Introduction
The circadian rhythm, an intrinsic biological
process with a period of ~24 h, regulates a wide array of
physiological functions such as sleep-wake cycles, metabolism,
hormone secretion and immune responses (1–3).
Disruptions in this rhythm, collectively termed circadian rhythm
disorders (CRDs), have been associated with various health issues,
including an increased risk of developing cancer. The impact of
CRDs on cancer is multifaceted, influencing not only the
proliferation and survival of cancer cells but also the tumor
immune microenvironment (TME) (4),
which serves a key role in tumor progression and response to
therapy (5–7).
F-box and leucine-rich repeat protein 22 (FBXL22),
an F-box protein-coding gene, is known to participate in the
ubiquitin-proteasome system, which is central to the regulation of
protein turnover and cellular homeostasis (8,9).
Emerging evidence suggests that FBXL22 also serves a role in cancer
development and progression, although the exact mechanism remains
unclear (8,10). Previous bioinformatics analysis has
indicated that FBXL22 is associated with breast cancer and acts as
a favorable factor for relapse-free survival in prostate cancer
(PCa) (11–13). However, a comprehensive pan-cancer
analysis of FBXL22, particularly in the context of the TME and its
interactions with the circadian rhythm, is lacking.
Due to the importance of circadian rhythms in
oncology and the potential role of FBXL22 in cancer, the present
study aimed to perform an extensive analysis of FBXL22 across
various cancer types. By leveraging data from The Cancer Genome
Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases, the
present study explored gene expression, genetic alterations and DNA
methylation patterns of FBXL22. The present analysis aimed to
reveal the correlation of these patterns with patient prognosis,
immune cell infiltration and TME, thereby providing insights into
the potential of FBXL22 as a biomarker and therapeutic target in
cancer.
Materials and methods
Data collection, normalization and
batch effect correction
Pan-cancer and normal tissue expression data for
FBXL22 were extracted from GTEx (https://www.gtexportal.org/home/datasets) and TCGA
datasets (TCGA-PANCAN, http://portal.gdc.cancer.gov/) of University of
California Santa Cruz XENA (https://xenabrowser.net/datapages/). University of
Alabama at Birmingham CANcer data analysis Portal (https://ualcan.path.uab.edu/) and cBioPortal
(https://www.cbioportal.org/). RNA
sequencing expression data from TCGA and GTEx were harmonized to
ensure cross-dataset comparability. Raw read counts from TCGA were
normalized to transcripts per million (TPM) using the ‘edgeR’ R
package (R Development Core Team, version 3.46.0; http://bioconductor.org/packages/release/bioc/html/edgeR.html).
GTEx data were processed using the same pipeline to maintain
consistency. Batch effects arising from differences in sequencing
platforms and study cohorts were corrected using the ComBat
algorithm (implemented via the ‘sva’ R package; version, 3.52.0;
http://bioconductor.org/packages/release/bioc/html/sva.html),
with ‘study cohort’ (TCGA vs. GTEx) as the batch variable.
Low-quality samples (read depth <10 million) and genes with zero
expression in >90% of the samples were excluded prior to
analysis.
Diagnostic receiver operating
characteristic (ROC) curve and immune infiltration analysis
The predictive accuracy of FBXL22 gene expression in
TCGA cancer types was assessed using ROC analysis with the ‘pROC’
package (Version, 1.18.0; http://cran.r-project.org/web/packages/pROC/) in R
software (R Development Core Team; Version, 4.4.2). To assess
immune cell infiltration using the ‘ssGSEA’ algorithm, predefined
gene sets were employed from the Molecular Signatures Database
(MSigDB; http://www.gsea-msigdb.org/gsea/msigdb/index.jsp)
hallmark collection, specifically the hallmark gene sets related to
the immune response. The cell-type marker genes used for immune
infiltration analysis were obtained from the ImmPort database
(version, DR35; http://www.immport.org/) and published literature,
ensuring their relevance and specificity for the immune cell types
under investigation. The ssGSEA algorithm was run with 1,000
permutations to assess statistical significance and normalized
enrichment scores were used to quantify the relative abundance of
immune cells in each sample. For the Tumor Immune Estimation
Resource 2.0 (TIMER2) tool (version 2.0; http://timer.cistrome.org), the present study used the
default settings with the immune gene module, which includes a
comprehensive set of immune-related genes and their corresponding
cell type-specific markers. Correlation analysis between FBXL22
expression and immune cell infiltration was performed using
Spearman's correlation coefficient to account for nonlinear
relationships.
Functional enrichment analysis of
FBXL22-related genes
Top 100 target genes associated with FBXL22
expression were identified using the ‘Similar Gene Detection’
module of Gene Expression Profiling Interactive Analysis 2
(http://gepia2.cancer-pku.cn/). Heatmap
data for the top 10 genes were generated using the ‘Gene_Corr’
module of TIMER2. Protein-protein interaction networks were
constructed using the Search Tool for the Retrieval of Interacting
Genes/Proteins database (STRING: functional protein association
networks; http://cn.string-db.org/). Functional
enrichment analysis was performed using the ‘clusterProfiler’
package in the R software (version 4.10.0; http://bioconductor.org/packages/clusterProfiler/).
Gene ontology (GO, http://www.geneontology.org/) and Kyoto Encyclopedia
of Genes and Genomes (KEGG, http://www.genome.jp/kegg) enrichment analyses
(14) were performed to explore the
biological processes and pathways associated with FBXL22.
Significant terms were defined as those with an adjusted P-value
<0.05 (Benjamini-Hochberg correction) and a minimum gene count
of 5 per term.
Subgroup survival prognosis analysis
for PCa and expression analysis in disease subtypes
RNA sequencing data from TCGA-prostate
adenocarcinoma (TCGA-PRAD; http://portal.gdc.cancer.gov/projects/TCGA-PRAD)
project were downloaded and organized using TCGA database
(https://portal.gdc.cancer.gov). Clinical
outcome data were obtained from a publication on general cancer
prognosis (15). The Prostate
Cancer Transcriptome Atlas (PCTA) database (version, 1.0.1;
http://www.thepcta.org/) was used to analyze
FBXL22 expression and its association with disease progression in
different subtypes of PCa. Patients were stratified based on
clinical features, such as Gleason score (GS) (16) and clinical stage and survival
analysis was performed using the Kaplan-Meier method with log-rank
tests (https://kmplot.com/analysis/).
Immuno-infiltration and immune
checkpoint gene co- expression analysis in PCa
Infiltration of the 24 immune cell types was
quantified using the ‘ssGSEA’ algorithm in the ‘GSVA’ R package (R
Development Core Team; version 1.52.0; http://bioconductor.org/packages/release/bioc/html/GSVA.html).
Spearman's correlation analysis was performed to examine the
relationship between FBXL22 expression and immune cell infiltration
matrix data as well as immune checkpoint protein expression in the
dataset.
Functional enrichment analysis of
FBXL22 in PCa
Patients were classified into high and low FBXL22
expression groups (low, 0–50%; high, 50–100%) for enrichment
analysis using the R package ‘clusterProfiler’ (version 4.10.0; R
Development Core Team; http://bioconductor.org/packages/clusterProfiler/).
Gene set enrichment analysis (GSEA) was performed using the
‘clusterProfiler’ and ‘msigdbr’ packages (version 7.5.1; http://cran.r-project.org/web/packages/msigdbr/index.html)
and the MSigDB gene collection database. For GO/KEGG analysis of
differentially expressed genes, significance thresholds were an
adjusted P-value <0.05 and a minimum gene count of 5. For GSEA,
gene sets with a false discovery rate <0.25 and a nominal
P-value <0.05 were considered statistically significant.
Cell culture and transfection
Human normal prostate epithelial cells (RWPE-1) and
PCa cell lines (DU145 and PC3) were obtained from Procell Life
Science and Technology Co., Ltd. and cultured in DMEM (cat. no.
D6429; MilliporeSigma) or F-12K medium (cat. no. 21127022; Thermo
Fisher Scientific, Inc.), supplemented with 10% FBS (cat. no.
A5670701; Thermo Fisher Scientific, Inc.) under standard conditions
(37°C; 5% CO2). Human natural killer (NK) cells (cat.
no. CP-H168), also from Procell Life Science and Technology Co.,
Ltd., were maintained in RPMI-1640 medium (cat. no. 11875093;
Thermo Fisher Scientific, Inc.) with 10% FBS under the same
conditions. The human FBXL22 gene was amplified using forward (F)
primer 5′-TACCGAGCTCGGATCCATGTGGCCACTCTGCACCATG-3′ and reverse (R)
primer 5′-GATATCTGCAGAATTCTCACTGCGGGAACGCATCCG-3′, then subcloned
into a pcDNA3.1 (+) mammalian expression vector (cat. no. V79020;
Invitrogen; Thermo Fisher Scientific, Inc.) to generate the
recombinant plasmid pcDNA3.1-FBXL22. The empty vector pcDNA3.1
served as a negative control (NC) and both were provided by
ABclonal Biotech Co., Ltd. (cat. no. RM09014P). For transfection,
cells were seeded into 24-well plates at a density of
5×104 cells per well and cultured until they reached 80%
confluence. Lipofectamine® 3000 (cat. no. L3000001;
Thermo Fisher Scientific, Inc.) was used as the transfection
reagent. A mixture of 25 µl Opti-MEM I medium and 0.5 µl
Lipofectamine® 3000 reagent was prepared. Meanwhile, 1
µl P3000TM reagent and 25 µl Opti-MEM I medium were added to 2 µg
pcDNA3.1-FBXL22 or pcDNA3.1-NC plasmid DNA. The two solutions were
subsequently combined and incubated at 25°C for 10 min. The
resulting 50 µl complex was added to the cell culture medium,
followed by a 48 h incubation at 37°C. After 48 h, the medium was
replaced with fresh complete medium. All functional assays were
performed 48 h post-transfection. Successful transfection was
confirmed using reverse transcription (RT)-quantitative (q)PCR and
western blotting.
Cell viability, invasion and migration
assays
At 48 h post-transfection, cell viability was
assessed using a Cell Counting Kit (CCK-8; cat. no. A50298; Thermo
Fisher Scientific, Inc.) assay following a 24 h incubation with the
CCK-8 reagent. Transwell invasion assays were performed using
24-well chambers (cat. no. 3378; Corning, Inc.,). The polycarbonate
membrane of the upper chamber was coated with 100 µl Matrigel (cat.
no. M8370; Beijing Solarbio Science & Technology Co., Ltd.)
diluted 1:8 in serum-free DMEM at 4°C and polymerized for 0.5–1 h
at 37°C. The upper chamber was seeded with ~5×105 cells
in 200 µl serum-free DMEM. The lower chamber contained 500 µl
medium with 20% FBS. Cells were incubated for 24 h at 37°C in 5%
CO2. The membrane was fixed with 600 µl of 4% tissue
cell fixative (cat. no. P1110; Beijing Solarbio Science &
Technology Co., Ltd.) for 20–30 min at room temperature. After
discarding the fixative, cells were stained with 0.1–0.2% crystal
violet (cat. no. G1062; Beijing Solarbio Science & Technology
Co., Ltd.) for 5–10 min at room temperature. Excess stain was
rinsed with PBS. Invasive cells in the lower membrane surface were
imaged using an inverted light microscope (model ICX41; Ningbo
Sunny Instruments Co., Ltd.) with a 50 µm scale bar. For the wound
healing assay, prostate cancer cell lines (DU145 and PC3) were
seeded in 6-well plates (Shanghai Kejin Biotechnology Co., Ltd.) at
a density of 5×105 cells per well and cultured until a
confluent monolayer had formed. Vertical scratches were created
across the monolayer using a sterile 20 µl pipette tip, aligned
with pre-marked horizontal lines on the back of the plate to ensure
consistent observation points. After scratching, cells were washed
with PBS (cat. no. C3580-0500; VivaCell; Shanghai XP Biomed Ltd.)
2–3 times to remove detached cells and then incubated in serum-free
DMEM. Images of the scratches were captured immediately at 0 h
using an inverted light microscope (model ICX41; Ningbo Sunny
Instruments) at 40× magnification. Cells were further cultured at
37°C in a 5% CO2 incubator, with additional images taken
at 24 h under the same 40× magnification. The healing rate was
quantified by measuring the change in scratch width using ImageJ
1.5.2a software (National Institutes of Health).
NK cell co-culture system with DU145
and PC3 PCa cell lines
NK cells were labeled with CellTracker™ Green CMFDA
(Thermo Fisher Scientific, Inc.) and co-cultured with
FBXL22-overexpressing or control DU145/PC3 cells in a Transwell
system (Corning, Inc.) with a pore size of 0.4 µm. The upper
chamber contained 1×105 NK cells in RPMI-1640 medium
without FBS and the lower chamber contained 2×105 PCa
cells in RPMI-1640 medium supplemented with 10% FBS. After 48 h of
co-culture, the Transwell membranes were first fixed with 600 µl of
4% tissue cell fixative (cat. no. P1110; Beijing Solarbio Science
& Technology Co., Ltd.) at room temperature for 20–30 min.
Following fixation, the membranes were stained with 0.1–0.2%
crystal violet (cat. no. G1062; Beijing Solarbio Science &
Technology Co., Ltd.) at room temperature for 5–10 min. Excess
stain was removed by rinsing with PBS three times. Analyzed for NK
cell migration and cytotoxic activity against PCa cells. For
Transwell assays assessing NK cell migration, the upper chamber
contained 1×105 NK cells in RPMI-1640 medium without
FBS, while the lower chamber contained RPMI-1640 medium with 10%
FBS. After 48 h, the membranes were processed for analysis.
Flow cytometry
Apoptosis was analyzed using an Annexin V-FITC/PI
Apoptosis Detection Kit (cat. no. CA1020; Solarbio Science &
Technology Co., Ltd.) according to the manufacturer's instructions.
Briefly, transfected DU145 and PC3 cells were harvested by
trypsinization with EDTA-free trypsin (cat. no. C0205; Beyotime
Institute of Biotechnology), centrifuged at 100 × g for 5 min at
4°C, and washed twice with cold PBS. The cell pellet was
resuspended in 1× binding buffer (10× binding buffer diluted 1:9
with deionized water) to a density of 1–5×106 cells/ml.
A 100 µl aliquot of the cell suspension was incubated with 5 µl
Annexin V-FITC in the dark at room temperature (25°C) for 15 min.
Subsequently, 5 µl PI solution and 400 µl PBS were added and
samples were immediately analyzed using a FACSCanto™ flow cytometer
(BD Biosciences). Data were analyzed using the FlowJo software
(v10.8.1; FlowJo LLC; BD Biosciences).
Gene expression
FBXL22 mRNA was quantified by reverse
transcription-quantitative PCR (RT-qPCR). RNA was extracted from
transfected DU145, PC3, and RWPE-1 cells using TRNzol (cat. no.
DP424; Tiangen Biotech Co., Ltd.). Concentration/purity was
measured via NanoDrop One/OneC (cat. no. 840-317400; Thermo Fisher
Scientific, Inc.; A260/280=1.8–2.0). cDNA was
synthesized with FastKing One-Step SuperMix (cat. no. KR118;
Tiangen Biotech Co., Ltd.; 20 µl; 4 µl 5× SuperMix; 2 µg RNA;
RNase-free water). The thermal cycling protocol was conducted as
follows: An initial step at 42°C for 15 min, followed by heat
inactivation at 95°C for 3 min and a final hold at 4°C for 30 min.
RT-qPCR was performed using a StepOnePlus™ Real-Time PCR System
(cat. no. 4376600; Thermo Fisher Scientific, Inc.) with SYBR™ Green
Master Mix (cat. no. G3320-15; Wuhan Servicebio Technology Co.,
Ltd.) or SuperReal Premix Plus (cat. no. FP205; Tiangen Biotech
Co., Ltd.). Each 20 µl reaction mixture contained 10 µl 2× Premix,
0.6 µl forward and reverse primer each (10 µM), 2 µl of cDNA
template, and was made up to the final volume with nuclease-free
water. The following thermocycling conditions were used: 95°C 15
min; 40× (95°C 10 sec; 60°C 32 sec). A melting curve analysis was
performed immediately after the amplification cycles to confirm the
specificity of the PCR products and the absence of primer-dimers.
Expression calculated via 2−ΔΔCq (17) with β-actin control. Primers
sequences were as follows: FBXL22, Forward
5′-CCATGCACATAACCCAGCTCA-3′; Reverse (R),
5′-CCGAGGTGATTTCGGTCCAAC-3′; β-actin F,
5′-CATGTACGTTGCTATCCAGGC-3′; R, 5′-CTCCTTAATGTCACGCACGAT-3′.
Western blotting
Protein levels were determined using western
blotting. Total protein was extracted from cells using RIPA lysis
buffer (cat. no. R0010; Beijing Solarbio Science & Technology
Co., Ltd.) and quantified with a BCA assay kit (cat. no. BL521A;
Biosharp Life Sciences). An equal amount (30 µg) of protein per
sample was separated by 10% SDS-PAGE and then electrophoretically
transferred to a PVDF membrane. The membrane was blocked with 5%
non-fat milk in TBST (containing 0.1% Tween 20), for 2 h at room
temperature. After primary antibody incubation (4°C; overnight;
Table SI) and three washes with
TBST, membranes were incubated with secondary antibodies: Goat
anti-rabbit IgG H and L (HRP) (cat. no. ab97051; Abcam) or
HRP-conjugated goat anti-mouse IgG (H+L) (cat. no. AS003; ABclonal
Biotech Co., Ltd.), both at a 1:10,000 dilution, for 1 h at room
temperature. Signals were detected using an enhanced
chemiluminescence kit (cat. no. P10300; Suzhou Xinsaimei
Biotechnology Co., Ltd) and visualized with a multi-functional
imaging system (SH-CUTE523; Zhehiang ICP; Table SII).
Co-immunoprecipitation (Co-IP)
An immunoprecipitation kit (cat. no. P2179S)
purchased from Beyotime Institute of Biotechnology was used for the
experiment. Transfected PC3 and DU145 cells (1×107) were
collected and lysed on ice with 500 µl lysis buffer (provided in
the kit) for 30 min. Lysates were centrifuged at 12,000 × g for 10
min at 4°C to pellet cellular debris and the protein concentration
of the supernatants was determined using a BCA assay (cat. no.
BL521A; Biosharp Life Sciences). A small amount of lysate (50 µl)
was incubated with 20 µl protein A/G agarose beads (pre-treated
with lysis buffer) at 4°C for 1 h to block non-specific binding.
For immunoprecipitation, 500 µl lysate (containing ~500 µg total
protein) was incubated with 1 µg anti-FBXL22 primary antibody (cat.
no. ab223059; Abcam) at 4°C overnight. Subsequently, 40 µl
pre-treated protein A/G agarose beads were added and incubated at
4°C for 2 h. The samples were centrifuged at 1,000 × g at 4°C for 3
min and the beads were collected and rinsed with elution buffer.
Finally, the bound proteins were eluted by boiling the beads in 2X
SDS-PAGE loading buffer at 95°C for 10 min. Western blotting was
performed on the eluent to analyze the co-precipitated
proteins.
Statistical analysis
Statistical analyses were performed using the R
software and GraphPad Prism 9.5.1(Dotmatics). Gene expression
differences were compared using Wilcoxon rank-sum (for two-group
comparisons) and Kruskal-Wallis tests (for multi-group
comparisons). For significant results using Kruskal-Wallis tests
(P<0.05), post hoc pairwise comparisons were performed using
Dunn's test with Bonferroni correction. Correlations were assessed
using Spearman's or Pearson's correlation analysis. Survival
characteristics were compared using Kaplan-Meier curves and Cox
regression analysis. For the KM plotter, mRNA expression data were
used; and patients were stratified into high and low FBXL22
expression groups using the 50th percentile (median) as the cut-off
(low, 0–50%; high, 50–100%). Data were represented as mean ±
standard deviation (SD) *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001 were considered to indicate a statistically
significant difference.
Results
Pan-cancer expression and epigenetic
regulation of FBXL22
FBXL22 was differentially expressed in 33 cancer
types when compared with corresponding normal tissues. FBXL22 was
significantly downregulated in 19 cancer types, including uterine
carcinosarcoma (P<0.0001), endometrial carcinoma (UCEC;
P<0.0001) and colon adenocarcinoma (COAD; P<0.0001), but
overexpressed in seven cancer types such as cholangiocarcinoma
(CHOL; P<0.0001) and acute myeloid leukemia (LAML; P<0.0001)
(Fig. 1A and B). When comparing
tumor tissues with the corresponding normal tissues from TCGA and
GTEx databases, significant variations in FBXL22 expression were
observed in 26 cancer types (P<0.05), with the exception of
kidney chromophobe, pheochromocytoma and paraganglioma and thyroid
carcinoma (THCA) (Fig. 1B). This
comparison highlights the distinct expression levels of FBXL22 in
neoplastic vs. non-neoplastic tissues and provides a foundation for
understanding its potential role in cancer. However, it should be
noted that expression levels in normal tissues were based on the
GTEx database and these data should be interpreted with caution
because of the potential differences in tissue collection and
processing methods between TCGA and GTEx.
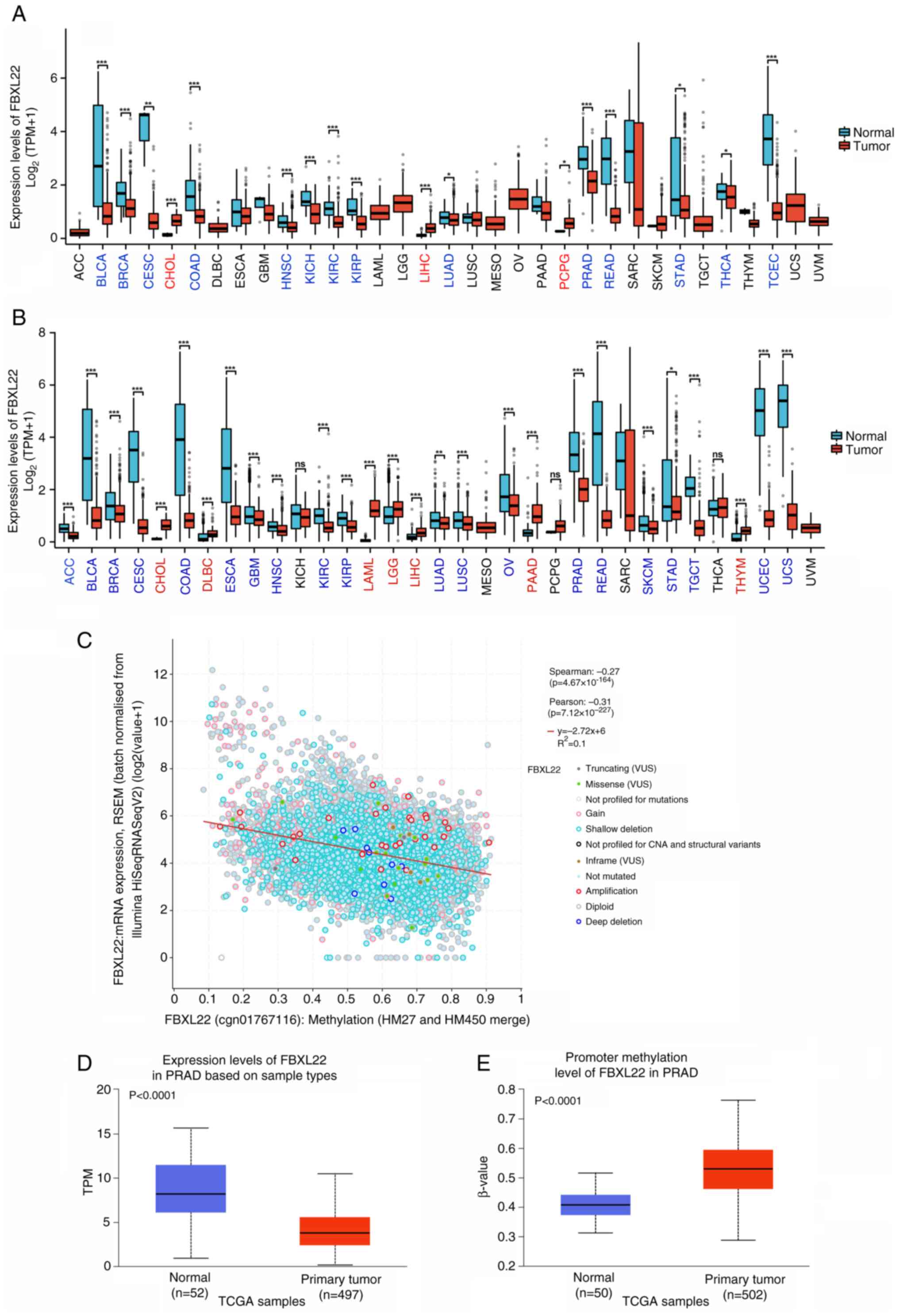 | Figure 1.Expression levels of FBXL22 in
pan-cancer. (A) The expression status of FBXL22 in different cancer
tissues and para-cancerous tissues in TCGA database. Error bars
represent standard deviation. (B) Expression status of FBXL22 in
matched cancer tissues and normal tissues in TCGA and GTEx
databases. Cancer names in red text indicate high FBXL22 expression
in this cancer, while blue text indicates low FBXL22 expression in
this cancer. (C) Correlation between FBXL22 expression and
methylation in TCGA database (D) Promoter methylation levels of
FBXL22 in PRAD samples. (E) Expression differences of FBXL22 in
PRAD samples based on sample types. *P<0.05; **P<0.01;
***P<0.001; ns, P≥0.05. FBXL22, F-box and leucine rich repeat
protein 22; ACC, adrenocortical carcinoma; BLCA, bladder urothelial
carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous
cell carcinoma and endocervical adenocarcinoma; CHOL,
cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid
neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma;
GBM, glioblastoma multiforme; HNSC, head and neck squamous cell
carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell
carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute
myeloid leukemia; LGG, brain lower grade glioma; LIHC, Liver
hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; MESO, mesothelioma; ns, not significant;
OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic
adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD,
prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC,
sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach
adenocarcinoma; TCGA, The Cancer Genome Atlas; TGCT, testicular
germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; TMB,
tumor mutation burden; TPM, transcripts per million; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma. |
A strong negative correlation between FBXL22
expression and promoter methylation was observed (P<0.0001,
Spearman's ρ=−0.27; Pearson's r=−0.31; Fig. 1C). Elevated methylation levels in
tumors vs. normal tissues were prominent in kidney renal papillary
cell carcinoma, breast invasive carcinoma (BRCA), THCA and PRAD
(P<0.0001), whereas reduced methylation was observed in
glioblastoma multiforme, liver hepatocellular carcinoma (LIHC) and
pancreatic adenocarcinoma (PAAD) (Figs.
1D and S1). In PCa, FBXL22
demonstrated significant promoter hypermethylation (P<0.0001)
and downregulation (P<0.0001) (Figs.
1D, E and S2).
Genomic alterations and mutation
frequency of FBXL22
Genomic alterations in FBXL22 were observed in 26
cases across 12 cancer types, with a somatic mutation frequency of
0.2% (Fig. S3). Among the various
cancer types, UCEC exhibited the highest mutation frequency of 1.7%
(9/529 cases), followed by stomach adenocarcinoma [STAD; 1.14%
(5/440 cases)] (Fig. S3). Missense
mutations were the predominant type of genetic alteration observed.
However, no significant correlation was found between FBXL22
expression levels and mutation types or copy number alteration
(Fig. S4; Table SIII; P>0.05). Although genomic
alterations of FBXL22 were identified across several cancer types,
the overall mutation frequency was relatively low. This suggested
that while genetic mutations can occur in FBXL22, they may not be
the primary driver of its expression changes in most cancer
types.
Survival analysis and prognostic
implications of FBXL22
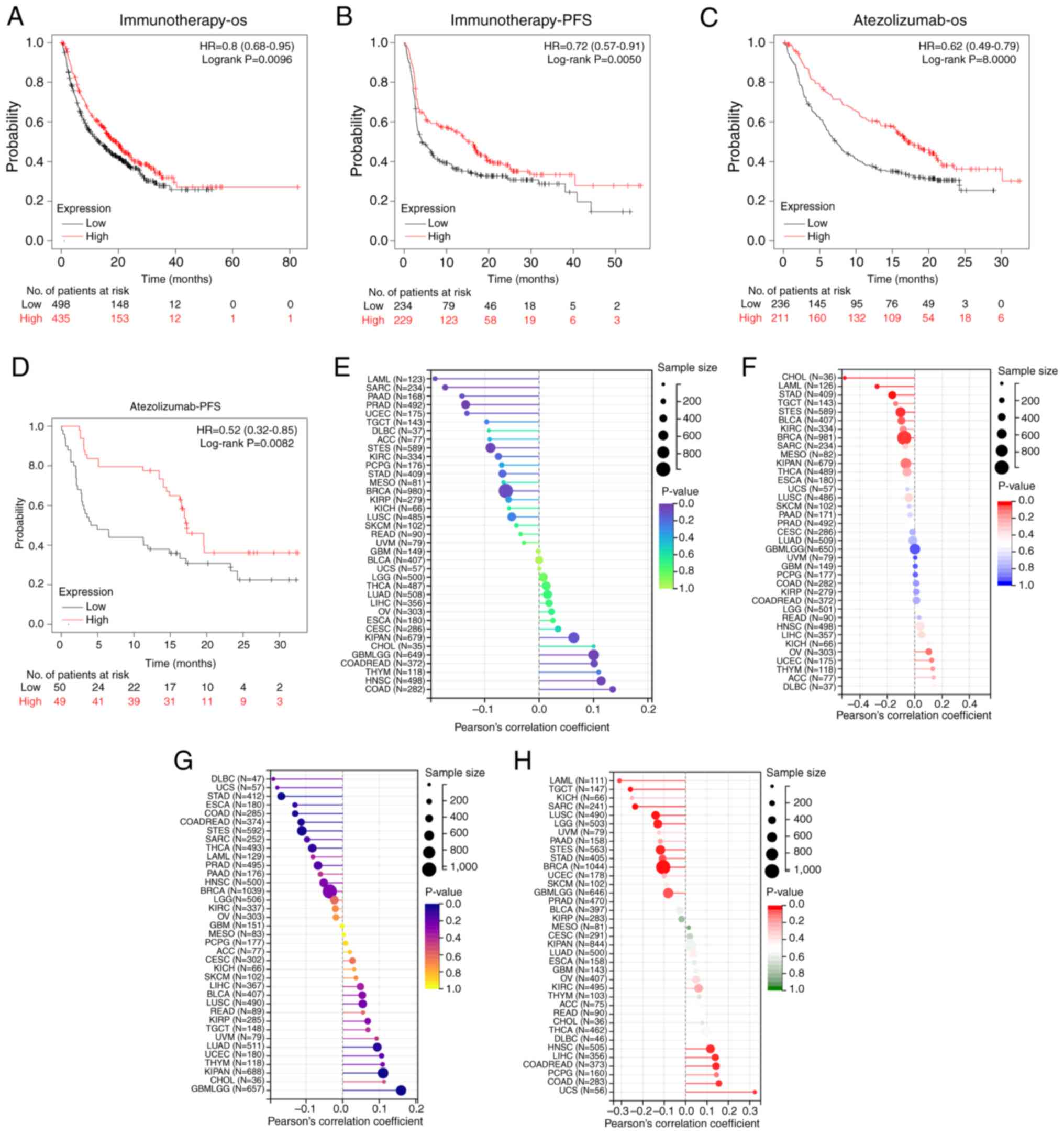
Survival analysis revealed that elevated FBXL22
expression was significantly associated with worse overall survival
(OS) in bladder cancer (BLCA; P<0.01) and STAD (P<0.05),
whereas low expression predicted unfavorable recurrence-free
survival in THCA (P<0.05) (Fig. 2A
and B). Kaplan-Meier validation across 21 cancer types
(n=7,489) further demonstrated that low FBXL22 expression
correlated with worse OS in esophageal adenocarcinoma (P<0.05)
and PAAD (P<0.05), whereas high expression remained a prognostic
factor for poor OS in BLCA (P<0.05) and STAD (P<0.05)
(Fig. S5). Notably, FBXL22
expression exhibited significant associations with PCa
clinicopathological features: Higher levels were observed in
patients with GS ≤7 (Fig. 2C and F;
P<0.001) and advanced clinical stages (Fig. 2D; P<0.05) and metastatic
castration-resistant PCa (Fig.
2E-G, P<0.001) and luminal B Prediction Analysis of
Microarray 50 subtypes (Fig. 2H,
P<0.001). Collectively, these findings underscore the key
relationship between FBXL22 expression and PCa progression markers
(GS, staging and molecular subtypes), which suggests it has a
potential role as a biomarker and functional driver in disease
progression.
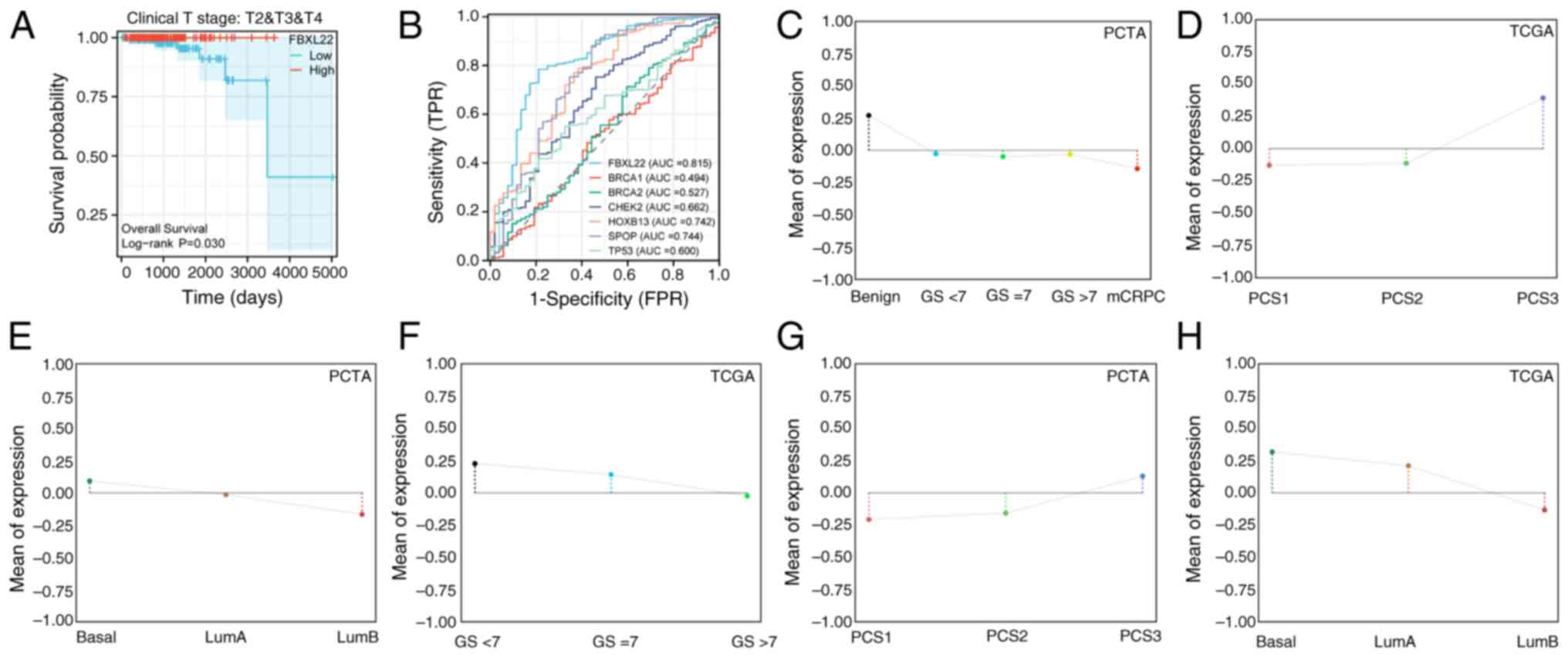 | Figure 2.Clinical diagnosis and prognostic
implications of FBXL22 in PCa. (A) Kaplan-Meier curve of PFI in
patients with intermediate to advanced disease stage (Clinical T
stage, T2, T3 and T4) in PCa by FBXL22-expression groups. (B)
Diagnostic ROC curves of FBXL22 and other genes such as SPOP,
HOXB13, CHEK2, TP53, BRCA2 and BRCA1 in PCa. (C) FBXL22 expression
across PCa disease course subtypes. (D) FBXL22 expression in PCS
subtypes. (E) FBXL22 expression in basal, LumA and LumB subtypes.
(F) FBXL22 expression across GS subgroups. (G) FBXL22 expression in
PCS subtypes. (H) FBXL22 expression in basal, LumA and LumB
subtypes. FBXL22, F-box and leucine rich repeat protein 22; PCa,
prostate cancer; PFI, progression-free interval; ROC, receiver
operating characteristic; SPOP, Speckle-type pox virus and zinc
finger protein; HOXB13, homeobox B13; CHEK2, checkpoint kinase 2;
PAM50, Prediction Analysis of Microarray 50; PCTA, Prostate Cancer
Transcriptome Atlas; TCGA, The Cancer Genome Atlas; PCS, PCa
subtypes; ROC, receiver operating characteristic; Lum, luminal; GS,
Gleason score; TPR, true positive rate; FPR, false positive rate;
AUC, area under the curve. |
FBXL22 immune activation and TME
Comprehensive analyses revealed a significant
association between FBXL22 expression and TME dynamics (Figs. 3 and S6). Violin plots demonstrated that
FBXL22-high tumors exhibited markedly elevated enrichment scores
across diverse immune cell populations (P<0.05; Wilcoxon test),
particularly in cytotoxic subsets, including NK and CD8+
T cells (Fig. 3A and I). Consistent
with this immune-activated phenotype, key immunoregulatory markers
[programmed cell death 1 (PDCD1), CD274 and PDCD1 ligand 2]
demonstrated 2.1- to 3.4-fold higher expression in FBXL22-high
samples [log2(TPM+1); P<0.01; Fig. 3B]. Heatmap analysis further
demonstrated strong co-expression patterns between FBXL22 and
immune checkpoint regulators (Pearson's r=0.62–0.71; Z-score
normalized; Fig. 3C). Quantitative
correlation analyses revealed dose-dependent relationships between
FBXL22 expression and infiltration densities of antitumor immune
subsets, including significant associations with NK cells
(Spearman's ρ=0.57; P<0.00001), CD8+ T cells (r=0.49;
P<0.001), Th1 cells (r=0.43; P<0.001) and dendritic cells
(r=0.38; P<0.01; Fig. 3D-H).
These multimodal findings collectively suggest that FBXL22 is a
potential immunomodulatory hub with high expression levels
correlating with enhanced immune infiltration and checkpoint
activation, which suggests it has functional involvement in shaping
antitumor immune responses.
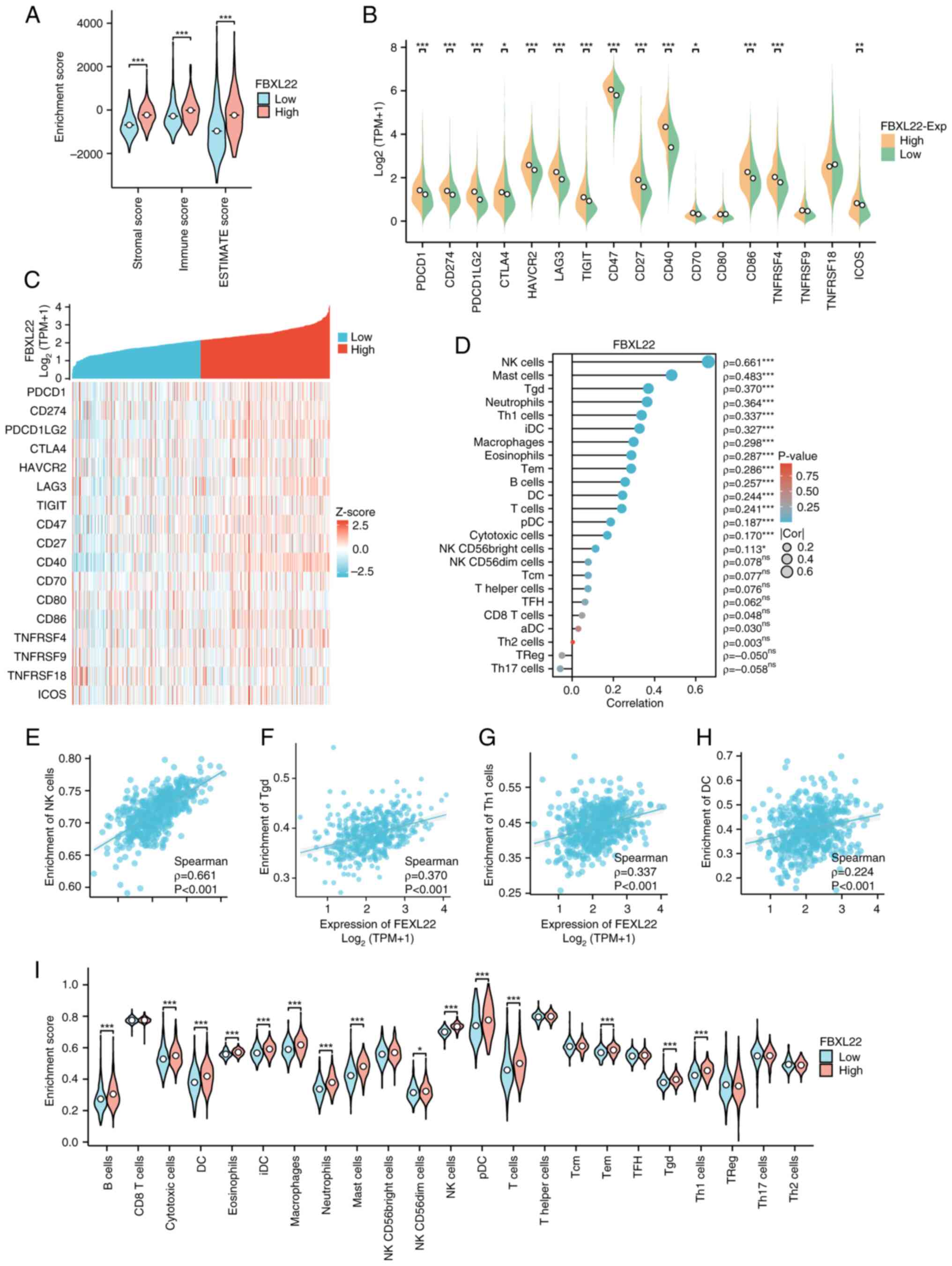 | Figure 3.Association of FBXL22 with
immune-infiltration in PCa. (A) The immune score of
FBXL22-expression groups. (B) Expression levels of the 17 immune
checkpoint proteins by FBXL22-expression group. (C) Heatmap
illustrating correlations between FBXL22 expression and 17 immune
checkpoint proteins. (D) Correlation between FBXL22 expression and
the infiltration levels of various immune cells. Scatter plot of
correlation between FBXL22 expression and the infiltration levels
of (E) NK, (F) Tgd, (G) Th1 and (H) DC cells. (I) Infiltration
levels of 23 types of immune cells were assessed by
FBXL22-expression group using ssGSEA. *P<0.05, **P<0.01 and
***P<0.001; ns, P≥0.05. FBXL22, F-box and leucine rich repeat
protein 22; PCa, prostate cancer; NK, natural killer; DC, dendritic
cells; Tgd cells, γδ T cells; Th1 cells, T helper type 1 cells;
Treg, regulatory T cells; ssGSEA, single-sample gene set enrichment
analysis; ESTIMATE, Estimation of STromal and Immune cells in
MAlignant Tumor tissues using Expression data; TPM, transcripts per
million; ns, not significant. |
Therapeutic response prediction based
on FBXL22 expression
Low FBXL22 expression was associated with worse OS
(P<0.01) and progression-free survival (PFS; P=0.005) in
immunotherapy-treated patients, whereas high expression predicted
improved outcomes in atezolizumab-treated cohorts (P<0.001 for
OS; P<0.01 for PFS) (Fig. 4A-D).
This suggests that FBXL22 expression may be a predictive biomarker
of therapeutic responses to specific cancer treatments. Notably,
FBXL22 expression was negatively correlated with tumor mutation
burden (TMB) in LAML, BRCA and STAD, and positively correlated with
homologous recombination deficiency (HRD) in COAD and LIHC
(Fig. 4E-H; all P<0.05). The
negative correlation between FBXL22 expression and TMB in certain
cancer types and the positive correlation with HRD in others
further emphasizes its potential as a biomarker for guiding
personalized treatment decisions.
Immune regulatory gene analysis
FBXL22 expression was significantly correlated with
immune checkpoint pathway genes across multiple cancer types,
including positive associations with inhibitory genes [for example,
programmed death-ligand 1 (PD-L1) and cytotoxic T-lymphocyte
associated protein 4 (CTLA4)] and stimulatory genes (for example,
CD40 and inducible T-cell co-stimulator) (Fig. S7; all P<0.05). Additionally,
FBXL22 correlated with immunomodulation-related genes such as
chemokines and major histocompatibility complex molecules in uveal
melanoma, READ and PRAD (Fig. S8;
all P<0.05). FBXL22 expression positively correlated with
cancer-associated fibroblast (CAF) and endothelial cell (EC)
infiltration in most cancer types, except brain lower grade glioma
(Fig. 5A-K). In THCA, FBXL22
demonstrated a negative correlation with CAFs (ρ=−0.1961;
P<0.0001) but a positive association with ECs (ρ=0.5977;
P<0.0001) (Fig. 5E, J and
K).
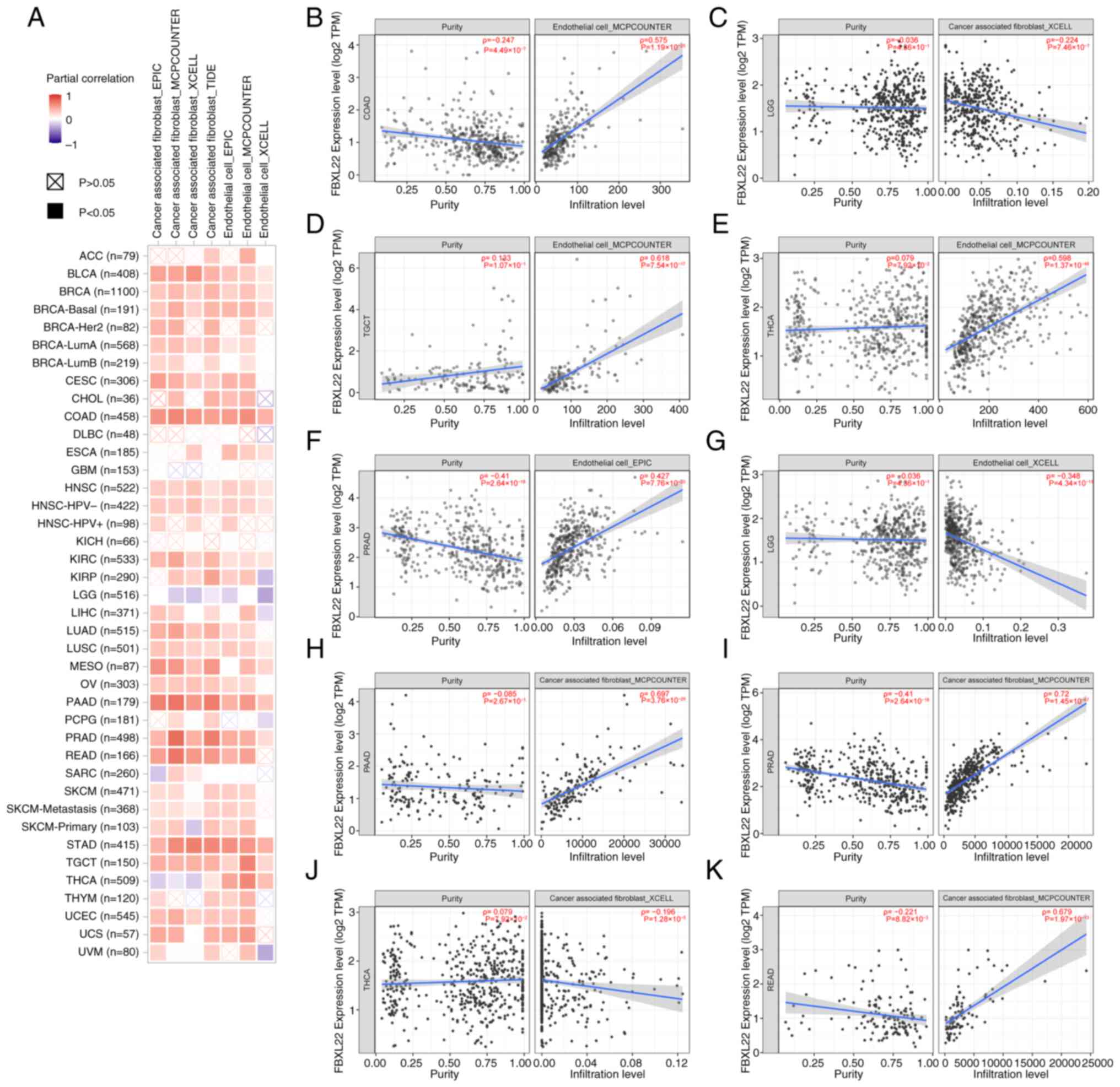 | Figure 5.Correlation analysis between FBXL22
expression and immune infiltration of CAFs and ECs across all types
of cancer in TCGA. (A) Corresponding heatmap and (B) COAD, (C) LGG,
(D) TGCT, (E) THCA, (F) PRAD, (G) LGG, (H) PAAD, (I) PRAD, (J)
THCA, (K) READ. Each scatter plot includes data points representing
FBXL22 expression levels (x-axis) and infiltration levels of CAFs
or ECs (y-axis), with a trend line indicating the correlation
direction. Spearman's correlation coefficient (ρ) and corresponding
P-value are annotated for each plot to quantify the strength and
significance of the association. FBXL22, F-box and leucine rich
repeat protein 22; CAFs, cancer-associated fibroblasts; ECs,
endothelial cells; TCGA, The Cancer Genome Atlas; TPM, transcripts
per million; COAD, colon adenocarcinoma; PRAD, prostate
adenocarcinoma; READ, rectum adenocarcinoma; TGCT, testicular germ
cell tumors; THCA, thyroid carcinoma; LGG, lower grade glioma. |
Functional enrichment analysis of
FBXL22-related genes
Protein-protein interaction analysis identified 50
FBXL22-binding partners, including filamin C (Fig. 6A). The top 100 FBXL22-correlated
genes, MRGPRF (r=0.83) and KCNMB1 (r=0.81), were enriched in
pathways such as ‘Circadian rhythm’, ‘Ubiquitin-mediated
proteolysis’ and ‘cGMP-PKG signaling’ (Figs. 6B, C and S9; Table
SIV and SV). These findings
provide insight into the molecular mechanisms by which FBXL22
exerts its effects on cancer cells and the TME.
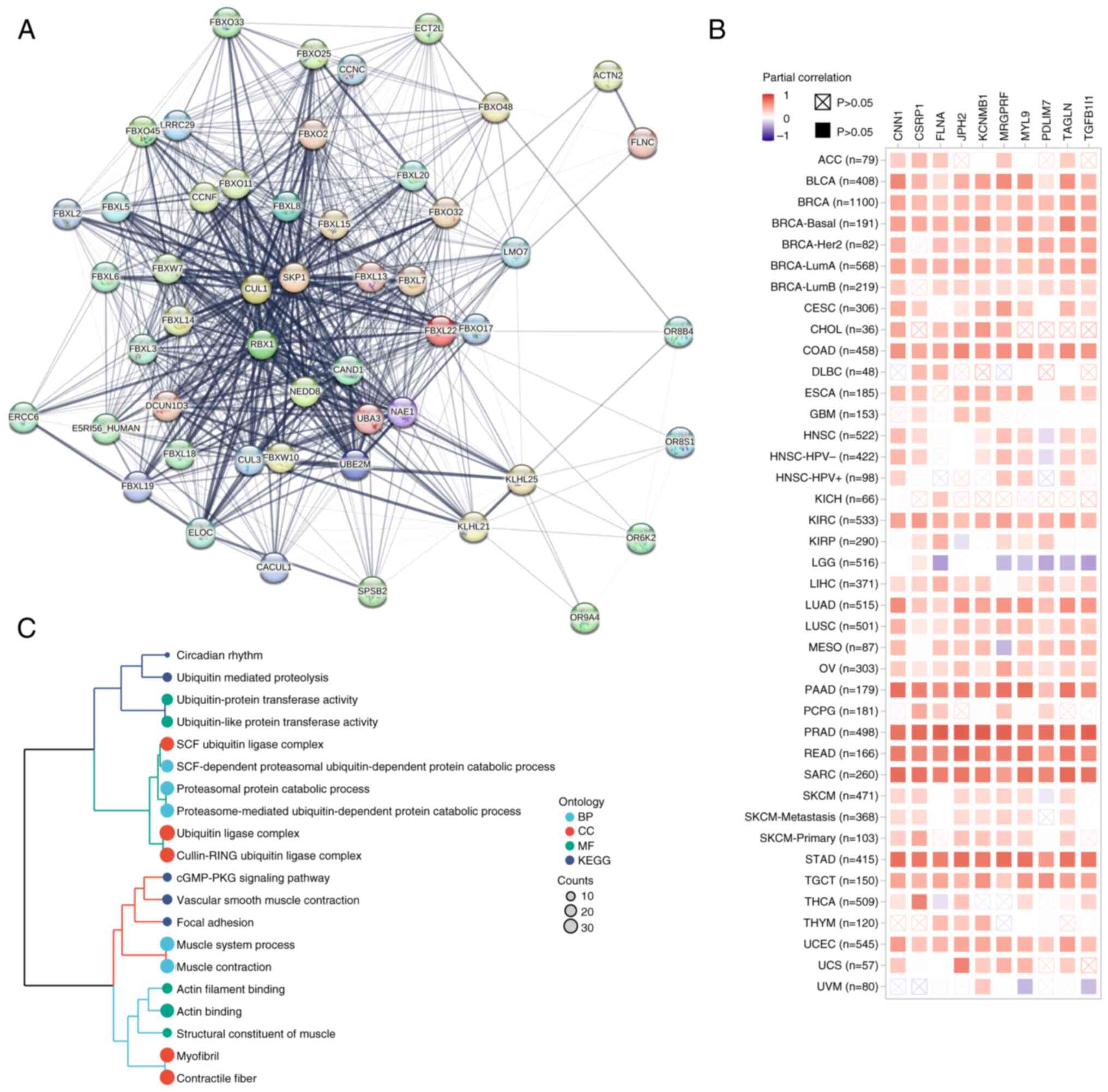 | Figure 6.FBXL22-related gene enrichment
analysis in pan-cancer. (A) Network of interactions among
FBXL22-binding proteins using the STRING tool. (B) Heatmap of
expression correlations between FBXL22 and the top 10
FBXL22-correlated genes in different cancer types of TCGA using the
GEPIA2 tools, including MRGPRF, KCNMB1, PDLIM7, TAGLN, TGFB1I1,
JPH2, CNN1, FLNA, CSRP1 and MYL9. (C) KEGG and GO enrichment
analyses are visualized by cluster tree diagram. FBXL22, F-box and
leucine rich repeat protein 22; STRING, Search Tool for the
Retrieval of Interacting Genes/Proteins; TCGA, The Cancer Genome
Atlas; GEPIA2, Gene Expression Profiling Interactive Analysis 2;
MRGPRF, Mas-related G protein-coupled receptor F; KCNMB1, potassium
calcium-activated channel subfamily M regulatory β subunit 1;
PDLIM7L, PDZ and LIM domain 7; TAGLN, Transgelin; TGFB1I1,
transforming growth factor β 1 induced 1; JPH2, junctophilin 2;
CNN1, calponin 1; FLNA, filamin A; CSRP1, cysteine and glycine rich
protein 1; MYL9, myosin light chain 9; KEGG, Kyoto Encyclopedia of
Genes and Genomes; GO, Gene ontology; BP, Biological process; CC,
Cellular component; MF, Molecular function. |
Differential expression and functional
enrichment of FBXL22 in PCa
In PCa, differential expression analysis identified
1,243 upregulated and 1,017 downregulated genes in the high-FBXL22
groups. These genes were enriched in pathways such as ‘Muscle
contraction’ and ‘cGMP-PKG signaling’ (Fig. 7A-D, Table SVI and SVII). GSEA revealed that FBXL22
negatively regulated pro-tumor pathways, such as ‘Myc active’,
‘E2F’ and ‘PLK1’, and were positively associated with
tumor-suppressive pathways, such as ‘Hippo signaling regulation’
and ‘Vitamin D receptor’ (Fig.
7E-F). These results suggested that FBXL22 serves a
tumor-suppressive role in PCa by modulating specific signaling
pathways.
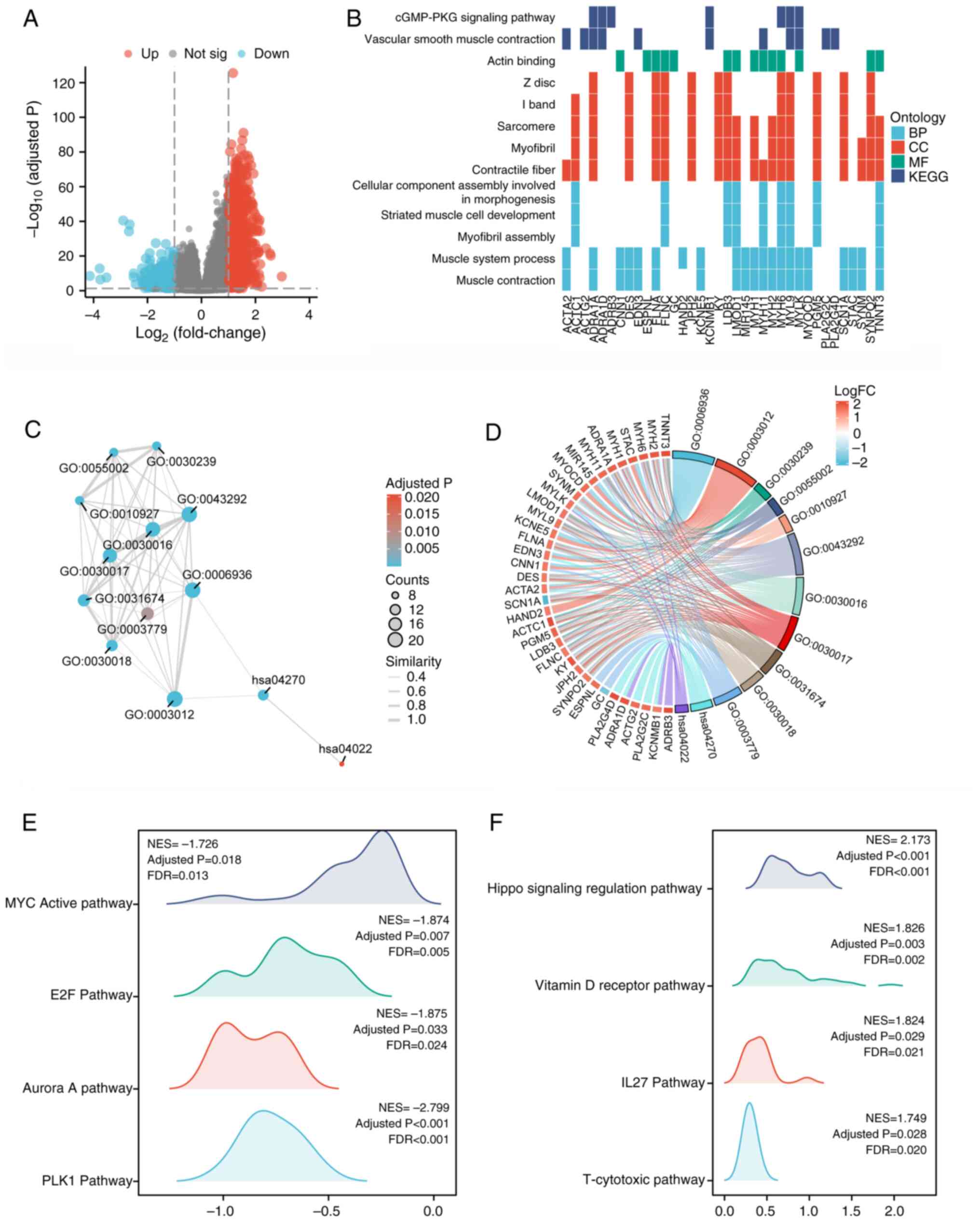 | Figure 7.Differential expression and
functional enrichment analysis of FBXL22-associated genes in
prostate cancer. (A) Volcano plot illustrating differential
expression analysis between high-FBXL22 and low-FBXL22 expression
groups in PCa, which highlights the genes that were significantly
upregulated or downregulated in association with FBXL22 expression
levels. The results of KEGG and GO enrichment analysis are depicted
using (B) heat map and (C) Enrichment Map Analysis Platform,
respectively. (D) The analysis of GO/KEGG combined log(fold change)
is represented by a chordal diagram. The mountain maps display the
pathways (E) significantly negative or (F) significantly positive
associated with expression levels of FBXL22 in GSEA analysis
results. Error bars represent standard deviation. FBXL22, F-box and
leucine rich repeat protein 22; PCa, prostate cancer; KEGG, Kyoto
Encyclopedia of Genes and Genomes; GO, Gene Ontology; GSEA, Gene
Set Enrichment Analysis; E2F, E2 DNA-binding factor; PLK1,
polo-like kinase 1; NES, nuclear export sequence; FDR, false
discovery rate; BP, biological process; CC, cellular component; MF,
molecular function. |
Inhibition of malignant behavior of
PCa cells by FBXL22
Analysis revealed that FBXL22 expression was
significantly decreased in PC3 and DU145 cells compared with normal
prostate epithelial cells (RWPE-1; Fig.
8A). Compared with NC cells, FBXL22 overexpression markedly
increased both mRNA and protein levels of FBXL22 in PC3 and DU145
cells (Fig. 8B-G). CCK-8 assays
revealed that FBXL22 overexpression significantly reduced the cell
viability of PC3 and DU145 cells (Fig.
8H and I). Transwell assays indicated that cell invasion was
significantly decreased in FBXL22-overexpressing cells compared
with NC cells (Fig. 8J-M). Wound
healing assays demonstrated that FBXL22 overexpression
substantially inhibited cell migration (Fig. 8N-Q). Flow cytometric analysis
revealed that FBXL22 overexpression significantly increased the
apoptosis rate in PC3 and DU145 cell lines (Fig. 8R-U). Co-culture experiments with NK
cells revealed enhanced migration and cytotoxic activity of NK
cells against FBXL22-overexpressing PCa cells (Fig. 8V and W). Co-IP experiments confirmed
the interaction between FBXL22 and PLK1 (Fig. 8X and Y). Collectively, these
findings indicate that FBXL22 overexpression suppresses
proliferation, invasion and migration of PCa cells while promoting
apoptosis and enhancing immune cell activity.
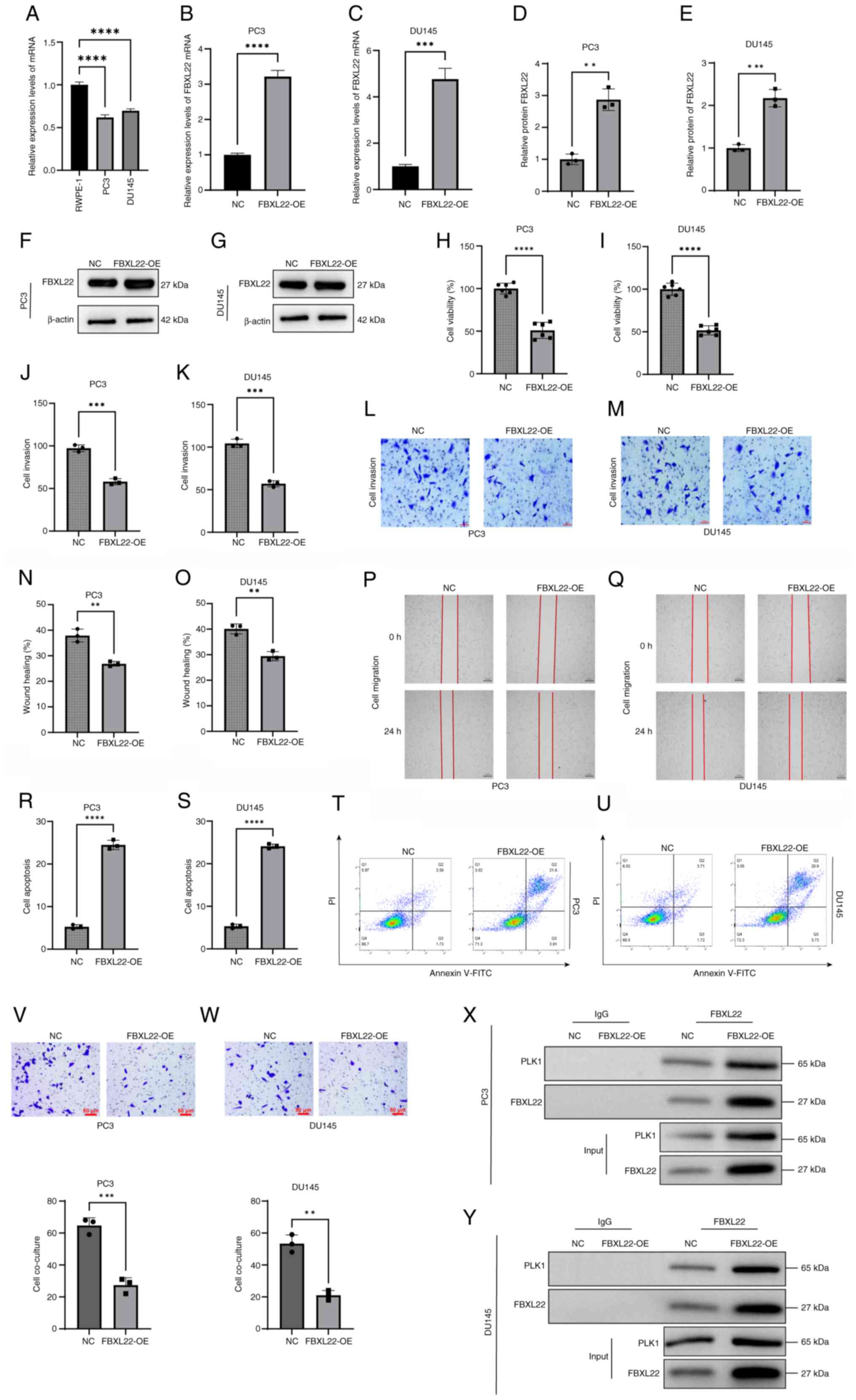 | Figure 8.Effects of FBXL22 on viability,
invasion, migration and apoptosis of PCa cells. (A) The relative
expression levels of FBXL22 in normal prostate epithelial cell
lines RWPE-1 and PCa cell lines PC3 and DU145. The relative
expression levels of FBXL22 in NC and FBXL22-OE in (B) PC3 and (C)
DU145 cell lines. Semi-quantification of relative protein levels of
FBXL22 in NC and FBXL22-OE groups in (D) PC3 and (E) DU145 cells.
Western blot of FBXL22 in NC and FBXL22-OE groups in (F) PC3 and
(G) DU145 cells. Cell viability was assessed using the CCK-8 assay
in (H) PC3 and (I) DU145 cells. (J-M) The Transwell chamber was
utilized to evaluate cell invasive ability (scale bar, 50 µm).
(N-Q) Cell migratory ability was examined using a wound healing
assay (scale bar, 250 µm). The histograms demonstrate the relative
percentage of wound healing. (R) Quantification of apoptosis rates
in PC3 cells transfected with FBXL22-OE compared with the NC group,
shown as a bar graph. (S) Quantification of apoptosis rates in
DU145 cells under the same conditions. (T) Representative flow
cytometry dot plots for PC3 cells, detailing the distribution of
cells in different apoptosis stages. (U) Representative flow
cytometry dot plots for DU145 cells, using annexin V-FITC and PI
staining to delineate cell viability and apoptosis stages. NK cell
migration and cytotoxicity against FBXL22-OE (V) PC3 and (W) DU145
cells in co-culture Transwell experiments (scale bar, 50 µm).
Co-immunoprecipitation analysis confirming the interaction between
FBXL22 and PLK1 in (X) PC3 and (Y) DU145 cells. Error bars
represent standard deviation. **P<0.01, ***P<0.001 and
****P<0.0001; ns, P≥0.05. PCa, prostate cancer; FBXL22, F-box
and leucine rich repeat protein 22; OE, overexpression; NC,
negative control; ns, not significant; NK, natural killer. |
Mechanism of FBXL22 in PCa
FBXL22 upregulation resulted in reduced levels of
phosphorylated PLK1, AR-V7 and PD-L1 (P<0.01), as well as
increased levels of cleaved caspases-3, −8 and −9 (P<0.01)
(Figs. 9A-I, S10). These findings suggest that the
modulation of the PLK1 pathway may be a key mechanism by which
FBXL22 exerts tumor-suppressive effects in PCa. These results
provide a basis for the further exploration of FBXL22 as a
therapeutic target in PCa.
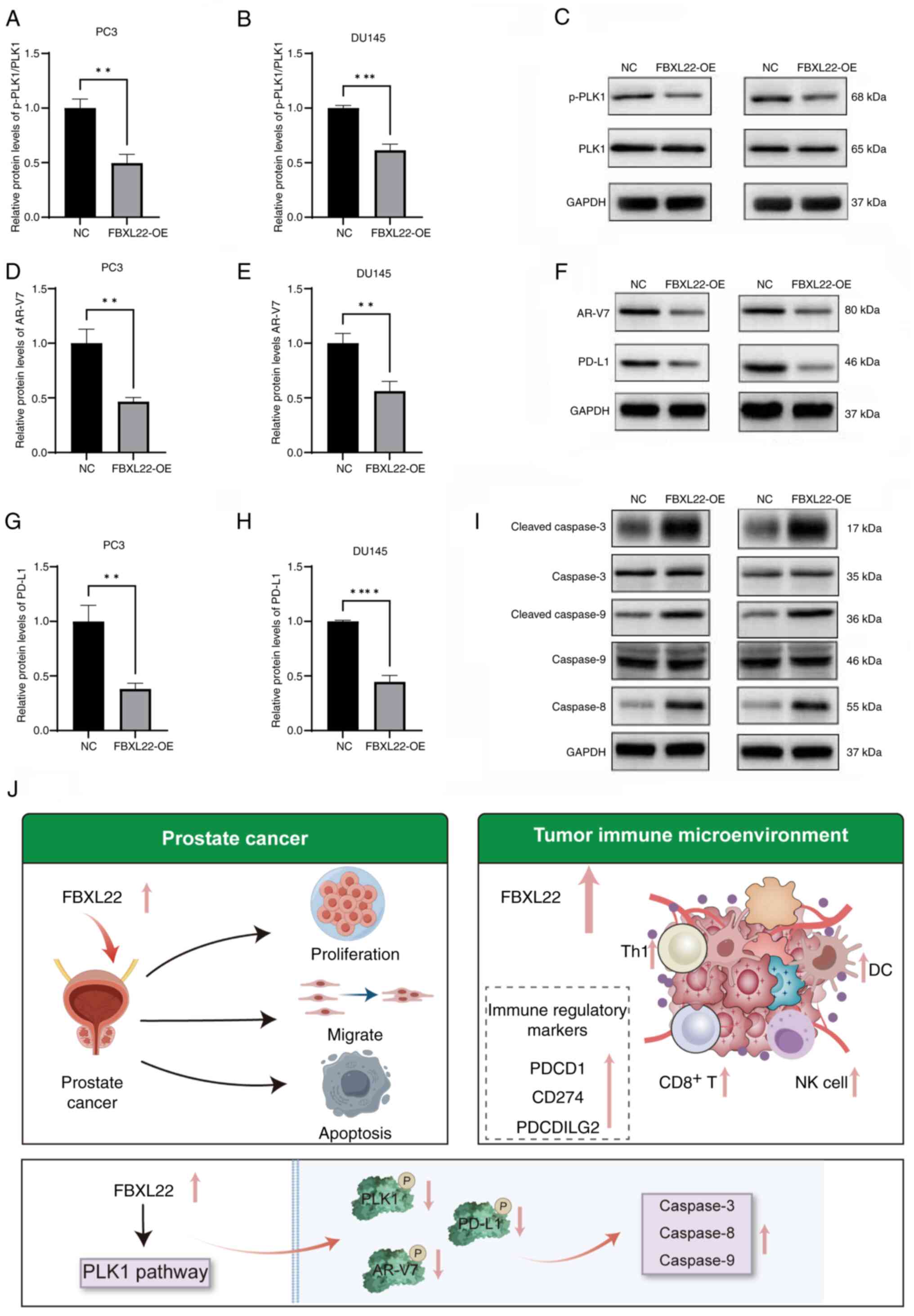 | Figure 9.Possible mechanism underlying the
impact of FBXL22 on PCa cell function. Western blot analysis
demonstrated the relative levels of p-PLK1 and PLK1 in (A) PC3 and
(B) DU145 cells transfected with FBXL22-OE compared with the NC
group. (C) Representative western blot images for p-PLK1, PLK1 and
GAPDH in PC3 and DU145 cells. Western blot analysis indicated the
relative levels of AR-V7 in (D) PC3 and (E) DU145 cells. (F)
Representative western blot images for AR-V7, PD-L1 and GAPDH in
PC3 and DU145 cells. Western blot analysis indicated the relative
levels of PD-L1 in (G) PC3 and (H) DU145 cells. (I) Representative
western blot images for cleaved-caspase-3, caspase-3,
cleaved-caspase-9, caspase-9, caspase-8 and GAPDH in PC3 and DU145
cells. (J) Mechanism by which FBXL22 functions in PCa cells. Error
bars represent standard deviation. The data indicates that FBXL22
overexpression reduces p-PLK1, AR-V7 and PD-L1 levels while
increasing cleaved caspase-3, −8 and −9 levels, which suggests
modulation of the PLK1 pathway and promotion of apoptosis.
**P<0.01, ***P<0.001 and ****P<0.0001. PCa, prostate
cancer; FBXL22, F-box and leucine rich repeat protein 22; OE,
overexpression; NC, negative control; PLK1, polo-like kinase 1;
AR-V7, androgen receptor splice variant 7; PD-L1, programmed
death-ligand 1; PDCD1, programmed cell death 1; PDCDILG2,
programmed cell death 1 Ligand 2. |
Discussion
The present study results demonstrated that FBXL22
upregulation suppresses cell viability, invasion and metastasis
while promoting apoptosis, potentially through the modulation of
the PLK1 pathway. Our comprehensive pan-cancer analysis of FBXL22
revealed its role as a circadian rhythm-regulated immune biomarker
with notable implications for cancer prognosis and therapy. These
findings indicate that FBXL22 is predominantly downregulated across
a multitude of cancer types and may serve as a prognostic and
diagnostic marker in specific cancer types, notably in PCa. This
downregulation was associated with poor OS in certain cancer types
such as BLCA and STAD, which highlights the potential utility of
FBXL22 as a prognostic biomarker (5–7).
Pan-cancer analysis of FBXL22 expression in the
present study revealed a complex and heterogeneous pattern.
Notably, FBXL22 was predominantly downregulated in most cancer
types compared with the corresponding normal tissues from the GTEx
database. However, significant overexpression was detected in CHOL
and LAML. This heterogeneity implies that FBXL22 may exert diverse
functions in different cancer contexts, which highlights the
importance of considering tissue-specific factors when interpreting
the role of FBXL22 in cancer. In cancer types such as BLCA and
STAD, elevated FBXL22 expression was associated with poor
prognosis, which appears to contradict its potential
tumor-suppressive role, as suggested by its low expression in other
cancer types. To reconcile this apparent discrepancy, we
hypothesized that the role of FBXL22 may be intricately associated
with the specific molecular landscape and TME of each cancer type.
For instance, in BLCA and STAD, FBXL22 overexpression might be
associated with the activation of alternative signaling pathways
that drive tumor progression, such as the PLK1 pathway, which has
been modulated by FBXL22 in PCa. In addition, tissue-specific
factors and genetic alterations can differentially influence the
function of FBXL22. In CHOL and LAML, the overexpression of FBXL22
might reflect its involvement in distinct pathophysiological
processes relevant to these cancer types. Further research is
warranted to delineate the specific mechanisms by which FBXL22
contributes to tumorigenesis in a cancer-specific manner, including
functional studies in CHOL and LAML models, as well as a more
detailed analysis of the molecular context in which FBXL22 exerts
its effects.
The present study results demonstrated a positive
correlation between high FBXL22 expression and immune checkpoint
genes such as PD-L1 and CTLA4, which suggests a potential role for
FBXL22 in immune suppression. However, high FBXL22 expression was
also associated with an improved response to immunotherapy, which
creates an apparent paradox. To address this contradiction, several
hypotheses were proposed that warrant further investigation. First,
the relationship between FBXL22 and immune checkpoint genes may be
context dependent. In some cancer types, FBXL22-induced immune
checkpoint expression may represent a feedback mechanism that
limits excessive immune activation, thereby maintaining immune
homeostasis while still permitting an overall immune-active
environment conducive to the immunotherapy response. This
hypothesis was supported by the observed co-expression patterns of
FBXL22 with both inhibitory and stimulatory immune checkpoint genes
across multiple cancer types, which suggests complex modulation of
immune responses. Second, the expression levels of immune
checkpoint genes, while correlated with FBXL22, may not solely
determine the therapeutic outcome but rather reflect a broader
immune landscape. High FBXL22 expression was also associated with
increased infiltration of antitumor immune cells, such as NK and
CD8+ T cells, which could outweigh the potential
immunosuppressive effects of checkpoint molecules, leading to
enhanced immunotherapy responses. Third, the TMB and the spectrum
of antigenic mutations present in tumors with high FBXL22
expression may enhance the efficacy of immune checkpoint blockade
therapy, even in the presence of elevated checkpoint expression.
This could be particularly relevant in cancer types in which FBXL22
expression is positively correlated with TMB, such as certain
subtypes of bladder and stomach cancer types. To fully understand
these interactions, future studies should incorporate in
vivo models to evaluate the direct impact of FBXL22 on immune
cell function within the TME and investigate how FBXL22 modulates
the balance between immune activation and suppression.
Additionally, analyzing the correlation among FBXL22 expression,
immune checkpoint usage and clinical outcomes across diverse cancer
types and immunotherapy regimens could provide further insights
into the role of FBXL22 as a biomarker of treatment response.
The mechanisms underlying the role of FBXL22 in
cancer appear to be multifaceted and involve circadian rhythm
regulation, ubiquitin-mediated proteolysis, focal adhesion
signaling and cGMP-PKG signaling (18). The present analysis also revealed
strong correlations between FBXL22 expression and immune checkpoint
genes, which suggests their involvement in the TME (19). This is further supported by the
observation that FBXL22 expression is associated with the
infiltration of CAFs and ECs, which may affect immunotherapy
outcomes (20,21).
In PCa, the present study provided evidence that
FBXL22 upregulation suppresses cell viability, invasion and
metastasis while promoting apoptosis, potentially by modulating the
PLK1 pathway (22). This suggests
that FBXL22 is a promising target for therapeutic interventions in
PCa.
The additional experiments further elucidate the
role of FBXL22 in PCa and the TME. The co-culture experiments with
NK cells provide evidence that FBXL22 overexpression enhances
immune cell activity, reinforcing its potential as an
immunomodulatory hub. This is supported by the observed increase in
NK cell migration and cytotoxicity, which suggests that FBXL22 may
promote antitumor immune responses by modulating the TME.
Furthermore, the Co-IP validation of the FBXL22-PLK1 interaction
strengthens the mechanistic basis for the tumor-suppressive effects
of FBXL22, likely mediated through PLK1 pathway modulation. These
findings collectively enhance current understanding of the dual
role of FBXL22 in regulating cancer cell behavior and immune cell
function, which underscores its therapeutic potential in PCa.
The correlation between FBXL22 expression and TMB,
microsatellite instability (MSI) and HRD across various cancer
types further underscores its potential as a predictive biomarker
of therapeutic response (23). The
findings of the present study indicate that FBXL22 can serve as a
biomarker to guide individualized cancer treatment decisions,
particularly in the context of immunotherapy and targeted therapies
(24).
TME, a complex network of cells and molecules that
surrounds a tumor, serves a key role in tumor biology (25). Based on TCGA data, the present
analysis suggested that FBXL22 was positively correlated with the
presence of CAFs and ECs in tumors (26). This correlation, along with the
notable associations between FBXL22 expression and genes associated
with immune checkpoint pathways and immunomodulation across
multiple cancer types, suggests that FBXL22 may function as an
oncogene involved in regulating the TME to promote an active state
or inflammatory response conducive to a robust response to immune
checkpoint inhibitor drugs (27).
Enrichment analysis integrating information about
FBXL22-binding protein genes and FBXL22 expression-related genes
across all tumor types revealed associations with processes such as
Skp1-Cullin-F-box-dependent, proteasomal, ubiquitin-dependent,
protein catabolic processes and muscle contraction (28). KEGG analysis suggested that the
pan-cancer role of FBXL22 may involve pathways, such as the
circadian rhythm, ubiquitin-mediated proteolysis, cGMP-PKG
signaling pathway and focal adhesion, which are closely associated
with tumor occurrence and development (29).
Therefore, to the best of our knowledge, the present
study provided the first comprehensive pan-cancer analysis of the
circadian rhythm gene FBXL22 and revealed its close association
with clinical prognosis, DNA methylation, immune regulation, immune
cell infiltration, TMB and MSI. The findings of the present study
suggest that FBXL22 is a promising novel marker of cancer
recurrence risk and immuno-infiltration in various tumor types,
particularly in PCa. These results contribute to current
understanding of the potential role and molecular mechanisms of
FBXL22 in tumor development and the TME based on clinical tumor
samples (30). Further research is
warranted to explore the mechanisms by which FBXL22 affects the
development and immune microenvironment of PCa and to validate its
potential as a therapeutic target.
In conclusion, the pan-cancer analysis of the
circadian rhythm gene FBXL22 revealed close associations between
its expression and clinical prognosis, DNA methylation, immune
regulation, immune cell infiltration, TMB and MSI. Furthermore,
preliminary investigations were performed into the mechanisms
underlying the potential ability of FBXL22 to inhibit PCa and
enhance the efficacy of immunotherapy. The aforementioned
comprehensive multi-omics analysis demonstrated that FBXL22 has
notable potential as a novel marker for cancer recurrence risk and
immuno-infiltration in various tumor types, particularly PCa. These
results contribute to current understanding of the potential role
and molecular mechanisms of FBXL22 in tumor development and TIM,
based on clinical tumor samples.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Hebei Province Medical
Science Research Program (grant no. 20240298) and Finance
Department of Hebei Province Government-Sponsored Program for
Cultivating Outstanding Clinical Medical Talents (grant no.
ZF2025162).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JL, WL and YS supervised the project and designed
the present study. JL, MF and JZ performed the data analysis and
prepared all the figures and tables. MF, SY and JZ performed the
experiments. JL and SY drafted the manuscript. WL and YS revised
the manuscript. JL obtained funding for the study. All authors read
and approved the final version of the manuscript. JL and WL confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BLCA
|
bladder urothelial carcinoma
|
|
BRCA
|
breast invasive carcinoma
|
|
CHOL
|
cholangiocarcinoma
|
|
COAD
|
colon adenocarcinoma
|
|
HRD
|
homologous recombination
deficiency
|
|
LAML
|
acute myeloid leukemia
|
|
LIHC
|
liver hepatocellular carcinoma
|
|
PAAD
|
pancreatic adenocarcinoma
|
|
PRAD
|
prostate adenocarcinoma
|
|
READ
|
rectum adenocarcinoma
|
|
STAD
|
stomach adenocarcinoma
|
|
THCA
|
thyroid carcinoma
|
|
TMB
|
tumor mutation burden
|
|
UCEC
|
uterine corpus endometrial
carcinoma
|
|
UVM
|
uveal melanoma
|
References
|
1
|
Martin RM, Donovan JL, Turner EL, Metcalfe
C, Young GJ, Walsh EI, Lane JA, Noble S, Oliver SE, Evans S, et al:
Effect of a low-intensity PSA-based screening intervention on
prostate cancer mortality: The CAP randomized clinical trial. JAMA.
319:883–895. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xuan W, Khan F, James CD, Heimberger AB,
Lesniak MS and Chen P: Circadian regulation of cancer cell and
tumor microenvironment crosstalk. Trends Cell Biol. 31:940–950.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu X, Maier G and Panda S: Learning from
circadian rhythm to transform cancer prevention, prognosis, and
survivorship care. Trends Cancer. 10:196–207. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI
|
|
5
|
Wang C, Barnoud C, Cenerenti M, Sun M,
Caffa I, Kizil B, Bill R, Liu Y, Pick R, Garnier L, et al:
Dendritic cells direct circadian anti-tumour immune responses.
Nature. 614:136–143. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang M, Jia K, Wang L, Li W, Chen B, Liu
Y, Wang H, Zhao S, He Y and Zhou C: Alterations of DNA damage
response pathway: Biomarker and therapeutic strategy for cancer
immunotherapy. Acta Pharm Sin B. 11:2983–2994. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shang S, Liu J, Verma V, Wu M, Welsh J, Yu
J and Chen D: Combined treatment of non-small cell lung cancer
using radiotherapy and immunotherapy: Challenges and updates.
Cancer Commun (Lond). 41:1086–1099. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spaich S, Will RD, Just S, Spaich S, Kuhn
C, Frank D, Berger IM, Wiemann S, Korn B, Koegl M, et al: F-box and
leucine-rich repeat protein 22 is a cardiac-enriched F-box protein
that regulates sarcomeric protein turnover and is essential for
maintenance of contractile function in vivo. Circ Res.
111:1504–1516. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hughes DC, Baehr LM, Driscoll JR, Lynch
SA, Waddell DS and Bodine SC: Identification and characterization
of Fbxl22, a novel skeletal muscle atrophy-promoting E3 ubiquitin
ligase. Am J Physiol Cell Physiol. 319:C700–C719. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Beisaw A, Kuenne C, Guenther S, Dallmann
J, Wu CC, Bentsen M, Looso M and Stainier DYR: AP-1 contributes to
chromatin accessibility to promote sarcomere disassembly and
cardiomyocyte protrusion during zebrafish heart regeneration. Circ
Res. 126:1760–1778. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haddad SA, Ruiz-Narváez EA, Haiman CA,
Sucheston-Campbell LE, Bensen JT, Zhu Q, Liu S, Yao S, Bandera EV,
Rosenberg L, et al: An exome-wide analysis of low frequency and
rare variants in relation to risk of breast cancer in African
American Women: the AMBER Consortium. Carcinogenesis. 37:870–877.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tekcham DS, Chen D, Liu Y, Ling T, Zhang
Y, Chen H, Wang W, Otkur W, Qi H, Xia T, et al: F-box proteins and
cancer: An update from functional and regulatory mechanism to
therapeutic clinical prospects. Theranostics. 10:4150–4167. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Tan Z, Yang S, Song X and Li W: A
circadian rhythm-related gene signature for predicting relapse risk
and immunotherapeutic effect in prostate adenocarcinoma. Aging
(Albany NY). 14:7170–7185. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2:1001412021.PubMed/NCBI
|
|
15
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–16.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ukimura O, Coleman JA, de la Taille A,
Emberton M, Epstein JI, Freedland SJ, Giannarini G, Kibel AS,
Montironi R, Ploussard G, et al: Contemporary role of systematic
prostate biopsies: Indications, techniques, and implications for
patient care. Eur Urol. 63:214–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nguyen L, J WMM, Van Hoeck A and Cuppen E:
Pan-cancer landscape of homologous recombination deficiency. Nat
Commun. 11:55842020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonzalez D and Stenzinger A: Homologous
recombination repair deficiency (HRD): From biology to clinical
exploitation. Genes Chromosomes Cancer. 60:299–302. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Antonarakis ES, Lu C, Wang H, Luber B,
Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et
al: AR-V7 and resistance to enzalutamide and abiraterone in
prostate cancer. N Engl J Med. 371:1028–1038. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng Z, Li J, Liu Y, Shi Z, Xuan Z, Yang
K, Xu C, Bai Y, Fu M, Xiao Q, et al: The crucial role of AR-V7 in
enzalutamide-resistance of castration-resistant prostate cancer.
Cancers. 14:48772022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
You S, Knudsen BS, Erho N, Alshalalfa M,
Takhar M, Al-Deen Ashab H, Davicioni E, Karnes RJ, Klein EA, Den
RB, et al: Integrated classification of prostate cancer reveals a
novel luminal subtype with poor outcome. Cancer Res. 76:4948–4958.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao SG, Chang SL, Erho N, Yu M, Lehrer J,
Alshalalfa M, Speers C, Cooperberg MR, Kim W, Ryan CJ, et al:
Associations of luminal and basal subtyping of prostate cancer with
prognosis and response to androgen deprivation therapy. JAMA Oncol.
3:1663–1672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Myers JA and Miller JS: Exploring the NK
cell platform for cancer immunotherapy. Nat Rev Clin Oncol.
18:85–100. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Xiang Y, Xin VW, Wang XW, Peng XC,
Liu XQ, Wang D, Li N, Cheng JT, Lyv YN, et al: Dendritic cell
biology and its role in tumor immunotherapy. J Hematol Oncol.
13:1072020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dhanasekaran R, Deutzmann A,
Mahauad-Fernandez WD, Hansen AS, Gouw AM and Felsher DW: The MYC
oncogene- the grand orchestrator of cancer growth and immune
evasion. Nat Rev Clin Oncol. 19:23–36. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu X, Boufaied N, Hallal T, Feit A, de
Polo A, Luoma AM, Alahmadi W, Larocque J, Zadra G, Xie Y, et al:
MYC drives aggressive prostate cancer by disrupting transcriptional
pause release at androgen receptor targets. Nat Commun.
13:25592022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arriaga JM, Panja S, Alshalalfa M, Zhao J,
Zou M, Giacobbe A, Madubata CJ, Kim JY, Rodriguez A, Coleman I, et
al: A MYC and RAS co-activation signature in localized prostate
cancer drives bone metastasis and castration resistance. Nat
Cancer. 1:1082–1096. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Faskhoudi MA, Molaei P, Sadrkhanloo M,
Orouei S, Hashemi M, Bokaie S, Rashidi M, Entezari M, Zarrabi A,
Hushmandi K, et al: Molecular landscape of c-Myc signaling in
prostate cancer: A roadmap to clinical translation. Pathol Res
Pract. 233:1538512022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han Z, Mo R, Cai S, Feng Y, Tang Z, Ye J,
Liu R, Cai Z, Zhu X, Deng Y, et al: Differential expression of E2F
transcription factors and their functional and prognostic roles in
human prostate cancer. Front Cell Dev Biol. 10:8313292022.
View Article : Google Scholar : PubMed/NCBI
|