Introduction
The National Cancer Center report on cancer
incidence and mortality in China in 2022 indicated that colorectal
cancer (CRC) is the second most prevalent tumor type, following
lung cancer (1). Notably, the liver
is the most common site of metastasis for CRC and liver metastasis
is a leading cause of mortality in patients (2). The management of liver metastases in
CRC poses a key challenge in current treatment practices. Numerous
patients with CRC have been reported to possess micrometastases in
surgically resected liver specimens, which are considered invisible
to the naked eye and imaging examinations before surgery, and these
micrometastases often develop into metachronous liver metastases
(MLM) (3–5). Consequently, MLM is prone to being
overlooked during follow-up, which leads to treatment failure
(6).
Compared with other treatments, such as radiotherapy
and chemotherapy, tumor ablation notably improves patient prognosis
and survival (7); however, current
imaging methods such as CT, MRI and ultrasound, can occasionally
miss microscopic lesions, which delay diagnosis and treatment.
Consequently, the establishment of robust screening protocols is
necessary to identify patients with CRC at an elevated risk of
developing MLM, thereby facilitating early detection, which would
enable timely surgical intervention, markedly improve prognosis and
enhance overall survival in patients (8). Previous studies have explored various
clinical factors, such as carcinoembryonic antigen (CEA) levels,
lymph node status, KRAS and primary tumor site, as potential
biomarkers for MLM; however, a clear consensus regarding these
biomarkers has not yet been established (9,10).
Radiomics is a novel technique that extracts
quantitative features from medical images and transforms the
features into mineable data. Through radiomics, key imaging
biomarkers that are undetectable to the naked eye can be identified
(11,12). A key advantage of radiomics is the
ability to analyze medical images across different modalities
(13). The proposed methodology
synergistically integrates multimodal data acquired from diverse
imaging modalities, which effectively overcome the constraints
inherent in single-modal analysis while facilitating a robust
evaluation of potential synergistic benefits. By adopting a
cross-modality approach, radiomics maximizes the use of extracted
imaging information and provides a more comprehensive understanding
of the underlying pathology (14,15).
We hypothesized that a change in the liver
microenvironment may occur before the appearance of imaging
manifestations. Previous studies that have investigated radiomics
in patients with CRC predominantly focused on a unimodal approach
or on multiple sequences with a single modality (8,16,17).
To the best of our knowledge, there are a limited number of
literature reports on the application of multimodality in patients
with CRC. Therefore, the present study aimed to develop an
integrated model combining CT and MRI modalities to improve the
prediction of MLM in patients with CRC. The primary objective of
the present study was to establish an imaging-based foundation for
clinical prediction and prevention of MLM in patients with CRC,
which may potentially prolong patient survival in the future.
Materials and methods
Subjects
The present study retrospectively analyzed a total
of 1,144 patients with CRC clinically diagnosed at Sir Run Run Shaw
Hospital (Zhejiang, China) between January 2010 and December 2020.
The present study received approval from the Ethical Review
Committee of Sir Run Run Shaw Hospital, Zhejiang University School
of Medicine (approval no. 0465-2022; Zhejiang, China) and a waiver
of consent was obtained due to the retrospective nature of the
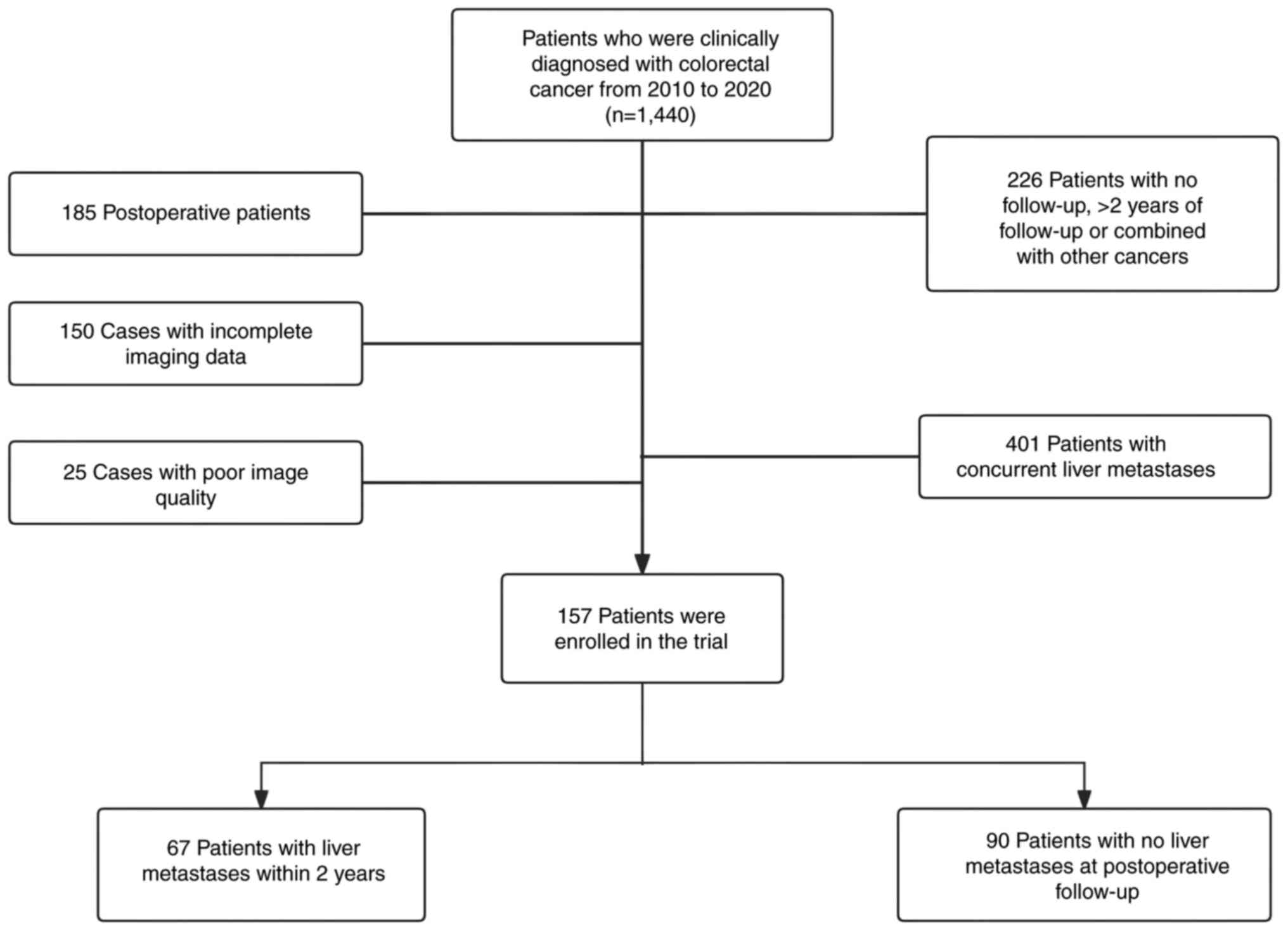
present study. A total of 157 cases were included in the present
study, which included 67 patients with liver metastases within 2
years of treatment and 90 patients without liver metastases at
postoperative follow-up. Clinical data of patients in terms of age,
sex, carcinoembryonic antigen (CEA), carbohydrate antigen 199
(CA199), anal distance, diameter, tumor stage, circumferential
resection margin (CRM) and extramural vascular invasion (EMVI),
were collected from both MLM and non-MLM groups. Tumor staging
followed the 8th American Joint Committee on Cancer
Tumor-Node-Metastasis classification, whereas EMVI and CRM were
assessed according to the 2017 European Society for Medical
Oncology guidelines (18,19). Tumor dimensions and location were
obtained from preoperative colonoscopy reports. All data analyzed
were the characteristics available in the medical records of
patients and were verified by an experienced radiologist (>10
years of experience).
The inclusion criteria for the present study were as
follows: i) Confirmation of CRC diagnosis through biopsy or
surgical pathology, ii) absence of primary tumors originating from
other sites, iii) absence of evident signs of liver metastasis upon
abdominal CT or MRI enhancement at the time of diagnosis and iv)
development of liver metastasis within 2 years of treatment,
confirmed through pathology or imaging review. The exclusion
criteria consisted of: i) Postoperative patients from other
hospitals; ii) combination of primary tumors from other sites; iii)
presence of liver metastasis at the time of diagnosis; iv) lack of
a 2-year follow-up period; and v) blurred image quality with
sequences missing, in which the tumor could not be identified
(Fig. 1).
Equipment
MRI acquisitions were performed using the following
3.0-T MRI scanners: Discovery™ MR 750 (GE Healthcare),
Signa™ HDx (GE Healthcare) and MAGNETOM Skyra (Siemens
Healthineers). The images of High-Resolution T2-weighted imaging
(HRT2) were specifically utilized for lesion outlining. No
intravenous contrast agents were administered.
The following CT scanning equipment were used:
LightSpeed VCT (GE Healthcare), Optima CT620 (GE Healthcare),
Revolution™ Maxima (GE Healthcare), Definition AS
(Siemens Healthineers), Definition AS 40 (Siemens Healthineers),
FORCE CT (Siemens Healthineers), Sensation16 (Siemens
Healthineers), SOMATOM Definition Flash (Siemens Healthineers),
SOMATOM go.Top® (Siemens Healthineers), UIHCT (United
Imaging) and Definition AS+ (Siemens Healthineers). Intravenous
contrast iohexol were administered.
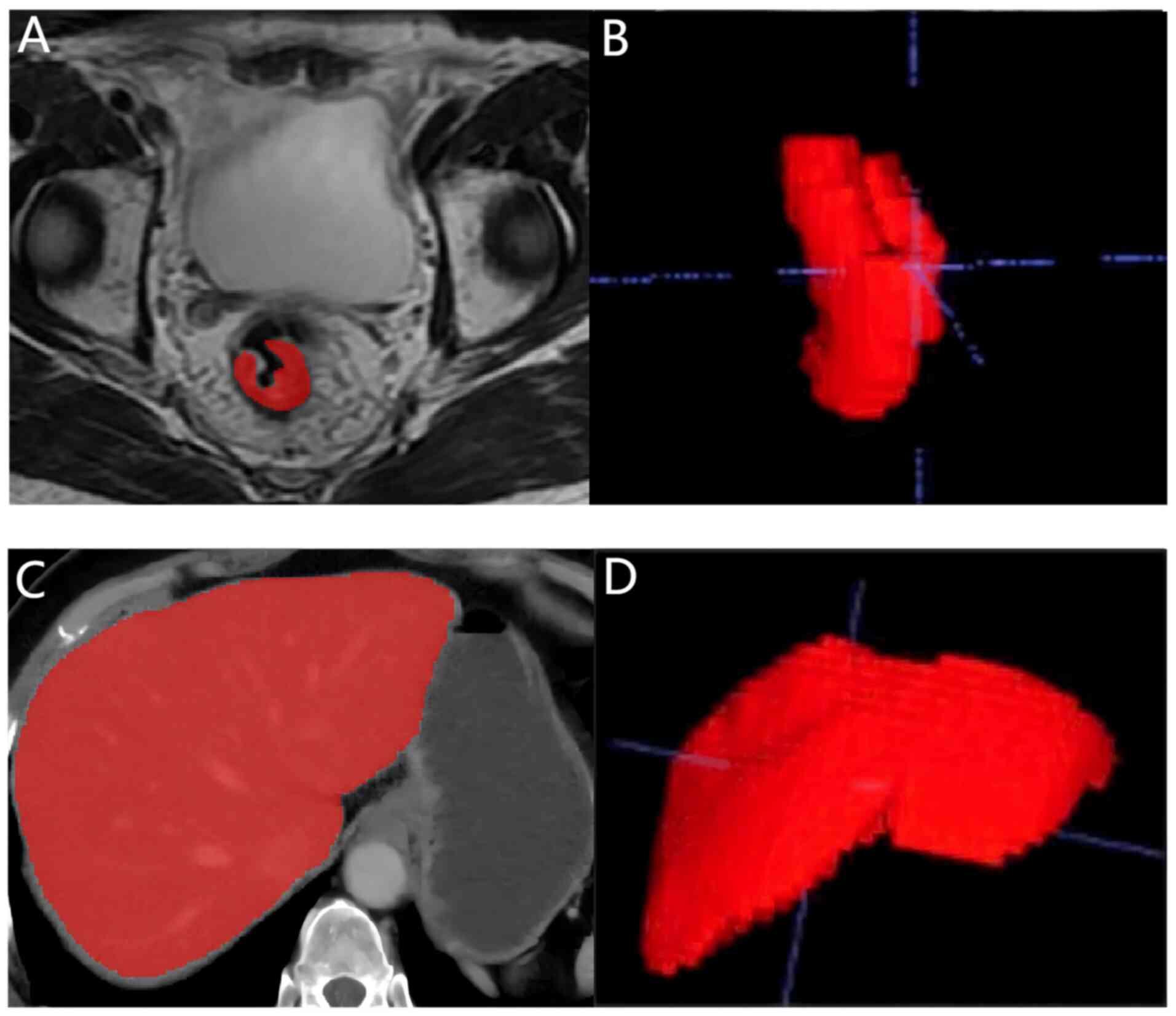
Image segmentation and feature
extraction
The liver was automatically outlined using the
Livermask package (version 1.4.1; http://github.com/andreped/livermask). ITK-SNAP
(version 3.8.0; http://www.itksnap.org) was then used to modify and
annotate the bowel MR images and the automatically outlined liver
CT images. All segmentation masks were reviewed by a junior
radiologist (with >5 years of experience in radiology) and
finally confirmed by another senior radiologist (with >10 years
of experience in radiology). Disagreements were resolved by
consensus decision making.
The International Biomarker Standardization
Initiative standard (https://theibsi.github.io/) was used during the
experimental procedure, z-score normalization was performed on the
signal intensity of HRT2 images and histogram normalization was
performed on CT images prior to feature extraction. The PyRadiomics
package (version 2.1.2; http://github.com/AIM-Harvard/pyradiomics) was
employed for radiomics feature extraction. To refine the feature
set, features with low variance (P<0.01) were eliminated by
conducting one-way ANOVA. An independent-sample t-test was
conducted to estimate the radiomics features. Features with
P<0.05 were considered significant for model development. The
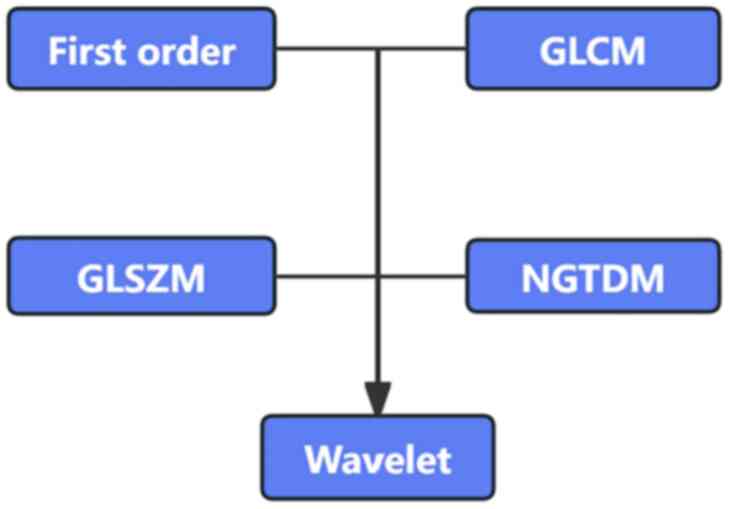
image acquisition and segmentation are shown in Fig. 2, and the radiomics feature
extraction is shown in Fig. 3.
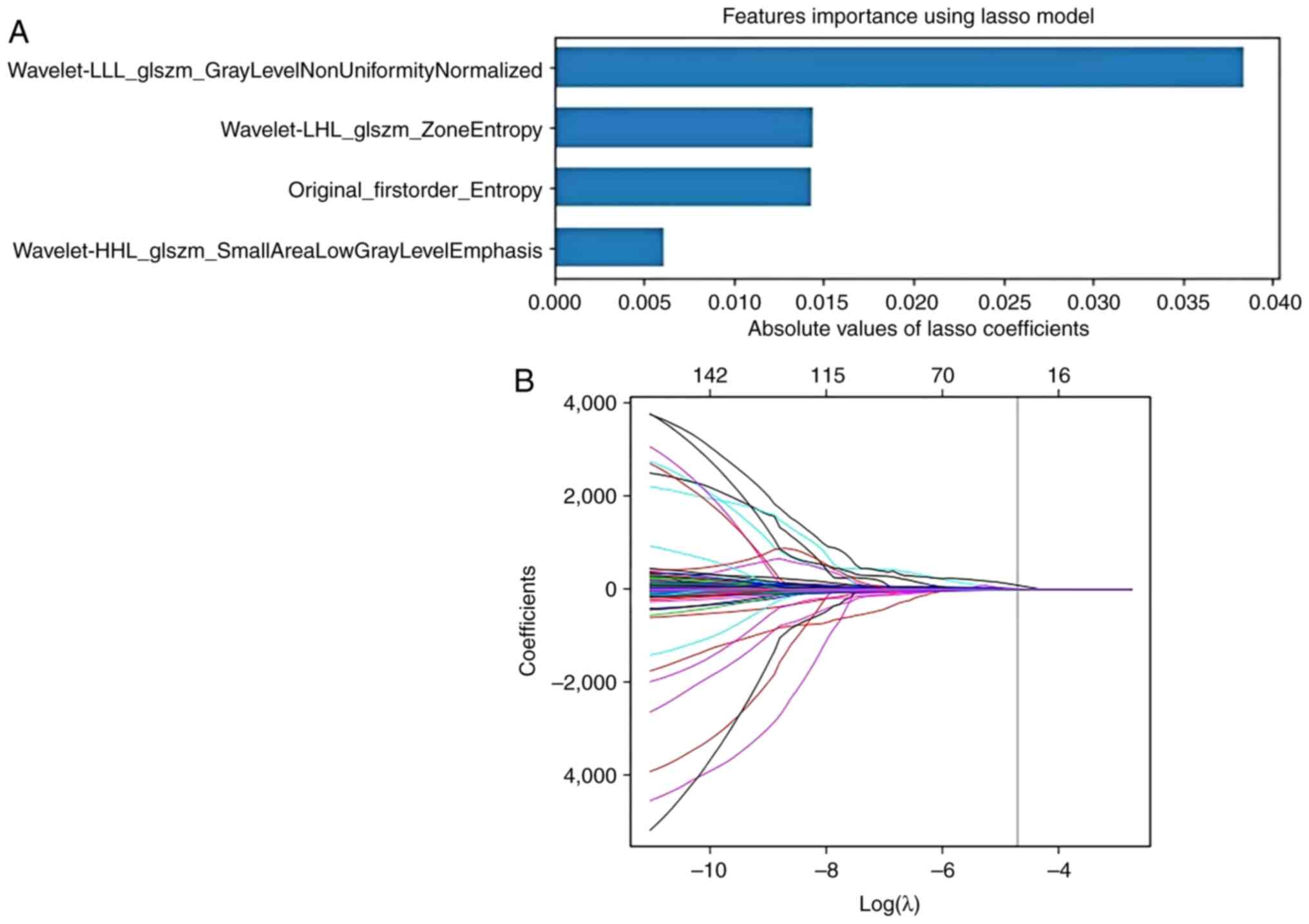
Feature selection and model
development
Since the imbalance in the original dataset (n=67
with MLM and n=90 without MLM) may lead to overfitting, a naive
random over-sampling method was employed within the open-source
Python package Imbalanced-learn (version 0.9.0, http://imbalanced-learn.org/) to create a balanced
dataset (n=90 with MLM and n=90 without MLM). Missing information
regarding the clinical characteristics (e.g., CEA and CA199) were
replaced with the mean value of the corresponding features. Then
bootstrap resampling was performed to derive confidence intervals
and improve the stability of the model. With a balanced dataset,
the Least Absolute Shrinkage and Selection Operator (LASSO) was
utilized to select the optimized subset of features from the
pre-processed features. For LASSO regression, α parameters of
0.093871 for CT, 0.061385 for MRI and 0.030567 for clinical
features were used. A prediction model was then built using the
Random Forest (RF) algorithm. The RF was built with 500 estimators,
a maximum depth of 20 and a minimum of 2 samples per leaf. To
ensure robustness and increase the generalizability of the models,
five-fold cross-validation was employed to assess the predictive
performance of each model. By performing feature selection
techniques on each cross-validation fold, the potential bias in the
prediction performance estimates were mitigated, which enhanced the
effective use of the available data. The feature selection and
model development is shown in Fig.
4.
A total of five models based on machine learning
technology were developed in the present study to predict MLM in
patients with CRC. Specifically, three radiomics models were
constructed using features from HRT2 images of CRC (MRI model), CT
images of the liver (CT model) and combined MRI-CT images (merged
model). Meanwhile, a clinical model (clinical model) was
independently developed using clinical data such as CEA, CA199 and
maximum tumor diameter, among others. A merged model combining
radiomics information and clinical features (clinical-merged model)
was also developed to enhance the predictive performance.
Model evaluation
The predictive performance of the models were
evaluated using the area under the receiver operating
characteristic (ROC) curve (AUC) over five-fold cross-validation.
McNemar's test and Delong test were conducted to compare the
classification models. The RF model generated individualized
prediction probabilities, which were subsequently subjected to
decision curve analysis (DCA) to measure the clinical utility of
the Clinical-Merged model for the prediction of MLM. Finally, a
visual nomogram was developed to estimate the probability of MLM
for each patient based on the clinical-merged model.
Statistical analysis
Statistical analysis of the clinical data was
performed using SPSS (version 26.0; IBM Corp.). Differences in
categorical characteristics between patients with CRC with and
without MLM were compared using Pearson's χ2 test and
Fisher's exact test. Continuous variables are presented as the mean
± standard deviation. For normally distributed continuous
variables, group comparisons were made using independent samples
t-test (for two groups) or one-way ANOVA (for multiple groups).
Differences in continuous characteristics between the MLM and
non-MLM groups of patients were compared using the Mann-Whitney U
test. McNemar's test and Delong test were conducted to compare the
classification models. For all statistical analyses, P<0.05
(two-tailed test) was considered to indicate a statistically
significant difference.
Results
Clinical characteristics of
patients
A total of 157 patients were included in the present
study, comprising 104 men with a mean age of 63.99±9.74 years and
53 women with a mean age of 60.53±12.51 years. The patients were
divided into MLM (n=67) and non-MLM (n=90) subgroups according to
imaging or pathological findings. These data indicate that 67
patients met the diagnostic criteria for MLM through either imaging
or pathological confirmation during the 2-year study period,
whereas 90 patients showed no clinical evidence of MLM. The
baseline characteristics of the patients are summarized in Table I.
 | Table I.Baseline demographics and clinical
characteristics of the patient cohort. |
Table I.
Baseline demographics and clinical
characteristics of the patient cohort.
|
Characteristics | MLM (n=67) | Non-MLM (n=90) | P-value |
|---|
| Age, years | 62.28±10.91 | 63.22±10.83 | 0.59 |
| Sex |
|
|
|
|
Men | 48 | 56 | 0.22 |
|
Women | 19 | 34 |
|
| CEA, ng/ml | 16.21±35.62 | 11.40±30.44 | 0.36 |
| CA199, IU/ml | 25.34±37.34 | 28.41±46.70 | 0.66 |
| Dis, mm | 66.61±33.55 | 69.02±66.61 | 0.65 |
| Dia, mm | 48.71±21.14 | 51.39±24.76 | 0.48 |
| T Stage at
diagnosis |
|
| 0.37 |
| T1 | 0 | 0 |
|
| T2 | 6 | 15 |
|
| T3 | 52 | 63 |
|
| T4 | 9 | 12 |
|
| N stage |
|
|
<0.01a |
| N0 | 9 | 31 |
|
| N1 | 21 | 34 |
|
| N2 | 35 | 1 |
|
| CRM |
|
| 0.07 |
|
Presence | 34 | 34 |
|
|
Absence | 33 | 56 |
|
| EMVI |
|
|
<0.01a |
|
Presence | 42 | 37 |
|
|
Absence | 25 | 53 |
|
Radiomics feature parameters
A total of 1,082 radiomics features were extracted
from CT images, which consisted of 14 shape features, 18
first-order intensity features and 1,050 texture features. From the
MRI images, 992 radiomics features were extracted, comprising 14
shape features, 18 first-order intensity features and 960 texture
features. The texture features included gray level co-occurrence
matrix, gray level size zone matrix (GLSZM) and neighboring gray
tone difference matrix. Finally, 11 features were selected by the
RF algorithm to construct the final radiomics models, with four
features from CT and seven features from MRI. The radiomics feature
selection results are listed in Table
II.
 | Table II.Radiomics feature selection
results. |
Table II.
Radiomics feature selection
results.
| A, CT
sequences |
|---|
|
|---|
| Feature name | Classification,
feature | Coefficient |
|---|
|
Wavelet-LHL_GLSZM_zoneentropy | Texture | −0.0144245 |
|
Original_firstorder_entropy | First-order
intensity | −0.0143472 |
|
Wavelet-HHL_GLSZM_smallarealowgraylevelemphasis | Texture | 0.00612166 |
|
Wavelet-LLL_GLSZM_graylevelnonuniformitynormalized | Texture | 0.03840038 |
|
| B, MRI
sequences |
|
| Feature
name | Classification,
feature |
Coefficient |
|
|
Wavelet-HHH_GLSZM_largeareahighgraylevelemphasis | Texture | −0.0419515 |
|
Wavelet-LHL_GLCM_clustershade | Texture | −0.0414815 |
|
Wavelet-LHH_NGTDM_coarseness | Texture | −0.0244287 |
|
Wavelet-HLH_GLSZM_largeareahighgraylevelemphasis | Texture | −0.0226032 |
|
Wavelet-HHH_GLSZM_Largeareaemphasis | Texture | −0.0194299 |
|
Log-sigma-0-5-mm-3D_firstorder_skewness | First-order
intensity | −0.0015288 |
|
Wavelet-HHH_GLSZM_zoneentropy | Texture | 0.01368654 |
Performance and evaluation of
different models
The merged model demonstrated the highest predictive
performance of the models analyzed, with sensitivity, specificity
and average AUC values of 0.71, 0.72 and 0.82, respectively. The
AUC values of the MRI, CT, clinical and clinical-merged models were
0.68, 0.80, 0.62 and 0.75, respectively. The predictive performance
of all models were summarized in Table III and Fig. 5. McNemar's test indicated that the
radiomics models were significantly different from the clinical
model (P=0.01), except for the MRI model (P=0.80). Additionally,
according to the Delong test results (z-score=0.52; P=0.60), no
significant differences were found between the merged model and the
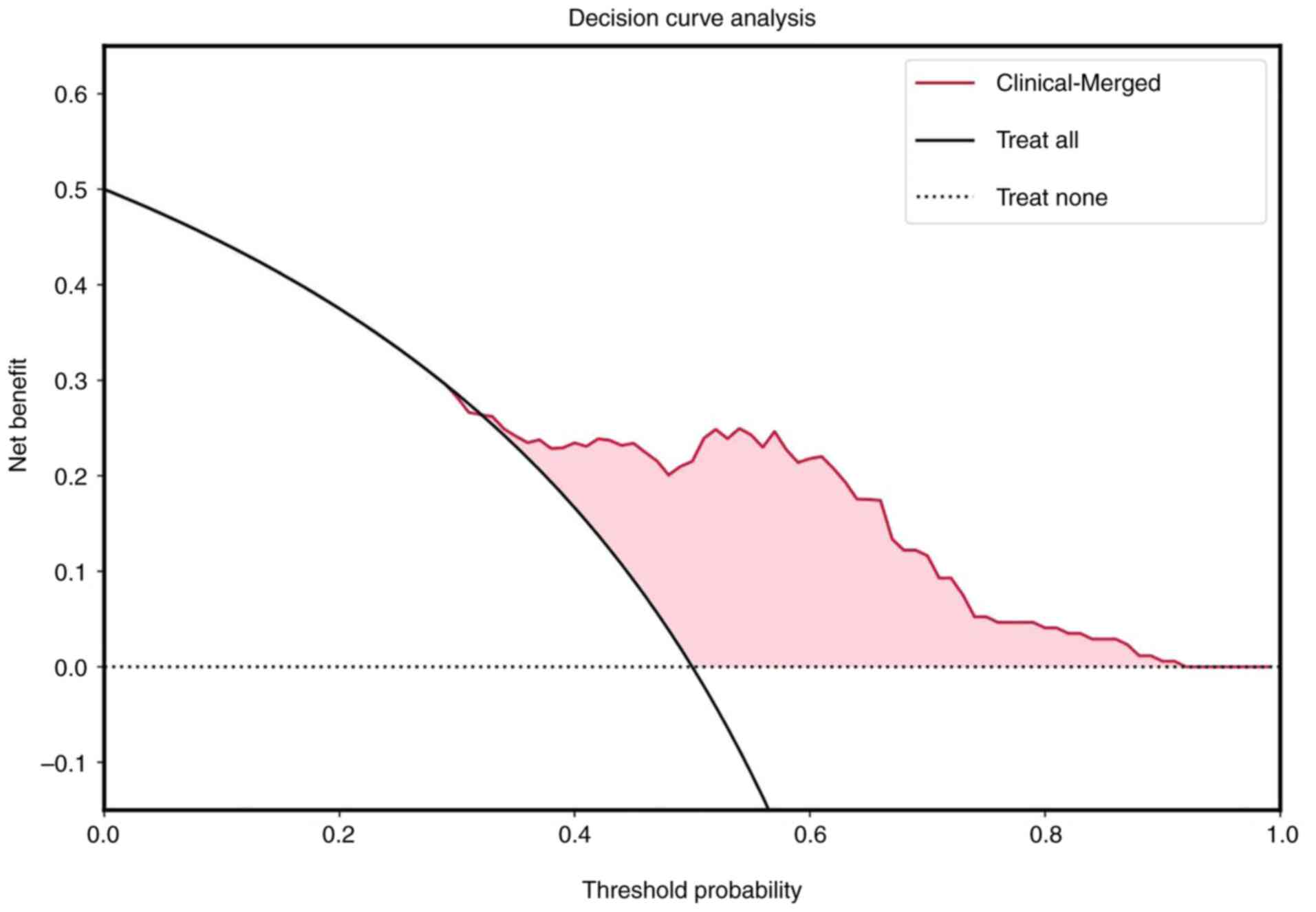
CT model. In terms of clinical utility, the DCA demonstrated that
the clinical-merged model had a higher net benefit compared with
the other four models. The calibrated probabilities served as
inputs to estimate net benefits across varying threshold
probabilities. When the threshold probability is <0.38 and
>0.93, the use of a radiomics nomogram may offer a higher net
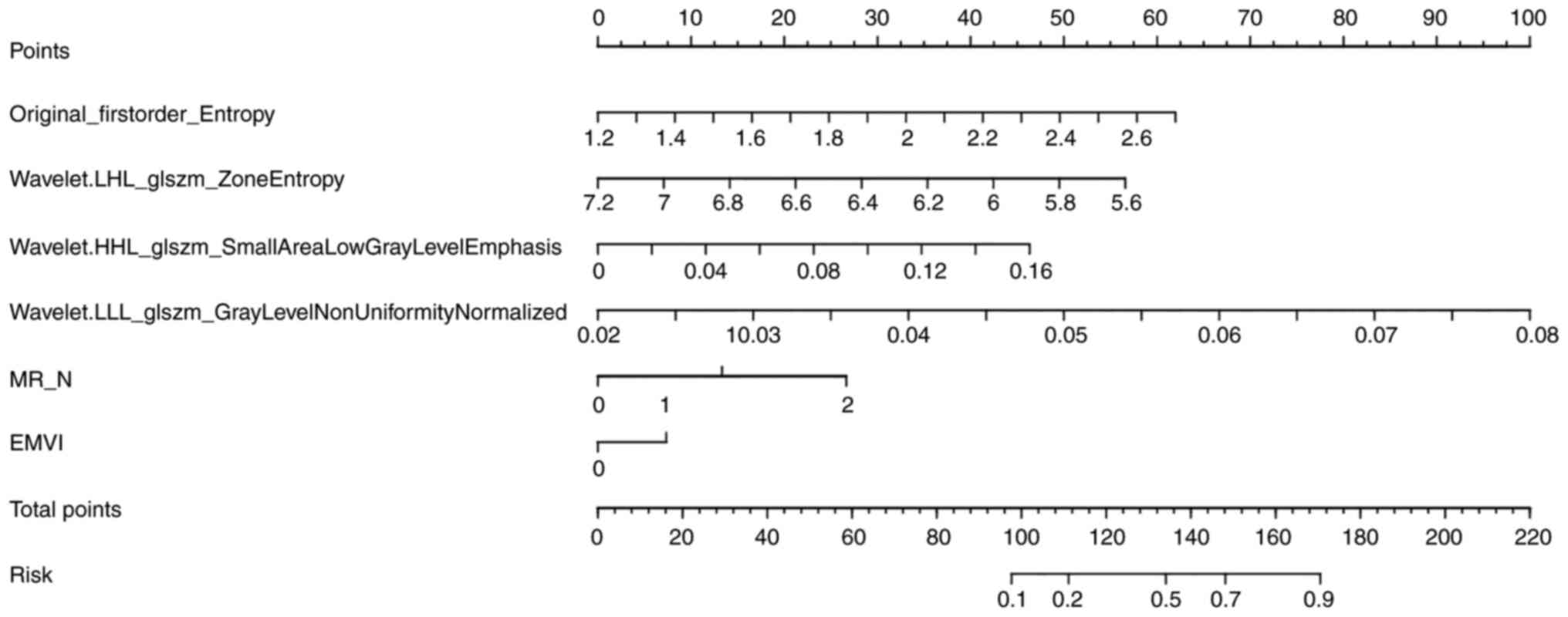
benefit compared with the other four models (Fig. 6). To generate quantitative,
individualized predictions incorporating key patient
characteristics (e.g., EVMI and MR_N status) predictive nomograms
were developed (Fig. 7).
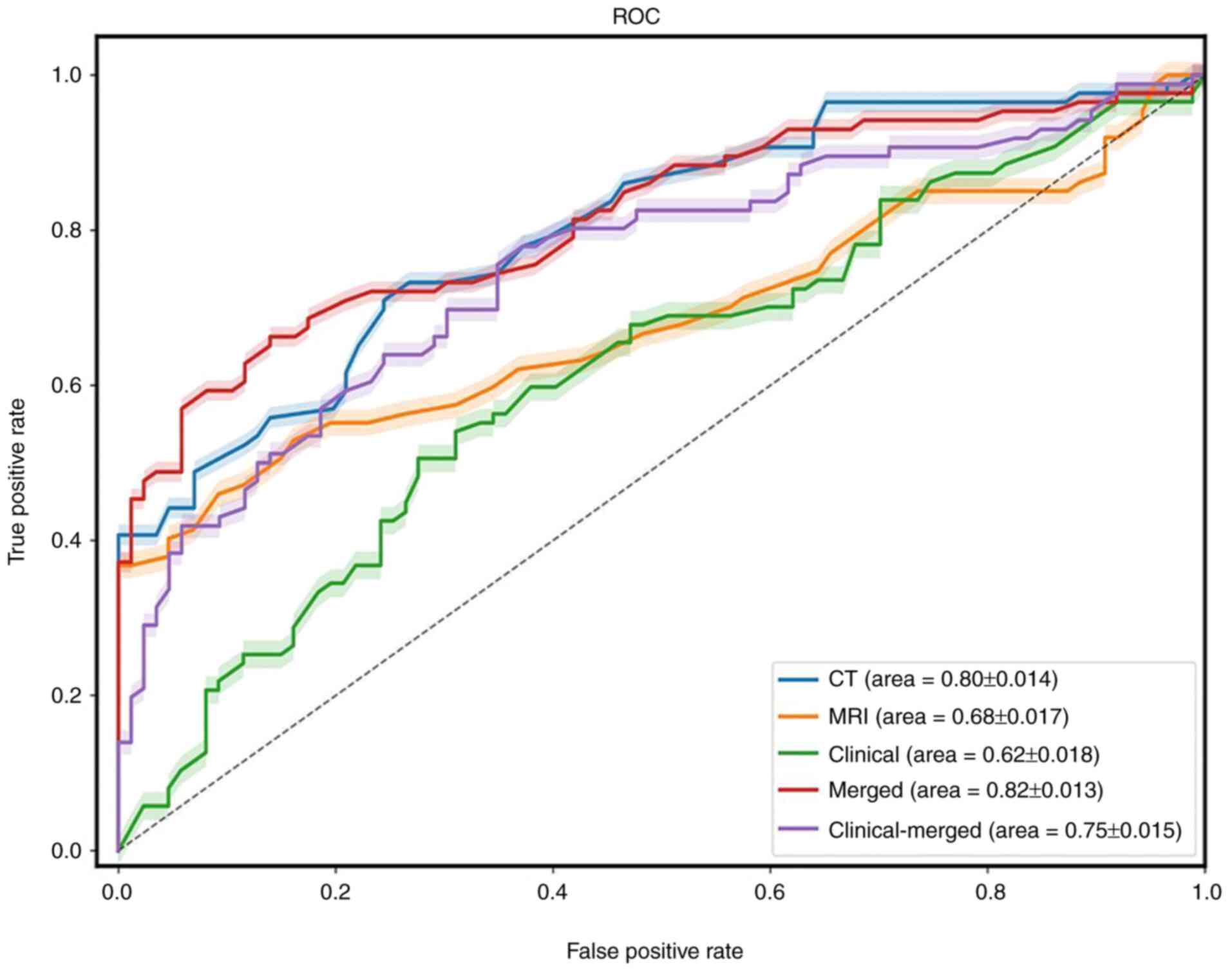 | Figure 5.ROC-AUC of different models for the
prediction of MLM. The models are represented as follows: Clinical,
green line; merged, red line; CT, blue line; MRI, orange line; and
clinical-merged, purple line. MLM, metachronous liver metastases;
ROC, receiver operating characteristic; AUC, area under the curve;
merged, the fusion radiomics signature based on features from CT
and MRI; clinical-merged, the combined model incorporating clinical
and radiological variables together. |
 | Table III.AUCs for all models. |
Table III.
AUCs for all models.
| Model | AUC | Specificity, % | Sensitivity, % |
|---|
| CT | 0.80 | 75 | 71 |
| MRI | 0.68 | 57 | 64 |
| Clinical | 0.62 | 46 | 68 |
| Merged | 0.82 | 71 | 72 |
|
Clinical-merged | 0.75 | 62 | 78 |
Discussion
Metastases serve a notable role in the mortality of
patients with cancer (20). A
substantial proportion of patients with CRC present with metastases
that are solely or predominantly localized to the liver at the time
of diagnosis. Liver metastases that are undetected during initial
diagnosis but emerge subsequently in the course of the disease are
referred to as MLM (21). In the
present study, 67 patients developed liver metastases within 2
years of treatment, with an incidence rate of ~42%, which was
higher compared with previous findings reported in the literature
(9,22,23).
CT is a non-invasive and reliable method of liver assessment and is
considered one of the standard imaging modalities for the
preoperative detection of liver metastases and postoperative
monitoring of patients with CRC (22,23).
However, by the time metastases are detected through imaging, it
may be too late to effectively intervene. A previous study reported
that prophylactic local adjuvant treatment of the liver parenchyma
in patients at risk of MLM can effectively reduce the incidence of
liver metastases after treatment (24). Therefore, it is important to predict
MLM before it manifests. The present study hypothesized that a
change in the liver microenvironment may occur before the
appearance of imaging manifestations. These changes cannot be
detected through conventional examinations; however, the present
study aimed to visualize and analyze the changes in the liver
microenvironment through radiomics. In patients diagnosed with CRC,
physicians assess systemic metastases, including the liver, to
determine the appropriate diagnosis and treatment (25). Therefore, the present study aimed to
develop a multimodal model using both CT and MRI to capture a
comprehensive view of CRC.
In the present study, a total of 11 features were
selected to build the radiomics model, which included nine
texture-related features (for example, wavelet) and two first-order
intensity features. Texture-related features reflect intratumoral
heterogeneity, while first-order intensity features extracted
through pixel-level analysis enable the quantification of intrinsic
image characteristics (8). Wavelet
transform extracts multi-frequency and multi-scale image
information, which enhances subtle contrast differences between
lesions and normal tissues. Wavelet transform is particularly
suitable for capturing complex clinical features in tumor images
that are difficult to describe with simple visual characteristics,
which indicates that images from patients with CRC contain
information that is difficult to discern with the naked eye
(26). These features provide
quantifiable information regarding texture patterns and tissue
distribution within the tumor, which may not be easily visually
discernible. Among these features, gray level non-uniformity
normalized was the best predictor, which measured the similarity of
gray-level intensity values in the image. Higher gray level
non-uniformity values have been associated with lower similarity,
which represent tumor heterogeneity (27). Apart from gray level non-uniformity,
small area low gray level emphasis and GLSZM_zoneentropy were also
illustrated in the present nomogram. Small area low gray level
emphasis has been potentially associated with pathological
characteristics such as tumor necrosis and fibrosis (28). Small area low gray level emphasis
quantifies the prevalence of small, low-intensity regions within
the lesion, which may correspond to necrotic areas or fibrotic
tissue deposition. GLSZM_zoneentropy reflects tumor heterogeneity,
which serves as a marker of intratumoral complexity. Higher values
of GLSZM_zoneentropy indicate greater variability in the size and
distribution of homogeneous gray-level zones, which suggest diverse
tissue subtype (29). The present
study combined traditional observation, radiomics and machine
learning to extract multidimensional imaging features of lesions
for predictive modeling. These features, whether easily
interpretable or not, capture the diversity within tumor regions
and reflect the biological variability of CRC. This provides a
crucial foundation for building a predictive model of MLM in
patients with CRC (30).
By adopting the aforementioned radiological features
combined with clinical information, five models were developed to
predict MLM in patients with CRC and the performance between these
five models were compared. Notably, the merged model exhibited high
performance with an AUC value of 0.82, compared with that of the
other models. The clinical model, which incorporated parameters
such as CEA, CA199, maximum tumor diameter, distance from the anus,
T-stage, N-stage, CRM and EMVI, yielded an AUC value of 0.62,
whereas the clinical-merged model achieved an AUC value of 0.75.
Among these models, the merged model demonstrated optimal
predictive performance and McNemar's test results provided
statistical validation of its significant diagnostic efficacy
(P<0.05). However, the Delong test demonstrated no significant
difference between the merged model and the CT model (P=0.60).
These findings indicated that the merged model and the CT model may
exhibit similar performance, although the merged model exhibited a
larger AUC value compared with that of the CT model. Notably, the
addition of clinical information failed to improve the performance
of the clinical-merged model in terms of AUC. This finding may be
attributed to the fact that the clinical data utilized in the
present study were obtained through subjective image assessments
and the small sample size could have exacerbated these biases. This
limitation could be mitigated in future studies involving larger
and more diverse cohorts. The present study concluded that
radiomics models may outperform clinical models in terms of
modelling and indicated that the construction of multimodal models
is feasible. However, the clinical significance of this observed
difference appears to be more limited than we hypothesized.
Previous radiomics studies have established the
prognostic value of CT and MRI features in CRC, which demonstrate
potential as predictive biomarkers for MLM (3,31,32).
In a multicenter study involving 91 patients with rectal cancer,
the radiomics, clinical and merged models achieved AUC values of
0.86, 0.71 and 0.86, respectively (10). However, the incorporation of
clinical features failed to significantly enhance the performance
of the clinical-merged model, a finding consistent with previous
studies and the present study findings (3). This phenomenon may be attributed to
the enhanced informational value of radiomics features regarding
liver parenchymal characteristics, which exert a notable influence
on model performance compared with conventional clinical predictors
such as CEA and CA199. Furthermore, a study conducted by Creasy
et al (23) emphasized the
importance of machine learning analysis of hepatic parenchyma in
the venous stage for the identification of patients at a high risk
of liver metastases. A number of studies have similarly
demonstrated the effectiveness of radiomics for the prediction of
MLM (22,33,34).
The present study not only expanded the sample size but also
extracted comprehensive three-dimensional information from the
liver parenchyma. In the present study, the merged model had the
best performance according to the results of AUC values, as well as
the results of the Delong test and McNemar's test. Therefore, the
radiomics model could potentially be used to predict MLM in
patients with CRC.
Liang et al (35) extracted features from T2-weighted
and venous phase sequence images of 108 patients with rectal
cancer, and used support vector machine and logistic regression
analysis to develop prediction models. The findings indicated that
MRI-based radiomics models derived from baseline rectal images may
hold potential for the prediction of MLM (35). Similarly, the present study also
tried to construct prediction models using RF algorithms and
logistic regression analysis, and a significant performance was
achieved using the RF algorithm. Li et al (36) utilized preoperative MRI sequences
diffusion-weighted imaging (DWI) and high-definition T2 for
predicting MLM in CRC, demonstrating marked efficacy, with the
fusion model achieving an impressive AUC value of 0.90. Compared
with the aforementioned study, the MRI model generated in the
present study performed worse with an AUC value of 0.68. This
discrepancy in performance could be attributed to the fact that the
present study exclusively selected HRT2 images without contrast,
which may have provided less clarity regarding the extent of the
lesion compared with enhanced images. Furthermore, the utilization
of a single sequence may have limited the amount of information
available for analysis, which contributed to the results from the
present study.
There were some limitations in the present study.
Firstly, the sample size was relatively small, which precludes the
application of certain artificial intelligence algorithms, such as
deep learning and neural networks. A larger sample size and
simultaneous improvement of the algorithms employed will serve a
key role in future studies. Secondly, as a single-center
retrospective study, external validation with independent datasets
is essential to confirm the findings and ensure reproducibility;
however, identifying a hospital with consistent examination
parameters and scan sequences among the multiple units has proven
to be challenging. Standardization of modalities will be a key
consideration for future multi-center studies. Thirdly, only the
HRT2 sequence of the tumor was selected for analysis, while other
sequences such as DWI were not included in the present study.
Practical experience demonstrated certain limitations in the
precise delineation of DWI sequences and notable interindividual
variability was observed. Functional sequences may contain
additional information that could be potentially valuable in future
studies.
In conclusion, five classification models were
developed to predict MLM by analyzing histological imaging and
clinical information in patients with CRC. The radiomics models
outperformed clinical models alone, which suggested that radiomics
may improve the ability to predict MLM in clinical practice. To the
best of our knowledge, the present study was the first attempt to
integrate two different modalities, CT and MRI, to predict MLM in
patients with CRC. Although the diagnostic performance did not
exhibit a significant improvement, the present study lays the
foundation for future research. Expanding the sample size and
incorporating multimodal testing will serve a key role in enhancing
the prediction accuracy of MLM in subsequent studies.
Acknowledgements
Not applicable.
Funding
The present study received funding from the Key Laboratory of
Functional Molecular Imaging of Tumor and Interventional Diagnosis
and Treatment of Shaoxing City, Zhejiang Province Public Welfare
Technology Application Social Development Field Project (grant no.
LGF20H180008), the Major Program Co-sponsored by Province and
Ministry (grant no. WKJ-ZJ-2210) and the General Plan for Medical
and Health Research in Zhejiang (grant no. 2020KY323, 2021KY371 and
2023KY1268).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JPW, ZNZ, DBS, YNH, WT, HBZ, ZHZ and JHS conceived
and designed the present study. JPW, ZNZ, DBS, YNH, WT, HBZ, ZHZ
and JHS acquired, analyzed and interpreted the data. JPW drafted
the manuscript. JPW, ZNZ, DBS, YNH, WT, HBZ, ZHZ and JHS revised
the manuscript. JPW and ZNZ performed the statistical analyses.
ZHZ, JHS, YNH and WT obtained funding for the present study. JHS
provided the materials for the present study. PW and ZNZ confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki (as revised in 2013). The present study
was approved by the Ethics Committee of Sir Run Shaw Hospital,
Zhenjiang University School of Medicine (approval no. 0465-2022;
Zhejiang, China). The need for informed consent was waived by the
ethics committee of The Sir Run Shaw Hospital, Zhenjiang University
School of Medicine, because of the retrospective nature of the
present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han B, Zheng R, Zeng H, Wang S, Sun K,
Chen R, Li L, Wei W and He J: Cancer incidence and mortality in
China, 2022. J Natl Cancer Cen. 4:47–53. 2024. View Article : Google Scholar
|
|
2
|
Wang Z, Kim SY, Tu W, Kim J, Xu A, Yang
YM, Matsuda M, Reolizo L, Tsuchiya T, Billet S, et al:
Extracellular vesicles in fatty liver promote a metastatic tumor
microenvironment. Cell Metab. 35:1209–1226.e13. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang M, Ma X, Wang L, Li D, Wang S, Zhang
H and Zhao X: Whole-liver enhanced CT radiomics analysis to predict
metachronous liver metastases after rectal cancer surgery. Cancer
Imaging. 22:502022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Horak J, Kubecek O, Siskova A, Honkova K,
Chvojkova I, Krupova M, Manethova M, Vodenkova S, García-Mulero S,
John S, et al: Differences in genome, transcriptome, miRNAome, and
methylome in synchronous and metachronous liver metastasis of
colorectal cancer. Front Oncol. 13:11335982023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu N, Gao Z, Wu D, Chen H, Zhang Z, Zhang
L, Wang Y, Lu X, Yao X, Liu X, et al: 5-hydroxymethylcytosine
features of portal venous blood predict metachronous liver
metastases of colorectal cancer and reveal phosphodiesterase 4 as a
therapeutic target. Clin Transl Med. 15:e701892025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Trailin A, Ali E, Ye W, Pavlov S,
Červenková L, Vyčítal O, Ambrozkiewicz F, Hošek P, Daum O, Liška V
and Hemminki K: Prognostic assessment of T-cells in primary
colorectal cancer and paired synchronous or metachronous liver
metastasis. Int J Cancer. 156:1282–1292. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Veen T, Kanani A, Zaharia C, Lea D and
Søreide K: Treatment-sequencing before and after index hepatectomy
with either synchronous or metachronous colorectal liver
metastasis: Comparison of recurrence risk, repeat hepatectomy and
overall survival in a population-derived cohort. Eur J Surg Oncol.
51:1095402025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inchingolo R, Maino C, Cannella R,
Vernuccio F, Cortese F, Dezio M, Pisani AR, Giandola T, Gatti M,
Giannini V, et al: Radiomics in colorectal cancer patients. World J
Gastroenterol. 29:2888–2904. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hao M, Wang K, Ding Y, Li H, Liu Y and
Ding L: Which patients are prone to suffer liver metastasis? A
review of risk factors of metachronous liver metastasis of
colorectal cancer. Eur J Med Res. 27:1302022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taghavi M, Trebeschi S, Simões R, Meek DB,
Beckers RCJ, Lambregts DMJ, Verhoef C, Houwers JB, van der Heide
UA, Beets-Tan RGH and Maas M: Machine learning-based analysis of CT
radiomics model for prediction of colorectal metachronous liver
metastases. Abdom Radiol (NY). 46:249–256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bera K, Braman N, Gupta A, Velcheti V and
Madabhushi A: Predicting cancer outcomes with radiomics and
artificial intelligence in radiology. Nat Rev Clin Oncol.
19:132–146. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bo Z, Song J, He Q, Chen B, Chen Z, Xie X,
Shu D, Chen K, Wang Y and Chen G: Application of artificial
intelligence radiomics in the diagnosis, treatment, and prognosis
of hepatocellular carcinoma. Comput Biol Med. 173:1083372024.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prelaj A, Miskovic V, Zanitti M, Trovo F,
Genova C, Viscardi G, Rebuzzi SE, Mazzeo L, Provenzano L, Kosta S,
et al: Artificial intelligence for predictive biomarker discovery
in immuno-oncology: A systematic review. Ann Oncol. 35:29–65. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valladares A, Beyer T, Papp L, Salomon E
and Rausch I: A multi-modality physical phantom for mimicking tumor
heterogeneity patterns in PET/CT and PET/MRI. Med Phys.
49:5819–5829. 2022. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalisvaart GM, van Velden FHP,
Hernández-Girón I, Meijer KM, Ghesquiere-Dierickx LMH, Brink WM,
Webb A, de Geus-Oei LF, Slump CH, Kuznetsov DV, et al: Design and
evaluation of a modular multimodality imaging phantom to simulate
heterogeneous uptake and enhancement patterns for radiomic
quantification in hybrid imaging: A feasibility study. Med Phys.
49:3093–3106. 2022. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie Z, Zhang Q, Wang X, Chen Y, Deng Y,
Lin H, Wu J, Huang X, Xu Z and Chi P: Development and validation of
a novel radiomics nomogram for prediction of early recurrence in
colorectal cancer. Eur J Surg Oncol. 49:1071182023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo X, Deng H, Xie F, Wang L, Liang J, Zhu
X, Li T, Tang X, Liang W, Xiang Z and He J: Prognostication of
colorectal cancer liver metastasis by CE-based radiomics and
machine learning. Transl Oncol. 47:1019972024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Glynne-Jones R, Wyrwicz L, Tiret E, Brown
G, Rödel C, Cervantes A and Arnold D; ESMO Guidelines Committee, :
Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 28 (Suppl_4):iv22–iv40. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vogel JD, Felder SI, Bhama AR, Hawkins AT,
Langenfeld SJ, Shaffer VO, Thorsen AJ, Weiser MR, Chang GJ,
Lightner AL, et al: The American society of colon and rectal
surgeons clinical practice guidelines for the management of colon
cancer. Dis Colon Rectum. 65:148–177. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Katipally RR, Martinez CA, Pugh SA,
Bridgewater JA, Primrose JN, Domingo E, Maughan TS, Talamonti MS,
Posner MC, Weichselbaum RR, et al: Integrated Clinical-molecular
classification of colorectal liver metastases: A biomarker analysis
of the phase 3 new EPOC randomized clinical trial. JAMA Oncol.
9:1245–1254. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siriwardena AK, Serrablo A, Fretland ÅA,
Wigmore SJ, Ramia-Angel JM, Malik HZ, Stättner S, Søreide K, Zmora
O, Meijerink M, et al: Multisocietal European consensus on the
terminology, diagnosis, and management of patients with synchronous
colorectal cancer and liver metastases: An E-AHPBA consensus in
partnership with ESSO, ESCP, ESGAR, and CIRSE. Br J Surg.
110:1161–1170. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim K, Kim S, Han K, Bae H, Shin J and Lim
JS: Diagnostic performance of deep learning-based lesion detection
algorithm in CT for detecting hepatic metastasis from colorectal
cancer. Korean J Radiol. 22:912–921. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Creasy JM, Cunanan KM, Chakraborty J,
McAuliffe JC, Chou J, Gonen M, Kingham VS, Weiser MR, Balachandran
VP, Drebin JA, et al: Differences in liver parenchyma are
measurable with CT radiomics at initial colon resection in patients
that develop hepatic metastases from stage II/III colon cancer. Ann
Surg Oncol. 28:1982–1989. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chandra P and Sacks GD: Contemporary
Surgical Management of Colorectal Liver Metastases. Cancers
(Basel). 16:9412024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ciracì P, Studiale V, Taravella A,
Antoniotti C and Cremolini C: Late-line options for patients with
metastatic colorectal cancer: A review and evidence-based
algorithm. Nat Rev Clin Oncol. 22:28–45. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng W, Mu R, Liu F, Qin X, Li X, Yang P,
Li X, Liang Y and Zhu X: Textural features of the frontal white
matter could be used to discriminate amnestic mild cognitive
impairment patients from the normal population. Brain Behav.
13:e32222023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Y, Wu D, Yan S, Xie Y, Zhang S, Lv W,
Qin Y, Liu Y, Liu C, Lu J, et al: Feasibility of a
Clinical-radiomics model to predict the outcomes of acute ischemic
stroke. Korean J Radiol. 23:811–820. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Grossmann P, Stringfield O, El-Hachem N,
Bui MM, Rios Velazquez E, Parmar C, Leijenaar RT, Haibe-Kains B,
Lambin P, Gillies RJ and Aerts HJ: Defining the biological basis of
radiomic phenotypes in lung cancer. Elife. 6:e234212017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Davnall F, Yip CS, Ljungqvist G, Selmi M,
Ng F, Sanghera B, Ganeshan B, Miles KA, Cook GJ and Goh V:
Assessment of tumor heterogeneity: An emerging imaging tool for
clinical practice? Insights Imaging. 3:573–589. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Liao M, Guo Q, Chen J, Wang S,
Liu S and Xiao F: Predicting N2 lymph node metastasis in
presurgical stage I–II non-small cell lung cancer using multiview
radiomics and deep learning method. Med Phys. 50:2049–2060. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Gong J, Shen X, Li M, Zhang H, Feng
F and Tong T: Assessment of primary colorectal Cancer CT radiomics
to predict metachronous liver metastasis. Front Oncol.
12:8618922022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hao M, Li H, Wang K, Liu Y, Liang X and
Ding L: Predicting metachronous liver metastasis in patients with
colorectal cancer: Development and assessment of a new nomogram.
World J Surg Oncol. 20:802022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma YQ, Wen Y, Liang H, Zhong JG and Pang
PP: Magnetic resonance imaging-radiomics evaluation of response to
chemotherapy for synchronous liver metastasis of colorectal cancer.
World J Gastroenterol. 27:6465–6475. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Radiya K, Joakimsen HL, Mikalsen K, Aahlin
EK, Lindsetmo RO and Mortensen KE: Performance and clinical
applicability of machine learning in liver computed tomography
imaging: A systematic review. Eur Radiol. 33:6689–6717. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang M, Cai Z, Zhang H, Huang C, Meng Y,
Zhao L, Li D, Ma X and Zhao X: Machine Learning-based analysis of
rectal cancer MRI Radiomics for prediction of metachronous liver
metastasis. Acad Radiol. 26:1495–1504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li ZF, Kang LQ, Liu FH, Zhao M, Guo SY, Lu
S and Quan S: Radiomics based on preoperative rectal cancer MRI to
predict the metachronous liver metastasis. Abdom Radiol (NY).
48:833–843. 2023.PubMed/NCBI
|