Introduction
Head and neck squamous cell carcinoma (HNSCC)
represents the most prevalent form of cancer in the head and neck
area, accounting for >90% of cases, with a 5-year overall
survival rate of 50–60% (1). Human
papillomavirus (HPV) serves as a primary risk-contributing factor
for HNSCC, contributing to ~70% of HNSCC cases in developed nations
(2,3). The pathogenesis and progression of
HPV+ HNSCC involve intricate molecular regulatory
mechanisms and signaling pathways, such as the regulation of
apoptosis, cell cycle progression and the immune response through
viral oncogenes or epigenetic silencing, leading to notable
differences in clinical characteristics, treatment responses and
prognosis compared with HPV− HNSCC (4,5).
Currently, there are no specific therapies for HPV+
HNSCC, which is still treated using standard HNSCC strategies such
as surgery, radiation and chemotherapy. These traditional
approaches frequently give rise to substantial side effects,
including toxicity, as well as malformation and dysfunction caused
by partial or complete resection of the jawbone or other
maxillofacial organs (6). Moreover,
the clinical application and efficacy of immunotherapies are
restricted, making it imperative to identify effective therapeutic
targets and treatments for HPV+ HNSCC (7).
Traditional Chinese medicine (TCM), notable for its
low toxicity, multi-component nature and multi-target effects, has
become a promising therapeutic option (8). With progress in network pharmacology,
exploring the components and mechanisms of TCM for anticancer
effects has become more efficient. For example, a study on Yinchen
Wuling San identified key compounds, such as cerevisterol,
polyporusterone E and genkwanin, which may exert anti-HNSCC effects
through pathways such as PI3K-Akt and MAPK signaling (9). This research underscores the potential
of TCM as a valuable source for developing novel anticancer
drugs.
Aloe vera (L.) Burm.f., a perennial succulent
herb, is widely used in TCM for its medicinal properties, including
immune modulation, anti-inflammatory, antiviral and hypoglycemic
effects (10,11). Previously, its antitumor efficacy
has gained attention, demonstrating activity against various types
of cancer. For example, aloe-emodin, a bioactive compound found in
Aloe vera, can trigger apoptosis in breast and lung cancer
by disrupting mitochondrial function (12). Additionally, ethanolic leaf extracts
of Aloe vera exhibit cytotoxic effects against cervical
cancer and lung adenocarcinoma cells (13). The anticancer effects of several
therapeutic ingredients have been identified, including
anthraquinones, Aloe vera polysaccharides and aloctin
(14); however, the therapeutic
effects of Aloe vera on HPV+ HNSCC remain
unclear.
Previous studies have underscored the potential of
serpin family member 1 (SERPINE1) as a promising target in HNSCC
therapy, particularly due to its role in regulating tumor
progression, metastasis and immune evasion (15,16).
In the context of HPV− HNSCC, emerging evidence has
indicated that SERPINE1 serves a critical role in modulating tumor
microenvironment dynamics and immune responses (17). Additionally, studies have
demonstrated that the expression of SERPINE1 is regulated by
multiple mechanisms, including m6A modifications mediated by the
α-ketoglutarate-dependent dioxygenase fat mass and
obesity-associated protein and circular RNAs, such as circPRMT5,
which stabilize SERPINE1 mRNA through interactions with
insulin-like growth factor 2 mRNA-binding protein 3 (15,18).
These findings collectively suggest that targeting SERPINE1 could
offer a novel therapeutic strategy for intervening in tumor biology
and enhancing antitumor immunity.
Based on network pharmacology analysis, the present
study aimed to identify potential therapeutic ingredients and
targets of Aloe vera against HPV+ HNSCC. Hub
genes were screened through topological analysis of protein-protein
interaction (PPI) networks and validated using The Cancer Genome
Atlas (TCGA) data for expression levels and prognostic importance.
Molecular docking was utilized to investigate the effective
ingredients and key targets. Subsequently, the antitumor effects
were evaluated through western blotting and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). The
research aimed to provide insights for the development of
anti-HPV+ HNSCC drugs derived from TCM.
Materials and methods
Ingredients and targets of Aloe
vera
The active ingredients and associated targets of
Aloe vera were explored through TCM Systems Pharmacology
Database and Analysis Platform (TCMSP; http://old.tcmsp-e.com/index.php) and
SwissTargetPrediction (http://www.swisstargetprediction.ch/) platforms. The
interaction between ingredients and targets was established using
Cytoscape v3.10.2 software (https://cytoscape.org/). A threshold of 30% for the
oral bioavailability and 0.18 for the drug likeness was set for
TCMSP, with a probability score of 1 for SwissTargetPrediction
(19,20).
Differential gene expression analysis
of HPV+ HNSCC
GSE53355 and GSE117973 were retrieved from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) using the key words
‘HPV positive HNSCC’, with GSE53355 containing 44 samples (26
HPV+ HNSCC, 18 HPV− HNSCC) and GSE117973
comprising 77 samples (54 HPV+ HNSCC, 23 HPV−
HNSCC) (21,22). Differential gene expression analysis
was performed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) (P<0.05;
logFC >1).
Identification of potential targets of
Aloe vera against HPV+ HNSCC
A Venn diagram was employed to analyze the overlap
between the predicted genes of Aloe vera and those
associated with HPV+ HNSCC, thereby identifying
potential therapeutic targets for this cancer subtype.
Functional enrichment analysis
Functional enrichment analysis was performed
employing WebGestalt v2024 (http://www.webgestalt.org). Top-ranked Gene Ontology
(GO) terms including biological processes (BPs), molecular
functions (MFs) and cellular components (CCs), as well as Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathways were visualized
as bar charts and bubble plots (23). The reference gene set was set to all
genome protein-coding genes, to serve as the background for
enrichment calculations.
Construction of a PPI network and hub
gene identification
PPI networks based on common targets between Aloe
vera and HPV+ HNSCC were created using the STRING
v12.0 platform (https://cn.string-db.org/) and were visualized in
Cytoscape. MCODE v2.0.3 (https://apps.cytoscape.org/apps/mcode) and Network
Analyzer plugins were employed to calculate the MCODE score,
betweenness centrality (BC), closeness centrality (CC) and degree
for each node, with the top 10 genes identified as hub genes
(24).
Expression validation and prognostic
analysis
The prognostic relevance of hub genes in a cohort of
patients with HNSCC (n=500) was analyzed using the Kaplan-Meier
Plotter (https://kmplot.com/analysis/) to
evaluate overall survival outcomes, employing the automatic
selection of the optimal cutoff based on percentile. To ensure the
robustness of the findings, the results of the survival analysis
were further validated using the Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) and UALCAN (https://ualcan.path.uab.edu/) platforms. These
platforms offer valuable insights into the association between gene
expression and survival based on TCGA datasets. Significant
associations were determined using a threshold of P<0.05.
Genomic alterations and immune
infiltrations
The cBioPortal (www.cbioportal.org/) and TIMER2.0 (http://timer.comp-genomics.org/) were employed to
examine the somatic copy number alterations (SCNAs) in key Aloe
vera targets against HPV+ HNSCC, and the correlation
between SCNAs and immune cell infiltrations. The infiltration
levels for each SCNA category were compared with normal controls
using a two-sided Wilcoxon rank-sum test.
Co-expression analysis and Gene Set
Enrichment Analysis (GSEA)
The LinkedOmics platform (www.linkedomics.org/) was employed to investigate the
co-expression of the hub gene, alongside GSEA to investigate their
functional implications (25).
Molecular docking
The core pharmacological compounds of Aloe
vera, along with the protein structures of the key genes, were
sourced from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) and Protein Data
Bank (PDB; http://www.rcsb.org/) databases.
Molecular docking was performed using PyRx software v0.8
(https://pyrx.sourceforge.io/) that was
integrated with AutoDock Vina v1.2.x (https://vina.scripps.edu/). This process identified
critical molecular interactions, with a binding energy threshold of
7 kcal/mol.
Experimental validation
Quercetin (Shanghai Yuanye Biotechnology Co., Ltd.)
was dissolved in dimethyl sulfoxide (MilliporeSigma) to create
solutions with concentrations of 5 and 10 µM. Post-sonication (30
min, 40 kHz, 25°C) and centrifugation (5 min, 13,500 × g, 4°C), the
supernatant was collected. Equivalent concentrations of dimethyl
sulfoxide without quercetin were used as a control.
Cell culture
UPCI:SCC154 cells (CRL-3241; American Type Culture
Collection) were cultured in DMEM supplemented with 10% FBS and 100
U/ml penicillin/streptomycin (all Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in 5% CO2.
Western blotting
After being treated for 24 h, total protein was
extracted using radioimmunoprecipitation assay lysis buffer
(Beyotime Institute of Biotechnology) containing protease and
phosphatase inhibitors. Protein concentration was determined using
a bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Equal amounts of protein (30 µg per lane) were then
separated by SDS-PAGE on 10% gels and transferred to polyvinylidene
fluoride membranes (Thermo Fisher Scientific, Inc.). The membranes
were blocked with 5% non-fat milk in TBS-0.1% Tween-20 (TBST) for 1
h at room temperature, and then incubated overnight at 4°C with the
following primary antibodies: Anti-SERPINE1 (1:1,000 dilution; cat.
no. 13801-1-AP; Proteintech Group, Inc.) and anti-β-actin (1:5,000
dilution; cat. no. 66009-1-Ig; Proteintech Group, Inc.). After
washing three times with TBST, the membranes were incubated with
HRP-conjugated secondary antibodies (goat anti-rabbit IgG, 1:5,000,
cat. no. SA00001-2; goat anti-mouse IgG, 1:5,000, cat. no.
SA00001-1; both from Proteintech Group, Inc.) for 1 h at room
temperature. Protein bands were visualized using an enhanced
chemiluminescence detection kit (MilliporeSigma), and imaged using
a ChemiDoc™ MP Imaging System (Bio-Rad Laboratories, Inc.)
Semi-quantification of protein bands was performed using ImageJ
software v1.54 (National Institutes of Health).
RT-qPCR
Cells were treated for 24 h before RNA extraction.
Total RNA was extracted using RNAiso Plus reagent (Takara Bio
Inc.), following the manufacturer's instructions. RT was performed
using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio
Inc.) according to the manufacturer's protocol. qPCR was conducted
using TB Green® Premix Ex Taq™ II (Takara Bio Inc.,) on
a StepOnePlus™ Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The primer sequences used were as
follows: SERPINE1: forward 5′-ACCGCAACGTGGTTTTCTCA-3′, reverse
5′-TTGAATCCCATAGCTGCTTGAAT-3′; and β-actin: forward
5′-AGAGCTACGAGCTGCCTGAC-3′, reverse 5′-GTCACCTTCACCGTTCCAGT-3′.
The qPCR cycling protocol included an initial
denaturation step at 95°C for 3 min, followed by 35 cycles of
denaturation at 95°C for 20 sec, annealing at 56°C for 20 sec and
extension at 72°C for 20 sec. Melting curve analysis was performed
with the following steps: 95°C for 15 sec, 60°C for 15 sec, 95°C
for 20 min and 95°C for 15 sec. All reactions were performed in
triplicate. Relative gene expression was quantified using the
2−ΔΔCq method, with β-actin used as the internal control
(26).
Statistical analysis
The experimental data were statistically analyzed
using GraphPad Prism v6.0 software (Dotmatics). Differences between
two groups were assessed using an unpaired Student's t-test, while
multi-group comparisons were performed using a one-way ANOVA
followed by Dunnett's test for post hoc analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Targets of Aloe vera against
HPV+ HNSCC
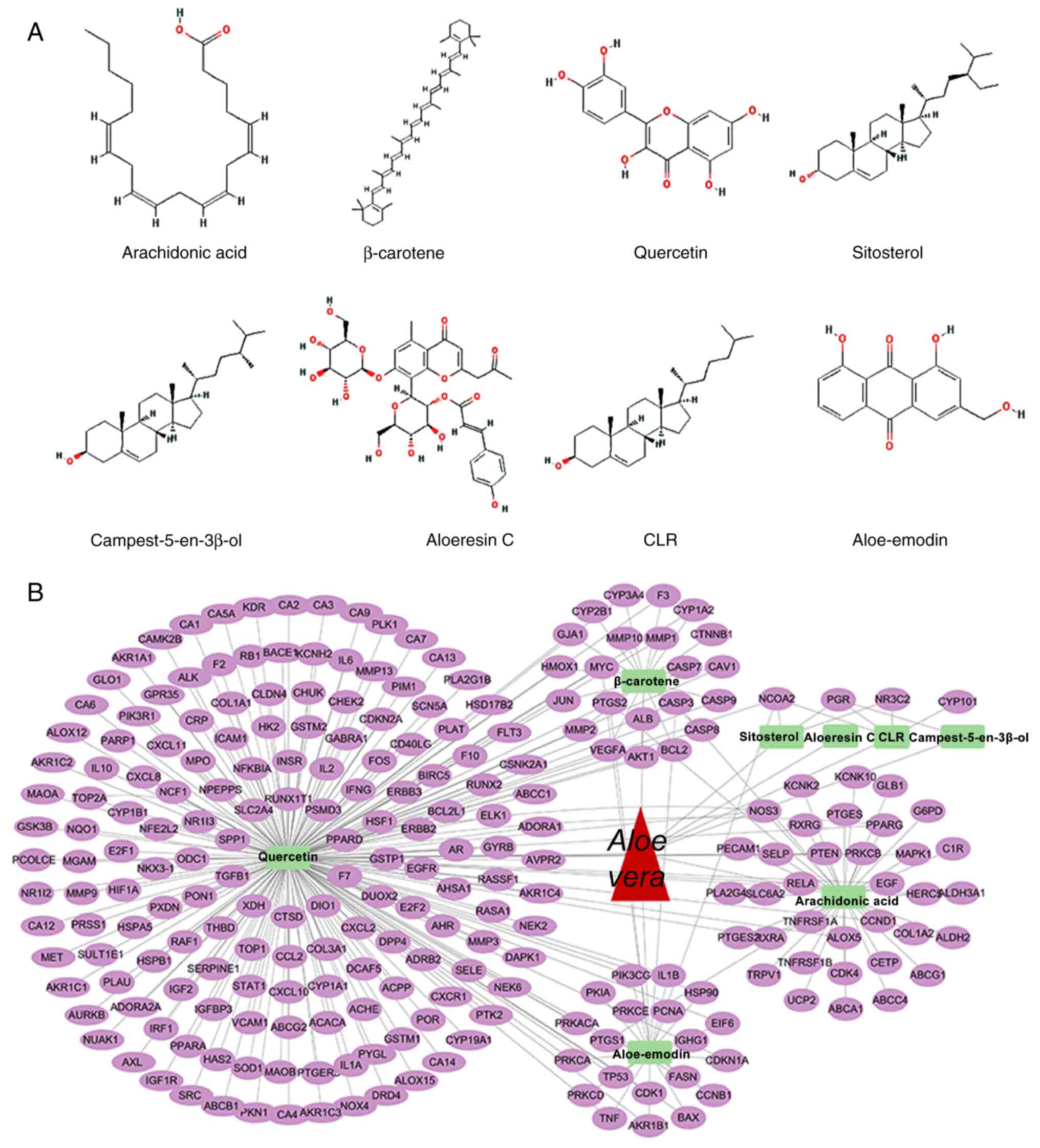
Using network pharmacology analysis, eight key
bioactive components of Aloe vera were identified:
Aloe-emodin, aloeresin C, arachidonic acid, β-carotene,
campest-5-en-3β-ol, cholesterol, quercetin and sitosterol (Fig. 1A). Concurrently, 244 potential
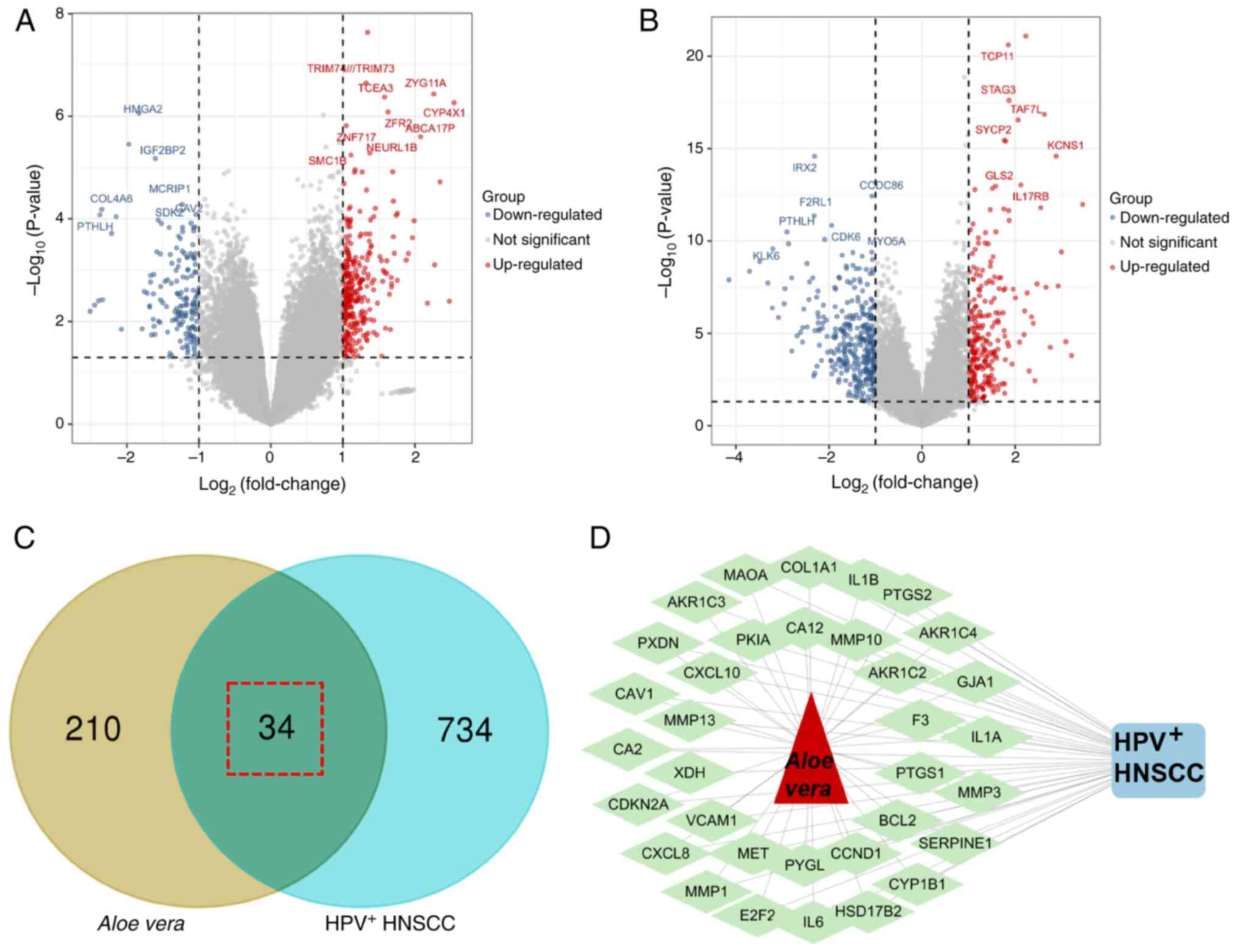
therapeutic targets for Aloe vera were identified (Fig. 1B). Differential gene expression
analysis of the GSE53355 dataset revealed 402 HPV+
HNSCC-specific genes, including 257 upregulated and 145
downregulated genes (Fig. 2A). From
the GSE117973 dataset, 461 HPV+ HNSCC-specific genes
were detected (194 upregulated and 267 downregulated; Fig. 2B). The integrated analysis of two
datasets identified a total of 768 genes specifically associated
with HPV+ HNSCC. Through the intersection of Aloe
vera targets and HPV+ HNSCC-specific genes, 34
potential therapeutic targets were identified as targets for
Aloe vera against HNSCC(Fig.
2C) and a drug-target-disease network was constructed (Fig. 2D).
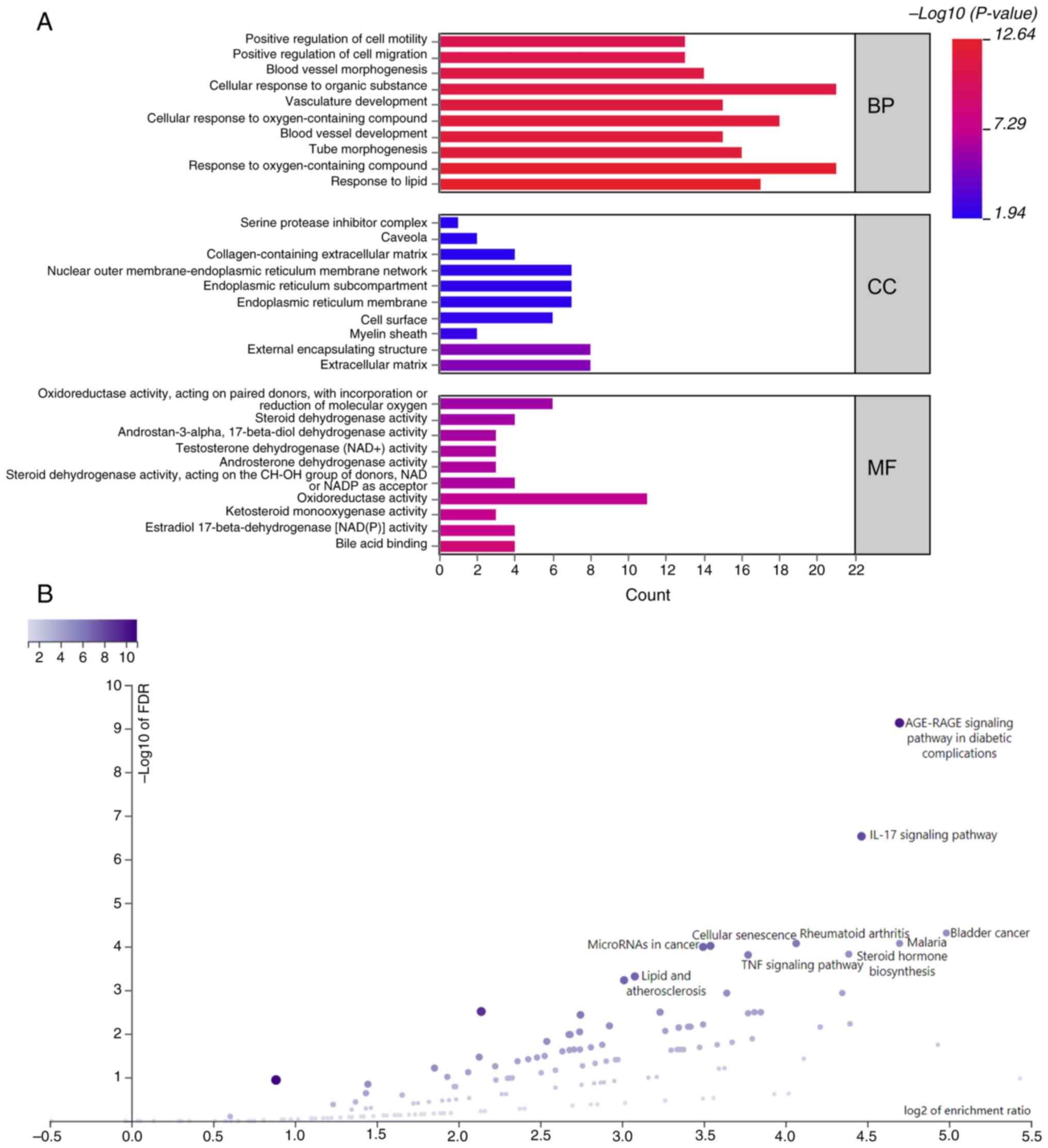
Functional enrichment analyses
GO analysis indicated that the target genes
primarily participated in BPs such as ‘response to lipid’ and
‘response to oxygen-containing compound’ (Fig. 3A and Table SI). At the MF level, they were
associated with ‘bile acid binding’ and ‘estradiol
17-beta-dehydrogenase [NAD(P)] activity’ and were localized in the
‘extracellular matrix’ and ‘external encapsulating structure’. KEGG
pathway analysis revealed their participation in pathways such as
‘AGE-RAGE signaling pathway in diabetic complications’ and ‘IL-17
signaling pathway’ (Fig. 3B).
Topological analysis and
identification of hub genes
A PPI network comprising 29 nodes was created using
STRING and Cytoscape (Fig. 4A). A
total of two significant clusters within the network were
identified by employing the MCODE plugin. By integrating
top-ranking nodes based on centrality measures such as degree, BC,
CC and MCODE scores, PTGS2, IL6, IL1B, CXCL8 and SERPINE1 were
recognized as the hub genes (Fig. 4B
and C).
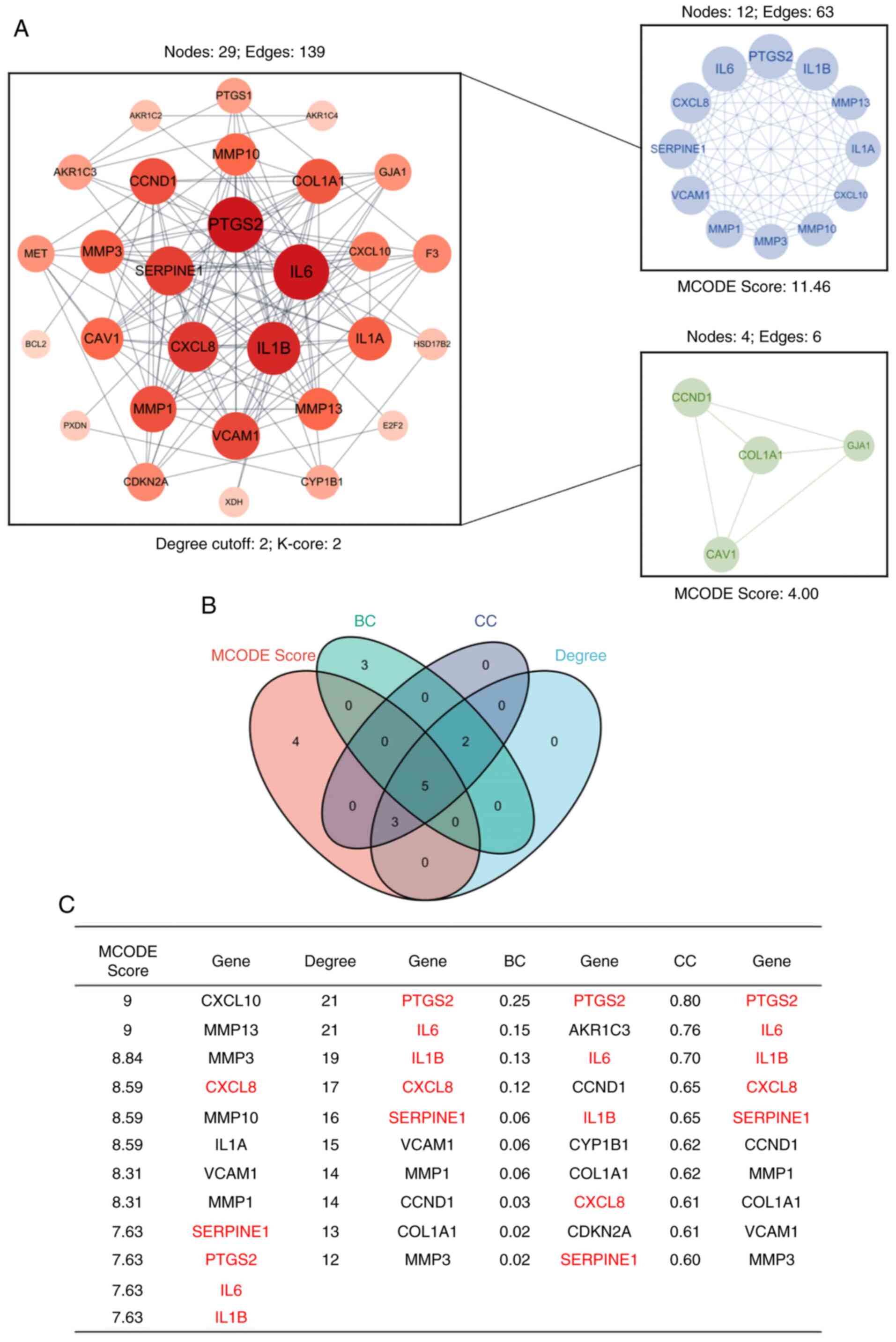 | Figure 4.PPI network analysis. (A) A PPI
network of Aloe vera- and human papillomavirus-positive head
and neck squamous cell carcinoma shared targets was constructed,
with two core subnetworks identified with degree cutoff=2 and
K-core=2. (B) Based on MCODE score, BC, CC and degree, a total of
five hub genes were identified, as shown in the Venn diagram. (C)
Five hub genes were selected including PTGS2, IL6, IL1B, CXCL8 and
SERPINE1, through the intersection of four gene sets. PPI,
protein-protein interaction; BC, betweenness centrality; CC,
closeness centrality; SERPINE1, serpin family member 1. |
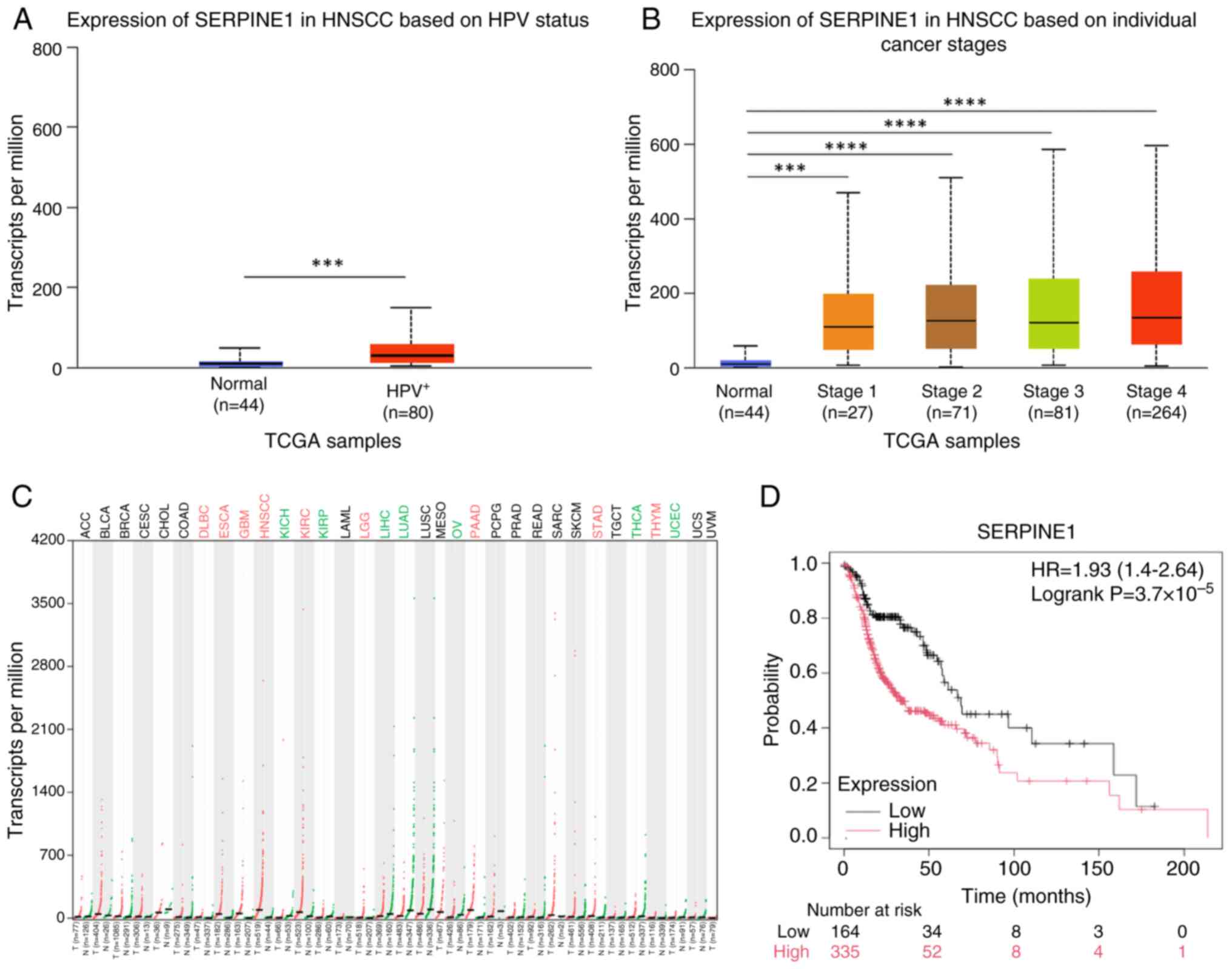
Validation of hub gene expression and
prognostic relevance
SERPINE1 expression was markedly upregulated in
HPV+ HNSCC tissues compared with that in normal tissues
from healthy individuals (Fig. 5A)
and was increased across all stages of HNSCC (Fig. 5B), as well as in eight other types
of cancer (Fig. 5C). Survival
analysis revealed that SERPINE1 expression was significantly
associated with overall survival in patients with HNSCC (Fig. 5D).
Genetic alterations and immune
infiltrations
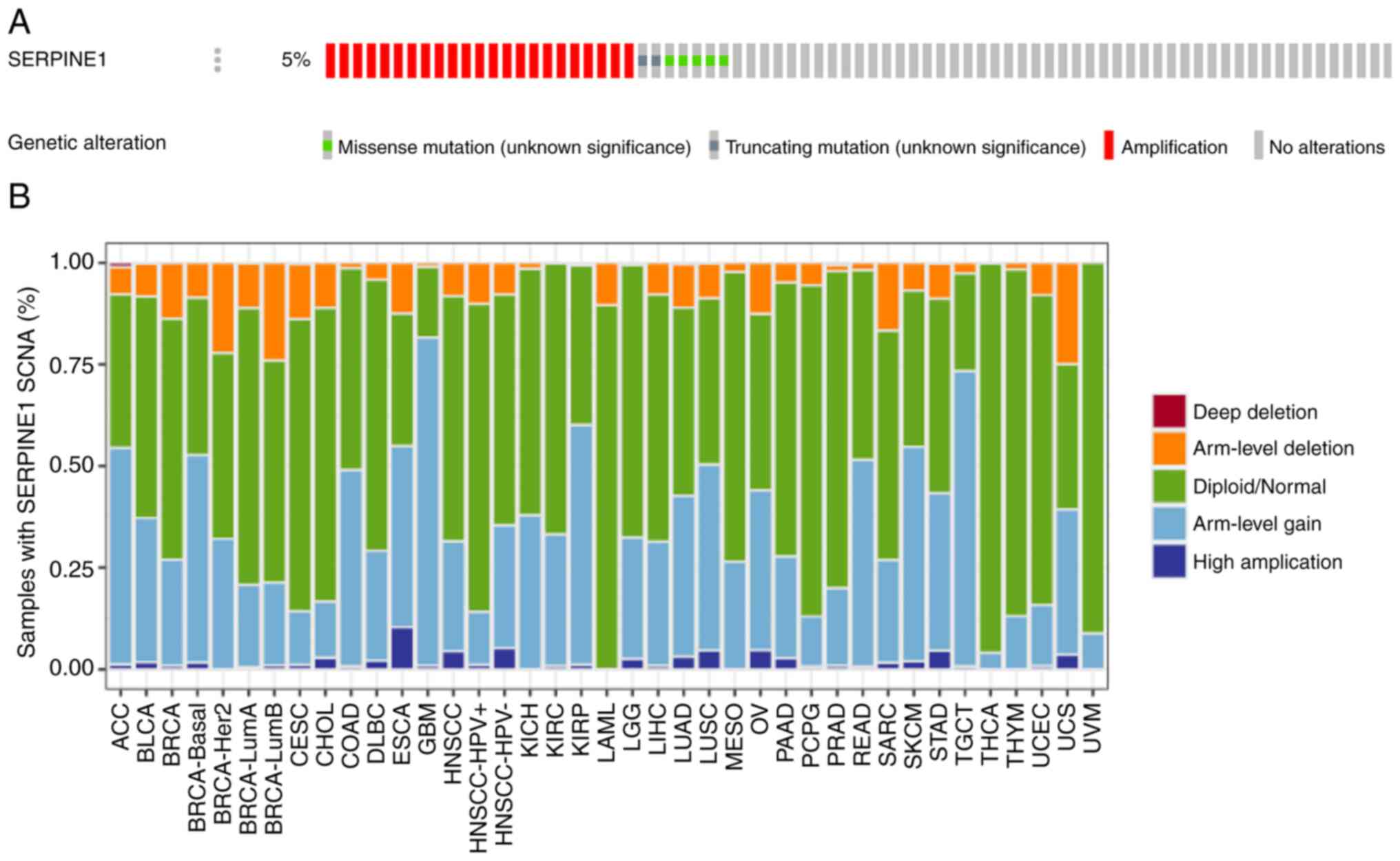
The overall frequency of genetic alterations in
SERPINE1 in HNSCC was ~5%, suggesting a potential genetic
predisposition associated with this gene (Fig. 6A). Amplifications were particularly
common and constituted a substantial proportion of these
alterations. By contrast, missense and truncating mutations
occurred at a lower frequency. In HPV+ HNSCC, SERPINE1
frequently underwent arm-level gains and deletions, suggesting that
gene dosage alterations may contribute to tumorigenesis in this
subgroup (Fig. 6B). These gains
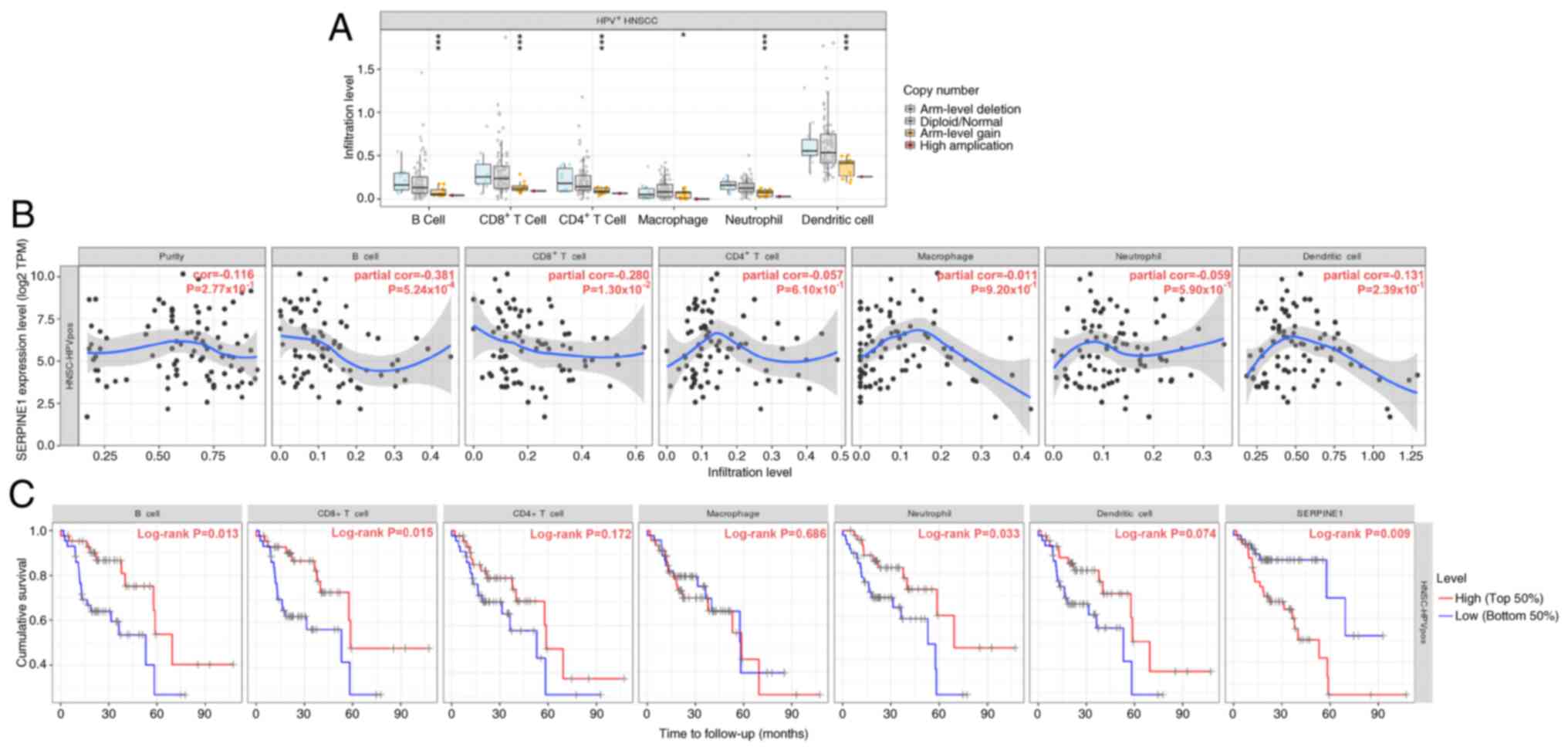
were associated with the infiltration of B cells, CD8+ T
cells, CD4+ T cells, macrophages, neutrophils and
dendritic cells (Fig. 7A).
Moreover, the increased expression of SERPINE1 was significantly
correlated with reduced B-cell infiltration, which in turn was
associated with worse survival outcomes in patients with
HPV+ HNSCC (Fig. 7B and
C). High levels of SERPINE1 expression were also associated
with diminished overall survival in patients with HPV+
HNSCC.
Co-expression analysis and GSEA of
SERPINE1 in HNSCC
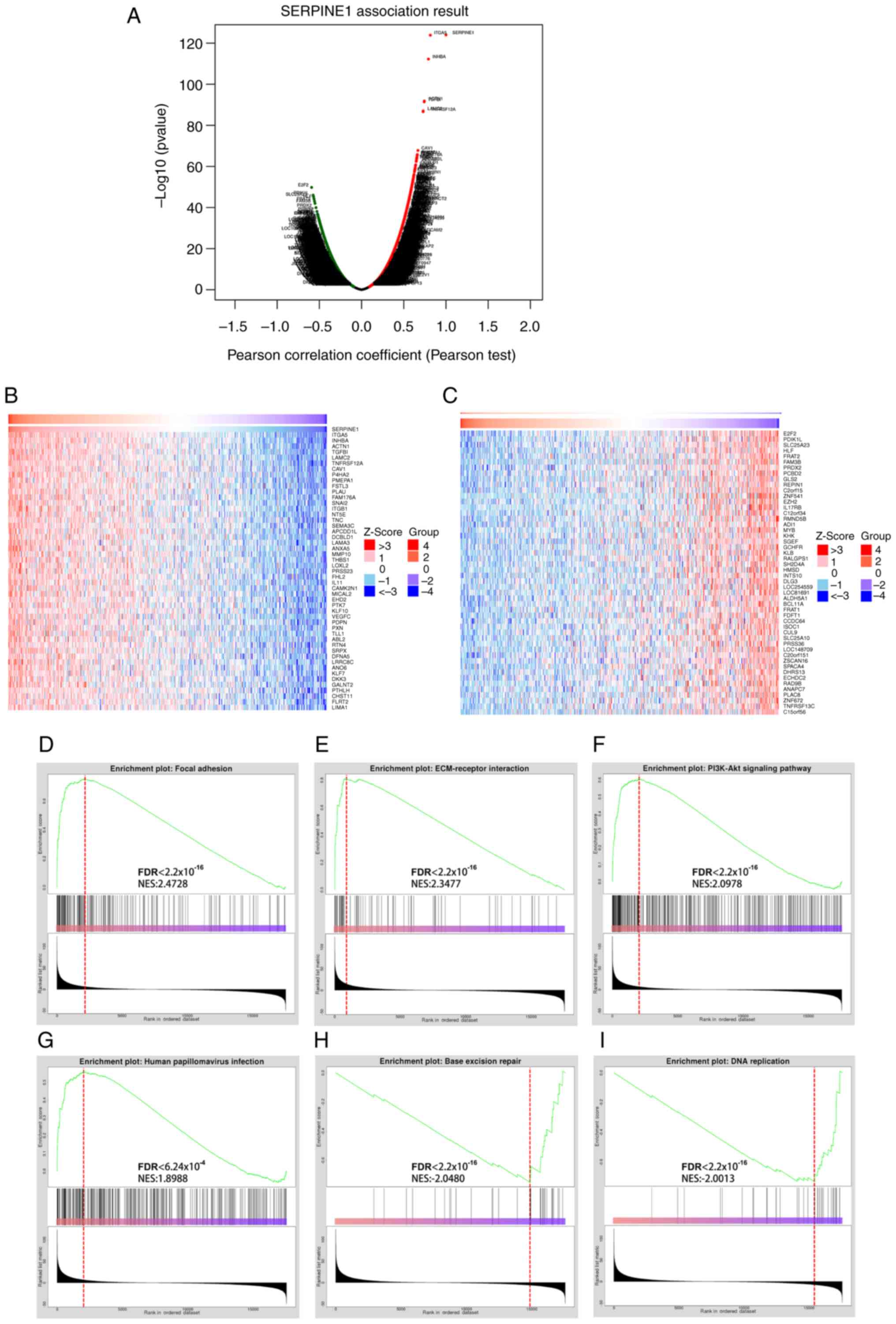
Co-expression analysis of SERPINE1 in HNSCC revealed
a significant association with several genes, including TGA5,
INHBA, ACTN1, E2F2 and PDIK1L (Fig.
8A). Heatmaps showed the top 100 genes associated with
SERPINE1, suggesting potential regulatory networks in HNSCC
(Fig. 8B and C). Furthermore, GSEA
revealed that key BPs were significantly associated with SERPINE1,
including ‘focal adhesion’, ‘ECM-receptor interaction’ and
‘PI3K-Akt signaling’, all crucial for cell-matrix interactions and
survival signaling (Fig. 8D-F).
Additionally, pathways associated with ‘human papillomavirus
infection’, ‘base excision repair’ and ‘DNA replication’ were
significantly enriched, highlighting the potential role of SERPINE1
in the oncogenic mechanisms of HPV+ HNSCC (Fig. 8G-I).
Molecular docking of quercetin with
SERPINE1 protein
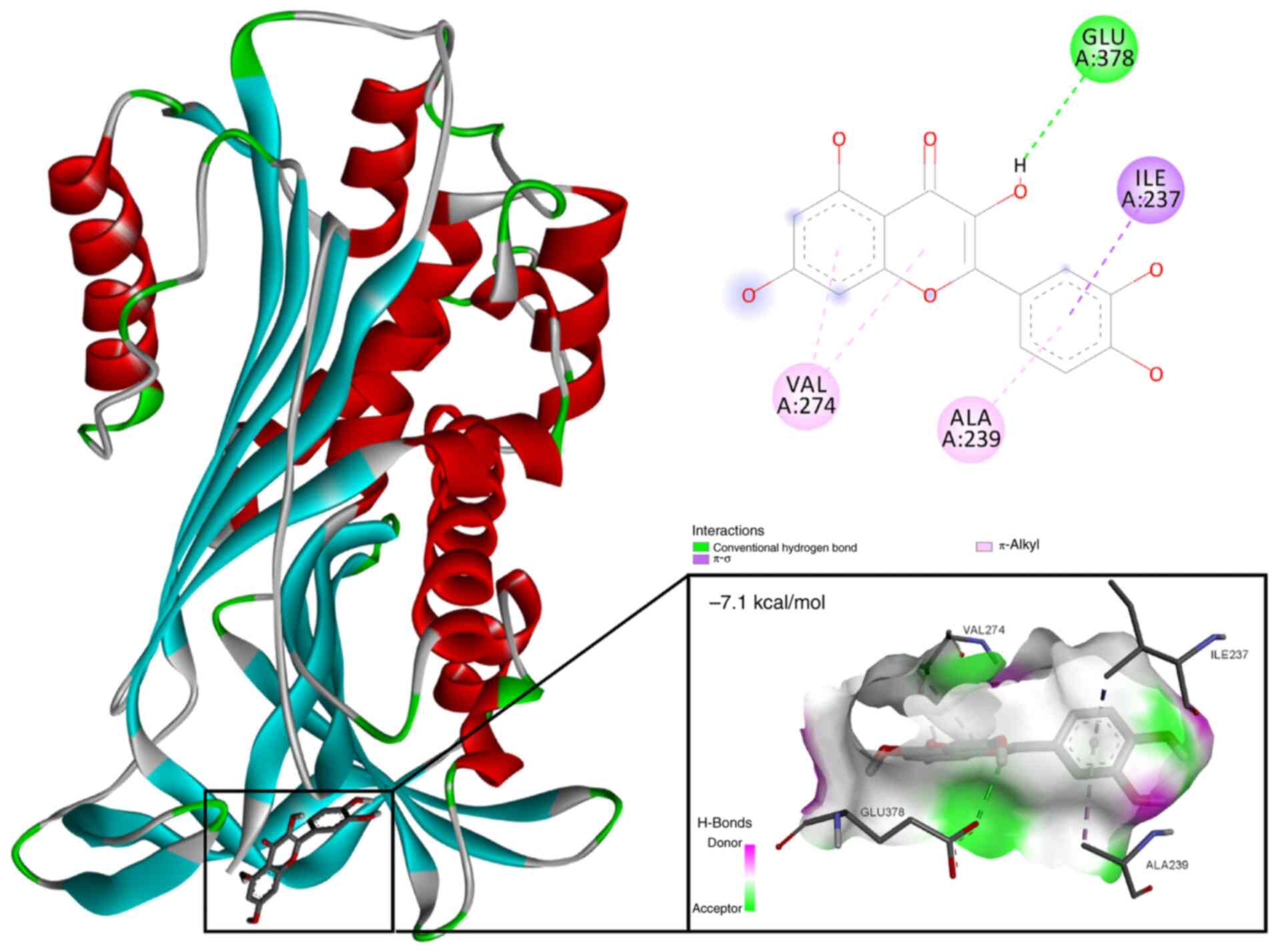
Based on the results of network pharmacological
analysis and the drug-component-target diagram (Fig. 1B), quercetin was identified as the
primary bioactive component in Aloe vera that specifically
targets SERPINE1. Subsequent molecular docking analysis revealed a
strong binding interaction between quercetin and the SERPINE1
protein (PDB identification, 1LJ5), with a binding energy
calculated at −7.1 kcal/mol (Fig.
9). The analysis revealed key interactions between quercetin
and critical residues within the SERPINE1 active site, including
hydrogen bonds with GLU378 and hydrophobic interactions involving
VAL274, ALA239 and ILE237. These interactions suggested that
quercetin may effectively inhibit the function of SERPINE1 by
stabilizing its active conformation.
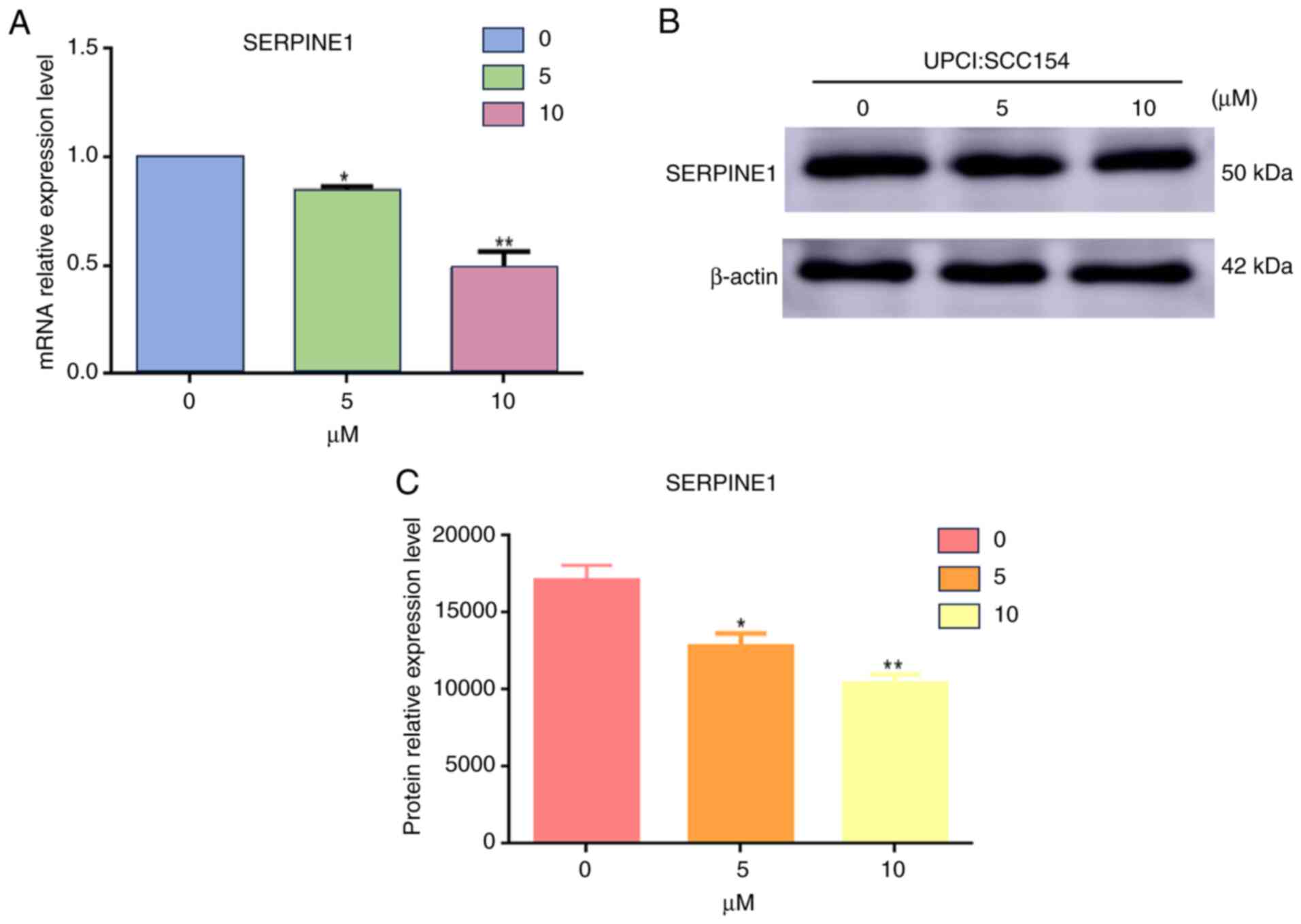
Downregulation of SERPINE1 by
quercetin in HPV+HNSCC Cells
The impact of quercetin on SERPINE1 expression was
further evaluated in HPV+ HNSCC cells. A dose-dependent
reduction in SERPINE1 expression was observed following quercetin
treatment. At the mRNA level, a significant decrease was detected
in response to 5 and 10 µM quercetin compared with 0 µM, with the
effect being more pronounced at 10 µM (Fig. 10A). This decrease in mRNA levels
was further validated by western blot analysis. This analysis
confirmed that the protein levels of SERPINE1 were downregulated
following treatment with quercetin, as depicted in Fig. 10B.
Discussion
HNSCC remains challenging to treat despite
advancements in therapeutic strategies, primarily due to its
complex molecular landscape. However, TCM has emerged as a
potential therapeutic option for this malignancy. A previous study
revealed that patients with oral cancer who received treatments
with TCM formulations, such as Xuán shēn, Shí hú, Mài mén dōng and
other herbal remedies, experienced a notable improvement in their
overall survival rates (6). Further
studies have suggested that TCM exerts antitumor effects by
triggering apoptosis and by regulating the programmed
death-1/programmed death-ligand 1 axis within the tumor immune
microenvironment (TIME), thereby enhancing CD8+ T-cell
activity and improving antitumor immunity (27,28).
Among various herbal formulations in TCM, Aloe
vera has shown notable anticancer potential. In the present
study, quercetin, a major bioactive compound in Aloe vera,
demonstrated promising molecular interactions with SERPINE1.
Quercetin (3,3′,4′,5,7-pentahydroxyflavone), broadly recognized for
its immune-protective, anti-inflammatory and anti-aging effects, is
a type of flavonol (29). Its
antiviral action is also notable, particularly in its ability to
bind to the HPV oncoprotein E6, interfering with the interaction
between E6 and E6-associated protein, leading to G2
phase arrest and apoptosis (30).
Furthermore, quercetin demonstrates extensive anticancer potential
by suppressing key signaling pathways such as mTOR and MAPK,
increasing reactive oxygen species levels and regulating
epithelial-mesenchymal transition markers together with matrix
metalloproteinases (31–34). These actions effectively inhibit the
migration, invasion and development of drug resistance in cancer
cells. Additionally, its interaction with key proteins such as
SERPINE1 indicates a targeted mechanism that holds promise for the
treatment of HPV+ HNSCC.
Previous research has indicated that the
upregulation of SERPINE1 is a marker of poor prognosis in several
types of cancer, including HPV+ oral squamous cell
carcinoma and gastric cancer (35,36).
Additionally, SERPINE1 serves a vital role in cancer-associated
pathways, such as ECM-receptor interaction and the PI3K-Akt
signaling pathway, which are essential for cancer cell survival,
migration and invasion. In HNSCC, SERPINE1 has also been associated
with the infiltration of immune cells such as CD8+ T
cells, CD4+ T cells and macrophages, thereby reshaping
the TIME (37,38).
GSEA demonstrated an association between SERPINE1
and HPV infection in the present study. In addition, survival
analysis highlighted SERPINE1 as a crucial gene associated with
poor prognosis in HPV+ HNSCC. Molecular docking showed
that quercetin binds strongly to SERPINE1 protein, with a binding
energy of −7.1 kcal/mol. Subsequent experiments demonstrated a
dose-dependent downregulation in the expression of SERPINE1 at both
the mRNA and protein levels in response to quercetin. These
findings indicated that quercetin may inhibit SERPINE1 by
stabilizing its active conformation, potentially reversing the
immunosuppressive TIME associated with elevated SERPINE1
expressions and SCNAs. Additionally, quercetin may exert anticancer
effects by directly binding and inhibiting the HPV oncoprotein E6,
further contributing to its potential as a multi-targeted
anticancer agent in HPV+ HNSCC. Therefore, the present
study presents a promising therapeutic strategy for ameliorating
the TIME and enhancing treatment efficacy in HPV+ HNSCC,
by targeting SERPINE1 with quercetin through a dual mechanism of
both inhibiting its expression and stabilizing its
conformation.
Although quercetin has been demonstrated to
downregulate SERPINE1 in HPV+ HNSCC cells, the current
study only confirmed the results through the use of in vitro
cell assays. Further investigations are needed to confirm its
therapeutic efficacy as a bioactive ingredient and to reveal its
related molecular mechanisms in complex tumor microenvironments
in vivo. Additionally, natural products are distinguished by
their multitarget and multi-pathway characteristics, implying their
potential to elicit synergistic effects through diverse
phytochemicals (39). The present
study investigated the regulatory effects of quercetin targeting
SERPINE1; however, further research is still needed to explore the
potential synergistic effects among different active components and
their combined regulatory impact on SERPINE1.
In summary, SERPINE1 is a highly promising
biological marker and therapeutic target in HPV+ HNSCC.
Quercetin has the potential to exert anticancer effects through
targeting and inhibiting SERPINE1, and it may also capitalize on
its antiviral properties. These findings indicate that quercetin,
along with Aloe vera, could be compelling candidates for
further investigation into the treatment of HPV+
HNSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Youth Science and Technology
Talent Program in Health and Wellness, Xinjiang Uygur Autonomous
Region (grant no. WJWY-202423).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
KZ, YH and KW conceived and designed the present
study. KZ, YH, YZ, XZ and SH collected the data. KZ, YH, YZ, XZ and
SH analyzed and interpreted the data. KZ and YH wrote the
manuscript. KW provided critical revisions important for the
intellectual content. KZ and KW confirm the authenticity of all the
raw data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barsouk A, Aluru JS, Rawla P, Saginala K
and Barsouk A: Epidemiology, risk factors, and prevention of head
and neck squamous cell carcinoma. Med Sci (Basel).
11:422023.PubMed/NCBI
|
|
2
|
Dickstein DR, Egerman MA, Bui AH, Doucette
JT, Sharma S, Liu J, Gupta V, Miles BA, Genden E, Westra WH, et al:
A new face of the HPV epidemic: Oropharyngeal cancer in the
elderly. Oral Oncol. 109:1046872020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Felice F, Bird T, Michaelidou A,
Thavaraj S, Odell E, Sandison A, Hall G, Morgan P, Lyons A,
Cascarini L, et al: Radical (chemo)radiotherapy in oropharyngeal
squamous cell carcinoma: Comparison of TNM 7th and 8th staging
systems. Radiother Oncol. 145:146–153. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aggarwal N, Yadav J, Thakur K, Bibban R,
Chhokar A, Tripathi T, Bhat A, Singh T, Jadli M, Singh U, et al:
Human papillomavirus infection in head and neck squamous cell
carcinomas: Transcriptional triggers and changed disease patterns.
Front Cell Infect Microbiol. 10:5376502020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abou Kors T, Hofmann L, Betzler A, Payer
K, Bens M, Truong J, von Witzleben A, Thomas J, Kraus JM, Kalaajieh
R, et al: INHBA is enriched in HPV-negative oropharyngeal squamous
cell carcinoma and promotes cancer progression. Cancer Res Commun.
4:571–587. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ben-Arie E, Lottering B, Inprasit C, Yip
HT, Ho WC, Ton G, Lee YC and Kao PY: Traditional Chinese medicine
use in patients with oral cancer: A retrospective longitudinal
cohort study in Taiwan. Medicine (Baltimore). 101:e307162022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang H, Zhao Q, Zhang Y, Zhang Q, Zheng Z,
Liu S, Liu Z, Meng L, Xin Y and Jiang X: Immunotherapy advances in
locally advanced and recurrent/metastatic head and neck squamous
cell carcinoma and its relationship with human papillomavirus.
Front Immunol. 12:6520542021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Qiu H, Li C, Cai P and Qi F: The
positive role of traditional Chinese medicine as an adjunctive
therapy for cancer. Biosci Trends. 15:283–298. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang B, Liu G, Wang X and Hu X:
Identification of molecular targets and potential mechanisms of
Yinchen Wuling San against head and neck squamous cell carcinoma by
network pharmacology and molecular docking. Front Genet.
13:9146462022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Catalano A, Ceramella J, Iacopetta D,
Marra M, Conforti F, Lupi FR, Gabriele D, Borges F and Sinicropi
MS: Aloe vera-an extensive review focused on recent studies.
Foods. 13:21552024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Riaz S, Hussain S, Syed SK and Anwar R:
Chemical characteristics and therapeutic potentials of Aloe vera.
RADS J Biol Res Appl Sci. 12:160–166. 2021. View Article : Google Scholar
|
|
12
|
Tong X, Li M, Li D, Lao C, Chen J, Xu W,
Du J, Zhang M, Yang X and Li J: Aloe vera gel extract: Safety
evaluation for acute and chronic oral administration in
Sprague-Dawley rats and anticancer activity in breast and lung
cancer cells. J Ethnopharmacol. 280:1144342021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karpagam T, Firdous J, Priya S,
Varalakshmi B, Gomathi S, Geetha S and Muhamad N: Anti-cancer
activity of Aloe vera ethanolic leaves extract against in vitro
cancer cells. Res J Pharm Technol. 12:2167–2170. 2019. View Article : Google Scholar
|
|
14
|
Akev N, Candoken E and Erdem Kuruca S:
Comparative study on the anticancer drug potential of a lectin
purified from aloe vera and aloe-emodin. Asian Pac J Cancer Prev.
21:99–106. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sa N, Liu X, Hao D, Lv Z, Zhou S, Yang L,
Jiang S, Tian J and Xu W: FTO-mediated m6A demethylation
of SERPINE1 mRNA promotes tumor progression in hypopharyngeal
squamous cell carcinoma. Transl Cancer Res. 14:595–612. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arroyo-Solera I, Pavón MÁ, León X, López
M, Gallardo A, Céspedes MV, Casanova I, Pallarès V, López-Pousa A,
Mangues MA, et al: Effect of serpinE1 overexpression on the primary
tumor and lymph node, and lung metastases in head and neck squamous
cell carcinoma. Head Neck. 41:429–439. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu C, Liu H, Li Z, Shi Y, Zhao J, Bai Y,
Chen Q and Li W: Prognostic significance and therapeutic potential
of SERPINE1 in head and neck squamous cell carcinoma. Cancer Med.
14:e706052025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang S, Ou L, Wang Y, Su K, Chen Z, He L,
Xu X, Cheng B, Xia J and Fan Z: CircPRMT5, a potential salivary
biomarker, facilitates the progression of head and neck squamous
cell carcinoma via the IGF2BP3-SERPINE1 pathway. Int J
Nanomedicine. 20:1597–1613. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ru J, Li P, Wang J, Zhou W, Li B, Huang C,
Li P, Guo Z, Tao W, Yang Y, et al: TCMSP: A database of systems
pharmacology for drug discovery from herbal medicines. J
Cheminform. 6:132014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Daina A, Michielin O and Zoete V:
SwissTargetPrediction: Updated data and new features for efficient
prediction of protein targets of small molecules. Nucleic Acids
Res. 47:W357–W364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mock A, Plath M, Moratin J, Tapken MJ,
Jäger D, Krauss J, Fröhling S, Hess J and Zaoui K: EGFR and PI3K
pathway activities might guide drug repurposing in HPV-negative
head and neck cancers. Front Oncol. 11:6789662021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Parker HS, Leek JT, Favorov AV, Considine
M, Xia X, Chavan S, Chung CH and Fertig EJ: Preserving biological
heterogeneity with a permuted surrogate variable analysis for
genomics batch correction. Bioinformatics. 30:2757–2763. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang C, Li D, Ko CN, Wang K and Wang H:
Active ingredients of traditional Chinese medicine for enhancing
the effect of tumor immunotherapy. Front Immunol. 14:11330502023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng H, Wang G, Liu M and Cheng H:
Traditional Chinese medicine inhibits PD-1/PD-L1 axis to sensitize
cancer immunotherapy: A literature review. Front Oncol.
13:1682262023.
|
|
29
|
Saeedi-Boroujeni A and Mahmoudian-Sani MR:
Anti-inflammatory potential of quercetin in COVID-19 treatment. J
Inflamm (Lond). 18:32021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clemente-Soto AF, Salas-Vidal E,
Milan-Pacheco C, Sánchez-Carranza JN, Peralta-Zaragoza O and
González-Maya L: Quercetin induces G2 phase arrest and apoptosis
with the activation of p53 in an E6 expression-independent manner
in HPV-positive human cervical cancer-derived cells. Mol Med Rep.
19:2097–2106. 2019.PubMed/NCBI
|
|
31
|
Zhuang Y, Coppock JD, Haugrud AB, Lee JH,
Messerli SM and Miskimins WK: Dichloroacetate and quercetin prevent
cell proliferation, induce cell death and slow tumor growth in a
mouse model of HPV-positive head and neck cancer. Cancers (Basel).
16:15252024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan CY, Hong SC, Chang CM, Chen YH, Liao
PC and Huang CY: Oral squamous cell carcinoma cells with acquired
resistance to erlotinib are sensitive to anti-cancer effect of
quercetin via pyruvate kinase M2 (PKM2). Cells. 12:1792023.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang CF, Liu SH, Ho TJ, Lee KI, Fang KM,
Lo WC, Liu JM, Wu CC and Su CC: Quercetin induces tongue squamous
cell carcinoma cell apoptosis via the JNK activation-regulated
ERK/GSK-3α/β-mediated mitochondria-dependent apoptotic signaling
pathway. Oncol Lett. 23:782022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Son HK and Kim D: Quercetin induces cell
cycle arrest and apoptosis in YD10B and YD38 oral squamous cell
carcinoma cells. Asian Pac J Cancer Prev. 24:283–289. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miranda-Galvis M, Carneiro Soares C,
Moretto Carnielli C, Ramalho Buttura J, Sales de Sá R, Kaminagakura
E, Marchi FA, Paes Leme AF, Lópes Pinto CA, Santos-Silva AR, et al:
New insights into the impact of human papillomavirus on oral cancer
in young patients: Proteomic approach reveals a novel role for
S100A8. Cells. 12:13232023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen S, Li Y, Zhu Y, Fei J, Song L, Sun G,
Guo L and Li X: SERPINE1 overexpression promotes malignant
progression and poor prognosis of gastric cancer. J Oncol.
2022:26478252022.PubMed/NCBI
|
|
37
|
Zhou Q, Yuan O, Cui H, Hu T, Xiao GG, Wei
J, Zhang H and Wu C: Bioinformatic analysis identifies HPV-related
tumor microenvironment remodeling prognostic biomarkers in head and
neck squamous cell carcinoma. Front Cell Infect Microbiol.
12:10079502022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao H, Wang F, Wang X, Zhao X and Ji J:
HPV-related prognostic signature predicts survival in head and neck
squamous cell carcinoma. J Oncol. 2022:73575662022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Tang J, Cao H, Wang C, Shen C and
Liu J: Effect and mechanism of magnolia officinalis in colorectal
cancer: Multi-component-multi-target approach. J Ethnopharmacol.
338:1190072025. View Article : Google Scholar : PubMed/NCBI
|