Introduction
Lung adenocarcinoma (LUAD) constitutes the
predominant subtype of lung cancer (LC), comprising ~40% of all LC
cases (1,2). The treatment of LUAD is tailored to
the stage and severity of the disease, typically involving a
combination of approaches such as surgery, radiotherapy,
chemotherapy, immunotherapy and targeted therapy (3). Despite these diverse treatment
strategies, the 5-year overall survival (OS) rate of patients with
LUAD remains low, at ~15%, and few patients achieve complete
remission (4,5). Accordingly, exploring the molecular
basis of LUAD and seeking novel biomolecules for therapeutic
purposes is a priority.
RNA helicases are essential regulators of several
aspects of RNA metabolism, including transcription, RNA splicing,
transport, storage, degradation, ribosome biogenesis and
translation (6–8). By modulating the expression levels of
oncogenes, RNA helicases contribute to tumorigenesis and
advancement (9). Additionally, they
are implicated in several pathological processes, such as
neurological disorders, viral infections and aging (10). Among these, the DEAD-box (DDX)
family stands out as a key group of human RNA helicases (11), with increasing evidence supporting
their involvement in human tumorigenesis. Specifically, DDX46,
located on chromosome 5q31.1, is a member of the DDX family. During
pre-mRNA splicing and in the assembly of the spliceosome, DDX46
functions as a component of the 17S U2 small nuclear
ribonucleoprotein complex (12).
Moreover, studies using zebrafish models have demonstrated that
DDX46 is crucial for the multilineage differentiation of
hematopoietic stem cells, as well as for the development of the
digestive organs and the brain (9).
Previous research has also reported that DDX46 is notably
overexpressed in esophageal squamous cell carcinoma, where its
silencing inhibits the Akt/IκBα/NF-κB signaling pathway, thereby
inducing apoptosis (13). In
osteosarcoma, DDX46 knockdown reduces tumor cell proliferation,
migration and invasion (14).
Furthermore, heightened expression of DDX46 has been reported in
gastric cancer, where it is activated via the Akt/GSK/β-catenin
pathway, promoting cancer cell proliferation and invasion (15). However, the relationship between
DDX46 expression and LUAD progression remains unclear, and further
investigation is needed to elucidate its potential role in
LUAD.
The present study aimed to systematically assess the
expression, pathological roles and the molecular basis of DDX46 in
LUAD. To achieve this, a synergistic approach was employed that
integrated bioinformatics analysis with experimental
techniques.
Materials and methods
Public databases
The gene expression and clinical data from 535 LUAD
samples (Table SI) and 59 normal
lung tissue samples (16) were
obtained from The Cancer Genome Atlas (TCGA) database (www.portal.gdc.cancer.gov/). Data processing was
performed using R (version 3.6.3, The R Foundation for Statistical
Computing; http://cran.r-project.org/src/base/R-3/R-3.6.3.tar.gz/)
and Strawberry Perl (version 5.40.0.1; http://strawberryperl.com/).
Tissue specimens
A total of 20 pairs of fresh LUAD samples with their
corresponding adjacent tissues, as well as 263 paraffin-embedded
LUAD tissue blocks, were collected from the Affiliated Hospital of
Nantong University (Nantong, China) between January 2012 and
December 2015. The inclusion criteria were as follows: i) Patients
pathologically diagnosed with LUAD; ii) no history of other
malignant tumors; iii) sufficient samples available from the first
visit for immunohistochemical analysis; iv) no prior anticancer
treatment before surgery. Patients who did not meet the
aforementioned criteria were not included in the present study.
Tissue microarrays were carefully prepared by integrating the
paraffin blocks with LUAD samples. The clinicopathological
characteristics of the patients from which the samples were
collected encompassed age, sex, smoking history, tumor
differentiation, clinical stage, tumor stage (T), lymph node
metastasis stage (N) and distant metastasis stage (M), along with
survival duration and outcome. Prior to surgery, all patients
provided written informed consent, confirming their clear
understanding of the risks and benefits associated with the
procedure. The study protocol and data collection were approved by
the Ethics Committee of the Affiliated Hospital of Nantong
University (approval no. 2022-L078).
Western blot analysis
Protein extraction was performed using RIPA buffer
(cat. no. P0013B; Beyotime Institute of Biotechnology), followed by
the measurement of total protein concentration in the samples. The
total protein concentration in the samples was measured using a
bicinchoninic acid protein assay kit (cat. no. P0010; Beyotime
Institute of Biotechnology), following the manufacturer's
instructions. This method ensures accurate quantification of
protein content before proceeding with western blot analysis. For
western blot analysis, equal amounts of total protein (20 µg/lane)
were mixed with appropriate volumes of 5X SDS-PAGE loading buffer
to ensure uniform loading volume across all lanes. Proteins were
separated by SDS-PAGE on 10% gels and were then transferred onto a
polyvinylidene difluoride (PVDF) membrane. The membrane was blocked
with 5% non-fat milk in Tris-buffered saline with 0.1% Tween-20 at
room temperature for 1 h to prevent non-specific binding. For
antibody incubation, the PVDF membrane was then incubated overnight
at 4°C with the following primary antibodies: Anti-DDX46 (1:1,000;
cat. no. ab72083; Abcam), anti-adenomatous polyposis coli (APC;
1:1,000, cat. no. ab40778; Abcam), anti-β-catenin (1:5,000; cat.
no. 51067-2-AP, Proteintech Group, Inc.) and anti-β-actin (1:4,000;
cat. no. 20536-1-AP; Proteintech Group, Inc.). Subsequently, the
membrane was incubated with a HRP-conjugated goat anti-rabbit IgG
secondary antibody (1:5,000; cat. no. SA00001-2; Proteintech Group,
Inc.) at room temperature for 1 h. Protein signals were visualized
using an enhanced chemiluminescence detection reagent (cat. no.
180-5001; Tanon Science and Technology Co., Ltd.). Densitometric
analysis was performed using ImageJ software (version 1.53;
National Institutes of Health).
Tissue chips immunohistochemical
staining and analysis
The. LUAD and adjacent tissue sections were fixed in
10% neutral buffered formalin at room temperature for 24 h,
embedded in paraffin and sectioned at a thickness of 4 µm. For
immunohistochemical staining and analysis of tissue chips, antigen
retrieval was performed using citrate buffer (pH 6.0) at 98°C for
20 min, followed by washing with phosphate-buffered saline (PBS)
and rehydration through a descending ethanol series (100, 95, 85
and 75%). Subsequently, the sections were incubated with 3%
hydrogen peroxide for 20 min to inhibit endogenous peroxidase
activity, followed by blocking with 5% normal rabbit serum (cat.
no. ab7487; Abcam) at room temperature for 30 min. The sections
were then incubated overnight at 4°C with anti-DDX46 antibodies
(1:200; cat. no. ab72083; Abcam), followed by incubation with an
HRP-conjugated goat anti-rabbit IgG secondary antibody (1:500
dilution; cat. no. ab6721; Abcam) at room temperature for 1 h to
enable signal detection. DDX46 staining was primarily localized in
the nucleus of the cells. Immunohistochemical staining was
evaluated and scored following the methodology described in our
previous study (17).
Cell culture
The present study employed the following cell lines:
BEAS-2B, PC-9, H1299, A549 and H1975, which were purchased from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. BEAS-2B and A549 cells were maintained in Dulbecco's
Modified Eagle Medium (cat. no. 11965092; Gibco; Thermo Fisher
Scientific, Inc.), whereas PC-9, H1299, and H1975 cells were
cultured in Roswell Park Memorial Institute (RPMI) 1640 medium
(cat. no. 11875093; Gibco; Thermo Fisher Scientific, Inc.) The
medium was supplemented with 10% fetal bovine serum (FBS; cat. no.
10099141; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (cat. no. 15140122; Gibco; Thermo Fisher
Scientific, Inc.) to prevent bacterial contamination. Cells were
maintained at 37°C in a humidified incubator with 5%
CO2.
Lentiviral transfection
Pre-packaged lentiviruses containing DDX46 short
hairpin RNA (shRNA) and negative control (NC) shRNA were purchased
from Shanghai GeneChem Co., Ltd. and were used directly for cell
infection according to the manufacturer's instructions. The shRNAs
were cloned into a GV248 plasmid backbone
(U6-MCS-Ubiquitin-EGFP-IRES-puromycin). The following sequences
were used: DDX46-shRNA#1,
5′-GATCCGCAGAAATCACCAGGCTCATACTCGAGTATGAGCCTGGTGATTTCTGCTTTTTG-3′
and
5′-AATTCAAAAAGCAGAAATCACCAGGCTCATACTCGAGTATGAGCCTGGTGATTTCTGCG-3′;
DDX46-shRNA#2,
5′-GATCCCCTGTGCTGGTCTATGATGTACTCGAGTACATCATAGACCAGCACAGGTTTTTG-3′
and
5′-AATTCAAAAACCTGTGCTGGTCTATGATGTACTCGAGTACATCATAGACCAGCACAGGG-3′;
and NC-shRNA, 5′-TTCTCCGAACGTGTCACGT-3′ and
5′-AAGAGGCTTGCACAGTGCA-3′. According to the manufacturer's
instructions, PC-9 and H1299 cells were infected with concentrated
lentiviruses containing either NC-shRNA or DDX46-shRNA at a
multiplicity of infection of 40 in serum-free RPMI-1640 medium
supplemented with 8 µg/ml polybrene. After 24 h, the supernatant
was replaced with RPMI-1640 medium containing 10% FBS. After 72 h
of infection, the cells were selected with puromycin at a
concentration of 2 µg/ml for 3 days, and were maintained in medium
containing 1 µg/ml puromycin for subsequent experiments. The
efficiency of NC-shRNA or DDX46-shRNA in infected cells was
assessed by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR).
Cell counting kit-8 (CCK8) assay
After transfecting NC-shRNA and DDX46-shRNA into
LUAD cells (PC-9 and H1299) for 48 h, the cells were collected,
resuspended and seeded at a concentration of 1×103
cells/well in a 96-well plate. The plate was subsequently incubated
at 37°C with 5% CO2 for 24–96 h. Following the
incubation period, 10 µl CCK-8 reagent (cat. no. C0042; Beyotime
Institute of Biotechnology) was added to each well, and the plate
was incubated for an additional 2.5 h to allow the reagent to react
with the viable cells. Microplate readers were used to measure the
absorbance (optical density) of each well at 450 nm, and cell
viability was calculated from the absorbance value.
5-ethynyl-2′-deoxyuridine (EdU)
assay
After transfecting NC-shRNA and DDX46-shRNA for 48
h, H1299 and PC-9 cells (4×104 cells/well) were seeded
in a 24-well plate and incubated with EdU solution (cat. no.
C0075S; Beyotime Institute of Biotechnology) for 2 h. Following
fixation with 4% paraformaldehyde for 15 min at room temperature,
cells were permeabilized for 15 min with 0.3% Triton X-100. The
cells were then incubated in the dark for 30 min at room
temperature. Subsequently, the cells were stained with Hoechst for
10 min at room temperature (Beyotime Biotechnology of
Biotechnology) and examined under a fluorescence microscope to
record the results.
Colony formation assay
Cells transfected with sh-NC and sh-DDX46 were
separately seeded at a density of 1,000 cells/well in a 6-well
plate and incubated at 37°C with 5% CO2 for 2 weeks.
Once colonies had formed, at room temperature the cells were fixed
with formaldehyde for 20 min and then stained with 0.5% crystal
violet solution for 10 min. Colonies were defined as cell clusters
containing ≥50 cells, indicating proliferation from a single cell.
Stained colonies were quantified using ImageJ (version 1.53) with
the Colony Counter plugin.
Detection of cell cycle
LUAD cells transfected with sh-NC and sh-DDX46 were
trypsinized, fixed with 70% ethanol at −20°C, cultured for 24 h,
washed with pre-chilled PBS at 4°C, and then incubated with 500 µl
PI/RNAse solution (cat. no. 550825; BD Biosciences) for 30 min at
37°C in the dark. Cell cycle analysis was performed using a BD
FACSCanto II (BD Biosciences), and the data were analyzed using
FlowJo v10.8 (BD Biosciences).
Wound healing assay
A wound was created by scratching the sh-NC- and
sh-DDX46-transfected cells in 6×-well plates using a 100-µl pipette
tip. After scratching, the cells were washed with PBS to remove
debris and then cultured in serum-free medium to minimize
proliferation during the assay. A microscope was used to observe
the healing progress at 0, 24 and 48 h and the wound healing area
percentage was determined using Image J software (version 1.53). At
the start of the study, cells on either side of the wound were at
100% confluence. Microscopic images were captured using an Olympus
IX73 inverted fluorescence microscope (Olympus Corporation).
Transwell assay
Cell migration was evaluated using Transwell
chambers (Corning, Inc.) with an 8-µm pore size. Initially, sh-NC-
and sh-DDX46-transfected cells were suspended in incomplete medium
and then seeded at a density of 2×104 cells/well in the
upper Transwell chamber. The lower chamber was filled with
RPMI-1640 and 10% FBS, and the cell culture was incubated at 37°C
with 5% CO2 for 24 h. After the cells migrated through
the chamber membrane, they were fixed with 4% paraformaldehyde at
room temperature for 15 min and stained with 0.1% crystal violet
solution for 10 min. Subsequently, cell images were captured using
an Olympus IX73 inverted fluorescence microscope, and the number of
cells in the lower chamber was counted.
RT-qPCR
LUAD cells transfected with sh-NC and sh-DDX46 were
lysed with TRIzol™ reagent (cat. no. 15596026CN; Thermo
Fisher Scientific, Inc.), following the manufacturer's
instructions. Chloroform, isopropanol and 75% ethanol were then
added to the cells, and the total RNA was extracted. A PCR machine
(Applied Biosystems 2720 Thermal Cycler; Thermo Fisher Scientific,
Inc.) was used to run at the following conditions to obtain cDNA
using RT (cat. no. R222-01; Vazyme Biotech Co., Ltd.): 42°C for 1 h
and 70°C for 10 min. SYBR Green (SYBR™ Green Master Mix;
Thermo Fisher Scientific, Inc.) was used for qPCR detection and the
qPCR thermocycling conditions included an initial denaturation step
at 95°C for 2–3 min, followed by 40 cycles at 95°C for 10–15 sec
and 60°C for 30 sec with fluorescence detection. Melt curve
analysis was performed at 95°C for 15 sec, 60°C for 1 min and a
gradual increase to 95°C (0.1°C/sec) with continuous fluorescence
acquisition. The forward and reverse sequences used were as
follows: DDX46 forward, 5′-AAGCTCTTGAATTGTCAGGGA-3′ and reverse,
5′-CCCTTACCAGAGAACCCACT-3′; and β-actin forward,
5′-AGTTGCGTTACACCCTTTCTTG-3′ and reverse,
5′-GCTGTCACCTTCACCGTTCC-3′. The 2−ΔΔCq method was used
for relative quantification of gene expression, with normalization
to β-actin as the internal reference gene (18).
TUNEL assay
The apoptotic rate of LUAD cells transfected with
sh-NC and sh-DDX46 was assessed using TUNEL staining. The control
and DDX46 knockdown groups were fixed with 4% paraformaldehyde for
30 min at room temperature, followed by permeabilization with 0.3%
Triton X-100 for 5 min. Subsequently, the cells were incubated with
TUNEL detection solution at 37°C for 60 min in the dark. Subsequent
Hoechst staining for 15 min at room temperature facilitated cell
imaging using a fluorescence microscope. The mounting medium used
for fluorescence microscopy was Fluoromount-G™ (Thermo
Fisher Scientific, Inc.) and 10 random fields per sample were
observed to ensure statistical reliability.
Single-cell analysis of DDX46
The Tumor Immune Single-cell Hub (TISCH; www.tisch.comp-generations.org/home/)
is a comprehensive database designed to facilitate several
single-cell analyses, containing nearly 190 samples (19). The present study specifically
focused on the distribution of DDX46 across different cell
subpopulations in LUAD; therefore, single-cell RNA sequencing data
was downloaded from the GSE117570 dataset (20) within the TISCH database to assess
DDX46 expression levels across different cell types. The expression
levels of DDX46 were quantified and visualized using scatter and
violin plots.
Immune infiltration analysis
Utilizing CIBERSORT (version 1.0; http://cibersort.stanford.edu/), an analysis was
performed to assess the correlation between DDX46 and
tumor-infiltrating immune cells (TIICs), evaluating the connection
between DDX46 expression levels and the prevalence of eight
distinct types of TIICs (21). To
analyze the correlation between DDX46 expression and immune
checkpoints and chemokines, gene expression levels were extracted
for seven immune checkpoints and 40 chemokines. Spearman
correlation analysis was performed and correlation heatmaps were
generated using the reshape2 (version 1.4.4; http://cran.r-project.org/package=reshape2),
corrplot (version 0.95; http://cran.r-project.org/package=corrplot), boot
(version 1.3–28.1; http://cran.r-project.org/package=boot), and ggplot2
packages (version 3.5.1; http://cran.r-project.org/package=ggplot2) in R
(version 4.1.3; http://www.r-project.org/).
Enrichment analysis
Enrichment analysis and the functional annotation of
genes was performed using the Gene Set Enrichment Analysis (GSEA)
website (www.software.broadinstitute:gsea/index.jsp). GSEA
is a widely utilized method for gene enrichment analysis that
assesses the enrichment levels of predefined gene sets within
specific biological processes, signaling pathways or diseases by
evaluating their correlation with these processes (22). GSEA enrichment analysis was
performed to annotate the sample genetics with Kyoto Encyclopedia
of Genes and Genomes (KEGG; http://www.kegg.jp/). The screening criteria were as
follows: P<0.05 and false discovery rate (FDR) <0.05.
Drug sensitivity analysis of
DDX46
The largest public pharmacogenomics resource, the
Cancer Drug Sensitivity Genomics website (www.cancerrxgene.org/), provided the anticancer drug
dataset used in the present study (23). To assess the association between
IC50 values of diverse anticancer drugs and DDX46
expression groups, the oncoPredict package 1.2 (https://cran.r-project.org/web/packages/oncoPredict/index.html)
was used. Subsequently, statistical analysis of drug sensitivity
within these groups was performed using the R package ‘pRRophetic’
0.5 (https://github.com/paulgeeleher/pRRophetic) to assess
differences in drug response levels among patients in different
risk groups.
Statistical analysis
Data are presented as the mean ± standard deviation
or mean ± standard error of the mean for continuous variables,
whereas categorical variables are presented as percentages or
frequencies. Three independent biological replicates (n=3) were
performed for in vitro experiments. Paired t-tests were used
to compare normal and malignant tissues from the same patient,
whilst unpaired t-tests were employed for comparisons between two
independent groups. One-way analysis of variance (ANOVA) was
performed to compare ≥3 groups. Dunnett's multiple comparison test
was subsequently applied to compare each experimental group with
the control, whilst controlling for the familywise error rate,
provided the ANOVA result revealed significant differences
(P<0.05). The Kaplan-Meier method and log-rank test were used to
evaluate the relationship between lung cancer prognosis and DDX46
expression levels, comparing both low and high expression groups.
P<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were performed using SPSS 26
(IBM Corp.) and GraphPad Prism 10 (Dotmatics).
Results
Increased expression of DDX46 in LUAD
samples
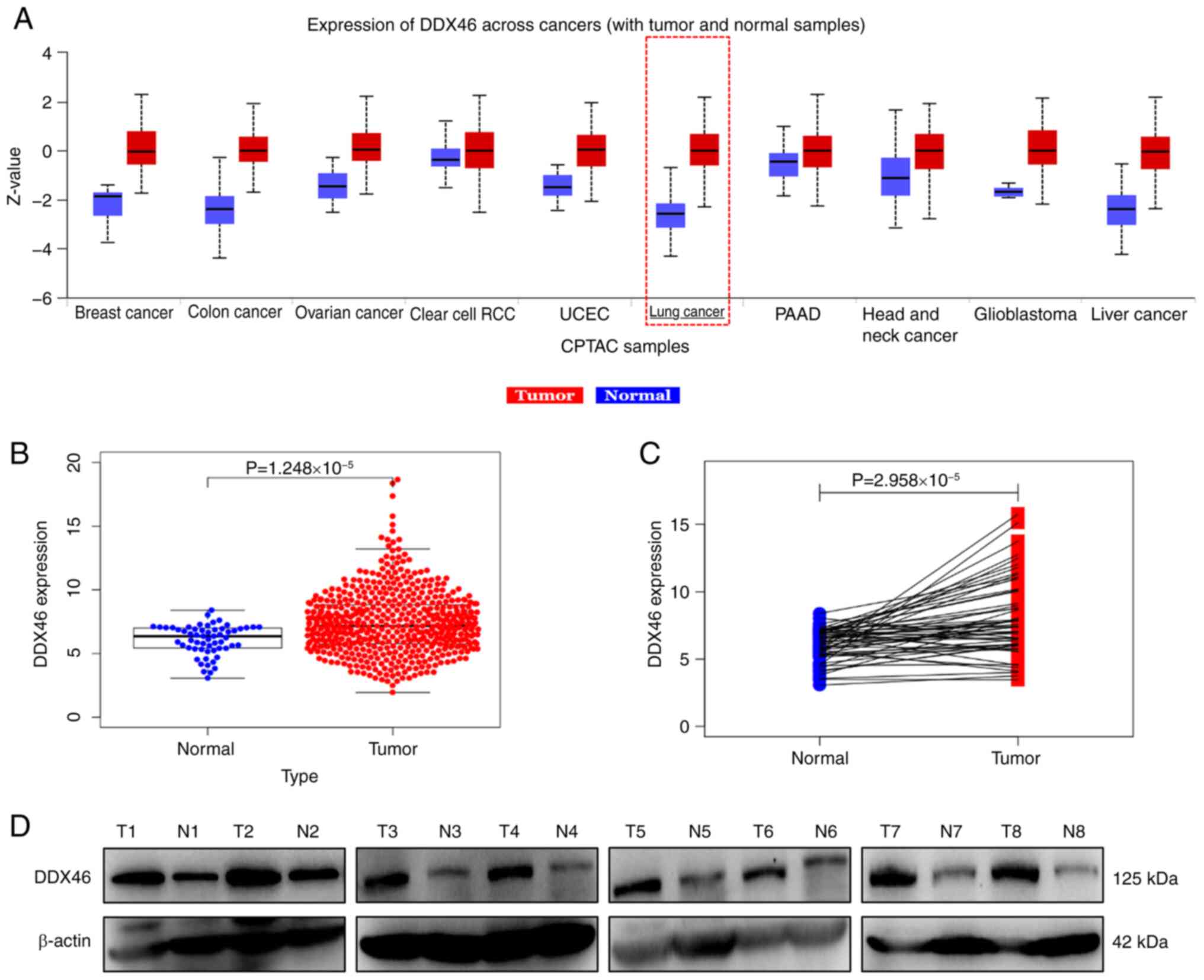
At the proteome level, DDX46 was demonstrated to be
significantly overexpressed in several tumor tissues, including
lung cancer, compared with that in normal tissues (Fig. 1A). Specifically in LUAD, DDX46
expression was revealed to be significantly higher in LUAD tissues
compared with that in normal lung tissues (Fig. 1B and C). Furthermore, western blot
analysis of eight LUAD samples and matched normal lung tissues
further demonstrated a marked increase in DDX46 protein levels in
LUAD tissues (Fig. 1D).
Association between DDX46 expression
and clinicopathological characteristics, along with the prognosis
of patients with LUAD
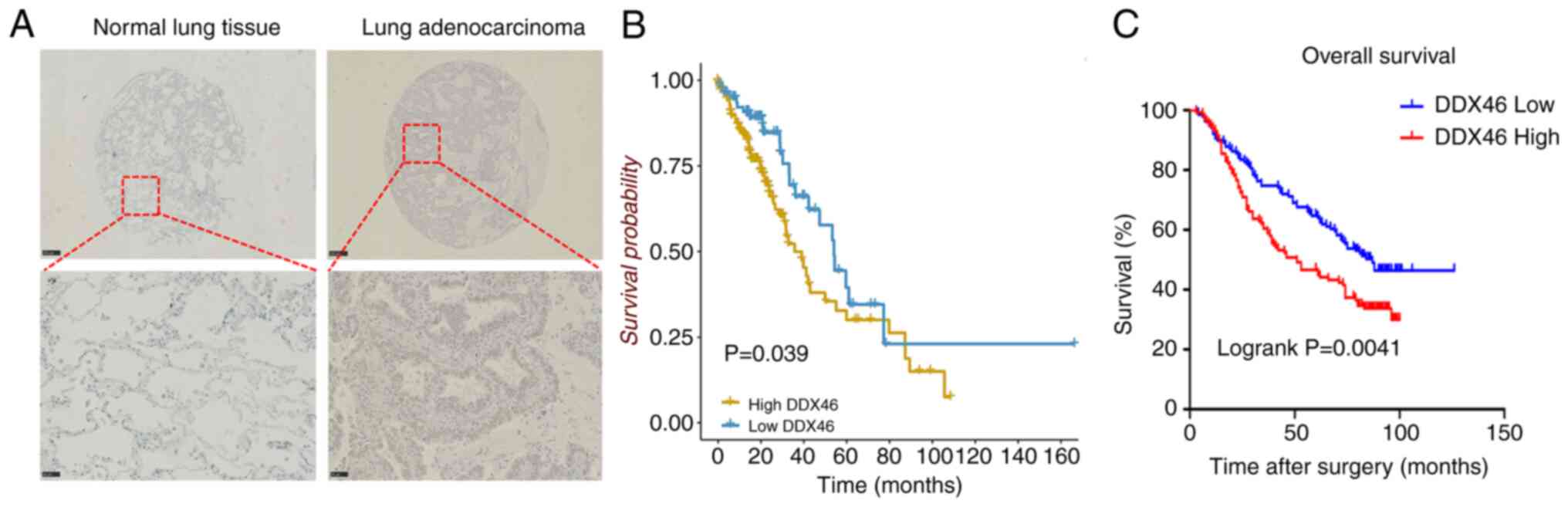
To assess the relationship between DDX46 expression
and clinicopathological characteristics, as well as patient
prognosis in LUAD, immunohistochemical staining was performed on
263 LUAD samples. The results revealed that DDX46 was predominantly
localized in the nucleus and had notably higher expression levels
in LUAD tissues than in adjacent non-tumor tissues (Fig. 2A). Moreover, DDX46 expression was
significantly associated with stage (P=0.007) and T classification
(P=0.036), whilst no statistically significant associations were
demonstrated for age, sex, smoking history, differentiation degree
or N classification (all P>0.05) (Table I).
 | Table I.Association between DEAD-box 46
expression and pathological parameters of lung adenocarcinoma. |
Table I.
Association between DEAD-box 46
expression and pathological parameters of lung adenocarcinoma.
|
|
| DDX46 expression,
n |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameter | Total, n | Low | High | P-value |
|---|
| Age |
|
|
| 0.403 |
| ≤60
years | 108 | 60 | 48 |
|
| >60
years | 155 | 78 | 77 |
|
| Sex |
|
|
| 0.210 |
|
Male | 160 | 79 | 81 |
|
|
Female | 103 | 59 | 44 |
|
| Smoking |
|
|
| 0.548 |
|
Nonsmoker | 212 | 113 | 99 |
|
|
Smoker | 51 | 25 | 26 |
|
| Stage |
|
|
| 0.007a |
| I +
II | 189 | 109 | 80 |
|
| III +
IV | 74 | 29 | 45 |
|
| Differentiated
degree |
|
|
| 0.871 |
| I +
II | 186 | 97 | 89 |
|
|
III | 77 | 41 | 36 |
|
| T
classification |
|
|
| 0.036b |
| 1 +
2 | 236 | 129 | 107 |
|
| 3 +
4 | 27 | 9 | 18 |
|
| N
classification |
|
|
|
|
| N0 | 149 | 84 | 65 | 0.147 |
| N1 +
N2 | 114 | 54 | 60 |
|
Kaplan-Meier analysis of data from the TCGA database
indicated that patients exhibiting high DDX46 expression
experienced significantly worse OS than those with low DDX46
expression (P=0.039; Fig. 2B).
Further analysis on tissue chips corroborated this finding,
revealing a significant negative association between DDX46
expression and OS in patients with LUAD (P=0.0041; Fig. 2C). Moreover, DDX46 expression was
demonstrated to be a significant independent prognostic factor in
LUAD (Table II). High DDX46
expression was significantly associated with a worse prognosis,
with a hazard ratio (HR) of 1.068 in univariate analysis (P=0.004)
and 1.056 in multivariate analysis (P=0.022), underscoring its
prognostic impact even when controlling for other variables.
Although tumor stage was also revealed to be a significant
independent prognostic factor (univariate HR=1.321; P=0.001 and
multivariate HR=1.567; P=0.009), the consistent association of
DDX46 with survival points to its unique role as a potential
standalone prognostic biomarker for LUAD.
 | Table II.Univariable and multivariable Cox
regression analyses. |
Table II.
Univariable and multivariable Cox
regression analyses.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathologic
parameter | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 1.017
(1.000–1.035) | 0.055 | 1.017
(0.999–1.035) | 0.062 |
| Stage | 1.321
(1.112–1.563) | 0.001a | 1.567
(1.121–2.191) | 0.009b |
| T
classification | 1.240
(1.043–1.475) | 0.015c | 1.012
(0.813–1.259) | 0.916 |
| N
classification | 1.166
(0.960–1.416) | 0.122 | 0.782
(0.564–1.085) | 0.142 |
| M
classification | 1.139
(0.503–2.580) | 0.754 | 0.528
(0.208–1.338) | 0.178 |
| DDX46
expression | 1.068
(1.02–1.117) | 0.004b | 1.056
(1.008–1.108) | 0.022c |
H1299 and PC-9 cells transfected with
lentivirus to knockdown DDX46 expression
Initially, the expression of DDX46 was assessed in
human normal lung epithelial BEAS-2B cells and LUAD cell lines,
including PC-9, H1299, H1975 and A549. DDX46 expression was
demonstrated to be significantly higher in PC-9, H1299, A549 and
H1975 cells compared with that in BEAS-2B cells (Fig. 3A). Based on these findings, H1299
and PC-9 cells were selected for transfection with lentiviral
vectors, including shNC, shDDX46#1 and shDDX46#2, and observation
of the fluorescence signal intensity confirmed successful viral
transfection (Fig. 3B). Subsequent
RT-qPCR and western blot analyses revealed a significant reduction
in DDX46 mRNA and protein levels in H1299 cells, in comparison with
the negative controls (Fig. 3C and
D). Similarly, DDX46 RNA and protein levels were significantly
decreased in PC-9 cells, in comparison with the negative controls
(Fig. 3E and F).
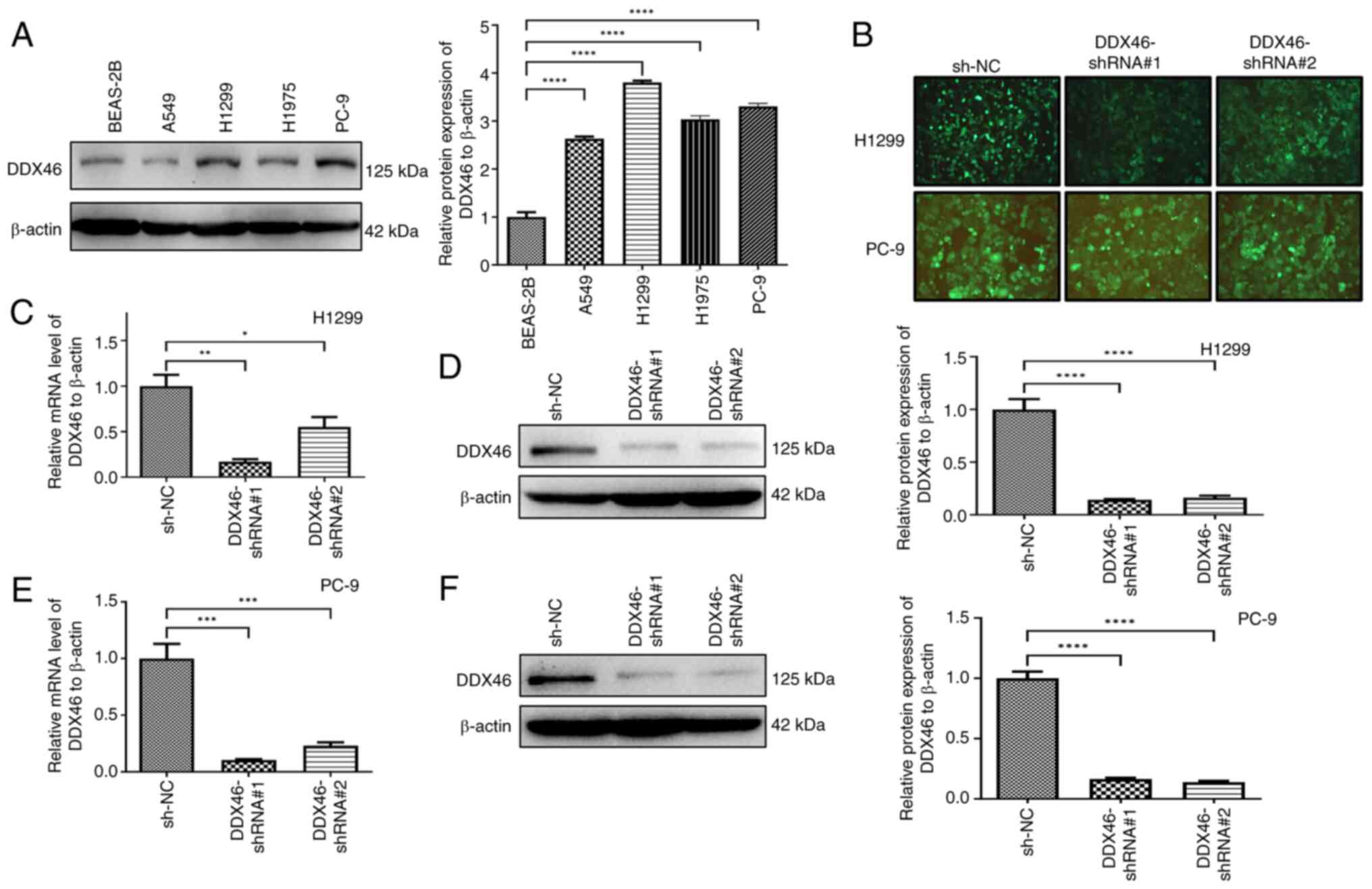 | Figure 3.Expression of DDX46 in several cell
lines and the effect of silencing DDX46 in vitro. (A) DDX46
protein levels in BEAS-2B, A549, H1975, H1299 and PC-9 cell lines.
(B) The knockdown DDX46 lentiviral vector was transfected into
H1299 and PC-9 cells, with the intensity of the green fluorescence
signal serves as an indicator of the efficiency of the
corresponding lentivirus transfection (magnification, ×200). The
effect of silencing DDX46 in H1299 cells was assessed using (C)
RT-qPCR and (D) western blotting, and the effect of silencing DDX46
in PC-9 cells was evaluated using (E) RT-qPCR and (F) western
blotting. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
DDX46, DEAD-box 4; RT-qPCR, reverse transcription-quantitative PCR;
ns, no significance; NC, negative control; shRNA, short hairpin
RNA. |
DDX46 knockdown inhibits the
proliferation potential of LUAD cells
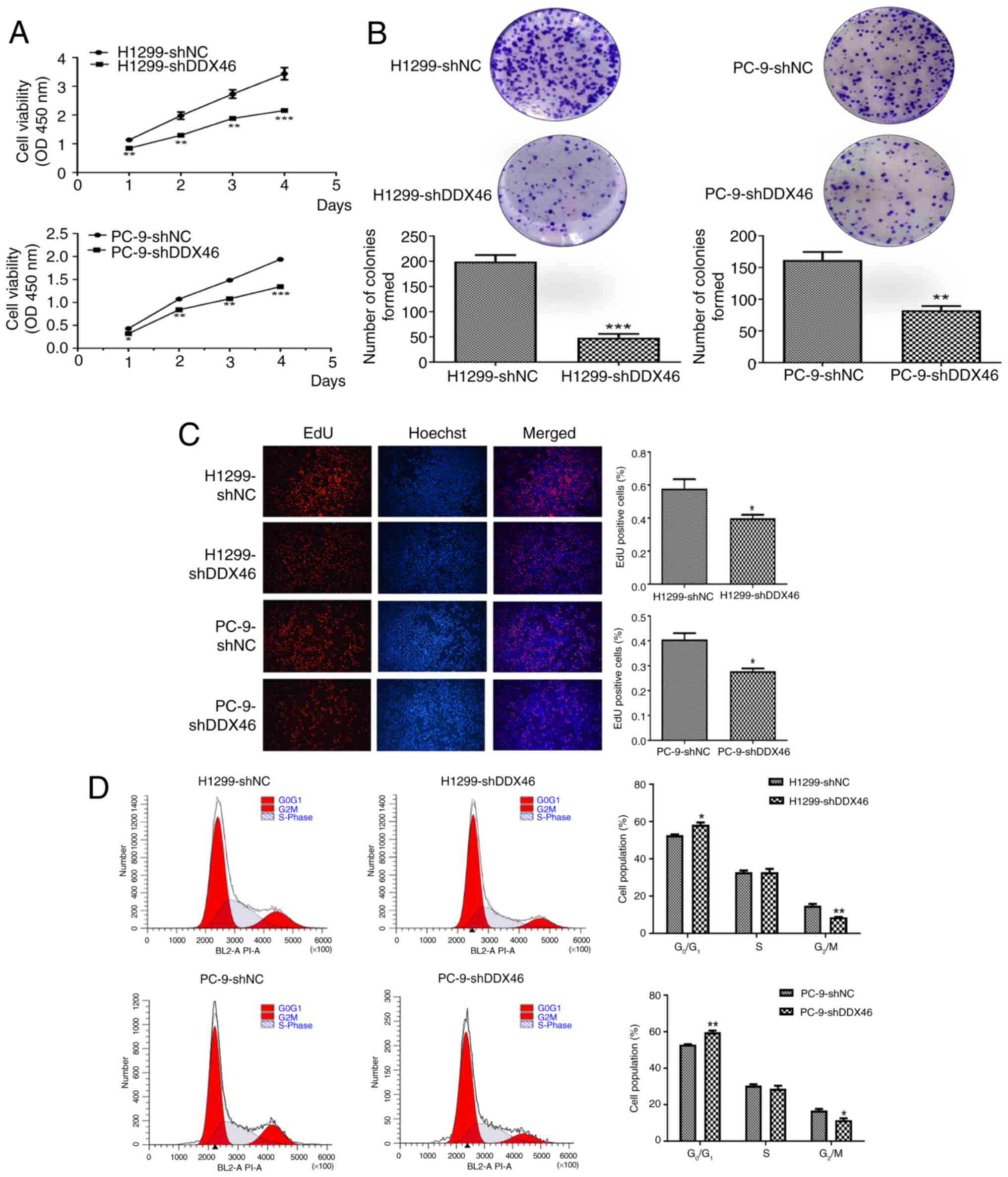
CCK-8 assays, colony formation analyses and EdU
experiments indicated a significantly lower proliferation activity
in the shDDX46 group compared with that in the NC group (Fig. 4A-C). Subsequently, flow cytometric
analysis of the cell cycle showed that knocking down the DDX46 gene
led to an increase in the proportion of G0/G1
phase cells and a decrease in G2/M phase cells,
indicating a cell cycle arrest in the G0/G1
phase (Fig. 4D).
Knockdown of DDX46 expression
suppresses the migration of LUAD cells
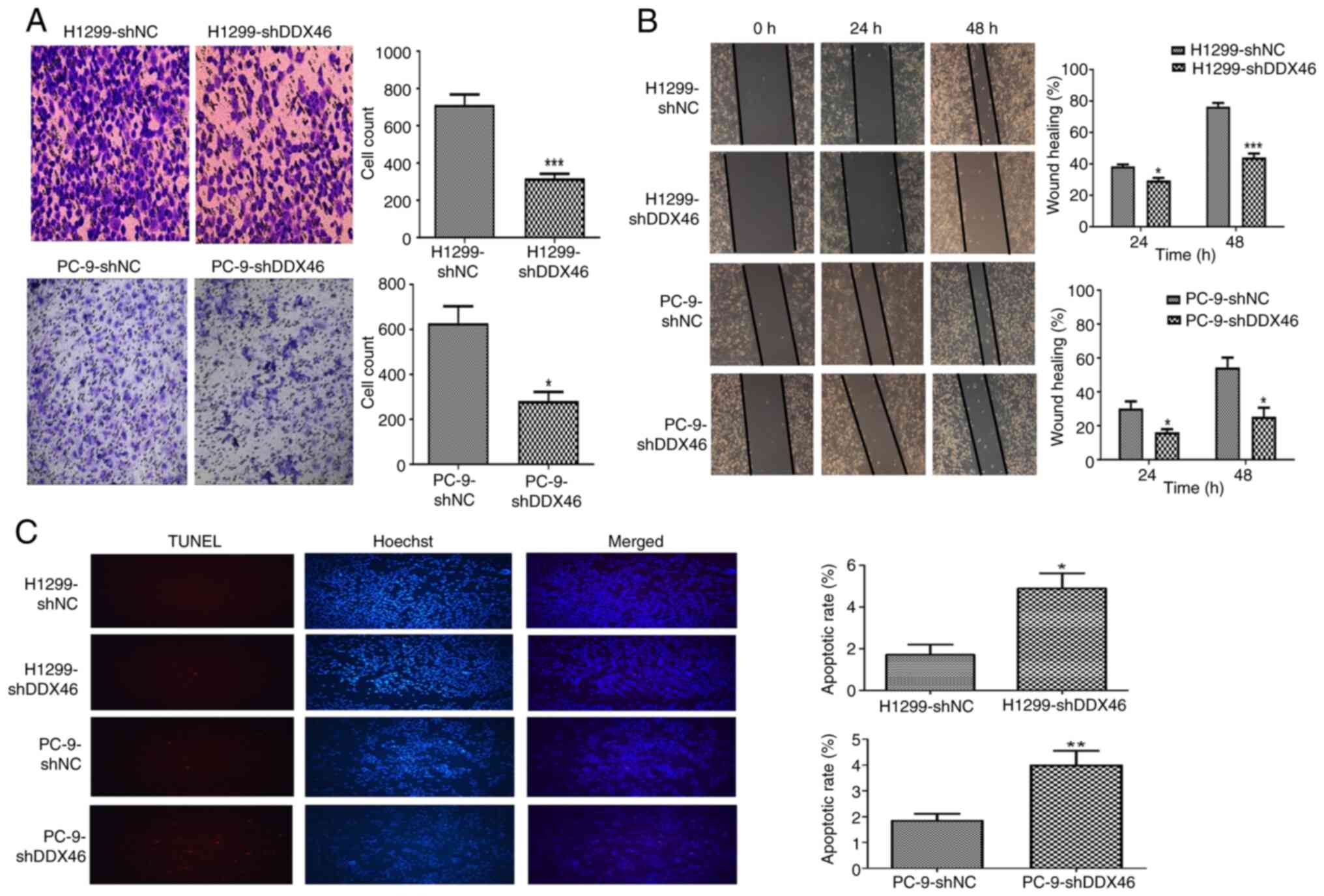
To assess the role of DDX46 in the migration of LUAD
cells, Transwell and wound healing assays were performed using
H1299 and PC-9 cells. The Transwell assay demonstrated a
significant reduction in the number of both DDX46-knockdown H1299
and PC-9 cells migrating through the polycarbonate membrane,
compared with in the control group (Fig. 5A). Similarly, the wound healing
assay revealed significantly reduced wound closure in H1299 and
PC-9 cells with DDX46 knockdown compared with in the control group
(Fig. 5B). Altogether, these
findings imply that knockdown of DDX46 expression attenuates the
migration of H1299 and PC-9 cells.
Knockdown of DDX46 expression promotes
apoptosis in LUAD cells
TUNEL staining was used to evaluate the impact of
DDX46 knockdown on the apoptosis of LUAD cells. The results
revealed that, relative to the control group, DDX46 knockdown was
associated with a significant increase in the rate of apoptosis in
both H1299 and PC-9 cells (Fig.
5C).
DDX46 regulates the Wnt signaling
pathway in LUAD cells
To further elucidate the potential mechanism of the
DDX46 gene in LUAD samples, TCGA-LUAD data was subjected to GSEA
analysis. In addition, KEGG pathway annotation was performed using
the data (Table SII). The
screening criteria were as follows: P<0.05 and FDR <0.05. The
KEGG analysis results indicate that DDX46 is closely associated
with the Wnt signaling pathway in LUAD (Fig. 6A). Subsequently, a list was compiled
of the top 10 genes enriched in the Wnt signaling pathway by DDX46
(Fig. 6B). The analysis of the
correlation between DDX46 and the top 10 genes in LUAD indicated
that the gene with the strongest correlation was APC, with a
correlation coefficient of 0.794 and a P-value of
6.28×10−113 (Fig. 6C).
Furthermore, through western Blot analysis, it was further
demonstrated that the expression of APC protein significantly
increased whilst the expression of β-catenin protein significantly
decreased in H1299 and PC-9 cells with DDX46 knockdown, in
comparison with control cells (Fig. 6D
and E). These results suggest that DDX46 is closely associated
with the Wnt signaling pathway and may regulate the malignant
progression LUAD through this pathway.
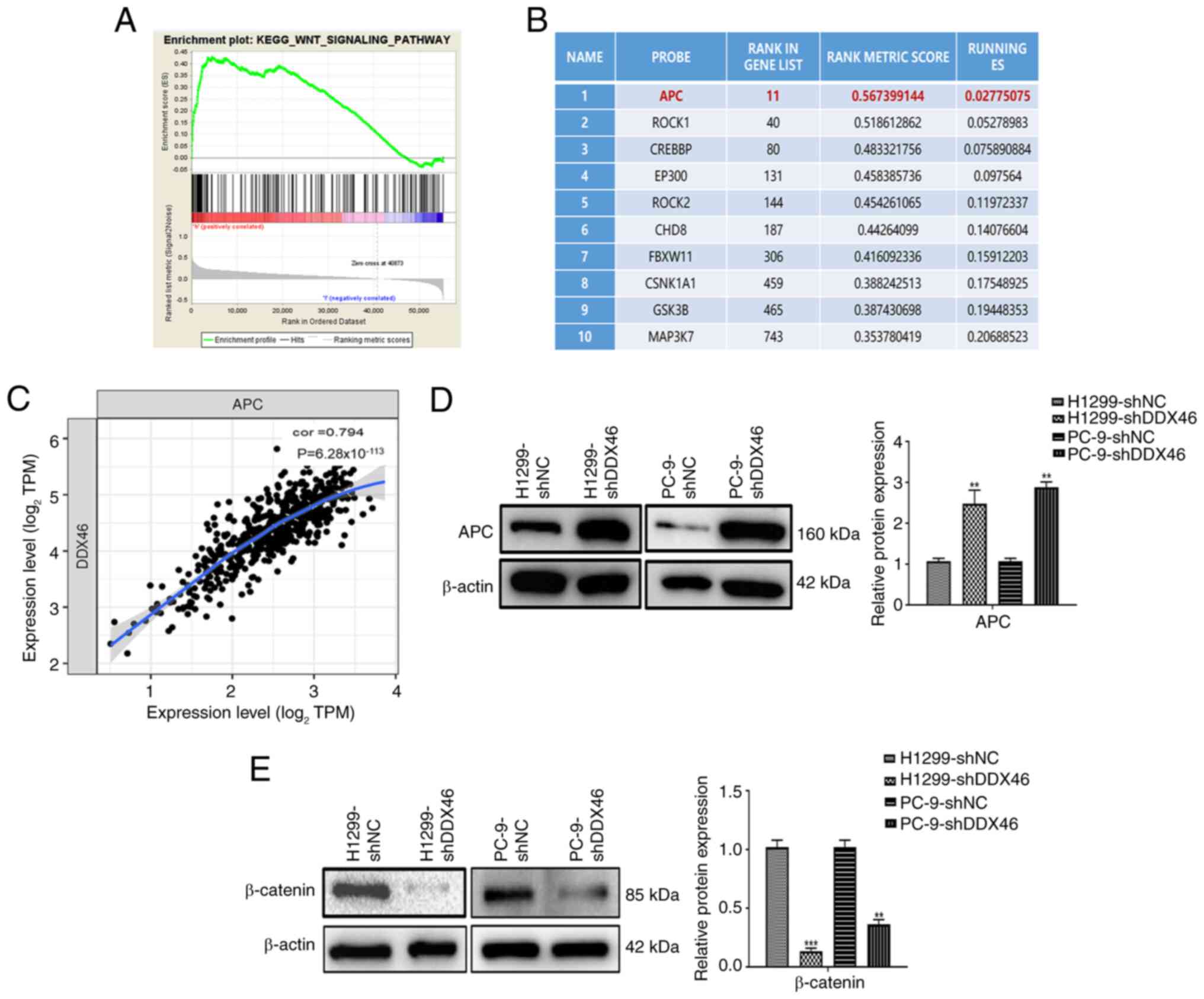 | Figure 6.DDX46 regulates the Wnt signaling
pathway in LUAD cells. (A) DDX46 is closely associated with the Wnt
signaling pathway in LUAD. (B) Top 10 genes enriched in the Wnt
signaling pathway by DDX46. (C) Correlation analysis between DDX46
and APC in LUAD. In (D) H1299 and (E) PC-9 cells, western blot
analysis revealed that APC protein expression increased after
sh-DDX46 treatment, whilst β-catenin protein expression decreased,
in comparison with sh-NC cells. **P<0.01; ***P<0.001. DDX46,
DEAD-box 4; LUAD, lung adenocarcinoma; APC, adenomatous polyposis
coli; sh, short hairpin; NC, negative control; KEGG, Kyoto
Encyclopedia of Genes and Genomes; TPM, transcripts per million;
cor, correlation coefficient; ES, enrichment score. |
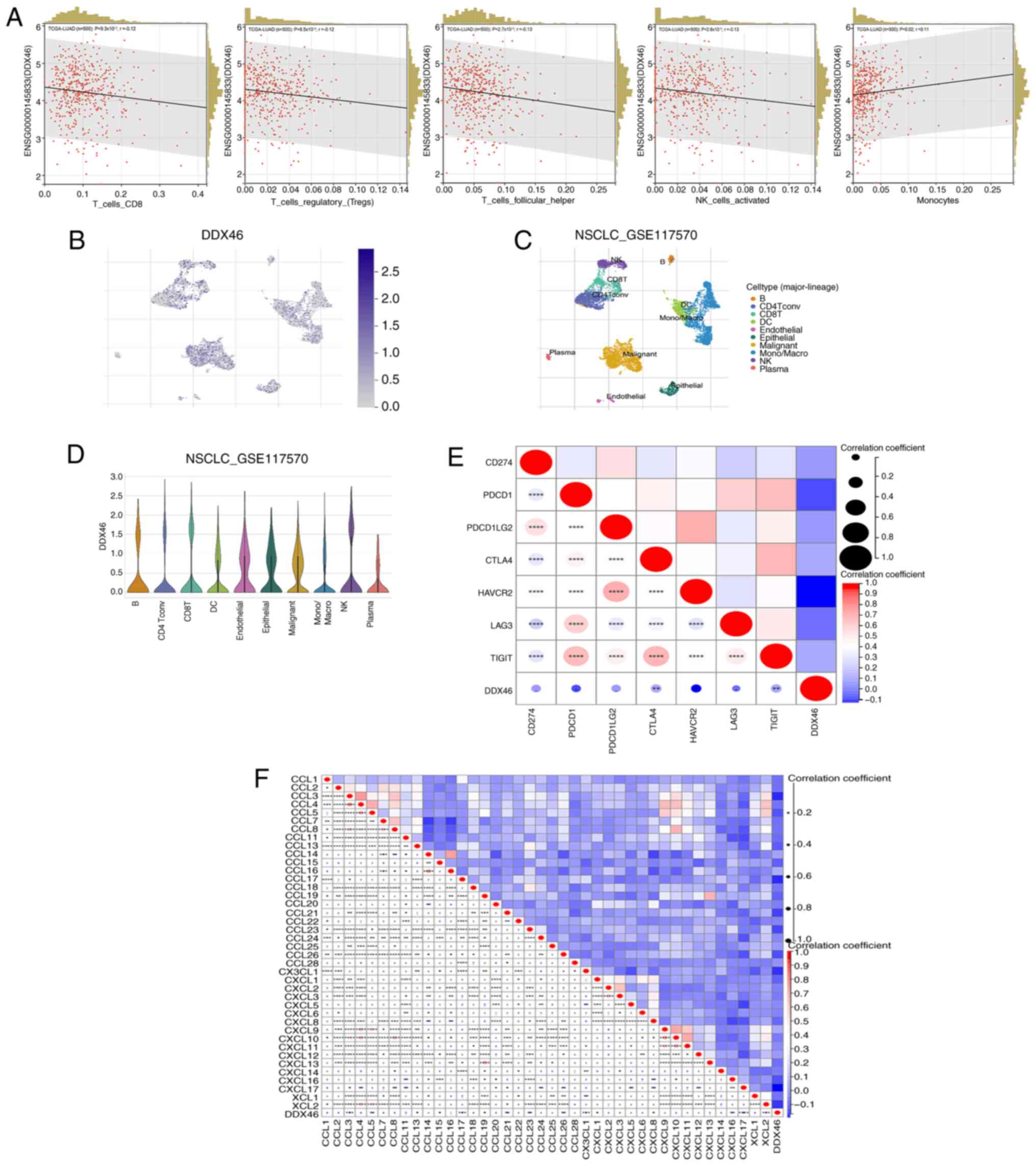
Association of DDX46 expression with
immune infiltration
Analysis of the correlation between DDX46 and 8
major immune cells in LUAD revealed that DDX46 expression is
significantly negatively correlated with CD8 T cells
(P=9.3×10−3; r=−0.12), regulatory T cells
(P=8.5×10−3; r=−0.12), follicular T cells
(P=2.7×10−3; r=−0.13) and natural killer (NK) cells
(P=2.6×10−3; r=−0.12), whilst it is significantly
positively correlated with monocytes (P=0.02; r=0.11) (Fig. 7A). However, no correlation between
the expression of DDX46 and B cells, CD4 T cells or macrophages was
identified (Fig. S1). Furthermore,
the GSE117570 dataset was used to determine the dominant cell types
associated with DDX46 gene expression. The results demonstrated
that the DDX46 gene is highly expressed in T cells, NK cells and
monocyte subsets within the non-small cell LC microenvironment
(Fig. 7B-D). Further correlation
analysis between DDX46 expression and several immune checkpoints
revealed that DDX46 is negatively correlated with cytotoxic
T-lymphocyte associated protein 4 (CTLA4), Hepatitis A virus
cellular receptor 2 (HAVCR2) and T cell immunoreceptor with Ig and
ITIM domains (TIGIT) (Fig. 7E).
Additionally, the relationship between DDX46 and several chemokines
was analyzed. DDX46 was correlated with the expression of several
chemokines, including CCL3, CCL5, CCL13, CCL15, CCL17, CCL19,
CCL21, CCL23, CCL26, CX3CL1, CXCL1, CXCL8, CXCL16, CXCL17 and XCL2
(Fig. 7F). The correlation data
between DDX46 and immune checkpoints and chemokines is presented in
Tables SIII and SIV.
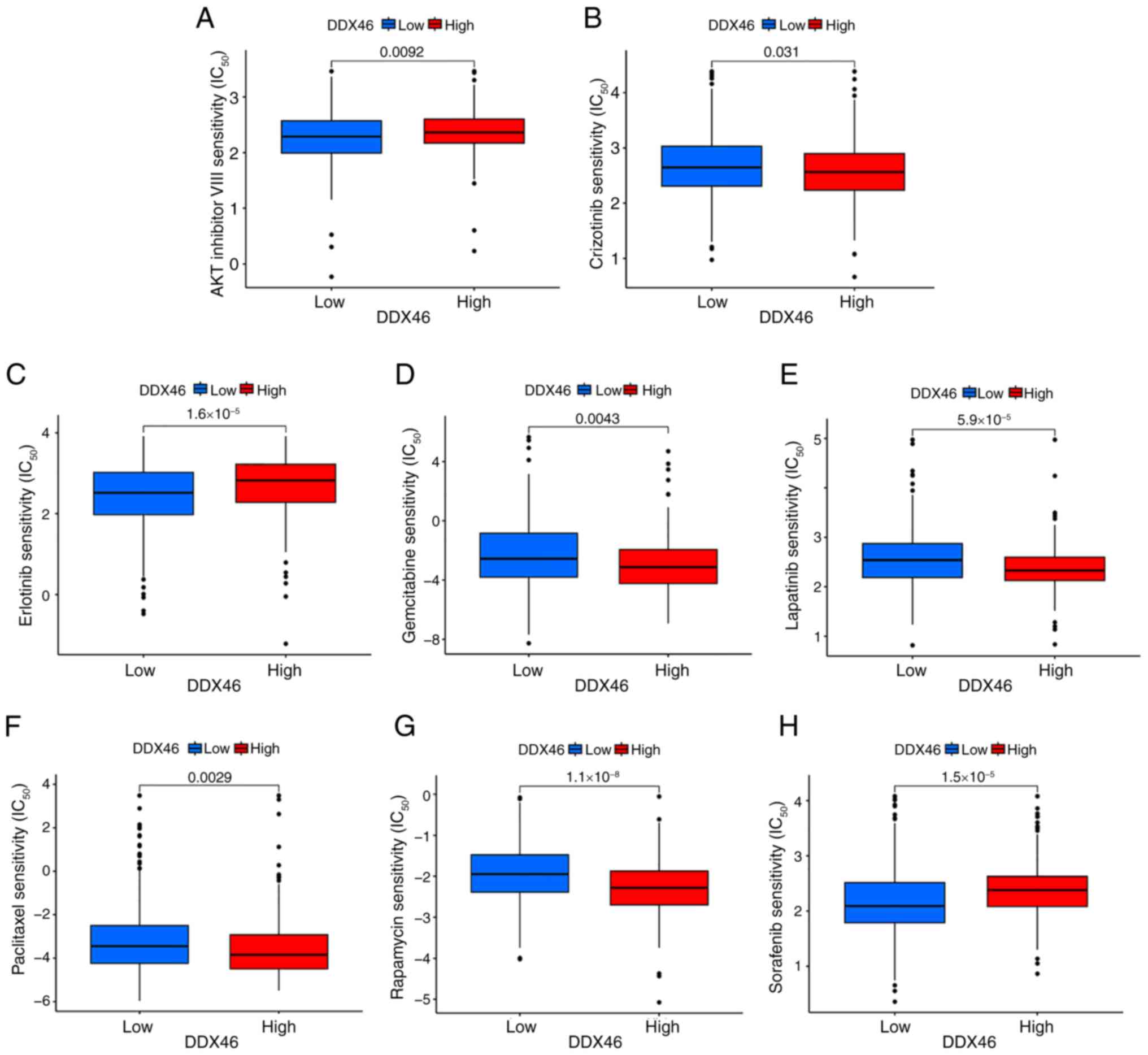
Analysis of DDX46 drug
susceptibility
The association between DDX46 gene expression and
the IC50 of commonly administered drugs in LUAD was
assessed to ascertain whether the DDX46 gene is suitable for the
personalized treatment of patients with LUAD. These commonly used
drugs for LUAD treatment include AKT inhibitors, paclitaxel,
crizotinib, erlotinib, gemcitabine, lapatinib, rapamycin and
sorafenib. The findings revealed that patients with LUAD with low
DDX46 expression demonstrated significantly increased sensitivity
to some drugs (crizotinib, gemcitabine, lapatinib, paclitaxel and
rapamycin), in comparison with those with high DDX46 expression.
However, high expression of DDX46 may be associated with increased
sensitivity to treatment with AKT inhibitors, erlotinib and
sorafenib, potentially due to its impact on key cellular survival
and proliferation pathways (Fig.
8).
Discussion
The oncogenic role of DDX46 has been reported in
several malignancies, including gastric cancer, colorectal cancer,
esophageal squamous cell carcinoma and breast cancer (13,24,25).
However, the specific biological function of DDX46 in LUAD
progression and its relationship with immune infiltration and drug
sensitivity remain poorly understood. The results of the present
study demonstrated a significant upregulation of DDX46 in LUAD,
which was associated with a poor prognosis. In LUAD cells, aberrant
expression of DDX46 affected cell proliferation, invasion and
migration. Bioinformatics analysis and experimental validation
suggested that DDX46 may regulate the Wnt signaling pathway.
Additionally, further bioinformatics analysis revealed that DDX46
is strongly linked to immune cell infiltration, immune checkpoints,
chemokines and drug sensitivity.
Research has shown that the overexpression of DDX46
in human breast cancer is related to elevated histological grade
and lymph node metastasis (25). In
a parallel study on esophageal squamous cell carcinoma, a notable
elevation in DDX46 expression was observed in both cancer cells and
tissues (13). Additionally,
colorectal cancer tissues have been reported to exhibit heightened
focal nuclear DDX46 staining compared with adjacent normal tissues
(24). Admoni-Elisha et al
(26) reported an association
correlation elevated DDX46 expression in patients with chronic
lymphocytic leukemia and reduced OS. In the present research, the
high expression of DDX46 in both LUAD tissues and cell lines was
initially demonstrated, revealing its association with a poor
prognosis. The findings suggest that DDX46 could serve as a
valuable biomarker for stratifying patients with LUAD based on
risk. By identifying patients with high DDX46 expression,
clinicians could improve the prediction of those at higher risk of
disease progression and poor outcomes. Functionally, abnormal
expression of DDX46 contributes to enhanced proliferation,
migration, invasion and suppressed apoptosis of esophageal squamous
cell carcinoma, colorectal cancer cells and cutaneous squamous cell
carcinoma in vitro and in vivo experiments (27). Additionally, in A549 cells, the
interaction between structural maintenance of chromosomes 4 and the
DDX46 gene was reported to impede cell M-phase progression, thereby
inhibiting cell proliferation and invasion (28). The findings of the study align with
the documented role of DDX46 in enhancing tumor cell proliferation,
invasion and migration.
Prior research has reported that DDX46 influences
the proliferation and migration of glioblastoma, gastric cancer and
preeclampsia through the MAPK-p38, Akt/glycogen synthase
kinase-3β/β-catenin and PI3K/Akt/mTOR signaling pathways (15,29,30).
In the mechanism, KEGG pathway enrichment analysis revealed that
DDX46 is strongly associated with the Wnt signaling pathway in
LUAD. Correlation analysis also revealed that DDX46 has a high
correlation coefficient with APC, in LUAD, reaching 0.794 with a
P-value of 6.28×10−113. APC serves a critical
suppressive role in the Wnt signaling pathway, and the interaction
between APC and β-catenin is crucial for cell signaling,
proliferation and fate determination. In cancer development,
mutations in APC lead to the abnormal activation of β-catenin, a
process that serves a critical role in tumorigenesis (31–34).
In the present study, western blot analysis further demonstrated
that, in H1299 and PC-9 cells with DDX46 knockdown, the expression
of APC protein increased whilst the expression of β-catenin protein
decreased. These findings offer fresh perspectives into the
downstream regulatory mechanisms of DDX46 in tumor progression.
The present study revealed a strong association
between DDX46 and diverse immune cells, immune checkpoints and
chemokines. Immune cells and chemokines within the tumor immune
microenvironment are pivotal in the pathogenesis of diverse tumors
(35), with the degree of immune
cell infiltration markedly impacting the prognosis of patients with
solid tumors (36,37). Research has also reported that
chemokines are involved in the circulation, homing, retention and
activation of immune active cells (38,39).
Notably, the findings of the present study indicate an inverse
relationship between DDX46 expression and T cells, NK cells and
immune checkpoints, CTLA4, HAVCR2 and TIGIT, suggesting a potential
role for DDX46 in immune evasion. This mechanism could contribute
to the progression of LUAD.
In the analysis of drug sensitivity associated with
DDX46, it was revealed that DDX46 expression is associated with the
sensitivity of cancer cells to several drugs, including AKT
inhibitors, paclitaxel, crizotinib, erlotinib, gemcitabine,
lapatinib, rapamycin and sorafenib. Numerous studies have reported
that the DDX helicase family proteins, in addition to their
critical roles in RNA biology, can stimulate and bind to several
protein kinases (40). This
provides a theoretical basis for the influence of DDX46 expression
on the sensitivity to multiple protein kinase inhibitors. AKT
inhibitors are a class of targeted antitumor drugs that inhibit the
AKT protein kinase (41). Previous
studies have reported that knockdown of DDX46 can notably reduce
the phosphorylation levels of AKT, thereby decreasing its kinase
activity (14,15). Erlotinib, a tyrosine kinase
inhibitor targeting the epidermal growth factor receptor (EGFR),
inhibits tumor cell proliferation, survival and migration by
blocking EGFR signaling pathways such as the Ras/Raf/MEK/ERK and
PI3K/AKT/mTOR pathways (42). A
study on preeclampsia also revealed that suppressing DDX46 inhibits
trophoblast proliferation and migration through the PI3K/AKT/mTOR
signaling pathway (30).
Furthermore, a study on human colorectal cancer reported that the
expression of DDX46 is associated with the apoptosis of tumor cells
(24). One of the mechanisms of
action of sorafenib is its ability to reduce the expression of
anti-apoptotic proteins through multiple pathways, thereby
promoting apoptosis (43). In
summary, the impact of DDX46 expression on drug sensitivity or
resistance in cancer cells is multifaceted, depending on the
interplay between the mechanism of action of each drug and the
cellular processes regulated by DDX46.
However, the present study has certain limitations.
Although bioinformatics provides valuable insights and preliminary
correlations, there is a lack of experimental validation to clarify
the association between DDX46 and the Wnt signaling pathway.
Similarly, the absence of in vivo experiments limits the
ability to confirm the biological and mechanistic roles of DDX46 in
LUAD. Future research should incorporate experiments targeting the
Wnt signaling pathway and utilize animal models to validate the
role of DDX46, establishing causal relationships with LUAD
progression and immune regulation.
In conclusion, elevated DDX46 levels in patients
LUAD may serve as a strong prognostic indicator and assist in
diagnosis. Furthermore, high DDX46 expression promotes cell
proliferation and survival in LUAD, highlighting its potential as a
therapeutic target for diagnosis and treatment.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was funded by the National Natural Science
Foundation of China (grant no. 82273422), the Nantong Basic
Research Plan Project (grant no. MS2023067), the 2022 Nantong Basic
Science Research and Social Livelihood Science and Technology Plan
Project (grant no. MS22022111), the Research Project on
Cutting-Edge Tumor Support Therapy (grant no. cphcf-2022-206), the
Jiangsu Provincial Research Hospital (grant nos. YJXYY202204-YSC01
and YJXYY202204-YSB01) and the Key Project of Nantong's 14th Five
Year Plan Science and Education Strengthening Health Project,
Oncology Clinical Medical Center (grant no. NTYXZX18).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
TB and MZ designed the study; TW, QZ and JZ
performed the experiments; TW analyzed the data; TB and MZ wrote
the paper and confirm the authenticity of all the raw data; YL and
WS designed the work, drafted the manuscript and contributed to its
revision. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki and was approved by the Ethics
Committee of the Affiliated Hospital of Nantong University
(approval no. 2022-L078). The data were anonymous and the
requirement for informed consent was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DDX46
|
DEAD-box 46
|
|
LC
|
lung cancer
|
|
LUAD
|
lung adenocarcinoma
|
|
TISCH
|
Tumor Immune Single Cell Hub
|
|
TIICs
|
tumor-infiltrating immune cells
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
APC
|
adenomatous polyposis coli
|
|
OS
|
overall survival
|
References
|
1
|
Collisson EA, Campbell JD, Brooks AN,
Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ,
Danilova L, et al: Comprehensive molecular profiling of lung
adenocarcinoma: The cancer genome atlas research network. Nature.
511:543–550. 2014. View Article : Google Scholar
|
|
2
|
Chen P, Liu Y, Wen Y and Zhou C: Non-small
cell lung cancer in China. Cancer Commun (Lond). 42:937–970. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zulfiqar B, Farooq A, Kanwal S and Asghar
K: Immunotherapy and targeted therapy for lung cancer: Current
status and future perspectives. Front Pharmacol. 13:10351712022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Denisenko TV, Budkevich IN and Zhivotovsky
B: Cell death-based treatment of lung adenocarcinoma. Cell Death
Dis. 9:1172018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hooper C and Hilliker A: Packing them up
and dusting them off: RNA helicases and mRNA storage. Biochim
Biophys Acta. 1829:824–834. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Linder P and Jankowsky E: From unwinding
to clamping-the DEAD box RNA helicase family. Nat Rev Mol Cell
Biol. 12:505–516. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Owttrim GW: RNA helicases: Diverse roles
in prokaryotic response to abiotic stress. RNA Biol. 10:96–110.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Robert F and Pelletier J: Perturbations of
RNA helicases in cancer. Wiley Interdiscip Rev RNA. 4:333–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steimer L and Klostermeier D: RNA
helicases in infection and disease. RNA Biol. 9:751–771. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abdelhaleem M, Maltais L and Wain H: The
human DDX and DHX gene families of putative RNA helicases.
Genomics. 81:618–622. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Will CL, Urlaub H, Achsel T, Gentzel M,
Wilm M and Lührmann R: Characterization of novel SF3b and 17S U2
snRNP proteins, including a human Prp5p homologue and an SF3b
DEAD-box protein. EMBO J. 21:4978–4988. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li B, Li YM, He WT, Chen H, Zhu HW, Liu T,
Zhang JH, Song TN and Zhou YL: Knockdown of DDX46 inhibits
proliferation and induces apoptosis in esophageal squamous cell
carcinoma cells. Oncol Rep. 36:223–230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang F, Zhang D, Li G and Wang X:
Knockdown of DDX46 inhibits the invasion and tumorigenesis in
osteosarcoma cells. Oncol Res. 25:417–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Xu M, Zhong W, Hu Y and Wang G:
Knockdown of DDX46 suppresses the proliferation and invasion of
gastric cancer through inactivating Akt/GSK-3β/β-catenin pathway.
Exp Cell Res. 399:1124482021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Jensen MA and Zenklusen JC: A
practical guide to the cancer genome atlas (TCGA). Methods Mol
Biol. 1418:111–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng M, Liu J, Bian T, Liu L, Sun H, Zhou
H, Zhao C, Yang Z, Shi J and Liu Y: Correlation between prognostic
indicator AHNAK2 and immune infiltrates in lung adenocarcinoma. Int
Immunopharmacol. 90:1071342021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun D, Wang J, Han Y, Dong X, Ge J, Zheng
R, Shi X, Wang B, Li Z, Ren P, et al: TISCH: A comprehensive web
resource enabling interactive single-cell transcriptome
visualization of tumor microenvironment. Nucleic Acids Res. 49(D1):
D1420–D1430. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lei Y, Zhou B, Meng X, Liang M, Song W,
Liang Y, Gao Y and Wang M: A risk score model based on lipid
metabolism-related genes could predict response to immunotherapy
and prognosis of lung adenocarcinoma: A multi-dataset study and
cytological validation. Discov Oncol. 14:1882023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ru B, Wong CN, Tong Y, Zhong JY, Zhong
SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, et al: TISIDB: An
integrated repository portal for tumor-immune system interactions.
Bioinformatics. 35:4200–4202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Komuro H, Shinohara S, Fukushima Y,
Demachi-Okamura A, Muraoka D, Masago K, Matsui T, Sugita Y,
Takahashi Y, Nishida R, et al: Single-cell sequencing on
CD8+ TILs revealed the nature of exhausted T cells
recognizing neoantigen and cancer/testis antigen in non-small cell
lung cancer. J Immunother Cancer. 11:e0071802023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang W, Soares J, Greninger P, Edelman EJ,
Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et
al: Genomics of drug sensitivity in cancer (GDSC): A resource for
therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.
41((Database Issue)): D955–D961. 2013.PubMed/NCBI
|
|
24
|
Li M, Ma Y, Huang P, Du A, Yang X, Zhang
S, Xing C, Liu F and Cao J: Lentiviral DDX46 knockdown inhibits
growth and induces apoptosis in human colorectal cancer cells.
Gene. 560:237–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma Z, Song J, Hua Y, Wang Y, Cao W, Wang H
and Hou L: The role of DDX46 in breast cancer proliferation and
invasiveness: A potential therapeutic target. Cell Biol Int.
47:283–291. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Admoni-Elisha L, Nakdimon I, Shteinfer A,
Prezma T, Arif T, Arbel N, Melkov A, Zelichov O, Levi I and
Shoshan-Barmatz V: Novel biomarker proteins in chronic lymphocytic
leukemia: Impact on diagnosis, prognosis and treatment. PLoS One.
11:e01485002016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin Q, Jin HJ, Zhang D and Gao L: DDX46
silencing inhibits cell proliferation by activating apoptosis and
autophagy in cutaneous squamous cell carcinoma. Mol Med Rep.
22:4236–4242. 2020.PubMed/NCBI
|
|
28
|
Zhang C, Kuang M, Li M, Feng L, Zhang K
and Cheng S: SMC4, which is essentially involved in lung
development, is associated with lung adenocarcinoma progression.
Sci Rep. 6:345082016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma J, Gao Z and Liu X: DDX46 accelerates
the proliferation of glioblastoma by activating the MAPK-p38
signaling. J BUON. 26:2084–2089. 2021.PubMed/NCBI
|
|
30
|
You X, Cui H, Yu N and Li Q: Knockdown of
DDX46 inhibits trophoblast cell proliferation and migration through
the PI3K/Akt/mTOR signaling pathway in preeclampsia. Open Life Sci.
15:400–408. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Disoma C, Zhou Y, Li S, Peng J and Xia Z:
Wnt/β-catenin signaling in colorectal cancer: Is therapeutic
targeting even possible? Biochimie. 195:39–53. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wan C, Mahara S, Sun C, Doan A, Chua HK,
Xu D, Bian J, Li Y, Zhu D, Sooraj D, et al: Genome-scale
CRISPR-Cas9 screen of Wnt/β-catenin signaling identifies
therapeutic targets for colorectal cancer. Sci Adv. 7:eabf25672021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hankey W, Frankel WL and Groden J:
Functions of the APC tumor suppressor protein dependent and
independent of canonical WNT signaling: Implications for
therapeutic targeting. Cancer Metastasis Rev. 37:159–172. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bai R, Yin P, Xing Z, Wu S, Zhang W, Ma X,
Gan X, Liang Y, Zang Q, Lei H, et al: Investigation of GPR143 as a
promising novel marker for the progression of skin cutaneous
melanoma through bioinformatic analyses and cell experiments.
Apoptosis. 29:372–392. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki
M, Kosuge T, Kanai Y and Hiraoka N: Immune cell infiltration as an
indicator of the immune microenvironment of pancreatic cancer. Br J
Cancer. 108:914–923. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schneider K, Marbaix E, Bouzin C, Hamoir
M, Mahy P, Bol V and Gregoire V: Immune cell infiltration in head
and neck squamous cell carcinoma and patient outcome: A
retrospective study. Acta Oncol. 57:1165–1172. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Franciszkiewicz K, Boissonnas A, Boutet M,
Combadiere C and Mami-Chouaib F: Role of chemokines and chemokine
receptors in shaping the effector phase of the antitumor immune
response. Cancer Res. 72:6325–6332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koizumi K, Hojo S, Akashi T, Yasumoto K
and Saiki I: Chemokine receptors in cancer metastasis and cancer
cell-derived chemokines in host immune response. Cancer Sci.
98:1652–1658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hirth A, Fatti E, Netz E, Acebron SP,
Papageorgiou D, Švorinić A, Cruciat CM, Karaulanov E, Gopanenko A,
Zhu T, et al: DEAD box RNA helicases are pervasive protein kinase
interactors and activators. Genome Res. 34:952–966. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shariati M and Meric-Bernstam F: Targeting
AKT for cancer therapy. Expert Opin Investig Drugs. 28:977–988.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu SG and Shih JY: Management of acquired
resistance to EGFR TKI-targeted therapy in advanced non-small cell
lung cancer. Mol Cancer. 17:382018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu CH, Lin KH, Fu BS, Hsu FT, Tsai JJ,
Weng MC and Pan PJ: Sorafenib induces apoptosis and inhibits
NF-κB-mediated anti-apoptotic and metastatic potential in
osteosarcoma cells. Anticancer Res. 41:1251–1259. 2021. View Article : Google Scholar : PubMed/NCBI
|