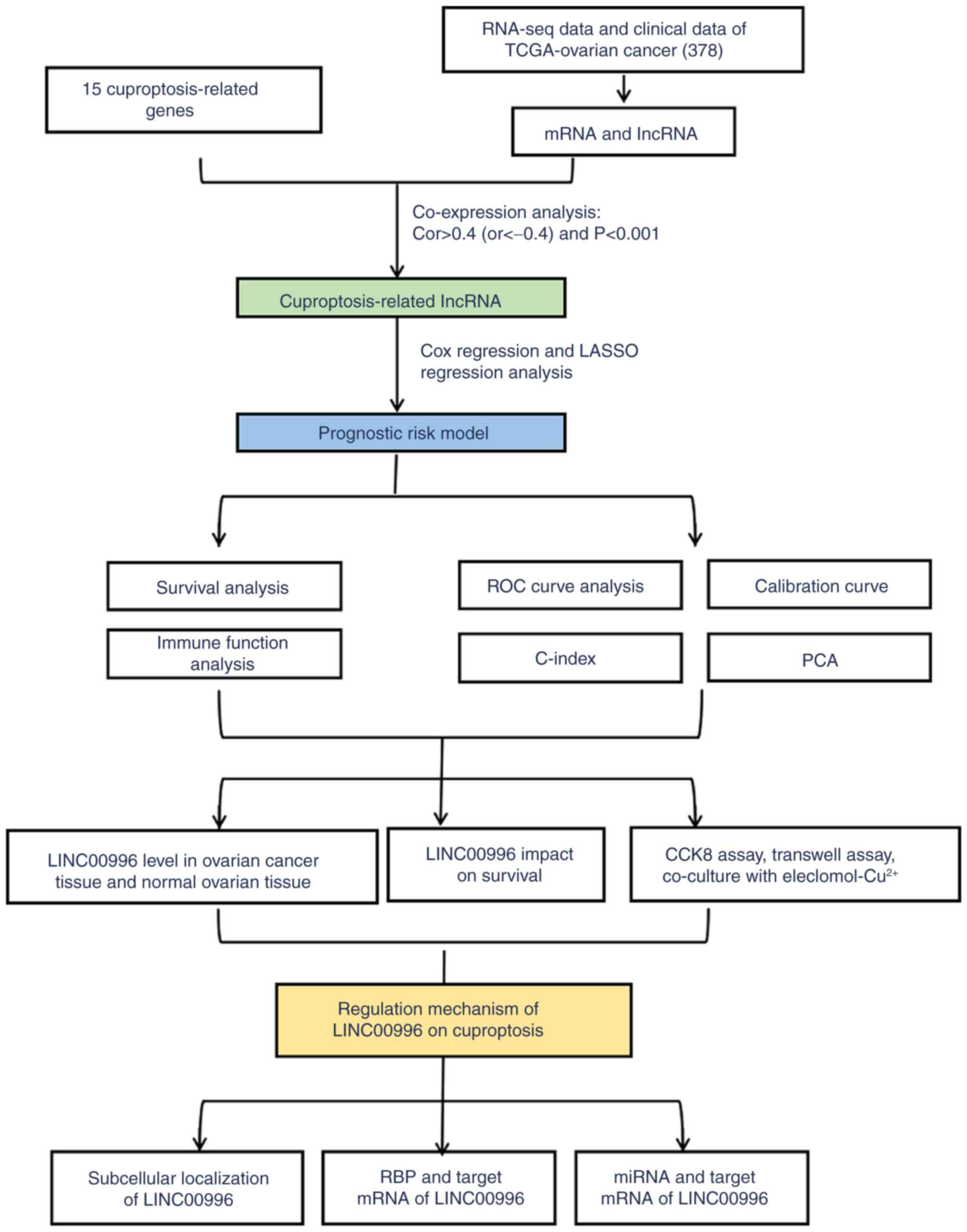
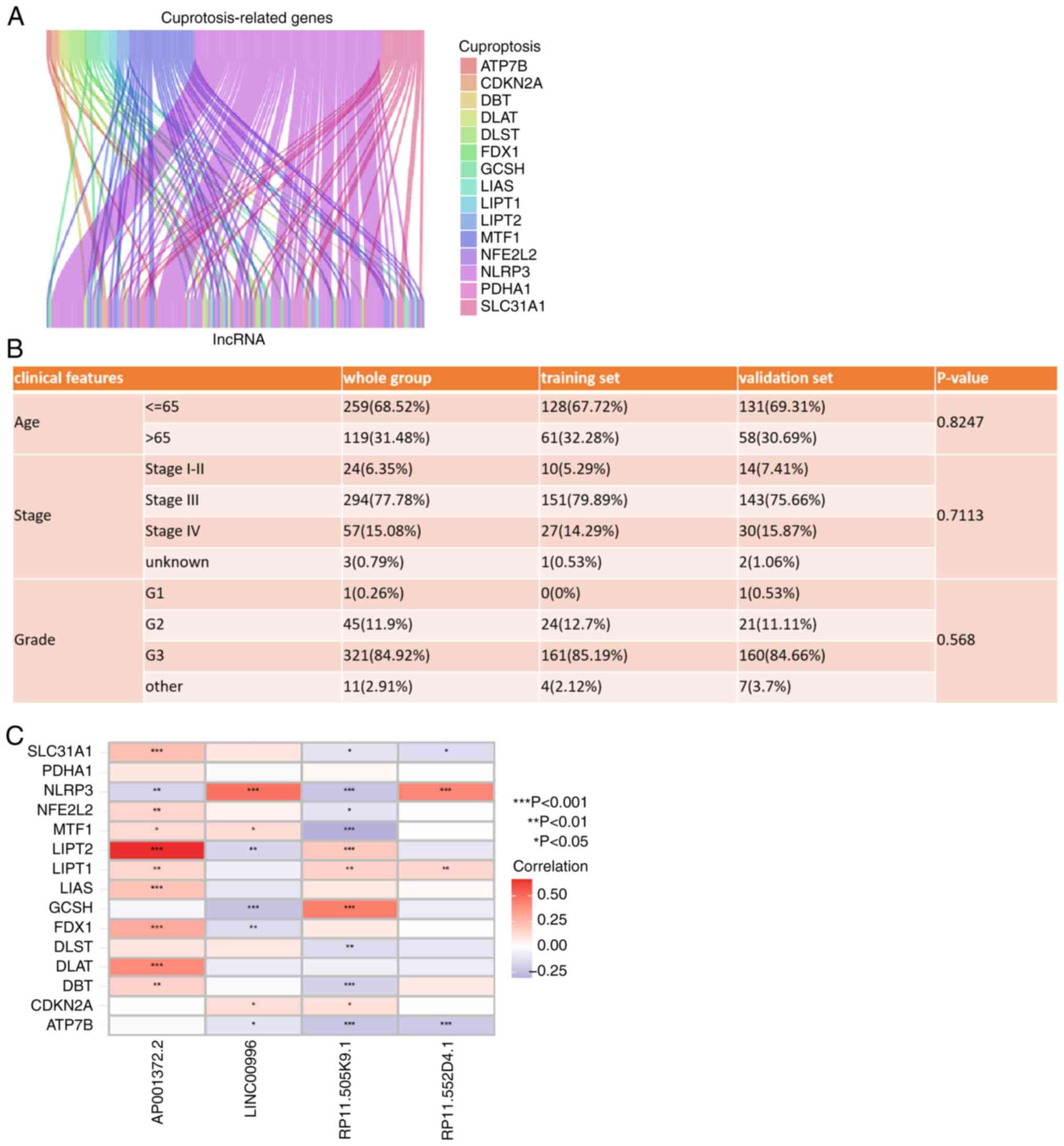
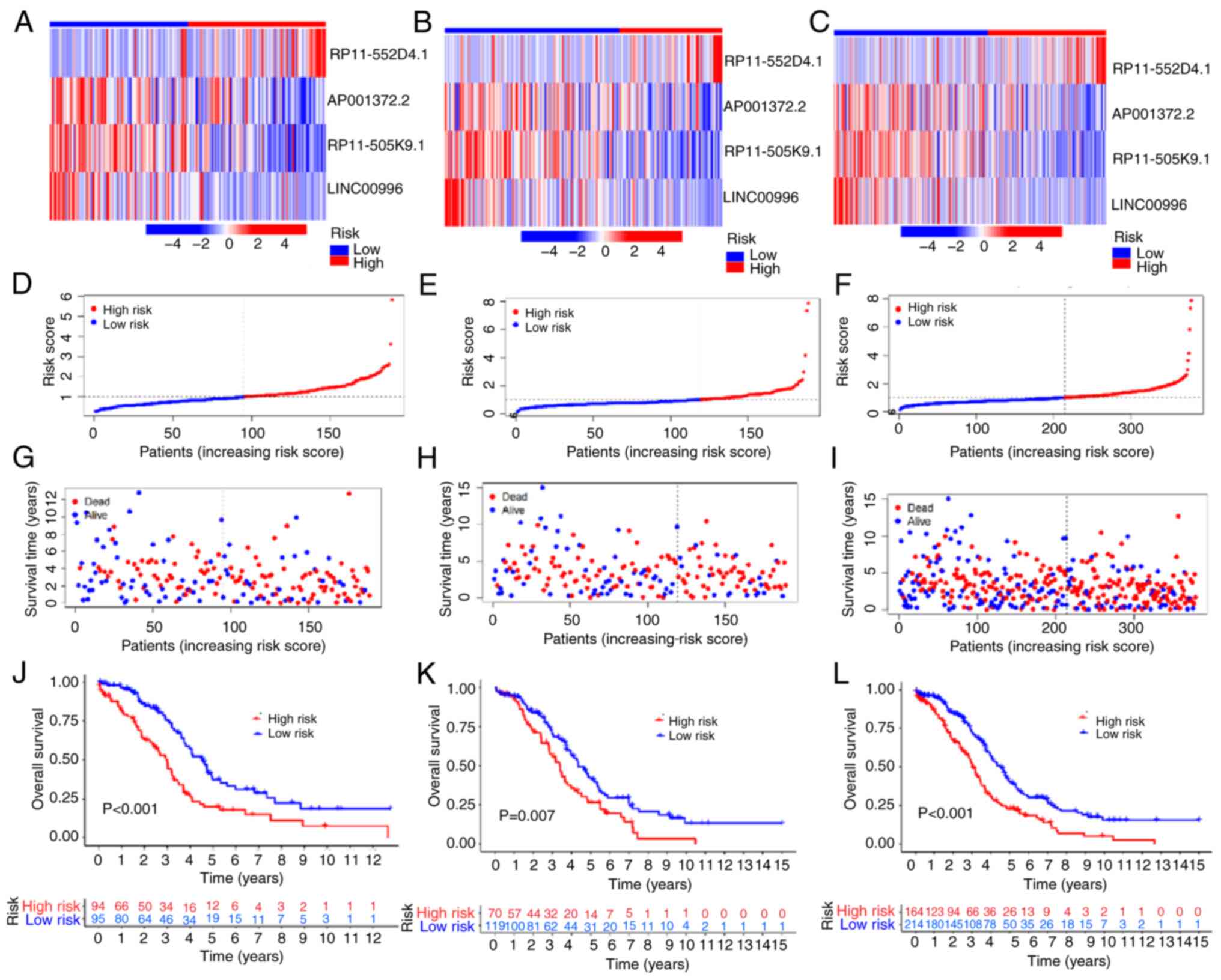
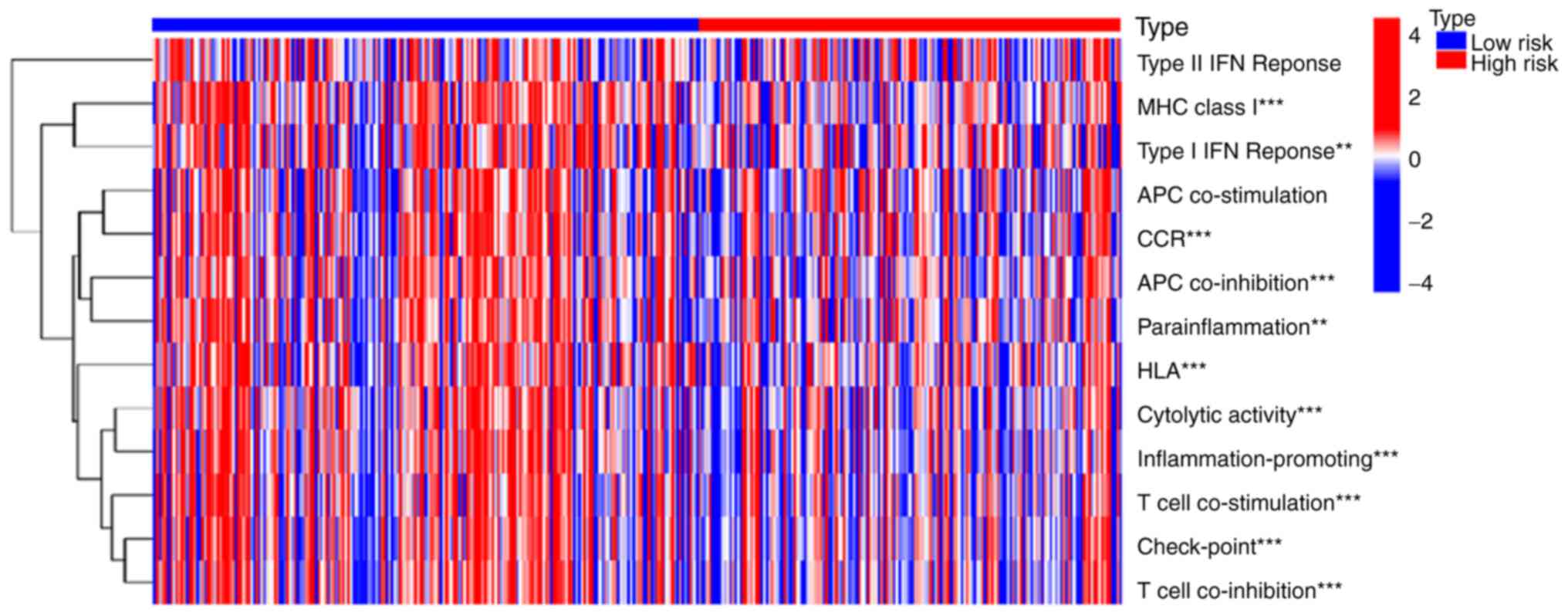
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Goding SA, Fedewa
SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal A:
Colorectal cancer statistics, 2020. CA Cancer J Clin. 70:145–164.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen S, Tang Y, Li Y, Huang M, Ma X, Wang
L, Wu Y, Wang Y, Fan W and Hou S: Design and application of prodrug
fluorescent probes for the detection of ovarian cancer cells and
release of anticancer drug. Biosens Bioelectron. 236:1154012023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li T, Wang X, Qin S, Chen B, Yi M and Zhou
J: Targeting PARP for the optimal immunotherapy efficiency in
gynecologic malignancies. Biomed Pharmacother. 162:1147122023.
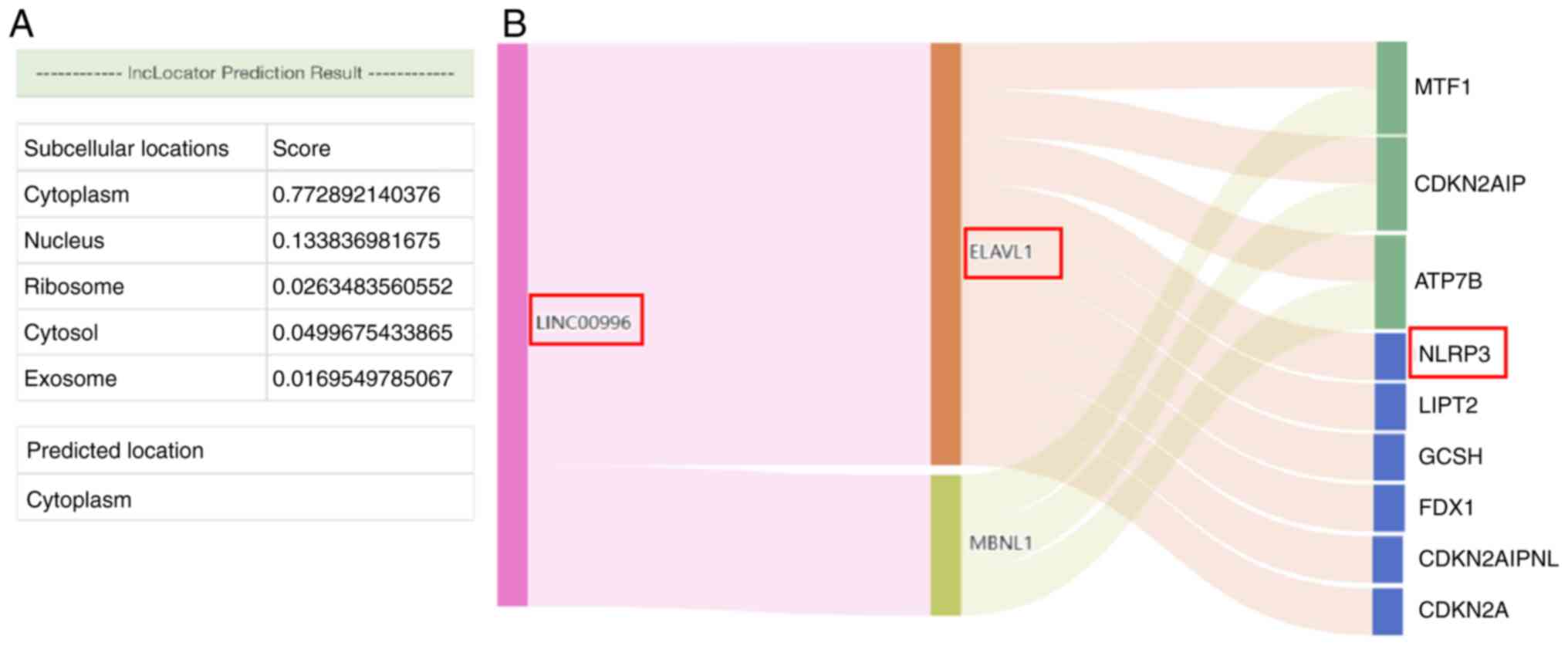
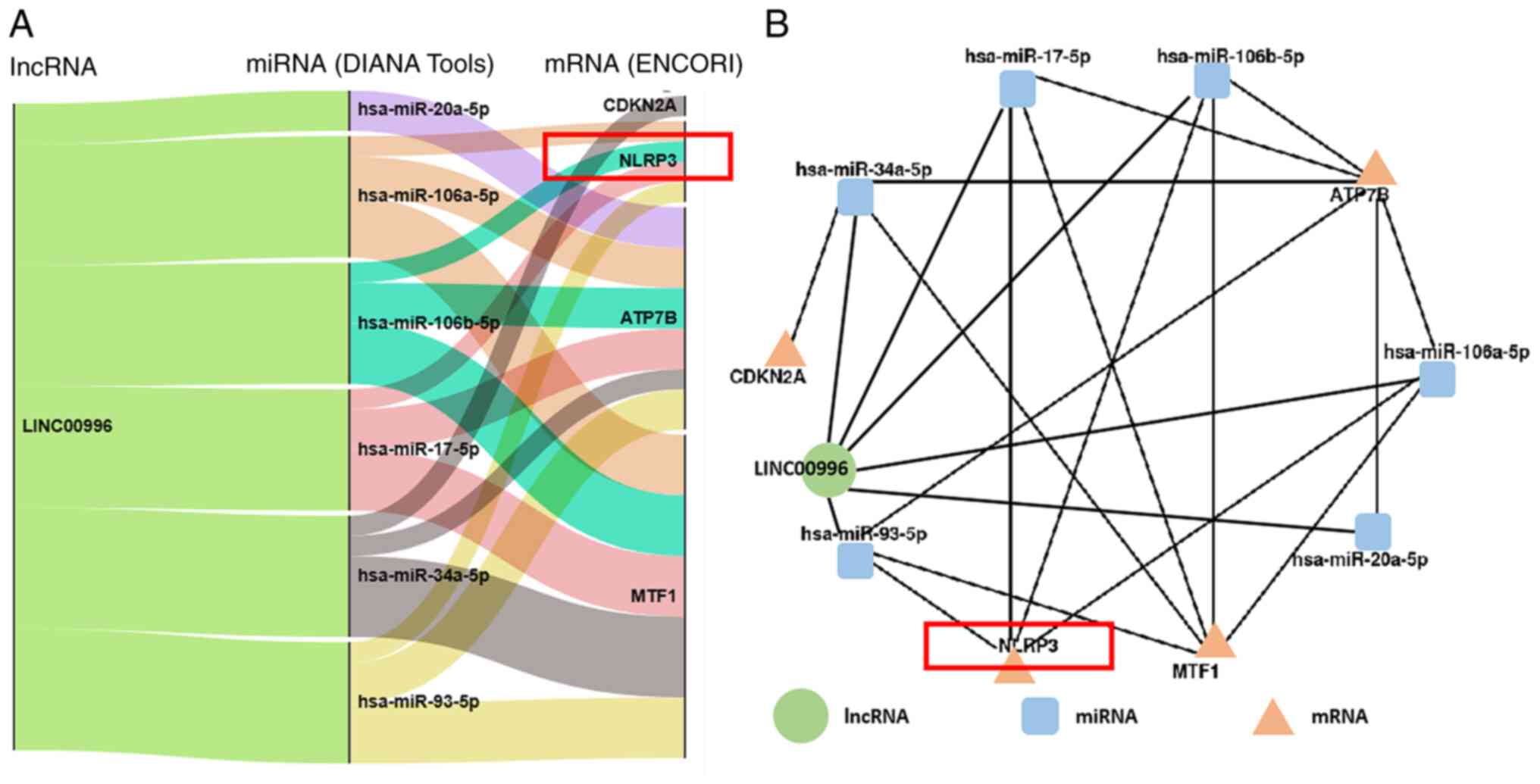
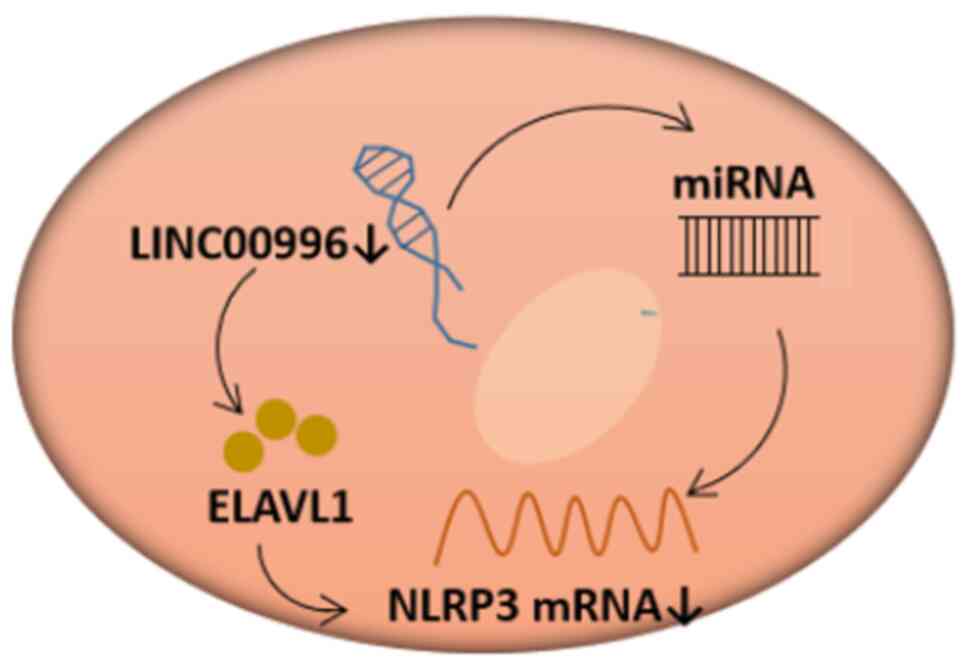
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kahn R, Filippova O, Gordhandas S, An A,
Straubhar AM, Zivanovic O, Gardner GJ, O'Cearbhaill RE, Tew WP,
Grisham RN, et al: Ten-year conditional probability of survival for
patients with ovarian cancer: A new metric tailored to long-term
survivors. Gynecol Oncol. 169:85–90. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh N, Jayraj AS, Sarkar A, Mohan T,
Shukla A and Ghatage P: Pharmacotherapeutic treatment options for
recurrent epithelial ovarian cancer. Expert Opin Pharmacother.
24:49–64. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perren TJ, Swart AM, Pfisterer J,
Ledermann JA, Pujade-Lauraine E, Kristensen G, Carey MS, Beale P,
Cervantes A, Kurzeder C, et al: A phase 3 trial of bevacizumab in
ovarian cancer. N Engl J Med. 365:2484–2496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bi R, Chen L, Huang M, Qiao Z, Li Z, Fan G
and Wang Y: Emerging strategies to overcome PARP inhibitors'
resistance in ovarian cancer. Biochim Biophys Acta Rev Cancer.
1879:1892212024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao J, Sun J, Shuai SC, Zhao Q and Shuai
J: Predicting potential interactions between lncRNAs and proteins
via combined graph auto-encoder methods. Brief Bioinform.
24:bbac5272023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang Y, Cheung BB, Atmadibrata B, Marshall
GM, Dinger ME, Liu PY and Liu T: The regulatory role of long
noncoding RNAs in cancer. Cancer Lett. 391:12–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao ZT, Yang YM, Sun MM, He Y, Liao L,
Chen KS and Li B: New insights into the interplay between long
non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun
(Lond). 42:117–140. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou HL, Luo G, Wise JA and Lou H:
Regulation of alternative splicing by local histone modifications:
Potential roles for RNA-guided mechanisms. Nucleic Acids Res.
42:701–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen XJ and An N: Long noncoding RNA ATB
promotes ovarian cancer tumorigenesis by mediating histone H3
lysine 27 trimethylation through binding to EZH2. J Cell Mol Med.
25:37–46. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu SS, Li JS, Xue M, Wu WJ, Li X and Chen
W: LncRNA UCA1 participates in De Novo synthesis of guanine
nucleotides in bladder cancer by recruiting TWIST1 to increase
IMPDH1/2. Int J Biol Sci. 19:2599–2612. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Haas R, Ganem NS, Keshet A, Orlov A,
Fishman A and Lamm AT: A-to-I RNA editing affects lncRNAs
expression after heat shock. Genes (Basel). 9:6272018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gordon MA, Babbs B, Cochrane DR, Bitler BG
and Richer JK: The long non-coding RNA MALAT1 promotes ovarian
cancer progression by regulating RBFOX2-mediated alternative
splicing. Mol Carcinog. 58:196–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma B, Wang S, Wu W, Shan P, Chen Y, Meng
J, Xing L, Yun J, Hao L, Wang X, et al: Mechanisms of
circRNA/lncRNA-miRNA interactions and applications in disease and
drug research. Biomed Pharmacother. 162:1146722023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou J, Xu Y, Wang L, Cong Y, Huang K, Pan
X, Liu G, Li W, Dai C, Xu P and Jia X: LncRNA IDH1-AS1
sponges miR-518c-5p to suppress proliferation of epithelial ovarian
cancer cell by targeting RMB47. J Biomed Res. 38:51–65. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalkavan H, Rühl S, Shaw JJP and Green DR:
Non-lethal outcomes of engaging regulated cell death pathways in
cancer. Nat Cancer. 6:795–806. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tong X, Tang R, Xiao M, Xu J, Wang W,
Zhang B, Liu J, Yu X and Shi S: Targeting cell death pathways for
cancer therapy: Recent developments in necroptosis, pyroptosis,
ferroptosis, and cuproptosis research. J Hematol Oncol. 15:1742022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsvetkov P, Coy S, Petrova B, Dreishpoon
M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R,
Spangler RD, et al: Copper induces cell death by targeting
lipoylated TCA cycle proteins. Science. 375:1254–1261. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deigendesch N, Zychlinsky A and Meissner
F: Copper regulates the canonical NLRP3 inflammasome. J Immunol.
200:1607–1617. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xue Q, Kang R, Klionsky DJ, Tang D, Liu J
and Chen X: Copper metabolism in cell death and autophagy.
Autophagy. 19:2175–2195. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu S, Ge J, Chu Y, Cai S, Wu J, Gong A
and Zhang J: Identification of hub cuproptosis related genes and
immune cell infiltration characteristics in periodontitis. Front
Immunol. 14:11646672023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun M, Zhan N, Yang Z, Zhang X, Zhang J,
Peng L, Luo Y, Lin L, Lou Y, You D, et al: Cuproptosis-related
lncRNA JPX regulates malignant cell behavior and epithelial-immune
interaction in head and neck squamous cell carcinoma via
miR-193b-3p/PLAU axis. Int J Oral Sci. 16:632024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou G, Chen C, Wu H, Lin J, Liu H, Tao Y
and Huang B: LncRNA AP000842.3 triggers the malignant progression
of prostate cancer by regulating cuproptosis related gene NFAT5.
Technol Cancer Res Treat. 23:153303382412555852024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai Y, Zhang Q, Liu F and Quan J: A novel
cuproptosis-related lncRNA signature predicts the prognosis and
immune landscape in bladder cancer. Front Immunol. 13:10274492022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang H, Chen G, Zhang Z, Wu G, Zhang Z,
Yu A, Wang J, Quan C, Li Y and Zhou M: Deciphering the role of
cuproptosis-related lncRNAs in shaping the lung cancer immune
microenvironment: A comprehensive prognostic model. J Cell Mol Med.
28:e185192024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu H, Lu X, Hu Y, Baatarbolat J, Zhang Z,
Liang Y, Zhang Y, Liu Y, Lv H and Jin X: Biomimic nanodrugs
overcome tumor immunosuppressive microenvironment to enhance
cuproptosis/chemodynamic-induced cancer immunotherapy. Adv Sci
(Weinh). 12:e24111222025. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guan M, Cheng K, Xie XT, Li Y, Ma MW,
Zhang B, Chen S, Chen W, Liu B, Fan JX and Zhao YD: Regulating
copper homeostasis of tumor cells to promote cuproptosis for
enhancing breast cancer immunotherapy. Nat Commun. 15:100602024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu X, Chen X, Lin C, Yi Y, Zhao S, Zhu B,
Deng W, Wang X, Xie Z, Rao S, et al: Elesclomol loaded copper oxide
nanoplatform triggers cuproptosis to enhance antitumor
immunotherapy. Adv Sci (Weinh). 11:e23099842024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen Z, Li X, Hu Z, Yang Y, Yang Z, Li S,
Zhou Y, Ma J, Li H, Liu X, et al: Linc00996 is a favorable
prognostic factor in LUAD: Results from bioinformatics analysis and
experimental validation. Front Genet. 13:9329732022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ge H, Yan Y, Wu D, Huang Y and Tian F:
Potential role of LINC00996 in colorectal cancer: a study based on
data mining and bioinformatics. Onco Targets Ther. 11:4845–4855.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Berek JS, Renz M, Kehoe S, Kumar L and
Friedlander M: Cancer of the ovary, fallopian tube, and peritoneum:
2021 update. Int J Gynaecol Obstet. 155 (Suppl):61–85. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kawase K, Kawashima S, Nagasaki J, Inozume
T, Tanji E, Kawazu M, Hanazawa T and Togashi Y: High expression of
MHC class I overcomes cancer immunotherapy resistance due to IFNγ
signaling pathway defects. Cancer Immunol Res. 11:895–908. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hiltner T, Szörenyi N, Kohlruss M,
Hapfelmeier A, Herz AL, Slotta-Huspenina J, Jesinghaus M, Novotny
A, Lange S, Ott K, et al: Significant tumor regression after
neoadjuvant chemotherapy in gastric cancer, but poor survival of
the patient? Role of MHC class I alterations. Cancers (Basel).
15:7712023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Colbert JD, Cruz FM, Baer CE and Rock KL:
Tetraspanin-5-mediated MHC class I clustering is required for
optimal CD8 T cell activation. Proc Natl Acad Sci USA.
119:e21221881192022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luda KM, Longo J, Kitchen-Goosen SM,
Duimstra LR, Ma EH, Watson MJ, Oswald BM, Fu Z, Madaj Z, Kupai A,
et al: Ketolysis drives CD8+ T cell effector function through
effects on histone acetylation. Immunity. 56:2021–2035.e8. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stanifer ML, Guo C, Doldan P and Boulant
S: Importance of type I and III interferons at respiratory and
intestinal barrier surfaces. Front Immunol. 11:6086452020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen L, Min J and Wang F: Copper
homeostasis and cuproptosis in health and disease. Signal Transduct
Target Ther. 7:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
O'Day S, Gonzalez R, Lawson D, Weber R,
Hutchins L, Anderson C, Haddad J, Kong S, Williams A and Jacobson
E: Phase II, randomized, controlled, double-blinded trial of weekly
elesclomol plus paclitaxel versus paclitaxel alone for stage IV
metastatic melanoma. J Clin Oncol. 27:5452–5458. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF,
Shan L, Zhang J, Wang S, Wang Y, Carmichael GG, et al: Distinct
processing of lncRNAs contributes to non-conserved functions in
stem cells. Cell. 181:621–636.e22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Q, Li G, Ma X, Liu L, Liu J, Yin Y,
Li H, Chen Y, Zhang X, Zhang L, et al: LncRNA TINCR impairs the
efficacy of immunotherapy against breast cancer by recruiting DNMT1
and downregulating MiR-199a-5p via the STAT1-TINCR-USP20-PD-L1
axis. Cell Death Dis. 14:762023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Priyanka P, Sharma M, Das S and Saxena S:
The lncRNA HMS recruits RNA-binding protein HuR to stabilize the
3′-UTR of HOXC10 mRNA. J Biol Chem. 297:1009972021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Latorre E, Carelli S, Raimondi I,
D'Agostino V, Castiglioni I, Zucal C, Moro G, Luciani A, Ghilardi
G, Monti E, et al: The ribonucleic complex HuR-MALAT1 represses
CD133 expression and suppresses epithelial-mesenchymal transition
in breast cancer. Cancer Res. 76:2626–2636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Brennan CM and Steitz JA: HuR and mRNA
stability. Cell Mol Life Sci. 58:266–277. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fu XD: RNA editing: New roles in feedback
and feedforward control. Cell Res. 33:495–496. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hildebrandt RP, Moss KR, Janusz-Kaminska
A, Knudson LA, Denes LT, Saxena T, Boggupalli DP, Li Z, Lin K,
Bassell GJ, et al: Muscleblind-like proteins use modular domains to
localize RNAs by riding kinesins and docking to membranes. Nat
Commun. 14:34272023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
González ÀL, Fernández-Remacha D, Borrell
JI, Teixidó J and Estrada-Tejedor R: Cognate RNA-binding modes by
the alternative-splicing regulator MBNL1 inferred from molecular
dynamics. Int J Mol Sci. 23:161472022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu Y, Wei W, Wang Y, Wan C, Bai Y, Sun X,
Ma J and Zheng F: TNF-α/calreticulin dual signaling induced NLRP3
inflammasome activation associated with HuR nucleocytoplasmic
shuttling in rheumatoid arthritis. Inflamm Res. 68:597–611. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jeyabal P, Thandavarayan RA, Joladarashi
D, Suresh Babu S, Krishnamurthy S, Bhimaraj A, Youker KA, Kishore R
and Krishnamurthy P: MicroRNA-9 inhibits hyperglycemia-induced
pyroptosis in human ventricular cardiomyocytes by targeting ELAVL1.
Biochem Biophys Res Commun. 471:423–429. 2016. View Article : Google Scholar : PubMed/NCBI
|