OM, Seashore-Ludlow B, Kaffenberger SD, Eaton JK, Shimada
K, Aguirre AJ, et al: Dependency of a therapy-resistant state of
cancer cells on a lipid peroxidase pathway. Nature. 547:453–457.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Mello SS, Valente LJ, Raj N, Seoane JA,
Flowers BM, McClendon J, Bieging-Rolett KT, Lee J, Ivanochko D,
Kozak MM, et al: A p53 super-tumor suppressor reveals a tumor
suppressive p53-ptpn14-yap axis in pancreatic cancer. Cancer Cell.
32:460–473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tombari C, Zannini A, Bertolio R, Pedretti
S, Audano M, Triboli L, Cancila V, Vacca D, Caputo M, Donzelli S,
et al: Mutant p53 sustains serine-glycine synthesis and essential
amino acids intake promoting breast cancer growth. Nat Commun.
14:67772023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fujihara KM, Corrales Benitez M, Cabalag
CS, Zhang BZ, Ko HS, Liu DS, Simpson KJ, Haupt Y, Lipton L, Haupt
S, et al: SLC7A11 Is a superior determinant of APR-246
(Eprenetapopt) response than TP53 mutation status. Mol Cancer Ther.
20:1858–1867. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Riscal R, Schrepfer E, Arena G, Cisse MY,
Bellvert F, Heuillet M, Rambow F, Bonneil E, Sabourdy F, Vincent C,
et al: Chromatin-bound MDM2 regulates serine metabolism and redox
homeostasis independently of p53. Mol Cell. 62:890–902. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Villalonga-Planells R, Coll-Mulet L,
Martinez-Soler F, Castano E, Acebes JJ, Gimenez-Bonafe P, Gil J and
Tortosa A: Activation of p53 by nutlin-3a induces apoptosis and
cellular senescence in human glioblastoma multiforme. PLoS One.
6:e185882011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Miyachi M, Kakazu N, Yagyu S, Katsumi Y,
Tsubai-Shimizu S, Kikuchi K, Tsuchiya K, Iehara T and Hosoi H:
Restoration of p53 pathway by nutlin-3 induces cell cycle arrest
and apoptosis in human rhabdomyosarcoma cells. Clin Cancer Res.
15:4077–4084. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Manfe V, Biskup E, Johansen P, Kamstrup
MR, Krejsgaard TF, Morling N, Wulf HC and Gniadecki R: MDM2
inhibitor nutlin-3a induces apoptosis and senescence in cutaneous
T-cell lymphoma: role of p53. J Invest Dermatol. 132:1487–1496.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xie Y, Lei X, Zhao G, Guo R and Cui N:
mTOR in programmed cell death and its therapeutic implications.
Cytokine Growth Factor Rev. 71–72. 66–81. 2023.PubMed/NCBI
|
|
76
|
Cordero OJ: CD26 and cancer. Cancers
(Basel). 14:51942022. View Article : Google Scholar : PubMed/NCBI
|