|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shanmugasundaram S and You J: Targeting
persistent human papillomavirus infection. Viruses. 9:2292017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu H, Ma H, Li Y and Zhao H: Advances in
epigenetic modifications and CC research. Biochim Biophys Acta Rev
Cancer. 1878:1888942023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao F, Dong Z, Li Y, Liu S, Guo P, Zhang
D and Li S: Comprehensive analysis of molecular clusters and
prognostic signature based on m7G-related LncRNAs in esophageal
squamous cell carcinoma. Front Oncol. 12:8931862022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo Y, Yao Y, Wu P, Zi X, Sun N and He J:
The potential role of N7-methylguanosine (m7G) in cancer. J Hematol
Oncol. 15:632022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alexandrov A, Martzen MR and Phizicky EM:
Two proteins that form a complex are required for 7-methylguanosine
modification of yeast tRNA. RNA. 8:1253–1266. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ying X, Liu B, Yuan Z, Huang Y, Chen C,
Jiang X, Zhang H, Qi D, Yang S, Lin S, et al: METTL1-m7
G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin
Transl Med. 11:e6752021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okamoto M, Fujiwara M, Hori M, Okada K,
Yazama F, Konishi H, Xiao Y, Qi G, Shimamoto F, Ota T, et al: tRNA
modifying enzymes, NSUN2 and METTL1, determine sensitivity to
5-fluorouracil in HeLa cells. PLoS Genet. 10:e10046392014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang M, Long J, Yao Z, Zhao Y, Zhao Y,
Liao J, Lei K, Xiao H, Dai Z, Peng S, et al: METTL1-Mediated m7G
tRNA modification promotes lenvatinib resistance in hepatocellular
carcinoma. Cancer Res. 83:89–102. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng W, Gao A, Lin H and Zhang W: Novel
roles of METTL1/WDR4 in tumor via m7G methylation. Mol Ther
Oncolytics. 26:27–34. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng Y, Zhou Z, Ji W, Lin S and Wang M:
METTL1-mediated m7G methylation maintains pluripotency in human
stem cells and limits mesoderm differentiation and vascular
development. Stem Cell Res Ther. 11:3062020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He J, Huang B, Zhang K, Liu M and Xu T:
Long non-coding RNA in CC: From biology to therapeutic opportunity.
Biomed Pharmacother. 127:1102092020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME, Sunkin SM, Mehler MF
and Mattick JS: Specific expression of long noncoding RNAs in the
mouse brain. Proc Natl Acad Sci USA. 105:716–721. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ravasi T, Suzuki H, Pang KC, Katayama S,
Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al:
Experimental validation of the regulated expression of large
numbers of non-coding RNAs from the mouse genome. Genome Res.
16:11–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brunner AL, Beck AH, Edris B, Sweeney RT,
Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al:
Transcriptional profiling of long non-coding RNAs and novel
transcribed regions across a diverse panel of archived human
cancers. Genome Biol. 13:R752012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y,
Brown M, Chen Y and Liu XS: Integrative genomic analyses reveal
clinically relevant long noncoding RNAs in human cancer. Nat Struct
Mol Biol. 20:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tomikawa C: 7-Methylguanosine
modifications in transfer RNA (tRNA). Int J Mol Sci. 19:40802018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Singh D, Vignat J, Lorenzoni V, Eslahi M,
Ginsburg O, Lauby-Secretan B, Arbyn M, Basu P, Bray F and
Vaccarella S: Global estimates of incidence and mortality of CC in
2020: A baseline analysis of the WHO Global CC elimination
initiative. Lancet Glob Health. 11:e197–e206. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
John RM and Rougeulle C: Developmental
epigenetics: Phenotype and the flexible epigenome. Front Cell Dev
Biol. 6:1302018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gibb EA, Becker-Santos DD, Enfield KS,
Guillaud M, van Niekerk D, Matisic JP, Macaulay CE and Lam WL:
Aberrant expression of long noncoding RNAs in cervical
intraepithelial neoplasia. Int J Gynecol Cancer. 22:1557–1563.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu XL, Huang XT, Zhang JN, Liu J, Wen LJ,
Xu X and Zhou JY: Long noncoding RNA MIR210HG is induced by
hypoxia-inducible factor 1α and promotes CC progression. Am J
Cancer Res. 12:2783–2797. 2022.PubMed/NCBI
|
|
33
|
Sharma S and Munger K: Expression of the
long noncoding RNA DINO in human papillomavirus-positive CC cells
reactivates the dormant TP53 tumor suppressor through ATM/CHK2
signaling. mBio. 11:e01190–e01120. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao H, Zheng GH, Li GC, Xin L, Wang YS,
Chen Y and Zheng XM: Long noncoding RNA LINC00958 regulates cell
sensitivity to radiotherapy through RRM2 by binding to
microRNA-5095 in CC. J Cell Physiol. 234:23349–23359. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J, Chew BL, Lai Y, Dong H, Xu L,
Balamkundu S, Cai WM, Cui L, Liu CF, Fu XY, et al: Quantifying the
RNA cap epitranscriptome reveals novel caps in cellular and viral
RNA. Nucleic Acids Res. 47:e1302019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ji H, Zhang JA, Liu H, Li K, Wang ZW and
Zhu X: Comprehensive characterization of tumor microenvironment and
m6A RNA methylation regulators and its effects on PD-L1 and immune
infiltrates in cervical cancer. Front Immunol. 13:9761072022.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen J, Li K, Chen J, Wang X, Ling R,
Cheng M, Chen Z, Chen F, He Q, Li S, et al: Aberrant translation
regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine
modification drives head and neck squamous cell carcinoma
progression. Cancer Commun (Lond). 42:223–244. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma J, Han H, Huang Y, Yang C, Zheng S, Cai
T, Bi J, Huang X, Liu R, Huang L, et al: METTL1/WDR4-mediated m7G
tRNA modifications and m7G codon usage promote mRNA translation and
lung cancer progression. Mol Ther. 29:3422–3435. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen Z, Zhu W, Zhu S, Sun K, Liao J, Liu
H, Dai Z, Han H, Ren X, Yang Q, et al: METTL1 promotes
hepatocarcinogenesis via m7 G tRNA modification-dependent
translation control. Clin Transl Med. 11:e6612021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Zhang Y, Chi Q, Wang Z and Sun B:
RETRACTED: Methyltransferase-like 1 (METTL1) served as a tumor
suppressor in colon cancer by activating 7-methyguanosine (m7G)
regulated let-7e miRNA/HMGA2 axis. Life Sci. 249:1174802020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qiu Z, He L, Yu F, Lv H and Zhou Y: LncRNA
FAM13A-AS1 regulates proliferation and apoptosis of cervical cancer
cells by targeting miRNA-205-3p/DDI2 axis. J Oncol.
2022:84119192022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang XJ, Li S, Fang J, Yan ZJ and Luo GC:
LncRNA FAM13A-AS1 promotes renal carcinoma tumorigenesis through
sponging miR-141-3p to upregulate NEK6 expression. Front Mol
Biosci. 9:7387112022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sugino RP, Ohira M, Mansai SP and Kamijo
T: Comparative epigenomics by machine learning approach for
neuroblastoma. BMC Genomics. 23:8522022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Roh J, Im M, Kang J, Youn B and Kim W:
Long non-coding RNA in glioma: Novel genetic players in
temozolomide resistance. Anim Cells Syst (Seoul). 27:19–28. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo X and Li M: LINC01089 is a
tumor-suppressive lncRNA in gastric cancer and it regulates
miR-27a-3p/TET1 axis. Cancer Cell Int. 20:5072020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang H, Zhang H, Li X, Huang S, Guo Q and
Geng D: LINC01089 functions as a ceRNA for miR-152-3p to inhibit
non-small lung cancer progression through regulating PTEN. Cancer
Cell Int. 21:1432021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li M and Guo X: LINC01089 blocks the
proliferation and metastasis of colorectal cancer cells via
regulating miR-27b-3p/HOXA10 axis. Onco Targets Ther. 13:8251–8260.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li X, Lv F, Li F, Du M, Liang Y, Ju S, Liu
Z, Zhou B, Wang B and Gao Y: LINC01089 inhibits tumorigenesis and
epithelial-mesenchymal transition of non-small cell lung cancer via
the miR-27a/SFRP1/Wnt/β-catenin axis. Front Oncol. 10:5325812020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li S, Han Y, Liang X and Zhao M: LINC01089
inhibits the progression of CC via inhibiting miR-27a-3p and
increasing BTG2. J Gene Med. 23:e32802021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu W, Zhao Y, Liu X, Zhang X, Ding J, Li
Y, Tian Y, Wang H, Liu W and Lu Z: A novel meiosis-related lncRNA,
Rbakdn, contributes to spermatogenesis by stabilizing Ptbp2. Front
Genet. 12:7524952021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qin R, Cao L, Ye C, Wang J and Sun Z: A
novel prognostic prediction model based on seven immune-related
RNAs for predicting overall survival of patients in early cervical
squamous cell carcinoma. BMC Med Genomics. 14:492021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tomoo Y: Prognostic factors of ovarian
cancer at our department. Igaku Kenkyu. 57:154–164. 1987.(In
Japanese). PubMed/NCBI
|
|
53
|
Xin Y, Shang X, Sun X, Xu G and Liu Y and
Liu Y: SLC8A1 antisense RNA 1 suppresses papillary thyroid cancer
malignant progression via the FUS RNA binding protein (FUS)/NUMB
like endocytic adaptor protein (Numbl) axis. Bioengineered.
13:12572–12582. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Y, Cao X and Li H: Identification and
validation of novel long non-coding RNA biomarkers for early
diagnosis of oral squamous cell carcinoma. Front Bioeng Biotechnol.
8:2562020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Komi DEA and Redegeld FA: Role of mast
cells in shaping the tumor microenvironment. Clin Rev Allergy
Immunol. 58:313–325. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu X, Li X, Wei H, Liu Y and Li N: Mast
cells in colorectal cancer tumour progression, angiogenesis, and
lymphangiogenesis. Front Immunol. 14:12090562023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shan F, Somasundaram A, Bruno TC, Workman
CJ and Vignali DAA: Therapeutic targeting of regulatory T cells in
cancer. Trends Cancer. 8:944–961. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tzankov A, Meier C, Hirschmann P, Went P,
Pileri SA and Dirnhofer S: Correlation of high numbers of
intratumoral FOXP3+ regulatory T cells with improved survival in
germinal center-like diffuse large B-cell lymphoma, follicular
lymphoma and classical Hodgkin's lymphoma. Haematologica.
93:193–200. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dolina JS, Van Braeckel-Budimir N, Thomas
GD and Salek-Ardakani S: CD8+ T cell exhaustion in cancer. Front
Immunol. 12:7152342021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Muthusami S, Sabanayagam R, Periyasamy L,
Muruganantham B and Park WY: A review on the role of epidermal
growth factor signaling in the development, progression and
treatment of CC. Int J Biol Macromol. 194:179–187. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Boromand N, Hasanzadeh M, ShahidSales S,
Farazestanian M, Gharib M, Fiuji H, Behboodi N, Ghobadi N,
Hassanian SM, Ferns GA and Avan A: Clinical and prognostic value of
the C-Met/HGF signaling pathway in cervical cancer. J Cell Physiol.
233:4490–4496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Guo K, Shelat AA, Guy RK and Kastan MB:
Development of a cell-based, high-throughput screening assay for
ATM kinase inhibitors. J Biomol Screen. 19:538–546. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hsieh MJ, Weng CC, Lin YC, Wu CC, Chen LT
and Cheng KH: Inhibition of β-catenin activity abolishes LKB1
loss-driven pancreatic cystadenoma in mice. Int J Mol Sci.
22:46492021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lu W, Ni K, Li Z, Xiao L, Li Y, Jiang Y,
Zhang J and Shi H: Salubrinal protects against cisplatin-induced
cochlear hair cell endoplasmic reticulum stress by regulating
eukaryotic translation initiation factor 2α signalling. Front Mol
Neurosci. 15:9164582022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
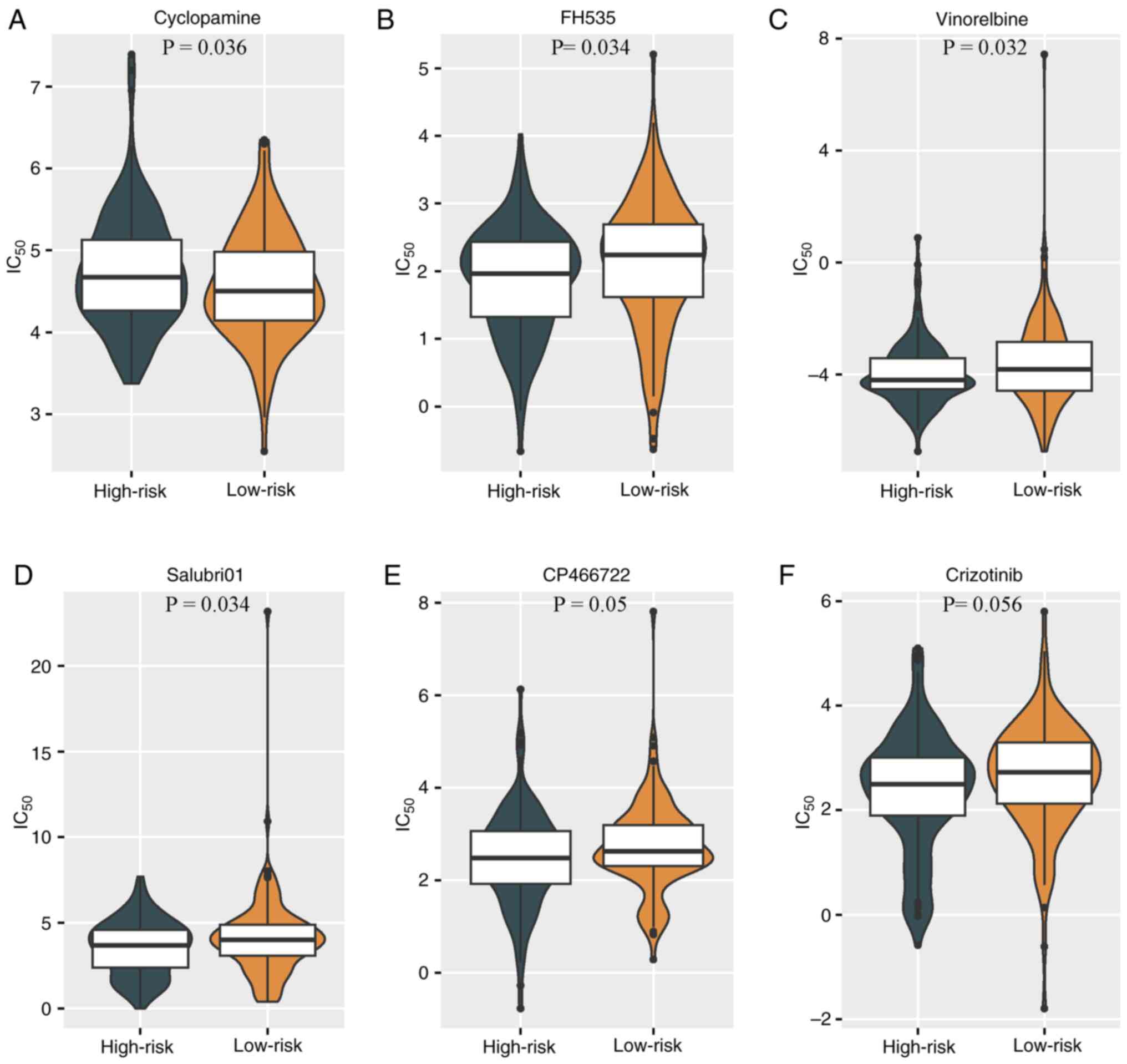
Frenel JS, Mathiot L, Cropet C, Borcoman
E, Hervieu A, Coquan E, De La Motte Rouge T, Saada-Bouzid E,
Sabatier R, Lavaud P, et al: Durvalumab and tremelimumab in
combination with metronomic oral vinorelbine for recurrent advanced
cervical cancer: An open-label phase I/II study. J Immunother
Cancer. 13:e0107082025. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Varma DA and Tiwari M: Crizotinib-induced
anti-cancer activity in human cervical carcinoma cells via
ROS-dependent mitochondrial depolarization and induction of
apoptotic pathway. J Obstet Gynaecol Res. 47:3923–3930. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jin MH and Oh DY: ATM in DNA repair in
cancer. Pharmacol Ther. 203:1073912019. View Article : Google Scholar : PubMed/NCBI
|