Introduction
Cancer ranks as one of the leading causes of death
worldwide, following cardiovascular and cerebrovascular disease
(1). In 2022, there were >20
million novel cancer cases and 9.7 million cancer-associated deaths
were reported worldwide (2),
accounting for nearly 16.8% of all global deaths and 22.8% of
fatalities from non-communicable disease (2), driven by population aging and global
development imbalances. Various factors, including environmental
pollution, viral infection, obesity, genetic variation and
unhealthy lifestyles, are implicated in cancer development
(3–6). However, the specific pathogens and
mechanisms remain to be elucidated.
Gene mutation, aberrant expression and epigenetic
modifications can disrupt normal biological functions, which leads
to tumorigenesis. Long non-coding RNAs (lncRNAs) are a type of
non-protein-coding RNA that include >200 nucleotides. lncRNAs
regulate gene expression at transcriptional, epigenetic and
post-transcriptional levels, and are involved in key processes such
as chromosomal silencing, genomic imprinting, transcriptional
interference and intranuclear transport (7). Aberrant lncRNA expression is observed
in various cancer types, such as oral cancer, lung cancer and
gastric cancer, which not only reflect clinical aspects but also
predict prognosis in patients with malignant disease. The
maternally expressed gene 3 (MEG3), a 32-kb imprinted gene located
on chromosome 14q32.3, encodes lncRNA MEG3 expression in diverse
tissues, such as cancerous tissues, normal tissues, and blood
(8). It was reported as the
ortholog of gene trap locus 2 in mice by Schuster-Gossler et
al (9) and identified in humans
in 2000 (10). To date, numerous
studies have demonstrated that MEG3 serves as a tumor suppressor
and is involved in cancer-associated processes such as cell
proliferation, differentiation, metastasis, immune response,
metabolism and apoptosis, which serve an important role in tumor
initiation and progression (11–13).
MEG3 is a key component of the p53 and mouse double minute 2 (MDM2)
signaling pathways, where it inhibits carcinogenesis by increasing
p53 expression (14). MEG3
expression is typically reduced or absent in a variety of cancer
types, including acute myeloid leukemia (15), serous ovarian cancer (16) and head and neck cancer (17), which suggests MEG3 may serve as a
tumor suppressor and target for cancer prevention and
treatment.
Genetic alterations, such as single nucleotide
polymorphisms (SNPs), serve a key role in cancer susceptibility. In
lncRNAs, SNPs lead to aberrant transcript splicing and structural
changes, which impairs lncRNA function. Cao et al (18) reported that individuals with the AA
genotype in the MEG3 rs7158663 G>A polymorphism have a markedly
higher risk of colorectal cancer compared with those with the GG
genotype. Since then, several case-control studies have examined
the association between MEG3 polymorphisms and cancer risk,
although the results remain inconclusive (19,20).
Therefore, the present meta-analysis aimed to clarify the potential
associations between MEG3 polymorphisms and cancer risk.
Materials and methods
Study design
The present meta-analysis was conducted in
accordance with the guidelines of PRISMA (21). All data were extracted from
previously published studies and no ethical concerns were
involved.
Literature search strategy
A total of five online databases (PubMed
(pubmed.ncbi.nlm.nih.gov/?Db=pubmed), Excerpta Medica Database
(embase.com/landing?status=grey), Web of Science (https://www.webofscience.com/wos/), China
National Knowledge Infrastructure (https://www.cnki.net/) and Wanfang (https://www.wanfangdata.com.cn/)) were mined for
studies that examined the association between MEG3 gene
polymorphisms and cancer susceptibility from inception up to
December 2024. Additionally, the references of all included studies
were reviewed to identify additional relevant studies (Table SI).
Eligibility criteria
Inclusion criteria were as follows: i) Observational
studies focusing on MEG3 gene polymorphisms; ii) studies with
sufficient genotype data on allele, homozygous and heterozygote for
each polymorphism locus; iii) studies published in English or
Chinese. The exclusion criteria were as follows: i) Case reports,
letters and reviews; ii) duplicate reports or those with
overlapping data; iii) studies lacking sufficient data to calculate
odds ratios (ORs) and 95% CI and iv) fundamental studies and others
using animal models or cell lines.
Data extraction
Two authors reviewed all included studies and
extracted the following data: Surname of first author, publication
year, country or region of study, ethnicity differences, control
group origin, sample sizes of patients and controls, frequency of
genotype distribution and genotyping method. Disagreements were
resolved through discussion with a third author.
Quality assessment
A total of two authors evaluated the quality of all
included studies with a modified version of the Newcastle-Ottawa
Quality Assessment Scale (NOS; Table
SII) (22). The NOS evaluates
six key aspects: Case representativeness, control source,
Hardy-Weinberg equilibrium (HWE) status in controls, genotyping
procedure, sample size and the assessment of the association.
Studies were scored on a scale of 0 to 11, with a score ≥8
indicating high quality.
Statistical analysis
Crude OR and 95% CIs were calculated to evaluate the
statistical strength of the associations between MEG3 polymorphisms
and cancer risk. A total of five genetic models were examined for
the rs7158663 G>A polymorphism: Allele contrast (A vs. G),
codominant models including heterozygote model (GA vs. GG) and
homozygote model (AA vs. GG), dominant model (AG + AA vs. GG) and
recessive model (AA vs. GG + GA). Similar genetic models were
applied to other loci (rs4081134 G>A, rs11160608 A>C,
rs3087918 T>G, rs3783355 G>A, rs2281511 G>A and rs12431658
T>C). Potential heterogeneity between the included studies was
assessed using the Cochran's χ2-based test (23). According to the recommendations of
Cochrane Handbook for Systematic Reviews of Interventions, a
random-effects model (DerSimonian and Laird method) was used to
guarantee the statistical power (24,25).
Subgroup analyses based on ethnicity, source of controls,
genotyping method and cancer subtypes were conducted when there
were multiple studies on the same theme. Cumulative analyses were
conducted to observe the trends in results when more studies were
added. Sensitivity analyses were conducted to assess the robustness
of the pooled results through gradual exclusion of studies.
Publication bias was evaluated with Egger's bias test and Begg's
funnel plots (26). All statistical
analyses were conducted using STATA (version 14.0; StataCorp LP).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overall results
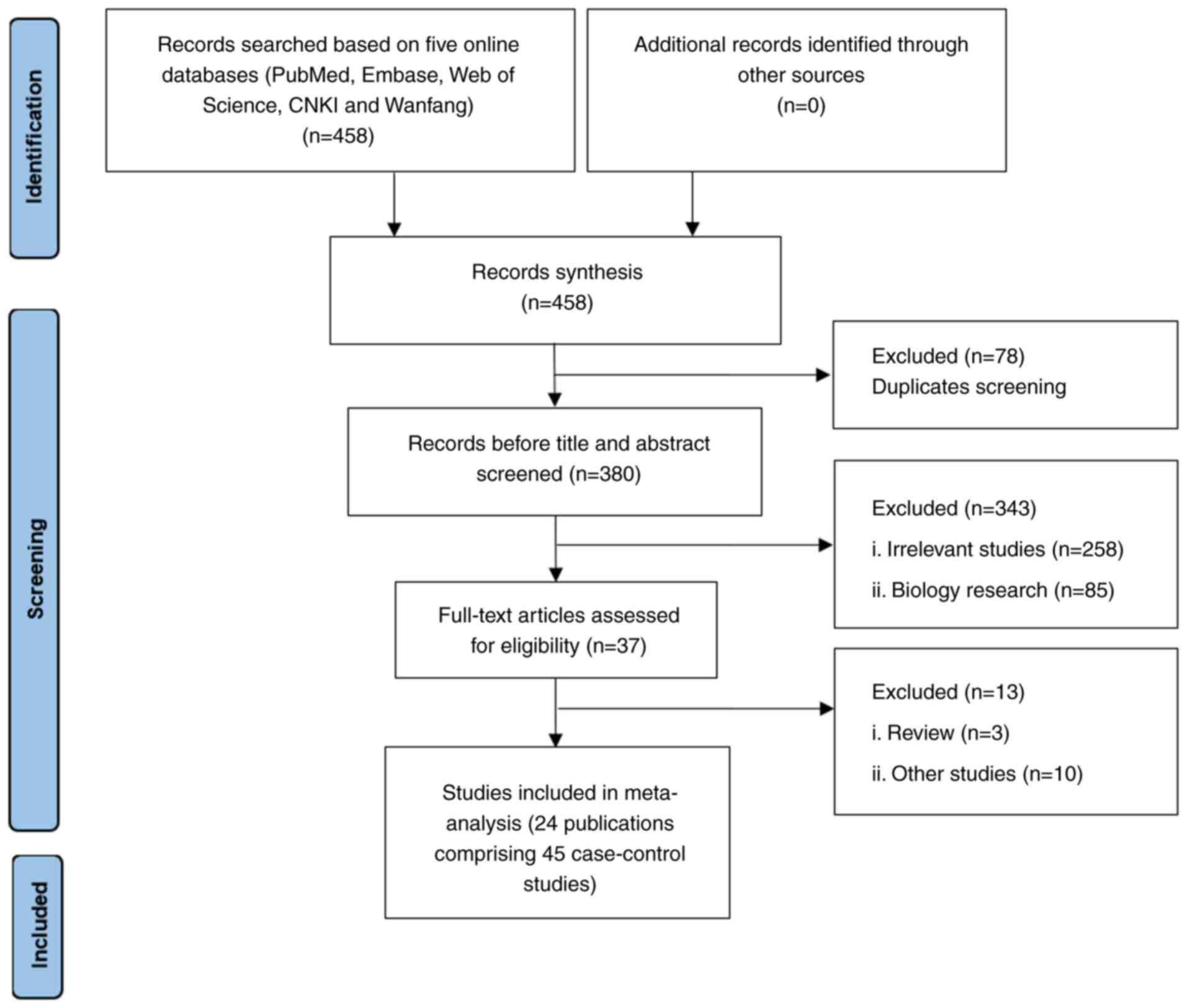
A total of 458 potential publications were
identified through a comprehensive search (Fig. 1). First, 78 duplicate articles were
excluded, 343 irrelevant articles were removed after title and
abstract screening and 13 articles were removed for reasons such as
reviews, fundamental research or lacking sufficient genotype data.
In total, 24 publications comprising 45 independent case-control
studies with 7,423 patients and 9,118 controls were included in
(18–20,27–47).
Of these, 16 studies were conducted in East Asia (based solely on a
Chinese population) and eight were conducted in the Middle East
(Egypt and Iran; Table I). There
were 21 studies that focused on rs7158663 G>A polymorphism
(18–20,27–38,41–46);
seven studies focused on rs4081134 G>A polymorphism (19,20,27,28,38–40),
six focused on s11160608 A>C polymorphism (30,33,38–40,47),
three focused on rs3087918 T>G polymorphism (30,33,38)
and two each focused on rs3783355 G>A (39,40),
rs2281511 G>A (39,40), rs12431658 T>C (39,40)
and rs10132552 T>C (32,42) polymorphisms, respectively. A total
of five case-control studies exhibited deviation from HWE across
multiple loci (Table I).
 | Table I.Characteristics of studies on MEG3
polymorphisms and cancer risk. |
Table I.
Characteristics of studies on MEG3
polymorphisms and cancer risk.
| A, rs7158663
G>A |
|---|
|
|---|
|
|
|
|
|
|
| Genotype
distribution |
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
|
| Case (%) | Control (%) |
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | Country | Ethnicity | Source of
controls | Case, n | Control, n | GG | GA | AA | GG | GA | AA | Genotyping
method | Cancer | P-value for
HWE | NOS | (Refs.) |
|---|
| Cao et
al, | China | EA | HB | 516 | 517 | 264 | 200 | 52 | 298 | 188 | 31 |
TaqMan™ | Colorectal | 0.85 | 9 | (18) |
| 2016 |
|
|
|
|
| (51) | (39) | (10) | (58) | (36) | (6) | cancer |
|
|
|
|
| Zhuo et
al, | China | EA | HB | 392 | 783 | 233 | 141 | 18 | 433 | 296 | 54 | TaqMan | Neuro- | 0.72 | 9 | (19) |
| 2018 |
|
|
|
|
| (59) | (36) | (5) | (55) | (38) | (7) |
| blastoma |
|
|
|
| Yang et
al, | China | EA | HB | 526 | 526 | 268 | 219 | 39 | 289 | 204 | 33 | TaqMan | Lung | 0.71 | 9 | (20) |
| 2018 |
|
|
|
|
| (51) | (42) | (7) | (55) | (39) | (6) |
| cancer |
|
|
|
| Zhang et
al, | China | EA | HB | 172 | 224 | 83 | 74 | 15 | 128 | 76 | 10 | TaqMan | Gastric | 0.76 | 7 | (27) |
| 2018 |
|
|
|
|
| (48) | (43) | (9) | (57) | (34) | (4) |
| cancer |
|
|
|
| Wei et
al, | China | EA | PB | 1,118 | 1,248 | 717 | 349 | 51 | 795 | 391 | 62 | TaqMan | Liver | 0.13 | 9 | (28) |
| 2019 |
|
|
|
|
| (64) | (31) | (5) | (64) | (31) | (5) |
| cancer |
|
|
|
| Ali et
al, | Egypt | ME | HB | 150 | 154 | 57 | 63 | 30 | 84 | 63 | 7 | TaqMan | Breast | 0.26 | 7 | (29) |
| 2020 |
|
|
|
|
| (38) | (42) | (20) | (55) | (41) | (5) |
| cancer |
|
|
|
| Zheng et
al, | China | EA | PB | 434 | 700 | 224 | 170 | 33 | 403 | 250 | 47 | Mass- | Breast | 0.33 | 9 | (30) |
| 2020 |
|
|
|
|
| (52) | (39) | (8) | (58) | (36) | (7) |
ARRAY® | cancer |
|
|
|
| Xu et
al, | China | EA | HB | 165 | 200 | 98 | 54 | 13 | 111 | 78 | 11 | TaqMan | Prostate | 0.57 | 9 | (31) |
| 2020 |
|
|
|
|
| (59) | (33) | (8) | (56) | (39) | (6) |
| cancer |
|
|
|
| Kong et
al, | China | EA | HB | 474 | 543 | 215 | 198 | 61 | 290 | 203 | 50 | TaqMan | Gastric | 0.10 | 9 | (32) |
| 2020 |
|
|
|
|
| (45) | (42) | (13) | (53) | (37%) | ( ) |
| cancer |
|
|
|
| Mazraeh et
al, | Iran | ME | HB | 100 | 100 | 43 | 36 | 21 | 16 | 48 | 36 | PCR- | Leukemia | 1.00 | 6 | (33) |
| 2020 |
|
|
|
|
| (43) | (36) | (21) | (16) | (48) | (36) | RFLP |
|
|
|
|
| Wang et
al, | China | EA | HB | 319 | 306 | 163 | 121 | 35 | 178 | 110 | 18 | PCR- | Lymphoma | 0.85 | 8 | (34) |
| 2020 |
|
|
|
|
| (51) | (38) | (11) | (58) | (36) | (6) | RFLP |
|
|
|
|
| Gao et
al, | China | EA | HB | 430 | 445 | 202 | 185 | 43 | 256 | 159 | 30 | TaqMan | Colorectal | 0.43 | 8 | (35) |
| 2021 |
|
|
|
|
| (47) | (43) | (10) | (58) | (36) | (7) |
| cancer |
|
|
|
| Shaker et
al, | Egypt | ME | HB | 180 | 150 | 63 | 70 | 47 | 93 | 24 | 33 | TaqMan | Breast | <0.01 | 6 | (36) |
| 2021 |
|
|
|
|
| (35) | (39) | (26) | (62) | (16) | (22) |
| cancer |
|
|
|
| Moham- | Egypt | ME | HB | 114 | 110 | 33 | 54 | 27 | 60 | 45 | 5 | TaqMan | Hepato | 0.34 | 7 | (37) |
| med et
al, |
|
|
|
|
| (29) | (47) | (24) | (55) | (41) | (5) |
| cellular |
|
|
|
| 2022 |
|
|
|
|
|
|
|
|
|
|
|
| cancer |
|
|
|
| Pei et al,
2022 | China | EA | HB | 266 | 266 | 118 | 119 | 29 | 153 | 96 | 17 | TaqMan | Leukemia | 0.71 | 10 | (38) |
|
|
|
|
|
|
| (44) | (45) | (11) | (58) | (36) | (6) |
|
|
|
|
|
| Mirzaz- | Iran | ME | HB | 175 | 175 | 46 | 111 | 18 | 69 | 95 | 11 | PCR Tetra- | Lymphoma | 0.04 | 7 | (41) |
| adeh et al,
2022 |
|
|
|
|
| (26) | (63) | (10) | (39) | (54) | (6) | ARMS |
|
|
|
|
| Wang et
al, | China | EA | HB | 118 | 345 | 72 | 39 | 7 | 184 | 135 | 26 | TaqMan | Cervical | 0.86 | 6 | (42) |
| 2022 |
|
|
|
|
| (61) | (33) | (6) | (53) | (39) | (8) |
| cancer |
|
|
|
| Elhelaly et
al, | Egypt | ME | HB | 160 | 160 | 78 | 50 | 32 | 96 | 56 | 8 | TaqMan | Colorectal | 0.96 | 7 | (43) |
| 2023 |
|
|
|
|
| (49) | (31) | (20) | (60) | (35) | (5) |
| cancer |
|
|
|
| Asadi et
al, | Iran | ME | HB | 230 | 240 | 75 | 132 | 23 | 104 | 127 | 9 | PCR-Tetra | Breast | <0.01 | 7 | (44) |
| 2024 |
|
|
|
|
| (33) | (57) | (10) | (43) | (53) | (4) | ARMS | cancer |
|
|
|
| Wang et
al, | China | EA | PB | 271 | 267 | 120 | 123 | 28 | 154 | 96 | 17 | PCR- | Hepato- | 0.60 | 8 | (45) |
| 2024 |
|
|
|
|
| (44) | (45) | (10) | (58) | (36) | (6) | sequencer | cellular |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| cancer |
|
|
|
| Lao et
al, | China | EA | HB | 200 | 200 | 87 | 76 | 37 | 108 | 71 | 21 | TaqMan | Nasopharyn | 0.08 | 7 | (46) |
| 2024 |
|
|
|
|
| (44) | (38) | (19) | (54) | (36) | (11) |
| geal cancer |
|
|
|
|
| B,
rs4081134G>A |
|
| Zhuo et
al, | China | EA | HB | 392 | 783 | 200 | 165 | 27 | 443 | 294 | 46 | TaqMan | Neuro- | 0.76 | 9 | (19) |
| 2018 |
|
|
|
|
| (51) | (42) | (7) | (57) | (38) | (6) |
| blastoma |
|
|
|
| Yang et
al, | China | EA | HB | 526 | 526 | 349 | 161 | 16 | 314 | 182 | 30 | TaqMan | Lung | 0.59 | 9 | (20) |
| 2018 |
|
|
|
|
| (66) | (31) | (3) | (60) | (35) | (6) |
| cancer |
|
|
|
| Hou et
al, | China | EA | HB | 444 | 984 | 243 | 169 | 16 | 598 | 332 | 54 |
Illumina® | Oral cancer | 0.38 | 7 | (39) |
| 2018 |
|
|
|
|
| (55) | (38) | (4) | (61) | (34) | (5) |
|
|
|
|
|
| Zhang et
al, | China | EA | HB | 172 | 224 | 102 | 58 | 12 | 135 | 73 | 16 | TaqMan | Gastric | 0.17 | 7 | (27) |
| 2018 |
|
|
|
|
| (59) | (34) | (7) | (60) | (33) | (7) |
| cancer |
|
|
|
| Yunho et
al, | China | EA | HB | 98 | 86 | 47 | 35 | 16 | 39 | 33 | 14 | PCR-RFLP | Oral cancer | 0.13 | 8 | (40) |
| 2019 |
|
|
|
|
| (48) | (36) | (16) | (45) | (38) | (16) |
|
|
|
|
|
| Wei et
al, | China | EA | HB | 1,118 | 1,248 | 533 | 461 | 120 | 604 | 537 | 103 | TaqMan | Liver | 0.28 | 9 | (28) |
| 2019 |
|
|
|
|
| (48) | (41) | (11) | (48) | (43) | (8) |
| cancer |
|
|
|
| Pei et al,
2022 | China | EA | HB | 266 | 266 | 141 | 107 | 18 | 149 | 101 | 16 | TaqMan | Leukemia | 0.84 | 10 | (38) |
|
|
|
|
|
|
| (53) | (40) | (7) | (56) | (38) | (6) |
|
|
|
|
|
|
| C. rs11160608
A>C |
|
| Hou et
al, | China | EA | HB | 444 | 984 | 112 | 237 | 93 | 308 | 485 | 191 | Illumina | Oral cancer | 1.00 | 7 | (39) |
| 2018 |
|
|
|
|
| (25) | (53) | (21) | (31) | (49) | (19) |
|
|
|
|
|
| Yunho et
al, | China | EA | HB | 98 | 86 | 55 | 30 | 13 | 44 | 28 | 14 | PCR-RFLP | Oral cancer | 0.02 | 8 | (40) |
| 2019 |
|
|
|
|
| (56) | (31) | (13) | (51) | (33) | (16) |
|
|
|
|
|
| Zheng et
al, | China | EA | PB | 434 | 700 | 126 | 218 | 80 | 227 | 341 | 132 | Mass- | Breast | 0.84 | 9 | (30) |
| 2020 |
|
|
|
|
| (29) | (50) | (18) | (32) | (49) | (19) | ARRAY | cancer |
|
|
|
| Mazraeh et
al, | Iran | ME | HeB | 100 | 100 | 28 | 58 | 14 | 39 | 48 | 13 | ARMS- | Leukemia | 0.77 | 6 | (33) |
| 2020 |
|
|
|
|
| (28) | (58) | (14) | (39) | (48) | (13) | PCR |
|
|
|
|
| Pei et al,
2022 | China | EA | HB | 266 | 266 | 80 | 137 | 49 | 85 | 131 | 50 | TaqMan | Leukemia | 0.97 | 10 | (38) |
|
|
|
|
|
|
| (30) | (52) | (18) | (32) | (49) | (19) |
|
|
|
|
|
| Abdi et
al, | Iran | ME | HB | 396 | 399 | 107 | 184 | 104 | 107 | 203 | 89 | Illumina | Gastric | 0.70 | 7 | (47) |
| 2024 |
|
|
|
|
| (27) | (46) | (26) | (27) | (51) | (22) |
| cancer |
|
|
|
|
| D, rs3087918
T>G |
|
| Zheng et
al, | China | EA | PB | 434 | 700 | 171 | 207 | 47 | 259 | 334 | 107 | Mass- | Breast | 0.97 | 9 | (30) |
| 2020 |
|
|
|
|
| (39) | (48) | (11) | (37) | (48) | (15) | ARRAY | cancer |
|
|
|
| Mazraeh et
al, | Iran | ME | HB | 100 | 100 | 52 | 36 | 12 | 41 | 52 | 7 | PCR-RFLP | Leukemia | 0.08 | 6 | (33) |
| 2020 |
|
|
|
|
| (52) | (36) | (12) | (41) | (52) | (7) |
|
|
|
|
|
| Pei et al,
2022 | China | EA | HB | 266 | 266 | 105 | 129 | 32 | 98 | 128 | 40 | TaqMan | Leukemia | 0.86 | 10 | (38) |
|
|
|
|
|
|
| (39) | (48) | (12) | (37) | (48) | (15) |
|
|
|
|
|
|
| E, rs3783355
G>A |
|
| Hou et
al, | China | EA | HB | 444 | 984 | 272 | 142 | 24 | 626 | 319 | 39 | Illumina | Oral cancer | 0.84 | 7 | (39) |
| 2018 |
|
|
|
|
| (61) | (32) | (5) | (64) | (32) | (4) |
|
|
|
|
|
| Yunho et
al, | China | EA | HB | 98 | 86 | 51 | 29 | 18 | 47 | 22 | 17 | PCR-RFLP | Oral cancer | <0.01 | 7 | (40) |
| 2019 |
|
|
|
|
| (52) | (30) | (18) | (55) | (26) | (20) |
|
|
|
|
|
|
| F, rs2281511
G>A |
|
| Hou et
al, | China | EA | HB | 444 | 984 | 299 | 134 | 11 | 673 | 269 | 42 | Illumina | Oral cancer | 0.03 | 7 | (39) |
| 2018 |
|
|
|
|
| (67) | (30) | (2) | (68) | (27) | (4) |
|
|
|
|
|
| Yunho et
al, | China | EA | HB | 98 | 86 | 63 | 23 | 12 | 58 | 16 | 15 | PCR-RFLP | Oral cancer | <0.01 | 8 | (40) |
| 2019 |
|
|
|
|
| (64) | (23) | (12) | (67) | (19) | (17) |
|
|
|
|
|
|
| G, rs12431658
T>C |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Hou et
al, | China | EA | HB | 444 | 984 | 390 | 50 | 3 | 839 | 139 | 6 | Illumina | Oral cancer | 0.93 | 7 | (39) |
| 2018 |
|
|
|
|
| (88) | (11) | (1) | (85) | (14) | (1) |
|
|
|
|
|
| Yunho et
al, | China | EA | HB | 98 | 86 | 74 | 19 | 5 | 67 | 16 | 3 | PCR-RFLP | Oral cancer | 0.12 | 8 | (40) |
| 2019 |
|
|
|
|
| (76) | (19) | (5) | (78) | (19) | (3%) |
|
|
|
|
|
|
| H. rs10132552
T>C |
|
| Kong et
al, | China | EA | HB | 474 | 543 | 239 | 207 | 28 | 278 | 219 | 46 | TaqMan | Gastric | 0.76 | 9 | (32) |
| 2020 |
|
|
|
|
| (50) | (44) | (6) | (51) | (40) | (8) |
| cancer |
|
|
|
| Wang et
al, | China | EA | HB | 118 | 345 | 82 | 32 | 4 | 187 | 142 | 16 |
TaqMan™ | Cervical | 0.09 | 6 | (42) |
| 2022 |
|
|
|
|
| (69) | (27) | (3) | (54) | (41) | (5) |
| cancer |
|
|
|
rs7158663 G>A polymorphism and
cancer risk
In total, 21 case-control studies involving 6,502
patients and 7,649 controls were included to investigate the
association between rs7158663 G>A polymorphism and cancer risk.
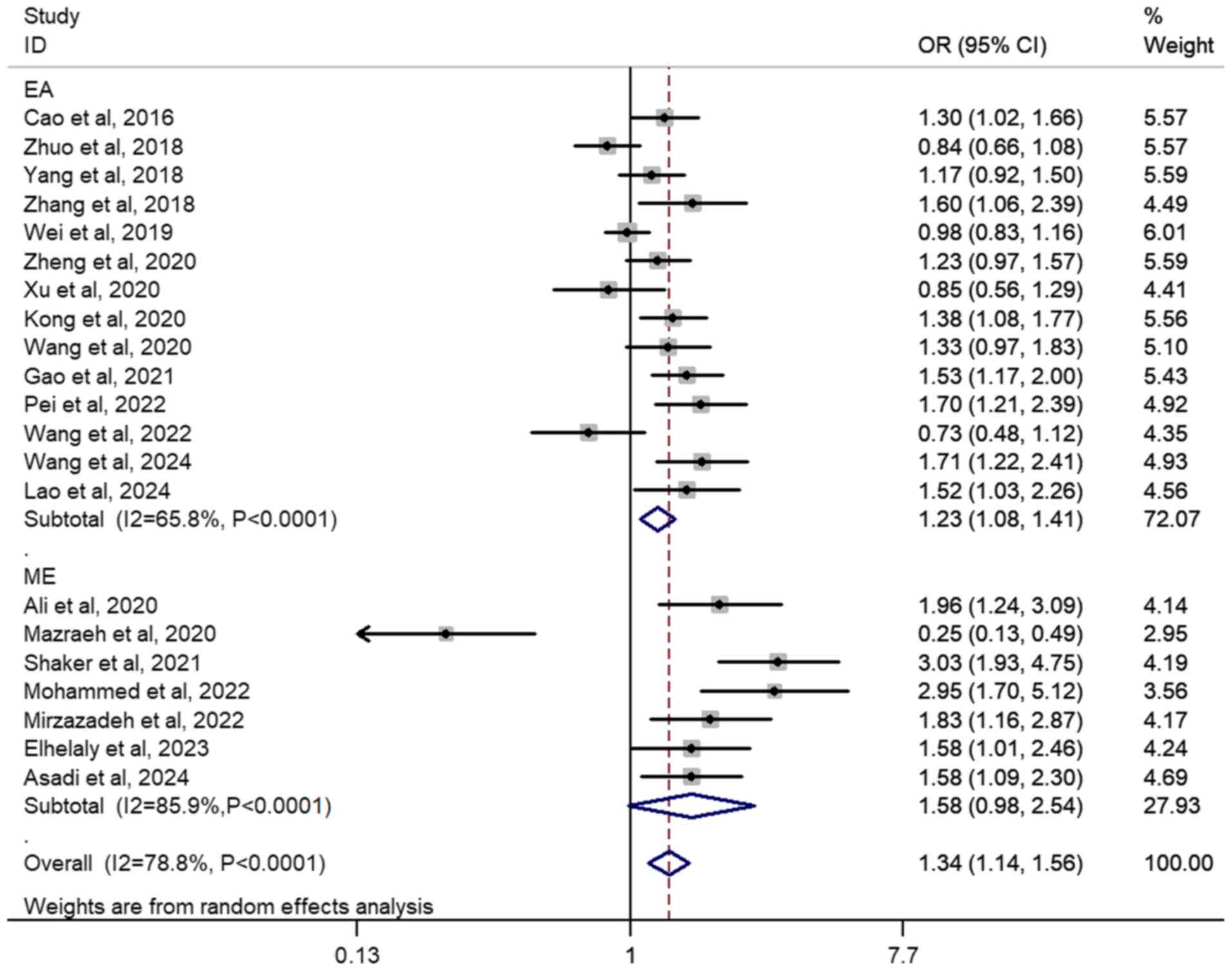
A significant association was observed in the overall population of
A vs. G (OR, 1.30; 95% CI, 1.14–1.48; P<0.01;
I2=82.3%), GA vs. GG (OR, 1.26; 95% CI, 1.09–1.45;
P<0.01; I2=72.0%) AA vs. GG (OR, 1.74; 95%CI,
1.31–2.32; P<0.01; I2=77.3%), GA + AA vs. GG (OR,
1.34; 95% CI, 1.14–1.56; P<0.01; I2=78.8%; Fig. 2) and AA vs. GG + GA (OR, 1.55; 95%
CI, 1.23–1.96; P<0.01; I2=67.9%; Table II). Subgroup analyses revealed
significantly elevated cancer risk associated with the rs7158663
G>A polymorphism in both HWE-yes groups [A vs. G (OR, 1.26; 95%
CI, 1.09–1.45; P<0.01; I2=83.3%); GA vs. GG (OR,
1.17; 95% CI, 1.03–1.34; P=0.02; I2=64.2%); AA vs. GG
(OR, 1.64; 95% CI, 1.20–2.25; P<0.01; I2=79.2%); AG +
AA vs. GG (OR,1.25; 95% CI, 1.07–1.47; P=0.01; I2=77.3%)
and AA vs. GG + GA (OR, 1.53; 95% CI, 1.18–1.98; P<0.01;
I2=71.1%)] and HWE-no groups [A vs. G (OR, 1.58; 95% CI,
1.31–1.91; P<0.01; I2=14.2%); GA vs. GG (OR, 2.15;
95% CI, 1.17–3.95; P=0.01; I2=80.4%); AA vs. GG (OR,
2.46; 95% CI, 1.65–3.68; P<0.01; I2=0%); AG + AA vs.
GG (OR, 2.05; 95% CI, 1.38–2.99; P<0.01; I2=59.5%)
and AA vs. GG + GA (OR, 1.69; 95% CI, 1.05–2.72; P=0.03;
I2=32.2%)]. For ethnic diversity, the similar increased
risks were observed across both East Asian and Middle Eastern
individuals (Table II).
Furthermore, similar results were found in the genotyping subgroup
using the TaqMan™ method (Table II). Increased tumorigenic risks
were observed in gastrointestinal tract cancer groups [A vs. G (OR,
1.40; 95% CI, 1.26–1.55; P<0.01; I2=0%); GA vs. GG
(OR, 1.32; 95% CI, 1.15–1.51; P<0.01; I2=0%); AA vs.
GG (OR, 2.04; 95% CI, 1.51–2.75; P<0.01; I2=29.6%);
GA + AA vs. GG (OR, 1.43; 95% CI, 1.25–1.63; P<0.01;
I2=0%); and AA vs. GG + GA (OR, 1.85; 95% CI, 1.33–2.56;
P<0.01; I2=43.1%)] and breast cancer groups [A vs. G
(OR, 1.59; 95% CI, 1.20–2.09; P<0.01; I2=75.3%); GA
vs. GG (OR, 1.75; 95% CI, 1.10–2.78; P=0.02; I2=81.2%);
AA vs. GG (OR, 2.56; 95% CI, 1.34–4.92; P=0.01;
I2=74.8%); GA + AA vs. GG (OR, 1.79; 95% CI, 2.22–2.63;
P<0.01; I2=76.9%) and AA vs. GG + GA (OR, 1.99; 95%
CI, 1.06–3.74; P=0.03; I2=75.4%; Table II)].
 | Table II.ORs and 95% CIs of MEG3 polymorphisms
and cancer risk. |
Table II.
ORs and 95% CIs of MEG3 polymorphisms
and cancer risk.
| A, rs7158663
G>A |
|---|
|
|---|
|
|
| A vs. G | GA vs. GG | AA vs. GG | GA + AA vs. GG | AA vs. GG + GA |
|---|
|
|
|
|
|
|
|
|
|---|
| Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|---|
| Total | 21 | 1.30 | 1.14- | <0.01 | 82.3 | 1.26 | 1.09- | <0.01 | 72.0 | 1.74 | 1.31- | <0.01 | 77.3 | 1.34 | 1.14- | <0.01 | 78.8 | 1.55 | 1.23- | <0.01 | 67.9 |
|
|
|
| 1.48 |
|
|
| 1.45 |
|
|
| 2.32 |
|
|
| 1.56 |
|
|
| 1.96 |
|
|
| HWE |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Yes | 18 | 1.26 | 1.09- | <0.01 | 83.3 | 1.17 | 1.03- | 0.02 | 64.2 | 1.64 | 1.20- | <0.01 | 79.2 | 1.25 | 1.07- | 0.01 | 77.3 | 1.53 | 1.18- | <0.01 | 71.1 |
|
|
|
| 1.45 |
|
|
| 1.34 |
|
|
| 2.25 |
|
|
| 1.47 |
|
|
| 1.98 |
|
|
| No | 3 | 1.58 | 1.33- | <0.01 | 14.2 | 2.15 | 1.17- | 0.01 | 80.4 | 2.46 | 1.65- | <0.01 | 0 | 2.05 | 1.38- | <0.01 | 59.5 | 1.64 | 1.13- | 0.03 | 32.2 |
|
|
|
| 1.88 |
|
|
| 3.95 |
|
|
| 3.68 |
|
|
| 2.99 |
|
|
| 2.38 |
|
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| EA | 14 | 1.21 | 1.08 | <0.01 | 70.4 | 1.18 | 1.05- | 0.01 | 51.7 | 1.47 | 1.18- | <0.01 | 53.1 | 1.23 | 1.08- | <0.01 | 65.8 | 1.33 | 1.16–1. | <0.01 | 34.7 |
|
|
|
| 1.35 |
|
|
| 1.33 |
|
|
| 1.83 |
|
|
| 1.41 |
|
|
| 53 |
|
|
| ME | 7 | 1.53 | 1.04- | 0.03 | 88.8 | 1.44 | 0.89- | 0.14 | 84.3 | 2.70 | 1.12- | 0.03 | 88.3 | 1.58 | 0.98- | 0.06 | 85.9 | 2.30 | 1.13- | 0.02 | 84.5 |
|
|
|
| 2.23 |
|
|
| 2.34 |
|
|
| 6.52 |
|
|
| 2.54 |
|
|
| 4.68 |
|
|
| Genotyping
method |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
TaqMan | 15 | 1.35 | 1.16- | <0.01 | 82.5 | 1.27 | 1.08- | <0.01 | 71.5 | 1.84 | 1.35- | <0.01 | 75.1 | 1.37 | 1.15- | <0.01 | 78.4 | 1.49 | 1.29- | <0.01 | 69.1 |
|
|
|
| 1.58 |
|
|
| 1.50 |
|
|
| 2.52 |
|
|
| 1.64 |
|
|
| 1.71 |
|
|
| PCR
series | 5 | 1.16 | 0.80- | 0.44 | 87.6 | 1.16 | 0.75- | 0.52 | 81.3 | 1.54 | 0.62- | 0.35 | 87.1 | 1.18 | 0.73- | 0.51 | 86.0 | 1.43 | 1.07- | 0.02 | 75.5 |
|
|
|
| 1.58 |
|
|
| 1.79 |
|
|
| 3.81 |
|
|
| 1.91 |
|
|
| 1.91 |
|
|
| Cancer type |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Gastroin- | 5 | 1.40 | 1.26- | <0.01 | 0 | 1.32 | 1.15- | <0.01 | 0 | 2.00 | 1.57- | <0.01 | 29.6 | 1.43 | 1.25- | <0.01 | 0 | 1.78 | 1.41- | <0.01 | 43.1 |
|
testinal |
|
| 1.55 |
|
|
| 1.51 |
|
|
| 2.54 |
|
|
| 1.63 |
|
|
| 2.25 |
|
|
|
tract |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Liver | 3 | 1.55 | 0.90- | 0.11 | 92.6 | 1.45 | 0.90- | 0.13 | 82.3 | 2.44 | 0.75- | 0.14 | 89.9 | 1.63 | 0.89- | 0.11 | 89.9 | 1.96 | 0.74- | 0.17 | 85.9 |
|
|
|
| 2.6 |
|
|
| 2.33 |
|
|
| 7.93 |
|
|
| 2.97 |
|
|
| 5.17 |
|
|
|
Breast | 4 | 1.59 | 1.20- | <0.01 | 75.3 | 1.75 | 1.10- | 0.02 | 81.2 | 2.56 | 1.34- | 0.01 | 74.8 | 1.79 | 2.22- | <0.01 | 76.9 | 1.99 | 1.06- | 0.03 | 75.4 |
|
|
|
| 2.09 |
|
|
| 2.78 |
|
|
| 4.92 |
|
|
| 2.63 |
|
|
| 3.74 |
|
|
|
Blood | 4 | 1.09 | 0.67- | 0.73 | 90.4 | 1.06 | 0.60- | 0.84 | 85.5 | 1.27 | 0.44- | 0.66 | 88.9 | 1.07 | 0.57- | 0.84 | 89.2 | 1.29 | 0.65- | 0.46 | 77.4 |
|
|
|
| 1.76 |
|
|
| 1.88 |
|
|
| 3.69 |
|
|
| 2.01 |
|
|
| 2.57 |
|
|
|
Other | 5 | 1.02 | 0.82- | 0.85 | 72.0 | 0.97 | 0.80- | 0.78 | 40.4 | 1.11 | 0.69- | 0.66 | 62.5 | 0.99 | 0.78- | 0.96 | 62.1 | 1.12 | 0.74- | 0.59 | 52.7 |
|
|
|
| 1.28 |
|
|
| 1.19 |
|
|
| 1.79 |
|
|
| 1.26 |
|
|
| 1.69 |
|
|
| B, rs4081134
G>A |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 7 | 1.02 | 0.91- | 0.70 | 47.7 | 1.04 | 0.94- | 0.44 | 35.5 | 0.99 | 0.74- | 0.93 | 44.6 | 1.04 | 0.91- | 0.57 | 41.6 | 0.97 | 0.74- | 0.86 | 43.6 |
|
|
|
| 1.15 |
|
|
| 1.15 |
|
|
| 1.32 |
|
|
| 1.19 |
|
|
| 1.29 |
|
|
| Oral cancer | 2 | 1.04 | 0.88- | 0.61 | 0 | 1.20 | 0.98- | 0.11 | 3.3 | 0.79 | 0.49- | 0.33 | 0 | 1.14 | 0.92- | 0.24 | 0 | 0.78 | 0.48- | 0.25 | 0 |
|
|
|
| 1.24 |
|
|
| 1.50 |
|
|
| 1.27 |
|
|
| 1.42 |
|
|
| 1.20 |
|
|
|
| C, rs11160608
A>C |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 6 | 1.09 | 1.00- | 0.57 | 0 | 1.15 | 1.10- | 0.05 | 0 | 1.17 | 0.94- | 0.10 | 0 | 1.16 | 1.01- | 0.04 | 2.7 | 1.08 | 0.92- | 0.35 | 0 |
|
|
|
| 1.19 |
|
|
| 1.33 |
|
|
| 1.40 |
|
|
| 1.33 |
|
|
| 1.26 |
|
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| EA | 4 | 1.08 | 0.98- | 0.13 |
| 1.19 | 1.01- | 0.04 | 0 | 1.15 | 0.93- | 0.20 | 0 | 1.18 | 1.01- | 0.04 | 0 | 1.03 | 0.86- | 0.77 | 0 |
|
|
|
| 1.20 |
|
|
| 1.41 |
|
|
| 1.42 |
|
|
| 1.37 |
|
|
| 1.23 |
|
|
| ME | 2 | 1.12 | 0.93- | 0.23 |
| 1.17 | 0.64- | 0.61 | 66.5 | 1.22 | 0.85- | 0.28 | 0 | 1.19 | 0.74- | 0.47 | 55.1 | 1.22 | 0.90- | 0.19 | 0 |
|
|
|
| 1.33 |
|
|
| 2.12 |
|
|
| 1.74 |
|
|
| 1.94 |
|
|
| 1.65 |
|
|
| Cancer type |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Oral | 2 | 1.05 | 0.77- | 0.76 | 50.0 | 1.26 | 0.99- | 0.07 | 36.5 | 1.24 | 0.91- | 0.17 | 37.4 | 1.13 | 0.71- | 0.61 | 57.1 | 1.07 | 0.82- | 0.68 | 0 |
|
|
|
| 1.43 |
|
|
| 1.61 |
|
|
| 1.68 |
|
|
| 1.79 |
|
|
| 1.39 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| .79 |
|
|
|
|
|
|
|
Leukemia | 2 | 1.09 | 0.89- | 0.40 | 0 | 1.25 | 0.90- | 0.18 | 0 | 1.13 | 0.73- | 0.57 | 0 | 1.22 | 0.90- | 0.20 | 24.3 | 1.11 | 0.68- | 1.00 | 0 |
|
|
|
| 1.35 |
|
|
| 1.74 |
|
|
| 1.75 |
|
|
| 1.67 |
|
|
| 1.47 |
|
|
|
| D, rs3087918
T>G |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 3 | 0.86 | 0.75- | 0.04 | 0 | 0.88 | 0.72- | 0.22 | 30.5 | 0.74 | 0.54- | 0.05 | 0 | 0.85 | 0.70- | 0.09 | 0 | 0.77 | 0.59- | 0.08 | 39.4 |
|
|
|
| 0.99 |
|
|
| 1.08 |
|
|
| 0.99 |
|
|
| 1.03 |
|
|
| 1.03 |
|
|
|
| E, rs3783355
G>A |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 2 | 1.08 | 0.91- | 0.37 | 0 | 1.04 | 0.83- | 0.71 | 0 | 1.25 | 0.81- | 0.31 | 0 | 1.07 | 0.86- | 0.52 | 0 | 1.22 | 0.79- | 0.37 | 0 |
|
|
|
| 1.30 |
|
|
| 1.31 |
|
|
| 1.94 |
|
|
| 1.33 |
|
|
| 1.86 |
|
|
|
| F, rs2281511
G>A |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 2 | 0.96 | 0.80- | 0.70 | 0 | 1.14 | 0.90- | 0.27 | 0 | 0.64 | 0.38- | 0.10 | 0 | 1.05 | 0.84- | 0.68 | 0 | 0.61 | 0.37- | 0.7 | 0 |
|
|
|
| 1.16 |
|
|
| 1.44 |
|
|
| 1.08 |
|
|
| 1.31 |
|
|
| 1.03 |
|
|
|
| G, rs12431658
T>C |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 2 | 0.88 | 0.67- | 0.37 | 16.5 | 0.82 | 0.60- | 0.22 | 0 | 1.27 | 0.47- | 0.64 | 0 | 0.84 | 0.63- | 0.30 | 0 | 1.28 | 0.47- | 0.64 | 0 |
|
|
|
| 1.16 |
|
|
| 1.12 |
|
|
| 3.44 |
|
|
| 1.14 |
|
|
| 3.50 |
|
|
|
| H, rs10132552
T>C |
|
|
|
| A vs. G | GA vs.
GG | AA vs.
GG | GA + AA vs.
GG | AA vs. GG +
GA |
|
|
|
|
|
|
|
|
|
Characteristic | n | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a | OR | 95% CI | P-value | I2,
%a |
|
| Total | 2 | 0.79 | 0.50- | 0.29 | 77.4 | 0.77 | 0.37- | 0.49 | 87.4 | 0.68 | 0.43- | 0.10 | 0 | 0.75 | 0.38- | 0.40 | 85.7 | 0.69 | 0.44- | 0.10 | 0 |
|
|
|
| 1.22 |
|
|
| 1.62 |
|
|
| 1.08 |
|
|
| 1.47 |
|
|
| 1.07 |
|
|
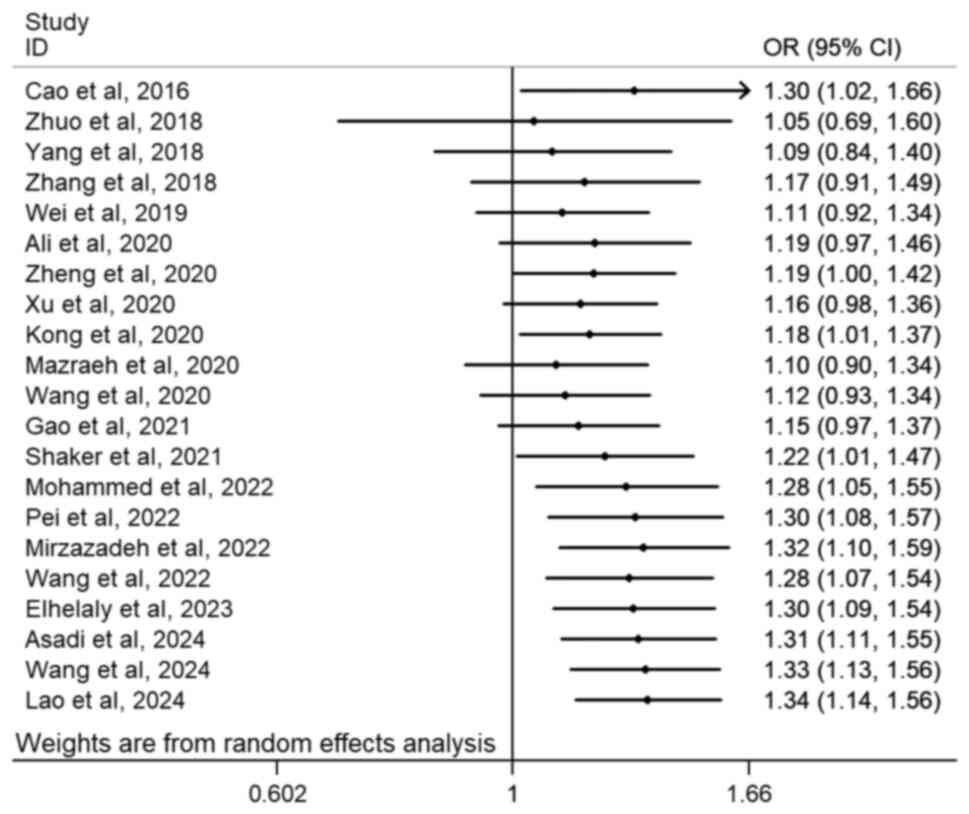
Cumulative analyses in the general population, which
revealed that the first statistically significant association
emerged when the study by Kong et al (2020) (32) was incorporated. This positive
finding specifically demonstrated an association between the
rs7158663 G>A polymorphism and cancer susceptibility (Fig. 3; GA + AA vs. GG). Furthermore,
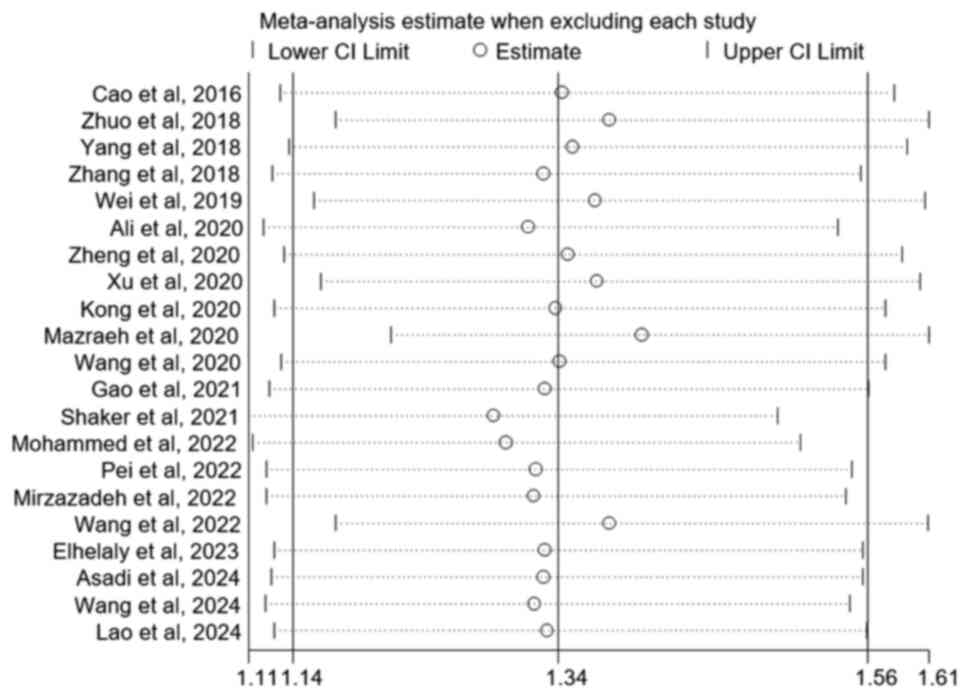
sensitivity analyses demonstrated a consistent and stable result
when each study was removed step-by-step (Fig. 4; GA + AA vs. GG).
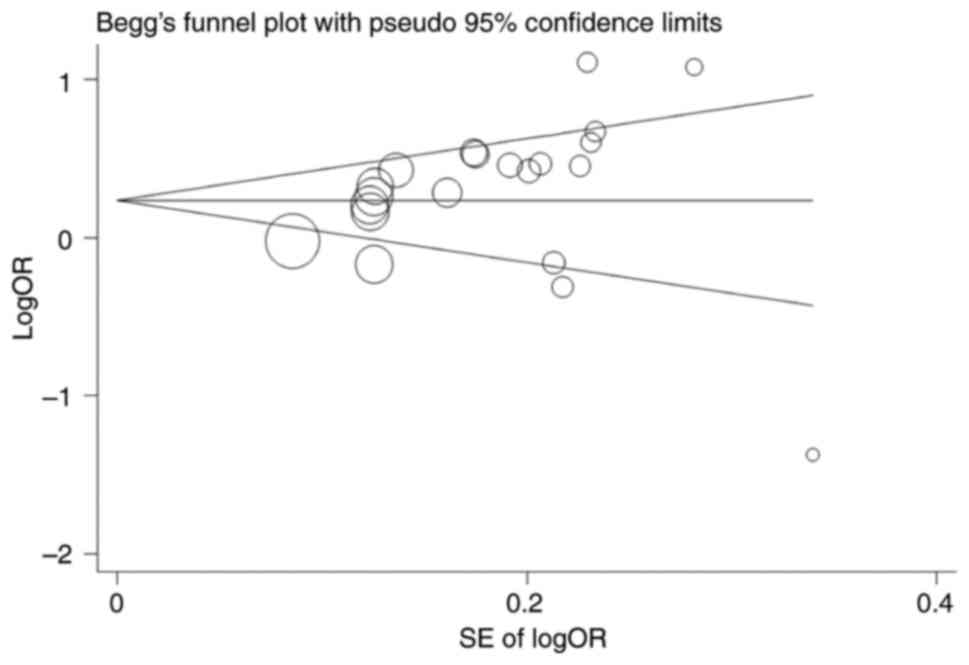
Funnel plot and Egger's test confirmed that there
was no significant publication bias in the general population [A
vs. G (P=0.13); GA vs. GG (P=0.30); AA vs. GG (P=0.10); GA + AA vs.
GG (P=0.18; Fig. 5) and AA vs. GG +
GA (P=0.03)].
rs4081134 G>A polymorphism and
cancer risk
A total of seven case-control studies involving
3,016 patients and 4,117 controls were included to examine the
association between the rs4081134G>A polymorphism and cancer
risk. No significant cancer risk was found overall [A vs. G (OR,
1.02; 95% CI, 0.91–1.15; P=0.70; I2=47.7%); GA vs. GG
(OR, 1.05; 95% CI, 0.91–1.20; P=0.52; I2=35.5%); AA vs.
GG (OR, 0.99; 95% CI, 0.74–1.32; P=0.93; I2=44.6%); AG +
AA vs. GG (OR, 1.04; 95% CI, 0.91–1.19; P=0.57;
I2=41.6%) and AA vs. GG + GA (OR, 0.97; 95% CI,
0.74–1.29; P=0.86; I2=43.6%)] or in the subgroup
analysis for oral cancer (Table
II).
Cumulative and sensitivity analyses demonstrated
fluctuations when studies were added or removed. No publication
bias was observed and the results were further supported by Egger's
test [A vs. G (P=0.61); GA vs. GG (P=0.95); AA vs. GG (P=0.19); AG
+ AA vs. GG (P=0.87) and AA vs. GG + GA (P=0.14); data not
shown].
rs11160608 A>C polymorphism and
cancer risk
A total of six case-control studies involving 1,725
patients and 2,535 controls were included to examine the
association between rs11160608 A>C polymorphism and cancer risk.
Overall, a significantly increased risk of tumorigenesis was
observed [AC + CC vs. AA (OR, 1.16; 95% CI, 1.01–1.33; P=0.04;
I2=2.7%); Fig. S1;
Table II].
Cumulative (Fig.
S2; AC + CC vs. AA model) and sensitivity analyses (Fig. S3; AC + CC vs. AA model)
demonstrated no significant differences when each study was added
or removed. Furthermore, no publication bias was observed and the
results were further supported by Egger's test [C vs. A (P=0.51);
AC vs. AA (P=0.84); CC vs. CC (P=0.48); AC + CC vs. AA (P=0.75); CC
vs. AA + AC (P=0.35; Fig.
S4)].
Other polymorphisms and cancer
risk
No significant association was observed between
rs3087918 T>G, rs3783355 G>A, rs2281511 G>A, rs12431658
T>C, rs10132552 T>C polymorphisms and cancer risk (Table II).
Discussion
Cancer is the leading cause of death globally and
imposes notable economic and emotional burdens on both society and
individuals (48). It is predicted
that the global cancer burden will rise to 28.4 million cases by
2040, which marks a 47% increase from 2020 (49). Despite advances in understanding
cancer, its etiology and pathogenesis remain incompletely
understood.
lncRNA is one of the most important ncRNAs, serve
key roles in various biological processes, including the regulation
of histone modification, DNA methylation, transcriptional
regulation and post-transcriptional regulation (50,51).
Increasing evidence highlights that aberrant expression of lncRNAs
is associated with metastasis, recurrence and prognosis across
multiple types of cancer (52,53).
MEG3, located in the imprinting region of the Δ-like
1 homolog-MEG3 locus on chromosome 14q32.3 in humans, has garnered
increasing attention as a tumor suppressor involved in
tumorigenesis (8,21). Dysregulated expression of MEG3
restricts the cancer cell proliferation, invasion and metastasis
while promoting apoptosis (54,55).
Previous studies have suggested that lower MEG3 expression in
patients with cancer is associated with worse pathological grade,
enhanced tumor invasion and worse prognosis (56,57).
Several signaling pathways, including p53, MDM2, VEFG,
Wnt/β-catenin and TGF-β, have been implicated in these processes
(14). MEG3 inhibits carcinogenesis
by interacting with microRNAs (miRNAs or miRs), such as miR-148a-3p
and miR-155, within both the intracellular and extracellular
microenvironment (3,4), which offers potential strategies for
cancer diagnosis and treatment.
SNPs are the most common forms of genetic mutations
and serve a notable regulatory role in tumorigenesis. SNPs alter
the secondary structure of lncRNAs and affect the binding affinity
between lncRNAs and other molecules. The nucleotide substitution in
the MEG3 rs7158663 G>A polymorphism modifies the folding
structures and minimum free energy (5,58).
Ghaedi et al (59) revealed
that the rs7158663 polymorphism disrupts binding sites for miR-4307
and miR-1265 within MEG3. These alterations impact miRNA-lncRNA
interactions, which modify binding sites for specific miRNAs and
indirectly regulate gene expression.
Cao et al (18) performed a first case-control study
in a Chinese population and reported a markedly increased risk for
colorectal cancer associated with the MEG3 rs7158663 G>A
polymorphism (OR, 1.31; 95% CI, 1.08–1.59; P=0.007). Numerous
studies have examined the association between the MEG3 rs7158663
G>A polymorphism and various cancer types (18–20),
however, the results were inconsistent. Ali et al (29) investigated the impact of the
rs7158663 G>A polymorphism on breast cancer and found that
individuals with the AA genotypes exhibited a markedly higher
cancer risk compared with those with the GG genotype (OR, 6.32; 95%
CI, 2.59–15.44; P<0.01). Kong et al (32) identified an increased susceptibility
to gastric cancer in patients carrying the A allele at rs7158663
(OR, 1.41; 95% CI, 1.14–1.74; P<0.01). Mazraeh et al
(33) reported that both the
mutated homozygous and heterozygous genotypes are more likely to
develop acute myeloid leukemia compared with the GG genotype.
However, other studies, such as those by Zhuo et al
(19), Yang et al (20) and Xu et al (31), found no notable association between
the rs7158663 G>A polymorphism and cancer risk. Similar
inconsistencies were observed for the rs4081134 G>A polymorphism
and other loci (38). These
discrepancies may be attributed to several factors, such as small
sample sizes for each polymorphism locus, ethnic differences
between studies, inconsistent quality of each study and deviation
of HWE in controls in some studies.
To the best of our knowledge, the present systematic
review and meta-analysis encompassed all relevant studies on the
association between multiple lncRNA MEG3 polymorphisms and cancer
susceptibility. The rs7158663 G>A polymorphism markedly
contributed to cancer development in the general population, as
well as in various subgroup analyses. The present findings
suggested that both East Asian and Middle Eastern populations
carrying the A allele are more susceptible to cancer: Ethnic
differences appear to influence cancer susceptibility associated
with these polymorphisms, with individuals from East Asia and the
Middle East exhibiting heightened cancer risk. The majority of
observational studies were conducted in East Asia and the Middle
East (19,20,43).
Furthermore, the present meta-analysis highlighted the potential
role of the rs11160608 A>C polymorphism as a tumorigenic factor;
however, the limited number of original studies warrants caution in
drawing definitive conclusions.
Numerous limitations in the present meta-analysis
should be addressed. First, ethnic bias could not be avoided, since
most studies were conducted in East Asia (based solely on Chinese
populations) and the Middle East. This raises concerns regarding
the generalizability of the present results to other ethnic groups.
Second, several interacting factors, such as viral infection,
environmental pollution, unhealthy lifestyle and dietary habits are
known to influence cancer susceptibility (60,61).
However, a comprehensive assessment of the impact of these factors
was not possible, as data on these variables were not extracted
from all included studies, which may have introduced deviation.
Third, although multiple loci in MEG3 were examined, the
interactive assessment with other genes was not conducted, which
may limit understanding of the tumorigenic mechanisms associated
with these polymorphism loci. Fourth, heterogeneity existed between
genetic models, which was alleviated in the subgroup analysis,
particularly based on ethnicity and cancer subtypes.
Despite these limitations, a comprehensive and
systematic search strategy was employed, yielding a large sample
size. Rigorous statistical methods, including cumulative and
sensitivity analysis and publication bias assessment, were used to
evaluate potential bias and subgroup analyses based on HWE status,
ethnicity, genotyping method and tumor subtypes were performed to
explore potential associations.
In conclusion, the present meta-analysis indicated
that MEG3 rs7158663 G>A polymorphism may serve a key role in the
development of cancer, particularly in East Asian and Middle
Eastern populations and numerous types of cancer. Furthermore,
future case-control studies with larger sample sizes across diverse
ethnic groups are warranted to confirm these associations in the
future.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research Grant for Health
Science and Technology of Pudong Health Commission (grant no.
PW2022A-63).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
YN and YH conceived and designed the study, analyzed
data and wrote and reviewed the manuscript. RC performed the
experiments and wrote the manuscript. XL analyzed data. All authors
have read and approved the final manuscript. YN and YH confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33.
2022.PubMed/NCBI
|
|
2
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
3
|
Pullella K and Kotsopoulos J: Arsenic
exposure and breast cancer risk: A Re-evaluation of the literature.
Nutrients. 12:33052020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosenberg AJ, Agrawal N, Pearson A, Gooi
Z, Blair E, Cursio J, Juloori A, Ginat D, Howard A, Chin J, et al:
Risk and response adapted de-intensified treatment for
HPV-associated oropharyngeal cancer: Optima paradigm expanded
experience. Oral Oncol. 122:1055662021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park S, Ahn S, Kim DG, Kim H, Kang SY and
Kim KM: High frequency of juxtamembrane domain ERBB2 mutation in
gastric cancer. Cancer Genomics Proteomics. 19:105–112. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tseng CM, Chen YT, Tao CW, Ou SM, Hsiao
YH, Li SY, Chen TJ, Perng DW and Chou KT: Adult narcoleptic
patients have increased risk of cancer: A nationwide
population-based study. Cancer Epidemiol. 39:793–797. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hrdlickova B, de Almeida RC, Borek Z and
Withoff S: Genetic variation in the non-coding genome: Involvement
of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys
Acta. 1842:1910–1922. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schuster-Gossler K, Bilinski P, Sado T,
Ferguson-Smith A and Gossler A: The mouse Gtl2 gene is
differentially expressed during embryonic development, encodes
multiple alternatively spliced transcripts, and may act as an RNA.
Dev Dyn. 212:214–228. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miyoshi N, Wagatsuma H, Wakana S,
Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino
T and Ishino F: Identification of an imprinted gene, Meg3/Gtl2 and
its human homologue MEG3, first mapped on mouse distal chromosome
12 and human chromosome 14q. Genes Cells. 5:211–220. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahmadi A, Rezaei A, Khalaj-Kondori M and
Khajehdehi M: A comprehensive bioinformatic analysis identifies a
tumor suppressor landscape of the MEG3 lncRNA in breast cancer.
Indian J Surg Oncol. 15:752–761. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Coan M, Haefliger S, Ounzain S and Johnson
R: Targeting and engineering long non-coding RNAs for cancer
therapy. Nat Rev Genet. 25:578–595. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu SJ, Dang HX, Lim DA, Feng FY and Maher
CA: Long noncoding RNAs in cancer metastasis. Nat Rev Cancer.
21:446–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Wei L, Jin Y, Fu H, Wu Y and Zheng X: Long Noncoding RNA MEG3
interacts with p53 protein and regulates partial p53 target genes
in hepatoma cells. PLoS One. 10:e01397902015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He C, Wang X, Luo J, Ma Y and Yang Z: Long
noncoding RNA maternally expressed gene 3 is downregulated, and its
insufficiency correlates with Poor-risk stratification, worse
treatment response, as well as unfavorable survival data in
patients with acute myeloid leukemia. Technol Cancer Res Treat.
19:15330338209458152020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Buttarelli M, De Donato M, Raspaglio G,
Babini G, Ciucci A, Martinelli E, Baccaro P, Pasciuto T, Fagotti A,
Scambia G and Gallo D: Clinical value of lncRNA MEG3 in High-grade
serous ovarian cancer. Cancers (Basel). 12:9662020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji Y, Feng G, Hou Y, Yu Y, Wang R and Yuan
H: Long noncoding RNA MEG3 decreases the growth of head and neck
squamous cell carcinoma by regulating the expression of miR-421 and
E-cadherin. Cancer Med. 9:3954–3963. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao X, Zhuang S, Hu Y, Xi L, Deng L, Sheng
H and Shen W: Associations between polymorphisms of long non-coding
RNA MEG3 and risk of colorectal cancer in Chinese. Oncotarget.
7:19054–19059. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhuo ZJ, Zhang R, Zhang J, Zhu J, Yang T,
Zou Y, He J and Xia H: Associations between lncRNA MEG3
polymorphisms and neuroblastoma risk in Chinese children. Aging
(Albany NY). 10:481–491. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Z, Li H, Li J, Lv X, Gao M, Bi Y,
Zhang Z, Wang S, Li S, Li N, et al: Association between long
noncoding RNA MEG3 polymorphisms and lung cancer susceptibility in
Chinese northeast population. DNA Cell Biol. 37:812–820. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: Preferred reporting items for systematic reviews and
meta-analyses: The PRISMA statement. J Clin Epidemiol.
62:1006–1012. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in Meta-analyses. Eur J Epidemiol.
25:603–605. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huedo-Medina TB, Sánchez-Meca J,
Marín-Martínez F and Botella J: Assessing heterogeneity in
meta-analysis: Q statistic or I2 index? Psychol Methods.
11:193–206. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
DerSimonian R: Meta-analysis in the design
and monitoring of clinical trials. Stat Med. 15:1237–1252. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
26
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Q, Ai L and Dai Y: Association
between polymorphism of long non-coding RNA maternally expressed
gene 3 and risk of gastric cancer. Chin J Bases Clin Gen Surg.
25:1323–1326. 2018.
|
|
28
|
Wei Z: Analysis of the Effect of Genetic
Variants of MEG3-P53-MDM2-PTEN Molecular Pathway on
Hepatocarcinogenesis. Master's degree dissertation. 2019.
|
|
29
|
Ali MA, Shaker OG, Alazrak M, AbdelHafez
MN, Khalefa AA, Hemeda NF, Abdelmoktader A and Ahmed FA:
Association analyses of a genetic variant in long non-coding RNA
MEG3 with breast cancer susceptibility and serum MEG3 expression
level in the Egyptian population. Cancer Biomark. 28:49–63. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng Y, Wang M, Wang S, Xu P, Deng Y, Lin
S, Li N, Liu K, Zhu Y, Zhai Z, et al: LncRNA MEG3 rs3087918 was
associated with a decreased breast cancer risk in a Chinese
population: A case-control study. BMC Cancer. 20:6592020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu B, Zhang M, Liu C, Wang C, You Z, Wang
Y and Chen M: Association of Long Non-coding RNA MEG3 polymorphisms
and risk of prostate cancer in chinese han population. Urol J.
18:176–180. 2020.PubMed/NCBI
|
|
32
|
Kong X, Yang S, Liu C, Tang H, Chen Y,
Zhang X, Zhou Y and Liang G: Relationship between MEG3 gene
polymorphism and risk of gastric cancer in Chinese population with
high incidence of gastric cancer. Biosci Rep. 40:BSR202003052020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mazraeh SA, Gharesouran J, Ghafouri-Fard
S, Ganjineh Ketab FN, Hosseinzadeh H, Moradi M, Javadlar M,
Hiradfar A, Rezamand A, Taheri M and Rezazadeh M: Association
between WT1 and MEG3 polymorphisms and risk of acute myeloid
leukemia. Meta Gene. 23:1006362020. View Article : Google Scholar
|
|
34
|
Wang XR: Association analyses of genetic
variants in long non-coding RNA MEG3 with diffuse large B-cell
lymphoma susceptibility in Chinese Han population. Master's degree
dissertation. 2020.
|
|
35
|
Gao X, Li X, Zhang S and Wang X: The
association of MEG3 gene rs7158663 polymorphism with cancer
susceptibility. Front Oncol. 11:7967742021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shaker O, Ayeldeen G and Abdelhamid A: The
impact of single nucleotide polymorphism in the Long Non-coding
MEG3 gene on MicroRNA-182 and MicroRNA-29 expression levels in the
development of breast cancer in egyptian women. Front Genet.
12:6838092021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mohammed SR, Shaker OG, Mohamed MM,
Abdelhafez Mostafa MN, Gaber SN, Ali DY, Hussein HA, Elebiary AM,
Abonar AA, Eid HM and El Sayed Ali HS: The emerging role of lncRNA
MEG3 and MEG3 rs7158663 in hepatocellular carcinoma. Eur Rev Med
Pharmacol Sci. 26:11–21. 2022.PubMed/NCBI
|
|
38
|
Pei JS, Chang WS, Chen CC, Mong MC, Hsu
SW, Hsu PC, Hsu YN, Wang YC, Tsai CW and Bau DT: Novel Contribution
of Long Non-coding RNA MEG3 genotype to prediction of childhood
leukemia risk. Cancer Genomics Proteomics. 19:27–34. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hou Y, Zhang B, Miao L, Ji Y, Yu Y, Zhu L,
Ma H and Yuan H: Association of long non-coding RNA MEG3
polymorphisms with oral squamous cell carcinoma risk. Oral Dis.
25:1318–1324. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jeong YH and Zhao ZH: Correlation between
long noncoding RNA Meg3 polymorphism and risk of oral squamous cell
carcinoma. J Modern Stomatol. 33:328–332. 2019.(In Chinese).
|
|
41
|
Mirzazadeh S, Sarani H, Nakhaee A, Hashemi
SM, Taheri M, Hashemi M and Bahari G: Association between PAX8AS1
(rs4848320 C > T, rs1110839 G > T, and rs6726151 T > G)
and MEG3 (rs7158663) gene polymorphisms and non-Hodgkin lymphoma
risk. Nucleosides Nucleotides Nucleic Acids. 41:1174–1186. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang CT, Wu HY, Xia W, Cheng YP, Yang S,
Feng YL, Lu XY, Liang GY and Xu M: The association between
polymorphisms in long non-coding RNA MEG3 and the different
cervical lesions. Modern Oncol. 30:2804–2810. 2022.(In
Chinese).
|
|
43
|
Elhelaly Elsherbeny M, Ramadan Elsergany A
and Gamil Shaker O: Association of lncRNA MEG3 Rs7158663
polymorphism and serum expression with colorectal cancer in
egyptian patients. Rep Biochem Mol Biol. 12:102–111.
2023.PubMed/NCBI
|
|
44
|
Asadi A, Barati F, Nakhaee A, Jahantigh D,
Hashemi SM, Taheri M and Bahari G: Association between PRNCR1,
PAX8AS1, MEG3, and PTENP1 gene polymorphisms and breast cancer
risk. Per Med. 21:373–383. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y, Gao F and Lu J: MEG3 rs7158663
genetic polymorphism is associated with the risk of hepatocellular
carcinoma. Nucleosides Nucleotides Nucleic Acids. 44:531–541. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lao X, Wang Y, Huang R, He Y, Lu H and
Liang D: Genetic variants of LncRNAs HOTTIP and MEG3 influence
nasopharyngeal carcinoma susceptibility and clinicopathologic
characteristics in the Southern Chinese population. Infect Agent
Cancer. 19:322024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Abdi E, Latifi-Navid S, Kholghi-Oskooei V,
Mostafaiy B, Pourfarzi F and Yazdanbod A: Roles of the lncRNAs
MEG3, PVT1 and H19 tagSNPs in gastric cancer susceptibility. BMC
Cancer. 24:14402024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 Countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
50
|
Mattick JS, Amaral PP, Carninci P,
Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME,
Fitzgerald KA, et al: Long non-coding RNAs: Definitions, functions,
challenges and recommendations. Nat Rev Mol Cell Biol. 24:430–447.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bridges MC, Daulagala AC and Kourtidis A:
LNCcation: LncRNA localization and function. J Cell Biol.
220:e2020090452021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nandwani A, Rathore S and Datta M: LncRNAs
in cancer: Regulatory and therapeutic implications. Cancer Lett.
501:162–171. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ahmad M, Weiswald LB, Poulain L, Denoyelle
C and Meryet-Figuiere M: Involvement of lncRNAs in cancer cells
migration, invasion and metastasis: Cytoskeleton and ECM crosstalk.
J Exp Clin Cancer Res. 42:1732023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ahadi A: Functional roles of lncRNAs in
the pathogenesis and progression of cancer. Genes Dis. 8:424–437.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
He Y, Luo Y, Liang B, Ye L, Lu G and He W:
Potential applications of MEG3 in cancer diagnosis and prognosis.
Oncotarget. 8:73282–73295. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang J, Lin Z, Gao Y and Yao T:
Downregulation of long noncoding RNA MEG3 is associated with poor
prognosis and promoter hypermethylation in cervical cancer. J Exp
Clin Cancer Res. 36:52017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ghaedi H, Zare A, Omrani MD, Doustimotlagh
AH, Meshkani R, Alipoor S and Alipoor B: Genetic variants in long
noncoding RNA H19 and MEG3 confer risk of type 2 diabetes in an
Iranian population. Gene. 675:265–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Virolainen SJ, VonHandorf A, Viel K,
Weirauch MT and Kottyan LC: Gene-environment interactions and their
impact on human health. Genes Immun. 24:1–11. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Su H and Xie H: Associations between
lifestyle habits, environmental factors and respiratory diseases: A
cross-sectional study from southwest China. Front Public Health.
13:15139262025. View Article : Google Scholar : PubMed/NCBI
|