Introduction
Kidney cancer remains one of the most prevalent and
deadliest tumors in urological oncology, with ~155,702 mortalities
and 434,419 new cases recorded worldwide in 2022 (1). Clear cell renal cell carcinoma (ccRCC)
accounts for >70% of all kidney cancer cases and is the most
common subtype (2). ccRCC typically
exhibits aggressive growth, frequent metastasis and substantial
immune infiltration, with ~30% of patients experiencing recurrence
after total nephrectomy (3,4). Conventional radiation therapy and
chemotherapy have demonstrated limited efficacy in the treatment of
all renal cell carcinoma subtypes (5). Immunotherapy and targeted therapy have
shown considerable potential in patients with ccRCC. However, owing
to low response rates, immunotherapy may not be suitable for all
individuals (6). In addition,
patients receiving targeted therapy are prone to resistance,
limiting the efficacy of existing treatments and complicating ccRCC
control (7). The outcomes of
patients with ccRCC are primarily affected by drug resistance and
metastasis (8). Therefore, to
effectively treat metastatic ccRCC, it is clinically important to
identify the most preferable biomarkers and immune-related
therapeutic targets.
Human guanylate-binding protein 5 (GBP5), which
belongs to the guanylate-binding protein (GBP) family, is
classified under the dynamin superfamily of large GTPases that are
induced by interferons (9,10). It is regarded as a key regulator of
immune responses in neoplastic diseases (9). Studies have shown that GBP5 exerts
antiviral activity and influences innate immunity and inflammation
(11,12). Due to the notable functions of other
members of the GBP family in proliferation and invasion, GBP5 may
also serve a major role in malignancy (13). Previous studies have supported the
key role of GBP5 in cancer progression. In gastric adenocarcinomas,
GBP5 expression is markedly upregulated, contributing to cancer
development through the Janus kinase 1 (JAK1)-STAT1/GBP5/C-X-C
motif chemokine ligand 8 (CXCL8) signaling circuit (14,15).
In addition, GBP5 enhances the migration, invasion and
proliferation of glioblastoma cells, promoting glioblastoma
progression through the proto-oncogene tyrosine-protein kinase
Src/ERK1/2/MMP3 pathway (16).
Furthermore, in triple-negative breast cancer cells exhibiting high
GBP5 expression, GBP5 knockdown notably reduces the number of
migrating cells, the activity of the IFN-γ/STAT1 and TNF-α/NF-κB
signaling axes and the expression of programmed death-ligand 1
(PD-L1) (17).
Previous reports have outlined the relevance of GBP5
in tumor immunity. Several studies have indicated that GBP5
expression is associated with the extent of immune cell
infiltration in breast cancer (17), melanoma (18), colon cancer (19), ovarian cancer (20), hepatocellular cancer (21) and small-cell lung cancer (22), emphasizing the role of GBP5 in
shaping the tumor immune microenvironment (TME). In oral squamous
cell carcinoma, GBP5 serves as an immune-related biomarker that
induces NF-κB activation and facilitates radioresistance, PD-L1
upregulation and tumor metastasis (23). In hepatocellular carcinoma and
ovarian cancer, GBP5 not only activates the PI3K-AKT signaling
pathway but also modulates tumor metabolism and immune evasion
through the regulation of DNA methylation and micro-RNA networks
(24,25). Furthermore, in cutaneous melanoma
and ovarian cancer, GBP5 induces pyroptosis through the
JAK2-STAT1-caspase-1 axis, therefore influencing cell death and
immune responses. Overall, GBP5 modulates the immune
microenvironment which further influences the prognosis of diverse
tumor types (20,26). In ccRCC, immune-suppressive cells,
such as regulatory T cells and myeloid-derived suppressor cells,
induce immune dysfunction, leading to poor therapeutic outcomes
(27). However, the precise
function of GBP5 is still not fully understood in ccRCC.
The present study employed R software together with
a variety of bioinformatics tools to integrate and analyze data,
aiming to comprehensively investigate the role of GBP5 in ccRCC.
Specifically, its differential expression patterns, associations
with clinicopathological features, prognostic and diagnostic
significance, protein-protein interaction networks, biological
functions, and potential involvement in tumor immune cell
infiltration were examined. Furthermore, in vitro cellular
experiments were performed to validate and further explore the
impact of GBP5 on the biological behavior of ccRCC. To the best of
our knowledge, the results of the present study demonstrate the
biological function and clinical significance of GBP5 and its
influence on tumor immunity in ccRCC for the first time. Fig. 1 illustrates the study design and
workflow, highlighting the step-by-step process from data
acquisition to experimental validation. The present study provides
novel insights into the function of GBP5 in ccRCC, potentially
offering new biomarkers and therapeutic targets for the diagnosis
and treatment of ccRCC.
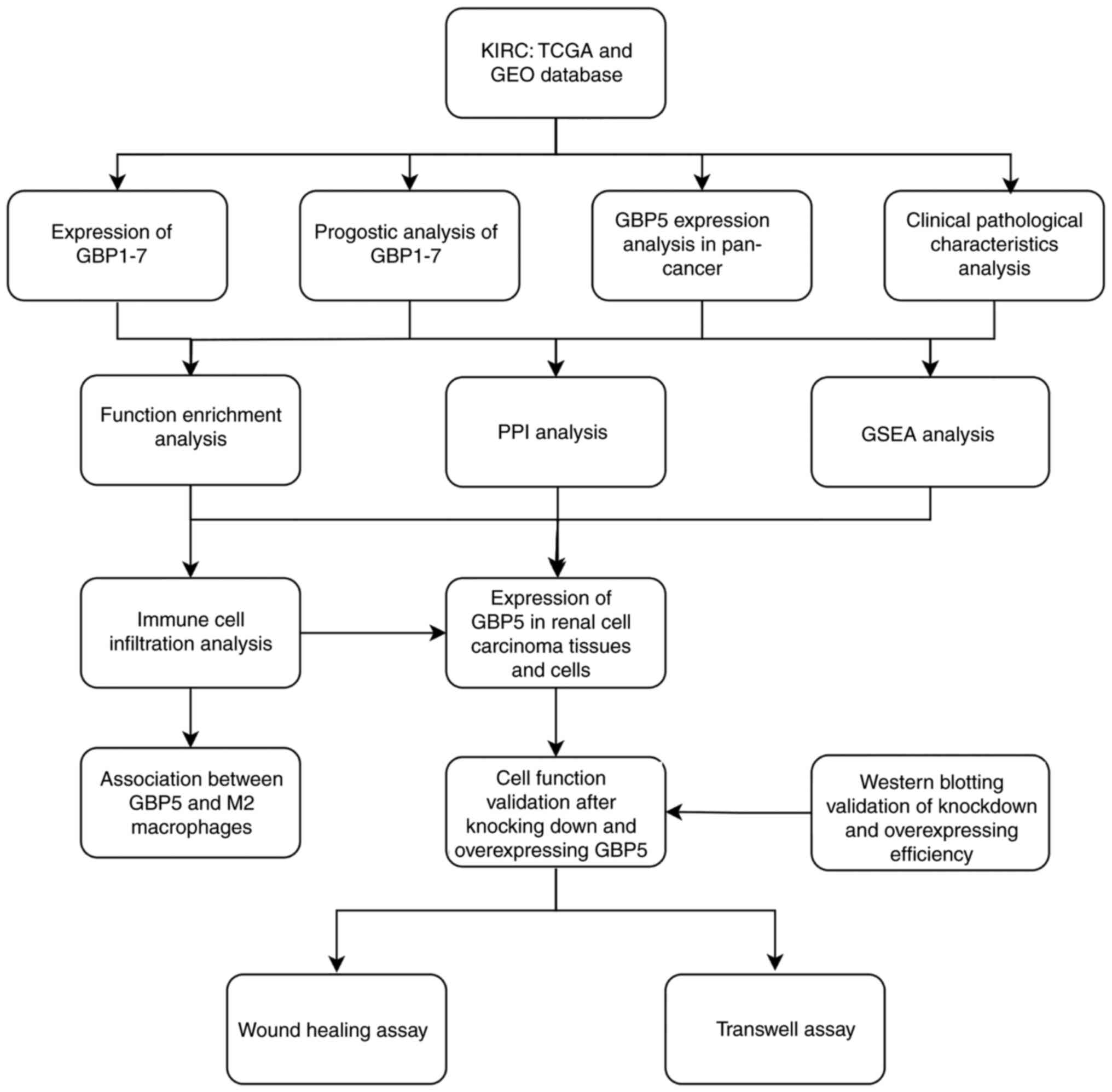 | Figure 1.Overall workflow of the study.
Schematic representation of the major steps and analytical methods
employed, offering a concise and visual summary of the entire
research process. KIRC datasets from TCGA and GEO databases were
utilized to perform differential gene expression and prognostic
analyses of GBP1-7 genes, with a focus on the clinical pathological
characteristics of GBP5 in ccRCC, as well as the gene function
enrichment analysis, PPI network analysis, GSEA analysis and immune
cell infiltration analysis conducted. Finally, wound-healing and
Transwell assays were conducted to evaluate the role of GBP5 in the
invasion and migration of ccRCC cells. KIRC, kidney renal clear
cell carcinoma; TCGA, The Cancer Genome Atlas; GEO, Gene Expression
Omnibus; GBP, guanylate-binding protein; PPI, protein-protein
interaction; GSEA, gene set enrichment analysis; ccRCC, clear cell
renal cell carcinoma. |
Materials and methods
Gene Expression Profiling Interactive
Analysis 2 (GEPIA2)
GEPIA2 (http://gepia2.cancer-pku.cn/) analyzes gene expression
differences based on tumor samples and normal samples from both The
Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov) and the Genotype-Tissue
Expression (https://www.gtexportal.org) databases (28). In the present study, this tool
facilitated the comparison of GBP5 expression between renal tumor
tissue and adjacent normal tissue.
Gene Expression Omnibus (GEO) dataset
selection
From the GEO database (http://www.ncbi.nlm.nih.gov/geo), two datasets were
acquired. GSE53757 (29) included
GBP1-6 mRNA data from 72 ccRCC and 72 normal tissues. GSE36895
(30) provided GBP1-6 mRNA data
from 23 ccRCC samples and 23 normal tissues. Analysis of GBP1-6
mRNA levels was conducted using these two datasets.
Kaplan-Meier (KM) plotter
As an online tool, the KM plotter (https://kmplot.com/analysis) provided survival
analysis capabilities for 21 tumor types by associating gene
expression profiles with clinical prognosis (31). Using this resource, the KIRC dataset
was selected in the pan-cancer module, and the ‘auto select best
cutoff’ option was applied for patient grouping to evaluate the
overall survival of GBP1-7.
GeneMANIA database analysis
GeneMANIA (http://www.genemania.org) is a publicly accessible
tool for analyzing genetic and protein interactions, co-expression,
pathways and gene co-localization (32). The potential interactions of GBP5
with related genes were assessed using this platform.
TIMER database analysis
TIMER (https://cistrome.shinyapps.io/timer/) is based on six
algorithms used to evaluate immune infiltration levels (33). The impact of GBP5 expression on
immune cell infiltration in kidney renal clear cell carcinoma
(KIRC) was analyzed in the present study. TIMER2.0 (http://timer.comp-genomics.org/) was also
employed to investigate GBP5 expression differences between tumor
tissue and adjacent tissue, alongside its association with immune
cell gene markers in KIRC (34).
Tumor Immune Single-cell Hub 2
(TISCH2) database analysis
Based on single-cell RNA sequencing, the TISCH2
database (http://tisch.comp-genomics.org/) is designed to
analyze the TME at single-cell resolution (35). The distribution and expression
patterns of GBP5 in the TME were analyzed using the GSE111360
(36) and GSE121636 (37) datasets.
Bioinformatics analysis
TCGA-KIRC RNA-sequencing data were downloaded from
TCGA database. Transcript per million format expression data and
the corresponding clinical information were extracted. Boxplots
illustrating GBP5 expression across different clinicopathological
features, including tumor-lymph node-metastasis (TNM) stages, were
generated using the ‘ggplot2 v3.3.5’ R package (RStudio, Inc.).
Receiver operating characteristic (ROC) curves were plotted to
assess the diagnostic value of GBP5 in ccRCC. Based on the criteria
of log2 fold change ≥2 and the adjusted P-value of
<0.05, GBP5-related differentially expressed genes (DEGs) were
identified and visualized through a volcano plot plotted using the
aforementioned ‘ggplot2’ R package. Gene Ontology (GO) biological
process enrichment analysis was performed using the DAVID database
(https://david.ncifcrf.gov/tools.jsp).
Gene set enrichment analysis (GSEA) was conducted to explore the
relevant molecular signaling pathways. Data from the Tracking Tumor
Immunophenotype (TIP) database (http://biocc.hrbmu.edu.cn/TIP) were used for immune
cell infiltration analysis (38).
The relationship between GBP5 expression and immune cell
infiltration and circulation was assessed using Spearman's rank
correlation analysis.
Clinical samples
A total of eleven pairs of clinical specimens were
obtained from the University of Hong Kong-Shenzhen Hospital
(Shenzhen, P.R. China) between August 2024 and November 2024.
Patients were eligible if no radiotherapy or chemotherapy had been
administered prior to biopsy, the pathological diagnosis confirmed
ccRCC and no other malignant tumors were present. The Ethics
Committee of the University of Hong Kong-Shenzhen Hospital approved
the collection of renal carcinoma tumor tissues and adjacent normal
tissues (approval no. 2024256), ensuring that all procedures
complied with the ethical guidelines of the Declaration of
Helsinki. All the donors signed an informed consent form.
Cell culture
Human ACHN, Caki-1, 786-O and human proximal tubular
epithelial (HK-2) cell lines were procured from Procell Life
Science & Technology Co., Ltd. RPMI 1640 complete medium
(Procell Life Science & Technology Co., Ltd.) was used to
culture the 786-O and ACHN cells. For Caki-1 cells, McCoy's 5A
complete medium (Procell Life Science & Technology Co., Ltd.)
was used. For HK-2 cells, Dulbecco's modified eagle medium (Gibco;
Thermo Fisher Scientific, Inc.) was utilized. A 10% fetal bovine
serum solution (FBS; Procell Life Science & Technology Co.,
Ltd.) and 1% antibiotics solution (100 µg/ml streptomycin and 100
U/ml penicillin) were added to prepare the complete medium.
Cultures were placed at 37°C in a 5% CO2 incubator,
Mycoplasma contamination was checked in all cell lines and
short tandem repeats analysis was used for authentication.
Transfection of lentivirus
Lentivirus overexpression, GBP5 overexpression (GBP5
OE; 47597-2; order of vector elements:
Ubi-MCS-3FLAG-SV40-puromycin), vector controls (CON254), GBP5 short
hairpin RNA (shRNA; order of vector elements:
hU6-MCS-CBh-gcGFP-IRES-puromycin) and negative control shRNAs
(shNC) were produced by Shanghai GeneChem Co., Ltd. The target
sequences of the shRNAs were as follows: shNC (CON313),
5′-TTCTCCGAACGTCACGT-3′; the GBP5-specific shRNA shGBP5#1
(103584–1), 5′-CTGGAAATAGATGGGCAACTT-3′ and the GBP5-specific shRNA
shGBP5#2 (103585–1), 5′-TGCCTCATCGAGAACTTTAAT-3′. Caki-1 and 786-O
cells were transfected according to manufacturer's instructions.
Cells infected with lentivirus were treated with puromycin (2
µg/ml) for 7 days, followed by evaluation of target gene knockdown
and overexpression efficiency using western blot analysis. The
stable cell lines obtained after successful selection and
maintained within 50 passages were used for subsequent
experiments.
Western blotting
Tissue and cellular protein lysates were prepared
using radioimmunoprecipitation assay buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology) supplemented with protease and
phosphatase inhibitors (cat. no. P1048, Beyotime Institute of
Biotechnology). Protein concentrations were determined using the
bicinchoninic acid method (cat. no. P0012; Beyotime Institute of
Biotechnology). The proteins were resolved using 8–10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis, the proteins
(10–50 µg) were then transferred on to nitrocellulose membranes
(cat. no. 66485; Pall Life Sciences). The membranes were blocked
with 5% skimmed milk for 1 h at 20–25°C, after which the membranes
were incubated at 4°C for 8–12 h with primary antibodies against
CD163 (1:1,000; cat. no. R381830; Chengdu Zen-BioScience Co.,
Ltd.), GAPDH (1:5,000; cat. no. D110016-0100; Sangon Biotech Co.,
Ltd.), β-actin (1:5,000; cat. no. D110022-0100; Sangon Biotech Co.,
Ltd.), Flag (1:5,000; cat. no. 20543-1-AP; Proteintech Group, Inc.)
and GBP5 (1:2,000; cat. no. 13220-1-AP; Proteintech Group, Inc.).
The membranes were incubated with secondary antibodies [horseradish
peroxidase (HRP)-conjugated goat anti-rabbit IgG; 1:5,000; cat. no.
511203; Chengdu Zen-BioScience Co., Ltd.] and HRP-conjugated goat
anti-mouse IgG (1:5,000; cat. no. 511103; Chengdu Zen-BioScience
Co., Ltd.) at 20–25°C for 2 h. Protein signals were detected using
the ChemiDoc™ MP enhanced chemiluminescence system
(Bio-Rad Laboratories, Inc.) and quantified using the ImageJ
software (version 1.40 g; National Institutes of Health).
Wound healing assay
786-O and Caki-1 cells were seeded in 12-well plates
and cultured until they reached full confluence. A 1,000 µl pipette
tip was used to create a linear scratch in the cell monolayer. The
wells were rinsed twice with PBS to remove detached cells and the
adherent cells were maintained in a medium containing 1% FBS.
Images of the wound area were captured at 0, 10, 12, 24 and 36 h
using time-lapse imaging under a light microscope (magnification,
×4).
Transwell assay
Transwell assay was performed to assess the
migratory potential of ccRCC cells. Cell migration was evaluated
using uncoated Transwell chambers (cat. no. 14341, 8 µm pore size;
LABSELECT®; Lanjieke Technology Co., Ltd.) and cell
invasion was assessed using Matrigel-coated inserts (Corning, Inc.)
pre-incubated at 37°C for 30 min. A total of 3×104 786-O
and Caki-1 cells were seeded into the upper chambers of 24-well
plates containing serum-free medium. Culture medium supplemented
with 15% FBS was added to the lower chambers, making up 500 µl as a
chemoattractant. After incubation at 37°C for 24 h for the invasion
assay and 12 h for the migration assay, the cells that had moved to
the lower surface of the membrane were fixed with 4%
paraformaldehyde at 20–25°C for 30 min, followed by crystal violet
staining at 20–25°C for 10 min. Following staining, the cells were
imaged using a light microscope (magnification, ×10) and cell
counts were performed in three or more randomly selected fields of
view.
Statistical analysis
GraphPad Prism (version 7.0; GraphPad; Dotmatics)
was used for statistical analyses. Each experiment was performed in
triplicate. Continuous variables are presented as mean ± standard
deviation. Both unpaired and paired two-tailed Student's t-tests
were used for comparisons between groups. For comparisons involving
multiple groups, Tukey's post hoc test was applied after one-way
ANOVA. Spearman's or Pearson's correlation coefficients were
determined, depending on suitability, to perform correlation
analyses. P<0.05 was considered to indicate a statistically
significant difference. Additionally, the following criteria
indicated statistical significance: *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001.
Results
Role of GBP expression and its
prognostic significance in ccRCC
Research suggests that the GBP gene family members
regulate the biological behavior of various cancers, indicating
their role in cancer initiation and progression (12). Therefore, the GEPIA2.0 tool was
initially applied to assess the expression of the GBP family
members in ccRCC. Results revealed that GBP1-7 exhibited a
consistent expression pattern, characterized by upregulation in
tumor tissues (Fig. 2A-G). Notably,
the upregulation of GBP1, GBP2 (39), GBP4 and GBP5 was statistically
significant. This observation was consistent with the results of
the analysis of ccRCC datasets from the GEO database, further
reinforcing their validity (Fig. 2H and
I). Furthermore, the Kaplan-Meier survival analysis results
indicated that elevated levels of GBP2 (39) and GBP5 were associated with a poor
prognosis, whereas low expression of GBP3, GBP4 and GBP7 was
associated with unfavorable outcomes (Fig. 2J-P). Collectively, these findings
support the hypothesis that the GBP gene family is critically
involved in ccRCC, with individual members exhibiting distinct
clinical relevance. Research specifically targeting GBP5 in ccRCC
has been limited. Therefore, the present study focused on
elucidating the function of GBP5 in ccRCC.
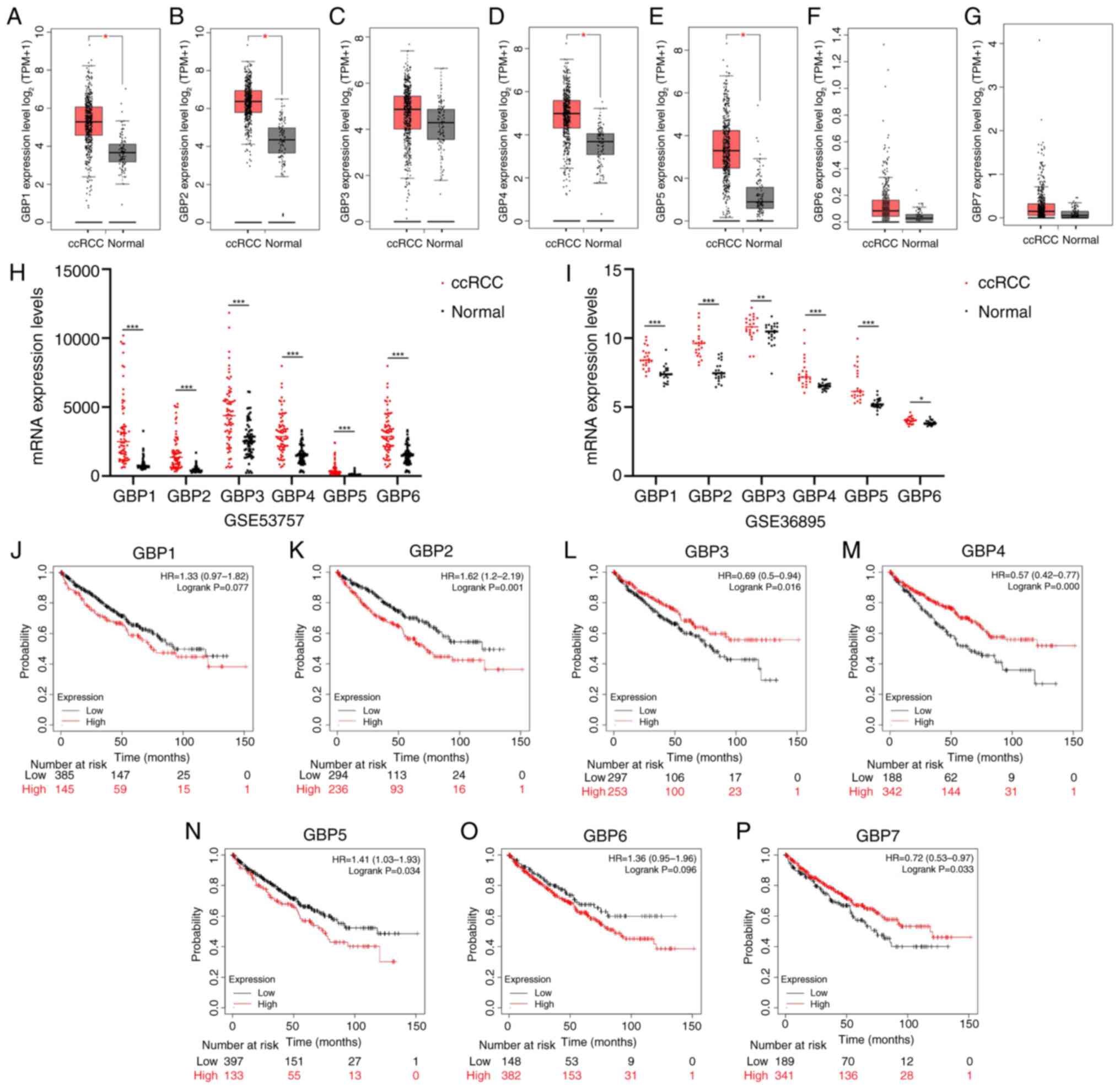 | Figure 2.Expression and prognostic analysis of
GBP1-7 in ccRCC. (A-G) The expression levels of GBP1-7 in KIRC and
matching adjacent tissues were assessed using the gene expression
profiling interactive analysis 2 online tool. Results indicate that
GBP1, GBP2, GBP4 and GBP5 are highly expressed in KIRC tissues. The
mRNA expression of GBP1-6 in ccRCC and adjacent tissues from Gene
Expression Omnibus databases (H) GSE53757 (n=72) and (I) GSE36895
(n=23) was analyzed. Results show that GBP1-6 are highly expressed
in ccRCC tissues. (J-P) Kaplan-Meier survival analysis results
indicate that elevated levels of GBP2 and GBP5 are associated with
a poor prognosis, whereas low expression of GBP3, GBP4 and GBP7 is
associated with unfavorable outcomes. *P<0.05, **P<0.01 and
***P<0.001. GBP, guanylate-binding protein; ccRCC, clear cell
renal cell carcinoma; KIRC, kidney renal clear cell carcinoma; TPM,
transcript per million; HR, hazard ratio. |
Clinical characteristics and
diagnostic value of GBP5 expression in ccRCC
To comprehensively assess the expression profile of
GBP5 across multiple tumor types, the TIMER database was employed.
Analysis showed a marked increase in GBP5 expression across a wide
range of malignancies, including KIRC and kidney renal papillary
cell carcinoma (Fig. 3A). To
investigate the clinical relevance of GBP5 in ccRCC, an in-depth
analysis was performed using transcriptomic and clinical data from
TCGA. Elevated GBP5 expression was significantly associated with
notable clinicopathological factors, including tumor size (T
stage), lymph node involvement (N stage), distant metastasis (M
stage) and both clinical and pathological stage classifications
(Fig. 3B-E). Notably, GBP5
expression increased progressively with advancing tumor stage. In
addition, univariate Cox regression analysis indicated that
elevated GBP5 expression was associated with unfavorable clinical
characteristics in patients with ccRCC (Fig. 3F). To evaluate the diagnostic
potential of GBP5 in ccRCC, ROC curve analysis was performed,
revealing a good diagnostic efficacy with an area under the curve
(AUC) value of 0.934 (Fig. 3G),
suggesting high sensitivity and specificity. These results imply
that GBP5 is a promising diagnostic biomarker involved in the
progression of ccRCC.
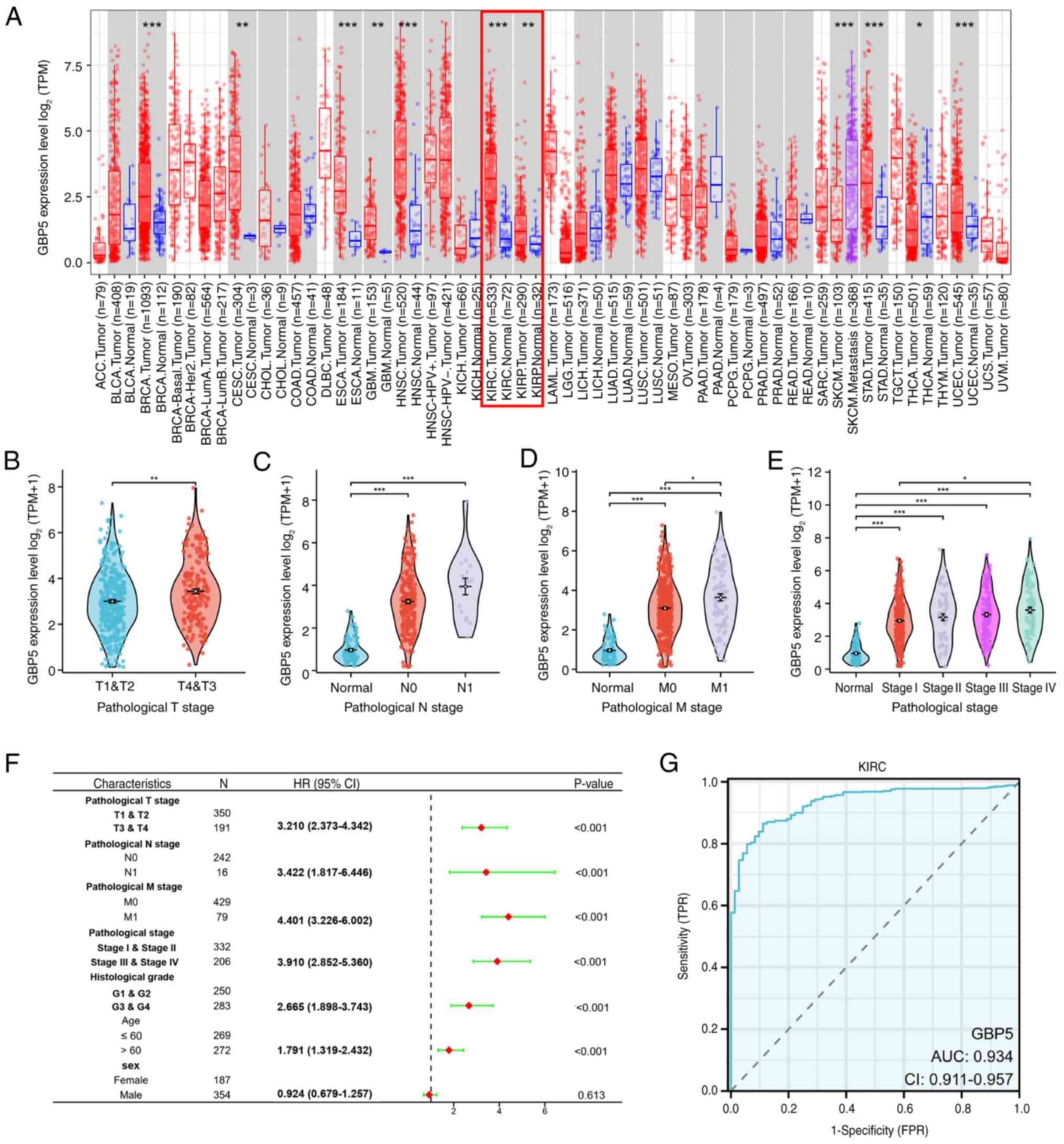 | Figure 3.Clinical characteristics of GBP5 in
ccRCC. (A) The analysis of GBP5 mRNA expression in tumor and normal
tissues was performed using the Tumor Immune Estimation Resource
2.0. Analysis showed a marked increase in GBP5 expression across a
wide range of malignancies, notably KIRC and KIRP. The association
of GBP5 mRNA expression with different clinical variables in
patients with KIRC, such as (B) T stage, (C) N stage, (D) M stage
and (E) pathological stage. (F) Univariate cox regression analysis
indicates that elevated GBP5 expression is linked to unfavorable
clinical characteristics in patients with ccRCC. (G) Receiver
operating characteristic curve analysis suggests that GBP5 has high
sensitivity and specificity in the diagnosis of ccRCC, with an AUC
of 0.934. *P<0.05, **P<0.01 and ***P<0.001. GBP5,
guanylate binding protein 5; ccRCC, clear cell renal cell
carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; T, tumor; N, lymph node; M,
metastasis; AUC, area under curve; TPM, transcript per million;
TPR, true-positive rate; FDR, false discovery rate. |
Functional enrichment of GBP5
interaction and expression related genes
To further investigate the potential biological
function of GBP5 in ccRCC, DEG analysis was performed and the
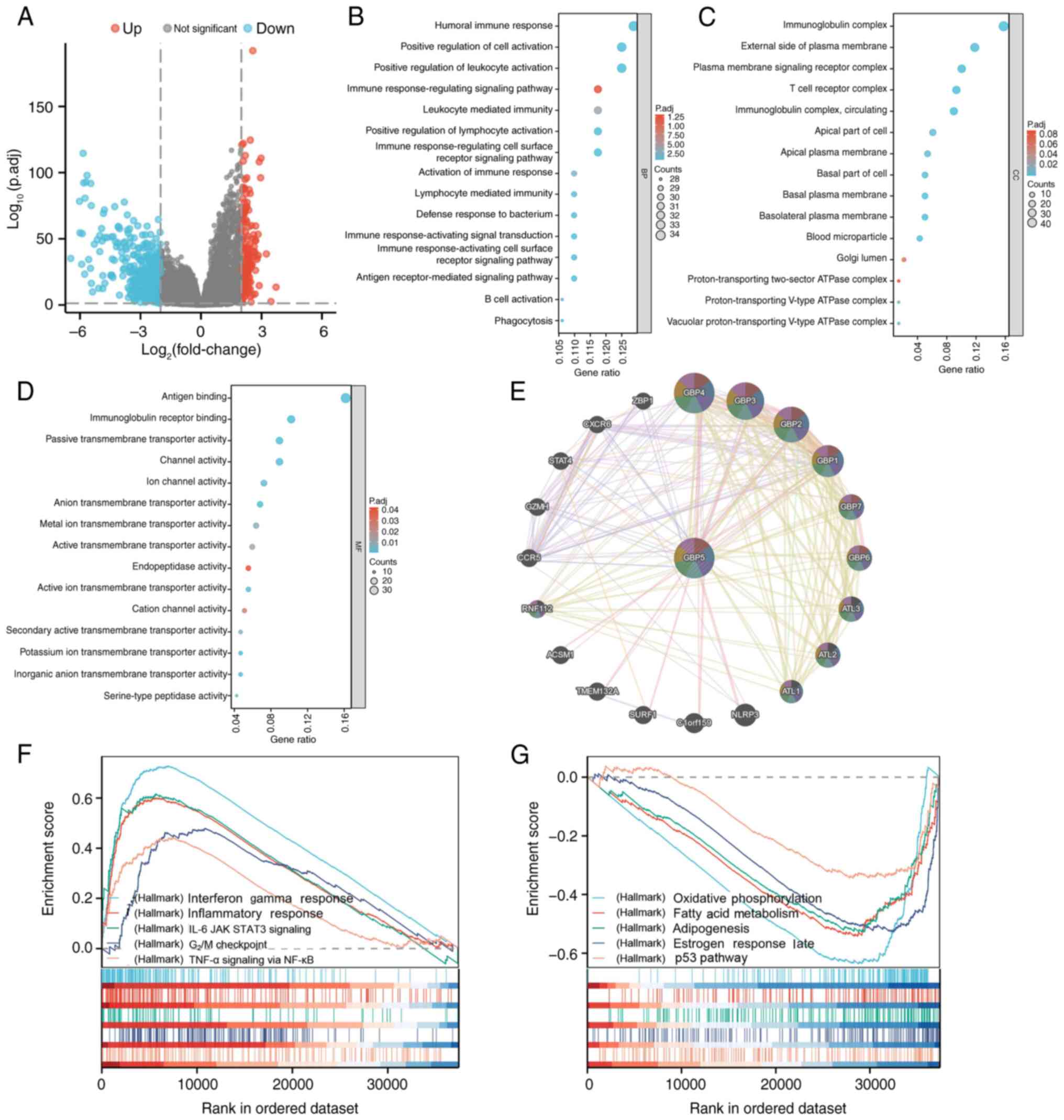
results were visualized using a volcano plot (Fig. 4A). Notably upregulated and
downregulated genes were identified using the criteria:
Log2 fold change >2 and adjusted P-value <0.05.
Enrichment analysis was performed on these DEGs, revealing
significant associations with various GO terms. Specifically, GO
biological process analysis revealed significant enrichment in
‘humoral immune response’, ‘positive regulation of cell
activation’, ‘positive regulation of leukocyte activation’ and
‘immune response-regulation signaling pathways’ (Fig. 4B). The enriched GO cellular
component terms were the ‘immunoglobulin complex’, ‘external side
of plasma membrane’, ‘plasma membrane signaling receptor complex’
and ‘T cell receptor complex’-related components (Fig. 4C). Regarding GO molecular functions,
enrichment was observed in functions such as ‘antigen binding’,
‘immunoglobulin receptor binding’, ‘passive transmembrane
transporter activity’ and ‘channel activity’-associated functions
(Fig. 4D).
Subsequently, GeneMANIA was used to build a
protein-protein interaction network for GBP5 in order to identify
interacting proteins potentially involved in tumorigenesis. Results
indicated that GBP5 exhibited strong co-expression with proteins
such as GBP1, GBP2, NLR family pyrin domain containing 3 (NLRP3)
and GBP4 (Fig. 4E), implying a
potential interaction between GBP5 and these genes, which could
promote tumor progression.
To further elucidate the biological pathways
regulated by GBP5 across the high and low expression groups, GSEA
was performed. As shown in Fig. 4F,
several biological pathways showed notable enrichment in the high
GBP5 expression group, including IFN-γ response, IL-6/JAK/STAT3
signaling, inflammatory response, G2/M damage checkpoint
and TNF-α signaling through NF-κB. By contrast, pathways such as
oxidative phosphorylation, fatty acid metabolism, adipogenesis,
estrogen response late and the p53 pathway were enriched in the low
GBP5 expression group (Fig. 4G).
According to these findings, GBP5 may influence ccRCC progression
through mechanisms related to tumor immunity, cell cycle regulation
and inflammation.
Immune cell infiltration is associated
with GBP5 in ccRCC
To investigate whether GBP5 affects immune cell
infiltration in ccRCC, the TIMER online tool was used to explore
the relationship between GBP5 expression and the abundance of
various immune cell types. In ccRCC, a strong positive correlation
was observed between GBP5 expression and the levels of immune cell
infiltration, including B cells, CD8+ T cells,
CD4+ T cells, macrophages, neutrophils and dendritic
cells (Fig. 5A). These findings
suggest that GBP5 serves a role in immune cell infiltration in
ccRCC. TIMER2.0 was then utilized to conduct a deeper analysis of
the association between GBP5 expression and marker genes of various
immune cells (Table I). The data
revealed strong positive correlations between GBP5 and marker genes
of T cells, CD8+ T cells, M2 macrophages and exhausted T
cells, further supporting the involvement of GBP5 in immune
regulation in ccRCC.
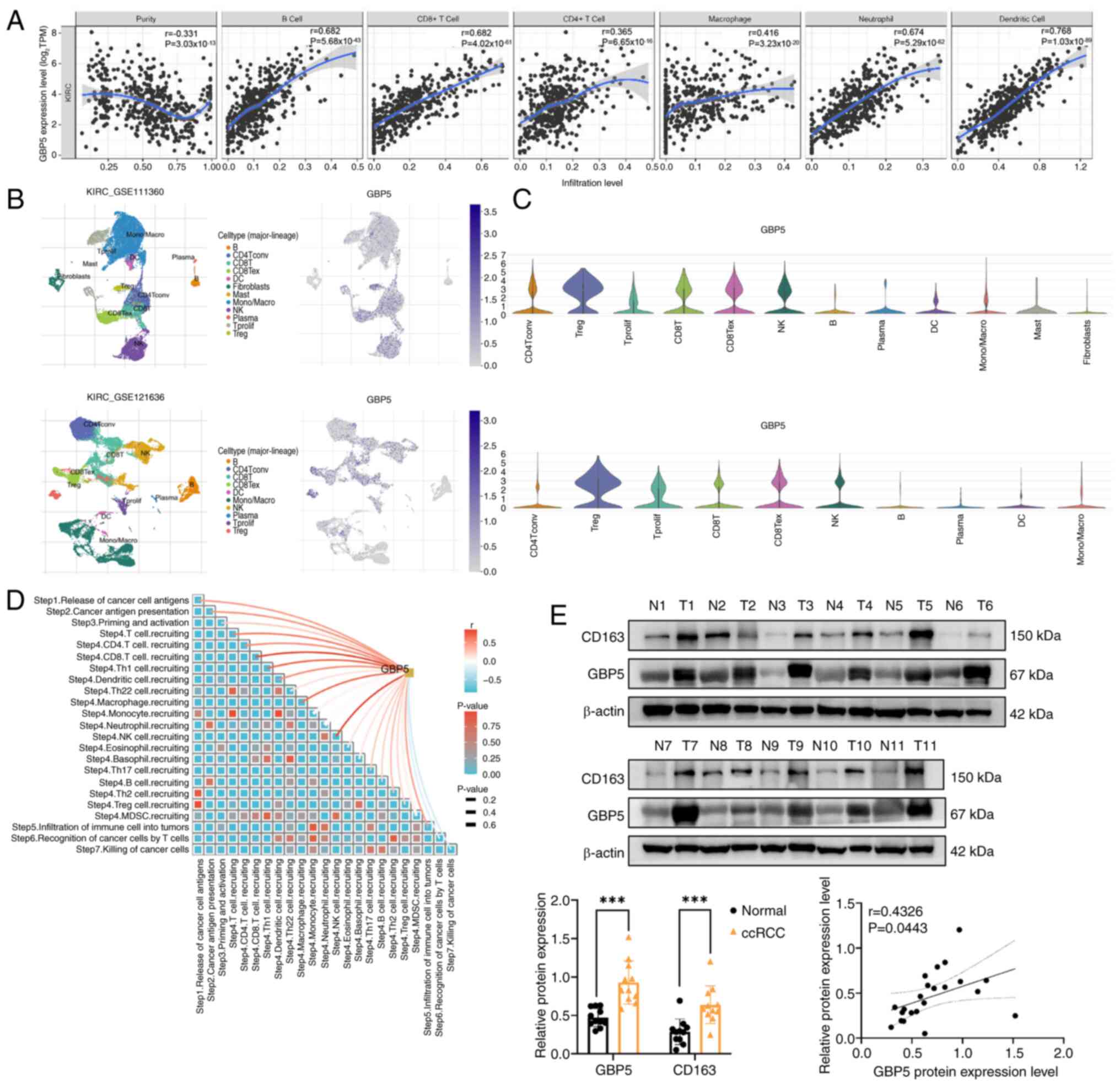 | Figure 5.Relationship between GBP5 expression
and immune cell infiltration in KIRC. (A) Analysis of the Tumor
Immune Estimation Resource database revealed a positive correlation
between GBP5 expression and infiltration levels of B cells,
CD8+ T cells, CD4+ T cells, macrophages,
neutrophils and DCs in KIRC. (B and C) The Tumor Immune Single-cell
Hub database was employed to assess the expression of GBP5 in
various tumor microenvironment-associated cellular subpopulations.
In both KIRC GSE111360 and GSE121636 datasets, GBP5 was found to be
highly expressed in CD4+ T cells, proliferating T cells,
regulatory T cells, CD8+ T cells and NK cells. (D) The
Tracking Tumor Immunophenotype database was employed to analyze the
immunological features of GBP5. Results indicate a strong positive
correlation between GBP5 and various immune cells, including
macrophages, NK cells and CD8+ T cells. (E) Western
blotting was used to measure GBP5 and CD163 protein levels in the
samples of patients with ccRCC. The levels of GBP5 and CD163 are
significantly higher in ccRCC tissues compared with adjacent normal
tissues and the relative expression levels of GBP5 and CD163
exhibit a positive correlation. ***P<0.001. GBP5,
guanylate-binding protein 5; KIRC, kidney renal clear cell
carcinoma; ccRCC, clear cell renal cell carcinoma; NK, natural
killer; DC, dendritic cells; TPM, transcript per million, N,
normal; T, tumor. |
 | Table I.Correlation analysis between
guanylate-binding protein 5 and immune cell markers in the Tumor
Immune Estimation Resource 2.0 database. |
Table I.
Correlation analysis between
guanylate-binding protein 5 and immune cell markers in the Tumor
Immune Estimation Resource 2.0 database.
|
|
| KIRC |
|---|
|
|
|
|
|---|
|
|
| None | Purity |
|---|
|
|
|
|
|
|---|
| Description | Gene markers | r | P-value | r | P-value |
|---|
| CD8+ T
cell | CD8A | 0.838 |
5.76×10−141 | 0.825 |
6.04×10−116 |
|
| CD8B | 0.785 |
3.79×10−119 | 0.767 |
5.25×10−95 |
| T cell
(general) | CD3D | 0.837 |
2.66×10−136 | 0.819 |
1.20×10−108 |
|
| CD3E | 0.852 |
6.79×10−152 | 0.836 |
1.73×10−122 |
|
| CD2 | 0.886 |
5.95×10−175 | 0.873 |
2.72×10−142 |
| B cell | CD19 | 0.474 |
2.97×10−34 | 0.439 |
4.92×10−26 |
|
| CD79A | 0.520 |
4.93×10−34 | 0.486 |
8.87×10−26 |
| Monocyte | CD86 | 0.750 |
2.52×10−95 | 0.726 |
4.68×10−76 |
|
| CD115 (CSF1R) | 0.604 |
2.19×10−54 | 0.556 |
1.39×10−39 |
| TAM | CCL2 | 0.108 |
1.14×10−2 | 0.052 | 0.263 |
|
| CD68 | 0.470 |
1.60×10−15 | 0.455 |
1.05×10−13 |
|
| IL10 | 0.589 |
3.28×10−50 | 0.523 |
2.33×10−34 |
| M1 Macrophage | INOS(NOS2) | 0.046 | 0.287 | −0.02 | 0.667 |
|
| IRF5 | 0.452 |
8.40×10−29 | 0.444 |
4.40×10−24 |
|
| COX2(PTGS2) | 0.068 | 0.114 | 0.028 | 0.548 |
| M2 Macrophage | CD163 | 0.512 |
4.58×10−39 | 0.483 |
0.85×10−30 |
|
| VSIG4 | 0.501 |
8.24×10−37 | 0.445 |
1.28×10−25 |
|
| MS4A4A | 0.563 |
4.87×10−46 | 0.526 |
5.76×10−35 |
| Neutrophils | CD66b
(CEACAM8) | 0.052 | 0.228 | 0.056 | 0.232 |
|
| CD11b (ITGAM) | 0.576 |
3.10×10−49 | 0.529 |
2.51×10−35 |
|
| CCR7 | 0.582 |
1.41×10−48 | 0.538 |
5.35×10−35 |
| Dendritic cell | HLA-DPB1 | 0.765 |
6.03×10−99 | 0.753 |
1.23×10−80 |
|
| HLA-DQB1 | 0.538 |
2.18×10−44 | 0.493 |
4.34×10−34 |
|
| HLA-DRA | 0.778 |
2.89×10−104 | 0.777 |
1.45×10−90 |
|
| HLA-DPA1 | 0.773 |
1.52×10−97 | 0.768 |
8.45×10−83 |
|
| BDCA-1 (CD1C) | 0.305 |
4.19×10−13 | 0.242 |
9.85×10−8 |
|
| BDCA-4 (NRP1) | 0.106 |
2.36×10−3 | 0.058 |
7.69×10−2 |
|
| CD11c (ITGAX) | 0.571 |
3.10×10−47 | 0.536 |
2.97×10−36 |
| Th1 | T-bet (TBX21) | 0.540 |
9.41×10−41 | 0.508 |
6.33×10−30 |
|
| STAT4 | 0.644 |
3.19×10−67 | 0.612 |
1.20×10−51 |
|
| STAT1 | 0.821 |
3.05×10−122 | 0.809 |
6.13×10−102 |
|
| IFN-γ (IFNG) | 0.841 |
3.15×10−147 | 0.823 |
7.02×10−117 |
|
| TNF-α (TNF) | 0.408 |
1.72×10−22 | 0.368 |
1.33×10−15 |
| Th2 | GATA3 | 0.346 |
2.95×10−16 | 0.363 |
1.18×10−15 |
|
| STAT6 | 0.142 |
1.21×10−5 | 0.152 |
3.70×10−5 |
|
| STAT5A | 0.652 |
6.78×10−64 | 0.621 |
1.01×10−49 |
|
| IL13 | 0.112 |
1.73×10−2 | 0.09 | 0.053 |
| Tfh | BCL6 | 0.092 | 0.035 | 0.089 | 0.059 |
|
| IL21 | 0.290 |
4.62×10−44 | 0.275 |
1.62×10−33 |
| Th17 | STAT3 | 0.268 |
4.13×10−11 | 0.242 |
3.34×10−8 |
|
| IL17A | 0.081 | 0.062 | 0.063 | 0.180 |
| Treg | FOXP3 | 0.678 |
2.03×10−68 | 0.651 |
4.99×10−55 |
|
| CCR8 | 0.734 |
3.03×10−96 | 0.719 |
4.66×10−77 |
|
| STAT5B | 0.168 |
2.28×10−5 | 0.179 |
4.09×10−5 |
|
| TGFβ | 0.194 |
2.65×10−6 | 0.157 |
3.51×10−4 |
| T cell
exhaustion | PDCD1 | 0.795 |
3.89×10−118 | 0.781 |
1.82×10−97 |
|
| CTLA4 | 0.773 |
1.90×10−113 | 0.760 |
8.78×10−95 |
|
| LAG3 | 0.815 |
2.78×10−119 | 0.798 |
6.42×10−99 |
|
| TIM-3 (HAVCR2) | 0.371 |
9.92×10−20 | 0.331 |
1.07×10−13 |
|
| GZMB | 0.489 |
1.35×10−33 | 0.444 |
7.90×10−23 |
To validate the relationship between GBP5 expression
and immune cell infiltration in ccRCC, two single-cell RNA datasets
of ccRCC were analyzed from the TISCH2 database. GBP5 expression
was assessed in different TME-associated cell subsets. The results
demonstrated that in both KIRC GSE111360 and GSE121636 datasets,
GBP5 was highly expressed in CD4+ T cells, proliferating
T cells, regulatory T cells, CD8+ T cells and natural
killer (NK) cells (Fig. 5B and
C).
Association between GBP5 and
tumor-associated macrophages
Further analysis of the immunological
characteristics of GBP5 in ccRCC was conducted. Results indicated a
strong positive correlation between GBP5 and various immune cells,
including macrophages, NK cells and CD8+ T cells
(Fig. 5D). To validate the findings
from public databases, western blotting of tumor and adjacent
normal tissues from 11 patients with ccRCC was performed. In ccRCC
tissues, the levels of GBP5 and CD163 were markedly higher compared
with those in adjacent normal tissues. The relative expression
levels of GBP5 and CD163 exhibited a positive correlation (Fig. 5E). These findings underscore the
association between GBP5 and M2 macrophages and suggest a potential
interaction between them.
Verification of GBP5 expression and
its effect on the biological behavior of ccRCC
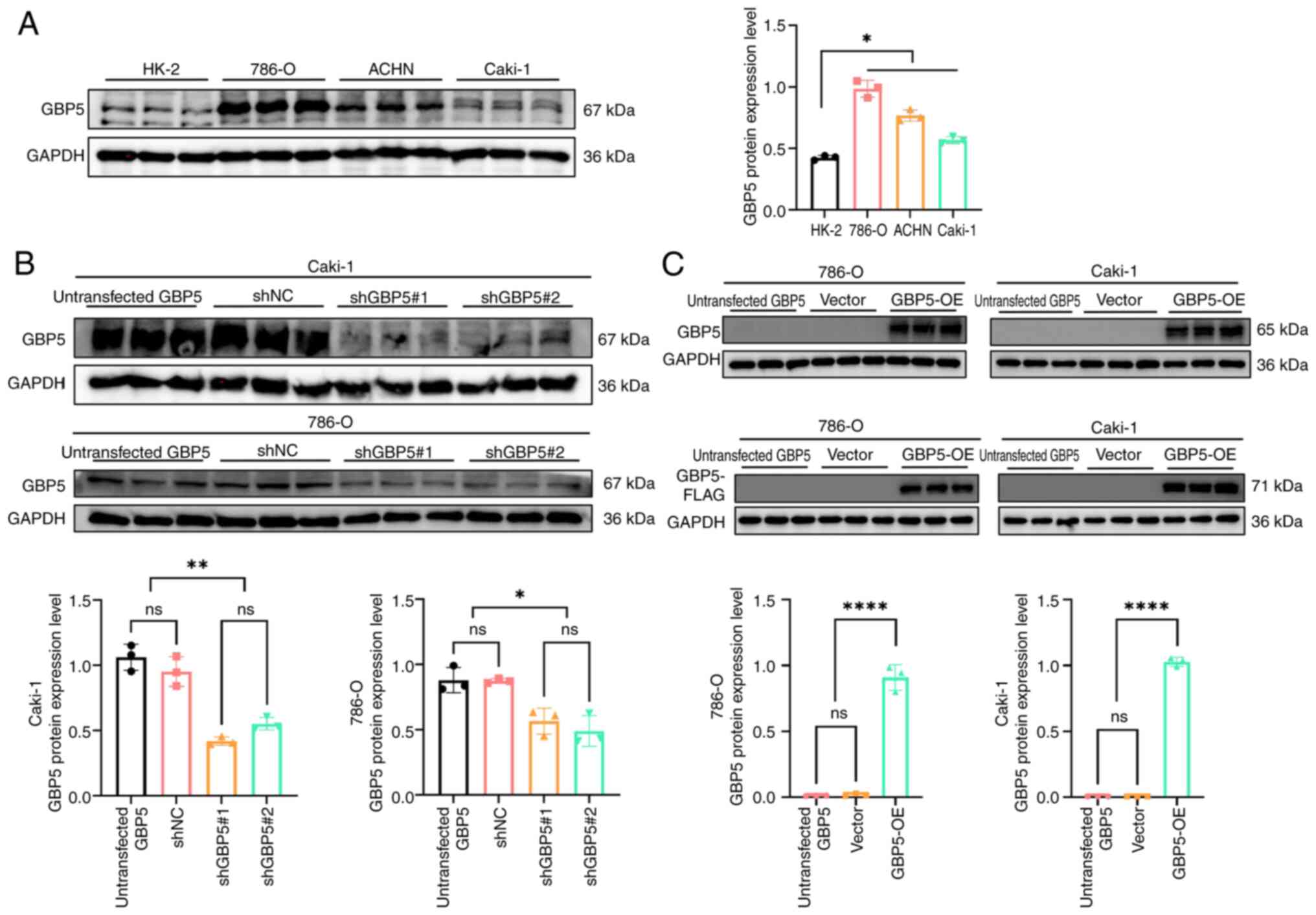
Several in vitro experiments were performed
to investigate the role of GBP5 in ccRCC cells. Western blotting
was performed to assess GBP5 expression in Caki-1, 786-O, ACHN and
HK-2 cells. Findings demonstrated that GBP5 protein levels were
markedly higher in renal cancer cell lines compared with HK-2 cells
(Fig. 6A). For subsequent
experiments, 786-O and Caki-1 cell lines were selected.
The efficiency of lentivirus-mediated gene silencing
was evaluated using western blotting. As shown in Fig. 6B, two lentiviral constructs carrying
different shRNA sequences targeting GBP5 were tested in Caki-1 and
786-O cells. Both shRNA constructs effectively knocked down GBP5
expression, with shGBP5#1 showing improved knockdown efficiency.
Thus, the shGBP5#1 lentivirus was selected to establish stable GBP5
knockdown cell lines. Similarly, GBP5 OE cell lines were generated
using a lentiviral system. Following the exogenous introduction of
GBP5 into 786-O and Caki-1 cells, western blotting analysis
confirmed a significant upregulation of GBP5 expression (Fig. 6C).
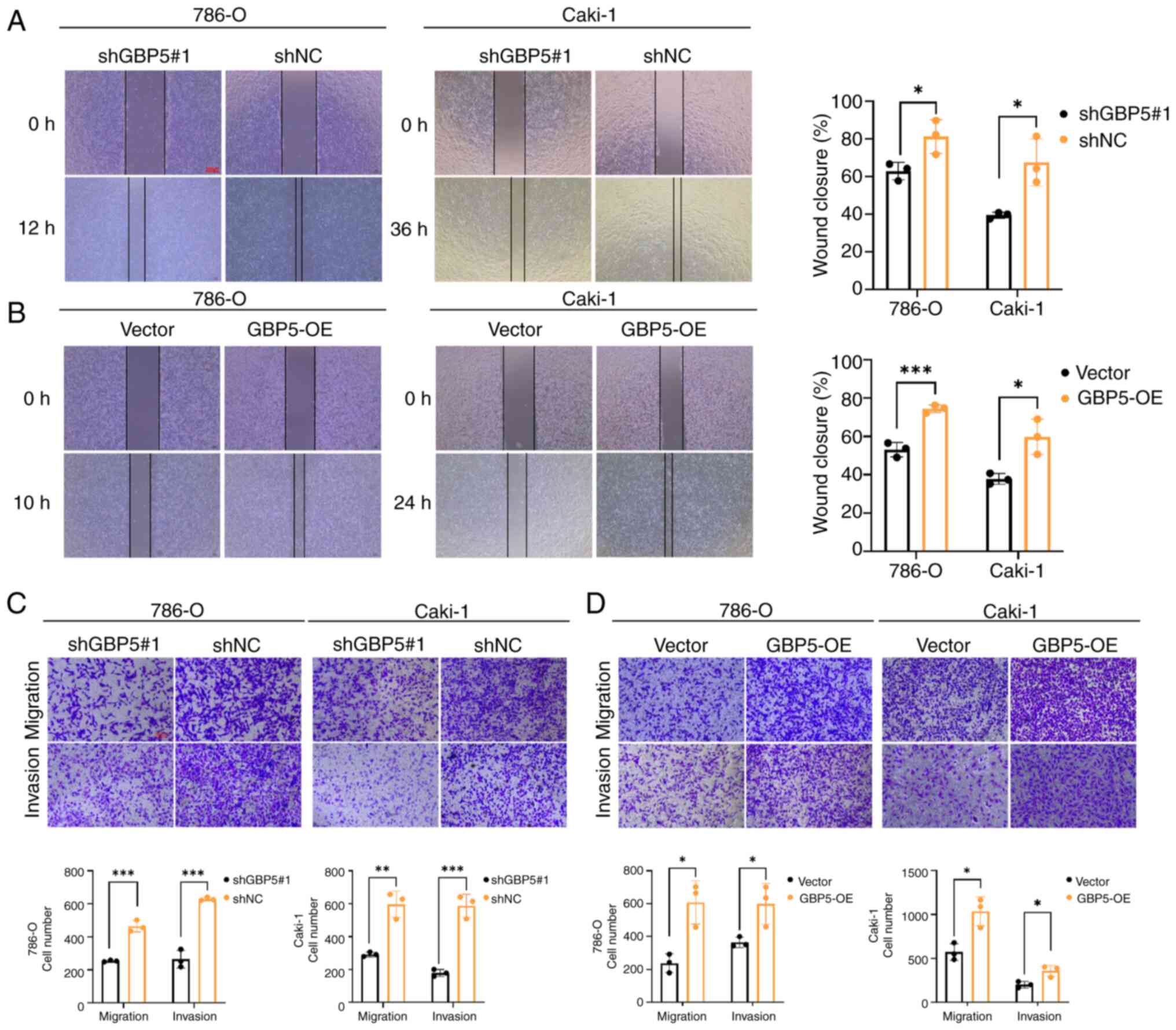
To explore the biological function of GBP5 in cancer
progression, a wound healing assay was performed. Notably, the
suppression of GBP5 expression significantly decreased the
migratory capacity of the cells (Fig.
7A). By contrast, GBP5 overexpression enhanced the
wound-closure rate (Fig. 7B). These
findings were consistently observed in Caki-1 and 786-O cell lines,
indicating that the GBP5-mediated regulation of cell migration is
not cell-type specific.
The Transwell migration assays demonstrated that
GBP5 knockdown inhibited cell migration, whereas GBP5
overexpression significantly promoted it (Fig. 7C and D). In addition, the
Matrigel-coated Transwell invasion assays demonstrated that GBP5
depletion markedly suppressed the invasive capabilities of Caki-1
and 786-O cells, whereas the overexpression of GBP5 exerted the
opposite effect, significantly enhancing cell invasion (Fig. 7C and D).
Discussion
ccRCC is the predominant subtype of RCC. It is
associated with a poor prognosis and also lacks effective
biomarkers. The primary cause of ccRCC-related mortality is the low
rate of early detection and scarcity of effective treatments for
patients with advanced or metastatic disease (40,41).
Consequently, it is key to identify reliable biomarkers for early
detection and prognosis, along with the development of innovative
therapeutic approaches to enhance the clinical outcomes of
ccRCC.
GBP5, which is induced by IFN-γ and classified
within the large GTPase family, is one of seven GBPs with strong
sequence homology that contribute to host defense mechanisms
(42). The function of GBP5 in
cancer, particularly ccRCC, remains largely unclear. In the present
study, GBP5 was recognized as a potential immunotherapeutic target,
exhibiting upregulation in several types of cancer. Previous
studies have highlighted the involvement of GBP5 in the progression
of oral squamous cell carcinoma (23), hepatocellular carcinoma (43), gastric cancer (15) and glioblastoma (16). In the present study, GBP5 was
characterized as a novel biomarker for ccRCC involved in the
tumorigenesis and progression of this malignancy. Findings show
that GBP5 expression was notably increased in ccRCC tissues and was
also associated with a poor prognosis. With this, the results of
the bioinformatics analysis demonstrated that GBP5 exhibited stage-
and grade-specific expression, making it a useful biomarker for
diagnosing and predicting the prognosis of patients with ccRCC.
However, GBP5 has been reported to serve an opposite role in
ovarian cancer (44), likely due to
the complexity of ovarian tumorigenesis and the organ-specific
characteristics, such as lineage-specific transcription factors
(45).
Cancer progression involves a complex array of
immune evasion mechanisms that collectively shape the immunological
characteristics of the TME. Tumor-associated immune cells have been
explored as potential therapeutic biomarkers for cancer (46). In response to pathogen- or
danger-associated molecular patterns, GBP5 serves a key role in the
activation of the NLRP3 inflammasome (47). It primarily mediates inflammation
and activates macrophages in the innate immune system (48). Previous studies have suggested that
GBP5 may be involved in tumor immunity (17–22).
The efficacy of both targeted therapies and immunotherapies is
influenced by the TME immune landscape (49). Various immune cell types such as T
cells, regulatory tumor-associated macrophages (TAMs), T regulatory
cells (Tregs), cancer-associated fibroblasts and myeloid-derived
suppressor cells, serve a role in the TME and contribute to immune
cell infiltration, which is regulated by tumor and
immune-modulating factors (50,51).
Associations between GBP5 expression and several immune cell
markers were observed, including CD8+ T cells, Tregs, T
helper type 1 cells, exhausted T cells, M2 macrophages and
monocytes. Patients with ccRCC showing elevated programmed cell
death protein 1 expression alongside CD8+ T cell and
Treg infiltration tended to have worse clinical outcomes (52–54).
Tregs may contribute to tumor progression and immune escape by
interacting with the TME and suppressing antigen-presenting cell
maturation (55). Although
CD8+ T cell infiltration is elevated in the ccRCC
microenvironment, it does not appear to enhance prognosis, most
likely because of functional T cell exhaustion (56). Moreover, analysis revealed a
positive association between GBP5 expression and CD163, an
established marker of M2-polarized TAMs. M2-like TAMs are commonly
found in tumors and facilitate tumor progression, cell
proliferation, angiogenesis, invasion and metastasis by secreting
diverse cytokines and chemokines (57–60).
These findings indicate that GBP5 is a possible driver of ccRCC
progression through TAMs.
Cancer metastasis is the primary cause of mortality
in patients with cancer (61).
Regarding cancer biology, GBP5 has been implicated in various
malignant behaviors, including tumor invasion, migration,
epithelial-mesenchymal transition, cell proliferation and
supporting properties of cancer stem cells (16,23).
Similarly, GBP5 was found to be involved in the regulation of renal
cancer cell invasion and migration, perhaps explaining why patients
with high GBP5 expression in ccRCC exhibit a poor prognosis. With
regards to the molecular mechanisms underlying the function of GBP5
in tumorigenesis, previous research has demonstrated that GBP5
might enhance tumor progression in breast and oral cancers by
upregulating PD-L1 expression and activating NF-κB signaling
(17,23). Additionally, GBP5 accelerates
gastric cancer development by engaging in a
JAK1-STAT1/GBP5/CXCL8-mediated positive feedback circuit (15). In the present study, through
bioinformatic approaches, GBP5 was found to associate with key
oncogenic signaling cascades such as the INF-γ response,
IL-6/JAK/STAT3 axis and the TNF-α/NF-κB network. These pathways are
recognized for their essential roles in both cancer development and
immune regulation (62,63). However, the precise molecular
mechanisms by which GBP5 modulates these pathways in RCC warrants
further investigation.
The present study has some limitations. Firstly, the
publicly available ccRCC datasets used in the analyses were limited
in scope, which may have introduced potential errors or biases.
Secondly, while the pro-migratory and pro-invasive roles of GBP5 in
renal cancer cells have been confirmed in vitro, the
underlying molecular mechanisms remain unexplored and no in
vivo validation has been performed. Finally, although GBP5
appears to be related to tumor immune cell infiltration, the
present study lacks supporting in vitro and in vivo
experimental evidence. These limitations must be considered when
interpreting the findings and they will be addressed in future
studies to provide a deeper understanding of the role of GBP5 in
immune cell infiltration and metastasis in ccRCC.
Overall, using bioinformatic analyses and in
vitro experiments, the upregulation of GBP5 in ccRCC was
confirmed. Moreover, immune cell infiltration in ccRCC is
associated with the upregulation of GBP5. In addition, GBP5
facilitates renal cancer cell migration and invasion, suggesting
its usefulness as a prognostic biomarker for poor clinical outcomes
and a promising immunotherapeutic target in ccRCC.
Acknowledgements
We gratefully acknowledge the Clinical,
Translational and Basic Research Laboratory of the University of
Hong Kong-Shenzhen Hospital for providing infrastructure support
and research resources essential to this study.
Funding
The present study was supported by the Medical Scientific
Research Foundation of Guangdong (grant no. B2023384).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding authors.
Authors' contributions
XC, SJY, XX and CL participated in the study design,
conducted the experiments, collected and analyzed the data and
drafted the initial manuscript. Bioinformatics analysis and some
experimentation was performed by XC, SJY, SY, WZ and ZL. SJY and CL
were also in charge of revising the manuscript, supervising the
study and offering technical experimental support. XC, SJY, XX and
CL confirm the authenticity of all the raw data. All authors were
involved in preparing, proofreading and submitting the manuscript.
All authors read, confirmed and approved the final version of the
manuscript, and are accountable for the manuscript's content.
Ethics approval and consent to
participate
The collection of renal carcinoma tumor tissues and
adjacent normal tissues was approved by the Ethics Committee of the
University of Hong Kong-Shenzhen Hospital (approval no. 2024256)
and was performed in accordance with the guidelines of the
Declaration of Helsinki. All donors signed informed consent
forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Shuch B, Amin A, Armstrong AJ, Eble JN,
Ficarra V, Lopez-Beltran A, Martignoni G, Rini BI and Kutikov A:
Understanding pathologic variants of renal cell carcinoma:
Distilling therapeutic opportunities from biologic complexity. Eur
Urol. 67:85–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Braun DA, Hou Y, Bakouny Z, Ficial M,
Sant' Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA,
Elagina L, et al: Interplay of somatic alterations and immune
infiltration modulates response to PD-1 blockade in advanced clear
cell renal cell carcinoma. Nat Med. 26:909–918. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choueiri TK and Motzer RJ: Systemic
therapy for metastatic renal-cell carcinoma. N Engl J Med.
376:354–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maines F, Caffo O, Veccia A, Trentin C,
Tortora G, Galligioni E and Bria E: Sequencing new agents after
docetaxel in patients with metastatic castration-resistant prostate
cancer. Crit Rev Oncol Hematol. 96:498–506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choueiri TK, Fishman MN, Escudier B,
McDermott DF, Drake CG, Kluger H, Stadler WM, Perez-Gracia JL,
McNeel DG, Curti B, et al: Immunomodulatory activity of nivolumab
in metastatic renal cell carcinoma. Clin Cancer Res. 22:5461–5471.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abah MO, Ogenyi DO, Zhilenkova AV, Essogmo
FE, Ngaha Tchawe YS, Uchendu IK, Pascal AM, Nikitina NM, Rusanov
AS, Sanikovich VD, et al: Innovative therapies targeting
drug-resistant biomarkers in metastatic clear cell renal cell
carcinoma (ccRCC). Int J Mol Sci. 26:2652024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Linehan WM and Ricketts CJ: Decade in
review-kidney cancer: Discoveries, therapies and opportunities. Nat
Rev Urol. 11:614–616. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Praefcke GJK and McMahon HT: The dynamin
superfamily: Universal membrane tubulation and fission molecules?
Nat Rev Mol Cell Biol. 5:133–147. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vestal DJ: The guanylate-binding proteins
(GBPs): Proinflammatory cytokine-induced members of the dynamin
superfamily with unique GTPase activity. J Interferon Cytokine Res.
25:435–443. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shenoy AR, Wellington DA, Kumar P, Kassa
H, Booth CJ, Cresswell P and MacMicking JD: GBP5 promotes NLRP3
inflammasome assembly and immunity in mammals. Science.
336:481–485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tretina K, Park ES, Maminska A and
MacMicking JD: Interferon-induced guanylate-binding proteins:
Guardians of host defense in health and disease. J Exp Med.
216:482–500. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fellenberg F, Hartmann TB, Dummer R,
Usener D, Schadendorf D and Eichmüller S: GBP-5 splicing variants:
New guanylate-binding proteins with tumor-associated expression and
antigenicity. J Invest Dermatol. 122:1510–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Patil PA, Blakely AM, Lombardo KA, Machan
JT, Miner TJ, Wang LJ, Marwaha AS and Matoso A: Expression of
PD-L1, indoleamine 2,3-dioxygenase and the immune microenvironment
in gastric adenocarcinoma. Histopathology. 73:124–136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao FY, Wang CH, Li X, Ma MZ, Tao GC, Yang
C, Li K, He XB, Tong SL, Zhao QC, et al: Guanylate binding protein
5 accelerates gastric cancer progression via the
JAK1-STAT1/GBP5/CXCL8 positive feedback loop. Am J Cancer Res.
13:1310–1328. 2023.PubMed/NCBI
|
|
16
|
Yu X, Jin J, Zheng Y, Zhu H, Xu H, Ma J,
Lan Q, Zhuang Z, Chen CC and Li M: GBP5 drives malignancy of
glioblastoma via the Src/ERK1/2/MMP3 pathway. Cell Death Dis.
12:2032021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng SW, Chen PC, Lin MH, Ger TR, Chiu HW
and Lin YF: GBP5 repression suppresses the metastatic potential and
PD-L1 expression in triple-negative breast cancer. Biomedicines.
9:3712021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng Z, Liu J, Yu YV and Jin YN: Machine
learning-based identification of an immunotherapy-related signature
to enhance outcomes and immunotherapy responses in melanoma. Front
Immunol. 15:14511032024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Elsayed I, Elsayed N, Feng Q, Sheahan K,
Moran B and Wang X: Multi-OMICs data analysis identifies molecular
features correlating with tumor immunity in colon cancer. Cancer
Biomark. 33:261–271. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zou C, Shen J, Xu F, Ye Y, Wu Y and Xu S:
Immunoreactive microenvironment modulator GBP5 suppresses ovarian
cancer progression by inducing canonical pyroptosis. J Cancer.
15:3510–3530. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiang S, Li J, Shen J, Zhao Y, Wu X, Li M,
Yang X, Kaboli PJ, Du F, Zheng Y, et al: Identification of
prognostic genes in the tumor microenvironment of hepatocellular
carcinoma. Front Immunol. 12:6538362021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tong Q, Li D, Yin Y, Cheng L and Ouyang S:
GBP5 expression predicted prognosis of immune checkpoint inhibitors
in small cell lung cancer and correlated with tumor immune
microenvironment. J Inflamm Res. 16:4153–4164. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chiu HW, Lin CH, Lee HH, Lu HW, Lin YHK,
Lin YF and Lee HL: Guanylate binding protein 5 triggers NF-κB
activation to foster radioresistance, metastatic progression and
PD-L1 expression in oral squamous cell carcinoma. Clin Immunol.
259:1098922024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen HQ, Zhao J, Li Y, He LX, Huang YJ,
Shu WQ, Cao J, Liu WB and Liu JY: Gene expression network regulated
by DNA methylation and microRNA during microcystin-leucine arginine
induced malignant transformation in human hepatocyte L02 cells.
Toxicol Lett. 289:42–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mitra A, Ghosh S, Porey S and Mal C: GBP5
and ACSS3: Two potential biomarkers of high-grade ovarian cancer
identified through downstream analysis of microarray data. J Biomol
Struct Dyn. 41:4601–4613. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi Z, Gu J, Yao Y and Wu Z:
Identification of a predictive gene signature related to pyroptosis
for the prognosis of cutaneous melanoma. Medicine (Baltimore).
101:e305642022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Diaz-Montero CM, Rini BI and Finke JH: The
immunology of renal cell carcinoma. Nat Rev Nephrol. 16:721–735.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res 47 (W1). W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
von Roemeling CA, Radisky DC, Marlow LA,
Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW and Copland JA:
Neuronal pentraxin 2 supports clear cell renal cell carcinoma by
activating the AMPA-selective glutamate receptor-4. Cancer Res.
74:4796–4810. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peña-Llopis S, Vega-Rubín-de-Celis S, Liao
A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L,
Sivanand S, Spence P, et al: BAP1 loss defines a new class of renal
cell carcinoma. Nat Genet. 44:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res. 23:e276332021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res 38 (Web Server Issue). W214–W220. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res 48 (W1). W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue
J, Wang H, Li T and Wang C: TISCH2: Expanded datasets and new tools
for single-cell transcriptome analyses of the tumor
microenvironment. Nucleic Acids Res. 51(D1): D1425–D1431. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Neal JT, Li X, Zhu J, Giangarra V,
Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, et
al: Organoid modeling of the tumor immune microenvironment. Cell.
175:1972–1988.e16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Borcherding N, Vishwakarma A, Voigt AP,
Bellizzi A, Kaplan J, Nepple K, Salem AK, Jenkins RW, Zakharia Y
and Zhang W: Mapping the immune environment in clear cell renal
carcinoma by single-cell genomics. Commun Biol. 4:1222021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu L, Deng C, Pang B, Zhang X, Liu W, Liao
G, Yuan H, Cheng P, Li F, Long Z, et al: TIP: A web server for
resolving tumor immunophenotype profiling. Cancer Res.
78:6575–6580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ye S, Li S, Qin L, Zheng W, Liu B, Li X,
Ren Z, Zhao H, Hu X, Ye N and Li G: GBP2 promotes clear cell renal
cell carcinoma progression through immune infiltration and
regulation of PD-L1 expression via STAT1 signaling. Oncol Rep.
49:492023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cerbone L, Cattrini C, Vallome G, Latocca
MM, Boccardo F and Zanardi E: Combination therapy in metastatic
renal cell carcinoma: Back to the future? Semin Oncol. 47:361–366.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Braun DA, Bakouny Z, Hirsch L, Flippot R,
Van Allen EM, Wu CJ and Choueiri TK: Beyond conventional
immune-checkpoint inhibition-novel immunotherapies for renal cell
carcinoma. Nat Rev Clin Oncol. 18:199–214. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Santos JC and Broz P: Sensing of invading
pathogens by GBPs: At the crossroads between cell-autonomous and
innate immunity. J Leukoc Biol. 104:729–735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fu J, Qin W, Tong Q, Li Z, Shao Y, Liu Z,
Liu C, Wang Z and Xu X: A novel DNA methylation-driver gene
signature for long-term survival prediction of hepatitis-positive
hepatocellular carcinoma patients. Cancer Med. 11:4721–4735. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zahra A, Dong Q, Hall M, Jeyaneethi J,
Silva E, Karteris E and Sisu C: Identification of potential
bisphenol A (BPA) exposure biomarkers in ovarian cancer. J Clin
Med. 10:19792021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Arner EN and Rathmell WK: Mutation and
tissue lineage lead to organ-specific cancer. Nature. 606:871–872.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu P, Ye L, Ren Y, Zhao G, Zhang Y, Lu S,
Li Q, Wu C, Bai L, Zhang Z, et al: Chemotherapy-induced phlebitis
via the GBP5/NLRP3 inflammasome axis and the therapeutic effect of
aescin. Br J Pharmacol. 180:1132–1147. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou L, Zhao H, Zhao H, Meng X, Zhao Z,
Xie H, Li J, Tang Y and Zhang Y: GBP5 exacerbates rosacea-like skin
inflammation by skewing macrophage polarization towards M1
phenotype through the NF-κB signalling pathway. J Eur Acad Dermatol
Venereol. 37:796–809. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wilkie KP and Hahnfeldt P: Tumor-immune
dynamics regulated in the microenvironment inform the transient
nature of immune-induced tumor dormancy. Cancer Res. 73:3534–3544.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Heidegger I, Pircher A and Pichler R:
Targeting the tumor microenvironment in renal cell cancer biology
and therapy. Front Oncol. 9:4902019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Najjar YG, Rayman P, Jia X, Pavicic PJ Jr,
Rini BI, Tannenbaum C, Ko J, Haywood S, Cohen P, Hamilton T, et al:
Myeloid-derived suppressor cell subset accumulation in renal cell
carcinoma parenchyma is associated with intratumoral expression of
IL1β, IL8, CXCL5, and Mip-1α. Clin Cancer Res. 23:2346–2355. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dai S, Zeng H, Liu Z, Jin K, Jiang W, Wang
Z, Lin Z, Xiong Y, Wang J, Chang Y, et al: Intratumoral
CXCL13+CD8+T cell infiltration determines
poor clinical outcomes and immunoevasive contexture in patients
with clear cell renal cell carcinoma. J Immunother Cancer.
9:e0018232021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Giraldo NA, Becht E, Pagès F, Skliris G,
Verkarre V, Vano Y, Mejean A, Saint-Aubert N, Lacroix L, Natario I,
et al: Orchestration and prognostic significance of immune
checkpoints in the microenvironment of primary and metastatic renal
cell cancer. Clin Cancer Res. 21:3031–3040. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Giraldo NA, Becht E, Vano Y, Petitprez F,
Lacroix L, Validire P, Sanchez-Salas R, Ingels A, Oudard S, Moatti
A, et al: Tumor-infiltrating and peripheral blood t-cell
immunophenotypes predict early relapse in localized clear cell
renal cell carcinoma. Clin Cancer Res. 23:4416–4428. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Paluskievicz CM, Cao X, Abdi R, Zheng P,
Liu Y and Bromberg JS: T Regulatory cells and priming the
suppressive tumor microenvironment. Front Immunol. 10:24532019.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Braun DA, Street K, Burke KP, Cookmeyer
DL, Denize T, Pedersen CB, Gohil SH, Schindler N, Pomerance L,
Hirsch L, et al: Progressive immune dysfunction with advancing
disease stage in renal cell carcinoma. Cancer Cell. 39:632–648.e8.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mantovani A, Marchesi F, Malesci A, Laghi
L and Allavena P: Tumour-associated macrophages as treatment
targets in oncology. Nat Rev Clin Oncol. 14:399–416. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pittet MJ, Michielin O and Migliorini D:
Clinical relevance of tumour-associated macrophages. Nat Rev Clin
Oncol. 19:402–421. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xiang X, Wang J, Lu D and Xu X: Targeting
tumor-associated macrophages to synergize tumor immunotherapy.
Signal Transduct Target Ther. 6:752021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zheng W, Ye S, Liu B, Liu D, Yan R, Guo H,
Yu H, Hu X, Zhao H, Zhou K and Li G: Crosstalk between GBP2 and M2
macrophage promotes the ccRCC progression. Cancer Sci.
115:3570–3586. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Meirson T, Gil-Henn H and Samson AO:
Invasion and metastasis: The elusive hallmark of cancer. Oncogene.
39:2024–2026. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018. View Article : Google Scholar : PubMed/NCBI
|