Introduction
Cervical neoplasia is the second most frequently
diagnosed malignancy in women worldwide and remains the foremost
cancer type in several low-income countries, imposing a notable
socio-economic burden (1). The
International Agency for Research on Cancer estimates that ~300,000
annual fatalities are attributed to cervical cancer (CC) in China
(2,3). Human papillomavirus (HPV)-18 is an
important strain responsible for initiating precancerous conditions
that frequently progress to cervical malignancies, accounting for
>90% of such cases globally. The occurrence of HPV-18 infections
among women is escalating worldwide; however, most of these
infections do not develop into cancer (4,5).
Consequently, it has become essential to explore supplementary
factors beyond mere infection that contribute to the onset of
carcinogenesis and CC progression.
YTH domain-containing protein 2 (YTHDC2) forms part
of the YTH protein unit and is instrumental in orchestrating an
array of mRNA metabolic functions, encompassing splicing, export,
dismantling and translation (6–8).
Members of the YTH protein group selectively interact with mRNAs
marked by N6-methyladenosine (m6A), a reversible and substantial
post-transcriptional alteration regulated by the activity of
methyltransferases and demethylases (9). In addition to its YTH domain, YTHDC2
has an RNA helicase domain, unique within this protein family that
is essential for efficiently translating specific mRNAs mediated by
YTHDC2 (10).
During tumorigenesis, YTHDC2 is pivotal for
synthesizing proteins that result in malignant traits. Moreover, it
suppresses colorectal cancer progression via its regulatory effects
on hypoxia-inducible factor 1α (HIF-1α) synthesis (11). YTHDC2 expression is also increased
in radiation-sensitive nasopharyngeal carcinoma, promoting
radioresistance by activating the insulin-like growth factor
1/AKT/S6 signaling cascade (12).
Furthermore, YTHDC2 expression is markedly increased in cancers
such as prostate cancer and glioblastoma (13–16),
but is diminished in pulmonary malignancies, as well as head and
neck squamous cell carcinomas (17,18).
However, the precise biological roles of YTHDC2 in CC remain poorly
defined, particularly in HPV-positive cases.
The present study aimed to assess the functional
effects of YTHDC2 on cervical carcinogenesis. The findings aimed to
demonstrate the association between YTHDC2 expression and HPV
prevalence across cervical neoplastic tissues and in vitro
cell models. The effect of YTHDC2 expression on cellular
proliferation and facilitated ferroptosis in CC was also
studied.
Materials and methods
Human tissue samples
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Henan University of
Science and Technology (Luoyang, China; approval no. 2023-0147).
Cervical tissue specimens were collected between July 2021 and
September 2023 from 25 women (age, 35.5–60.1 years) histologically
diagnosed with stage IA CC featuring lymphovascular space
involvement, or classified as IA2, as detailed in our previous
research (19,20). The selection criteria for patients
with cervical cancer were as follows: i) Pathologically confirmed
patients with cervical cancer; and (2) the patients had no history of other
cancers. No patients had preoperative chemotherapy, radiotherapy,
or other treatment history or other inflammatory diseases. Patient
conditions were staged according to the criteria of the
International Federation of Gynecology and Obstetrics (21). Additionally, samples were collected
from women with high-risk (HR)-HPV-negative typical CC (n=25) and
HR-HPV-positive cervical malignancies (n=25) who had undergone
HR-HPV screening and ThinPrep® cytology (Hologic, Inc.)
at the First Affiliated Hospital of Henan University of Science and
Technology. Written informed consent was obtained from every
participant involved.
Immunohistochemistry (IHC)
IHC was performed on cervical cancer tissue sections
prepared as formalin-fixed paraffin-embedded samples. The tissues
were fixed in 10% neutral-buffered formalin at room temperature for
24 h, embedded in paraffin wax, and sectioned at a 4-µm thickness.
Fresh frozen tissues were snap-frozen in liquid nitrogen-cooled
isopentane at approximately −150°C and sectioned at 7 µm. The
sections were permeabilized with 0.2% Triton X-100 (Sigma-Aldrich;
Merck KGaA) for 10 min at room temperature, followed by blocking
with 5% normal goat serum (cat. no. 31873; Thermo Fisher
Scientific, Inc.) in PBS for 1 h at room temperature. Primary
antibody incubation was performed using anti-YTHDC2 (cat. no.
ab220160; Abcam) at a 1:100 dilution overnight at 4°C. After
washing, sections were incubated with HRP-conjugated goat
anti-rabbit IgG secondary antibody (cat. no. 7074; Cell Signaling
Technology) at a 1:500 dilution for 1 h at room temperature. Signal
detection was performed using 3,3′-diaminobenzidine chromogen (cat.
no. K3468; Dako; Agilent Technologies, Inc.). Slides were
counterstained with hematoxylin and examined using a bright-field
microscope (Olympus BX53; Olympus Corporation).
Cell culture
Cell lines (purchased from ATCC), including HFF-1
(HPV-negative human epithelial foreskin fibroblasts), C33A and
DoTc2 4510 (both from HPV-negative cervical carcinomas), SiHa and
CaSKi (from HPV16-positive cervical squamous carcinomas), SW756
(from an HPV18-positive squamous carcinoma), HeLa (from an
HPV18-positive cervical epithelial adenocarcinoma) and H8 (from
HPV-positive, immortalized cervical cells), were maintained in
RPMI-1640 medium (cat. no. 11875093; Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (cat. no. A5256701;
HyClone; Cytiva). All cell lines were incubated at 37°C in a 5%
CO2 atmosphere with humidity.
Transfection procedures
SiHa and CaSKi cells (5×106/well) were
transfected with 1.5 µg YTHDC2-overexpression vectors (pCMV-YTHDC2)
and/or 1.5 µg control vectors (pCMV-empty), and with 2 µg
YTHDC2-specific small interfering (si)RNAs (#1,
5′-ATATAAGAGATGTGACGAGGG-3′; #2, 5′-CTTTAGTCGAAGTTCTGACTA-3′; and
#3, 5′-GGAAGCTAAATCGAGCCTT-3′), and control siRNAs
(5′-CACAGGGUAAGGAACUCGUCUCUCA-3′) using Lipofectamine™ 2000 (cat.
no. 11668027; Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature for 36 h according to the manufacturer's protocols. The
vector and siRNA were constructed and purchased from Genscript
Biotech Corporation. The effect of overexpression or knockdown was
evaluated using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR), as described below, at 48 h post-transfection
Subsequently, siRNA #1 was randomly selected for further
experiments as the knockdown efficacies of all three siRNAs were
similar (Fig. S1). SiHa and CaSKi
cells were co-transfected with pCMV1-YTHDC2 along with either
pcDNA3.1-empty or pcDNA3.1-SLC7A11 for 48 h.
Additionally, SiHa and CaSKi cells were transfected
with 2 µg SLC7A11-overexpression vector pcDNA3-SLC7A11 or 2 µg
SLC7A11-specific siRNA (5′-CTGGAGTTATGCAGCTAAT-3′), or
co-transfected with pCMV1-YTHDC2 along with either pcDNA3.1-empty
or pcDNA3.1-SLC7A11, using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) at room temperature for 36 h according to the
manufacturer's instructions, to upregulate or downregulate SLC7A11
expression, respectively. Concentrations of 10 nM for vectors and
20 nM for siRNAs were used for the transfections. Subsequent
experiments were performed 48 h after transfection.
Cell viability assay
Cell viability was assessed using Cell Counting
Kit-8 (CCK-8) reagent (cat. no. CK04; Dojindo Laboratories, Inc.)
at 48 h following transfection, according to the manufacturer's
guidelines. A total of 10 µl CCK-8 solution was added to each
designated well (5×106/well). Subsequently, the plate
was incubated for 2 h and absorbance readings were then taken at
450 nm using the Tecan Infinite M200 microplate (Tecan Group,
Inc.).
Colony formation assay
SiHa and CaSKi cells were cultured in a series of
12-well plates, with each well containing 3,000 cells/ml. These
cells were cultured for 7–9 days at 37°C, after which they were
fixed using 10% neutral-buffered formalin for ≥4 h at room
temperature, stained using crystal violet for 30 min at room
temperature (cat. no. C0121; Beyotime Institute of Biotechnology)
and visually assessed using an advanced optical system, and counted
manually (Olympus CX23; Olympus Corporation). A colony was
typically defined as: i) A group containing at least 50 cells; ii)
a group clearly separated from neighboring colonies; and iii) a
group of cells that must be stained.
Flow cytometry
Following transfection, the cell specimens were
incubated in 6-well plates for 48 h and then trypsinized and
preserved in 75% ethanol at −20°C overnight. Following
stabilization, these specimens were treated with 0.5 µg/ml RNase A
(Thermo Fisher Scientific, Inc.) and 100 µg/ml propidium iodide
(PI) for 30 min. Subsequently, apoptotic indices were evaluated
using a dual staining Annexin V-FITC/PI apoptosis kit (Abcam) with
analytical procedures performed using a BD FACScan flow cytometer
(BD Biosciences) equipped with BD CellQuest™ software (version 5.1;
BD Biosciences). Specimens unreactive to FITC/PI were utilized as
negative controls in these assays.
Reactive oxygen species (ROS)
measurements
A total of 5×104 SiHa and CaSKi cells
were added to the wells of a 96-well white plate optimized for
luminescence studies. Following cell adhesion, cells were cultured
for 24 h. After thoroughly rinsing with PBS (cat. no. 10010023;
Thermo Fisher Scientific, Inc.), a solution of 20 µM
carboxy-H2-DCFDA (cat. no. C400; Invitrogen; Thermo Fisher
Scientific, Inc.) was added to the cellular milieu and incubated at
37°C for 1 h to facilitate ROS-associated fluorescence development.
Fluorescence detection was performed using the 1420 Multi-label
Counter (PerkinElmer, Inc.). Furthermore, cells were resuspended in
1 ml PBS and incubated for 1 h under identical temperature
conditions after applying a second dose of 20 µM carboxy-H2-DCFDA.
The cells were rinsed with PBS to remove extraneous dye, and the
fluorescence intensity was quantified using a microplate reader at
485 and 535 nm excitation and emission wavelengths, respectively,
to determine the ROS levels within the samples accurately.
RT-qPCR
RNA isolation from SiHa and CaSKi cell or tissue
samples (10 mg) was performed using TRIzol™ Reagent (cat. no.
15596018CN; Invitrogen; Thermo Fisher Scientific, Inc.), with the
quantitative assessment of yields performed using a NanoDrop™ 2000
Spectrophotometer (Thermo Fisher Scientific, Inc.) at an optical
density of 260 nm. Subsequent reverse transcription to generate
cDNA was performed using the SuperScript™ IV First-Strand Synthesis
System (cat. no. 18091050; Invitrogen; Thermo Fisher Scientific,
Inc.) with oligo(dT) 20 primers. The thermal conditions for reverse
transcription PCR were as follows: 37°C for 2 min, and 55°C for 5
min. For the quantitative analysis, the prepared cDNA was subjected
to amplification using SYBR™ Select Master Mix (cat. no. 4472918;
Invitrogen; Thermo Fisher Scientific, Inc.), adhering to the
protocols specified by the supplier. The internal standard GAPDH
mRNA was used to calibrate the reaction, which was initiated with a
10 min denaturation step at 95°C, followed by 40 cycles of 15 sec
denaturation at the same temperature, and a 40 sec extension phase
at 60°C. The abundance of the target mRNA was quantitatively
evaluated using the comparative 2−ΔΔCq method (22). Each experimental sequence was
performed in triplicate to ensure the reproducibility and
reliability of the results. The sequences for primers used in this
experiment were as follows: YTHDC2-forward (F),
5′-CCAGGCCGAGCAGCGTCTCC-3′; YTHDC2-reverse (R),
5′-ACAGTTAATCAGTATGGGAGCC-3′; GAPDH-F, 5′-AACAGCGACACCCACTCCTC-3′;
GAPDH-R, 5′-CATACCAGGAAATGAGCTTGACAA-3′; SLC7A11-F,
5′-TCCTGCTTTGGCTCCATGAACG-3′; and SLC7A11-R,
5′-AGAGGAGTGTGCTTGCGGACAT-3′. The thermocycling conditions for qPCR
were as follows: 95°C for 2 min, 95°C for 10 sec, 55°C for 30 sec
and 72°C for 30 sec, for 40 cycles.
Western blotting (WB)
SiHa and CaSKi cells were disrupted in a lysis
buffer comprising HEPES (0.02 M), NaCl (0.15 M), EDTA (1 mM, pH
7.4), glycerol (10%), Triton X-100 (1%), a comprehensive protease
inhibitor mixture and Na3VO4 (5 mM). The
resulting lysate was incubated at 4°C for 1 h, followed by
centrifugation at 12,000 × g at 4°C for 15 min. After separation
via 5–12% sodium dodecyl sulphate-polyacrylamide gel
electrophoresis, proteins (10 µg/lane; determined by BCA method)
were transferred onto polyvinylidene difluoride membranes. The
membranes were then blocked for 2 h at 4°C in PBS supplemented with
5% skim milk powder (cat. no. LP0033B; Invitrogen; Thermo Fisher
Scientific, Inc.) and 0.5% Tween 20. Subsequently, proteins were
probed with appropriate primary antibodies, including anti-YTHDC2
(1:1,000; cat. no. ab220160; Abcam), anti-actin (1:5,000; cat. no.
ab8227; Abcam), anti-SLC7A11 (1:2,000; cat. no. ab37185; Abcam),
anti-p53 (1:2,000; cat. no. ab32389; Abcam), anti-ACSL4 (1:1,000;
cat. no. ab155282; Abcam) and anti-GPX4 (1:1,000; cat. no. ab41787;
Abcam) antibodies for 1 h at room temperature, and detected using
horseradish peroxidase-conjugated secondary antibodies derived from
rabbits or mice, including goat anti-mouse HRP antibody (1:10,000;
cat. no. ab6708; Abcam) and goat anti-rabbit antibody (1:10,000;
cat. no. ab6721; Abcam) for another 1 h at room temperature.
Finally, protein expression was quantified using Image-Pro Plus
software (version 6.0; Media Cybernetics, Inc.).
Dual-luciferase reporter assay
Fragments of SLC7A11 wild-type (−WT) or SLC7A11
mutant (−Mut) (where m6A was substituted with C) were inserted into
the pRP (Exp)-Puro-EF1A-Luciferase (mSLC7A11_3′UTR_1862-4861bp:
SNP) (VectorBuilder) plasmid to create pcDNA3-SLC7A11-WT and
pcDNA3-SLC7A11-Mut plasmids as aforementioned. At 72 h
post-transfection using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions,
luciferase activity was measured using the Dual-Luciferase Reporter
Assay System (Promega Corporation). Relative firefly luciferase
(Fluc)/Renilla luciferase (Rluc) activity was calculated by
normalizing the activity of firefly luciferase to that of
Renilla luciferase. Each experiment was conducted in
triplicate for each group.
Statistical analysis
Data were statistically analyzed using SPSS version
18.0 software (IBM Corp.). Data are expressed as the mean ±
standard deviation. Comparisons between two groups were assessed
using an unpaired t-test, whereas comparisons between multiple
groups were analyzed using one-way analysis of variance with
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
YTHDC2 levels are increased in
HPV-positive CC specimens and cell lines
CC specimens were first classified as HPV- positive
or -negative. YTHDC2 mRNA levels in HPV-positive CC specimens were
significantly lower than those in their HPV-negative counterparts,
according to RT-qPCR, WB and immunohistochemistry results (Fig. 1A-C). To further evaluate these
observations based on clinical specimens, YTHDC2 expression in
HPV-negative (HFF-1, C33A and DoTc2 4510) and HPV-positive CC cell
lines (SiHa, CaSKi, SW756, HeLa and H8) were assessed. At the
cellular level, the significant downregulation of YTHDC2 mRNA and
protein expression was detected in HPV-positive CC cells compared
with levels in HPV-negative CC cells (Fig. 1D and E). These data indicate that
HPV-positive CC specimens and cell lines consistently have lower
YTHDC2 levels.
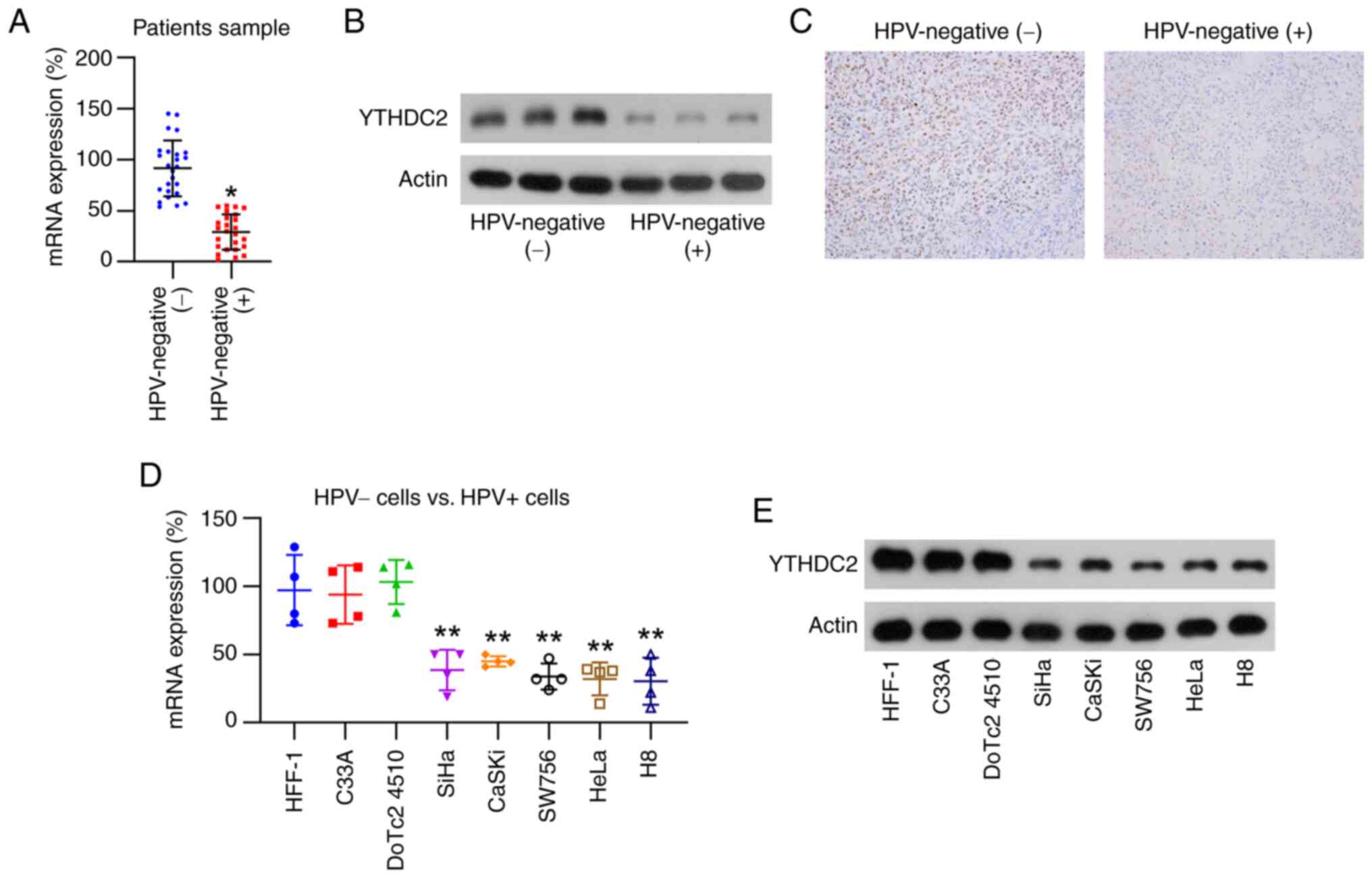 | Figure 1.Analysis of YTHDC2 expression in
clinical and cell samples of HPV-positive cervical cancer. (A)
RT-qPCR results demonstrating YTHDC2 expression in tissue samples
from HPV-positive (n=25) and HPV-negative (n=25) cervical cancer.
(B) Western blot analysis of the expression of YTHDC2 in tissue
samples from HPV-positive (n=3) and HPV-negative (n=3) cervical
cancer from the aforementioned 25 HPV-positive tissues and 25
HPV-negative CC samples. (C) Immunohistochemistry assessment of the
expression of YTHDC2 in the tissue samples from HPV-positive and
HPV-negative cervical cancer. Magnification, ×40. Analysis of
YTHDC2 mRNA and protein levels in cervical cancer cell lines
(CaSKi, HFF-1, SiHa, SW756, HeLa, DoTc2 4510, C33A and H8) using
(D) RT-qPCR and (E) western blotting, respectively. *P<0.05;
**P<0.01. YTHDC2, YTH N6-methyladenosine RNA-binding protein C2;
HPV, human papillomavirus; RT-qPCR, reverse
transcription-quantitative PCR. |
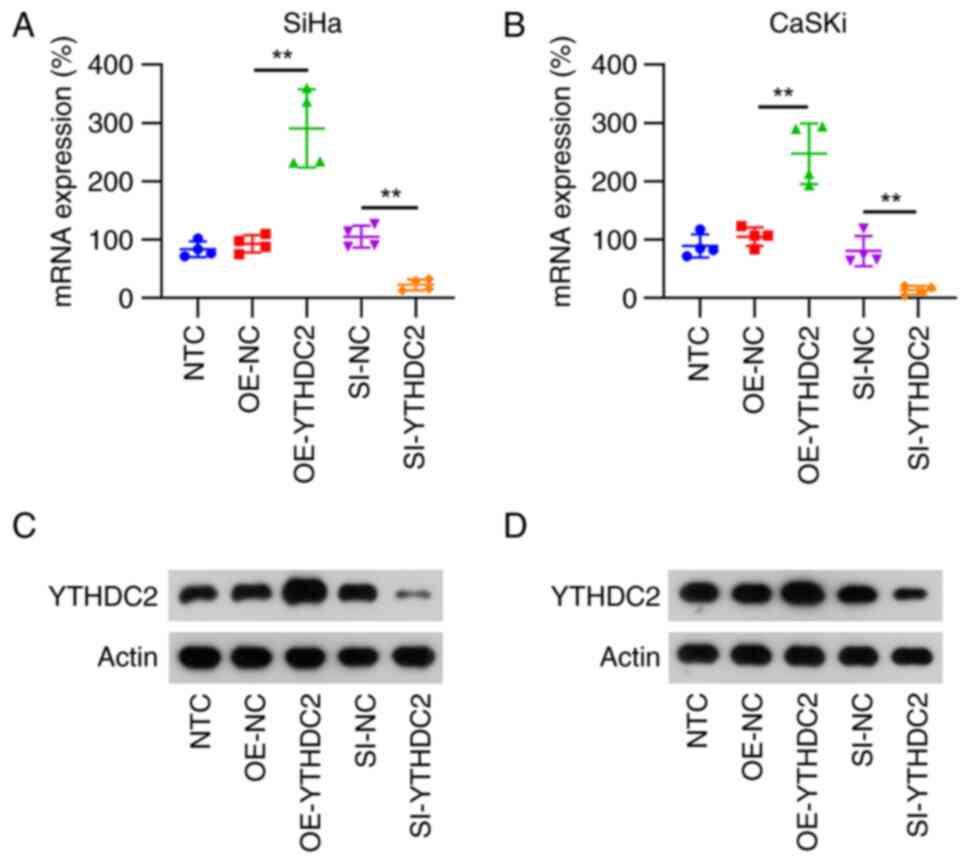
YTHDC2 silencing and overexpression in
HPV-positive CC cells (SiHa and CaSKi)
YTHDC2 expression was modulated in SiHa and CaSKi
cells through overexpression and silencing techniques to further
evaluate the function of YTHDC2 in HPV-positive CC cells. After
transfection with the overexpression vector, both RT-qPCR and WB
analyses revealed a significant increase in YTHDC2 mRNA and protein
levels compared with those in the control group. Conversely,
silencing YTHDC2 expression via siRNA transfection significantly
diminished its mRNA and protein levels in both SiHa and CaSKi
cells, compared with those in the control group (Fig. 2).
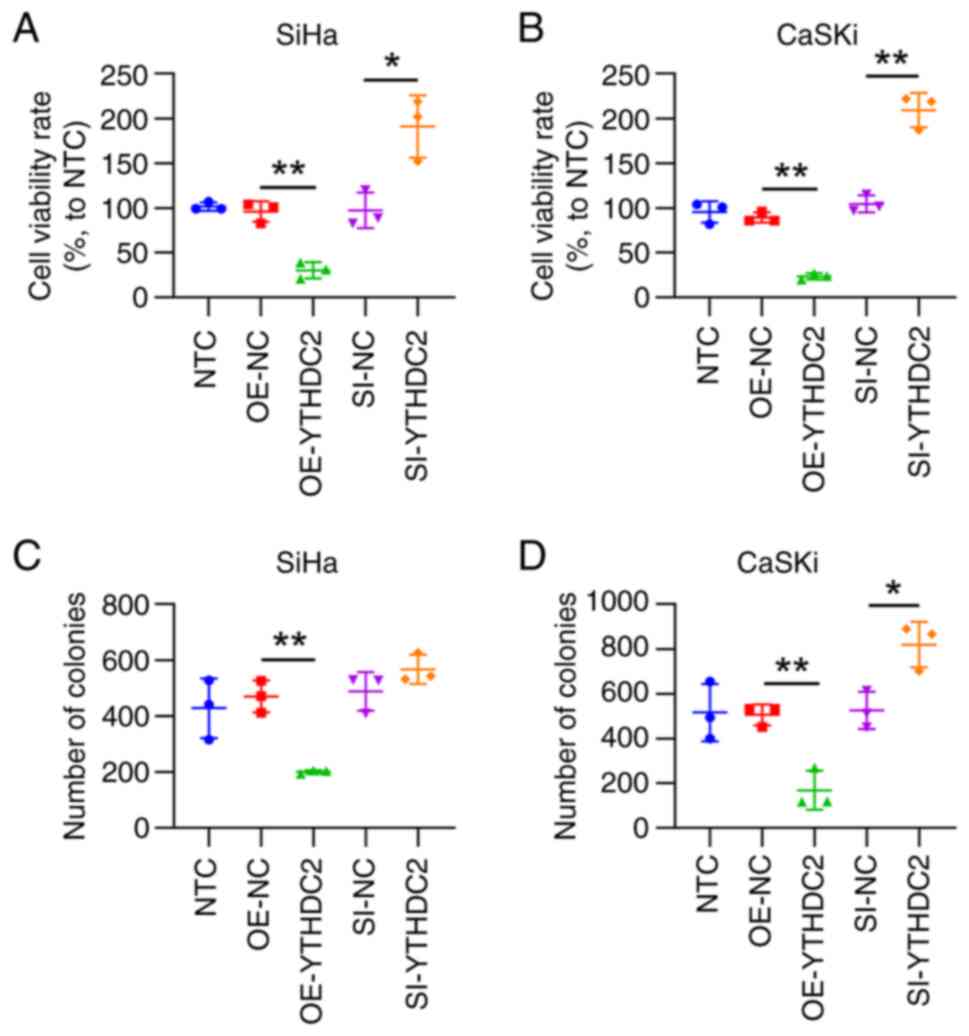
Upregulated YTHDC2 expression impedes
cellular proliferation and induces ferroptosis in SiHa and CaSKi
cells
YTHDC2 overexpression led to a significant decrease
in cell viability in both cell lines at 48 h post-treatment,
compared with that in the control group. Meanwhile, YTHDC2
silencing significantly increased cell viability, compared with
that in the control group, according to CCK-8 assays (Fig. 3A and B). The results of the colony
formation assays indicated that the cell proliferation rate was
significantly decreased after YTHDC2 overexpression and
significantly elevated after YTHDC2 knockdown, compared with that
in the controls groups (Fig. 3C and
D). These data suggest that YTHDC2 overexpression attenuates
HPV-positive cell proliferation.
Moreover, flow cytometry was used to assess the role
of YTHDC2 in cell apoptosis, especially ferroptosis in SiHa and
CaSKi cells. The numbers of apoptotic cells for both cell lines
overexpressing YTHDC2 were significantly elevated, whereas cells
with YTHDC2 knockdown exhibited a significantly decreased cell
apoptosis rate, compared with that of controls (Fig. 4A and B). Ferroptosis is a newly
identified type of apoptosis, and excessive ROS production is a
predominant marker of this process (23). Therefore, the present study assessed
the ROS levels in cell lines after overexpressing or silencing
YTHDC2. Compared with that of controls, increased ROS levels were
observed in cells overexpressing YTHDC2, whereas they were
significantly reduced in cells with silenced YTHDC2 expression
(Fig. 4C and D). Furthermore, the
protein expression of the ferroptosis markers SLC7A11, p53,
acyl-CoA synthetase long chain family member 4 (ACSL4) and
glutathione peroxidase 4 (GPX4) in cells with varying YTHDC2
expression levels were assessed. WB analyses demonstrated that GPX4
and ACSL4 levels were negatively associated with the expression of
YTHDC2 in SiHa and CaSKi cells, whereas SLC7A11 and p53 were
positively associated with YTHDC2 levels in these cells (Fig. 4E and F). These findings indicate
that YTHDC2 promotes ferroptosis in SiHa and CaSKi cells.
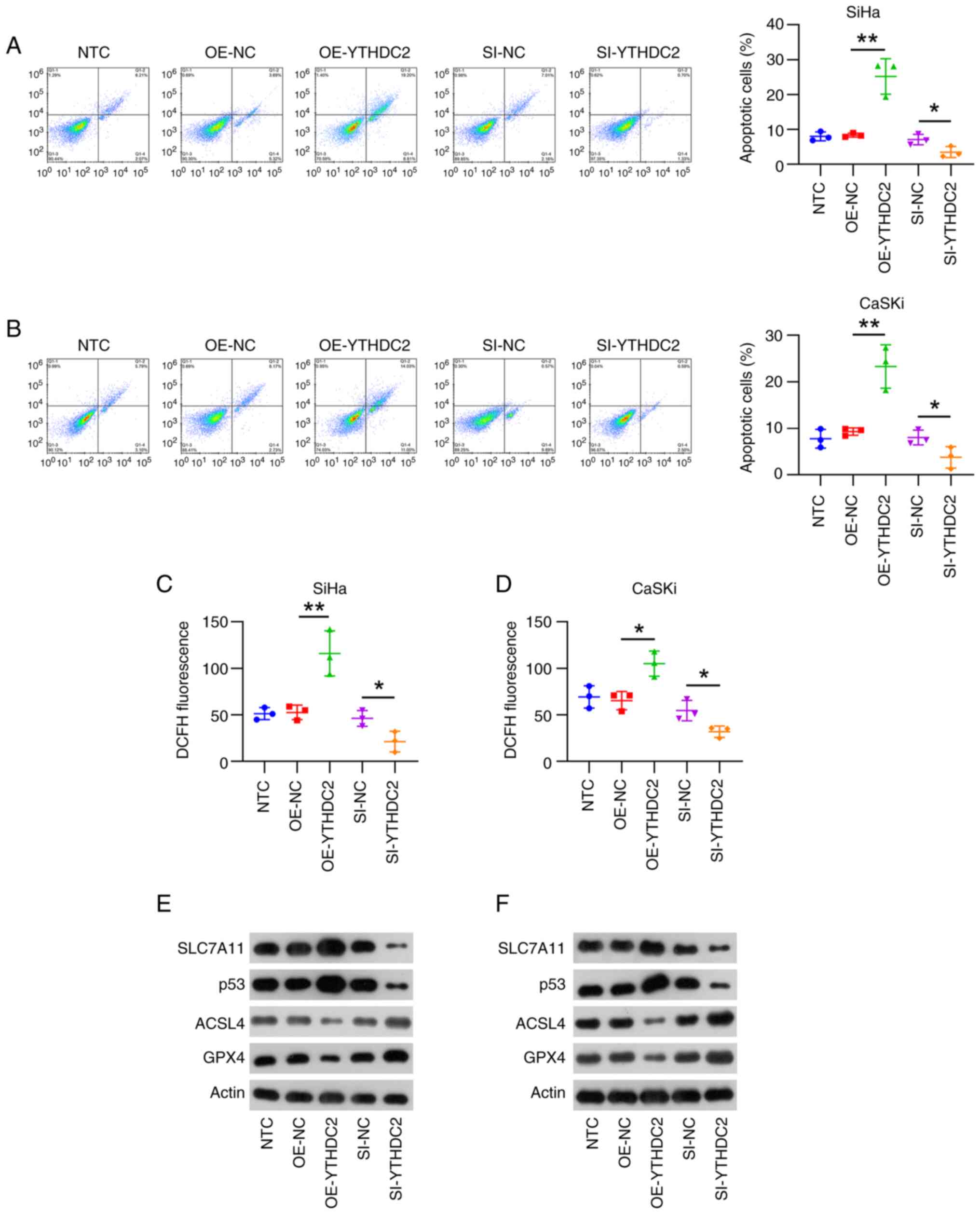 | Figure 4.Effect of YTHDC2 on ferroptosis in
SiHa and CaSKi cells. SiHa and CaSKi cells were either untreated or
transfected with pCMV1-empty, pCMV1-YTHDC2, control siRNA or YTHDC2
siRNA for 48 h. Flow cytometry was performed to assess the
apoptosis rates in (A) SiHA and (B) CaSKi cells. ROS levels were
assessed after a 10 min exposure to the ROS-sensitive fluorescent
dye H2DCF-DA (5 µM) in (C) SiHa and (D) CaSKi cells. Western
blotting was performed to assess the levels of proteins involved in
ferroptosis, including p53, ACSL4, GPX4 and SLC7A11, in (E) SiHa
and (F) CaSKi cells. *P<0.05; **P<0.01. YTHDC2, YTH
N6-methyladenosine RNA-binding protein C2; si, small interfering;
OE, overexpression; NC, negative control; ROS, reactive oxygen
species; ACSL4, acyl-CoA synthetase long chain family member 4;
GPX4, glutathione peroxidase 4; SLC7A11, solute carrier family 7
member 11; NTC, non-transfected control. |
SLC7A11 expression is regulated by
YTHDC2 through m6A methylation
Based on previous publications (24,25),
we hypothesized that SLC7A11 could be modulated by YTHDC2 (an RNA
m6A reader) in an m6A-dependent manner. Therefore, the present
study aimed to assess this hypothesis. The results revealed that,
compared with controls, SLC7A11 mRNA expression was significantly
downregulated after YTHDC2 overexpression and significantly
upregulated after YTHDC2 silencing in both cell lines, according to
RT-qPCR analysis (Fig. 5A and B).
Subsequently, luciferase reporter assays were performed to assess
SLC7A11 mRNA 5′-UTR methylation and gene expression. SiHa and CaSKi
cells demonstrating either upregulated or downregulated YTHDC2
expression were transfected with reporter plasmids that included
the full-length SLC7A11 5′-UTR immediately upstream of the
luciferase gene and incubated for 36 h. Compared with in controls,
luciferase activity was significantly diminished in cells with
elevated YTHDC2 levels, whereas cells with suppressed YTHDC2
expression exhibited a significant increase in luciferase activity
(Fig. 5C and D). Subsequently, the
G residue within the SLC7A11 5′-UTR consensus sequence was mutated
(5′-GGCUGC-3′ to 5′-AACUAC- 3′ in mRNA), and wild-type and mutant
reporter-driven luciferase activities were compared using
luciferase assays. The A residue mutation reduced the luciferase
activity by ~70% in SiHa and CaSKi cells (Fig. 5E and F). Collectively, these data
suggest that YTHDC2 contributes to SLC7A11 mRNA 5′-UTR m6A
methylation, thereby inhibiting SLC7A11 expression and protein
translation.
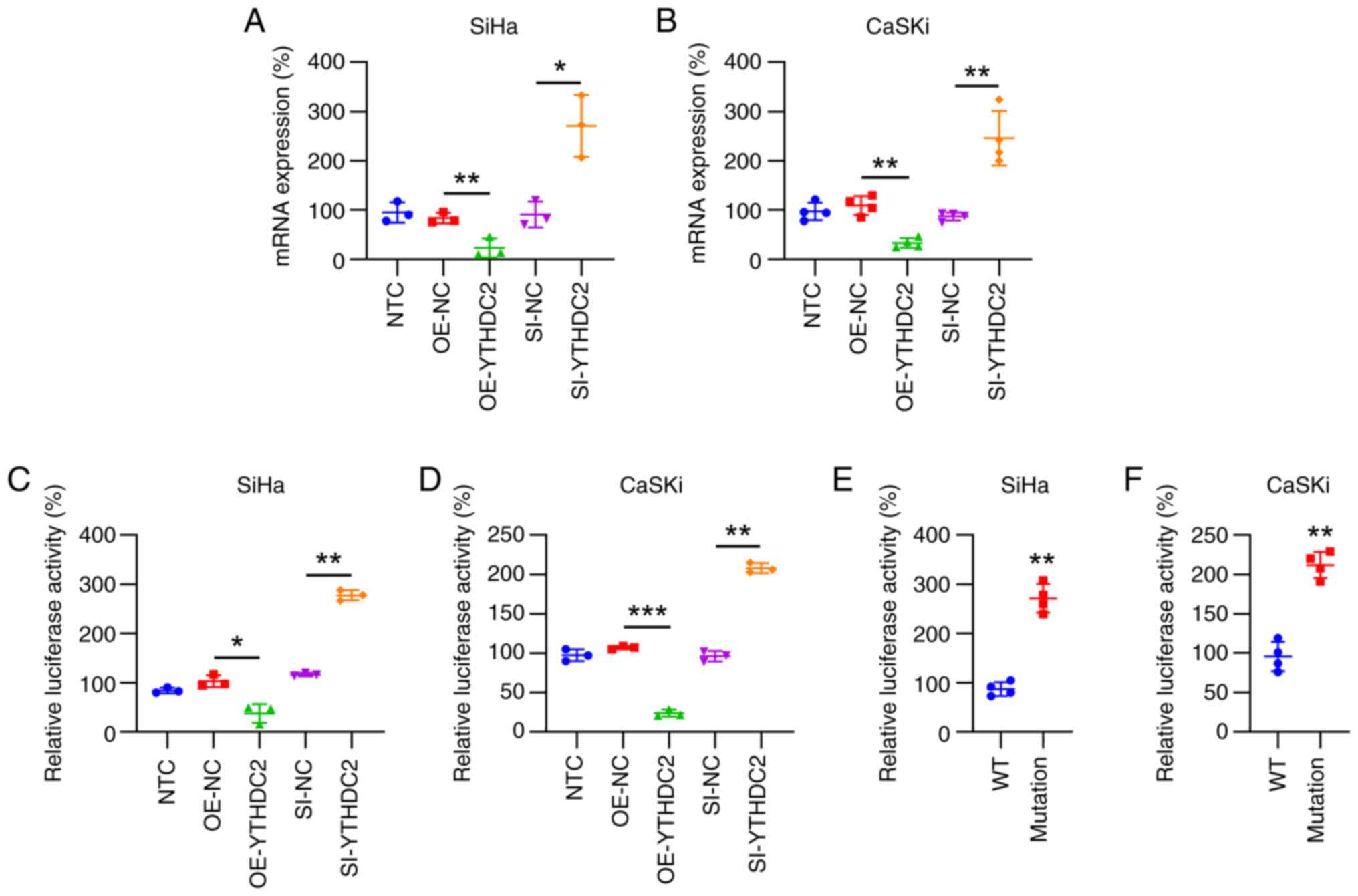 | Figure 5.Role of YTHDC2 in N6-methyladenosine
methylation and suppression of SLC7A11 mRNA expression. CaSKi cells
were either untreated or transfected with pCMV1-empty,
pCMV1-YTHDC2, control siRNA or YTHDC2 siRNA for 48 h. Reverse
transcription-quantitative PCR of (A) SiHA and (B) CaSKi cells
cells revealed the changes in SLC7A11 expression levels. Luciferase
assays were performed on (C) SiHa and (D) CaSKi cells transfected
with either YTHDC2 overexpression or silencing vectors alongside a
reporter plasmid, including the SLC7A11 5′-UTR associated with
luciferase sequences. Luciferase activity ratios corresponding with
the SLC7A11 5′-UTR were normalized to controls. Additional
luciferase assays involved (E) SiHa and (F) CaSKi cells transfected
with either WT or mutant SLC7A11 5′-UTR luciferase reporters.
*P<0.05; **P<0.01; ***P<0.001. YTHDC2, YTH
N6-methyladenosine RNA-binding protein C2; SLC7A11, solute carrier
family 7 member 11; UTR, untranslated region; si, small
interfering; OE, overexpression; NC, negative control; WT,
wild-type; NTC, non-transfected control. |
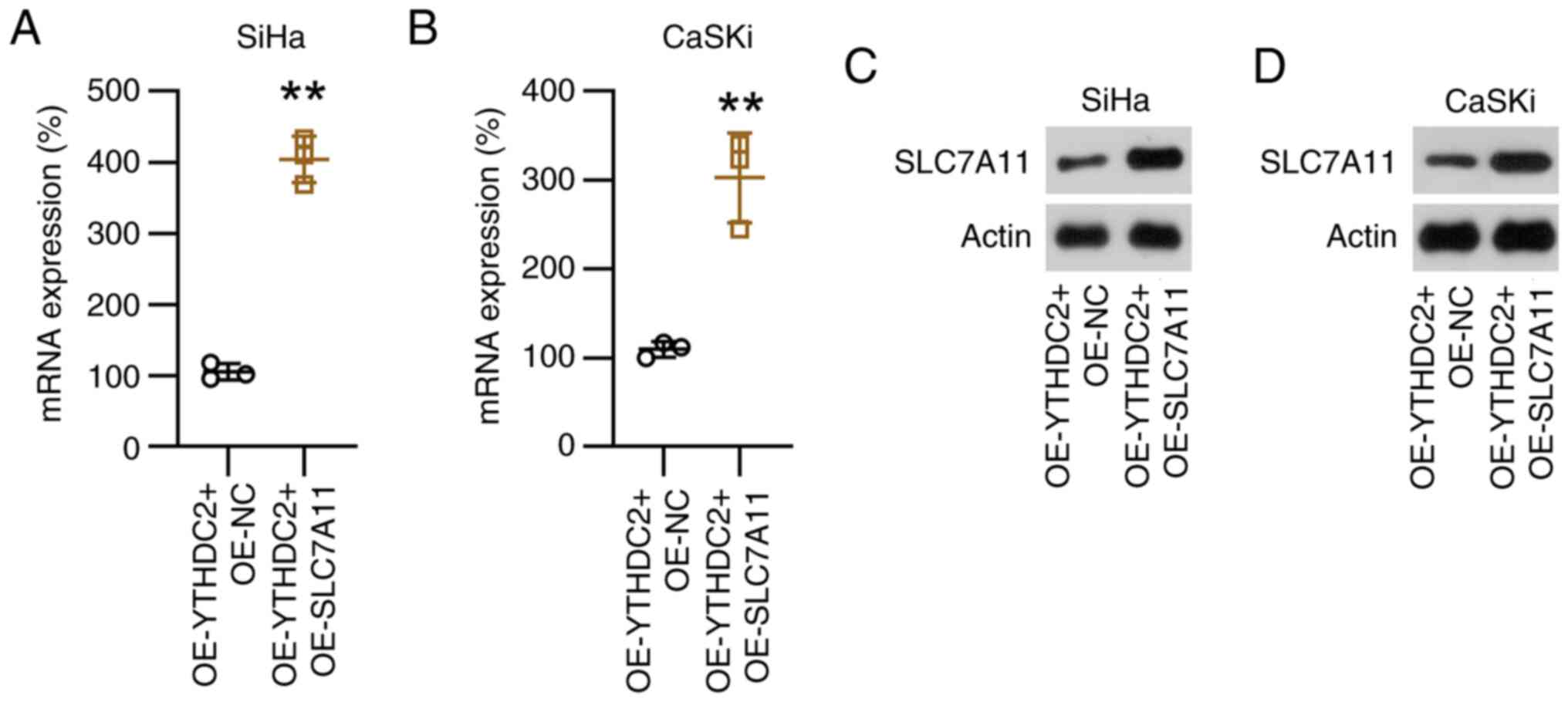
SLC7A11 overexpression counteracts
YTHDC2-mediated SiHa and CaSKi cell proliferation and
ferroptosis
Subsequently, the present study evaluated the
function of SLC7A11 in YTHDC2-regulated cell proliferation and
ferroptosis in SiHa and CaSKi cells by overexpressing it in these
cell lines, both with and without YTHDC2 overexpression. Compared
with the controls, SLC7A11 mRNA and protein levels were
significantly upregulated in SiHa and CaSKi cells, independent of
YTHDC2 overexpression (Fig. 6).
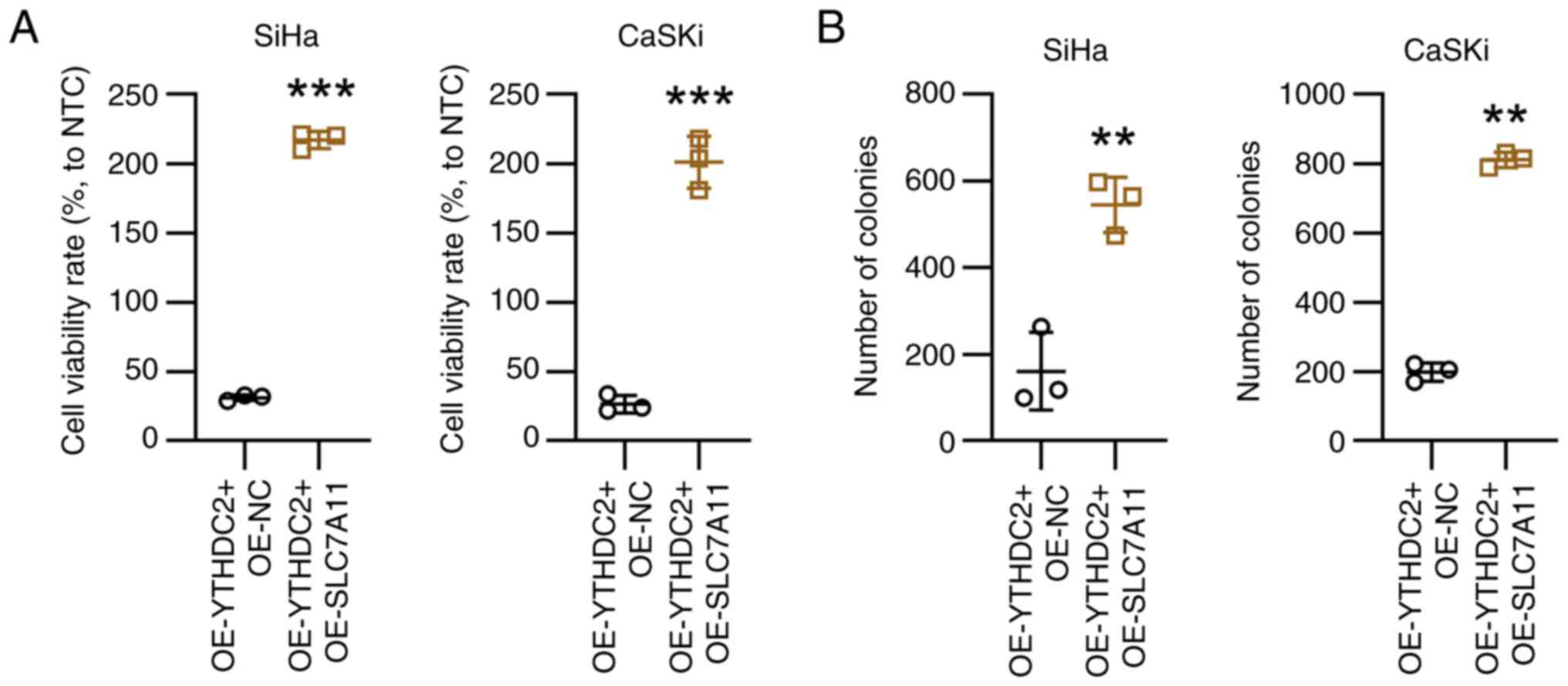
Subsequently, proliferation and ferroptosis in SiHa and CaSKi cells
was assessed following YTHDC2 overexpression and/or SLC7A11
overexpression. Compared with the controls, increased SLC7A11
expression was associated with a significant increase in cell
viability and colony formation in SiHa and CaSKi cells (Fig. 7).
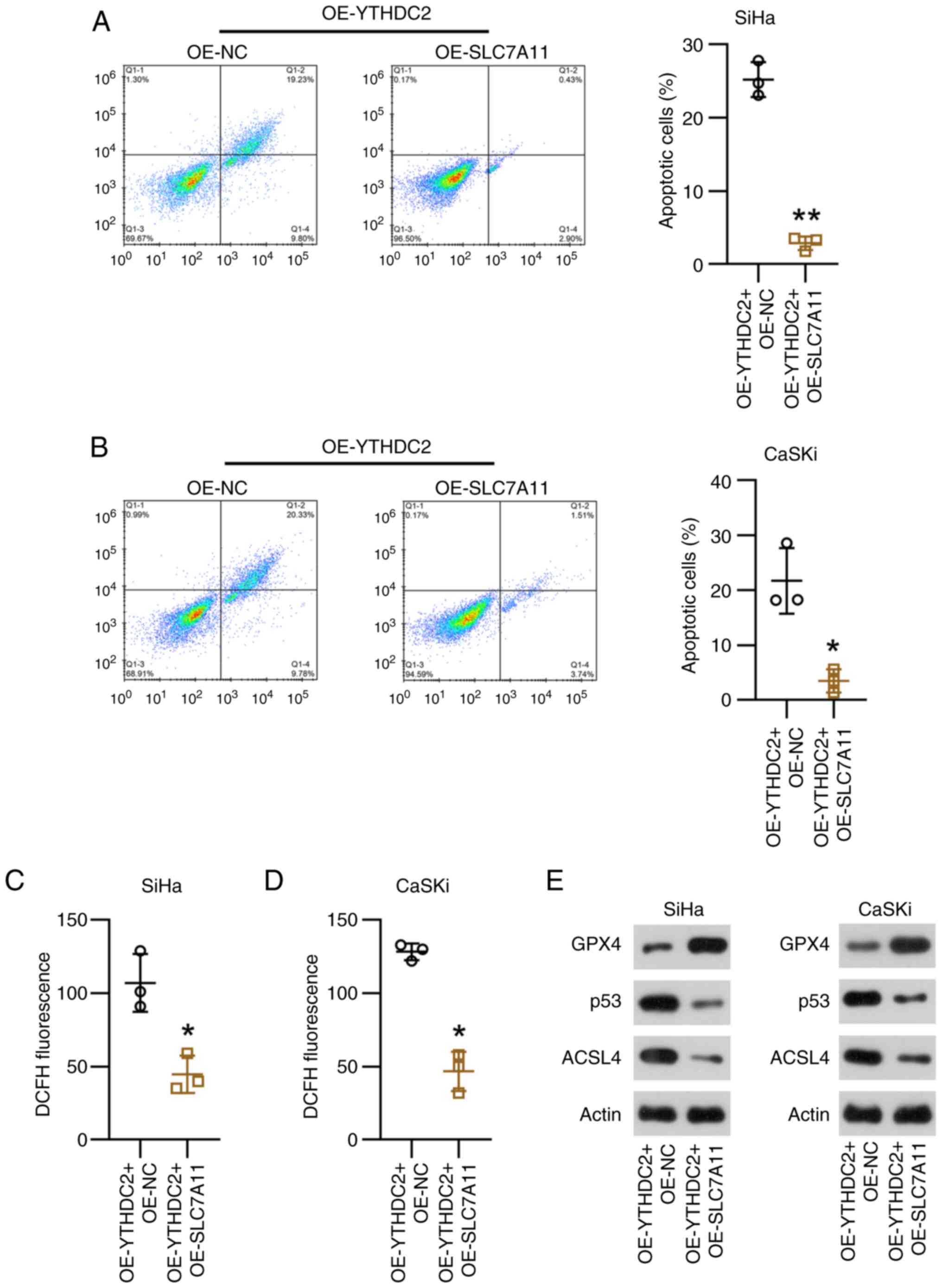
Finally, regarding ferroptosis, SLC7A11
overexpression was demonstrated to significantly reduce apoptosis
in SiHa and CaSKi cells with silenced YTHDC2 expression, compared
with controls (Fig. 8A and B).
Moreover, the generation of ROS in cells overexpressing YTHDC2 with
elevated SLC7A11 levels was significantly reduced compared with the
control group (Fig. 8C and D).
Meanwhile, SLC7A11 and p53 markedly promoted GPX expression, and
ACSL levels were notably reduced by SLC7A11, according to WB
analysis (Fig. 8E). These results
suggest that SLC7A11 overexpression is associated with
YTHDC2-mediated proliferation and ferroptosis in SiHa and CaSKi
cells.
Discussion
CC associated with HPV infection is a malignancy
that is largely preventable through immunization strategies, with
vaccines currently available and subject to ongoing refinement
(26). Findings from the present
study indicate the suppression of YTHDC2 expression in CC samples
and cell cultures positive for HPV. The enhanced expression of
YTHDC2 in HPV-positive CC cell lines (SiHa and CaSKi) curtailed
cellular proliferation and increased ferroptosis in these cells,
whereas YTHDC2 knockdown reversed these effects on proliferation
and ferroptosis. Furthermore, SLC7A11 (a notable ferroptosis
marker) was conversely controlled by YTHDC2 in an m6A
modification-dependent manner. SLC7A11 overexpression in
YTHDC2-overxpressing cells mitigated the effect of YTHDC2 on cell
proliferation and the progression of ferroptosis. However, as the
present analysis was limited to two cell lines, additional
empirical studies using animal models are imperative to corroborate
these findings.
Extensive research has demonstrated the role of m6A
methylation in several pathologies, including acute myeloid
leukemia and type 2 diabetes (27–30).
Subsequent investigations have delved deeper into the functionality
of m6A-associated proteins in CC. For example,
methyltransferase-like 3 enhances the methylation-dependent
stability of hexokinase 2 in CC cells, facilitating the Warburg
effect and cellular proliferation (31). Furthermore, elevated levels of YTH
N6-methyladenosine RNA binding protein F1 (YTHDF1), a protein from
the YTH protein family, are associated with a worse clinical
prognosis in CC. The attenuation of YTHDF1 expression markedly
curtails the proliferation, migration and invasion of CC cells,
whilst increasing apoptosis rates (32). In vivo experiments using nude
mice have demonstrated the role of YTHDF1 in accelerating CC cell
oncogenesis (32). Moreover, YTHDC2
was suggested to augment metastasis in colon cancer by promoting
HIF-1α translation (11).
Conversely, another study reported reduced YTHDC2 expression in
lung cancer based on tissue and cellular models. Functionality
assays reported that YTHDC2 overexpression can inhibit the
proliferation and migration of lung cancer cells, indicating that
it is a vital prognostic factor (18). Therefore, these results highlight
controversy regarding the function of YTHDC2 in multiple types of
cancers. Furthermore, YTHDC2 is implicated in breast cancer
progression by modulating transcription factors responsible for
cellular stemness, indicating its viability as a target for
therapeutic interventions (33). In
the present study, YTHDC2 overexpression alleviated HPV-positive CC
cell proliferation and cell viability, as demonstrated by the CCK-8
and colony formation assay results. Additionally, the
manifestations of ferroptosis, including apoptosis, ROS generation
and biomarker (SLC7A11, ACSL4, GPX4 and p53) dysregulation, was
induced after YTHDC2 overexpression in SiHa and CaSKi cells. These
observations indicate that YTHDC2 exerts a notable
tumor-suppressing effect on HPV-positive CC, with multifaceted
roles across several cancer types.
Pertaining to the dynamics between HPV and YTHDC2,
the findings of the present study indicate that m6A modifications
may serve a pivotal role in the advancement of HPV-positive CC.
Enhancing the expression levels of YTHDC2 within CC raises
questions about the potential regulatory effects of HPV on YTHDC2
or other m6A modifying enzymes. Further evaluating the connections
between HPV and m6A regulatory mechanisms with a particular focus
on YTHDC2 could provide critical insights into the molecular
processes driving the progression of CC induced by HPV.
The results of the present study also demonstrated
that SLC7A11 is conversely controlled by YTHDC2 in an m6A
modification-dependent manner. Previous studies of the occurrence
of ferroptosis in CC have not specifically targeted HPV-positive CC
(34,35). Ferroptosis is distinguished by a
non-apoptotic, programmed cell death process initiated by the
deactivation of GPX4 and SLC7A11, followed by iron-dependent lipid
peroxidation (36). Glutathione
(GSH) is a crucial GPX4-reducing agent that is pivotal for
maintaining cellular redox homeostasis and shielding cells from
oxidative damage by curtailing ROS accumulation. Inhibition of the
SLC7A11 system markedly reduces cystine levels within the cell,
curtails GSH metabolism and consequently triggers ferroptosis
(37). In the analysis in the
present study, a negative association between SLC7A11 expression
and YTHDC2 was identified in HPV-positive CC cells. Therefore,
reduced SLC7A11 expression inhibited cell proliferation and induced
ferroptosis in cells, which is consistent with previous studies
(38). Increasing SLC7A11 levels
partially negates the effects of YTHDC2 overexpression on
proliferation and ferroptosis in SiHa and CaSKi cells.
Consequently, the present study elucidated a novel pathway wherein
YTHDC2 diminishes SLC7A11 expression, thereby amplifying
ferroptosis.
In conclusion, the present study identified and
assessed the interaction between YTHDC2 and the 5′-UTR of SLC7A11,
based on results from luciferase assays and phenotypic changes. The
present study also demonstrated that SLC7A11-associated ferroptosis
is regulated by YTHDC2. However, future studies should focus on
further experiments and an in-depth analysis of the underlying
molecular interaction and signal pathway. This lack of an in-depth
analysis is considered a limitation of the present study. Another
limitation is that sample size used in the patient sample analysis
was too low to reach a solid conclusion. In summary, the present
research demonstrates that YTHDC2 facilitates cell proliferation
and suppresses ferroptosis in HPV-positive CC cells. Additionally,
SLC7A11 was identified as a direct target influenced by YTHDC2. The
regulatory effect of YTHDC2 on SLC7A11 occurs through an
m6A-mediated translational mechanism, which is critical for the
pathology of HPV-positive CC. These insights suggest that targeting
YTHDC2 could offer a promising therapeutic avenue for treating
HPV-positive CC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LR designed the study. JZ, JY and JG performed the
research. LR and LJ analyzed the data. LR and JZ wrote the paper.
All authors read and approved the final manuscript. LR and JZ
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the First Affiliated Hospital of Henan University of
Science and Technology (approval no. 2023-0147). Written informed
consent was obtained from every participant involved.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kumar S, Malviya R and Meenakshi DU:
Overview of Women's Health. Women's Health: A Comprehensive Guide
to Common Health Issues in Women. Bentham Science Publishers; pp.
1–21. 2024
|
|
2
|
Sun D, Li H, Cao M, He S, Lei L, Peng J
and Chen W: Cancer burden in China: Trends, risk factors and
prevention. Cancer Biol Med. 17:879–895. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou L, Li Y, Wang H, Qin R, Han Z and Li
R: Global cervical cancer elimination: Quantifying the status,
progress, and gaps. BMC Med. 23:672025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kombe Kombe AJ, Li B, Zahid A, Mengist HM,
Bounda GA, Zhou Y and Jin T: Epidemiology and burden of human
papillomavirus and related diseases, molecular pathogenesis, and
vaccine evaluation. Front Public Health. 8:5520282021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schiffman M, Castle PE, Jeronimo J,
Rodriguez AC and Wacholder S: Human papillomavirus and cervical
cancer. Lancet. 370:890–907. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liao S, Sun H and Xu C: YTH domain: A
family of N6-methyladenosine (m6A) readers.
Genomics Proteomics Bioinformatics. 16:99–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi R, Ying S, Li Y, Zhu L, Wang X and Jin
H: Linking the YTH domain to cancer: The importance of YTH family
proteins in epigenetics. Cell Death Dis. 12:3462021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma C, Liao S and Zhu Z: Crystal structure
of human YTHDC2 YTH domain. Biochem Biophys Res Commun.
518:678–684. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B
and Qian SB: m6A in mRNA coding regions promotes
translation via the RNA helicase-containing YTHDC2. Nat Commun.
10:53322019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanabe A, Tanikawa K, Tsunetomi M, Takai
K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, et al:
RNA helicase YTHDC2 promotes cancer metastasis via the enhancement
of the efficiency by which HIF-1α mRNA is translated. Cancer Lett.
376:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He JJ, Li Z, Rong ZX, Gao J, Mu Y, Guan
YD, Ren XX, Zi YY, Liu LY, Fan Q, et al: m6A reader YTHDC2 promotes
radiotherapy resistance of nasopharyngeal carcinoma via activating
IGF1R/AKT/S6 signaling axis. Front Oncol. 10:11662020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Sun B, Xia Y, Sun S and He C: RNA
N6-methyladenosine-related gene contribute to clinical prognostic
impact on patients with liver cancer. Front Genet. 11:3062020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu J, Wang D, Zhou J, Wang L, Zhang N,
Zhou L, Zeng J, Liu J and Yang M: N6-methyladenosine reader YTHDC2
and eraser FTO may determine hepatocellular carcinoma prognoses
after transarterial chemoembolization. Arch Toxicol. 95:1621–1629.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang W, Li J, Lin F, Guo J and Zhao J:
Identification of N 6-methyladenosine-related lncRNAs for patients
with primary glioblastoma. Neurosurg Rev. 44:463–470. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Q, Xie X, Huang Y, Meng S, Li Y, Wang H
and Hu Y: N6-methyladenosine RNA methylation regulators contribute
to the progression of prostate cancer. J Cancer. 12:682–692. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao X and Cui L: Development and
validation of a m6A RNA methylation regulators-based signature for
predicting the prognosis of head and neck squamous cell carcinoma.
Am J Cancer Res. 9:2156–2169. 2019.PubMed/NCBI
|
|
18
|
Sun S, Han Q, Liang M, Zhang Q, Zhang J
and Cao J: Downregulation of m6A reader YTHDC2 promotes tumor
progression and predicts poor prognosis in non-small cell lung
cancer. Thorac Cancer. 11:3269–3279. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Benevolo M, Mottolese M, Marandino F,
Vocaturo G, Sindico R, Piperno G, Mariani L, Sperduti I, Canalini
P, Donnorso RP and Vocaturo A: Immunohistochemical expression of
p16INK4a is predictive of HR-HPV infection in cervical low-grade
lesions. Mod Pathol. 19:384–391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Fan P, Yang Y, Xu C, Huang Y, Li D,
Qing Q, Sun C and Zhou H: Human papillomavirus and human telomerase
RNA component gene in cervical cancer progression. Sci Rep.
9:159262019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wright JD, Matsuo K, Huang Y, Tergas AI,
Hou JY, Khoury-Collado F, St Clair CM, Ananth CV, Neugut AI and
Hershman DL: Prognostic performance of the 2018 international
federation of gynecology and obstetrics cervical cancer staging
guidelines. Obstet Gynecol. 134:49–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan HF, Zou T, Tuo QZ, Xu S, Li H, Belaidi
AA and Lei P: Ferroptosis: Mechanisms and links with diseases. Sig
Transduct Target Ther. 6:492021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu
K, Xu X, Niu Y, Guo S, Zhang C, et al: The m(6)A reader YTHDC2
inhibits lung adenocarcinoma tumorigenesis by suppressing
SLC7A11-dependent antioxidant function. Redox Biol. 38:1018012021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang K, Yang Z, Yang Z, Du L, Zhou Y, Fu
S, Wang X, Li X, Liu D and He X: The m6A reader YTHDC2 promotes the
pathophysiology of temporal lobe epilepsy by modulating
SLC7A11-dependent glutamate dysregulation in astrocytes.
Theranostics. 14:5551–5570. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen SH, Song YY, Gan N, Wang PT, Yan K,
Wang SF, Zu YE and Peng XW: Human papillomavirus infection and
screening strategies. World J Clin Oncol. 16:1050552025. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su H, Wang G, Wu L, Ma X, Ying K and Zhang
R: Transcriptome-wide map of m6A circRNAs identified in
a rat model of hypoxia mediated pulmonary hypertension. BMC
Genomics. 21:392020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mo X-B, Zhang Y-H and Lei S-F: Genome-wide
identification of N6-methyladenosine (m6A)
SNPs associated with rheumatoid arthritis. Front Genet. 9:2992018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
De Jesus DF, Zhang Z, Kahraman S, Brown
NK, Chen M, Hu J, Gupta MK, He C and Kulkarni RN: m6A
mRNA methylation regulates human β-cell biology in physiological
states and in type 2 diabetes. Nat Metab. 1:765–774. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paris J, Morgan M, Campos J, Spencer GJ,
Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA,
Sepulveda C, et al: Targeting the RNA m6A reader YTHDF2
selectively compromises cancer stem cells in acute myeloid
leukemia. Cell Stem Cell. 25:137–148.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Q, Guo X, Li L, Gao Z, Su X, Ji M and
Liu J: N6-methyladenosine METTL3 promotes cervical
cancer tumorigenesis and Warburg effect through YTHDF1/HK2
modification. Cell Death Dis. 11:9112020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Luo Q, Kang J, Wei Q, Yang Y, Yang
D, Liu X, Liu T and Yi P: YTHDF1 aggravates the progression of
cervical cancer through m6A-mediated up-regulation of
RANBP2. Front Oncol. 11:6503832021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tanabe A, Nakayama T, Kashiyanagi J,
Yamaga H, Hirohashi Y, Torigoe T, Satomi F, Shima H, Maeda H,
Kutomi G, et al: YTHDC2 promotes malignant phenotypes of breast
cancer cells. J Oncol. 2022:91889202022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qi X, Zhou J, Wang X, Shen Y, Cao Y, Jiang
L, Shen M, Zhang H, Wang T, Wei P, et al: HPV E6/E7-induced
acetylation of a peptide encoded by a long non-coding RNA inhibits
ferroptosis to promote the malignancy of cervical cancer. Adv Sci
(Weinh). 12:e24140182025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wei E: Heterogeneity analysis of low-risk
HPV infection and high-risk HPV infection, HPV-positive and
HPV-negative cancers. Dissertation; LMU München: pp. 1–88. 2023
|
|
36
|
Song X, Zhu S, Chen P, Hou W, Wen Q, Liu
J, Xie Y, Liu J, Klionsky DJ, Kroemer G, et al: AMPK-mediated BECN1
phosphorylation promotes ferroptosis by directly blocking system
Xc-activity. Curr Biol. 28:2388–2399.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen H, Cao L, Han K, Zhang H, Cui J, Ma
X, Zhao S, Zhao C, Yin S, Yin S, et al: Patulin disrupts
SLC7A11-cystine-cysteine-GSH antioxidant system and promotes renal
cell ferroptosis both in vitro and in vivo. Food Chem Toxicol.
166:1132552022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Angeli JPF, Shah R, Pratt DA and Conrad M:
Ferroptosis inhibition: Mechanisms and opportunities. Trends
Pharmacol Sci. 38:489–498. 2017. View Article : Google Scholar : PubMed/NCBI
|