Introduction
Among gynecological malignancies, ovarian cancer
(OV) exhibits the highest mortality rate and constitutes a
significant hazard to the health of female patients (1). Specifically, there were 21,750 new OV
cases and 13,940 deaths from OV in the United States in 2020
(2), and there were 57,090 new
cases of OV and 39,306 deaths from OV in China in 2022 (3). Due to its deep pelvic location, lack
of typical symptoms and lack of early screening tools,
approximately two-thirds of patients have progressed to the
advanced stage of disease at diagnosis, increasing the difficulty
of treatment (4,5). Patients with stage I/II OV have a
10-year survival rate of 29–75%, whereas the 10-year survival rate
drops to 6.9–22% for those with stage III–IV OV (6). Furthermore, >70% of patients with
OV relapse within 2–3 years (7,8).
At present, the treatment of OV is multidisciplinary
and comprehensive, which includes surgery, chemotherapy, targeted
therapy and other therapeutic methods (5,9).
However, the clinical effectiveness of these treatments is often
compromised by drug resistance, cancer recurrence and immune escape
(4,9). Consequently, there is an urgent need
to identify novel molecular targets to improve the assessment of
the progression of OV and enhance the available therapies.
Long non-coding (lnc)RNA refers to a category of RNA
molecules whose transcript length is >200 nt (10). Due to the lack of conservative open
reading frames, most lncRNAs are incapable of direct translation
into proteins (11–13). However, lncRNA can recruit
chromatin-remodeling complexes to regulate chromosome structure and
modification (for instance, DNA methylation and histone
modification), thus affecting gene expression levels (14,15).
Chen and An (16) reported that the
expression of lncRNA activated by TGF-β (ATB) is elevated in OV
tissues compared with adjacent normal tissues, and it is linked to
unfavorable outcomes. Furthermore, ATB facilitated the
proliferation, invasion and migration of OV cells by mediating
histone H3 lysine 27 trimethylation through binding to enhancer of
zeste homolog 2 (16). lncRNAs also
possess the capacity to regulate the transcription of target genes
through interacting with transcriptional regulatory molecules
(17–19). Moreover, lncRNA can bind RNA binding
protein (RBP) to participate in alternative splicing, mRNA
metabolism and transport (17,18,20).
Gordon et al (21) reported
that high metastasis associated lung adenocarcinoma transcript 1
(MALAT1) expression is associated with increased stage, recurrence
and decreased survival in OV, and that MALAT1 promotes OV
progression by promoting the expression of RNA binding fox-1
homolog 2 and inhibiting preferential splicing of the pro-apoptotic
isoform of kinesin family member 1b. Furthermore, lncRNA can
function as a micro (mi)RNA sponge that reduces the activity of
miRNA, thus enhancing the expression level of target genes
(22). Zhou et al (23) reported that the expression levels of
lncRNA isocitrate dehydrogenase 1 antisense RNA 1 (IDH1-AS1) are
lower in epithelial OV cells than in normal ovarian epithelial
cells, and IDH1-AS1 expression indicates a favorable prognosis.
Additionally, IDH1-AS1 serves as a sponge for miR-518c-5p to
suppress the proliferation of epithelial OV cells by targeting RNA
binding motif protein 47 (23). In
conclusion, lncRNAs serve a vital role in the progression of
malignant tumors.
Regulated cell death (RCD) is a mode of cell death
based on precise signaling networks and molecular mechanisms, and
includes apoptosis, pyroptosis, necroptosis and ferroptosis
(24,25). Recently, Tsvetkov et al
(26) reported a new
copper-dependent RCD, termed cuproptosis. Excess intracellular
copper can interact with lipid-acylated proteins, which triggers
irregular clustering of lipid-acylated proteins and depletion of
iron-sulfur (Fe-S) clusters, ultimately culminating in cuproptosis
(26). In addition, elesclomol (a
potent copper ionophore) combined with copper can induce
cuproptosis, whilst tetrathiopolybdate (a copper chelator) can
inhibit cuproptosis, and cells with higher acylated protein level
are more sensitive to cuproptosis (26). Ferrodoxin 1 functions as a vital
positive regulator in cuproptosis, and other genes have also been
identified as being closely related to cuproptosis (26–29).
For instance, NLR family pyrin domain containing 3 (NLRP3) can be
activated by intracellular copper, which can lead to cell death,
whilst tetrathiopolybdate can inhibit NLRP3 activation (27). Furthermore, certain studies have
reported that certain lncRNAs can regulate cuproptosis-related
genes and are thereby considered novel potential targets in
cuproptosis (30–33). For instance, LINC00996 has been
identified as the key cuproptosis-related lncRNA in lung and
bladder cancer (32,33). An in-depth study of cuproptosis
could provide the ideal risk model and an accurate biomarker for
malignant tumors; however, the function and underlying regulatory
pathways of cuproptosis in OV are still poorly studied.
Therefore, the present study aimed to construct a
prognostic risk model based on lncRNAs associated with cuproptosis
in OV and to explore the regulatory mechanism of LINC00996 in
cuproptosis in OV.
Materials and methods
Data collection and processing
The present study accessed the RNA sequencing
(RNA-seq) data and clinical records of patients with OV from The
Cancer Genome Atlas (TCGA)-OV dataset (https://portal.gdc.cancer.gov). The RNA-seq data was
in fragments per Kilobase per Million format. The clinical records
obtained included the age, stage, grade, survival status and
survival time of the patients. A total of 378 patients with OV with
complete data were randomly allocated to two groups, a training set
(n=189) and a validation set (n=189), based on
‘createDataPartition’ package
(rdocumentation.org/packages/caret/versions/6.0–94/topics/createDataPartition)
in R programming (version 4.1.1). ‘createDataPartition’ uses the
following basic syntax: createDataPartition (y, p=0.5, list=FALSE,
…), with y representing the vector of outcomes and prepresenting
percentage of data to use in the training set. The remaining data
formed the validation set.
Identification of cuproptosis-related
lncRNAs
Using the keywords ‘cuproptosis,’ ‘copper’ and
‘regulated cell death’ to search the PubMed (https://pubmed.ncbi.nlm.nih.gov/) and Web of Science
(webofscience.com/) databases, articles related to cuproptosis were
screened. Subsequently, by carefully reading the relevant articles,
the cuproptosis-related genes and their functions were summarized.
In addition, relevant reviews were consulted to identify other
cuproptosis-related genes and their functions. Finally, 15
cuproptosis-related genes were obtained from previous studies
(26–29). ENSEMBL (Release 111) (ensembl.org)
was used to extract lncRNAs from the RNA-seq data. Subsequently,
using the ‘limma’ packages (version 3.48.3) in R programming
(version 4.1.1) and the Wilcoxon rank sum test, the Pearson
correlation coefficients between lncRNA and cuproptosis-related
genes were calculated.. The cut-off criteria included correlation
coefficient (r) of >0.4 or <-0.4) and P<0.001. Additional
notes that correlation coefficient |r| <0.4 indicated a weak
correlation, 0.4 ~ 0.7 indicated a moderate correlation, and
>0.7 indicated a strong correlation. Finally, a Sankey diagram
was drawn based on the ‘dplyr (version 1.0.7)’, ‘ggplot2 (version
3.4.0)’ and ‘ggalluvial (version 0.12.3)’ packages in R programming
(version 4.1.1).
Construction of the risk model
There was no discernible difference in the clinical
characteristics between the training and the validation sets.
Univariate Cox regression was performed to identify
cuproptosis-related lncRNAs associated with overall survival (OS)
in the training set. Among these lncRNAs, the risk model was
constructed using least absolute shrinkage selection operator
(LASSO) regression analysis and multivariate Cox regression. The
risk score was calculated as follows: Risk score =
∑nk=1 coef(lncRNAk) ×
expr(lncRNAk), where coef(lncRNA) represents the link
between the lncRNA and the survival of the patient with OV, and
expr(lncRNA) represents the expression level of the lncRNA. The
analysis was performed using R programming (version 4.1.1).
Identification of the prognostic
signature
Patients with a risk score >1 were considered
high-risk, while those with a risk score ≤1 were considered
low-risk, and the survival status and immune function in the low-
and high-risk groups were analyzed. Univariate and multivariate Cox
regression analysis were performed to identify independent
predictors. Kaplan-Meier survival curves were then produced to
compare the difference in the survival rate between the low- and
high-risk groups using the ‘survival (version 3.4.0)’ and
‘survminer (version 0.4.9)’ packages. Furthermore, receiver
operating characteristic (ROC) curves, calibration curves, C-index
and principal component analysis (PCA) were performed. The analysis
was carried out by R programming (version 4.1.1).
Cell culture
OVCAR3 cells were acquired from the American Type
Culture Collection and cultured in Roswell Park Memorial
Institute-1640 medium with 10% fetal bovine serum (FBS; both Gibco,
Thermo Fisher Scientific Corporation). The cells were incubated in
an environment that mimicked physiological conditions, with an
atmosphere containing 5% CO2 to maintain an optimal pH
level and ensure proper gas exchange, while being maintained at a
constant temperature of 37°C.
Transfection of small interference
(si)RNA
The LINC00996 siRNA (siLINC00996) and control siRNA
(siControl) were purchased from Shanghai GenePharma Co., Ltd. When
the cell density reached 60–75%, the transfection solution was
configured as follows: Solution A, 250 µl OPTI-MEM (Gibco, Thermo
Fisher Scientific, Inc.) with 5 µl Lipo3000 (Invitrogen; Thermo
Fisher Scientific Corporation); and solution B, 250 µl OPTI-MEM
with 50 nM siRNA (Genepharma Corporation), which were both
incubated at room temperature for 5 min. Subsequently, solutions A
and B were mixed and incubated at room temperature for 20 min.
Then, the medium was removed from a 6-well plate containing OVCAR3
cells, and 1500 µl serum-free medium and 500 µl mixed solution was
added at room temperature. Finally, After incubation for 12 h, it
was replaced with fresh medium and further cultured in an incubator
with 5% CO2 at 37°C for 48 h. The sequences of the siRNAs used were
as follows: siLINC00996-1, 5′-GCUGUGUGAAAGGGUUUAATT-3′ and
5′-UUAAACCCUUUCACACAGCTT-3′; siLINC00996-2,
5′-CCGGCCUUAUUGUUUCUAUTT-3′ and 5′-AUAGAAACAAUAAGGCCGGTT-3′; and
siControl, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
5′-ACGUGACACGUUCGGAGAATT-3′.
Extraction of RNA and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Extraction of total RNA from OVCAR3 cells was
performed using the RNA Extraction Kit (Vazyme Biotech Co., Ltd.)
according to the manufacturer's protocol. RT was performed to
convert RNA into cDNA with HiScript® II Q RT SuperMix
(Vazyme Biotech Co., Ltd.) according to the manufacturer's
protocol. RNA expression was measured by ChamQ Universal SYBR qPCR
Master Mix (Vazyme Biotech Co., Ltd.) with ABI ViiA™ 7 real-time
fluorescence quantitative PCR instrument equipped with ViiATM7
system. Thermocycling conditions are as follows: Pre-denaturation
at 95°C for 3 min, then 95°C for 10 sec, 60°C for 30 sec and 95°C
for 15 sec as 1 cycle for 40 cycles. The relative gene expression
was calculated by 2−∆∆Cq method with GAPDH as the
control (34). The primers (Tsingke
Corporation) used were as follows: LINC00996 forward,
5′-CTCTGCCACATCGTTCGGTTC-3′ and reverse
5′-CTTCTTACGCTGCCAACTGCTAA-3′; and GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Cell proliferation and migration
assay
A Cell Counting Kit-8 (CCK-8) assay was performed to
measure proliferative ability. OVCAR3 cells were cultured in a
96-well plate. After adding CCK-8 solution (Beyotime Institute of
Biotechnology) for 1 h, the absorbance was measured at 490 nm based
on microplate reader.
A Transwell assay was the performed to evaluate
migration ability. A cell suspension including 2×10^5 cells without
FBS was added to the upper chamber and medium with 20% FBS was
added to the lower chamber of the Transwell plate. The cells were
incubated at 37°C for 24 h. When the cells passed through the
membrane, 4% paraformaldehyde and crystal violet were added to the
upper chamber for 15 min at room temperature. The results were
observed under an inverted optical microscope (Shanghai Optical
Instrument Factory) and counted using ImageJ 1.8.0 software
(National Institutes of Health).
Sensitivity to cuproptosis
Elesclomol can facilitate the transport of
Cu2+ into cells. After entering the cells,
Cu2+ directly binds to lipoylated enzymes and prompts
the aggregation of lipoylated proteins and the dissipation of Fe-S
cluster proteins, leading to proteotoxic stress and ultimately
cuproptosis (26). Therefore,
cuproptosis was induced by adding elesclomol-CuCl2
(Medchemexpress Corporation; 1×10−9 M to 10−4
M into the medium of OVCAR3 cells in a 5% CO2 incubator
at 37°C for 4 h. CCK-8 solution (Beyotime Institute of
Biotechnology) was added for 1 h to measure the number of living
cells and thus assess the sensitivity of cells to cuproptosis, in
which the absorbance was measured at 490 nm based on microplate
reader.
Prediction of RBPs and target
mRNA
The RBPs that can interact with LINC00996 were
predicted using the ENCORI database (https://rnasysu.com/encori/). The filtering condition
was CLIP Data ≥1 (results were supported by at least one
cross-linking immunoprecipitation. Based on ENCORI, the target
mRNAs of the aforementioned RBPs were predicted. Because of the
large number of predicted target mRNAs, we set more stringent
filtering criteria as follows: CLIP Data was ≥4 and the number of
pan-cancer was ≥10, which meant the results were supported by ≥4
cross-linking immunoprecipitation and were validated in at least 10
cancer types. In addition, expression of target lncRNA in ovarian
cancer patients was analyzed using the Gene Expression Profiling
Interactive Analysis (GEPIA; gepia.cancer-pku.cn/), and locations
of target lncRNA are predicted by lncLocator
(csbio.sjtu.edu.cn/bioinf/lncLocator/).
Prediction of miRNA and target
mRNA
The miRNAs that can interact with LINC00996 were
identified using DIANA Tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php).
Furthermore, ENCORI was used to predict the target genes of the
aforementioned miRNAs.
Statistical analysis
Statistical analyses were performed using R
programming (version 4.1.1) (https://www.r-project.org/about.html) and GraphPad
Prism 9 (version 9.0.0; Dotmatics). Statistical differences for
three-group comparisons were determined using one-way ANOVA with
Tukey's post hoc test. The CCK-8 assay data were analyzed two-way
ANOVA with Tukey's post hoc test. All P-values were two-sided and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of cuproptosis-related
lncRNAs with prognostic significance
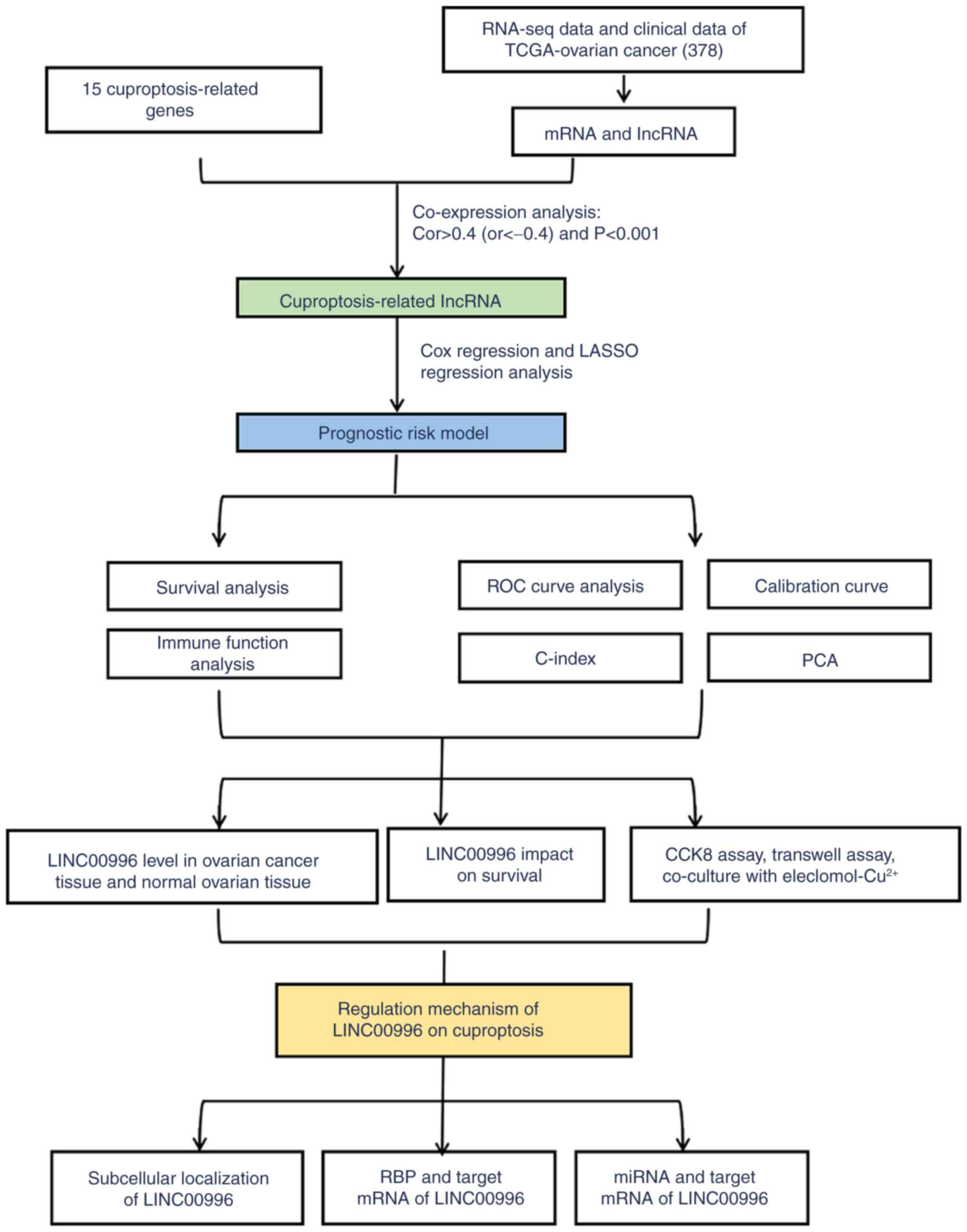
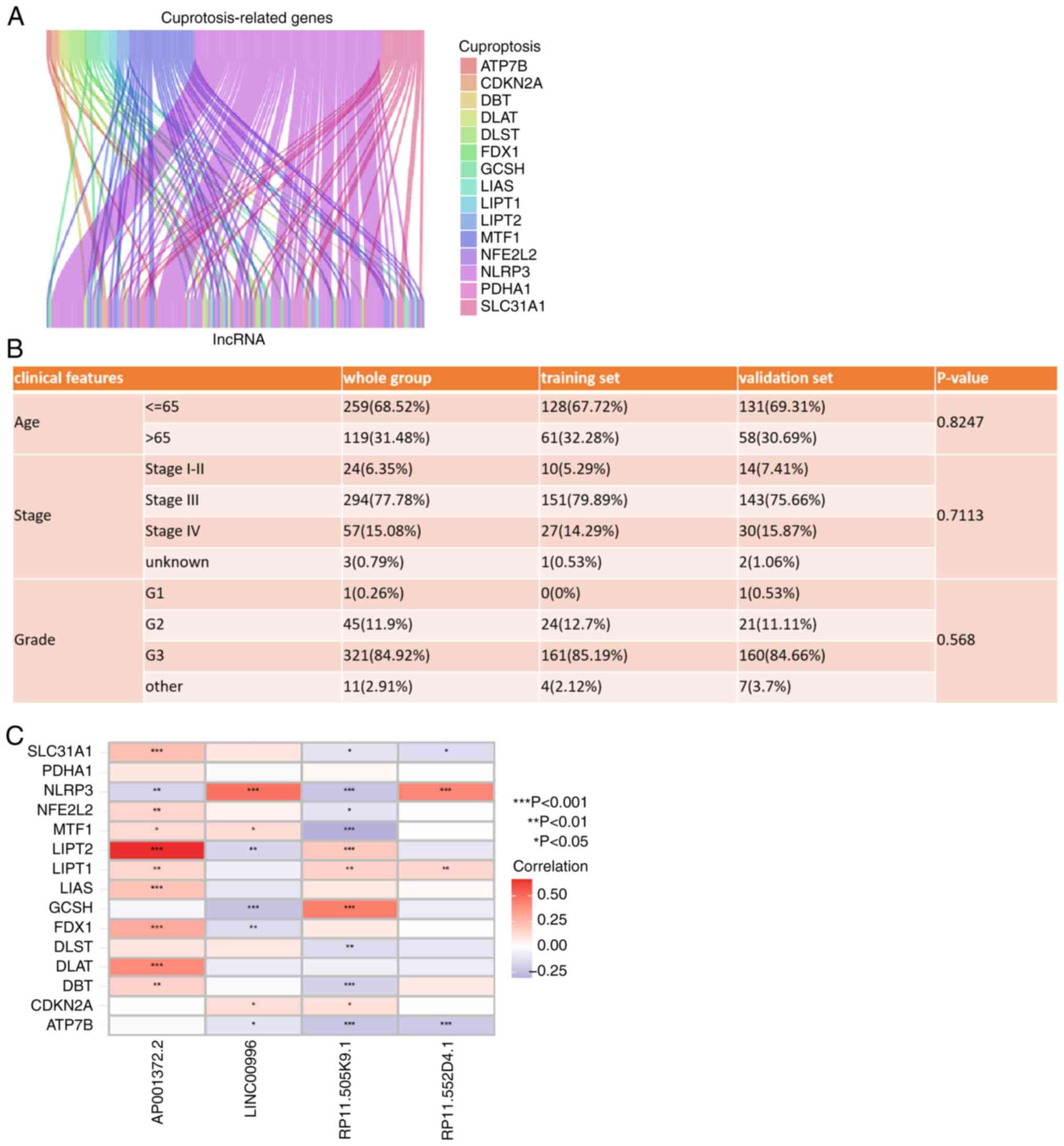
The study flowchart is shown in Fig. 1. Based on previous articles and
reviews related to cuproptosis, 15 cuproptosis-related genes were
identified (Table SI) (26–29).
Subsequently, 14,831 lncRNAs were obtained from TCGA-OV cohort
through the ENSEMBL database. Pearson correlation analysis was
performed to assess the correlation between cuproptosis-related
genes and lncRNAs based on the ‘limma (version 3.48.3)’ packages
and the Wilcoxon rank sum test. The criteria for
cuproptosis-related lncRNAs were r>0.4 (or <-0.4) and
P<0.001. Finally, 140 lncRNAs were identified as
cuproptosis-related lncRNAs (Fig.
2A). The RNA-seq data and clinical records of 378 patients with
OV were collected from TCGA-OV dataset. These patients with OV were
randomly divided into a training set (n=189) and a validation set
(n=189), both of which had similar clinical characteristics
(Fig. 2B). Univariate Cox
regression analysis was performed to identify the
cuproptosis-related lncRNAs with prognostic significance in the
training set. Among these lncRNAs, a risk model was constructed
using LASSO regression and multivariate Cox regression. The risk
model was calculated as the following formula: Risk = (0.687927022
× RP11-552D4.1) - (0.659783022 × AP001372.2) - (0.652465319 ×
RP11-505K9.1) - (1.627006889 × LINC00996). Among the lncRNAs
identified, RP11-552D4.1 was found to be a risk factor, while
AP001372.2, RP11-505K9.1 and LINC00996 were protective factors. The
correlation between these 4 lncRNAs and the identified
cuproptosis-related genes is shown in Fig. 2C. LINC00996 with the highest
correlation coefficient in the risk model is significantly
positively correlated with NLRP3 (P<0.001).
Predictive ability of the risk
model
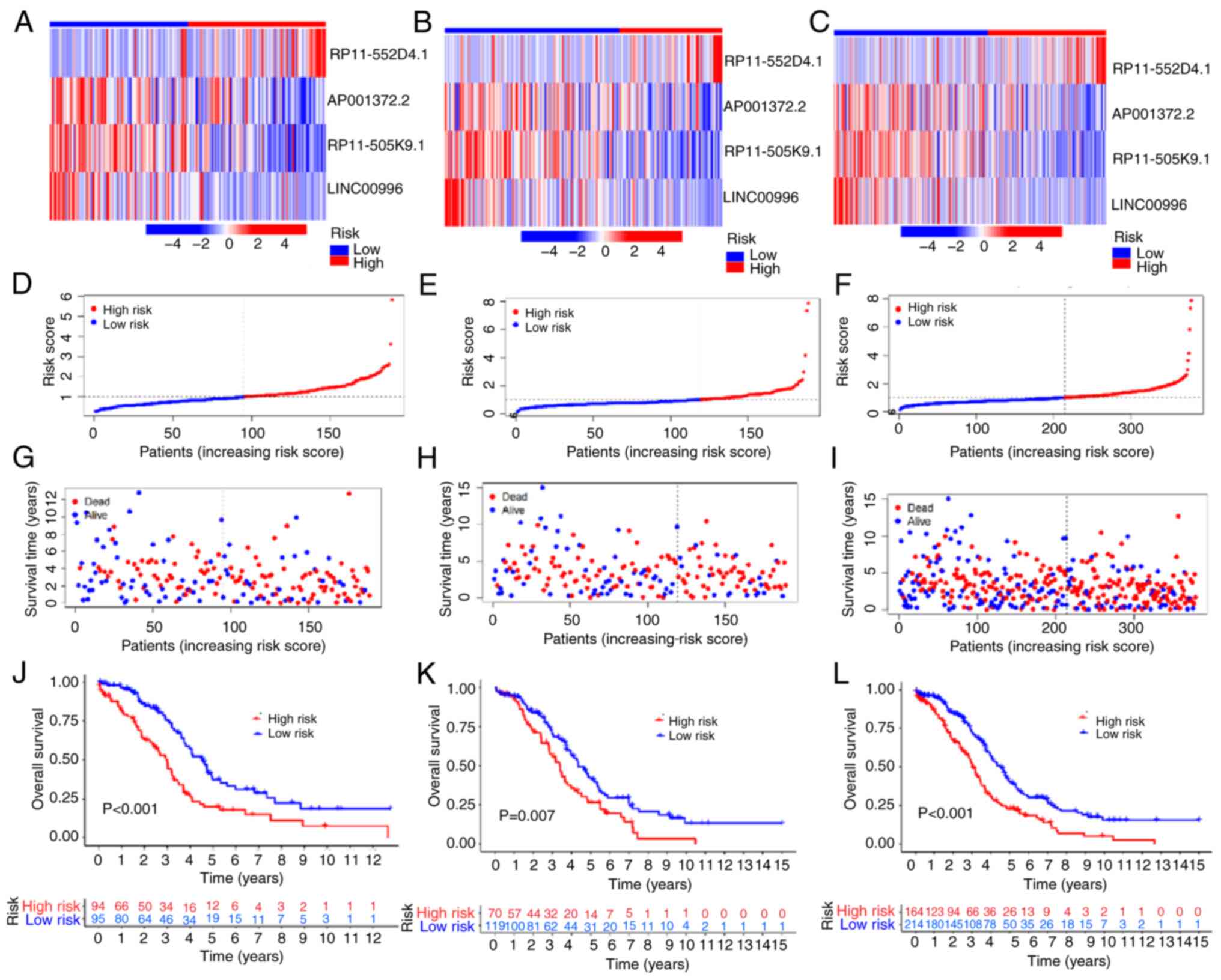
The expression of 4 lncRNAs (RP11.552D4.1,
AP001372.2, RP11.505K9.1 and LINC00996) in the training set,
validation set and whole group are shown using heat maps in
Fig. 3A-C. Patients with a risk
score of >1 were considered high-risk, while those with a risk
score of ≤1 were considered low-risk. (Fig. 3D-F). Compared with the low-risk
group, more deaths were observed in the high-risk group (Fig. 3G-I) in the training and validation
set and whole group. Kaplan-Meier survival curves demonstrated that
the prognosis of patients with OV in the low-risk group was
superior compared with the high-risk group (training set,
P<0.001; validation set, P=0.007; whole group, P<0.001;
Fig. 3J-L). These results suggested
that the risk model is reliable in predicting the prognosis of
patients with OV.
Immune function analysis of the risk
model
There is an association between cuproptosis and
immune function (35). Excessive
intracellular copper can lead to cuproptosis, which subsequently
triggers immunogenic cell death and activates antitumor immune
responses, thereby reprogramming the immunosuppressive tumor
microenvironment (35,36). Moreover, cuproptosis-inducing drugs
can markedly inhibit tumor growth and hold promising prospects for
broad clinical application (35,36).
Recent research has developed nanomedicines that can induce
cuproptosis, which can notably inhibit tumor growth and promote
immune responses, and in combination with programmed cell death 1
immunotherapy, they can markedly enhance antitumor efficacy
(37). Thus, a thorough
investigation into the association between cuproptosis and immune
function holds promise for refining immunotherapy and enhancing
antitumor efficacy in OV.
Based on immune function analysis, the generated
heat map demonstrated that the high- and low-risk groups differed
significantly in their immune functions (Fig. 4). The high-risk group had poorer
immune functions in major histocompatibility complex (MHC)-class-I,
type I interferon (IFN) response, chemokine receptor (CCR),
antigen-presenting cell (APC) co-inhibition, parainflammation,
human leukocyte antigen (HLA), cytolytic activity,
inflammation-promoting, T cell co-stimulation, check-point and T
cell co-inhibition. Therefore, the changes in immune function in
the high-risk group may result in the progression of patients with
OV.
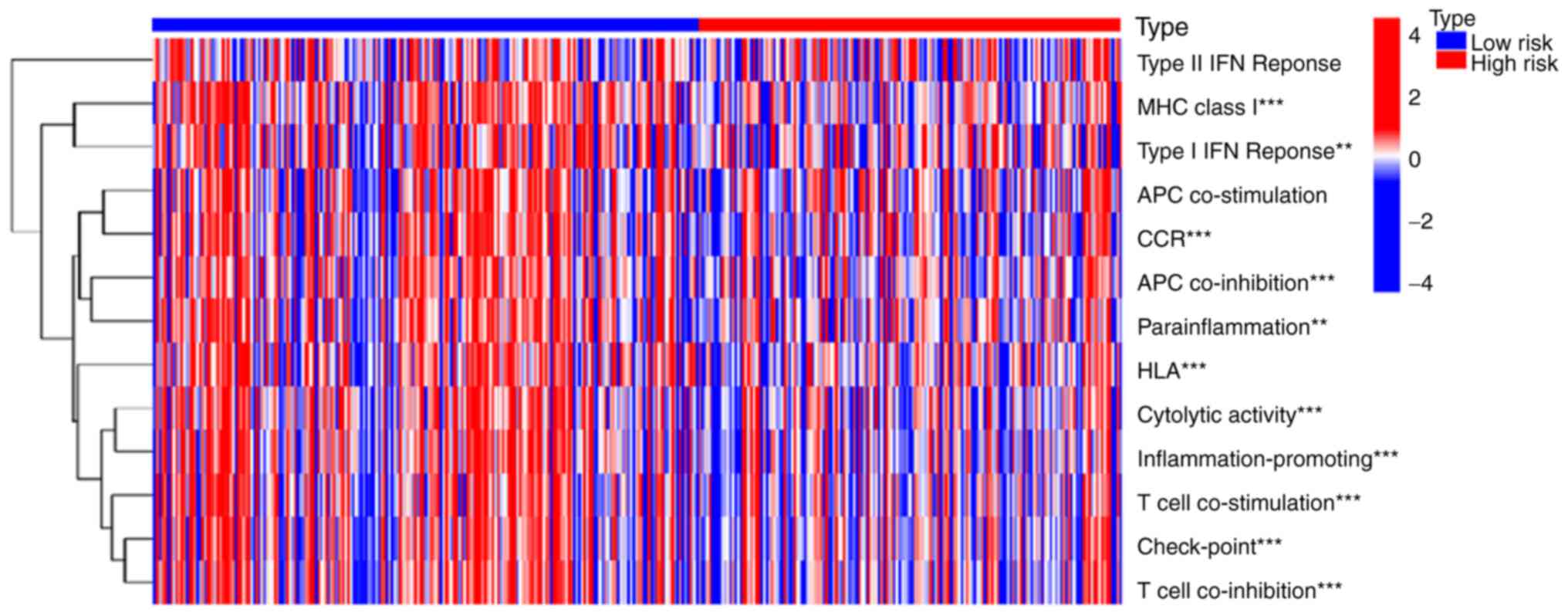 | Figure 4.Immune function analysis of the risk
model. The results showed that the high-risk group had poorer
immune functions in MHC-class-I, type I IFN response, CCR, APC
co-inhibition, parainflammation, HLA, cytolytic activity,
inflammation-promoting, T cell co-stimulation, check-point and T
cell co-inhibition. **P<0.01 and ***P<0.001. MHC, major
histocompatibility complex; IFN, interferon; CCR, chemokine
receptor; APC, antigen-presenting cell; HLA, human leukocyte
antigen. All the original data came from TCGA-OV cohort. |
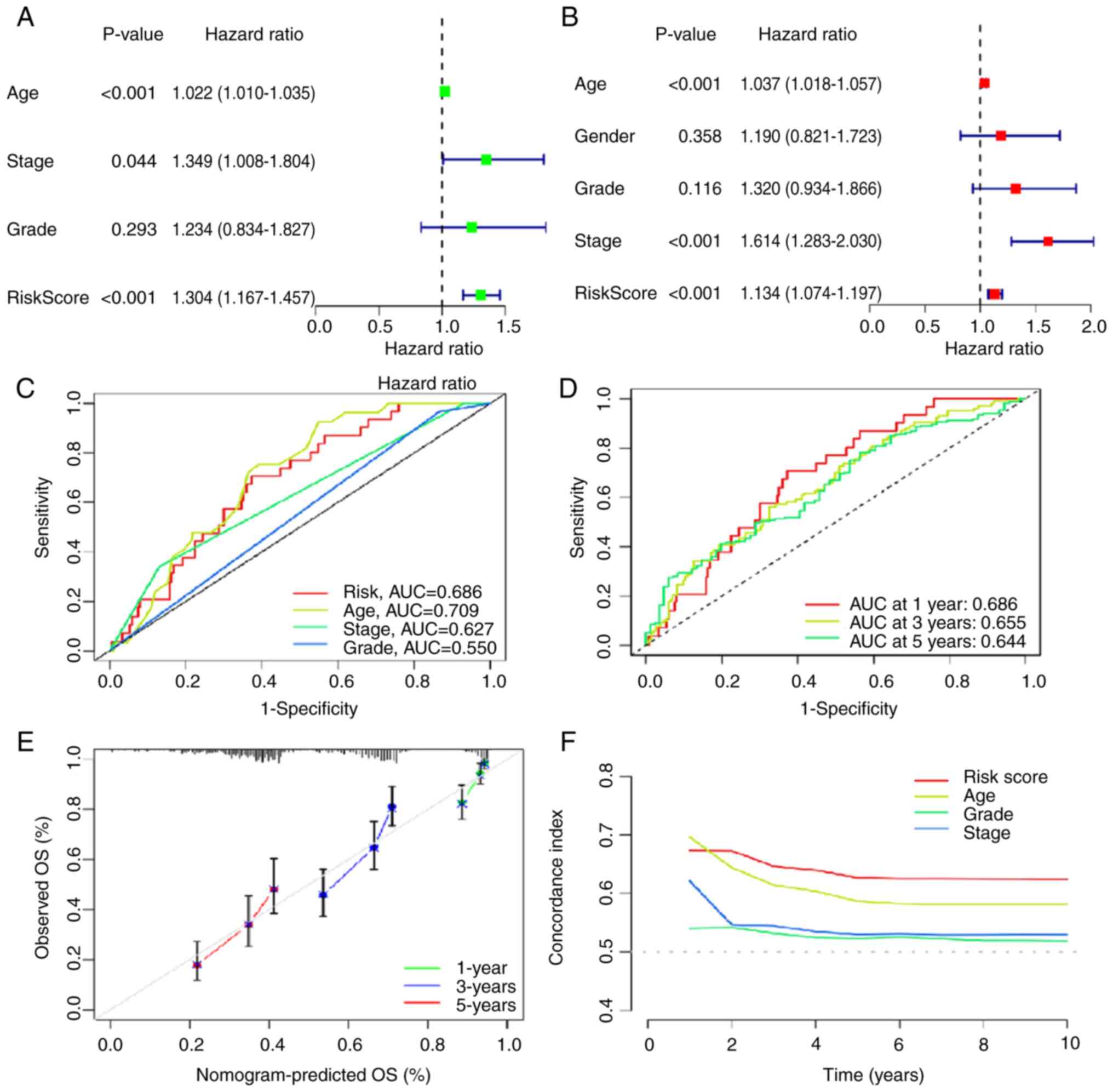
Clinical significance of the risk
model
Using univariate and multivariate Cox regression,
the risk model was identified as an independent prognostic risk
factor in OV (Fig. 5A and B).
Additionally, the area under curve (AUC) of the ROC was calculated,
which measured the prediction accuracy of the risk model. The
closer the AUC value was to 1, the better the differentiation of
the risk model and the higher the prediction accuracy of the risk
model. As shown in Fig. 5C, the AUC
of the risk model was 0.686, which demonstrated good
differentiation and prediction accuracy. Moreover, the prognostic
predictions of the risk model displayed good predictive performance
for OS in patients with OV (1-year, AUC=0.686; 3-year, AUC=0.655;
and 5-year, AUC=0.644; Fig. 5D).
The calibration curve for 1-, 3- and 5-year OS also showed reliable
predictive ability of the risk model (Fig. 5E) and the C-index of the risk model
outranked that of other factors (Fig.
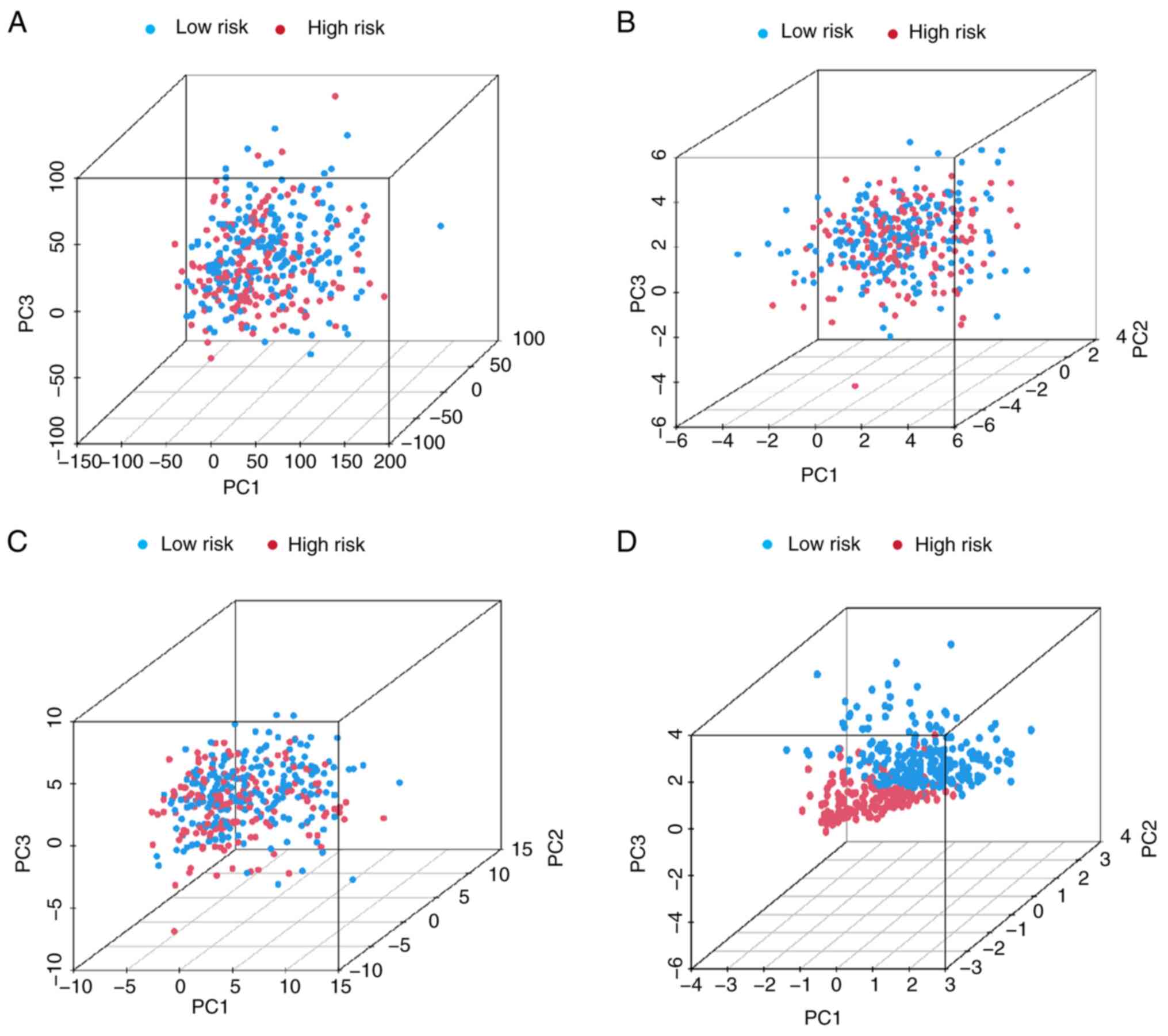
5F). Furthermore, PCA was performed based on all genes
(Fig. 6A), the cuproptosis-related
gene set (Fig. 6B), the
cuproptosis-related lncRNA set (Fig.
6C) and the risk model (Fig.
6D). There were markedly different distribution of the high-
and low-risk groups in the PCA based on the risk model, suggesting
that the risk model may serve as a promising biomarker for risk
stratification. In summary, the risk model could accurately predict
prognosis and has notable clinical value.
Effect of LINC00996 in patients with
OV
In the risk model, the correlation coefficient of
LINC00996 was the highest compared with the other lncRNAs,
indicating that it may serve a crucial role in cuproptosis in OV.
Certain studies have reported the function of LINC00996 in several
malignant tumors, such as colorectal cancer and lung
adenocarcinoma, yet its potential role and importance in OV remain
incompletely understood (38,39).
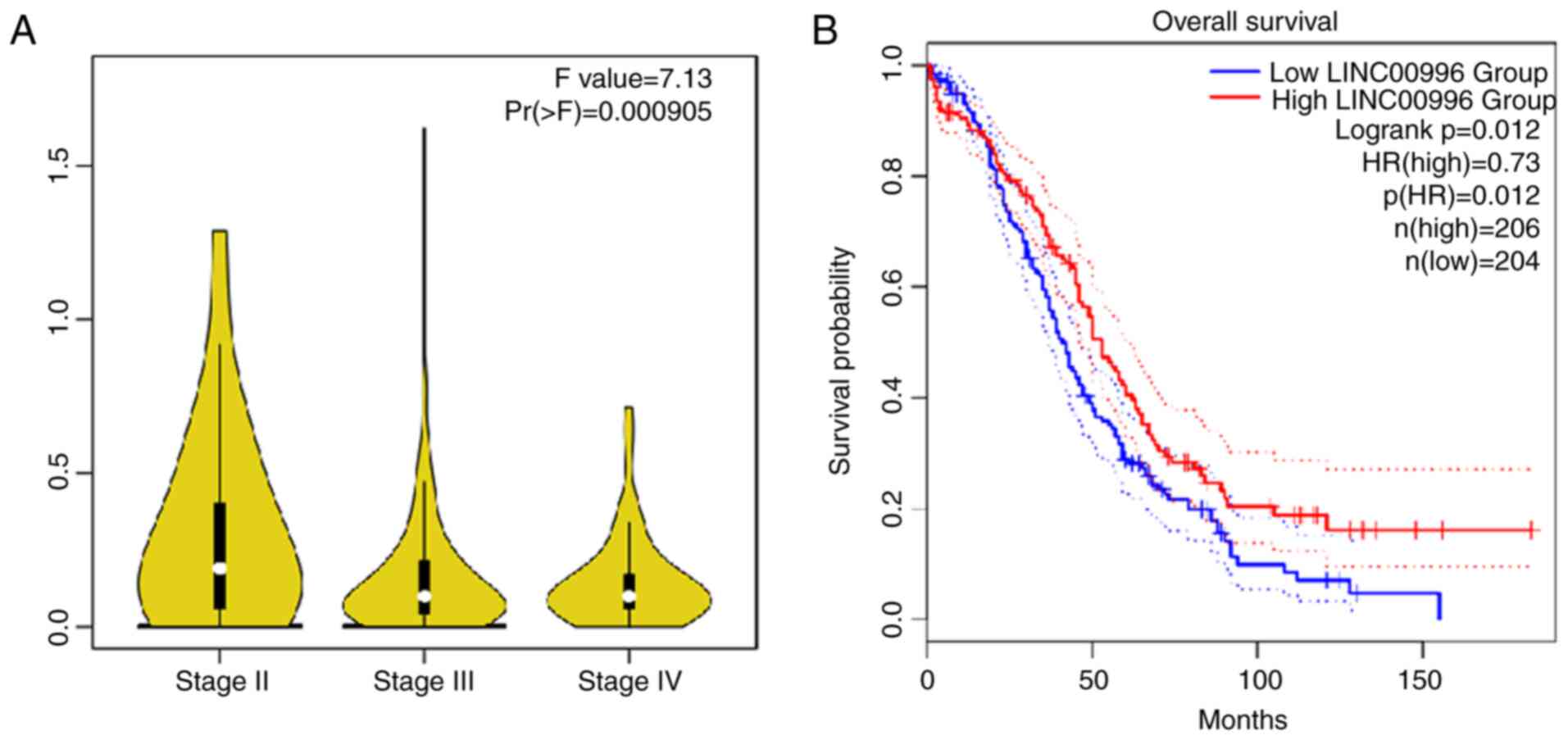
Furthermore, data from GEPIA suggested that the LINC00996
expression level is lower in patients with OV with a higher
International Federation of Gynecology and Obstetrics (FIGO) stage
compared with those with a lower FIGO stage (40) (Fig.
7A). Kaplan-Meier survival curves also demonstrated that the
lower the expression of LINC00996, the lower the survival rate of
patients with OV (Fig. 7B). These
results suggest that LINC00996 may serve an inhibitory role in
OV.
Effect of LINC00996 on the biological
behavior of OV cells
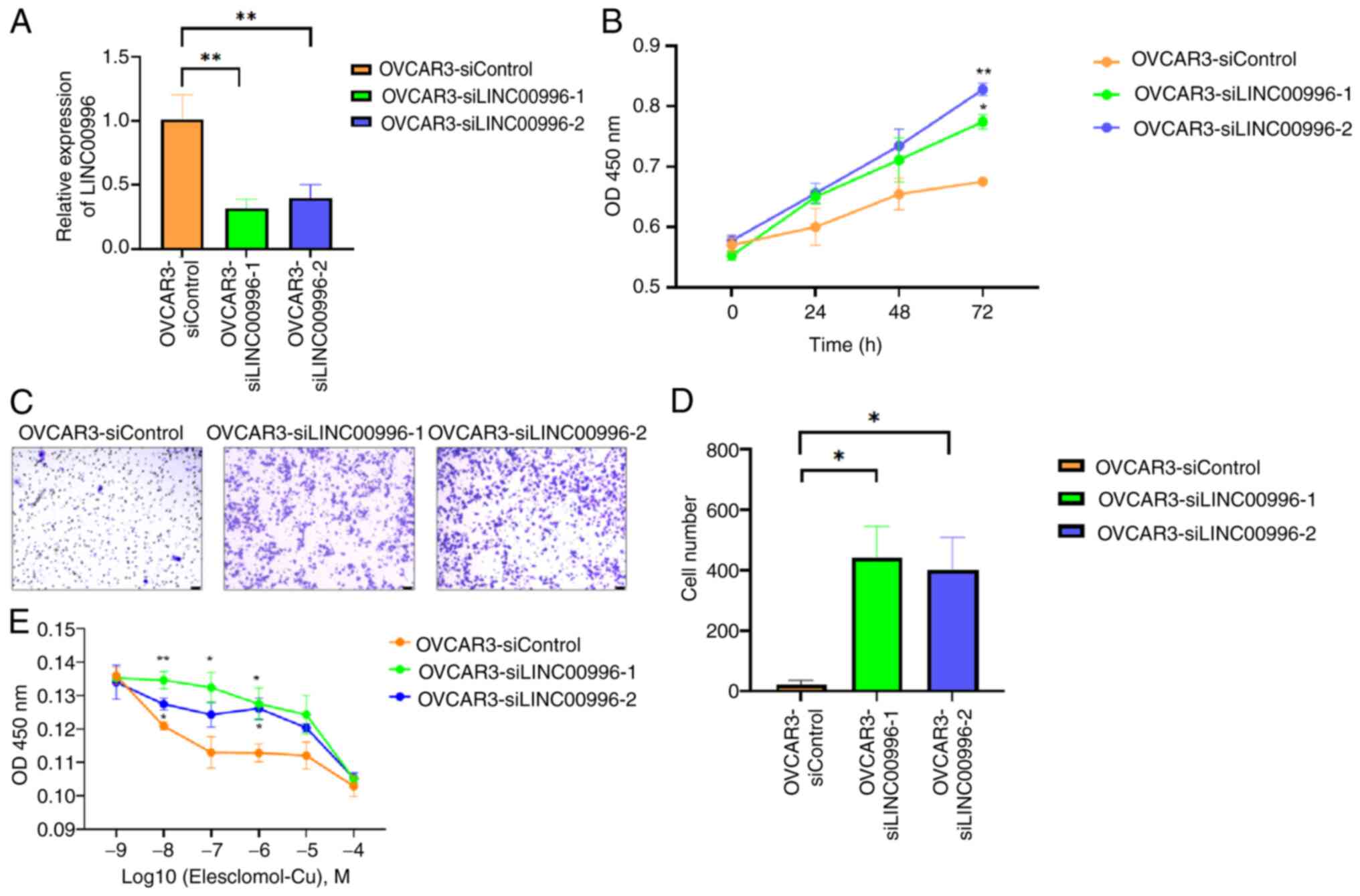
After transfecting siLINC00996-1 and siLINC00.996-2
into OVCAR3 cells to knockdown LINC00996 expression, the role of
LINC00996 in OV cells was further explored (Fig. 8A). The results of the CCK-8 assay
revealed that knockdown of LINC00996 improved the proliferative
ability of OV cells (Fig. 8B).
Moreover, the results of the Transwell assay demonstrated that the
knockdown of LINC00996 enhanced the migration ability of OV cells
(Fig. 8C and D). Elesclomol
facilitates the intracellular translocation of Cu2+,
thereby inducing mitochondrial dysfunction and triggering
cuproptosis (26). After culturing
the cells with different concentrations of
elesclomol-Cu2+ solution (ranging from 10−9
to 10−4 mol/l), a CCK-8 assay was performed to assess
the sensitivity of the cells to cuproptosis. The results suggested
that the knockdown of LINC00996 reduced the sensitivity of OV cells
to cuproptosis (Fig. 8E).
Regulatory mechanism of LINC00996
binding to ELAV-like RNA binding protein 1 (ELAVL1) in
cuproptosis
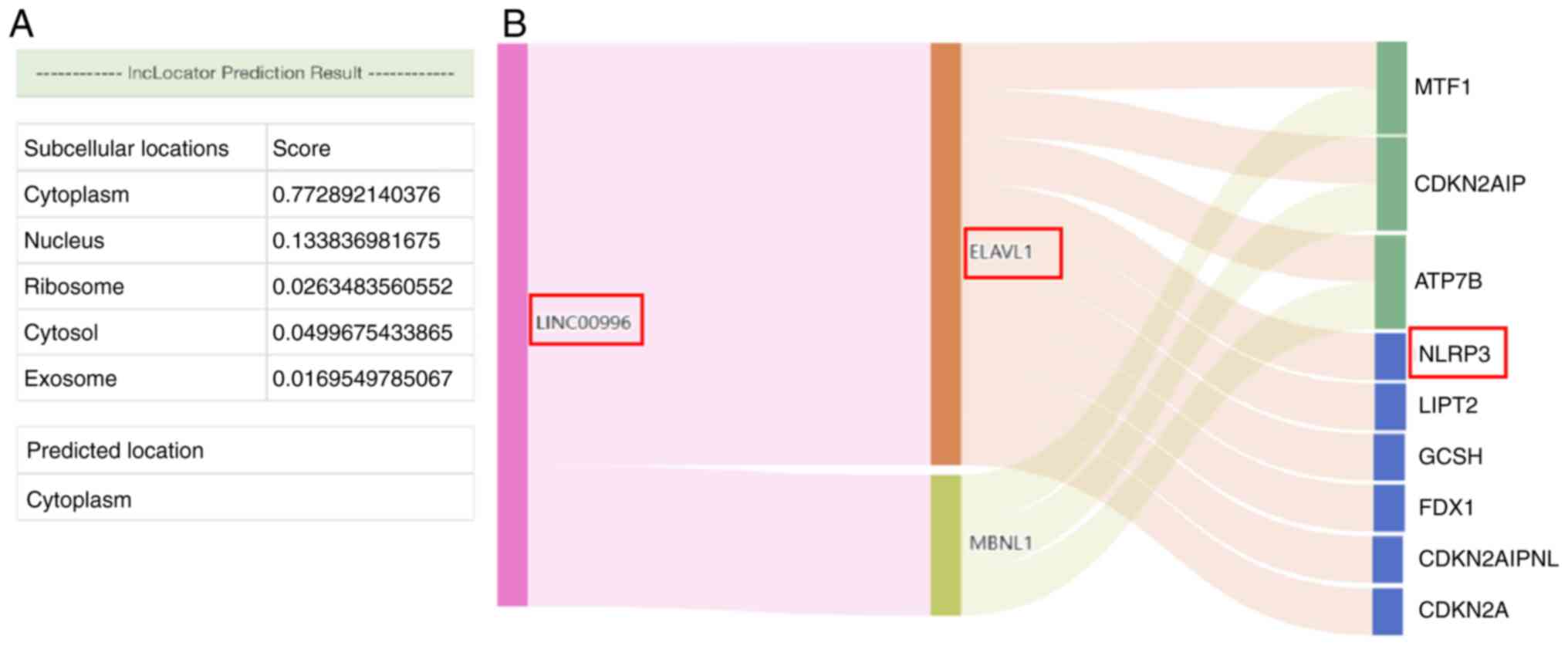
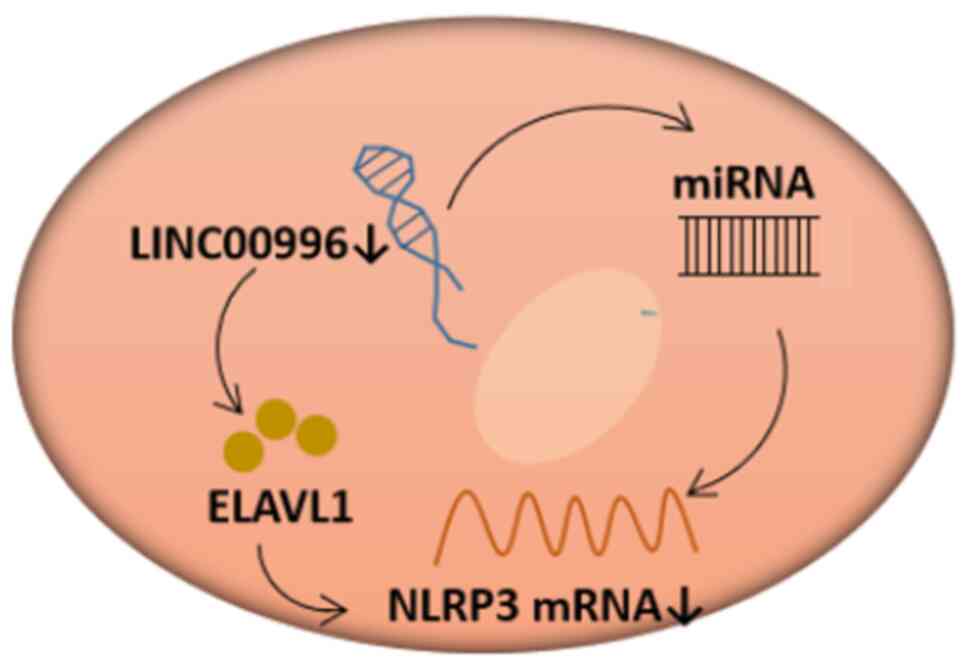
Based on the results from lncLocator, LINC00996 was
determined to be primarily located in the cytoplasm (Fig. 9A). This subcellular localization
suggested that LINC00996 may bind to RBPs or act as an miRNA
sponge. According to the correlation between LINC00996 and genes
associated with cuproptosis, LINC00996 was significantly positively
correlated with NLRP3 (P<0.001) (Fig. 2C). Moreover, to predict the RBPs
that can bind to LINC00996 as well as the target mRNAs, stringent
filtering conditions were set in ENCORI. RBPs interacting with
LINC00996 were predicted using ENCORI, filtered by CLIP Data ≥1.
The results showed that ELAVL1 and muscleblind-like splicing
regulator 1 may bind to LINC00996. Subsequently, the target mRNAs
of the aforementioned two RBPs were predicted, filtered by CLIP
Data ≥4 and pan-cancer count ≥10. The results suggested that ELAVL1
could interact with NLRP3 mRNA (Fig.
9B).
Regulatory mechanism of LINC00996 as
an miRNA sponge in cuproptosis
Previous studies have suggested that miRNA recognize
and bind to target mRNA, typically promoting mRNA degradation or
inhibiting its translation. Additionally, lncRNAs can bind to
specific miRNAs and modulate the expression levels of the target
mRNAs, thereby forming a regulatory network consisting of
lncRNA-miRNA-mRNA (22). In the
present study, the miRNAs that interact with LINC00996 were
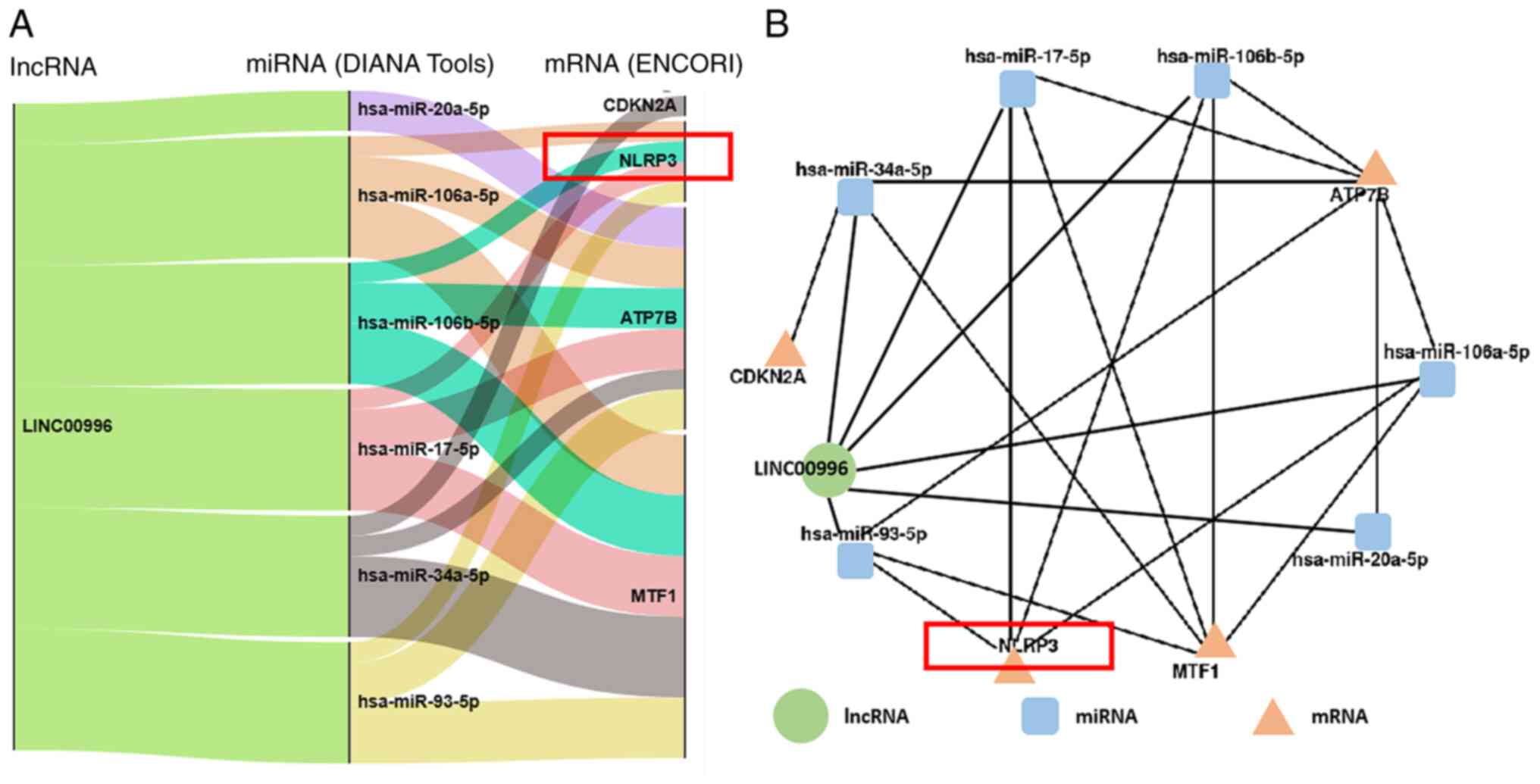
predicted utilizing the DIANA tools. The findings revealed that
hsa-miR-20a-5p, hsa-miR-106a-5p, hsa-miR-106b-5p, hsa-miR-17-5p,
hsa-miR-34a-5p and hsa-miR-93-5p could interact with LINC00996.
Based on ENCORI, the target mRNAs of these 6 miRNAs were then
predicted (Fig. 10A).
Consequently, a comprehensive lncRNA-miRNA-mRNA regulatory network
pertinent to cuproptosis was constructed (Fig. 10B). Notably, among the
aforementioned 6 miRNAs, hsa-miR-106a-5p, hsa-miR-106b-5p,
hsa-miR-17-5p and hsa-miR-93-5p may act on NLRP3 mRNA. This
comprehensive lncRNA-miRNA-mRNA regulatory network will be further
verified in subsequent studies.
Discussion
In the present study, after analyzing the TCGA-OV
cohort, a prognostic risk model based on cuproptosis-associated
lncRNAs was constructed. Survival analysis, ROC curves, calibration
curves, C-index and PCA demonstrated the reliability of the
constructed risk model. The potential mechanism of LINC00996 in the
aforementioned risk model was then predicted using ENCORI and DIANA
tools. The prognostic risk model can divide patients with OV into
high- and low-risk groups. Furthermore, the immune function
analysis demonstrated that MHC-class-I, type I IFN response, CCR,
APC co-inhibition, parainflammation, HLA, cytolytic activity,
inflammation-promoting, T cell co-stimulation, check-point and T
cell co-inhibition were improved in the low-risk group (41–45).
MHC-class-I can present intracellular peptides, which are
recognized by CD8+ T cells, thus inducing an antitumor
immune response (41–43). The findings of the present study
suggested that the low-risk group had a more optimal immune
function in MHC-class-I, indicating that the improved prognosis in
the low-risk group may be associated with the antitumor immune
response induced by MHC-class-I. However, the effect of different
immune functions between groups on the prognosis of patients with
OV needs further analysis and experimental verification.
Excess intracellular copper can interact with lipid
acylated proteins, and this process triggers irregular clustering
of lipid-acylated proteins and depletion of Fe-S clusters,
ultimately culminating in cuproptosis (26,46).
However, the role of cuproptosis in tumor progression and its
regulatory mechanism are poorly understood. The present study
constructed a prognostic risk model of lncRNAs associated with
cuproptosis and demonstrated that patients with OV in low-risk
group had an improved prognosis. We hypothesized that patients with
OV in the low-risk group would be more sensitive to cuproptosis
than those in the high-risk group. Cuproptosis provides new
perspectives on the treatment approach of OV, and the induction of
cuproptosis may be an emerging treatment option. Moreover, O'Day
et al (47) reported that
combination therapy of elesclomol and paclitaxel could markedly
increase the survival rate of patients with stage IV metastatic
melanoma. In the present study, after culturing with
elesclomol-Cu2+, the sensitivity of OV cells to
cuproptosis was positively associated with the level of LINC00996.
Therefore, LINC00996 could be a new prognostic biomarker for OV,
and the induction of cuproptosis may be a potent treatment for
patients with OV.
The key lncRNA of the prognostic risk model in the
present study was LINC00996, which was significantly positively
correlated with the cuproptosis-related gene, NLRP3. According to
previous reports, lncRNAs that are located in the nucleus can
regulate chromatin structure and histone modifications and affect
gene transcription, while lncRNAs that are localized in the
cytoplasm can regulate mRNA translation and degradation (48,49).
LINC00996 is mainly localized in the cytoplasm, suggesting that
LINC00996 regulates cuproptosis in OV through binding RBPs or
acting as an miRNA sponge (Fig.
11). Data from ENCORI suggested that the RBP, ELAVL1, can bind
to LINC00996. ELAVL1 can increase mRNA stability and serves a vital
role in tumor progression (50,51).
Additionally, NLRP3 was the target mRNA of ELAVL1 based on ENCORI.
ELAVL1 can bind to adenine and uridine-rich stability elements to
improve mRNA stability (52–55).
Liu et al (56) reported
that in rheumatoid arthritis fibroblast-like synoviocytes and human
umbilical vein endothelial cells, the nucleocytoplasmic shuttling
of ELAVL1 is initiated and ELAVL1 expression is notably elevated in
the cytoplasm, which increases the stability of NLRP3 mRNA and
promotes NLRP3 expression, after TNF-α and calreticulin dual
stimulation. Furthermore, in high glucose-treated cardiomyocytes
and in human diabetic hearts, Jeyabal et al (57) also reported that ELAVL1 knockdown
reduced the expression level of NLRP3. Therefore, we hypothesized
that LINC00996 can increase the stability of NLRP3 mRNA by binding
to ELAVL1 in OV cells, thereby regulating sensitivity to
cuproptosis. The present study found that LINC00996, as a sponge,
can bind to hsa-miR-20a-5p, hsa-miR-106a-5p, hsa-miR-106b-5p,
hsa-miR-17-5p, hsa-miR-34a-5p and hsa-miR-93-5p. Among these miRNA,
hsa-miR-106a-5p, hsa-miR-106b-5p, hsa-miR-17-5p and hsa-miR-93-5p
can act on NLRP3 mRNA.
The present study has the following limitations:
First, it used a dataset of 378 patients with OV from TCGA to
explore a cuproptosis-related risk model, but further validation of
the risk model in cohorts from other institutions is needed to
ensure broader applicability. Second, while preliminary validation
was performed using cell experiments, further in vitro and
in vivo experiments are required to verify the interactions
and regulatory mechanisms.
In conclusion, the present study constructed a
prognostic risk model based on lncRNAs associated with cuproptosis
in OV and explored the regulatory mechanism of LINC00996 in
cuproptosis. The results provide new perspectives to assess the
role of cuproptosis in OV; however, more validation experiments are
required to further explore this role.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by Natural Science Foundation of Zhejiang
Province (grant no. LQ23H160033), Zhejiang Medical and Health
Science and Technology program (grant no. 2024KY116) and Key
R&D Program of Zhejiang Province (grant no. 2022C03013).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
BC and HW proposed the conception and implementation
of the study. BC and SY were responsible for data download and the
interpretation of data. BC was a major contributor in the
preparation of the manuscript. BC and HW performed the final
proofreading. BC, SY and HW confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Goding SA, Fedewa
SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal A:
Colorectal cancer statistics, 2020. CA Cancer J Clin. 70:145–164.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen S, Tang Y, Li Y, Huang M, Ma X, Wang
L, Wu Y, Wang Y, Fan W and Hou S: Design and application of prodrug
fluorescent probes for the detection of ovarian cancer cells and
release of anticancer drug. Biosens Bioelectron. 236:1154012023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li T, Wang X, Qin S, Chen B, Yi M and Zhou
J: Targeting PARP for the optimal immunotherapy efficiency in
gynecologic malignancies. Biomed Pharmacother. 162:1147122023.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kahn R, Filippova O, Gordhandas S, An A,
Straubhar AM, Zivanovic O, Gardner GJ, O'Cearbhaill RE, Tew WP,
Grisham RN, et al: Ten-year conditional probability of survival for
patients with ovarian cancer: A new metric tailored to long-term
survivors. Gynecol Oncol. 169:85–90. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh N, Jayraj AS, Sarkar A, Mohan T,
Shukla A and Ghatage P: Pharmacotherapeutic treatment options for
recurrent epithelial ovarian cancer. Expert Opin Pharmacother.
24:49–64. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perren TJ, Swart AM, Pfisterer J,
Ledermann JA, Pujade-Lauraine E, Kristensen G, Carey MS, Beale P,
Cervantes A, Kurzeder C, et al: A phase 3 trial of bevacizumab in
ovarian cancer. N Engl J Med. 365:2484–2496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bi R, Chen L, Huang M, Qiao Z, Li Z, Fan G
and Wang Y: Emerging strategies to overcome PARP inhibitors'
resistance in ovarian cancer. Biochim Biophys Acta Rev Cancer.
1879:1892212024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao J, Sun J, Shuai SC, Zhao Q and Shuai
J: Predicting potential interactions between lncRNAs and proteins
via combined graph auto-encoder methods. Brief Bioinform.
24:bbac5272023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang Y, Cheung BB, Atmadibrata B, Marshall
GM, Dinger ME, Liu PY and Liu T: The regulatory role of long
noncoding RNAs in cancer. Cancer Lett. 391:12–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao ZT, Yang YM, Sun MM, He Y, Liao L,
Chen KS and Li B: New insights into the interplay between long
non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun
(Lond). 42:117–140. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou HL, Luo G, Wise JA and Lou H:
Regulation of alternative splicing by local histone modifications:
Potential roles for RNA-guided mechanisms. Nucleic Acids Res.
42:701–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen XJ and An N: Long noncoding RNA ATB
promotes ovarian cancer tumorigenesis by mediating histone H3
lysine 27 trimethylation through binding to EZH2. J Cell Mol Med.
25:37–46. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu SS, Li JS, Xue M, Wu WJ, Li X and Chen
W: LncRNA UCA1 participates in De Novo synthesis of guanine
nucleotides in bladder cancer by recruiting TWIST1 to increase
IMPDH1/2. Int J Biol Sci. 19:2599–2612. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Haas R, Ganem NS, Keshet A, Orlov A,
Fishman A and Lamm AT: A-to-I RNA editing affects lncRNAs
expression after heat shock. Genes (Basel). 9:6272018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gordon MA, Babbs B, Cochrane DR, Bitler BG
and Richer JK: The long non-coding RNA MALAT1 promotes ovarian
cancer progression by regulating RBFOX2-mediated alternative
splicing. Mol Carcinog. 58:196–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma B, Wang S, Wu W, Shan P, Chen Y, Meng
J, Xing L, Yun J, Hao L, Wang X, et al: Mechanisms of
circRNA/lncRNA-miRNA interactions and applications in disease and
drug research. Biomed Pharmacother. 162:1146722023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou J, Xu Y, Wang L, Cong Y, Huang K, Pan
X, Liu G, Li W, Dai C, Xu P and Jia X: LncRNA IDH1-AS1
sponges miR-518c-5p to suppress proliferation of epithelial ovarian
cancer cell by targeting RMB47. J Biomed Res. 38:51–65. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalkavan H, Rühl S, Shaw JJP and Green DR:
Non-lethal outcomes of engaging regulated cell death pathways in
cancer. Nat Cancer. 6:795–806. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tong X, Tang R, Xiao M, Xu J, Wang W,
Zhang B, Liu J, Yu X and Shi S: Targeting cell death pathways for
cancer therapy: Recent developments in necroptosis, pyroptosis,
ferroptosis, and cuproptosis research. J Hematol Oncol. 15:1742022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsvetkov P, Coy S, Petrova B, Dreishpoon
M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R,
Spangler RD, et al: Copper induces cell death by targeting
lipoylated TCA cycle proteins. Science. 375:1254–1261. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deigendesch N, Zychlinsky A and Meissner
F: Copper regulates the canonical NLRP3 inflammasome. J Immunol.
200:1607–1617. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xue Q, Kang R, Klionsky DJ, Tang D, Liu J
and Chen X: Copper metabolism in cell death and autophagy.
Autophagy. 19:2175–2195. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu S, Ge J, Chu Y, Cai S, Wu J, Gong A
and Zhang J: Identification of hub cuproptosis related genes and
immune cell infiltration characteristics in periodontitis. Front
Immunol. 14:11646672023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun M, Zhan N, Yang Z, Zhang X, Zhang J,
Peng L, Luo Y, Lin L, Lou Y, You D, et al: Cuproptosis-related
lncRNA JPX regulates malignant cell behavior and epithelial-immune
interaction in head and neck squamous cell carcinoma via
miR-193b-3p/PLAU axis. Int J Oral Sci. 16:632024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou G, Chen C, Wu H, Lin J, Liu H, Tao Y
and Huang B: LncRNA AP000842.3 triggers the malignant progression
of prostate cancer by regulating cuproptosis related gene NFAT5.
Technol Cancer Res Treat. 23:153303382412555852024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai Y, Zhang Q, Liu F and Quan J: A novel
cuproptosis-related lncRNA signature predicts the prognosis and
immune landscape in bladder cancer. Front Immunol. 13:10274492022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang H, Chen G, Zhang Z, Wu G, Zhang Z,
Yu A, Wang J, Quan C, Li Y and Zhou M: Deciphering the role of
cuproptosis-related lncRNAs in shaping the lung cancer immune
microenvironment: A comprehensive prognostic model. J Cell Mol Med.
28:e185192024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu H, Lu X, Hu Y, Baatarbolat J, Zhang Z,
Liang Y, Zhang Y, Liu Y, Lv H and Jin X: Biomimic nanodrugs
overcome tumor immunosuppressive microenvironment to enhance
cuproptosis/chemodynamic-induced cancer immunotherapy. Adv Sci
(Weinh). 12:e24111222025. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guan M, Cheng K, Xie XT, Li Y, Ma MW,
Zhang B, Chen S, Chen W, Liu B, Fan JX and Zhao YD: Regulating
copper homeostasis of tumor cells to promote cuproptosis for
enhancing breast cancer immunotherapy. Nat Commun. 15:100602024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu X, Chen X, Lin C, Yi Y, Zhao S, Zhu B,
Deng W, Wang X, Xie Z, Rao S, et al: Elesclomol loaded copper oxide
nanoplatform triggers cuproptosis to enhance antitumor
immunotherapy. Adv Sci (Weinh). 11:e23099842024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen Z, Li X, Hu Z, Yang Y, Yang Z, Li S,
Zhou Y, Ma J, Li H, Liu X, et al: Linc00996 is a favorable
prognostic factor in LUAD: Results from bioinformatics analysis and
experimental validation. Front Genet. 13:9329732022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ge H, Yan Y, Wu D, Huang Y and Tian F:
Potential role of LINC00996 in colorectal cancer: a study based on
data mining and bioinformatics. Onco Targets Ther. 11:4845–4855.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Berek JS, Renz M, Kehoe S, Kumar L and
Friedlander M: Cancer of the ovary, fallopian tube, and peritoneum:
2021 update. Int J Gynaecol Obstet. 155 (Suppl):61–85. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kawase K, Kawashima S, Nagasaki J, Inozume
T, Tanji E, Kawazu M, Hanazawa T and Togashi Y: High expression of
MHC class I overcomes cancer immunotherapy resistance due to IFNγ
signaling pathway defects. Cancer Immunol Res. 11:895–908. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hiltner T, Szörenyi N, Kohlruss M,
Hapfelmeier A, Herz AL, Slotta-Huspenina J, Jesinghaus M, Novotny
A, Lange S, Ott K, et al: Significant tumor regression after
neoadjuvant chemotherapy in gastric cancer, but poor survival of
the patient? Role of MHC class I alterations. Cancers (Basel).
15:7712023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Colbert JD, Cruz FM, Baer CE and Rock KL:
Tetraspanin-5-mediated MHC class I clustering is required for
optimal CD8 T cell activation. Proc Natl Acad Sci USA.
119:e21221881192022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luda KM, Longo J, Kitchen-Goosen SM,
Duimstra LR, Ma EH, Watson MJ, Oswald BM, Fu Z, Madaj Z, Kupai A,
et al: Ketolysis drives CD8+ T cell effector function through
effects on histone acetylation. Immunity. 56:2021–2035.e8. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stanifer ML, Guo C, Doldan P and Boulant
S: Importance of type I and III interferons at respiratory and
intestinal barrier surfaces. Front Immunol. 11:6086452020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen L, Min J and Wang F: Copper
homeostasis and cuproptosis in health and disease. Signal Transduct
Target Ther. 7:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
O'Day S, Gonzalez R, Lawson D, Weber R,
Hutchins L, Anderson C, Haddad J, Kong S, Williams A and Jacobson
E: Phase II, randomized, controlled, double-blinded trial of weekly
elesclomol plus paclitaxel versus paclitaxel alone for stage IV
metastatic melanoma. J Clin Oncol. 27:5452–5458. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF,
Shan L, Zhang J, Wang S, Wang Y, Carmichael GG, et al: Distinct
processing of lncRNAs contributes to non-conserved functions in
stem cells. Cell. 181:621–636.e22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Q, Li G, Ma X, Liu L, Liu J, Yin Y,
Li H, Chen Y, Zhang X, Zhang L, et al: LncRNA TINCR impairs the
efficacy of immunotherapy against breast cancer by recruiting DNMT1
and downregulating MiR-199a-5p via the STAT1-TINCR-USP20-PD-L1
axis. Cell Death Dis. 14:762023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Priyanka P, Sharma M, Das S and Saxena S:
The lncRNA HMS recruits RNA-binding protein HuR to stabilize the
3′-UTR of HOXC10 mRNA. J Biol Chem. 297:1009972021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Latorre E, Carelli S, Raimondi I,
D'Agostino V, Castiglioni I, Zucal C, Moro G, Luciani A, Ghilardi
G, Monti E, et al: The ribonucleic complex HuR-MALAT1 represses
CD133 expression and suppresses epithelial-mesenchymal transition
in breast cancer. Cancer Res. 76:2626–2636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Brennan CM and Steitz JA: HuR and mRNA
stability. Cell Mol Life Sci. 58:266–277. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fu XD: RNA editing: New roles in feedback
and feedforward control. Cell Res. 33:495–496. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hildebrandt RP, Moss KR, Janusz-Kaminska
A, Knudson LA, Denes LT, Saxena T, Boggupalli DP, Li Z, Lin K,
Bassell GJ, et al: Muscleblind-like proteins use modular domains to
localize RNAs by riding kinesins and docking to membranes. Nat
Commun. 14:34272023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
González ÀL, Fernández-Remacha D, Borrell
JI, Teixidó J and Estrada-Tejedor R: Cognate RNA-binding modes by
the alternative-splicing regulator MBNL1 inferred from molecular
dynamics. Int J Mol Sci. 23:161472022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu Y, Wei W, Wang Y, Wan C, Bai Y, Sun X,
Ma J and Zheng F: TNF-α/calreticulin dual signaling induced NLRP3
inflammasome activation associated with HuR nucleocytoplasmic
shuttling in rheumatoid arthritis. Inflamm Res. 68:597–611. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jeyabal P, Thandavarayan RA, Joladarashi
D, Suresh Babu S, Krishnamurthy S, Bhimaraj A, Youker KA, Kishore R
and Krishnamurthy P: MicroRNA-9 inhibits hyperglycemia-induced
pyroptosis in human ventricular cardiomyocytes by targeting ELAVL1.
Biochem Biophys Res Commun. 471:423–429. 2016. View Article : Google Scholar : PubMed/NCBI
|