Introduction
Osteosarcoma (OS), a typical malignant bone tumor,
predominantly affects children and young adults. Notably progress
in the prevention and treatment of OS has been achieved through the
introduction of potential treatment approaches. However, the global
annual incidence of OS is ~3.4 cases per million population, with a
slight male predominance. Despite advancements in treatment, the
5-year survival rate for patients with localized disease is 60–70%,
which drops to 15–30% for those with metastatic or recurrent
disease. Mortality trends have shown an annual increase of
approximately 1.4% in certain populations (1–4). It
has also been reported that ~80% of patients with OS have high
programmed cell death ligand 1 (PD-L1) mRNA expression (5). Programmed cell death protein 1
(PD-1)/PD-L1 pathway inhibitors have been evaluated in clinical
trials for the treatment of bone and soft-tissue sarcomas; however,
there has been no notable responses recorded in patients with
advanced OS (6). The success of
immunotherapy is highly dependent on the level of immune cells
within the microenvironment of the host (7,8).
Therefore, assessing the immunoreactivity of OS is crucial in
making decisions about individualized treatment.
Ferroptosis, a novel type of cell death, is
typically accompanied by high levels of lipid peroxidation and iron
build-up (9). It serves a role in
numerous key pathological processes, including the development and
incidence of cancer (10).
Moreover, a previous study reported that ferroptosis serves a core
role in modulating genes involved in tumor immune escape (11). For example, the absence of
glutathione peroxidase 4, a key regulator of ferroptosis, prevents
an increase in the levels of CD8+ and CD4+ T
cells (11). Hence, determination
of immune infiltration-related ferroptosis genes for cancer
immunoreactivity could reveal novel insights into OS treatment.
In the present study, a single-sample Gene Set
Enrichment Analysis (ssGSEA) was used to assign patients with OS
into two clusters. A risk model was constructed using two immune
infiltration-related ferroptosis genes.
Materials and methods
Data abstraction
Clinical and pathological data from 87 patients with
OS were retrieved from the Therapeutically Applicable Research to
Generate Effective Treatments (TARGET) database (accession no.
phs000468.v22.p8) (https://www.cancer.gov/ccg/research/genome-sequencing/target)
(12). Furthermore, the expression
profile of the GSE21257 dataset (13) was downloaded from the Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), which contains data
from 53 human OS tissues. The gene probes of the platform were
transformed to gene names by referencing the GPL10295 platform
(14).
OS subgroup determination based on
immune gene sets
The 29 immune-related gene sets encompass several
types of immune cells, functions and pathways, were obtained from a
previously published study by Nguyen et al (15). For each overall survival dataset
(TARGET-OS database, phs000468.v22.p8), the level of immune-cell
infiltration in cancers was estimated using ssGSEA with the
GSVApackage (bioconductor.org/packages/release/bioc/html/GSVA.html)
in R language (R 4.2.2 with Bioconductor 3.16.). Subsequently,
tumor purity, immune score and stromal score were inferred using
the ESTIMATE R package [v1.0.13 Kosuke Yoshihara Lab (Academic)].
Consensus clustering and molecular subtyping were performed using
the ‘ConsensusClusterPlus’ R package
[bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.htmlv1.68.0(Bioconductor
release 3.19)], based on the ssGSEA scores. K-means clustering,
spectral clustering and PCA-k-means clustering were each replicated
50 times, and the optimal clustering outcome was utilized.
Differential expression analysis
Gene expression data from RNA-sequencing (RNA-seq)
were normalized using the voom transformation prior to differential
expression analysis. The ‘limma’ R package was used to screen the
differentially expressed genes (DEGs) between high immunity
(Immunity_H) and low immunity (Immunity_L) samples. DEGs were
identified based adjusted P<0.05 and (|log2FC|) >0.5. This
threshold was selected to capture moderate transcriptional changes
relevant to immune modulation in OS, where subtle gene expression
variations may markedly impact biological pathways. Whilst
stringent cutoffs (such as |log2FC| >1) are common, studies
indicate that immune-related genes often exhibit smaller yet
functionally critical expression shifts in tumor microenvironments
(16–18).
Moreover, the ‘pheatmap’ R package was used to plot
heatmaps, whilst ‘ggpubr’ was used to generate boxplots. FerrDb
(http://www.zhounan.org/ferrdb/) and
PubMed (https://pubmed.ncbi.nlm.nih.gov/) were used to obtain
data on the 283 ferroptosis genes. For each of the DEGs and
ferroptosis genes, a Venn diagram was used for overlapping.
Development of a prognostic model
based on ferroptosis genes
Prognosis-related ferroptosis genes were identified
using the univariate Cox regression (UCR) model. Subsequently, the
expression levels of these genes were combined, and the estimated
regression coefficients were used to calculate prognostic scores
for each patient. The following formula was utilized to determine
the risk score for each patient: Risk score=β1×1 + β2×2 + … + βixi.
Subsequently, based on the median risk score as the cut-off value,
samples were divided into high- and low-risk groups. The
Kaplan-Meier (K-M) method was employed to determine the survival
rates in each group. Variables were included in both UCR and
multivariate Cox regression (MCR) analyses to assess the
independence of the risk variables. Receiver operating
characteristic (ROC) curves were used to evaluate the accuracy and
specificity of the signature associated with immune
infiltration-related ferroptosis genes.
Gene ontology (GO) annotation and
pathway enrichment analysis
RStudio software (version 4.2; Posit Software, PBC;
rstudio.com/) was utilized to perform mRNA GO enrichment and Kyoto
Encyclopedia of Genes and Genomes (KEGG) functional analyses.
CIBERSORT estimation
CIBERSORT estimates the abundance ratios based on
gene expression data from several cell types within a mixed cell
population (19,20). In the present study, the CIBERSORT
algorithm was utilized to measure the percentage of immune cells.
Subsequently, patients were categorized into low- and high-risk
groups based on prognostic risk score for overall survival.
Gene set enrichment analysis
(GSEA)
GSEA was used to elucidate the different signaling
pathways between the two risk subgroups, and all datasets were used
for a GSEA using c2.cp.kegg.v7.0.symbols.gmt. Nominal P<0.05,
|normalized ES| Enrichment Score)>1 and false Discovery Rate)
q<0.25 were selected out.
Drug sensitivity analysis
Drug sensitivity data were retrieved from the
CellMiner database (https://discover.nci.nih.gov/cellminer/). The R
packages, ‘impute’, ‘limma’, ‘ggplot2’ and ‘ggpubr’ were used for
data processing, statistical analysis and result visualization.
Quantification of gene expression
using reverse transcription-quantitative PCR (RT-qPCR)
The relative mRNA expression levels of IFNG and
TLR4were measured in human OS cells lines, MG-63, 143B and U2OS,
and in the normal human osteoblast hFOB1.19 cell line. The hFOB1.19
cell line was maintained in DMEM/F12 medium (Gibco; Thermo Fisher
Scientific, Inc.) at 34°C with 5% CO2 in a humidified
atmosphere. Moreover, The human OS cell lines MG-63 and 143B cells
(iCell Bioscience Inc.) were cultured in MEM (Gibco; Thermo Fisher
Scientific, Inc.), while U2OS cells (iCell Bioscience Inc.) were
cultured in McCoy's 5A medium (cat. no. 16600082; Gibco; Thermo
Fisher Scientific, Inc.). Both culture media were supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 U/ml streptomycin (100 µg/ml) at 37°C with 5%
CO2 in a humidified environment.
Total RNA was extracted using TRIzol reagent (Thermo
Fisher Scientific, Inc.) and subsequently used to synthesize cDNA
with SuperScript™ II Reverse Transcriptase
(Invitrogen™; Thermo Fisher Scientific, Inc.),
incorporating 5 µg oligo (dT) primers per sample. RT was performed
under the following temperature conditions: 25°C for 10 min (primer
annealing), followed by 42°C for 50 min (cDNA synthesis), and 70°C
for 15 min (enzyme inactivation). qPCR was performed using
SYBR® Green PCR Master Mix (Applied
Biosystems®; Thermo Fisher Scientific, Inc.) in a total
volume of 20 µl with the 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions were set at 95°C for 5 min, followed by 40 cycles of
95°C for 30 sec and 60°C for 45 sec. Melt-curve analysis was used
to confirm the specificity of the amplification, and GAPDH served
as the endogenous control for normalization of the amount of total
RNA in each group. The relative gene expression was calculated in
fold change according to the 2−ΔΔCq method (21), repeated independently in triplicate.
The primer sequences were designed as follows: Interferon γ (IFNG)
forward, 5′-TGCCTGCAATCTGAGCCAGT-3′ and reverse,
5′-TGGAAGCACCAGGCATGAAA-3′; toll-like receptor 4 (TLR4) forward,
5′-TAGCGAGCCACGCATTCACA-3′ and reverse, 5′-TAGGAACCACCTCCACGCAG-3′;
and GAPDH forward, 5′-GACCTGACCTGCCGTCTA-3′ and reverse,
5′-AGGAGTGGGTGTCGCTGT-3′.
Authentication testing of the MG-63, 143B, U2OS and
hFOB1.19 cell lines was performed (Shanghai Biowing Applied
Biotechnology Co., Ltd.) via short tandem repeat (STR) profiling.
The STR profiles matched the standards recommended for the
authentication of the MG-63, 143B, U2OS and hFOB1.19 cell
lines.
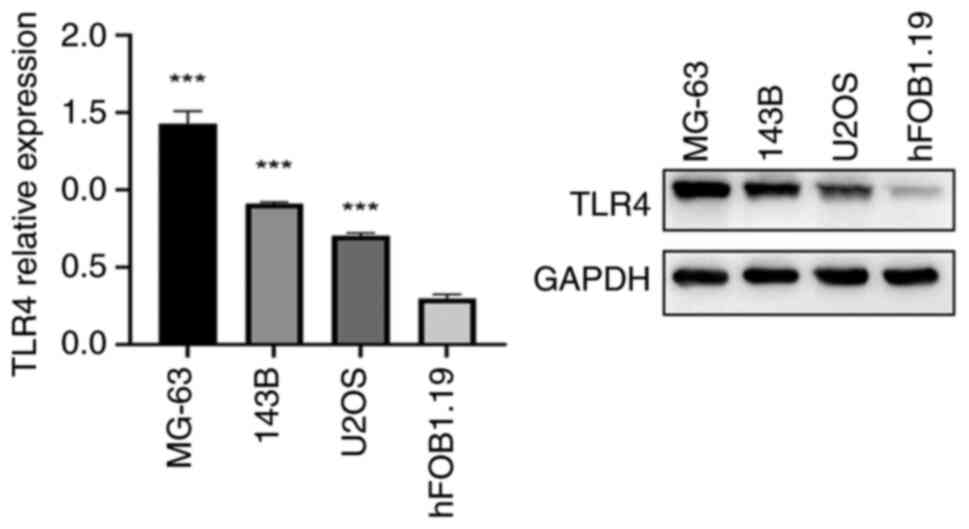
Semi-quantification of gene expression
using western blot analysis
Total proteins extracted from human OS (MG-63, 143B
and U2OS) and the normal human osteoblast hFOB1.19 cell line were
harvested using ice-cold radioimmunoprecipitation lysis buffer
(Thermo Fisher Scientific, Inc.) supplemented with
phenylmethanesulfonyl fluoride for 1 h. Following the
manufacturer's instructions, a bicinchoninic acid protein assay kit
(Sigma-Aldrich; Merch KGaA) was used to determine the amount of
protein. A total of 20 µg protein/lane was separated on 12%
SDS-PAGE (Beyotime Biotechnology) and transferred onto PVDF
membranes (MilliporeSigma; Merck KGaA). The membranes were blocked
with 5% skimmed milk at room temperature for 1 h, and washed in
Tris-buffered saline Tween-20 [150 mmol/l NaCl (pH 7.5), 20 mmol/l
Tris-HCl and 0.1% Tween 20] at room temperature three times.
Primary monoclonal antibodies against TLR4 (1:1,000; cat. no.
BA1717; Wuhan Boster Biological Technology, Ltd.) and the loading
control, GAPDH (1:10,000; cat. no. AB0037; Shanghai Abways
Biotechnology Co., Ltd.) were incubated with the membranes
overnight at 4°C. Membranes were incubated with horseradish
peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody
(1:10,000 dilution; cat. no. BE0101; bioeasy) for 1 h at room
temperature. Protein bands were visualized using ECL) reagent (cat.
no. T4600; Tianneng Technologies) according to the manufacturer's
instructions.
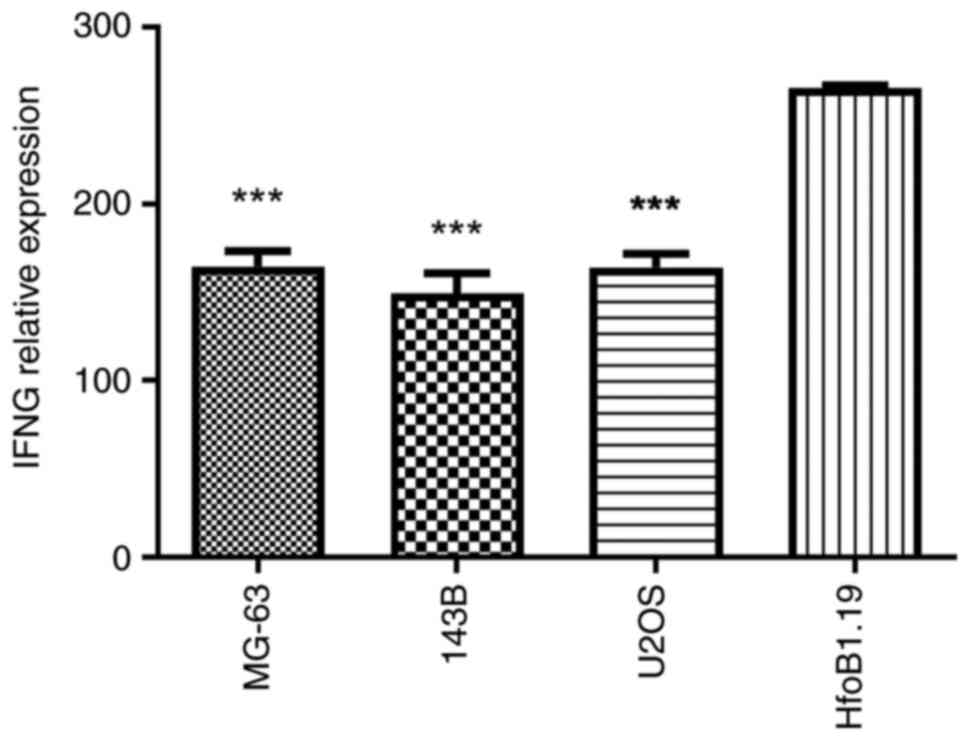
Detection of IFNG expression using
ELISA
The levels of the anti-fibrotic factor IFNG in
MG-63, 143B, U2OS and normal human osteoblasts were assessed using
the Human Interferon γ ELISA Kit (cat. no. H0108c; Wuhan
Elabscience Biotechnology Co., Ltd.) according to the
manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism (version 8.0; Dotmatics) or SPSS (version 24.0; IBM Corp.).
Data are presented as the mean ± standard deviation of ≥3
independent experimental repeats. For linear correlation, Pearson's
correlation coefficient was used. Comparisons between multiple
groups were performed using one-way analysis of variance followed
by Tukey's post hoc test. Overall survival was determined using K-M
survival curves along with the log-rank test. P<0.05 was
considered to indicate a statistically significant difference. For
comparisons between two groups, statistical significance was
assessed using the Mann-Whitney U test for non-normally distributed
data (immune cell infiltration proportions and immune checkpoint
gene expression). P<0.05 was considered to indicate a
statistically significant difference.
Results
Immune subtypes identified in OS
A total of 87 patients with OS were included in the
present study. ssGSEA was performed on each sample using gene sets
comprised of mRNA transcripts specific to most subpopulations of
both innate and adaptive immune cells. All tumor samples were
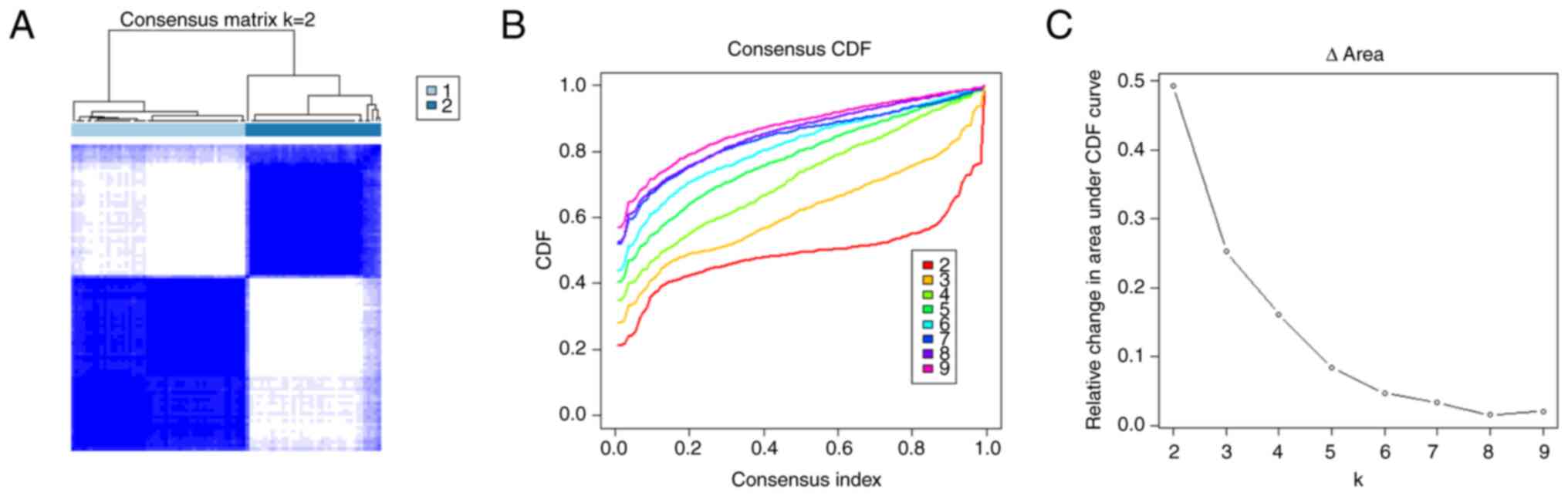
categorized into k subtypes, with k-values of 2–9. Based on the
consensus score of the cumulative distribution function curve, k=2
was determined to be the optimal number of subtypes (referred to as
Immunity_H and Immunity_L; Fig.
1).
Immune features of the three immune
subtypes
The ssGSEA clustering analysis indicated that the
extent of immune infiltration was higher in the Immunity_H group
than in the Immunity_L group (Fig.
2A). The overall survival cases in the Immunity_H group
exhibited significantly lower tumor purity and higher ESTIMATE,
stromal and immune scores compared with those in the Immunity_L
group (Fig. 2A; P<0.005).
Additionally, the expression of most human leukocyte antigens was
significantly higher in the high immune cell infiltration cluster
(Immunity_H group) compared with the low immune cell infiltration
cluster (Immunity_L group). (P<0.05; Fig. 2B). The Immunity_H subtype showed
significantly increased infiltration of CD8+ T cells,
memory activated CD4+ T cells, M2 macrophages and M1
macrophages compared with the Immunity_L subtype (P<0.05;
Fig. 2C). However, the Immunity_H
subtype exhibited significantly lower infiltration of M0
macrophages and natural killer (NK) cells resting compared with the
Immunity_L subtype (P<0.05; Fig.
2C). Furthermore, the expression of the following eight immune
checkpoint genes associated with immune escape were assessed:
lymphocyte-activation gene 3 (LAG3), V-set immunoregulatory
receptor, CD274, hepatitis A virus cellular receptor 2 (HAVCR2),
killer cell immunoglobulin like receptor (KIR)-two Ig domains and
long cytoplasmic tail 1, killer cell lectin like receptor C1,
5′-nucleotidase ecto, cytotoxic T-lymphocyte associated protein 4
(CTLA4), T cell immunoreceptor with Ig and ITIM domains (TIGIT),
KIR-three Ig domains and long cytoplasmic tail 2, KIR-two Ig
domains and long cytoplasmic tail 3 and programmed cell death 1
(PDCD1). The expression levels of these genes were significantly
higher in the Immunity_H group than in Immunity_L group (P<0.05;
Fig. 2D). Finally, K-M analysis
indicated that patients with OS in the Immunity_H cluster had a
significantly improved survival rate compared with those in the
Immunity_L cluster (P=0.0171; Fig.
2E).
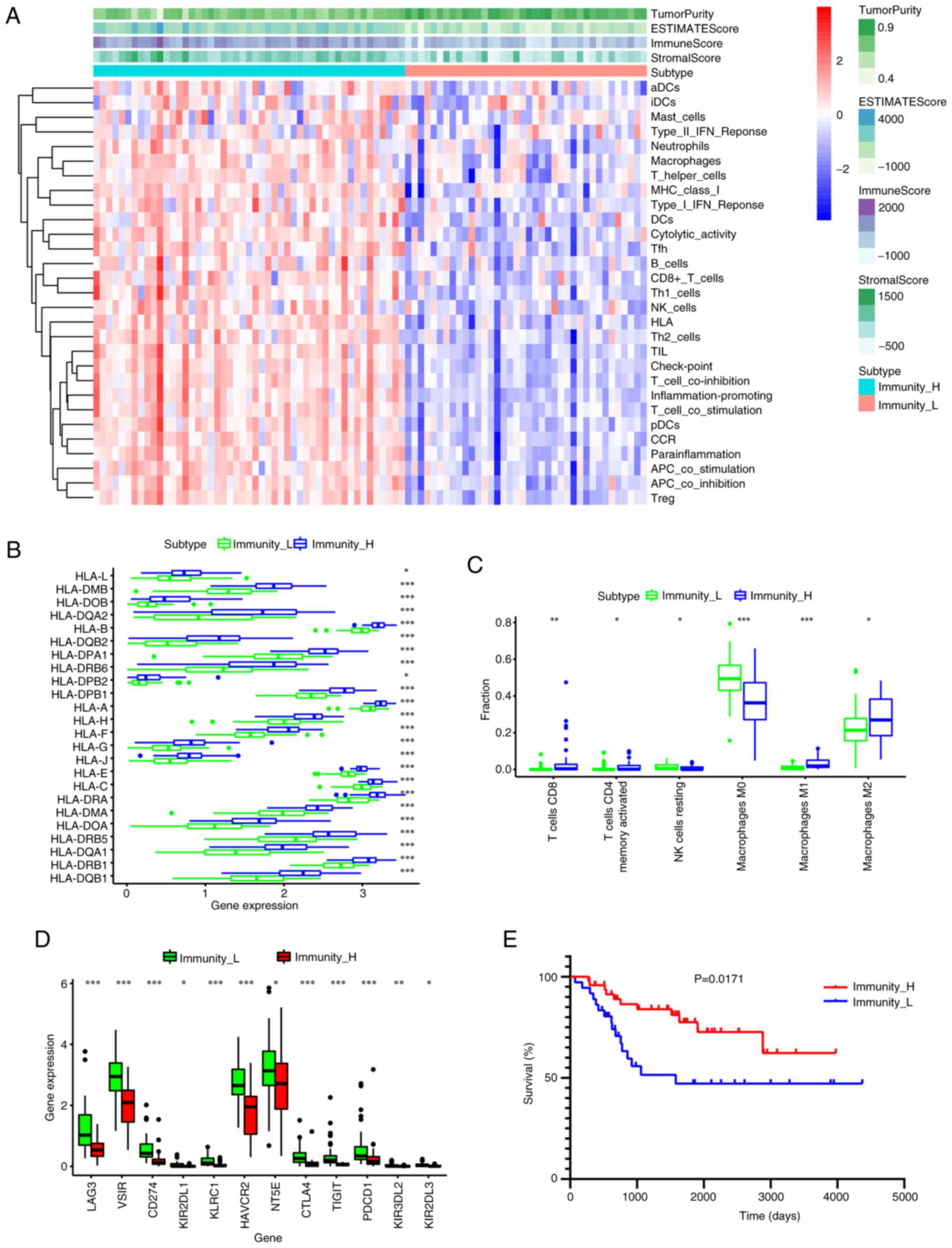 | Figure 2.Determination of the two immune
subtypes in osteosarcoma. (A) Heatmap of single-sample Gene Set
Enrichment Analysis scores. (B) There was a significant difference
in the expression of most HLAs between the high- and low-immune
cell infiltration clusters. (C) Difference in the expression levels
of infiltrating immune cells between the two clusters. (D)
Difference in the expression levels of the immune checkpoint genes,
LAG3, VSIR, CD274, HAVCR2, NT5E, CTLA4, TIGIT and PDCD1, among the
two subtypes. (E) Patients in low immune cell infiltration cluster
showed worse overall survival than those in high immune cell
infiltration cluster. *P<0.05; **P<0.01; ***P<0.005. LAG3,
lymphocyte-activation gene 3; VSIR, V-set immunoregulatory
receptor; HAVCR2, hepatitis A virus cellular receptor 2; NT5E,
5′-nucleotidase ecto; CTLA4, cytotoxic T-lymphocyte associated
protein 4; TIGIT, T cell immunoreceptor with Ig and ITIM domains;
PDCD1, programmed cell death 1. |
Identification of DEGs and functional
enrichment analyses
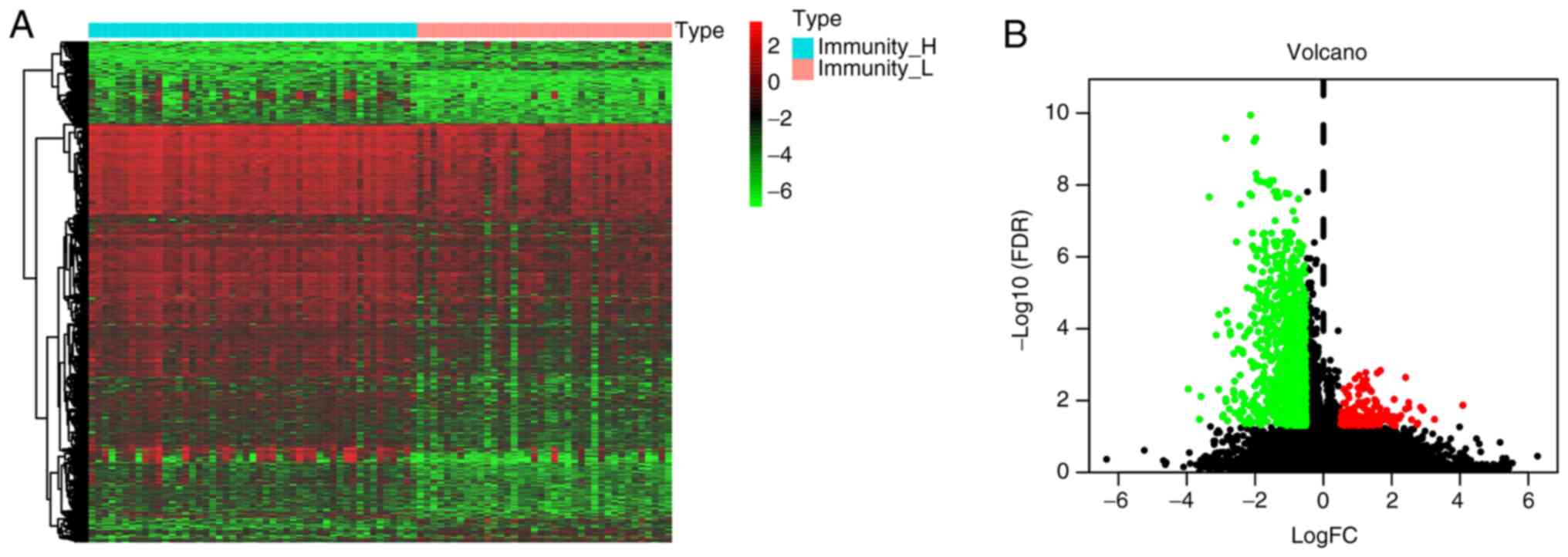
The results revealed that Immunity_H represents an
immune-infiltration cluster, whilst Immunity_L signifies a cluster
with low immune cell infiltration. Consequently, differentially
expressed immune infiltration-related genes were identified from
both Immunity_H and Immunity_L groups. In total, 1,240 DEGs were
identified, comprising 159 upregulated genes and 1,081
downregulated genes (Fig. 3).
Immune infiltration-related
ferroptosis genes and functional enrichment analyses
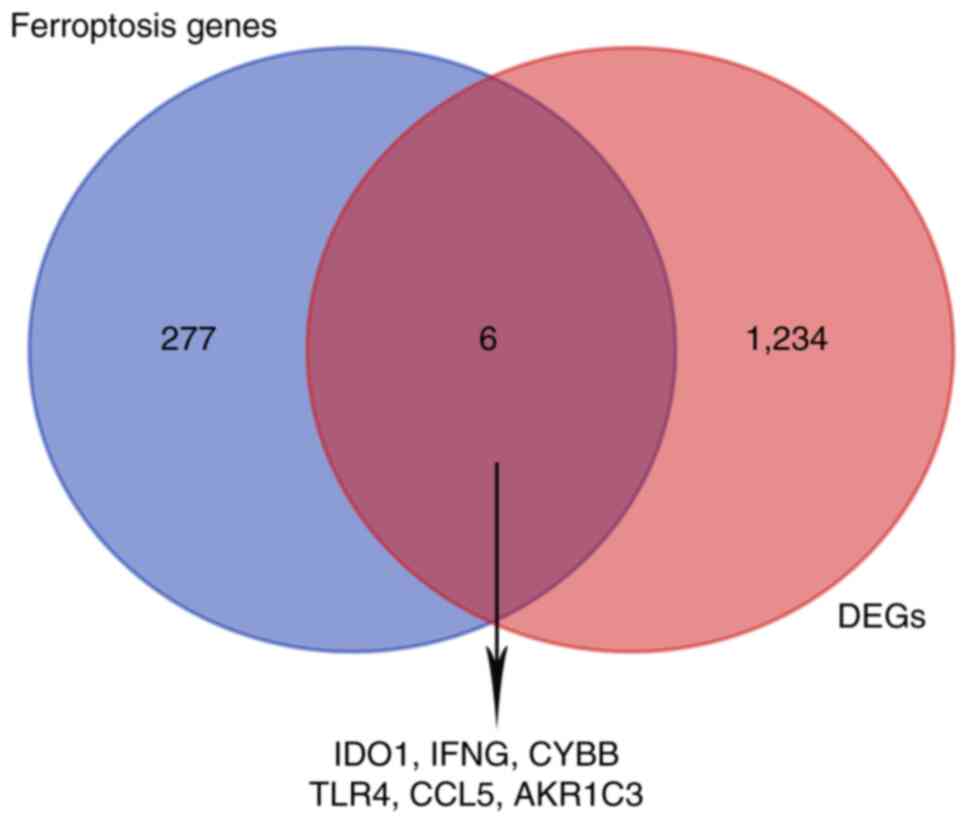
Immune infiltration-related genes that were
differentially expressed were intersected with 283 ferroptosis
genes. A total of six overlapping genes were screened for
subsequent analyses (Fig. 4).
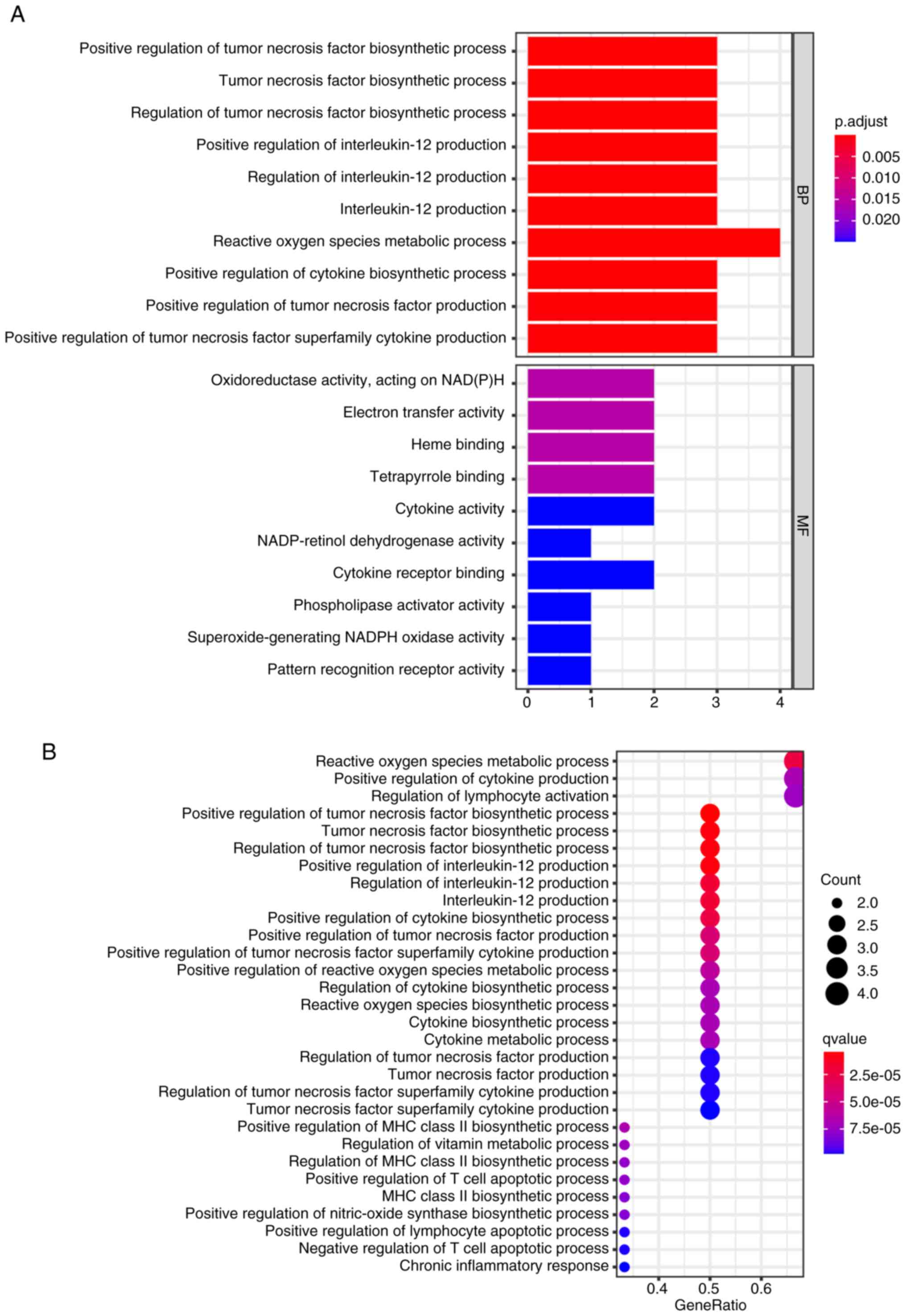
Additional investigations were performed into the potential
functions of the genes by employing GO and KEGG analyses. The GO
terms were mainly enriched in positive regulation of the tumor
necrosis factor biosynthetic process, reactive oxygen species
metabolic process, regulation of tumor necrosis factor biosynthetic
process and positive regulation of interleukin-12 production
(Fig. 5A). Additionally, the KEGG
pathway analysis (Fig. 5B)
demonstrated that the hypoxia-inducible factor 1 signaling pathway,
nucleotide-binding and oligomerization domain-like receptor
signaling pathway in cancer, PD-L1 expression and PD-1 checkpoint
pathway were also enriched in the PD-L1-positive and toll-like
receptor-signaling pathways (Fig.
5B). The results indicate that the inflammatory and
immunological responses may be involved in the development of OS
tumorigenesis.
Construction of the diagnostic and
prognosis model based on two ferroptosis genes
UCR analysis was performed with OS prognosis as the
dependent variable, and P<0.05 was considered to indicate a
statistically significant difference. TLR4 was revealed to have a
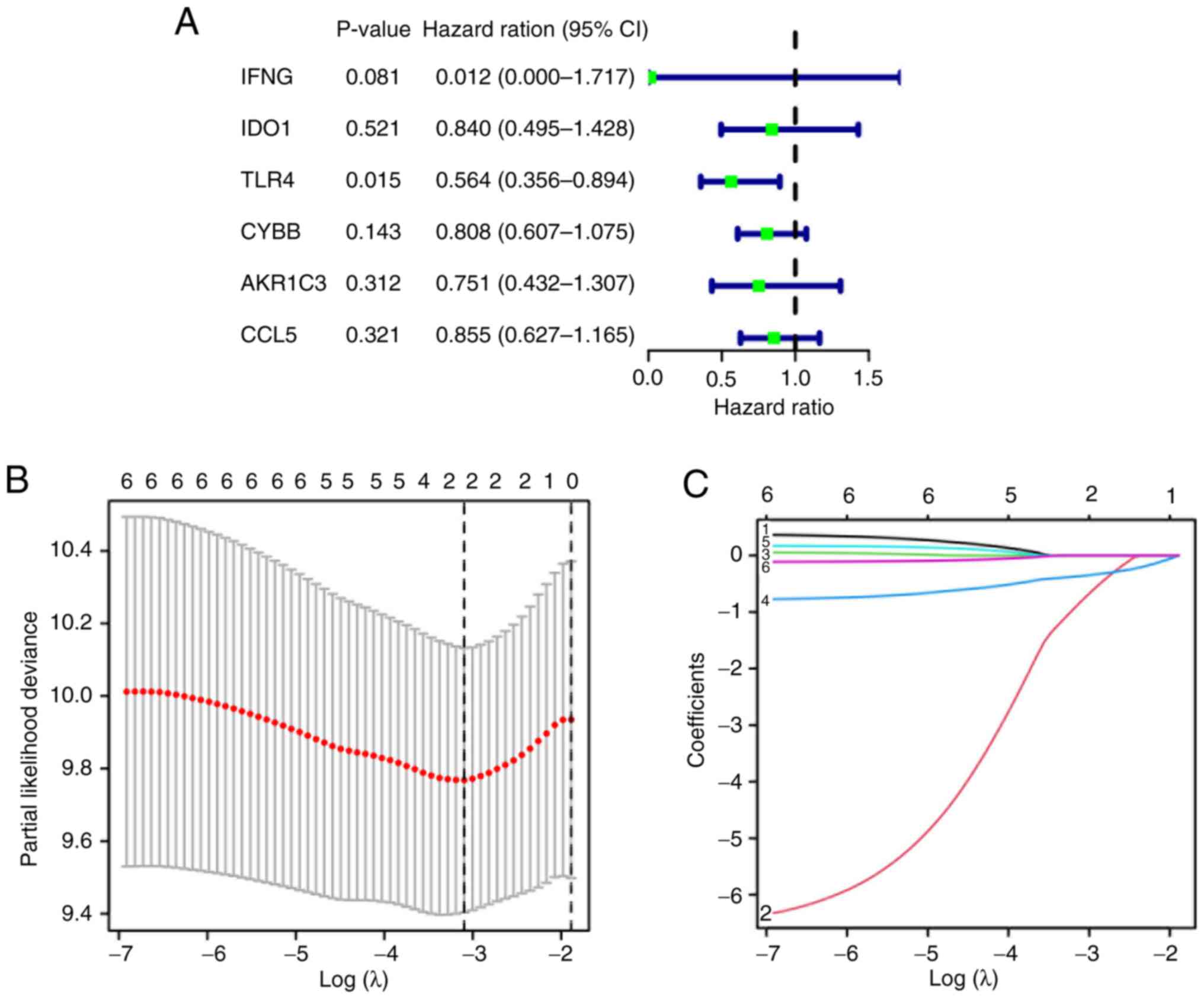
notable association with overall survival (P<0.05; Fig. 6A). Subsequently, Least Absolute
Shrinkage and Selection Operator (LASSO) regression analysis
identified two genes, IFNG and TLR4 (Fig. 6B and C). After which, a prognostic
signature was developed based on the following formula: Risk
score=(−0.3623 × TLR4 expression value) + (−0.8072 × IFNG
expression value). Patients were stratified into high-(n=43) and
low-(n=44) risk groups. The high-risk group exhibited a
significantly worse prognosis compared with the low-risk group
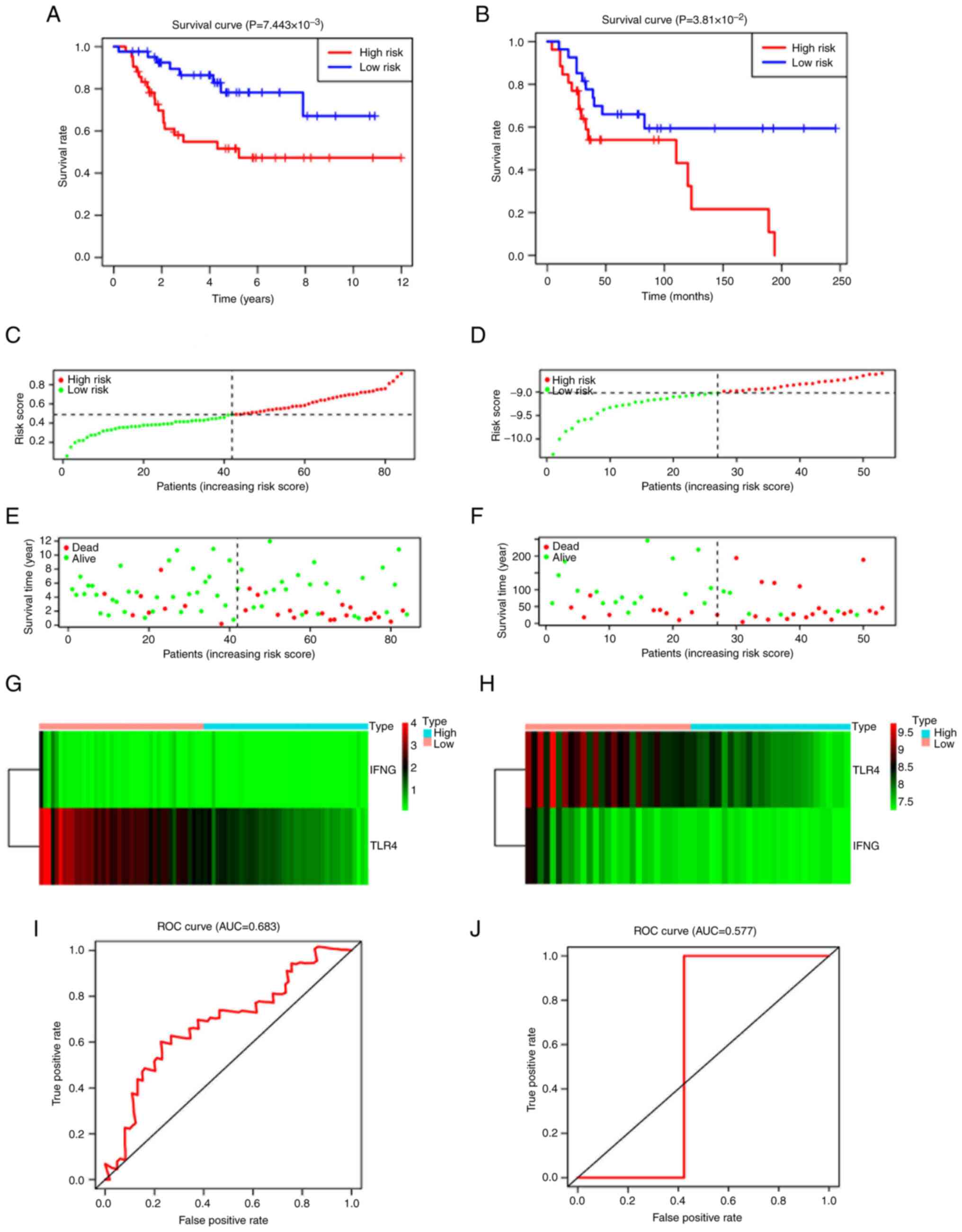
(P=7.443×10−3; Fig. 7A).
Moreover, ROC curve analysis demonstrated that the Cox prediction
model achieved an area under the curve (AUC) of 0.683 for survival
prediction at 3 years (Fig. 7I),
showing improved accuracy and suggesting moderate predictive
capability. The riskScore plot, status plot and the expression of
the two signature genes are presented in Fig. 7C, E and G. The GEO dataset was then
used to assess the robustness of the model. Compared with patients
in the low-risk group, patients in the higher risk group had a
shorter survival period, which was in-line with the previous
results (P=7.443×10−3; Fig.
7B); the AUC of ROC was 0.577 (Fig.
7J). The detailed risk scores, status plot and
ferroptosis-related gene expression are presented in Fig. 7B, D, F and H.
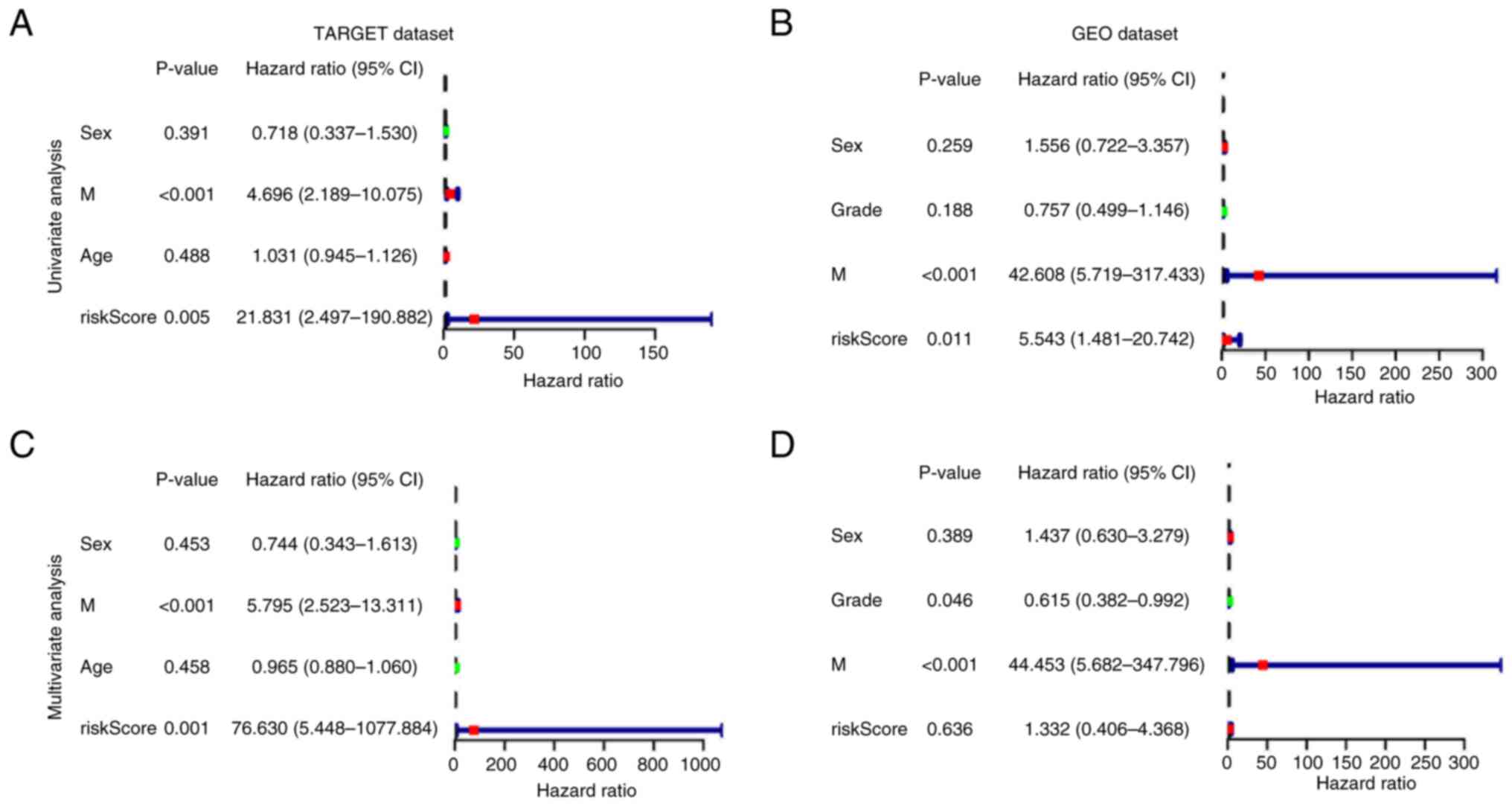
Additionally, to assess whether the signature was
independent of other clinical characteristics, UCR and MCR analyses
were performed in two cohorts. UCR indicated that the metastasis
status and risk score had a significant influence on prognosis in
both datasets (Fig. 8A and B). In
MCR analysis, the risk score derived from the signature was an
independent prognostic factor in the TARGET cohort [hazard ratio
(HR), 76.630; 95% confidence interval (CI), 5.448–1077.884;
P<0.001]. However, the risk score was not an independent
prognostic factor in the GEO cohort (HR, 1.332; 95% CI,
0.406–4.368; P=0.636; Fig. 8C and
D); however, this may be due to the limited number of samples
used (n=53).
Assessment of the diverse
tumor-infiltrating immune cell (TIIC) types between low- and
high-risk cohorts
Using CIBERSORT, the immune infiltration percentage
of each cell type was calculated using the gene matrix as a basis.
In patients in the high-risk group, there was a significantly
increased number of M0 macrophages (P=0.00074; Fig. 9A). By contrast, the low-risk group
exhibited significantly higher levels of M1 macrophages (P=0.006;
Fig. 9B) and M2 macrophages
(P=0.017; Fig. 9C). Furthermore,
the expression of immune checkpoint genes in the low-risk group,
including LAG3, CD274, CTLA4, HAVCR2, TIGIT, HAVCR2 and PDCD1 was
higher than that in the high-risk group (P<0.05; Fig. 9G). Additionally, consistent results
were obtained using the GEO cohort (Fig. 9D-F and H; P<0.005).
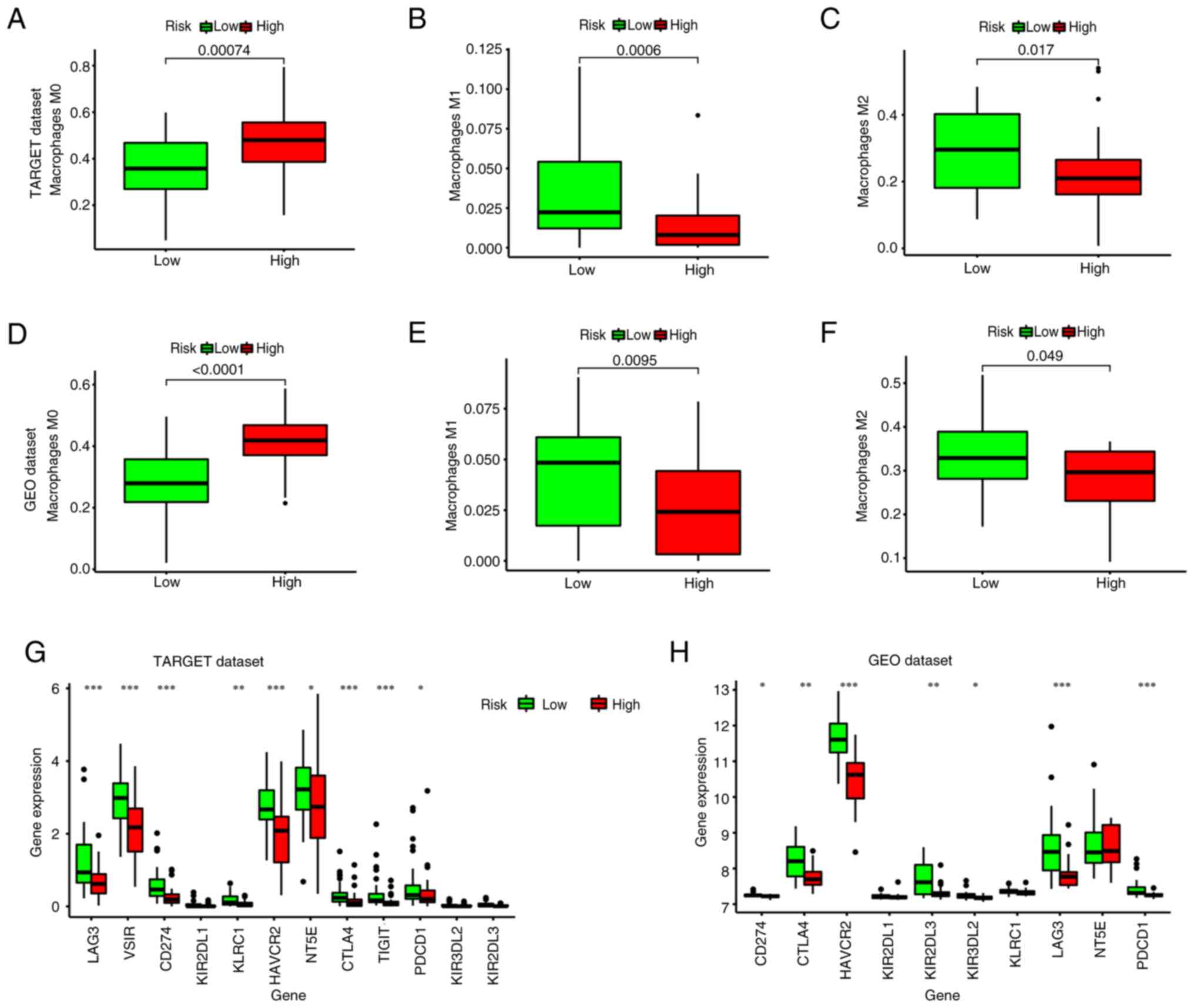 | Figure 9.Analysis of immune cell infiltration
in patients with osteosarcoma. Differences in the number of (A) M0,
(B) M1 and (C) M2 macrophages between the high- and low-risk groups
in the TARGET dataset. Differences in the number of (D) M0, (E) M1
and (F) M2 macrophages between the high- and low-risk groups in the
GEO dataset. Differences in the expression of immune checkpoint
genes among the high- and low-risk groups in the (G) TARGET and (H)
GEO datasets. *P<0.05; **P<0.01; ***P<0.005. TARGET,
Therapeutically Applicable Research to Generate Effective
Treatments; GEO, Gene Expression Omnibus; LAG3,
lymphocyte-activation gene 3; VSIR, V-set immunoregulatory
receptor; KIR2DL1, killer cell Immunoglobulin-like Receptor, two Ig
domains, Long cytoplasmic tail, inhibitory receptor 1; KLRC1,
Killer cell Lectin-like Receptor subfamily C member 1; HAVCR2,
hepatitis A virus cellular receptor 2; NT5E, 5′-nucleotidase ecto;
CTLA4, cytotoxic T-lymphocyte associated protein 4; TIGIT, T cell
immunoreceptor with Ig and ITIM domains; PDCD1, programmed cell
death 1. |
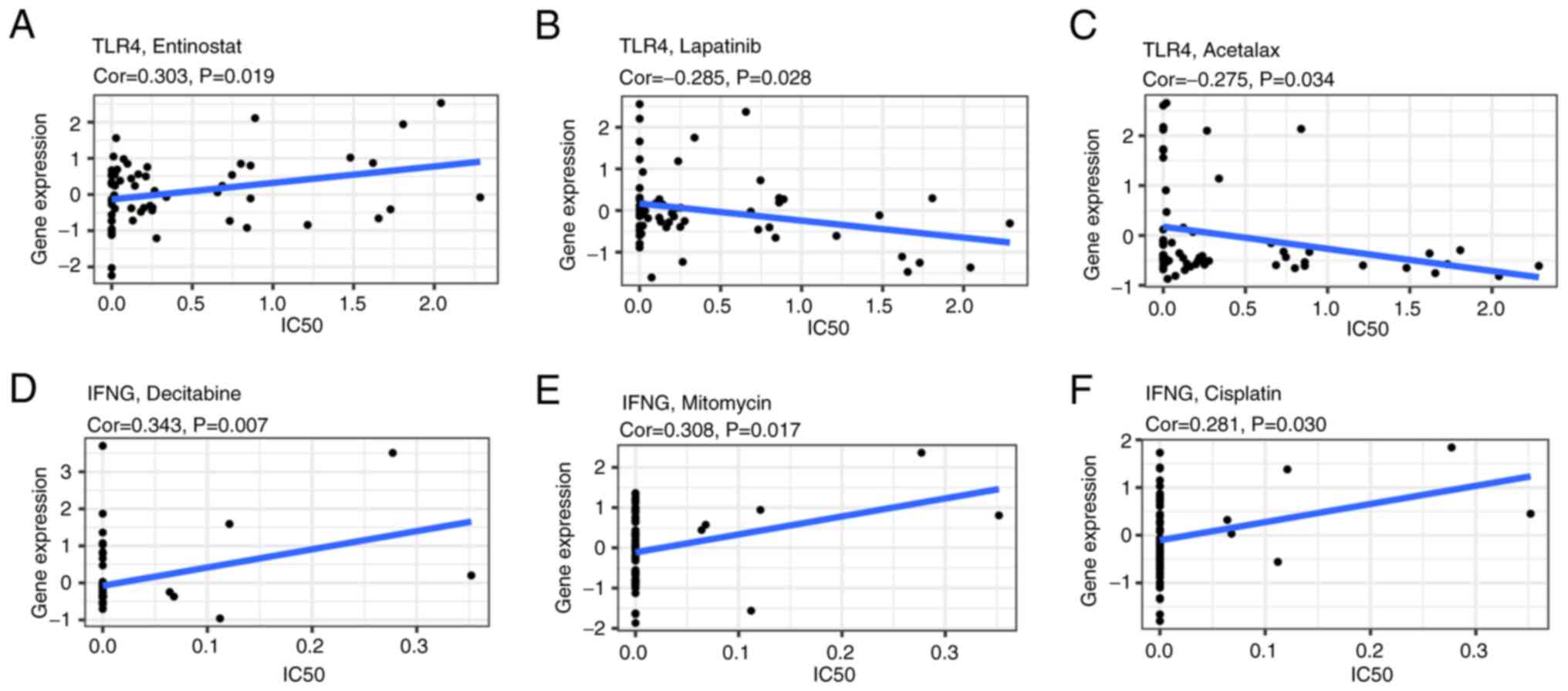
Drug sensitivity analysis of IFNG and
TLR4
To assess the underlying correlation between IFNG
and TLR4 gene expression and drug sensitivity in several human
cancer cell lines from the CellMiner database, a correlation
analysis was performed. TLR4 was demonstrated to be significantly
correlated with the drug sensitivity of entinostat (correlation
coefficient=0.303; P=0.019; Fig.
10A), lapatinib (correlation coefficient=−0.285; P=0.028;
Fig. 10B) and acetalax
(correlation coefficient=−0.275; P=0.034; Fig. 10C). Additionally, IFNG expression
was significantly correlated with the drug sensitivity of
decitabine (correlation coefficient=0.343; P=0.007; Fig. 10D), mitomycin (correlation
coefficient=0.308; P=0.017; Fig.
10E) and cisplatin (correlation coefficient=0.281; P=0.030;
Fig. 10F).
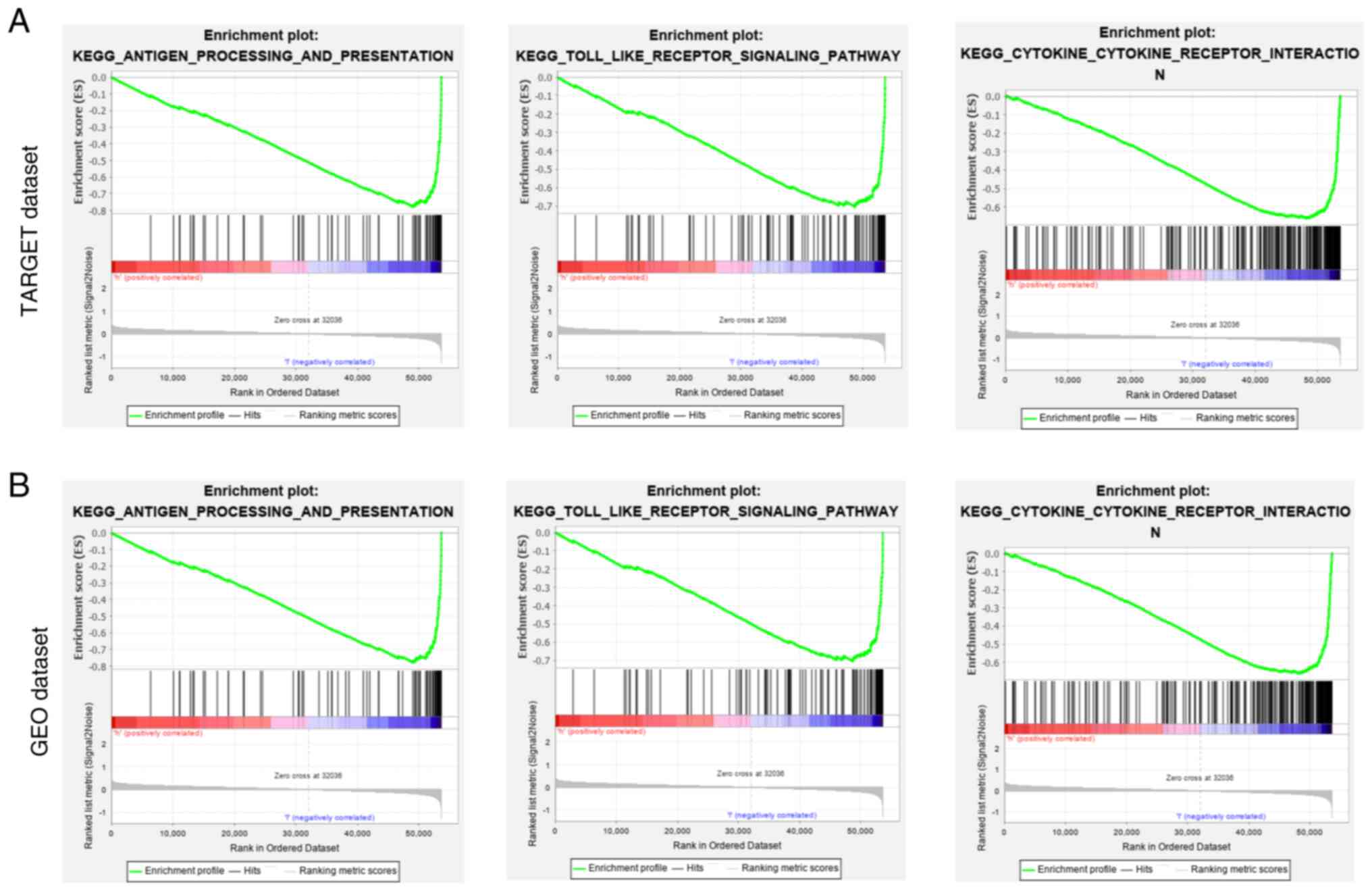
Association between risk and
immune-related pathways
GSEA indicated that, compared with the low-risk
group, the high-risk group was negatively associated with
immune-related pathways, such as antigen processing and
presentation, the Toll-like receptor signaling pathway and
cytokine-cytokine receptor interaction (Fig. 11).
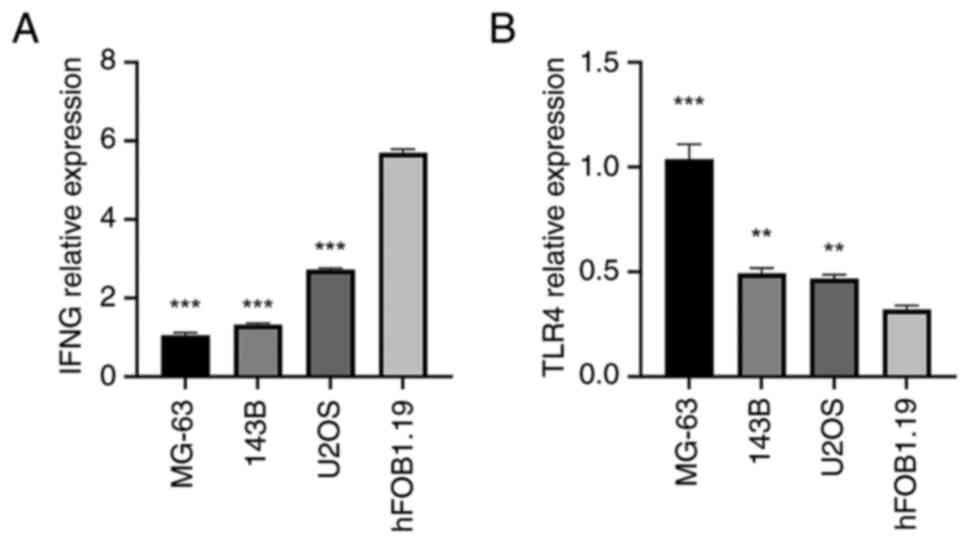
Validation of the expression level of
immune infiltration-related ferroptosis genes (IFNG and TLR4) in
hFOB1.19, MG-63, 143B and U2OS cell lines
To confirm that the two immune infiltration-related
ferroptosis genes, IFNG and TLR4, are specifically expressed in OS
cells, RT-qPCR was performed using the MG-63, 143B, U2OS and normal
human osteoblast (hFOB1.19) cell lines. Both IFNG and TLR4 were
expressed in MG-63, 143B and U2OS cells, with TLR4 expression
significantly higher in these OS cell lines than in hFOB1.19 cells.
By contrast, IFNG expression in the MG-63, 143B and U2OS cells was
significantly lower than in hFOB1.19 cells than in the OS cell
lines (Fig. 12). The same pattern
was observed in the results of the western blot analysis and ELISA
(Figs. 13 and 14). Consequently, this cellular
experiment corroborates the reliability of the bioinformatics
analysis results.
Discussion
Human OS, a primary malignant bone tumor, is most
prevalent during adolescence and in children (22). With the rapid development of
molecular biology technology, there has been a growing interest in
anticancer immunotherapies, including immune modulators, immune
checkpoint inhibitors and genetically modified T cells (23,24).
Patients with OS lacking immune cell infiltration present with high
rates of metastasis and poor clinical outcomes (25). Notably, immune reconstitution has
been reported to suppress OS recurrence and improve survival in
metastatic OS (26).
Recent studies have reported that
ferroptosis-related genes exhibit cell-type-specific expression in
immune cells, directly influencing their functional roles. TLR4 was
reported to be predominantly expressed in myeloid cells
(macrophages and dendritic cells). Moreover, its upregulation in M2
macrophages was reported to promote iron retention via ferritin
heavy chain 1 and suppress lipid peroxidation, enhancing
anti-inflammatory functions (27,28).
IFNG is secreted by activated CD8+ T cells and NK cells.
It induces tumor ferroptosis by downregulating the solute carrier
family 7 member 11/glutathione peroxidase 4 pathway whilst
upregulating acyl-CoA synthetase long-chain family member 4
(ACSL4) (29,30). In OS, TLR4 likely stabilizes iron
homeostasis in stromal macrophages, whilst IFNG from infiltrating T
cells sensitizes tumor cells to ferroptosis. This spatial crosstalk
creates a feed-forward loop where ferroptotic cells release
damage-associated molecular patterns [DAMPs; such as high-mobility
group box 1 (HMGB1)], further recruiting dendritic cells and
amplifying antitumor immunity (30–32).
Based on the immune signature score, patients with
OS were categorized into two clusters. Significant differences were
observed in tumor purity, ESTIMATE scores, stromal scores, immune
scores and the expression of immune checkpoint markers between the
Immunity_H and Immunity_L groups. Patients in the Immunity_H group
exhibited greater immune infiltration compared with those in the
Immunity_L group.
A prognosis model incorporating two
ferroptosis-related genes was utilized to develop a risk model for
predicting the overall survival of patients with OS. The risk score
was then used to classify patients into two distinct risk groups.
The reliability of the identified immune subtypes (Immunity_H/L)
was supported by the internal consensus clustering stability within
the TARGET cohort (Fig. 1). Whilst
the identical immunotyping procedure was not replicated on the
external GEO validation cohort due to platform heterogeneity, the
successful validation of the downstream immunotype-derived
ferroptosis prognostic model (IFNG/TLR4 risk score) in this
independent dataset (Fig. 7F-J)
provides indirect support for the biological relevance captured by
the initial subtyping approach. Furthermore, the K-M curve, score
plot, survival status plot and ROC curve indicated that the two
immune-related risk levels had a favorable predictive potential for
prognosis.
Notably, the multivariate Cox regression analysis,
incorporating clinical factors such as age, sex and metastasis
status, demonstrated that the ferroptosis-based risk score remained
an independent prognostic predictor in the primary cohort (TARGET),
suggesting its value beyond these potential confounders. However,
in the smaller GEO cohort, the risk score did not reach independent
significance in the multivariate model (P=0.636), which may be due
to the limited sample size (n=53).
To address how ferroptosis influences the tumor
immune microenvironment, the following mechanistic insights are
proposed based on recent studies: i) Ferroptosis modulates
myeloid-derived suppressor cells (MDSCs) and T-cell activity; Lnk
deficiency in MDSCs triggers ferroptosis via the FMS-like tyrosine
kinase 3/STAT1/interferon regulatory factor-1/arachidonate
(12S)-lipoxygenase axis, reducing Arginase-1 expression and
increasing TNF-α, thereby weakening MDSC-mediated immunosuppression
(33). This aligns with the results
of the Immunity_H cluster (enriched in CD8+ T cells/M1
macrophages) and improved survival rates; ii) with a dual role in
T-cell function, CD8+ T cells promote tumor ferroptosis
via IFN-γ-mediated xCT suppression, but are themselves
vulnerable to ferroptosis due to ROS accumulation (34,35).
This may explain the protective role of IFNG in the model in
the present study, and its downregulation in OS cells; iii)
macrophage polarization: M1 macrophages exhibit ferroptosis
sensitivity via ACSL4, whilst M2 macrophages resist it via
ferroptosis suppressor protein 1 (35). The elevated M1/M2 infiltration in
the low-risk group in the present study may reflect
ferroptosis-driven plasticity, supported by enriched HIF-1/TLR
pathways in KEGG; iv) immunogenic cell death: Ferroptotic cells
release DAMPs (such as HMGB1) that activate dendritic cells and
T-cell priming (36,37). This may have driven the compensatory
immune checkpoint upregulation (PD-L1/CTLA4/LAG3) in the low-risk
group; and v) TLR4/IFNG bridge ferroptosis and immunity: TLR4
inhibits ferroptosis via ROS/p38-MAPK suppression, potentially
explaining its high-risk association. Conversely, IFNG
enhances tumor ferroptosis but risks T-cell depletion (38), creating a feedback loop evident in
the cellular data in the present study.
IFNG was excessively expressed in the low-risk
group. Moreover, the circulating concentrations of IFNG have been
reported to be markedly associated with overall survival in
patients with OS (39).
Furthermore, IFNG serves a crucial role in the antitumor immune
response by enhancing antigen presentation, T-lymphocyte
differentiation and maturation across several cancers (40). For example, IFN-γ, encoded by the
IFNG gene, acts on tumor cells, enhancing their recognition by
CD8+ T cells as well as by CD4+ T cells.
IFN-γ can promote tumor cell apoptosis by upregulating the
expression of caspase-1, −3 and −8. An important aspect is the
ability of IFN-γ to induce PD-L1 expression in cancer, stromal and
myeloid cells, which impairs effector tumor immunity (41,42).
The aforementioned research indicates that IFNG is involved in
tumor immunity, which aligns with the findings of the present
study.
TLR4 is known for its function in recognizing
pathogen-associated antigens and activating the production of
pro-inflammatory factors (43). It
has previously been reported to serve a role in lung tumorigenesis
induced by chemically induced pulmonary inflammation (44). Furthermore, the inhibition of TLR4
has been reported to mitigate oxygen-glucose deprivation-induced
ferroptosis by suppressing oxidative stress and p38-MAPK signaling,
ultimately enhancing neuronal cell viability (38). Moreover, the present study
investigated the association between IFNG/TLR4 expression and drug
sensitivity using publicly available pharmacogenomic databases
(GDSC or CTRP). Spearman correlation analysis revealed that higher
IFNG expression was significantly associated with increased
sensitivity to entinostat (r=0.32, P<0.05) and decitabine
(r=0.28, P<0.05), both epigenetic modulators. Conversely,
elevated TLR4 expression correlated with resistance to cisplatin
(r=−0.35, P<0.05), a conventional chemotherapeutic agent
commonly used in OS treatment. These findings suggest that our
ferroptosis-based risk signature may not only have prognostic value
but also potential implications for predicting treatment
response.
Considering the pivotal role of TIICs in the
progression of cancers, the diverse types of TIICs were explored
between low- and high-risk cohorts in the present study. A previous
investigation reported that resting memory CD4+ T cells
are among the most enriched TIICs in gastric cancer samples
(45). In addition, studies have
reported the presence of follicular helper T cells in tertiary
lymphoid structures of numerous tumors, suggesting that they serve
a role in generating effective and sustained antitumor immune
responses (46). Macrophages are
considered to serve as both antitumor agents (M1 macrophages) and
protumor agents (M2 macrophages) (47). In the present study, patients in the
high-risk group exhibited an elevated level of M0 macrophages. By
contrast, patients in the low-risk group showed increased
proportions of both M0 and M2 macrophages. Immune checkpoint
molecules, including CD274, CTLA4 and LAG3, have been demonstrated
to serve a role in tumor immunology (48). The targeted blockade of two
molecules, CD274 and CTLA-4, resulted in clinical benefits for
individuals with several solid tumors (49,50).
The present study revealed that the expression of immune checkpoint
genes in the low-risk group was higher than in the high-risk group.
Furthermore, GSEA based on RNA-Seq data revealed that the risk
score was associated with the antigen processing and presentation
pathway, the toll-like receptor signaling pathway and the
cytokine-cytokine receptor interaction. In the tumor
microenvironment, numerous chemokines are released to facilitate
the migration of immune cells, thereby mediating the balance
between protumor and antitumor responses (51). The toll-like receptor pathway was
originally recognized as a critical mediator regulating immune
responses (52). Therefore, the
toll-like receptor signaling pathway could potentially be used to
predict responses to immunotherapy. In summary, the results of the
present study indicate that a high risk of mortality (poor overall
survival), as predicted by ferroptosis-based prognostic signature,
may influence immune infiltration-related biological processes.
Several limitations of the present study warrant
consideration. Firstly, the analysis was retrospective and relied
on publicly available datasets (TARGET and GEO). Whilst the
prognostic model was validated in an independent cohort (GEO), the
inherent biases and unmeasured confounders associated with
retrospective data cannot be fully eliminated. Prospective
validation in larger, uniformly treated cohorts is essential.
Secondly, data heterogeneity between the TARGET (RNA-seq) and GEO
(microarray) datasets, including differences in platform
technology, normalization methods and sample processing, poses
challenges for direct integration and validation. Mitigation of
this was attempted using standardized methods (such as ssGSEA,
CIBERSORT and consistent bioinformatic pipelines) and focusing on
relative risk stratification rather than absolute expression
cut-offs. However, this heterogeneity likely contributed to the
reduced statistical significance of the risk score as an
independent prognostic factor in the smaller GEO cohort during MCR
(Fig. 8D). Thirdly, the statistical
models have inherent limitations. The LASSO Cox regression used for
model construction helps prevent overfitting but may not capture
all relevant biological complexity. The sample size, particularly
for subgroup analyses and the GEO validation cohort, limits
statistical power. Furthermore, whilst standard significance
thresholds (P<0.05) were used for certain comparative analyses
(such as immune cell infiltration and checkpoint expression),
formal adjustment for multiple testing was not applied in all
exploratory comparisons, potentially increasing the risk of Type I
errors. Fourthly, whilst the cellular experiments demonstrated
differential expression of IFNG and TLR4 in OS cell lines compared
with in normal osteoblasts (Fig.
12, Fig. 13, Fig. 14), these in vitro models
cannot fully recapitulate the complexity of the in vivo
tumor microenvironment and its immune interactions. Therefore,
functional studies are needed to definitively establish the causal
roles of these genes in modulating ferroptosis and immune
infiltration within OS. Finally, the precise mechanistic links
between ferroptosis (as defined by the specific genes IFNG and
TLR4) and the observed immune cell profiles remain correlative
based on the bioinformatic and cellular expression data. Dedicated
experiments investigating how modulation of IFNG or TLR4 affects
ferroptosis sensitivity and immune cell behavior in OS models are
crucial next steps.
In conclusion, in the present study, two immune
subtypes in OS (Immunity_H and Immunity_L) with differing immune
infiltration and patient outcomes were identified. A prognostic
model based on ferroptosis genes (IFNG and TLR4) stratified
patients into high- and low-risk groups, where high-risk was
characterized by increased M0 macrophages and reduced immune
checkpoint expression. The findings highlight the interplay between
ferroptosis and immune regulation in OS. Future studies should
validate these genes as therapeutic targets and explore their
mechanisms in larger cohorts and preclinical models.
Acknowledgements
Not applicable.
Funding
Funding was received from the Hunan Provincial Key Laboratory of
Pediatric Orthopedics (grant no. 2023TP1019), Science and
Technology Project of Furong Laboratory (grant no. 2023SK2111),
Hunan Provincial Clinical Medical Research Center for Pediatric
Limb Deformities (grant no. 2019SK4006) and Hunan Provincial Health
Research Project (grant no. 20256815).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LRZ and GY conceived and designed the study. LRZ
collected data and wrote the manuscript. GHZ and HBM analyzed data
and constructed the figures. GY revised the manuscript, LRZ and GY
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
OS
|
osteosarcoma
|
|
DEG
|
differentially expressed gene
|
|
UCR
|
univariate Cox regression
|
|
MCR
|
multivariate Cox regression
|
|
ssGSEA
|
single-sample Gene Set Enrichment
Analysis
|
|
TARGET
|
Therapeutically Applicable Research to
Generate Effective Treatments
|
|
ROC
|
receiver operating characteristic
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GO
|
Gene Ontology
|
|
TIIC
|
tumor-infiltrating immune cell
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33.
2022.PubMed/NCBI
|
|
2
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar
|
|
3
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pei Y, Yao Q, Li Y, Zhang X and Xie B:
microRNA-211 regulates cell proliferation, apoptosis and
migration/invasion in human osteosarcoma via targeting EZRIN. Cell
Mol Biol Lett. 24:482019. View Article : Google Scholar
|
|
5
|
Shen JK, Cote GM, Choy E, Yang P, Harmon
D, Schwab J, Nielsen GP, Chebib I, Ferrone S, Wang X, et al:
Programmed cell death ligand 1 expression in osteosarcoma. Cancer
Immunol Res. 2:690–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tawbi HA, Burgess M, Bolejack V, Van Tine
BA, Schuetze SM, Hu J, D'Angelo S, Attia S, Riedel RF, Priebat DA,
et al: Pembrolizumab in advanced soft-tissue sarcoma and bone
sarcoma (SARC028): A multicentre, two-cohort, single-arm,
open-label, phase 2 trial. Lancet Oncol. 18:1493–1501. 2017.
View Article : Google Scholar
|
|
7
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Daud AI, Wolchok JD, Robert C, Hwu WJ,
Weber JS, Ribas A, Hodi FS, Joshua AM, Kefford R, Hersey P, et al:
Programmed death-ligand 1 expression and response to the
anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin
Oncol. 34:4102–4109. 2016. View Article : Google Scholar
|
|
9
|
Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao
N, Sun B and Wang G: Ferroptosis: Past, present and future. Cell
Death Dis. 11:882020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He GN, Bao NR, Wang S, Xi M, Zhang TH and
Chen FS: Ketamine induces ferroptosis of liver cancer cells by
targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des Devel Ther.
15:3965–3978. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang W, Green M, Choi JE, Gijón M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al:
CD8+ T cells regulate tumour ferroptosis during cancer
immunotherapy. Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Bahrami A, Pappo A, Easton J,
Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, et al:
Recurrent somatic structural variations contribute to tumorigenesis
in pediatric osteosarcoma. Cell Rep. 7:104–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen W, Liao Y, Sun P, Tu J, Zou Y, Fang
J, Chen Z, Li H, Chen J, Peng Y, et al: Construction of an ER
stress-related prognostic signature for predicting prognosis and
screening the effective anti-tumor drug in osteosarcoma. J Transl
Med. 22:662024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pradervand S, Weber J, Thomas J, Bueno M,
Wirapati P, Lefort K, Dotto GP and Harshman K: Impact of
normalization on miRNA microarray expression profiling. RNA.
15:493–501. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nguyen CB, Kumar S, Zucknick M, Kristensen
VN, Gjerstad J, Nilsen H and Wyller VB: Associations between
clinical symptoms, plasma norepinephrine and deregulated immune
gene networks in subgroups of adolescent with chronic fatigue
syndrome. Brain Behav Immun. 76:82–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang T, Wang S, Hua D, Shi X, Deng H, Jin
S and Lv X: Identification of ZIP8-induced ferroptosis as a major
type of cell death in monocytes under sepsis conditions. Redox
Biol. 69:1029852024. View Article : Google Scholar
|
|
17
|
Yu D, Hu H, Zhang Q, Wang C, Xu M, Xu H,
Geng X, Cai M, Zhang H, Guo M, et al: Acevaltrate as a novel
ferroptosis inducer with dual targets of PCBP1/2 and GPX4 in
colorectal cancer. Signal Transduct Target Ther. 10:2112025.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou Y, Palte MJ, Deik AA, Li H, Eaton JK,
Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dančík V, et al: A
GPX4-dependent cancer cell state underlies the clear-cell
morphology and confers sensitivity to ferroptosis. Nat Commun.
10:16172019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bashur L and Zhou G: Cancer stem cells in
osteosarcoma. Case Orthop J. 10:38–42. 2013.PubMed/NCBI
|
|
23
|
Wang SD, Li HY, Li BH, Xie T, Zhu T, Sun
LL, Ren HY and Ye ZM: The role of CTLA-4 and PD-1 in anti-tumor
immune response and their potential efficacy against osteosarcoma.
Int Immunopharmacol. 38:81–89. 2016. View Article : Google Scholar
|
|
24
|
Tsukahara T, Emori M, Murata K, Mizushima
E, Shibayama Y, Kubo T, Kanaseki T, Hirohashi Y, Yamashita T, Sato
N and Torigoe T: The future of immunotherapy for sarcoma. Expert
Opin Biol Ther. 16:1049–1057. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scott MC, Temiz NA, Sarver AE, LaRue RS,
Rathe SK, Varshney J, Wolf NK, Moriarity BS, O'Brien TD, Spector
LG, et al: Comparative transcriptome analysis quantifies immune
cell transcript levels, metastatic progression, and survival in
osteosarcoma. Cancer Res. 78:326–337. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Merchant MS, Melchionda F, Sinha M, Khanna
C, Helman L and Mackall CL: Immune reconstitution prevents
metastatic recurrence of murine osteosarcoma. Cancer Immunol
Immunother. 56:1037–1046. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morgan PK, Pernes G, Huynh K, Giles C,
Paul S, Smith AAT, Mellett NA, Liang A, van Buuren-Milne T, Veiga
CB, et al: A lipid atlas of human and mouse immune cells provides
insights into ferroptosis susceptibility. Nat Cell Biol.
26:645–659. 2024. View Article : Google Scholar
|
|
28
|
Ma L, Chen C, Zhao C, Li T, Ma L, Jiang J,
Duan Z, Si Q, Chuang TH, Xiang R and Luo Y: Targeting carnitine
palmitoyl transferase 1A (CPT1A) induces ferroptosis and synergizes
with immunotherapy in lung cancer. Signal Transduct Target Ther.
9:642024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bell HN, Stockwell BR and Zou W: Ironing
out the role of ferroptosis in immunity. Immunity. 57:941–956.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng Y, Sun L, Guo J and Ma J: The
crosstalk between ferroptosis and anti-tumor immunity in the tumor
microenvironment: Molecular mechanisms and therapeutic controversy.
Cancer Commun (Lond). 43:1071–1096. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Li Y, Tuerxun H and Zhao Y, Liu X
and Zhao Y: Firing up ‘cold’ tumors: Ferroptosis causes immune
activation by improving T cell infiltration. Biomed Pharmacother.
179:1172982024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng Z, Wang K, Wang Y, Liu T, Li J, Wang
Y, Chen W, Awuti R, Zhou H, Tong W, et al: Ferroptosis mediated by
the IDO1/Kyn/AhR pathway triggers acute thymic involution in
sepsis. Cell Death Dis. 16:5622025. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou J, Yin H, Pan J, Yin R, Wei X, Shen
M, Cai L, Liu Z, Zhao J, Chen W, et al: Lnk deficiency attenuates
the immunosuppressive capacity of MDSCs via ferroptosis to suppress
tumor development. Cell Death Dis. 16:6102025. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li S, Ouyang X, Sun H, Jin J, Chen Y, Li
L, Wang Q, He Y, Wang J, Chen T, et al: DEPDC5 protects
CD8+ T cells from ferroptosis by limiting
mTORC1-mediated purine catabolism. Cell Discov. 10:532024.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu J, Cui L, Hou B, Ding X, Liu H, Sun W,
Mi Y, Chen Y and Zou Z: Ferroptosis in tumor associated immune
cells: A double-edged sword against tumors. Crit Rev Oncol Hematol.
212:1048182025. View Article : Google Scholar
|
|
36
|
Gupta T, Wu SR, Chang LC, Lin FC, Shan YS,
Yeh CS and Su WP: Radiocleavable rare-earth nanoactivators
targeting over-expressed folate receptors induce mitochondrial
dysfunction and remodel immune suppressive microenvironment in
pancreatic cancer. J Nanobiotechnology. 23:5622025. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li P, Huang C, He Y, Lei X, Shen X, Xu J,
Mo Y, Sun X, Zheng L and Niu Y: M2 macrophage-laden vascular grafts
orchestrate the optimization of the inflammatory microenvironment
for abdominal aorta regeneration. Acta Biomater. 204:323–339. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu K, Zhu X, Sun S, Yang W, Liu S, Tang
Z, Zhang R, Li J, Shen T and Hei M: Inhibition of TLR4 prevents
hippocampal hypoxic-ischemic injury by regulating ferroptosis in
neonatal rats. Exp Neurol. 345:1138282021. View Article : Google Scholar
|
|
39
|
Flores RJ, Kelly AJ, Li Y, Nakka M,
Barkauskas DA, Krailo M, Wang LL, Perlaky L, Lau CC, Hicks MJ and
Man TK: A novel prognostic model for osteosarcoma using circulating
CXCL10 and FLT3LG. Cancer. 123:144–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Castro F, Cardoso AP, Gonçalves RM, Serre
K and Oliveira MJ: Interferon-gamma at the crossroads of tumor
immune surveillance or evasion. Front Immunol. 9:8472018.
View Article : Google Scholar
|
|
41
|
Shtrichman R and Samuel CE: The role of
gamma interferon in antimicrobial immunity. Curr Opin Microbiol.
4:251–259. 2001. View Article : Google Scholar
|
|
42
|
Garcia-Diaz A, Shin DS, Moreno BH, Saco J,
Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X,
et al: Interferon receptor signaling pathways regulating PD-L1 and
PD-L2 expression. Cell Rep. 19:1189–1201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rosadini CV, Zanoni I, Odendall C, Green
ER, Paczosa MK, Philip NH, Brodsky IE, Mecsas J and Kagan JC: A
single bacterial immune evasion strategy dismantles both MyD88 and
TRIF signaling pathways downstream of TLR4. Cell Host Microbe.
18:682–693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bauer AK, Dixon D, DeGraff LM, Cho HY,
Walker CR, Malkinson AM and Kleeberger SR: Toll-like receptor 4 in
butylated hydroxytoluene-induced mouse pulmonary inflammation and
tumorigenesis. J Natl Cancer Inst. 97:1778–1781. 2005. View Article : Google Scholar
|
|
45
|
Li L, Ouyang Y, Wang W, Hou D and Zhu Y:
The landscape and prognostic value of tumor-infiltrating immune
cells in gastric cancer. PeerJ. 7:e79932019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gu-Trantien C, Loi S, Garaud S, Equeter C,
Libin M, de Wind A, Ravoet M, Le Buanec H, Sibille C,
Manfouo-Foutsop G, et al: CD4+ follicular helper T cell
infiltration predicts breast cancer survival. J Clin Invest.
123:2873–2892. 2013. View Article : Google Scholar
|
|
47
|
Gambardella V, Castillo J, Tarazona N,
Gimeno-Valiente F, Martínez-Ciarpaglini C, Cabeza-Segura M, Roselló
S, Roda D, Huerta M, Cervantes A and Fleitas T: The role of
tumor-associated macrophages in gastric cancer development and
their potential as a therapeutic target. Cancer Treat Rev.
86:1020152020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Harrington BK, Wheeler E, Hornbuckle K,
Shana'ah AY, Youssef Y, Smith L, Hassan Q II, Klamer B, Zhang X,
Long M, et al: Modulation of immune checkpoint molecule expression
in mantle cell lymphoma. Leuk Lymphoma. 60:2498–2507. 2019.
View Article : Google Scholar
|
|
49
|
Wolchok JD, Kluger H, Callahan MK, Postow
MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K,
et al: Nivolumab plus ipilimumab in advanced melanoma. N Engl J
Med. 369:122–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Motzer RJ, Rini BI, McDermott DF, Redman
BG, Kuzel TM, Harrison MR, Vaishampayan UN, Drabkin HA, George S,
Logan TF, et al: Nivolumab for metastatic renal cell carcinoma:
Results of a randomized phase II trial. J Clin Oncol. 33:1430–1437.
2015. View Article : Google Scholar
|
|
51
|
Goralski KB, Jackson AE, McKeown BT and
Sinal CJ: More than an adipokine: The complex roles of chemerin
signaling in cancer. Int J Mol Sci. 20:47782019. View Article : Google Scholar
|
|
52
|
Tran TH, Tran TTP, Truong DH, Nguyen HT,
Pham TT, Yong CS and Kim JO: Toll-like receptor-targeted particles:
A paradigm to manipulate the tumor microenvironment for cancer
immunotherapy. Acta Biomater. 94:82–96. 2019. View Article : Google Scholar : PubMed/NCBI
|