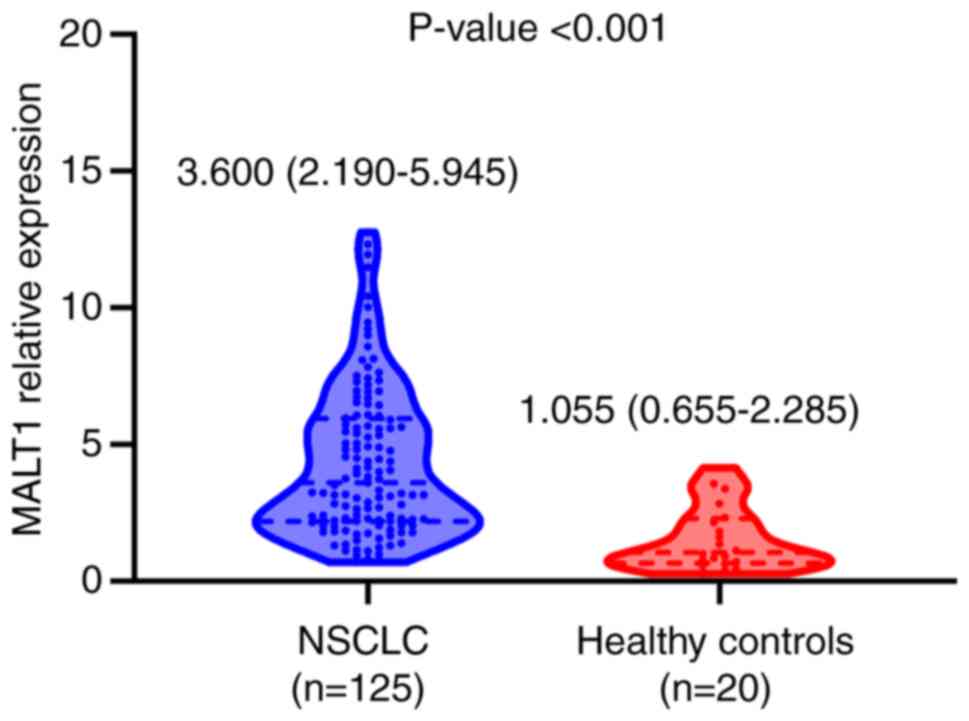
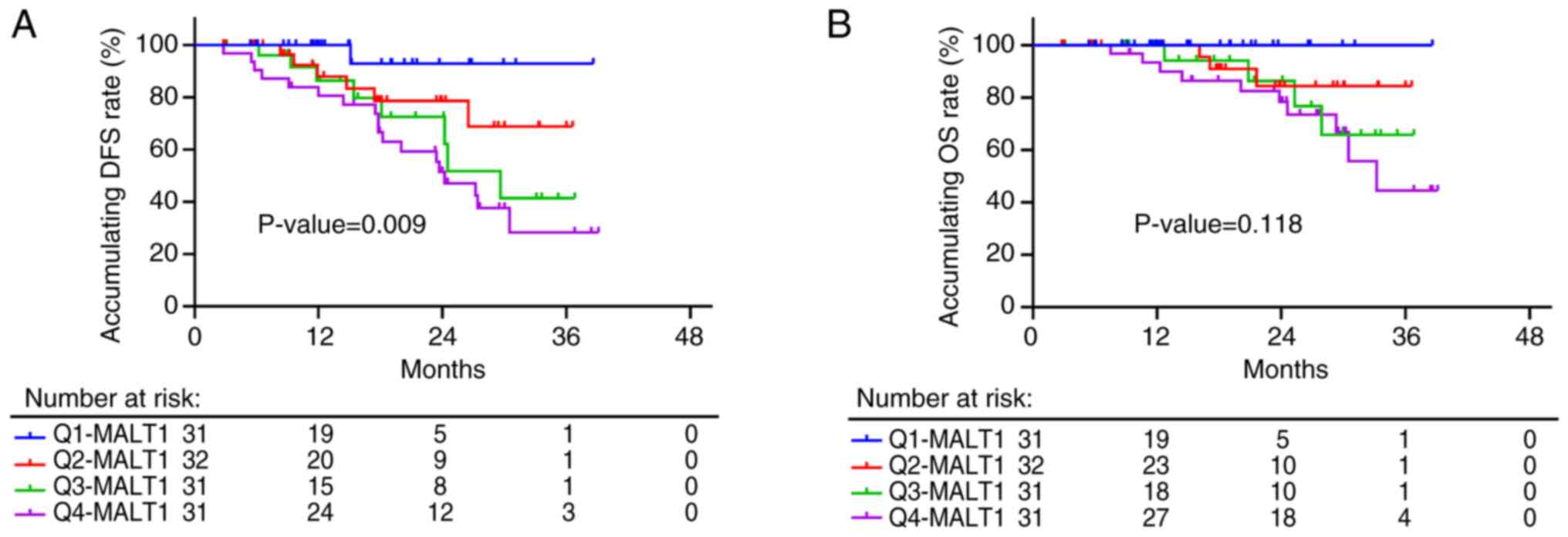
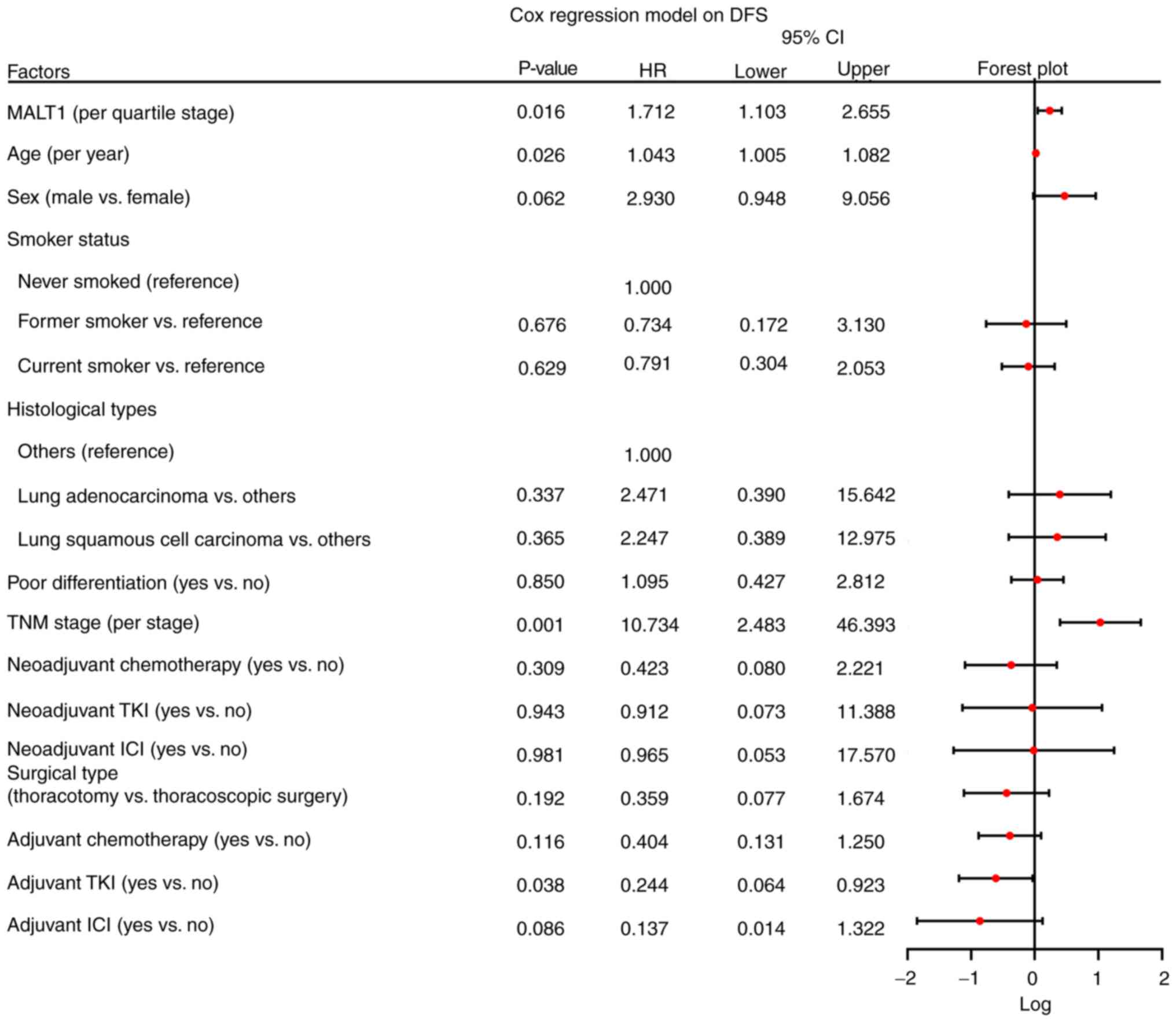
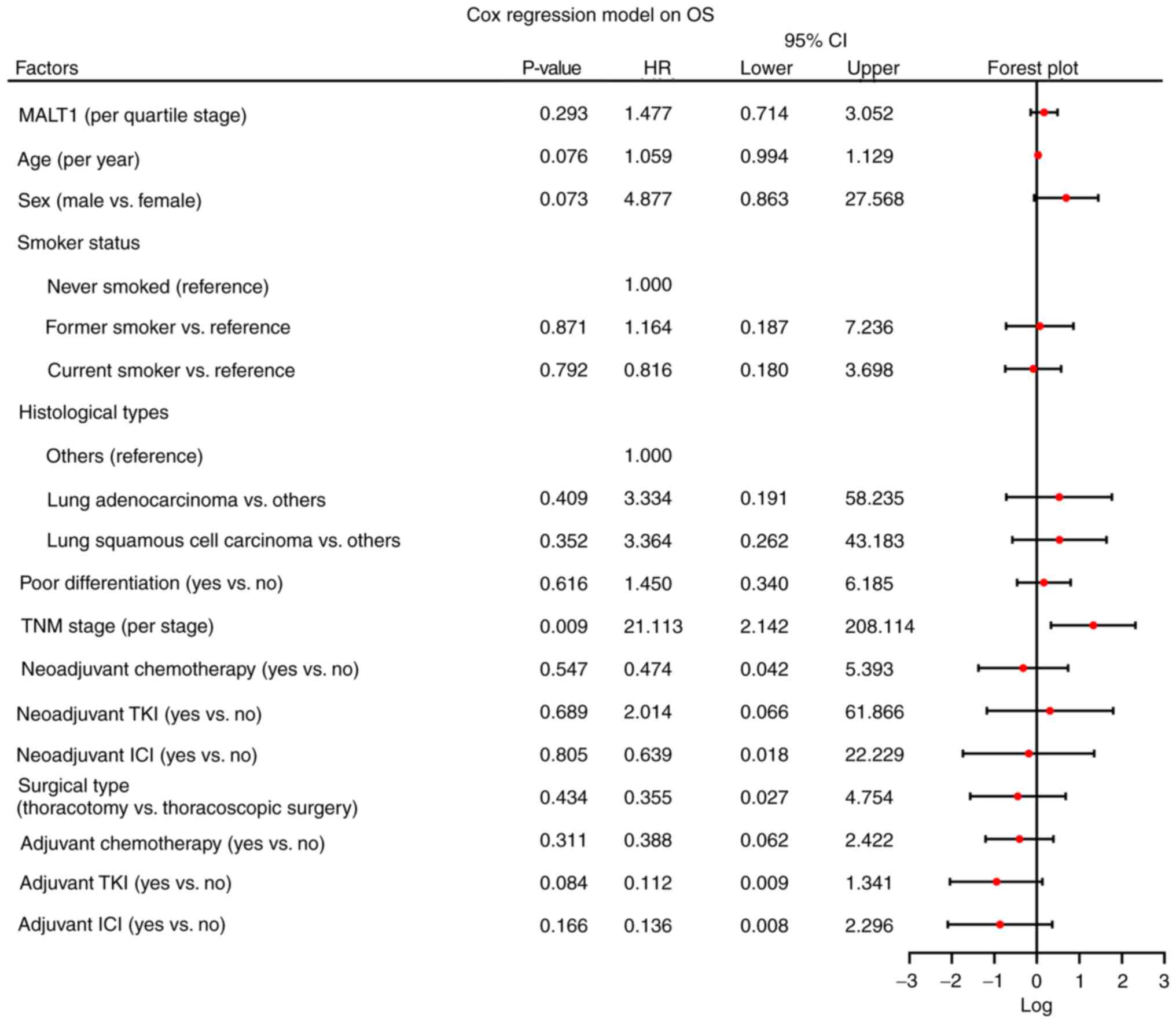
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng RS, Zhang SW, Sun KX, Chen R, Wang
SM, Li L, Zeng HM, Wei WW and He J: Cancer statistics in China,
2016. Zhonghua Zhong Liu Za Zhi. 45:212–220. 2023.(In Chinese).
PubMed/NCBI
|
|
3
|
Leiter A, Veluswamy RR and Wisnivesky JP:
The global burden of lung cancer: Current status and future trends.
Nat Rev Clin Oncol. 20:624–639. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qi J, Li M, Wang L, Hu Y, Liu W, Long Z,
Zhou Z, Yin P and Zhou M: National and subnational trends in cancer
burden in China, 2005-20: An analysis of national mortality
surveillance data. Lancet Public Health. 8:e943–e955. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen P, Liu Y, Wen Y and Zhou C: Non-small
cell lung cancer in China. Cancer Commun (Lond). 42:937–970. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang SR, Schultheis AM, Yu H, Mandelker D,
Ladanyi M and Buttner R: Precision medicine in non-small cell lung
cancer: Current applications and future directions. Semin Cancer
Biol. 84:184–198. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Restrepo JC, Duenas D, Corredor Z and
Liscano Y: Advances in genomic data and biomarkers: Revolutionizing
NSCLC diagnosis and treatment. Cancers (Basel). 15:34742023.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tostes K, Siqueira AP, Reis RM, Leal LF
and Arantes L: Biomarkers for immune checkpoint inhibitor response
in NSCLC: Current developments and applicability. Int J Mol Sci.
24:118872023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
O'Neill TJ, Tofaute MJ and Krappmann D:
Function and targeting of MALT1 paracaspase in cancer. Cancer Treat
Rev. 117:1025682023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gomez Solsona B, Schmitt A,
Schulze-Osthoff K and Hailfinger S: The paracaspase malt1 in
cancer. Biomedicines. 10:3442022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou B, Mo Z, Lai G, Chen X, Li R, Wu R,
Zhu J and Zheng F: Targeting tumor exosomal circular RNA cSERPINE2
suppresses breast cancer progression by modulating MALT1-NF-.
B-IL-6 axis of tumor-associated macrophages. J Exp Clin Cancer Res.
42:482023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vanneste D, Staal J, Haegman M, Driege Y,
Carels M, Van Nuffel E, De Bleser P, Saeys Y, Beyaert R and Afonina
IS: CARD14 signalling ensures cell survival and cancer associated
gene expression in prostate cancer cells. Biomedicines.
10:20082022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kurden-Pekmezci A, Cakiroglu E, Eris S,
Mazi FA, Coskun-Deniz OS, Dalgic E, Oz O and Senturk S: MALT1
paracaspase is overexpressed in hepatocellular carcinoma and
promotes cancer cell survival and growth. Life Sci. 323:1216902023.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Minderman M, Lantermans HC, Gruneberg LJ,
Cillessen SAGM, Bende RJ, van Noesel CJM, Kersten MJ, Pals ST and
Spaargaren M: MALT1-dependent cleavage of CYLD promotes NF-κB
signaling and growth of aggressive B-cell receptor-dependent
lymphomas. Blood Cancer J. 13:372023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Di Pilato M, Gao Y, Sun Y, Fu A, Grass C,
Seeholzer T, Feederle R, Mazo I, Kazer SW, Litchfield K, et al:
Translational studies using the MALT1 inhibitor (S)-Mepazine to
induce treg fragility and potentiate immune checkpoint therapy in
cancer. J Immunother Precis Oncol. 6:61–73. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li C, Yu F and Xu W: Early low blood MALT1
expression levels forecast better efficacy of PD-1 inhibitor-based
treatment in patients with metastatic colorectal cancer. Oncol
Lett. 26:3292023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Zhang G, Jin J, Degan S, Tameze Y
and Zhang JY: MALT1 promotes melanoma progression through JNK/c-Jun
signaling. Oncogenesis. 6:e3652017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tibiletti MG, Martin V, Bernasconi B, Del
Curto B, Pecciarini L, Uccella S, Pruneri G, Ponzoni M,
Mazzucchelli L, Martinelli G, et al: BCL2, BCL6, MYC, MALT 1, and
BCL10 rearrangements in nodal diffuse large B-cell lymphomas: A
multicenter evaluation of a new set of fluorescent in situ
hybridization probes and correlation with clinical outcome. Hum
Pathol. 40:645–652. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan D, Jiang C, Ma Z, Blonska M, You MJ
and Lin X: MALT1 is required for EGFR-induced NF-κB activation and
contributes to EGFR-driven lung cancer progression. Oncogene.
35:919–928. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Israel L, Gluck A, Berger M, Coral M, Ceci
M, Unterreiner A, Rubert J, Bardet M, Ginster S, Golding-Ochsenbein
AM, et al: CARD10 cleavage by MALT1 restricts lung carcinoma growth
in vivo. Oncogenesis. 10:322021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mempel TR and Krappmann D: Combining
precision oncology and immunotherapy by targeting the MALT1
protease. J Immunother Cancer. 10:e0054422022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Huang Q and He F: Aberrant blood
MALT1 and its relevance with multiple organic dysfunctions, T
helper cells, inflammation, and mortality risk of sepsis patients.
J Clin Lab Anal. 36:e243312022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qian R, Niu X, Wang Y, Guo Z, Deng X, Ding
Z, Zhou M and Deng H: Targeting MALT1 suppresses the malignant
progression of colorectal cancer via miR-375/miR-365a-3p/NF-κB
axis. Front Cell Dev Biol. 10:8450482022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saba NS, Wong DH, Tanios G, Iyer JR,
Lobelle-Rich P, Dadashian EL, Liu D, Fontan L, Flemington EK,
Nichols CM, et al: MALT1 inhibition is efficacious in both naive
and ibrutinib-resistant chronic lymphocytic leukemia. Cancer Res.
77:7038–7048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Yue C, Shi L, Liu G, Cao Q, Shan Q,
Wang Y, Chen X, Li H, Wang J, et al: MALT1 is a potential
therapeutic target in glioblastoma and plays a crucial role in
EGFR-induced NF-κB activation. J Cell Mol Med. 24:7550–7562. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Flynn SM, Chen C, Artan M, Barratt S,
Crisp A, Nelson GM, Peak-Chew SY, Begum F, Skehel M and de Bono M:
MALT-1 mediates IL-17 neural signaling to regulate C. elegans
behavior, immunity and longevity. Nat Commun. 11:20992020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu JW, Hoffman S, Beal AM, Dykon A,
Ringenberg MA, Hughes AC, Dare L, Anderson AD, Finger J, Kasparcova
V, et al: MALT1 protease activity is required for innate and
adaptive immune responses. PLoS One. 10:e01270832015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng L, Deng N, Yang N, Zhao X and Lin X:
Malt1 protease is critical in maintaining function of regulatory T
cells and may be a therapeutic target for antitumor immunity. J
Immunol. 202:3008–3019. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kingeter LM and Schaefer BC: Malt1 and
cIAP2-Malt1 as effectors of NF-kappaB activation: Kissing cousins
or distant relatives? Cell Signal. 22:9–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen W, Li Z, Bai L and Lin Y: NF-kappaB
in lung cancer, a carcinogenesis mediator and a prevention and
therapy target. Front Biosci (Landmark Ed). 16:1172–1185. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pathan J, Mondal S, Sarkar A and
Chakrabarty D: Daboialectin, a C-type lectin from Russell's viper
venom induces cytoskeletal damage and apoptosis in human lung
cancer cells in vitro. Toxicon. 127:11–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo B, Chu X, Yu P and Tian J: The TCR
repertoire diversity and its application in the prevention and
treatment of lung cancer. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
38:939–943. 2022.(In Chinese). PubMed/NCBI
|
|
36
|
Che T, You Y, Wang D, Tanner MJ, Dixit VM
and Lin X: MALT1/paracaspase is a signaling component downstream of
CARMA1 and mediates T cell receptor-induced NF-kappaB activation. J
Biol Chem. 279:15870–15876. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shah K, Al-Haidari A, Sun J and Kazi JU: T
cell receptor (TCR) signaling in health and disease. Signal
Transduct Target Ther. 6:4122021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taniuchi I: CD4 Helper and CD8 Cytotoxic T
Cell Differentiation. Annu Rev Immunol. 36:579–601. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Chen X, Wang Z, Zhao D, Chen H, Chen
W, Zhou Z, Zhang J, Zhang J, Li H and Chen C: The HECTD3 E3
ubiquitin ligase suppresses cisplatin-induced apoptosis via
stabilizing MALT1. Neoplasia. 15:39–48. 2013. View Article : Google Scholar : PubMed/NCBI
|