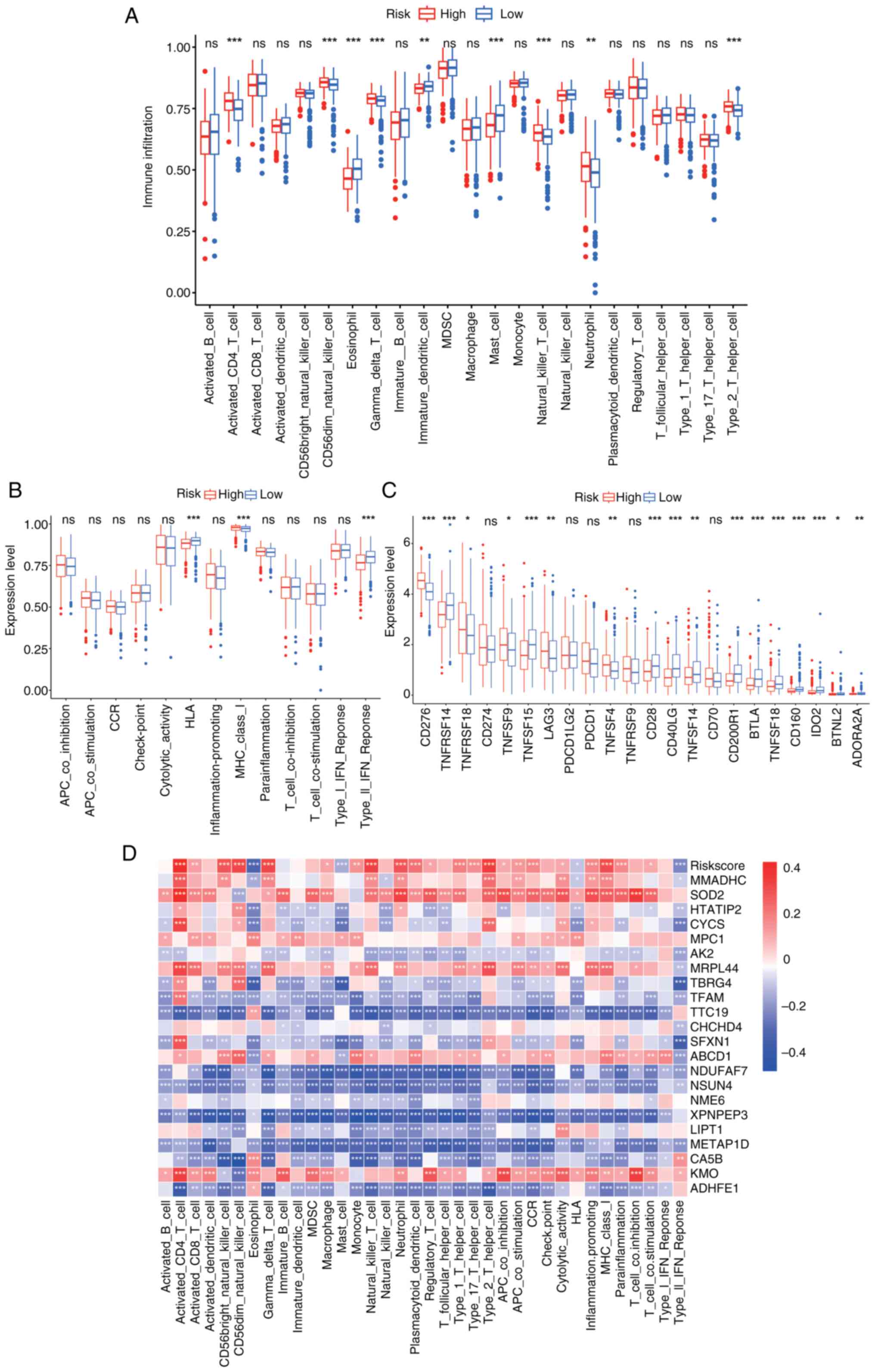
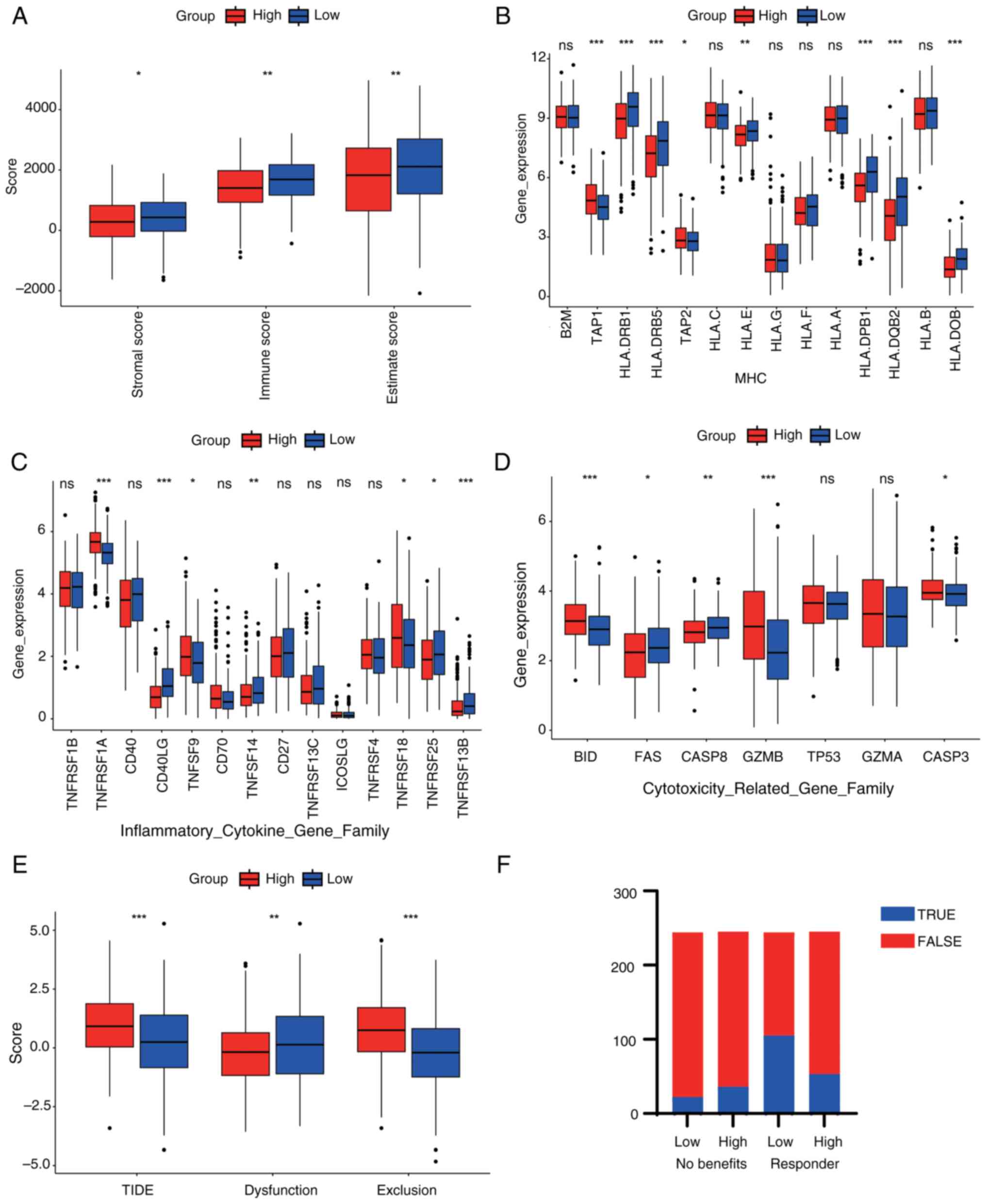
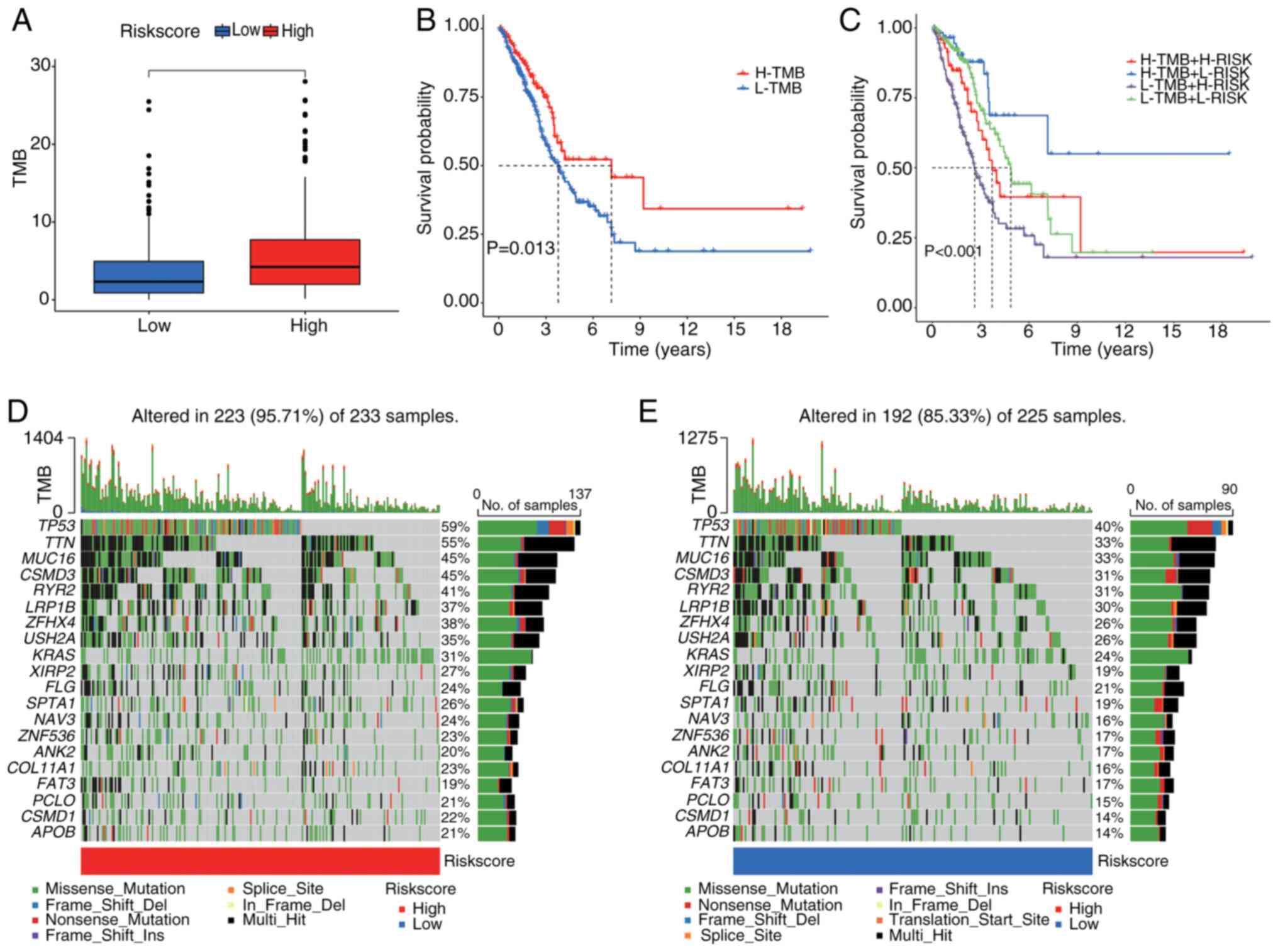
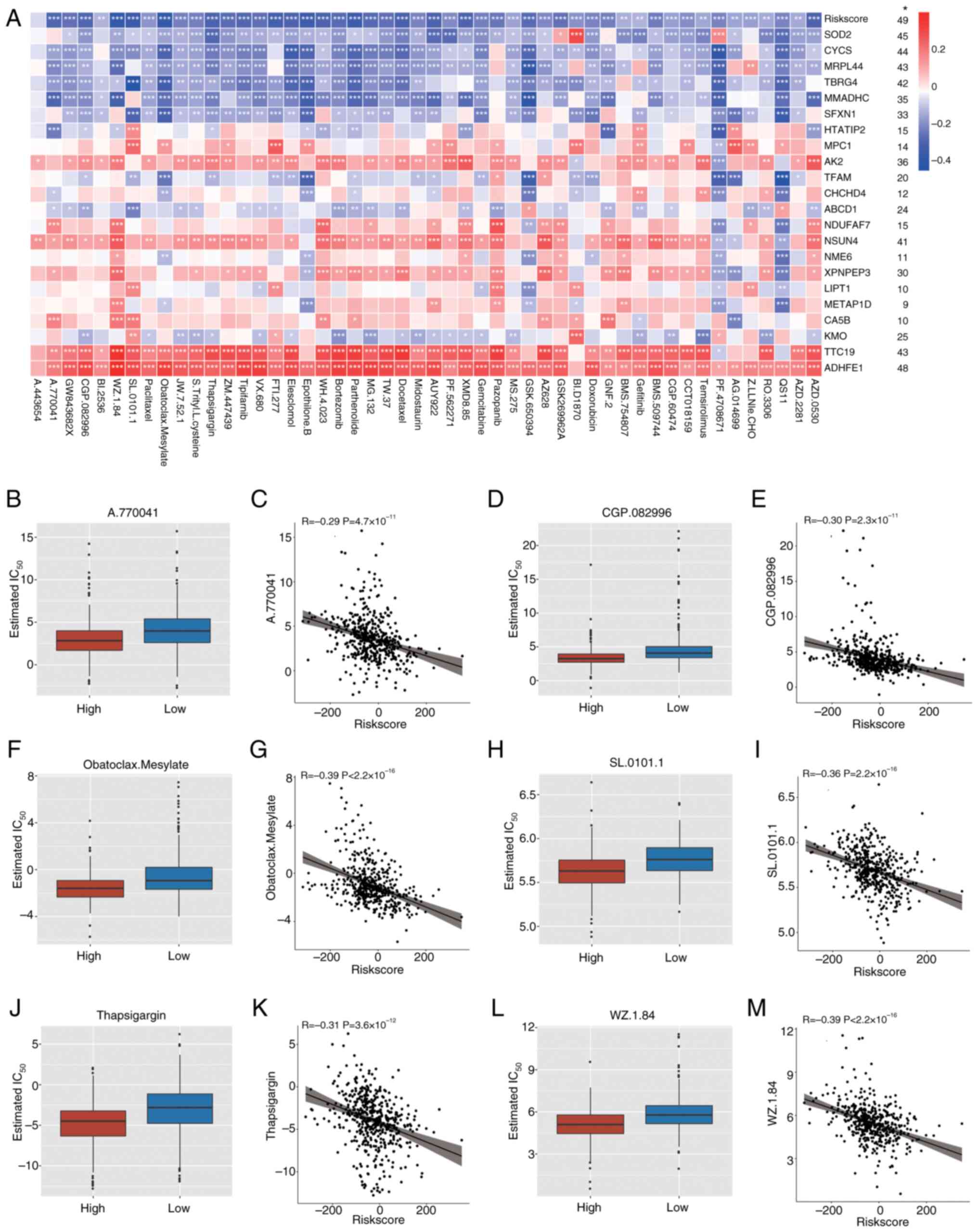
|
1
|
Zhang Y, Vaccarella S, Morgan E, Li M,
Etxeberria J, Chokunonga E, Manraj SS, Kamate B, Omonisi A and Bray
F: Global variations in lung cancer incidence by histological
subtype in 2020: A population-based study. Lancet Oncol.
24:1206–1218. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Florez N, Kiel L, Riano I, Patel S,
DeCarli K, Dhawan N, Franco I, Odai-Afotey A, Meza K, Swami N, et
al: Lung cancer in women: The past, present, and future. Clin Lung
Cancer. 25:1–8. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carter-Harris L: Lung cancer stigma as a
barrier to medical help-seeking behavior: Practice implications. J
Am Assoc Nurse Pract. 27:240–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu HI, Chiang CJ, Su SY, Jhuang JR, Tsai
DR, Yang YW, Lin LJ, Wang YC and Lee WC: Incidence trends and
spatial distributions of lung adenocarcinoma and squamous cell
carcinoma in Taiwan. Sci Rep. 13:16552023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakagawa K, Yoshida Y, Yotsukura M and
Watanabe SI: Minimally invasive open surgery (MIOS) for clinical
stage I lung cancer: Diversity in minimally invasive procedures.
Jpn J Clin Oncol. 51:1649–1655. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duma N, Santana-Davila R and Molina JR:
Non-small cell lung cancer: Epidemiology, screening, diagnosis, and
treatment. Mayo Clin Proc. 94:1623–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doval DC, Desai CJ and Sahoo TP:
Molecularly targeted therapies in non-small cell lung cancer: The
evolving role of tyrosine kinase inhibitors. Indian J Cancer. 56
(Suppl 1):S23–S30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laerum D, Strand TE, Brustugun OT,
Gallefoss F, Falk R, Durheim MT and Fjellbirkeland L: Evaluation of
sex inequity in lung-cancer-specific survival. Acta Oncol.
63:343–350. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lyu G, Dai L, Deng X, Liu X, Guo Y, Zhang
Y, Wang X, Huang Y, Wu S, Guo JC and Liu Y: Integrative analysis of
cuproptosis-related mitochondrial depolarisation genes for
prognostic prediction in non-small cell lung cancer. J Cell Mol
Med. 29:e704382025. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tong X, Tang R, Xiao M, Xu J, Wang W,
Zhang B, Liu J, Yu X and Shi S: Targeting cell death pathways for
cancer therapy: Recent developments in necroptosis, pyroptosis,
ferroptosis, and cuproptosis research. J Hematol Oncol. 15:1742022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang W, Lu K, Jiang X, Wei Q, Zhu L, Wang
X, Jin H and Feng L: Ferroptosis inducers enhanced cuproptosis
induced by copper ionophores in primary liver cancer. J Exp Clin
Cancer Res. 42:1422023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Min J and Wang F: Copper
homeostasis and cuproptosis in health and disease. Signal Transduct
Target Ther. 7:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Faubert B, Li KY, Cai L, Hensley CT, Kim
J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al:
Lactate metabolism in human lung tumors. Cell. 171:358–371. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He Y, Ji Z, Gong Y, Fan L, Xu P, Chen X,
Miao J, Zhang K, Zhang W, Ma P, et al: Numb/Parkin-directed
mitochondrial fitness governs cancer cell fate via metabolic
regulation of histone lactylation. Cell Rep. 42:1120332023.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Elnaggar GN, El-Hifnawi NM, Ismail A,
Yahia M and Elshimy RAA: Micro RNA-148a targets Bcl-2 in patients
with non-small cell lung cancer. Asian Pac J Cancer Prev.
22:1949–1955. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ran XM, Xiao H, Tang YX, Jin X, Tang X,
Zhang J, Li H, Li YK and Tang ZZ: The effect of
cuproptosis-relevant genes on the immune infiltration and
metabolism of gynecological oncology by multiply analysis and
experiments validation. Sci Rep. 13:194742023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang H, Shi Z, Li Y, Zhu G, Chen C, Zhang
Z, Shi R, Su L, Cao P, Pan Z, et al: Pyroptosis-related LncRNA
signatures correlate with lung adenocarcinoma prognosis. Front
Oncol. 12:8509432022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilkerson MD, Yin X, Walter V, Zhao N,
Cabanski CR, Hayward MC, Miller CR, Socinski MA, Parsons AM, Thorne
LB, et al: Differential pathogenesis of lung adenocarcinoma
subtypes involving sequence mutations, copy number, chromosomal
instability, and methylation. PLoS One. 7:e365302012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schabath MB, Welsh EA, Fulp WJ, Chen L,
Teer JK, Thompson ZJ, Engel BE, Xie M, Berglund AE, Creelan BC, et
al: Differential association of STK11 and TP53 with KRAS
mutation-associated gene expression, proliferation and immune
surveillance in lung adenocarcinoma. Oncogene. 35:3209–3216. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao W, Huang H, Zhao Z, Ding C, Jia C,
Wang Y, Wang G, Li Y, Liu H and Chen J: Identification of hypoxia
and mitochondrial-related gene signature and prediction of
prognostic model in lung adenocarcinoma. J Cancer. 15:4513–4526.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Z, Zhang P, Xie J, Cui Y, Shuo W and
Yue D: Five-gene prognostic model based on autophagy-dependent cell
death for predicting prognosis in lung adenocarcinoma. Sci Rep.
14:264492024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang J, Zhang J, Zhang F, Lu S, Guo S,
Shi R, Zhai Y, Gao Y, Tao X, Jin Z, et al: Identification of a
disulfidptosis-related genes signature for prognostic implication
in lung adenocarcinoma. Comput Biol Med. 165:1074022023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ritchie ME: limma: Linear models for
microarray and RNA-Seq data. The R Foundation for Statistical
Computing. 2024.
|
|
25
|
He Y, Jiang Z, Chen C and Wang X:
Classification of triple-negative breast cancers based on
Immunogenomic profiling. J Exp Clin Cancer Res. 37:3272018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang G, Song C, Wang X, Xu Y, Li H, He Z,
Cai Y, Zheng M and Mao W: The multi-omics analysis identifies a
novel cuproptosis-anoikis-related gene signature in prognosis and
immune infiltration characterization of lung adenocarcinoma.
Heliyon. 9:e140912023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang B, Yin Y, Wang A, Liu W, Chen J and
Li T: SMR-guided molecular subtyping and machine learning model
reveals novel prognostic biomarkers and therapeutic targets in
non-small cell lung adenocarcinoma. Sci Rep. 15:16402025.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Li J, Xiong C, Sun X and Shen T:
Development of a prognostic model for NSCLC based on differential
genes in tumour stem cells. Sci Rep. 14:209382024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Z, Zhang J, Zhang H, Dai Z, Liang X,
Li S, Peng R, Zhang X, Liu F, Liu Z, et al: CMTM family genes
affect prognosis and modulate immunocytes infiltration in grade
II/III glioma patients by influencing the tumor immune landscape
and activating associated immunosuppressing pathways. Front Cell
Dev Biol. 10:7408222022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Z and Zhang Y: Development of an
immune-related gene signature applying ridge method for improving
immunotherapy responses and clinical outcomes in lung
adenocarcinoma. PeerJ. 13:e191212025. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ghisai SA, Barin N, van Hijfte L, Verhagen
K, de Wit M, van den Bent MJ, Hoogstrate Y and French PJ:
Transcriptomic analysis of EGFR co-expression and activation in
glioblastoma reveals associations with its ligands. Neurooncol Adv.
7:vdae2292025.PubMed/NCBI
|
|
32
|
Han AX, Long BY, Li CY, Huang DD, Xiong
EQ, Li FJ, Wu GL, Liu Q, Yang GB and Hu HY: Machine learning
framework develops neutrophil extracellular traps model for
clinical outcome and immunotherapy response in lung adenocarcinoma.
Apoptosis. 29:1090–1108. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen T, Yang Y, Huang Z, Pan F, Xiao Z,
Gong K, Huang W, Xu L, Liu X and Fang C: Prognostic risk modeling
of endometrial cancer using programmed cell death-related genes: A
comprehensive machine learning approach. Discov Oncol. 16:2802025.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng Q, Lu H and Wu L: Identification of
M2-like macrophage-related signature for predicting the prognosis,
ecosystem and immunotherapy response in hepatocellular carcinoma.
PLoS One. 18:e02916452023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu L, Wu J, Tian J, Zhang B, Zhao Y, Zhao
Z, Luo Y and Li Y: Machine Learning unveils sphingolipid
Metabolism's role in tumour microenvironment and immunotherapy in
lung cancer. J Cell Mol Med. 29:e704352025. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meng WJ, Guo JM, Huang L, Zhang YY, Zhu
YT, Tang LS, Wang JL, Li HS and Liu JY: Anoikis-related long
non-coding RNA signatures to predict prognosis and immune
infiltration of gastric cancer. Bioengineering (Basel). 11:8932024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Therneau TM: Survival: Survival analysis.
The R Foundation for Statistical Computing. 2024.
|
|
38
|
Kassambara A: Survminer: Drawing survival
curves using ‘ggplot2’. The R Foundation for Statistical Computing.
2024.
|
|
39
|
Wickham H: ggplot2: Create elegant data
visualisations using the grammar of graphics. The R Foundation for
Statistical Computing. 2025.
|
|
40
|
Harrell FE Jr: rms: Regression modeling
strategies. The R Foundation for Statistical Computing. 2024.
|
|
41
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aran D, Hu Z and Butte AJ: xCell:
Digitally portraying the tissue cellular heterogeneity landscape.
Genome Biol. 18:2202017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mayakonda A: maftools: Summarize, analyze
and visualize MAF files. The R Foundation for Statistical
Computing. 2025.
|
|
46
|
Maeser D: oncoPredict: Drug response
modeling and biomarker discovery. The R Foundation for Statistical
Computing. 2024.
|
|
47
|
Kassambara A: ggpubr: ‘ggplot2’-Based
Publication Ready Plots. The R Foundation for Statistical
Computing. 2025.
|
|
48
|
Kolde R: pheatmap: Pretty Heatmaps. The R
Foundation for Statistical Computing. 2025.
|
|
49
|
Revelle W: Psych: Procedures for
psychological, psychometric, and personality research. The R
Foundation for Statistical Computing. 2025.
|
|
50
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pan J, Liu F, Xiao X, Xu R, Dai L, Zhu M,
Xu H, Xu Y, Zhao A, Zhou W, et al: METTL3 promotes colorectal
carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp
Clin Cancer Res. 41:192022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li J, Xie H, Ying Y, Chen H, Yan H, He L,
Xu M, Xu X, Liang Z, Liu B, et al: YTHDF2 mediates the mRNA
degradation of the tumor suppressors to induce AKT phosphorylation
in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer.
19:1522020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wen H, Qu C, Wang Z, Gao H, Liu W, Wang H,
Sun H, Gu J, Yang Z and Wang X: Cuproptosis enhances docetaxel
chemosensitivity by inhibiting autophagy via the DLAT/mTOR pathway
in prostate cancer. FASEB J. 37:e231452023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu S, Wu H, Cui H, Guo H, Ouyang Y, Ren
Z, Deng Y, Geng Y, Ouyang P, Wu A, et al: Induction of mitophagy
via ROS-dependent pathway protects copper-induced hypothalamic
nerve cell injury. Food Chem Toxicol. 181:1140972023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tsvetkov P, Detappe A, Cai K, Keys HR,
Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, et al:
Mitochondrial metabolism promotes adaptation to proteotoxic stress.
Nat Chem Biol. 15:681–689. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Su Y, Zhang X, Li S, Xie W and Guo J:
Emerging roles of the copper-CTR1 axis in tumorigenesis. Mol Cancer
Res. 20:1339–1353. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu Z, Zhang W and Kang YJ: Copper affects
the binding of HIF-1α to the critical motifs of its target genes.
Metallomics. 11:429–438. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zheng P, Zhou C, Lu L, Liu B and Ding Y:
Elesclomol: A copper ionophore targeting mitochondrial metabolism
for cancer therapy. J Exp Clin Cancer Res. 41:2712022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li Q, Wang T, Zhou Y and Shi J:
Cuproptosis in lung cancer: Mechanisms and therapeutic potential.
Mol Cell Biochem. 479:1487–1499. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li H, Fu Y, Xu Y, Miao H, Wang H, Zhang T,
Mei X, He Y, Zhang A and Ge X: Cuproptosis associated cytoskeletal
destruction contributes to podocyte injury in chronic kidney
disease. Am J Physiol Cell Physiol. 327:C254–C269. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao W, Ding C, Zhao M, Li Y, Huang H, Li
X, Cheng Q, Shi Z, Gao W, Liu H and Chen J: Identification and
validation of a hypoxia and glycolysis prognostic signatures in
lung adenocarcinoma. J Cancer. 15:1568–1582. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu Y, Lin W, Yang Y, Shao J, Zhao H, Wang
G and Shen A: Role of cuproptosis-related gene in lung
adenocarcinoma. Front Oncol. 12:10809852022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang J, Liu K, Yang L, Ji J, Qin J, Deng H
and Wang Z: Identification and validation of a novel
cuproptosis-related stemness signature to predict prognosis and
immune landscape in lung adenocarcinoma by integrating single-cell
and bulk RNA-sequencing. Front Immunol. 14:11747622023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
You J, Yu Q, Chen R, Li J, Zhao T and Lu
Z: A prognostic model for lung adenocarcinoma based on cuproptosis
and disulfidptosis related genes revealing the key prognostic role
of FURIN. Sci Rep. 15:60572025. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liang P, Chen J, Yao L, Hao Z and Chang Q:
A deep learning approach for prognostic evaluation of lung
adenocarcinoma based on cuproptosis-related genes. Biomedicines.
11:14792023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang W, Qu H, Ma X, Li L, Wei Y, Wang Y,
Zeng R, Nie Y, Zhang C, Yin K, et al: Identification of cuproptosis
and immune-related gene prognostic signature in lung
adenocarcinoma. Front Immunol. 14:11797422023. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang C, Zhang S, Liu J, Tian Y, Ma B, Xu
S, Fu Y and Luo Y: Secreted pyruvate kinase M2 promotes lung cancer
metastasis through activating the integrin Beta1/FAK signaling
pathway. Cell Rep. 30:1780–1797. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yin L, Shi J, Zhang J, Lin X, Jiang W, Zhu
Y, Song Y, Lu Y and Ma Y: PKM2 is a potential prognostic biomarker
and related to immune infiltration in lung cancer. Sci Rep.
13:222432023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu Y, Liang X, Zhang H, Dong J, Zhang Y,
Wang J, Li C, Xin X and Li Y: ER stress-related genes EIF2AK3,
HSPA5, and DDIT3 polymorphisms are associated with risk of lung
cancer. Front Genet. 13:9387872022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cheuk IW, Siu MT, Ho JC, Chen J, Shin VY
and Kwong A: ITGAV targeting as a therapeutic approach for
treatment of metastatic breast cancer. Am J Cancer Res. 10:211–223.
2020.PubMed/NCBI
|
|
72
|
Wu D, Jin J, Qiu Z, Liu D and Luo H:
Functional analysis of O-GlcNAcylation in cancer metastasis. Front
Oncol. 10:5852882020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Crawford A, Fassett RG, Geraghty DP, Kunde
DA, Ball MJ, Robertson IK and Coombes JS: Relationships between
single nucleotide polymorphisms of antioxidant enzymes and disease.
Gene. 501:89–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Isomoto K, Haratani K, Hayashi H, Shimizu
S, Tomida S, Niwa T, Yokoyama T, Fukuda Y, Chiba Y, Kato R, et al:
Impact of EGFR-TKI treatment on the tumor immune microenvironment
in EGFR mutation-positive non-small cell lung cancer. Clin Cancer
Res. 26:2037–2046. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen S, Ren Y and Duan P: Biomimetic
nanoparticle loading obatoclax mesylate for the treatment of
non-small-cell lung cancer (NSCLC) through suppressing bcl-2
signaling. Biomed Pharmacother. 129:1103712020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bhat V, Narayanan DL and Shukla A: Report
of rapid diagnosis and precise management of MMADHC-related
intracellular cobalamin defect. BMJ Case Rep. 14:e2397552021.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu D and Casey PJ: GPCR-Gα13 involvement
in mitochondrial function, oxidative stress, and prostate cancer.
Int J Mol Sci. 25:71622024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Mukherjee S, Forde R, Belton A and
Duttaroy A: SOD2, the principal scavenger of mitochondrial
superoxide, is dispensable for embryogenesis and imaginal tissue
development but essential for adult survival. Fly (Austin).
5:39–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hernandez-Saavedra D and McCord JM:
Association of a new intronic polymorphism of the SOD2 gene
(G1677T) with cancer. Cell Biochem Funct. 27:223–227. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhao C, Zhang Z, Wang Z and Liu X:
Circular RNA circRANGAP1/miR-512-5p/SOD2 axis regulates cell
proliferation and migration in non-small cell lung cancer (NSCLC).
Mol Biotechnol. 66:3608–3617. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li P, Sun Q, Bai S, Wang H and Zhao L:
Combination of the cuproptosis inducer disulfiram and anti-PD-L1
abolishes NSCLC resistance by ATP7B to regulate the HIF-1 signaling
pathway. Int J Mol Med. 53:192024. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang X, Liu Z and Lin C: Metal
ions-induced programmed cell death: How does oxidative stress
regulate cell death? Life Sci. 374:1236882025. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Guo Z, Chen D, Yao L, Sun Y, Li D, Le J,
Dian Y, Zeng F, Chen X and Deng G: The molecular mechanism and
therapeutic landscape of copper and cuproptosis in cancer. Signal
Transduct Target Ther. 10:1492025. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ning X, Chen X, Li R, Li Y, Lin Z and Yin
Y: Identification of a novel cuproptosis inducer that induces ER
stress and oxidative stress to trigger immunogenic cell death in
tumors. Free Radic Biol Med. 229:276–288. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun W, Lu H, Zhang P, Zeng L, Ye B, Xu Y,
Chen J, Xue P, Yu J, Chen K, et al: Localized propranolol delivery
from a copper-loaded hydrogel for enhancing infected burn wound
healing via adrenergic β-receptor blockade. Mater Today Bio.
30:1014172025. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ge EJ, Bush AI, Casini A, Cobine PA, Cross
JR, DeNicola GM, Dou QP, Franz KJ, Gohil VM, Gupta S, et al:
Connecting copper and cancer: From transition metal signalling to
metalloplasia. Nat Rev Cancer. 22:102–113. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Oliveri V: Selective targeting of cancer
cells by copper ionophores: An overview. Front Mol Biosci.
9:8418142022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Tsvetkov P, Coy S, Petrova B, Dreishpoon
M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R,
Spangler RD, et al: Copper induces cell death by targeting
lipoylated TCA cycle proteins. Science. 375:1254–1261. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Bagherpoor AJ, Shameem M, Luo X, Seelig D
and Kassie F: Inhibition of lung adenocarcinoma by combinations of
sulfasalazine (SAS) and disulfiram-copper (DSF-Cu) in cell line
models and mice. Carcinogenesis. 44:291–303. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liu X, Wang L, Cui W, Yuan X, Lin L, Cao
Q, Wang N, Li Y, Guo W, Zhang X, et al: Targeting ALDH1A1 by
disulfiram/copper complex inhibits non-small cell lung cancer
recurrence driven by ALDH-positive cancer stem cells. Oncotarget.
7:58516–58530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu D, Cao J, Ding X, Xu W, Yao X, Dai M,
Tai Q, Shi M, Fei K, Xu Y and Su B: Disulfiram/copper complex
improves the effectiveness of the WEE1 inhibitor Adavosertib in p53
deficient non-small cell lung cancer via ferroptosis. Biochim
Biophys Acta Mol Basis Dis. 1870:1674552024. View Article : Google Scholar : PubMed/NCBI
|