|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rutegård M, Charonis K, Lu Y, Lagergren P,
Lagergren J and Rouvelas I: Population-based esophageal cancer
survival after resection without neoadjuvant therapy: An update.
Surgery. 152:903–910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Hagen P, Hulshof MC, van Lanschot JJ,
Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ,
Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al: Preoperative
chemoradiotherapy for esophageal or junctional cancer. N Engl J
Med. 366:2074–2084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liang H, Fan JH and Qiao YL: Epidemiology,
etiology, and prevention of esophageal squamous cell carcinoma in
China. Cancer Biol Med. 14:33–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li H, Ding C, Zeng H, Zheng R, Cao M, Ren
J, Shi J, Sun D, He S, Yang Z, et al: Improved esophageal squamous
cell carcinoma screening effectiveness by risk-stratified
endoscopic screening: Evidence from high-risk areas in China.
Cancer Commun (Lond). 41:715–725. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li P, Liu X, Dong ZM and Ling ZQ:
Epigenetic silencing of HIC1 promotes epithelial-mesenchymal
transition and drives progression in esophageal squamous cell
carcinoma. Oncotarget. 6:38151–38165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu T, Ding H, Chen J, Lei J, Zhao M, Ji B,
Chen Y, Qin S and Gao Q: Research progress of DNA methylation in
endometrial cancer. Biomolecules. 12:9382022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Sheng H, Hu C, Li F, Cai B and Ma
Y, Wang Y and Ma Y: Effects of DNA methylation on gene expression
and phenotypic traits in cattle: A review. Int J Mol Sci.
24:118822023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peneder P, Stütz AM, Surdez D, Krumbholz
M, Semper S, Chicard M, Sheffield NC, Pierron G, Lapouble E, Tötzl
M, et al: Multimodal analysis of cell-free DNA whole-genome
sequencing for pediatric cancers with low mutational burden. Nat
Commun. 12:32302021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miyoshi J, Zhu Z, Luo A, Toden S, Zhou X,
Izumi D, Kanda M, Takayama T, Parker IM, Wang M, et al: A
microRNA-based liquid biopsy signature for the early detection of
esophageal squamous cell carcinoma: A retrospective, prospective
and multicenter study. Mol Cancer. 21:442022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiao G, Zhuang W, Dong B, Li C, Xu J, Wang
G, Xie L, Zhou Z, Tian D, Chen G, et al: Discovery and validation
of methylation signatures in circulating cell-free DNA for early
detection of esophageal cancer: A case-control study. BMC Med.
19:2432021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin Y, Wu CW, Taylor WR, Sawas T, Burger
KN, Mahoney DW, Sun Z, Yab TC, Lidgard GP, Allawi HT, et al:
Discovery, validation, and application of novel methylated DNA
markers for detection of esophageal cancer in plasma. Clin Cancer
Res. 25:7396–7404. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xue Y, Wang K, Jiang Y, Dai Y, Liu X, Pei
B, Li H, Xu H and Zhao G: An ultrasensitive and multiplexed miRNA
one-step real time RT-qPCR detection system and its application in
esophageal cancer serum. Biosens Bioelectron. 247:1159272024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu D, Wu X, Wu W, Wu S, Li H, Zhang Y, Yan
X, Zhai J, Dong X, Feng S, et al: Plasma cell-free DNA
5-hydroxymethylcytosine and whole-genome sequencing signatures for
early detection of esophageal cancer. Cell Death Dis. 14:8432023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang X, Suo C, Zhang T, Yin X, Man J, Yuan
Z, Yu J, Jin L, Chen X, Lu M and Ye W: Targeted proteomics-derived
biomarker profile develops a multi-protein classifier in liquid
biopsies for early detection of esophageal squamous cell carcinoma
from a population-based case-control study. Biomark Res. 9:122021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv J, Wang J, Shen X, Liu J, Zhao D, Wei
M, Li X, Fan B, Sun Y, Xue F, et al: A serum metabolomics analysis
reveals a panel of screening metabolic biomarkers for esophageal
squamous cell carcinoma. Clin Transl Med. 11:e4192021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chan KC, Jiang P, Chan CW, Sun K, Wong J,
Hui EP, Chan SL, Chan WC, Hui DS, Ng SS, et al: Noninvasive
detection of cancer-associated genome-wide hypomethylation and copy
number aberrations by plasma DNA bisulfite sequencing. Proc Natl
Acad Sci USA. 110:18761–18768. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hattori N and Ushijima T: Compendium of
aberrant DNA methylation and histone modifications in cancer.
Biochem Biophys Res Commun. 455:3–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang M, Zhou J, Xie X, Huang Z, Liu R and
Lv Y: Single nanoparticle counting-based liquid biopsy for cancer
diagnosis. Anal Chem. 94:15433–15439. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Llueca A, Canete-Mota S, Jaureguí A,
Barneo M, Ibañez MV, Neef A, Ochoa E, Tomas-Perez S, Mari-Alexandre
J, Gilabert-Estelles J, et al: The impact of liquid biopsy in
advanced ovarian cancer care. Diagnostics (Basel). 14:18682024.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weinberg DN, Rosenbaum P, Chen X, Barrows
D, Horth C, Marunde MR, Popova IK, Gillespie ZB, Keogh MC, Lu C, et
al: Two competing mechanisms of DNMT3A recruitment regulate the
dynamics of de novo DNA methylation at PRC1-targeted CpG islands.
Nat Genet. 53:794–800. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nie K, Jia Y and Zhang X: Cell-free
circulating tumor DNA in plasma/serum of non-small cell lung
cancer. Tumour Biol. 36:7–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang
F, Wang N, Yang H, Zheng Y and Zhang J: Identification of a DNA
methylome profile of esophageal squamous cell carcinoma and
potential plasma epigenetic biomarkers for early diagnosis. PLoS
One. 9:e1031622014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
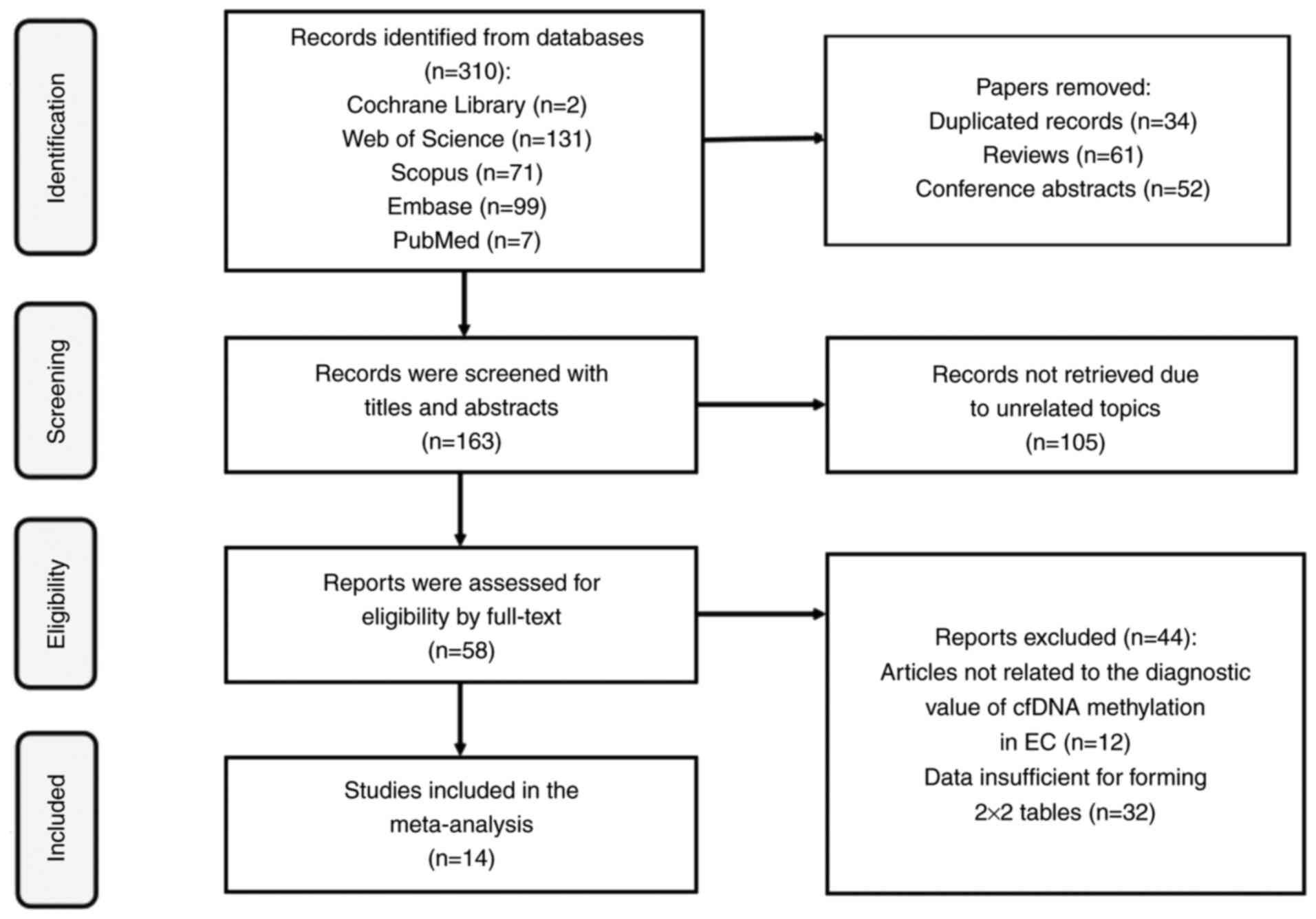
Page MJ, McKenzie JE, Bossuyt PM, Boutron
I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan
SE, et al: The PRISMA 2020 statement: an updated guideline for
reporting systematic reviews. BMJ. 372:n712021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
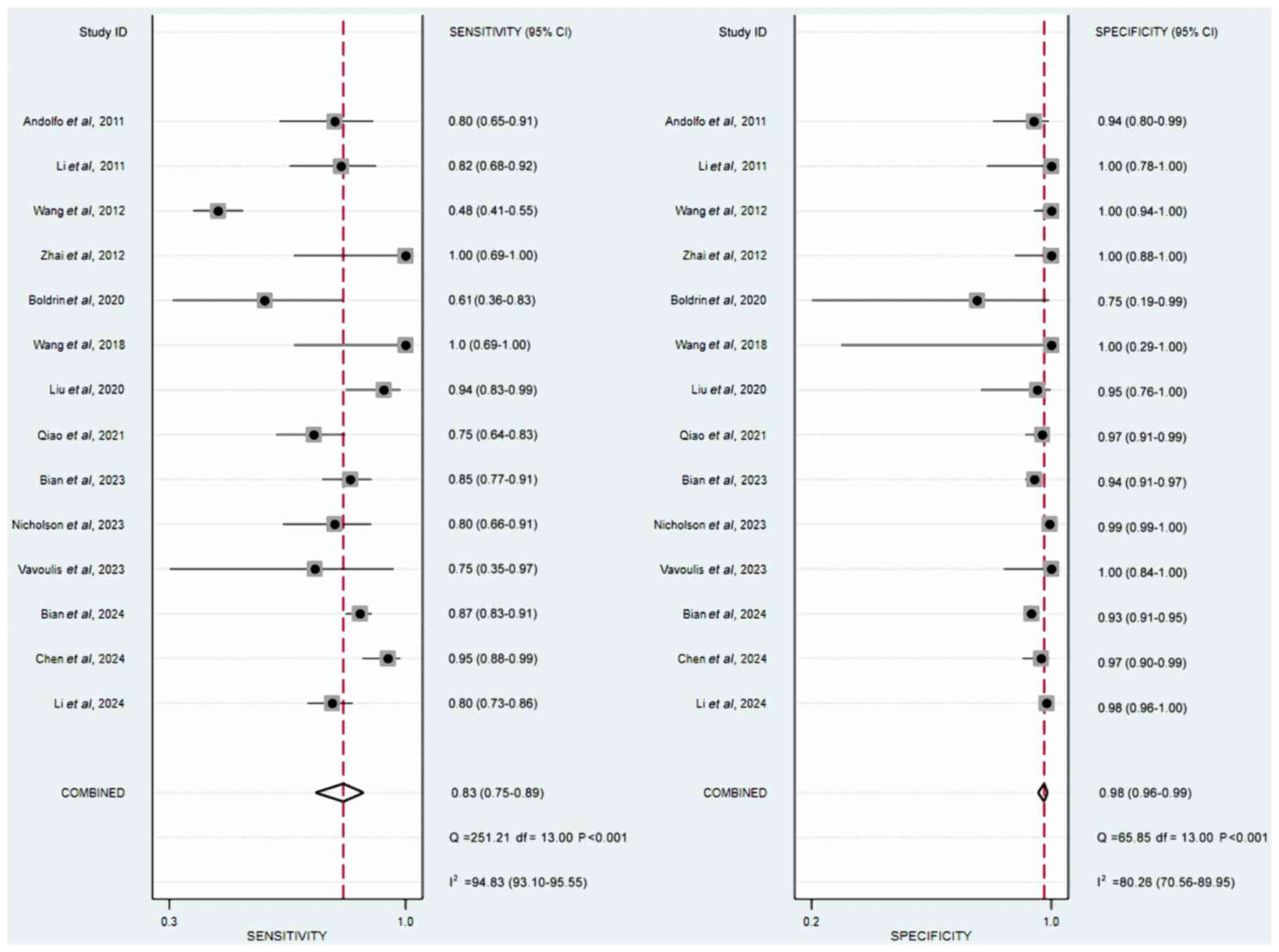
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM; QUADAS-2 Group, : QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
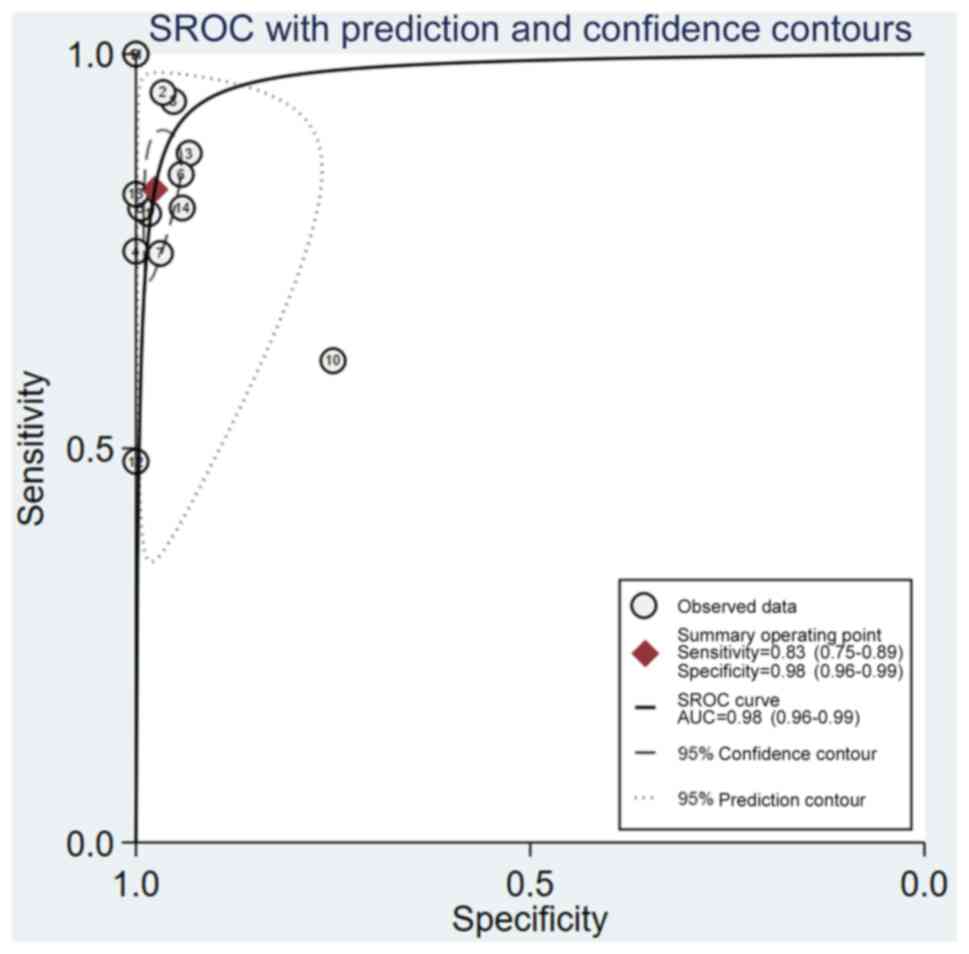
Zamora J, Abraira V, Muriel A, Khan K and
Coomarasamy A: Meta-DiSc: A software for meta-analysis of test
accuracy data. BMC Med Res Methodol. 6:312006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
StataCorp. Stata Statistical Software, .
Release 16. College Station, TX: StataCorp LLC; 2019, https://www.stata.comJuly 25–2024
|
|
29
|
Review Manager (RevMan) [Computer
program], . Version 5.4. The Cochrane Collaboration. 2020.Available
from. https://training.cochrane.org/online-learning-materials/revman
|
|
30
|
Li Y, Liu B, Zhou X, Yang H, Han T, Hong
Y, Wang C, Huang M, Yan S, Li S, et al: Genome-Scale multimodal
analysis of cell-free DNA whole-methylome sequencing for
noninvasive esophageal cancer detection. JCO Precis Oncol.
8:e24001112024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Li M, Li Y, Aiolfi A, Bonavina L,
Lerut T, Wu X and Zhang Q: Combining non-invasive liquid biopsy and
a methylation analysis to assess surgical risk for early esophageal
cancer. Transl Cancer Res. 13:3075–3089. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bian Y, Gao Y, Lin H, Sun C, Wang W, Sun
S, Li X, Feng Z, Ren J, Chen H, et al: Non-invasive diagnosis of
esophageal cancer by a simplified circulating cell-free DNA
methylation assay targeting OTOP2 and KCNA3: A double-blinded,
multicenter, prospective study. J Hematol Oncol. 17:472024.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vavoulis D, Cutts A, Thota N, Brown J,
Sugar R, Rueda A, Ardalan A, Matos Santo F, et al: Multimodal
cell-free DNA whole-genome analysis combined with TET-Assisted
Pyridine Borane Sequencing is sensitive and reveals specific cancer
signals. MedRxiv. DOI:. https://doi.org/10.1101/2023.09.29.23296336
|
|
34
|
Nicholson BD, Oke J, Virdee PS, Harris DA,
O'Doherty C, Park JE, Hamady Z, Sehgal V, Millar A, Medley L, et
al: Multi-cancer early detection test in symptomatic patients
referred for cancer investigation in England and Wales (SYMPLIFY):
a large-scale, observational cohort study. Lancet Oncol.
24:733–743. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bian Y, Gao Y, Lu C, Tian B, Xin L, Lin H,
Zhang Y, Zhang X, Zhou S, Wan K, et al: Genome-wide methylation
profiling identified methylated KCNA3 and OTOP2 as promising
diagnostic markers for esophageal squamous cell carcinoma. Chin Med
J (Engl). 137:1724–1735. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu MC, Oxnard GR, Klein EA, Swanton C and
Seiden MV; CCGA Consortium, : Sensitive and specific multi-cancer
detection and localization using methylation signatures in
cell-free DNA. Ann Oncol. 31:745–759. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang HQ, Yang CY, Wang SY, Wang T, Han JL,
Wei K, Liu FC, Xu JD, Peng XZ and Wang JM: Cell-free plasma
hypermethylated CASZ1, CDH13 and ING2 are promising biomarkers of
esophageal cancer. J Biomed Res. 32:424–433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Boldrin E, Curtarello M, Dallan M, Alfieri
R, Realdon S, Fassan M and Saggioro D: Detection of LINE-1
hypomethylation in cfDNA of esophageal adenocarcinoma patients. Int
J Mol Sci. 21:15472020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhai R, Zhao Y, Su L, Cassidy L, Liu G and
Christiani DC: Genome-wide DNA methylation profiling of cell-free
serum DNA in esophageal adenocarcinoma and Barrett esophagus.
Neoplasia. 14:29–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang CC, Mao WM and Ling ZQ: DNA
methylation status of RARβ2 and p16(INK4α) in peripheral blood and
tumor tissue in patients with esophageal squamous cell carcinoma.
Zhonghua Zhong Liu Za Zhi. 34:441–445. 2012.(In Chinese).
PubMed/NCBI
|
|
41
|
Li B, Wang B, Niu LJ, Jiang L and Qiu CC:
Hypermethylation of multiple tumor-related genes associated with
DNMT3b up-regulation served as a biomarker for early diagnosis of
esophageal squamous cell carcinoma. Epigenetics. 6:307–316. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Andolfo I, Petrosino G, Vecchione L, De
Antonellis P, Capasso M, Montanaro D, Gemei M, Troncone G, Iolascon
A, Orditura M, et al: Detection of erbB2 copy number variations in
plasma of patients with esophageal carcinoma. BMC Cancer.
11:1262011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Constantin N, Sina AA, Korbie D and Trau
M: Opportunities for early cancer detection: The rise of ctDNA
methylation-based pan-cancer screening technologies. Epigenomes.
6:62022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nikanjam M, Kato S and Kurzrock R: Liquid
biopsy: Current technology and clinical applications. J Hematol
Oncol. 15:1312022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lo YM, Chan KC, Sun H, Chen EZ, Jiang P,
Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR and Chiu RW: Maternal
plasma DNA sequencing reveals the genome-wide genetic and
mutational profile of the fetus. Sci Transl Med. 2:61ra912010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Heitzer E, Auinger L and Speicher MR:
Cell-Free DNA and apoptosis: How dead cells inform about the
living. Trends Mol Med. 26:519–528. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jones PA: Functions of DNA methylation:
islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dor Y and Cedar H: Principles of DNA
methylation and their implications for biology and medicine.
Lancet. 392:777–786. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Esfahani MS, Hamilton EG, Mehrmohamadi M,
Nabet BY, Alig SK, King DA, Steen CB, Macaulay CW, Schultz A,
Nesselbush MC, et al: Inferring gene expression from cell-free DNA
fragmentation profiles. Nat Biotechnol. 40:585–597. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lo YM, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW and Wainscoat JS: Presence of fetal DNA in
maternal plasma and serum. Lancet. 350:485–487. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu SC, Lee SW, Jiang P, Leung TY, Chan KC,
Chiu RW and Lo YM: High-resolution profiling of fetal DNA clearance
from maternal plasma by massively parallel sequencing. Clin Chem.
59:1228–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Feng H, Jin P and Wu H: Disease prediction
by cell-free DNA methylation. Brief Bioinform. 20:585–597. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Azad TD, Chaudhuri AA, Fang P, Qiao Y,
Esfahani MS, Chabon JJ, Hamilton EG, Yang YD, Lovejoy A, Newman AM,
et al: Circulating tumor DNA analysis for detection of minimal
residual disease after chemoradiotherapy for localized esophageal
cancer. Gastroenterology. 158:494–505.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Egyud M, Tejani M, Pennathur A, Luketich
J, Sridhar P, Yamada E, Ståhlberg A, Filges S, Krzyzanowski P,
Jackson J, et al: Detection of circulating tumor DNA in plasma: A
potential biomarker for esophageal adenocarcinoma. Ann Thorac Surg.
108:343–349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fleischhacker M and Schmidt B: Circulating
nucleic acids (CNAs) and cancer-a survey. Biochim Biophys Acta.
1775:181–232. 2007.PubMed/NCBI
|
|
58
|
Frommer M, McDonald LE, Millar DS, Collis
CM, Watt F, Grigg GW, Molloy PL and Paul CL: A genomic sequencing
protocol that yields a positive display of 5-methylcytosine
residues in individual DNA strands. Proc Natl Acad Sci USA.
89:1827–1831. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang J and Wang L: Cell-Free DNA
methylation profiling analysis-technologies and bioinformatics.
Cancers (Basel). 11:17412019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhu H, Ma X, Ye T, Wang H, Wang Z, Liu Q
and Zhao K: Esophageal cancer in China: Practice and research in
the new era. Int J Cancer. 152:1741–1751. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu T, Chen D, Wang Y, Sun X, Li S, Miao S,
Wo Y, Dong Y, Leng X, Du W and Jiao W: Identification of DNA
methylation-driven genes in esophageal squamous cell carcinoma: a
study based on The Cancer Genome Atlas. Cancer Cell Int. 19:522019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xu Y, Wang Z, Pei B, Wang J, Xue Y and
Zhao G: DNA methylation markers in esophageal cancer. Front Genet.
15:13541952024. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bixby B, Vrba L, Lenka J, Oshiro M, Watts
GS, Hughes T, Erickson H, Chopra M, Knepler JL, Knox KS, et al:
Cell-Free DNA methylation analysis as a marker of malignancy in
pleural fluid. Sci Rep. Feb 5–2023.(Epub ahead of print).
|
|
64
|
Zhou X, Cheng Z, Dong M, Liu Q, Yang W,
Liu M, Tian J and Cheng W: Tumor fractions deciphered from
circulating cell-free DNA methylation for cancer early diagnosis.
Nat Commun. 13:76942022. View Article : Google Scholar : PubMed/NCBI
|