Introduction
Chronic myeloid leukemia (CML) is classified as a
tumor-related disease characterized by clonal expansion of
hematopoietic stem cells from the bone marrow. The global incidence
is 1–2 cases per 100,000 adults, representing 15% of newly
diagnosed leukemia cases in adults (1). Notably, ~50% of patients with CML lack
typical symptoms in the early stage and are often incidentally
discovered through routine physical examinations or blood tests.
Bone marrow biopsy is the gold standard for confirming hyperplasia
of bone marrow cells and abnormal maturation of the myeloid cell
line. Molecular diagnosis relies on the detection of the BCR-ABL1
fusion gene, which can be identified through the translocation of
chromosomes t(9;22)(q34;q11) using fluorescence in situ
hybridization (FISH) or quantitative PCR (2).
It is noteworthy that 5–10% of cases negative for
the Philadelphia (Ph) chromosome require further verification of
BCR-ABL1 mutations through next-generation sequencing (3). Over 95% of CML cases are linked to the
Ph chromosome, whereas ~5% display variant Ph chromosomes
characterized by complex translocations (4). These chromosomal rearrangements lead
to the fusion of the 3′ end of the ABL1 gene on chromosome 9 with
the 5′ end of the BCR gene on chromosome 22, resulting in the
BCR-ABL1 fusion gene (5). For
numerous years, traditional karyotyping has served as the primary
diagnostic approach for identifying t(9;22)(q34; q11.2); however,
this method possesses significant limitations, potentially leading
to the oversight of BCR-ABL1 rearrangements in ~5% of patients with
CML. Over 95% of CML cases exhibit BCR-ABL1 e13a2 or e14a2 fusion
transcripts at the time of diagnosis. Approximately 1% of patients
possess a breakpoint between exons 1 and 2 of the BCR gene,
resulting in the production of the e1a2 transcript (6).
The prognosis of CML has significantly improved with
the application of tyrosine kinase inhibitors (TKIs), with the
5-year survival rate increasing from 50 to >90% (7). However, ~9% of patients may progress
to blast crisis or accelerated phase, leading to a sharp reduction
in survival. Regular assessment of hematological, cytogenetic and
molecular indicators is necessary to monitor treatment response
(8). Therefore, individualized
treatment and lifelong monitoring are crucial for optimizing the
management of CML.
The current report presents a case of a patient with
CML who tested negative for both reverse transcription (RT)-PCR and
cytogenetic analysis during the initial diagnostic phase.
Subsequent FISH revealed a concealed insertion of the ABL1 gene
into the BCR gene on chromosome 22. The e13a3 BCR-ABL1 fusion
transcript was subsequently validated via DNA sequencing employing
alternative primer sets. The patient received a diagnosis of CML
with the e13a3 atypical fusion gene and underwent treatment with
nilotinib.
Case report
In September 2021, a 39-year-old male patient was
referred to the Department of Hematology, (Shandong Provincial
Hospital Affiliated to Shandong First Medical University, Jinan,
China) after abnormal blood parameters were identified during a
routine health examination. The patient had exhibited splenomegaly
for 3 years. A CT scan of the abdomen conducted upon admission
verified the existence of splenomegaly. Laboratory analyses
indicated a white blood cell count of 95.94×109/l
(reference range,3.5–9.5×109/l), a hemoglobin
concentration of 97 g/l (reference range, 130–175 g/l) and a
platelet count of 929×109/l (reference range,
125–350×109/l). The absolute neutrophil count was
73.49×109/l (reference range, 1.8–6.3×109/l).
Analysis of the peripheral blood smear revealed notable
leukocytosis, comprising ~1% blasts (reference range, 0–1%), 1%
promyelocytes (reference range, 1.0–2.2%), 23% myelocytes and
metamyelocytes (reference range, 4.5–8.5%), and 16% basophils
(reference range, 0–0.5%). Platelets were often observed in a
scattered arrangement. The analysis of the bone marrow smear
indicated active hematopoietic proliferation, exhibiting a
granulocyte-to-erythrocyte ratio of 11.19:1.00. Granulopoiesis was
markedly heightened, characterized by an augmented ratio of
neutrophilic myelocytes and metamyelocytes. Prominent nucleoli were
identified in 74% of cells, a significant number of which displayed
excessive granulation. The levels of eosinophils and basophils were
elevated. The neutrophil alkaline phosphatase (NAP) score was 8
(The NAP activity test employs the Fast Blue B staining method.
Under an oil immersion microscope, 100 mature neutrophils are
observed, and cytoplasmic granules are scored from 0 to 4 based on
staining intensity and density: 0 points, no granules; 1 point,
<25% of cytoplasm lightly stained; 2 points, 25–50% moderately
stained; 3 points, 50–75% deeply stained; 4 points: >75% densely
and deeply stained; the individual scores are summed to yield a
total score ranging from 0 to 400. The normal reference range is
30–130 points), indicating a significant likelihood of CML. Flow
cytometry of the bone marrow demonstrated an increased percentage
of granulocytes at early, intermediate, and late maturation stages.
Myelocytes comprised 83.0% of nucleated cells and exhibited
expression levels of CD33, CD64, CD13 and CD15, alongside partial
expression of CD10, CD11b and CD16, indicative of the chronic phase
of CML.
Molecular analysis (9) for the BCR-ABL1 fusion gene
(p190/p210/p230) yielded negative results, as did tests for JAK2
V617F, CALR exon 9 and MPL exon 10 mutations. Subsequent analysis
of atypical BCR-ABL1 fusion transcripts revealed a positive result
for the e13a3 fusion variant. Chromosomal karyotype analysis
verified the existence of t(9;22)(q34; q11.2). Following these
findings, the patient received a diagnosis of chronic-phase CML
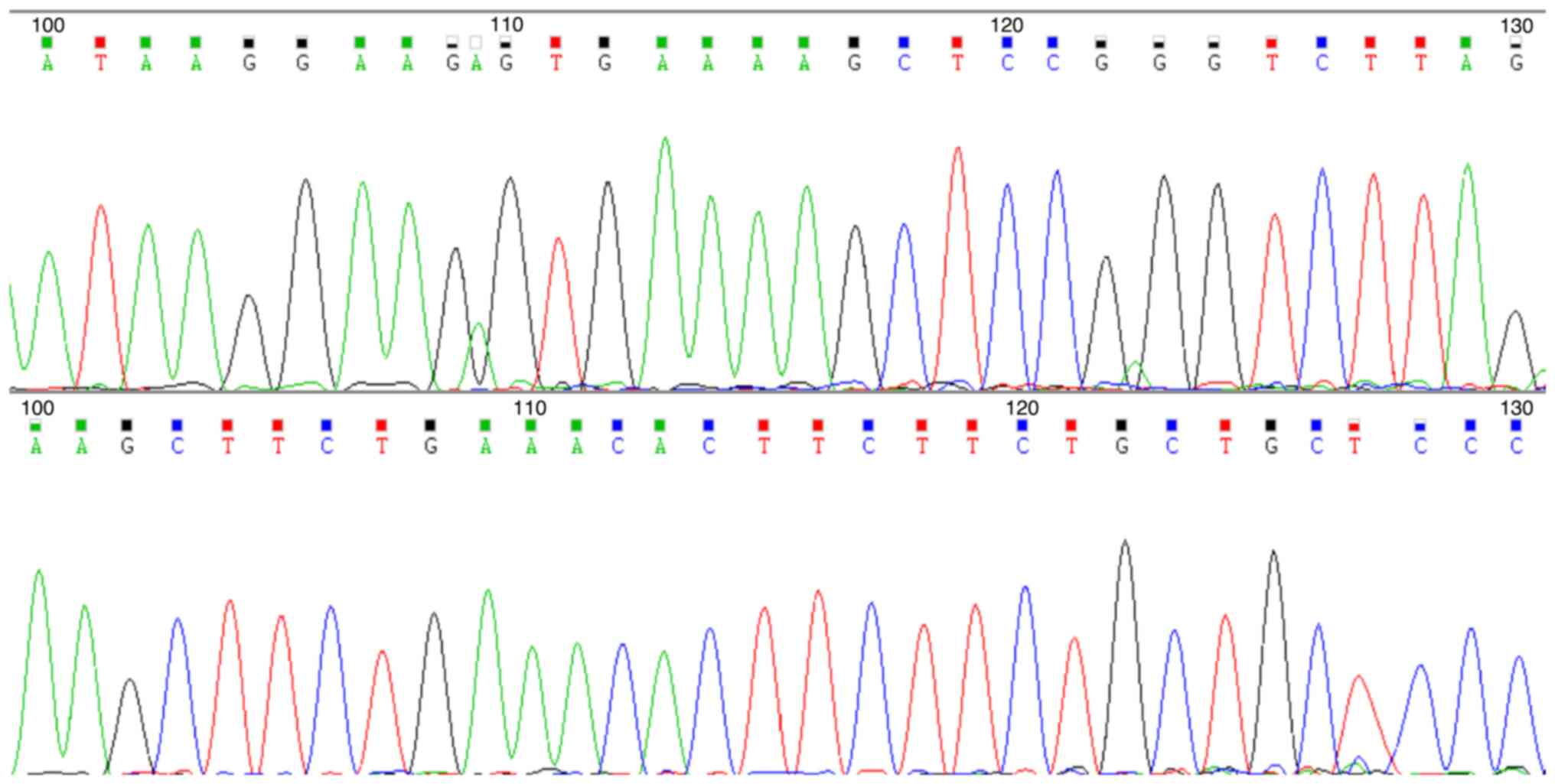
(e13a3 type) (Fig. 1). To elucidate
the characteristics of the fusion gene, RT-PCR was used to amplify
the BCR/ABL e13a3 sequence. The amplified product was subjected to
Sanger sequencing to confirm the precise breakpoint of the e13a3
fusion transcript. RNA was extracted from cells using the Lab-Aid
896 Blood Total RNA Kit (Xiamen Zeesan Biotech Co., Ltd.). RT was
performed using the LF Enzyme 03 Reverse Transcription Kit (Xiamen
Zeesan Biotech Co., Ltd.) under the following thermal conditions:
37°C for 15 min (cDNA synthesis) and 85°C for 5 sec (reverse
transcriptase inactivation). The BCR:ABL1 fusion gene was amplified
using the following five primers: ABL-3 reverse,
CCATTGTGATTATAGCCTAAGACCCGGAG; BCR-1 forward,
CTCCAGCGAGGAGGACTTCTCCT; BCR-6 forward,
CCTGAGAGCCAGAAGCAACAAAGATGCC; BCR-12 forward,
AGAACATCCGGGAGCAGCAGAAGAA; and BCR-19 forward,
ACTGAAGGCAGCCTTCGACGTC.
The reaction was performed with the following
thermal profile: Initial denaturation at 95°C for 3 min, followed
by 39 cycles of denaturation at 95°C for 10 sec, annealing at 63°C
for 20 sec and extension at 72°C for 30 sec, with a final extension
at 72°C for 5 min. The reaction was terminated by holding at 4°C.
Amplified products were analyzed using agarose gel electrophoresis.
The reaction system consisted of 1 µl each of forward and reverse
primers, 3 µl of cDNA template, 10 µl of PCR Mix, and ddH2O to make
up the total volume to 20 µl. After purification of the amplified
products, sequencing reactions were performed using the BigDye
Terminator v3.1 sequencing kit (Thermo Fisher Scientific, Inc.),
and sequence analysis was ultimately completed with the 3130×l
Genetic Analyzer (Thermo Fisher Scientific, Inc.). Treatment
commenced with nilotinib at a dosage of 300 mg administered twice
daily. After 6 months of consistent therapy, the atypical BCR-ABL1
fusion gene became undetectable, and the patient proceeded with
regular monthly follow-up. At 2 years post-therapy initiation, the
patient exhibited skin induration in the limbs, diagnosed as
cutaneous amyloidosis, attributed to a drug-related adverse effect
of nilotinib. The treatment with the TKI was sustained as the
patient could endure the condition. The patient remains in profound
molecular remission as of the last check-up inn June 2024.
Discussion
The BCR-ABL1 fusion gene serves as a critical
molecular marker in CML, with the e13a2 (b2a2) and e14a2 (b3a2)
transcripts representing >95% of all instances. Nonetheless,
infrequent BCR-ABL1 transcripts, specifically e13a3 (b2a3) and
e14a3 (b3a3), are observed in a minor fraction of patients, with an
occurrence rate of 0.1–0.3% (10,11).
In a substantial cohort study involving 2,331 patients with CML,
only 4 cases (0.1%) exhibited e13a3 or e14a3 transcripts (12). Despite infrequency of occurrence,
recent evidence indicates that the e13a3 transcript may impart
distinct clinical and molecular attributes. The e13a3 transcript is
produced by the direct fusion of exon e13 (b2) of the BCR gene with
exon a3 of the ABL1 gene, resulting in the exclusion of exon a2 of
ABL1 (13). This exon encodes the
SH3 domain, which is essential for the negative regulation of
BCR-ABL1 kinase activity, suggesting that the e13a3 transcript may
modify BCR-ABL1-mediated signaling pathways (14). The absence of the SH3 domain may
impact downstream signaling, including the STAT5 pathway, thereby
affecting the proliferative characteristics of leukemic cells
(15). However, the specific
biological implications of this structural modification require
additional examination.
In terms of clinical manifestations, patients
possessing the e13a3 transcript may display unique characteristics.
An increased basophil count at initial presentation has been
documented in certain cases (16).
Moreover, these patients typically exhibit a positive response to
TKI, frequently attaining molecular responses more swiftly, with
some achieving deep molecular remission (DMR) (17). In a study of 33 patients with CML
with uncommon BCR-ABL1 transcripts, all 4 e13a3 cases attained a
minimum 1-log reduction in BCR-ABL1 transcripts within 3 months,
and 50% achieved undetectable transcript levels within 2 years
(18).
At present, RT-PCR is the established molecular
method for identifying prevalent CML-associated transcripts,
chiefly e13a2 and e14a2. Nonetheless, the lack of the ABL1 a2 exon
in e13a3 may result in conventional RT-PCR assays failing to detect
this variant, thereby producing false-negative outcomes (12). A report indicated that a patient
with CML initially tested negative for BCR-ABL1 via RT-PCR.
However, FISH later identified BCR-ABL1 gene rearrangement, which
was subsequently corroborated by Sanger sequencing that confirmed
the presence of the e13a3 transcript (13). This highlights the inadequacy of
depending exclusively on RT-PCR in these instances. FISH is an
essential adjunctive diagnostic method for identifying BCR-ABL1
rearrangements in cases of negative RT-PCR results (16), whereas Sanger sequencing can
accurately delineate the atypical transcript (17). Therefore, when CML is clinically
suspected but RT-PCR results are negative, the incorporation of
FISH and Sanger sequencing is advised to reduce the risk of
misdiagnosis or overlooked diagnosis. In recent years,
next-generation sequencing (NGS) has emerged as an alternative to
traditional PCR, providing the ability to detect rare transcripts
such as e13a3 and facilitating dynamic monitoring of molecular
responses (18). Therefore, for
patients possessing the e13a3 transcript, refining RT-PCR primer
design to focus on the a3 exon, alongside FISH, Sanger sequencing
and NGS, can significantly improve diagnostic sensitivity and
specificity.
While data on e13a3 patients is scarce, current
studies indicate that this transcript may affect TKI response,
molecular remission rates and treatment-free remission (TFR)
outcomes following TKI cessation (19). The specific BCR-ABL1 transcript
variant influences the rate and magnitude of molecular responses
elicited by TKIs. Patients with the e13a2 transcript demonstrate a
protracted molecular remission and diminished optimal response
rates at 12 months during imatinib therapy in contrast to those
with e14a2 (11). The e13a3
transcript, which omits the ABL1 a2 exon, may similarly undermine
the structural integrity of the BCR-ABL1 protein and its
interaction with TKIs, potentially resulting in a protracted
molecular remission compared to e13a2 or e14a2 instances.
Second-generation TKIs (2G TKIs), such as nilotinib and dasatinib,
typically elicit more profound and rapid molecular responses
compared to imatinib. Patients with the e14a2 transcript attain
molecular remission more swiftly under 2G TKI therapy compared with
those with the e13a2 variant (19).
Despite the limited clinical data on e13a3, its structural
similarity to e13a2 implies that patients with e13a3 may exhibit
inadequate responses to imatinib, rendering 2G TKIs a more suitable
treatment option. The transcript type affects both the initial
response to TKI and the long-term disease-free survival (DFS) and
overall survival (OS). Patients with the e14a2 transcript exhibit
elevated rates of attaining stable DMR and sustaining TFR following
TKI cessation in comparison to those with e13a2 (11). The long-term prognostic implications
of e13a3 remain ambiguous. However, its molecular characteristics
and response patterns indicate that DFS and OS may resemble those
of e13a2 patients, potentially exhibiting slower molecular
remission and reduced TFR maintenance, while not significantly
impacting OS (20).
Adverse events linked to TKI therapy are essential
factors influencing long-term outcomes. Nilotinib is associated
with an increased occurrence of cutaneous adverse reactions,
including keratosis pilaris, which, while generally mild, may
impact treatment adherence (20).
In addition, certain patients may necessitate dose modifications or
therapeutic transitions owing to TKI-associated toxicities, which
could indirectly affect molecular remission and prognosis.
In CML, 1–2% of patients with CML possess atypical
BCR-ABL1 transcripts, such as e13a3, e14a3, e1a2 and e19a2
(21). These uncommon transcripts
influence diagnostic accuracy, treatment response, precision in
minimal residual disease monitoring, and long-term prognosis
(22). Standardized quantitative
PCR assays predominantly focus on e13a2 and e14a2, which diminishes
detection sensitivity for rare transcripts, potentially leading to
an underestimation of relapse risk (23). A current therapeutic objective in
CML management is to enable TFR after the successful cessation of
TKI treatment. The viability of TFR in patients with atypical
transcripts remains predominantly uncertain. Research indicates
that e14a2 patients are more likely to sustain TFR than e13a2
patients; however, information regarding e13a3 cases is limited
(11). Generally, patients with
e14a2 typically show more pronounced DMR, resulting in elevated TFR
success rates, while individuals with e13a2 or atypical transcripts
generally experience suboptimal TFR maintenance (24). Considering the structural attributes
of the e13a3 BCR-ABL1 fusion protein, there may be a diminished
likelihood of maintaining TFR following the cessation of treatment
(19). Patients contemplating TFR
should undergo a meticulous evaluation of molecular response
kinetics. Significantly, 2G TKIs may enhance the durability of
treatment-free remission in comparison to imatinib. Nilotinib is
associated with an increased probability of maintaining
treatment-free remission after cessation, likely attributable to
its capacity to elicit more rapid and significant molecular
responses (19). Patients attaining
Molecular Response (MR)4.5 (25) or
more profound responses through prolonged nilotinib or dasatinib
treatment may demonstrate improved TFR outcomes. Nonetheless,
patients exhibiting atypical transcripts such as e1a2 have shown
elevated rates of treatment discontinuation failure, indicating
that TFR strategies may be inappropriate for this subgroup
(26). In e13a3 patients, the early
achievement of MR4 (BCR-ABL1/ABL1 ≤0.01%) or more profound
responses may improve TFR success. However, additional research is
required to validate this hypothesis. Future research should
concentrate on clarifying remission patterns, determining optimal
TKI regimens, and developing discontinuation strategies for e13a3
patients to refine treatment protocols, enhance quality of life and
improve disease management (24).
The diagnosis and management of the present study's
rare patient with e13a3 BCR-ABL1 CML highlights the complexities
and strategic considerations involved in treating atypical
transcript cases. The patient's diagnosis was initially overlooked
by routine RT-PCR and was subsequently validated through Sanger
sequencing. To enhance molecular diagnostics, RT-PCR protocols must
be refined to identify atypical transcripts such as e13a3 and
integrated with FISH and NGS to improve diagnostic sensitivity and
specificity. The present patient attained complete cytogenetic
remission within 6 months on a 2G TKI (nilotinib), indicating that
e13a3 patients may exhibit favorable early responses to TKIs. The
emergence of drug-related adverse events, such as skin sclerosis,
underscores the necessity for rigorous safety monitoring during TKI
therapy in patients with atypical BCR-ABL1 transcripts.
Consequently, management must prioritize disease remission and
proactive surveillance of treatment-related toxicities,
facilitating personalized therapeutic modifications to enhance
adherence and tolerance.
The transition of treatment objectives for CML from
prolonged medication use to TFR renders the prognosis for achieving
TFR in patients with atypical BCR-ABL1 transcripts markedly
uncertain. This case illustrates that patients with e13a3
transcripts can attain rapid DMR during TKI therapy, indicating the
viability of pursuing TFR with rigorous molecular surveillance.
However, it is essential to underscore that the results of this
single-case study possess inherent limitations regarding
generalization. The emergence of drug-related adverse reactions,
such as skin hardening, in this patient highlights the potential
intricacies of safety profiles during prolonged TKI administration
in atypical transcript carriers.
The present report offers clinical insights into TKI
efficacy in e13a3 CML, yet the molecular mechanisms influencing
signaling dynamics have not been fully elucidated. Structural
analyses reveal that the e13a3 transcript truncates the SH3 domain
of ABL1, which typically suppresses kinase activity via allosteric
interactions with the SH1 domain (27). The lack of SH3 disrupts the SH3-SH2
complex, destabilizing the kinase's inactive conformation. The
absence of SH3 residues critical for its inhibitory role (such as
Tyr-89 and Tyr-134) leads to persistent kinase activation (12). This structural modification also
hinders the binding of allosteric inhibitors such as asciminib to
the myristoyl pocket, thereby facilitating resistance. Conversely,
ATP-competitive TKIs retain efficacy as their interaction with the
active kinase conformation remains unaltered (28). Kinase assays and molecular dynamics
simulations elucidate that SH3 truncation affects the spatial
configuration of the ATP-binding cleft and modifies the allosteric
signaling network (26), accounting
for the varied TKI resistance profiles in BCR-ABL1
leukemogenesis.
The present single-case report lacks the statistical
power to establish definitive clinical correlations, and the
observed outcomes may not represent the wider population of
patients with atypical transcripts. Consequently, clinical
management should implement a dual-faceted strategy: Pursuing
molecular responses via personalized TKI regimens while remaining
alert to the onset of treatment-related toxicities, thereby
facilitating prompt dose adjustments or therapeutic shifts to
enhance adherence and efficacy. In summary, the long-term
management of patients with e13a3 BCR-ABL1 CML requires the
incorporation of accurate molecular diagnostics, customized
therapeutic approaches and vigilant monitoring for adverse events.
Future multicenter collaborative studies and systematic reviews
that consolidate data from analogous rare cases are essential to
substantiate these preliminary findings and to formulate
evidence-based clinical guidelines. Until such data are accessible,
clinicians should regard the current findings as
hypothesis-generating and exercise prudence when extrapolating
these results to other instances involving atypical
transcripts.
Acknowledgements
Not applicable.
Funding
The work was supported by a grant from the Taishan Youth Scholar
Foundation of Shandong Province (grant no. tsqn201812140).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XZ and NS confirm the authenticity of all the raw
data. XZ designed the study. ML performed the acquisition, analysis
and interpretation of data. NS performed the analysis and
interpretation of data, and contributed to manuscript drafting and
critical revisions of the intellectual content. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
The patient provided written informed consent for
publication, authorizing the use of their imaging, pathological and
clinical data for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arzoun H, Srinivasan M, Thangaraj SR,
Thomas SS and Mohammed L: The progression of chronic myeloid
leukemia to myeloid sarcoma: A systematic review. Cureus.
14:e210772022.PubMed/NCBI
|
|
2
|
Tsuchiya K, Tabe Y, Ai T, Ohkawa T, Usui
K, Yuri M, Misawa S, Morishita S, Takaku T, Kakimoto A, et al:
Eprobe mediated RT-qPCR for the detection of leukemia-associated
fusion genes. PLoS One. 13:e02024292018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luatti S, Baldazzi C, Marzocchi G, Ameli
G, Bochicchio MT, Soverini S, Castagnetti F, Tiribelli M, Gugliotta
G, Martinelli G, et al: Cryptic BCR-ABL fusion gene as variant
rearrangement in chronic myeloid leukemia: Molecular cytogenetic
characterization and influence on TKIs therapy. Oncotarget.
8:29906–29913. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abdulmawjood B, Costa B, Roma-Rodrigues C,
Baptista PV and Fernandes AR: Genetic biomarkers in chronic myeloid
leukemia: What have we learned so far? Int J Mol Sci. 22:125162021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Soverini S, Mancini M, Bavaro L, Cavo M
and Martinelli G: Chronic myeloid leukemia: The paradigm of
targeting oncogenic tyrosine kinase signaling and counteracting
resistance for successful cancer therapy. Mol Cancer. 17:492018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abdullah MA, Amer A, Nawaz Z, Abdullah AS,
Al-Sabbagh A, Kohla S, Nashwan AJ and Yassin MA: An uncommon case
of chronic myeloid leukemia with variant cytogenetic. Acta Biomed.
89:28–32. 2018.PubMed/NCBI
|
|
7
|
Al-Bayati AMS, Al-Bayti AAH and Husain VI:
A short review about chronic myeloid leukemia. J Life Bio Sci Res.
4:15–19. 2023. View Article : Google Scholar
|
|
8
|
Iqbal Z, Absar M, Akhtar T, Aleem A,
Jameel A, Basit S, Ullah A, Afzal S, Ramzan K, Rasool M, et al:
Integrated genomic analysis identifies ANKRD36 gene as a novel and
common biomarker of disease progression in chronic myeloid
leukemia. Biology (Basel). 10:11822021.PubMed/NCBI
|
|
9
|
Burmeister T and Reinhardt R: A multiplex
PCR for improved detection of typical and atypical BCR-ABL fusion
transcripts. Leuk Res. 32:579–585. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue M, Wang Q, Huo L, Wen L, Yang X, Wu Q,
Pan J, Cen J, Ruan C, Wu D and Chen S: Clinical characteristics and
prognostic significance of chronic myeloid leukemia with rare
BCR-ABL1 transcripts. Leuk Lymphoma. 60:3051–3057. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Molica M, Abruzzese E and Breccia M:
Prognostic significance of transcript-Type BCR-ABL1 in chronic
myeloid leukemia. Mediterr J Hematol Infect Dis. 12:e20200622020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schäfer V, White HE, Gerrard G, Möbius S,
Saussele S, Franke GN, Mahon FX, Talmaci R, Colomer D, Soverini S,
et al: Assessment of individual molecular response in chronic
myeloid leukemia patients with atypical BCR-ABL1 fusion
transcripts: Recommendations by the EUTOS cooperative network. J
Cancer Res Clin Oncol. 147:3081–3089. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jean J, Sukhanova M, Dittmann D, Gao J and
Jennings LJ: A novel BCR::ABL1 variant detected with multiple
testing modalities. Case Rep Hematol. 2024:84862672024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crampe M, Kearney L, O'Brien D, Bacon CL,
O'Shea D and Langabeer SE: Molecular monitoring in adult
philadelphia chromosome-positive acute lymphoblastic leukemia with
the variant e13a3 BCR-ABL1 fusion. Case Rep Hematol.
2019:96350702019.PubMed/NCBI
|
|
15
|
Hu LH, Pu LF, Yang DD, Zhang C, Wang HP,
Ding YY, Li MM, Zhai ZM and Xiong S: How to detect the rare BCR-ABL
(e14a3) transcript: A case report and literature review. Oncol
Lett. 14:5619–5623. 2017.PubMed/NCBI
|
|
16
|
Li Y, Zhang Y, Meng X, Chen S, Wang T,
Zhang L and Ma X: Chronic myeloid leukemia with two rare fusion
gene transcripts of atypical BCR::ABL: A case report and literature
review. Medicine (Baltimore). 103:e367282024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pecciarini L, Brunetto E, Grassini G, De
Pascali V, Ogliari FR, Talarico A, Marra G, Magliacane G, Redegalli
M, Arrigoni G, et al: Gene fusion detection in NSCLC routine
clinical practice: Targeted-NGS or FISH? Cells. 12:11352023.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee T, Clarke JM, Jain D, Ramalingam S and
Vashistha V: Precision treatment for metastatic non-small cell lung
cancer: A conceptual overview. Cleve Clin J Med. 88:117–127. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Su YJ, Kuo MC, Chen TY, Wang MC, Yang Y,
Ma MC, Lin TL, Lin TH, Chang H, Teng CJ, et al: Comparison of
molecular responses and outcomes between BCR::ABL1 e14a2 and e13a2
transcripts in chronic myeloid leukemia. Cancer Sci. 113:3518–3527.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leong WMS and Aw CWD: Nilotinib-induced
keratosis pilaris. Case Rep Dermatol. 8:91–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cross NCP, Ernst T, Branford S, Cayuela
JM, Deininger M, Fabarius A, Kim DDH, Machova Polakova K, Radich
JP, Hehlmann R, et al: European LeukemiaNet laboratory
recommendations for the diagnosis and management of chronic myeloid
leukemia. Leukemia. 37:2150–2167. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kearney L, Crampe M and Langabeer SE:
Frequency and spectrum of atypical BCR-ABL1 transcripts in chronic
myeloid leukemia. Exp Oncol. 42:78–79. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee SE, Choi SY, Kim SH, Song HY, Yoo HL,
Lee MY, Kang KH, Hwang HJ, Jang EJ and Kim DW: BCR-ABL1 transcripts
(MR4.5) at post-transplant 3 months as an early
predictor for long-term outcomes in chronic myeloid leukemia.
Korean J Intern Med. 32:125–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pai HL, Liu CY and Yeh MH:
Scleroderma-like lesions in a patient undergoing combined
pembrolizumab and routine chemotherapy: A case report and
literature review. Medicina (Kaunas). 60:10922024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang YN, Chu WY, Chen JS, Yeh YH and Cheng
CN: Long-term outcomes of chronic myeloid leukemia in children and
adolescents-real world data from a single-institute in Taiwan. J
Formos Med Assoc. S0929-6646(25)00014-2. 2025.(Epub ahead of
print). View Article : Google Scholar
|
|
26
|
Lang F, Wunderle L, Pfeifer H, Schnittger
S, Bug G and Ottmann OG: Dasatinib and azacitidine followed by
haploidentical stem cell transplant for chronic myeloid leukemia
with evolving myelodysplasia: A case report and review of treatment
options. Am J Case Rep. 18:1099–1109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leske IB and Hantschel O: The e13a3 (b2a3)
and e14a3 (b3a3) BCR::ABL1 isoforms are resistant to asciminib.
Leukemia. 38:2041–2045. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hijiya N and Mauro MJ: Asciminib in the
treatment of philadelphia chromosome-positive chronic myeloid
leukemia: Focus on patient selection and outcomes. Cancer Manag
Res. 15:873–891. 2023. View Article : Google Scholar : PubMed/NCBI
|