Digestive system neoplasms are the most common
malignant tumor in China, mainly including esophageal squamous cell
carcinoma (ESCC), gastric cancer (GC), colorectal cancer (CRC),
hepatocellular carcinoma (HCC) and pancreatic cancer (PC) (1,2).
Notably, CRC is the second leading cause of global cancer
mortality, and GC ranks fourth. Furthermore, the incidence rates of
both cancers remain persistently high. A pivotal reason for the
high mortality is metastasis, which accounts for ~90% of cancer
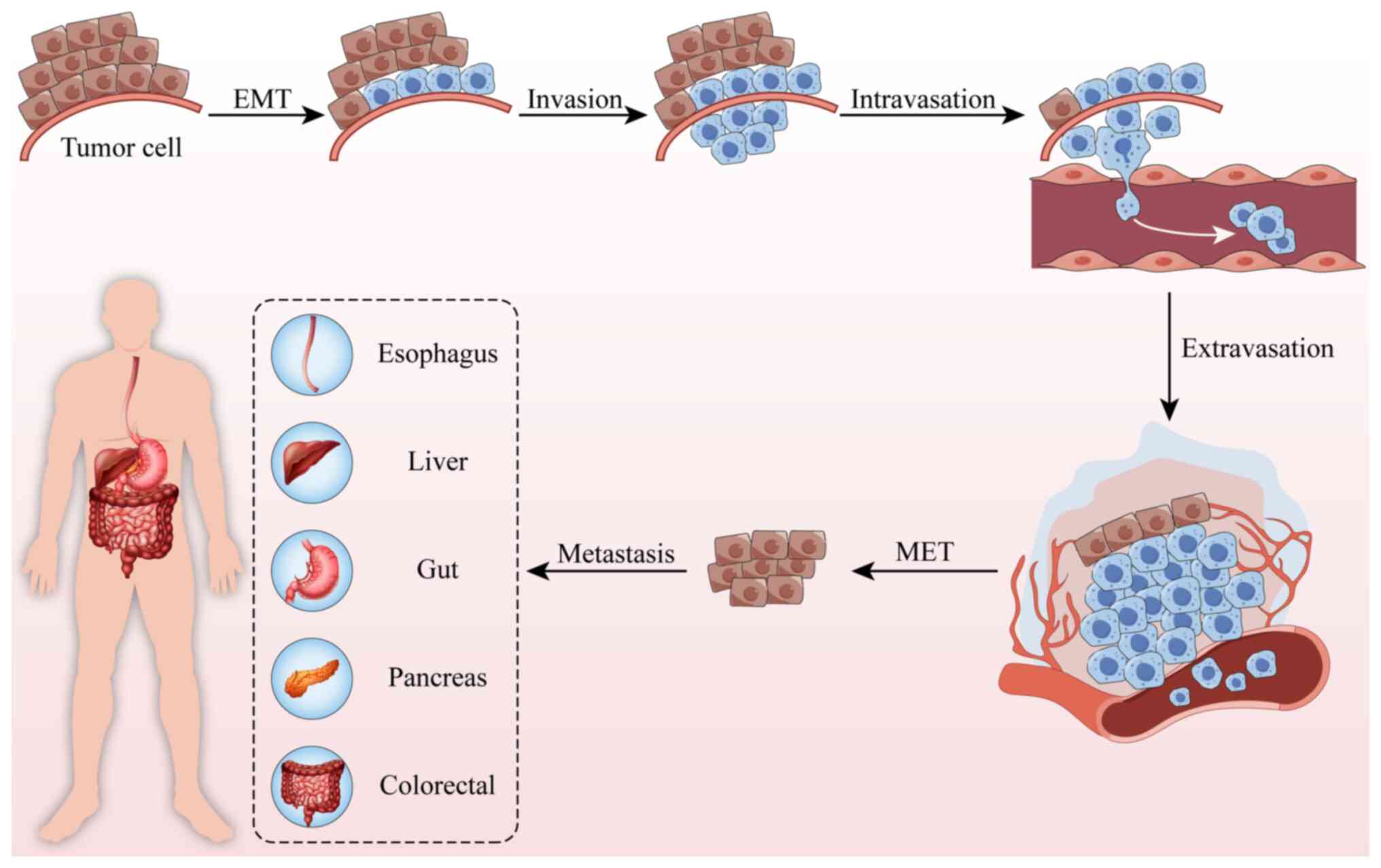
deaths. The metastatic process entails essential steps such as
epithelial-mesenchymal transition (EMT), tumor invasion,
intravasation, extravasation and mesenchymal-epithelial transition
(MET), culminating in the establishment of distant metastases
(Fig. 1). In the present review,
‘invasion’ specifically refers to the ability of cells to degrade
and penetrate the extracellular matrix or basement membrane,
emphasizing its destructive nature; ‘migration’ describes the
motility of cells such as tumor cells; and ‘metastasis’ denotes the
multistep process by which tumor cells disseminate from the primary
site to distant organs and form secondary tumors (3,4).
Emerging evidence indicates that metabolic
reprogramming of tumor cells serves as a critical driver of tumor
metastasis. Malignant tumor cells alter metabolic reprogramming
through the modulation of conventional activities of key enzymes,
as well as the regulation of their non-canonical roles (5–7).
Meanwhile, several metabolic pathways have been reported to be
involved in metabolic reprogramming to meet increased bioenergetic
and biosynthetic needs to support cancer cell proliferation,
invasion and metastasis (8). With
the growing identification of aberrantly expressed non-coding
(nc)RNAs in tumors, these regulatory molecules have been reported
to induce extensive metabolic crosstalk through targeting
rate-limiting enzymes, ultimately forming sophisticated regulatory
networks. The present review summarizes abnormal substance
metabolism in digestive system tumor metastasis and the related
metabolic pathway networks.
A hallmark of cancer is aberrant metabolism. Cells
often metabolize glucose, and cancer cells preferentially use
glycolysis over mitochondrial oxidative phosphorylation to create
glucose-dependent ATP and glycoferment intermediates for
macromolecular biosynthesis, even in conditions where oxygen supply
is sufficient. This is referred to as the Warburg effect (9–11).
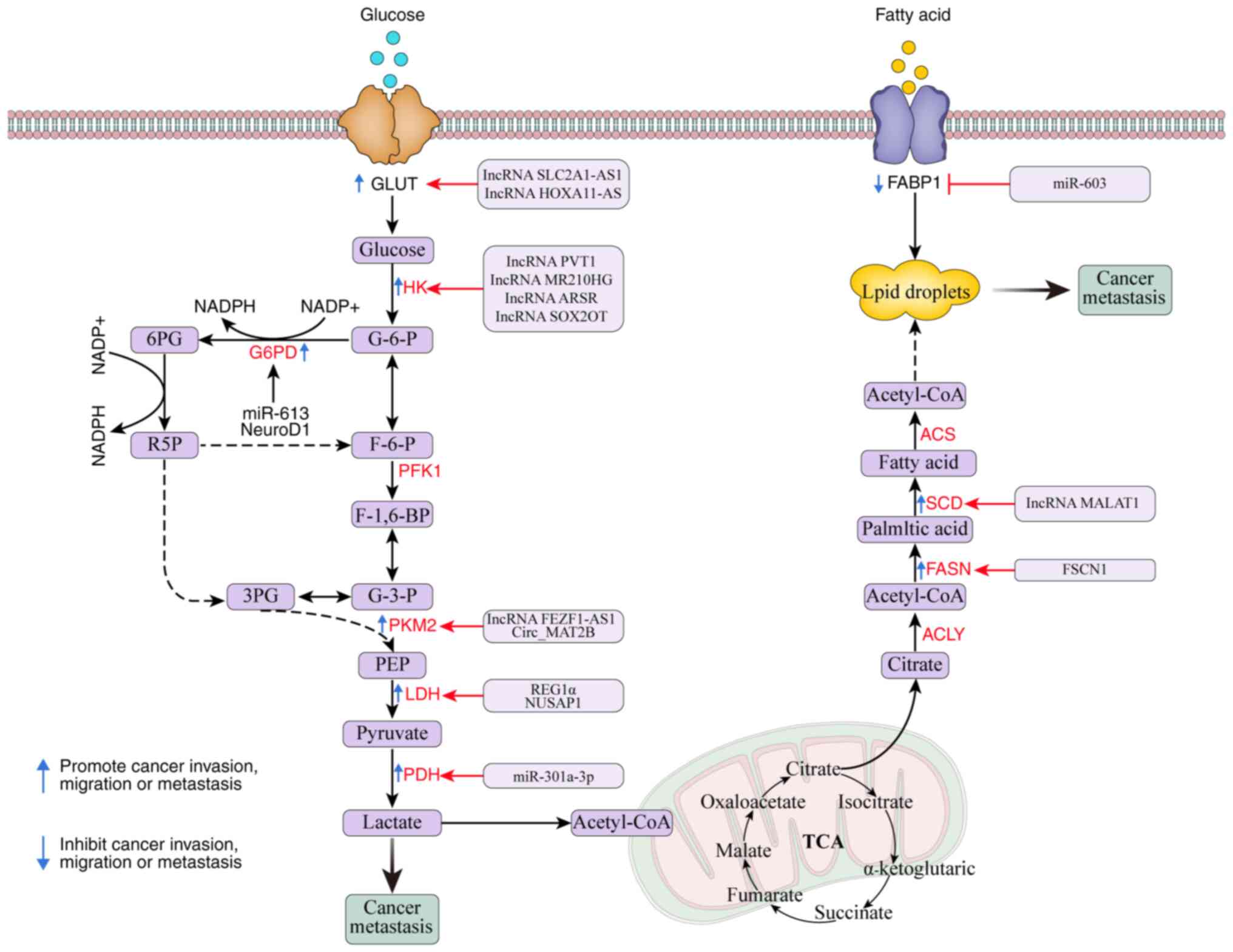
Important players in glycolytic pathways include pyruvate kinase
(PK), phosphofructokinase (PFK) and HK. Moreover, research has
reported that a number of coding RNAs and ncRNAs are notable
regulators of the metabolic enzymes and signaling pathways involved
in the metabolism of glucose in cancer cells (12,13)
(Table I and Fig. 2).
HK phosphorylates glucose to G6P in an ATP-dependent
manner, catalyzing the first irreversible step in the glycolytic
pathway. The four primary isoenzymes, HK1, HK2, HK3 and HK4, are
encoded by several genes (12). The
primary isoenzyme, HK2, is engaged in the first and rate-limiting
phases of glycolysis, which transforms glucose into
glucose-6-phosphate. Several cancer types show elevated levels of
this compound due to its ability to enhance glucose uptake in cells
and the Warburg effect (13,14).
Guo et al (15) identified a
putative competing endogenous (ce)RNA regulatory network in which
the lysyl oxidase like 1 (LOXL1)/microRNA (miR)-1224-5p/miR-761/HK2
axis was shown to promote cell proliferation and metastasis by
regulating aerobic glucose metabolism in CRC HCT-116 and SW480
cells, wherein the long non-coding (lnc)RNA LOXL1 modulated HK2
expression through competitively binding to endogenous miR-1224-5p
and miR-761. Similarly, lncRNA MIR210HG was reported to be
abnormally upregulated in PC, and promote lactate accumulation
through the miR-125b-5p/HK2/PKM2 axis, ultimately affecting
malignant phenotypes, including proliferation, invasion and
migration in PC PANC-1 and MIA PaCa-2 cells (16). Additionally, Chen et al
(17) reported that sponging of
miR-125b reduced the HK2-mediated Warburg effect in HCC Hep3B
cells, and sponging of miR-125b downregulated hsa_circ_0001806
expression, thereby suppressing the HK2-mediated Warburg effect,
primarily through inhibiting lactate accumulation and ATP
production. This ultimately impeded tumor proliferation and
migration. Several studies have reported that circPRMT5,
hsa_circ_0045932 and circRPS19 actively regulate the expression of
HK2 via sponge miRNAs, thereby accelerating glycolysis and
promoting tumor development (18–20).
Another isoenzyme of hexokinase, HK1, is reported to be positively
regulated by the lncRNA ARSR. This lncRNA binds to miR-34a-5p
through a sponge adsorption mechanism, thereby promoting glucose
uptake, lactate and ATP production in CRC Caco-2 cells, and
ultimately enhancing the invasion and migration capabilities of
tumor cells (21).
The rate-limiting enzyme PK catalyzes the last stage
of glycolysis, which is the conversion of phosphoenol pyruvate to
pyruvate (22). There are several
subtypes of mammalian PK, of which PKM2 is directly involved in
cancer aerobic glycolysis. Although mitochondrial oxidative
phosphorylation generates more net energy than aerobic glycolysis
in tumors, cancer cells use the Warburg effect to digest glucose
more quickly in response to higher energy demands. PKM2 is thought
to be a crucial metabolic enzyme for the Warburg effect in cancer
cells, and the overexpression of this enzyme is primarily
associated with an increased use of glucose and altered redox
balance in cells (23). Increasing
evidence points to the possible involvement of PKM2 in tumor
metastasis (24–26). PKM2 proteins are bound by the lncRNA
FEZF1-antisense RNA (AS)1 (FEZF1-AS1), which enhances their
stability and raises the amounts of PKM2 in the cytoplasm and
nucleus. Whilst FEZF1-AS1 regulates PKM2/STAT3 signaling and
glycolysis to increase the proliferation and migration of CRC LoVo
and HCT116 cells, an increase in cytosolic PKM2 stimulates PK
activity and lactate generation (27). Notably, under hypoxic conditions,
miR-338-3p interacts with circRNA MAT2B, thereby upregulating PKM2
expression levels and enhancing glycolysis in HCC Huh7 and HepG2
cells. The ATP generated from this metabolic reprogramming directly
fuels cytoskeletal remodeling and membrane fluidity, whilst lactate
accumulation contributes to creating an acidic microenvironment
conducive to invasion, collectively promoting tumor progression
(28).
One of the major glycolytic enzymes, lactate
dehydrogenase A (LDHA) is highly associated with thyroid cancer,
CRC and pancreatic ductal adenocarcinoma (PDAC) malignancies due to
its aberrant expression and overexpression (29–31).
Regenerating islet-derived protein 1-α (REG1α), for instance, was
reported to be upregulated in CRC serum and tissues. By enhancing
glycolysis mediated by the β-catenin/MYC axis, REG1α promoted
invasion in CRC DLD-1 and HCT-116 cells, and lung metastasis in
BALB/c nude mouse models. Furthermore, the Wnt/β-catenin/MYC axis
or glycolysis pathway could be silenced to successfully reverse the
malignant phenotype controlled by REG1α. Furthermore, REG1α
expression was moderated by methyltransferase-like 3 (METTL3). The
METTL3/REG1α/β-catenin/MYC axis in CRC suggests that REG1α may
serve as a novel biomarker and a possible target for treatment in
patients with CRC. REG1α drives glycolysis to provide both the
energy and carbon sources required for the synthesis and activation
of matrix metalloproteinases, which are essential for extracellular
matrix degradation during tumor cell invasion (30). Nucleolar and spindle associated
protein 1 (NUSAP1) was markedly overexpressed in PDAC, and in
several PDAC cell lines, NUSAP1 expression was consistent with that
of LDHA. Additionally, NUSAP1 facilitated PDAC metastasis and
LDHA-mediated glycolysis in BALB/c nude mouse models (31). This suggests that NUSAP1 promotes
the formation of distant metastases by regulating lactate
production and consequently influencing the pH of the TME.
Phosphoglycerate kinase 1 (PGK1) and
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB), are
key components of the glycolytic pathway. Furthermore, PFKFB has
four isoenzymes (PFKFB1, PFKFB2, PFKFB3 and PFKFB4), and studies
have reported that PFKFB3 and PFKFB4 are involved in EMT and
associated with tumor metastasis (32). Moreover, linc01572 modulated HCC
Huh7 and SNU-449 cells invasion and migration by increasing PFKFB4
levels by sponging miR-195-5p and subsequently enhancing glycolysis
and PI3K-AKT signaling activation (33).
The Warburg effect begins with the transfer of
glucose from the extracellular milieu. Using simple diffusion, the
transport capacity of glucose is determined by its hydrophilicity.
Consequently, certain membrane transporters are needed for this
process. In this mechanism, the GLUT family is crucial (34). Out of the 14 GLUT family members,
GLUT1 is most frequently expressed, which is also recognized as the
basal switch for glycolysis in tumor cells (35). Research has revealed that low
survival rates in human cancer are associated with elevated GLUT1
expression, potentially due to its association with the presence of
metastases (36). The promotion of
cancer cell proliferation and metastatic behavior is closely
associated with GLUT1 overexpression (37). Shang et al (38) reported that lncRNA SLC2A1-AS1 is
downregulated in liver cancer tissues. This lncRNA was shown to
inhibit aerobic glycofermentation and the progression of HCC
MHCC97-H and Huh7 cells by inactivating GLUT1, which is encoded by
the opposite-strand-encoding gene SLC2A1. Meanwhile, lncRNA
SLC2A1-AS1 enhanced GLUT1 expression through sponging miR-378a-3p,
thereby promoting aerobic glycolysis in ESCC EC9706, TE1 and
KYSE180 cells, and ultimately facilitating tumor cell invasion and
migration through the EMT pathway (39). Furthermore, FOXD1 was upregulated in
PC, and it activated SLC2A1 and the lncRNA HOXA11-AS by sponging
miR-148b. Reduced FOXD1 inhibited MIA PaCa-2 and PANC-1 PC cells
invasion, migration and aerobic glycolysis. Furthermore, a positive
correlation was observed between FOXD1 and GLUT1, suggesting their
synergistic role in driving oncogenesis (40). According to recent research, GLUT3
stimulates the EMT process via the TGF-β/JNK/ATF2 signaling
pathway, suggesting that this system may be a target for the
treatment of metastatic CRC (41).
Moreover, by controlling aerobic glycolysis to stimulate GC tumor
growth and migration, the oncogenic role of GLUT12 may be targeted
by the let-7a-5p/GLUT12 pathway to amplify the anticancer
properties of everolimus, suggesting a promising and viable
therapeutic strategy (42).
Solid tumors are characterized by hypoxia, which is
a diminished oxygen supply that promotes tumor growth. Hypoxic
conditions induce molecular responses in both normal and tumor
cells, driving the activation of the key transcription factor HIF
(43). As a crucial regulator of
oxygen balance, HIF-1 is a heterodimer transcription factor made up
of stable HIF-1β subunits and unstable HIF-1α subunits (44). HIF-1α is a transcription factor that
is frequently activated by the hypoxic milieu found in tumors and
is intimately associated with the growth, metastasis and
angiogenesis of tumors (45). It
has been established that lncRNA SNHG11 stimulates HIF-1α
downstream targets to enhance CRC HCT-116 and RKO cell invasion and
migration (46). It has also been
reported that upregulation of TMEM161B-AS1 promoted the expression
of HIF1AN via absorbing miR-23a-3p, inhibiting Eca109 and KYSE30
proliferation, invasion and glycolysis, further inducing the
downregulation of glycoenzyme-associated proteins, and ultimately
leading to the inhibition of ESCC progression (47). Furthermore, research has reported
that circRNF13 expression increased in PC and was associated with a
low rate of survival. The signaling pathway activated by HIF-1α and
RNF13 used the miR-654/PDK3 axis to increase PC cell invasion in
vitro and metastasis in BALB/c nude mouse lung and liver
metastasis models (48). By
enhancing PDK3 activity, this axis strengthens glycolytic
metabolism, thereby providing the energy required for the formation
of invaders and the activation of proteases, which mediate the
degradation of extracellular matrix during distant local invasion
and extravasation processes. Exosome release was stimulated by
hypoxia, and miR-301 expression was raised. Exosomes may carry
enhanced miR-301a-3p from one GC cell to another. The transferred
miR-301a-3p subsequently helped to block HIF-1α degradation by
focusing on prolyl hydroxylase domain-containing protein 3, which
could hydroxylate HIF-1α subunits and ubiquitinate their
destruction. HIF-1α and miR-301a-3p created a positive feedback
loop that promoted GC invasion, migration, proliferation and EMT in
BALB/c nude mouse models of lung metastasis (49).
An important glucose catabolic route is the PPP. The
PPP involves numerous enzymes and is essential in controlling the
proliferation of cancer cells (50).
One of the essential enzymes in the PPP oxidation
branch, 6PGD is involved in redox and nucleotide biosynthesis
(51). In several studies, elevated
G6PD levels have been reported to enhance the cancer progression in
several tumor types (52–54). Through the modulation of important
enzymes, ncRNAs perform their roles in PPP regulation. In HCC
MHCC97H and HCCLM3 cells, G6PD promotes migration and invasion by
activating the STAT3 pathways, which in turn causes EMT (52). circ_0003215 inhibits invasion and
migration in CRC SW480 and HT29 cells by suppressing the PPP
through DLG4-mediated, K48-linked ubiquitination and degradation of
G6PD (54). Additionally, circNOLC1
interacts with AZGP1 to activate the mTOR/SREBP1 signaling pathway,
or functions as a sponge for miR-212-5p to upregulate c-Met
expression. Collectively, these mechanisms induce G6PD to enhance
the PPP in CRC HCT116 and LoVo cells, promoting liver metastasis in
CRC (55). Furthermore, it has been
reported that NeuroD1 positively regulates G6PD in CRC and alters
tumor cell metabolism by stimulating the PPP, therapy enhancing CRC
tumorigenesis (56).
In the non-oxidized phase of PPP, TKT is an
essential enzyme that helps to maintain the levels of ribose
5-phosphate. The transketase family includes three human genes, TKT
and TKT-like genes 1 and 2 (57).
TKT has been reported to be upregulated in HCC and CRC (58,59),
and it has been demonstrated to have a positive association with
HCC metastasis and proliferation. Previous research reported that
TKT controlled metastasis in CRC via attaching to GRP78, which
changed AKT phosphorylation to influence cell glycolysis (59). In the meantime, ncRNAs took part in
networks connected to TKT. Through its interactions with TKT and
STAT3, the lncRNA TSLNC8 inactivated the IL6/STAT3 signaling
pathway, suppressing STAT3 phosphorylation and transcriptional
activity in HCC Huh-7 cells. This in turn prevented the growth of
tumors (60). Zeng et al
(61) reported that downregulation
of S100A11 suppressed the malignant progression of PDAC and
inhibited the PPP via TKT expression.
The TCA cycle satisfies the needs of cells for
bioenergetics, biosynthesis and redox equilibrium, and is a key
route for oxidative phosphorylation. Research indicates that
certain cancer cells, particularly those with dysregulated oncogene
and tumor suppressor gene expression, may heavily depend on the TCA
cycle to produce energy and synthesize macromolecules, despite the
earlier hypothesis that cancer cells primarily used aerobic
glycolysis and avoided the TCA cycle (62).
There are four isoenzymes that make up pyruvate
dehydrogenase kinases (PDKs). The pyruvate dehydrogenase complex is
phosphorylated and rendered inactive by PDKs, preventing
mitochondria from oxidatively metabolizing pyruvate. This procedure
may stimulate anabolic pathways, which in turn may stimulate the
growth of cancer cells. By raising pyruvate dehydrogenase (PDH)
activity and thus increasing ROS generation, inhibition of PDK
causes cell death (63).
A gatekeeper enzyme called PDK1 is involved in the
growth, survival and angiogenesis of tumors in cancer. PDK1 is a
regulator of PDH, a key rate-limiting enzyme of cellular
glycolysis, and has been widely confirmed to be involved in the
regulation of cancer progression (64). Research has reported that
circ_0000284 serves an oncogene role in intrahepatic
cholangiocarcinoma (ICC), which facilitates ICC HCCC-9810 and
HUCCT1 cell growth and metastasis through sponging miR-152-3p to
increase PDK1 expression (65).
According to another study, by sponging miR-599, lncRNA PTPRG-AS1
upregulated PDK1 expression, thereby promoting migration and
proliferation in ESCC TE-8 and KYSE-150 cells (66). In GC SGC7901 and MGC803 cells, to
restore the activity of PDH, the gatekeeping enzyme that catalyzes
the decarboxylation of pyruvate to create acetyl-CoA, miR-422a was
reported to suppress PDK2. This process reduced NADPH generation,
thereby compromising the ability of the cell to counteract
oxidative damage incurred during migration (67).
A class of intracellular lipid chaperones known as
FABPs binds to long-chain fatty acids and related lipids to help
move them into different parts of the cell, including the nucleus
(71). Humans have been found to
exhibit 10 distinct FABP subtypes. FABP5, a diminutive constituent
of the cytoplasmic FABP family, exhibits a strong propensity for
binding to long-chain FAs. Research suggests that FABP5, through
triggering EMT and controlling angiogenic responses, serves a
critical role in the invasion, metastasis and development of cancer
(72,73). In addition, FABP1 is overexpressed
in the cytoplasm of HCC cells (74). However, IL-6 has been reported to
increase the expression of miR-603 and then suppresses the
production of FABP1, which promotes lipid metabolism and the
manufacture of related proteins, ultimately raising the levels of
cellular oxidative stress and causing HCC Huh-7 cells migration and
invasion (75).
SREBPs are the most important transcription factors
for regulating lipid homeostasis. Previous studies have reported
that SREBP is highly upregulated and promotes tumor growth in
several cancers (70). The lncRNA
ZFAS1 has been reported to rewire fat metabolism to accelerate the
invasion of CRC SW480 and RKO cells by binding to PABP2 and
promoting the interaction between PABP2 and SREBP1 (76). Moreover, the lncRNA SNHG16 was
reported to promote invasion and migration by directly controlling
the miR-195/SREBP2 axis, which in turn sped up the development of
PC PANC-1 and AsPC-1 cells. By increasing cholesterol synthesis,
SREBP2 enhances plasma membrane fluidity, which promotes
deformability and motility for cell migration. Concurrently, it
supplies the essential lipid components required by metastatic
cells to preserve membrane integrity (77). Furthermore, through mRNA splicing
and transcription, lncRNA MALAT1 stimulates the expression of
several genes in AMPK signaling and unsaturated fatty acid
metabolism pathways, leading to the upregulation of glucose uptake
and adipogenesis, which indicates the function of MALAT1 in tumor
cell invasion and migration (78).
Increased expression of the SCD1 enzyme has been
reported to accelerate the development of several malignancies.
SCD1 is involved in the synthesis of monounsaturated fatty acids,
including oleic acid (79). The
orthotopic CRC model demonstrated that SULT2B1 promotes metastatic
progression of colorectal cancer. Furthermore, experiments in
HCT116 and SW480 cells revealed that SULT2B1 directly interacts
with SCD to enhance lipid metabolism, thereby facilitating the
metastasis of CRC (80). In
addition, low expression of miR-4310 is associated with a poor
prognosis. By inhibiting SCD1 and FASN-mediated lipid synthesis,
miR-4310 has been reported to inhibit HCC cell migration and
invasion in vitro and HCC tumor growth and metastasis in
vivo (81). Furthermore, Ly1
antibody reactive (LYAR) and fascin actin-bundling protein 1
(FSCN1) has been reported to participate in CRC progression and
promote cell metastasis, and LYAR partly regulates FSCN1 expression
in CRC. Additionally, FSCN1 is associated with lipid metabolism,
and FSCN1 silencing reduces the expression levels of FASN and SCD1
in CRC progression (82).
Serum deprivation-response protein (SDPR) has been
reported to participate in TGF-β-induced GC metastasis. It has been
shown that overexpression of SDPR inhibits TGF-β-induced GC MKN45
and MGC803 cells invasion and migration by suppressing ERK and
enhancing the expression of peroxisome proliferator-activated
receptor α, thereby upregulating the expression of carnitine
palmitoyltransferase 1A (CPT1A), promoting fatty acid oxidation
(FAO) and inducing GC metastasis. By driving FAO, CPT1A supports
the TCA cycle function through acetyl-CoA production, which in turn
furnishes the persistent energy required for cell migration
(83). Zhao et al (84) reported that hexokinase domain
containing 1 (HKDC1) expression was upregulated in GC.
Additionally, lipid metabolism serves a critical role in the role
of PRKDC as a downstream effector of HKDC1-mediated GC
carcinogenesis. HKDC1 can mechanically regulate the well-known
oncoprotein G3BP stress granule assembly factor 1 (G3BP1), and
HKDC1 and G3BP1 collaborate to support PRKDC stability. The
HKDC1/G3BP1-PRKDC axis reprograms lipid metabolism to promote GC
metastasis and chemoresistance (84).
Glutamine is mostly utilized for the synthesis of
energy and is regarded as a non-essential amino acid. However, when
certain cancer cells proliferate quickly, when there is stress, or
when proliferating cells grow quickly, glutamine becomes a
conditional necessary amino acid (85,86).
SLC1A5 is a sodium-coupled transporter of alanine, serine, cysteine
and glutamine. It is often referred to as alanine-serine-cysteine
transporter 2. The expression of circ_0001273 is upregulated in
esophageal cancer (EC) tissues and cell lines. As circ_0001273
regulates miR-622 and downstream SLC1A5, it partially explains why
silencing EC TE1 and ECA109 cells was reported to reduce their
survival, proliferation, migration and rate of glutamine metabolism
(87). Moreover, circ_0001093 has
been reported to function as ceRNA by sponging miR-579-3p, raising
the expression of glutaminase, enhancing glutamine metabolism and
advancing malignancy in ESCC (88).
Building on the established role of glutamic-oxaloacetic
transaminase 1 (GOT1) in glutamate metabolism (89), further research demonstrates that
silencing circ_MBOAT2 inhibits glutamine catabolism and progression
in PC PANC-1 and SW1990 cells via the circ_MBOAT2/miR-433-3p/GOT1
axis (90). Furthermore, HIF-2α is
markedly upregulated in PDAC Panc-1 and Capan-2 cells and promotes
glutamine metabolism by targeting GOT1 via activation of the
PI3K/mTOR complex 2 pathways (91).
Low GOT2 expression is also associated with glutamine metabolic
reprogramming to glutathione synthesis, and promotes HCC
progression by activating the PI3K/AKT/mTOR pathway (92) (Table
III).
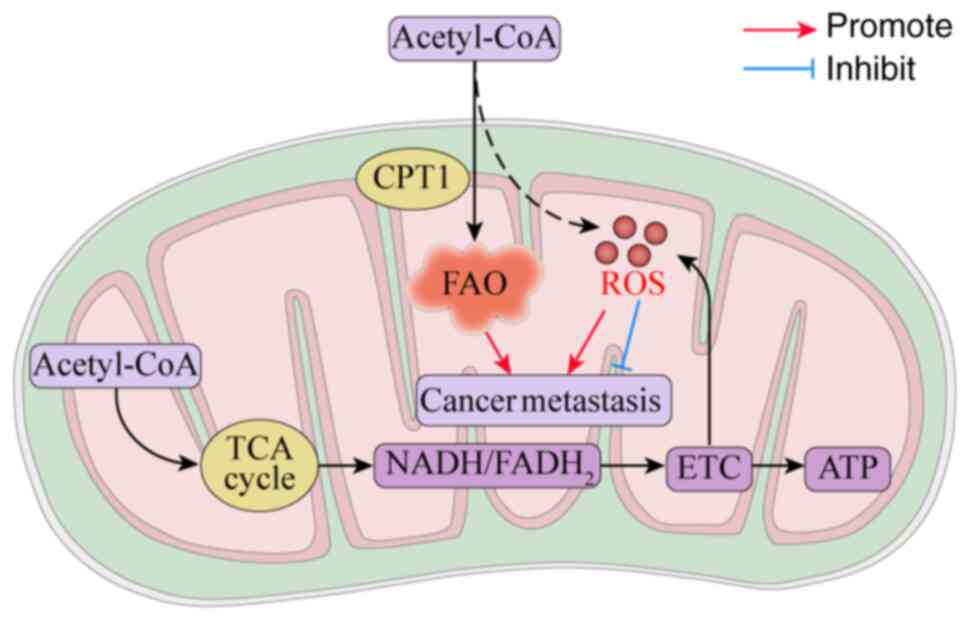
Mitochondria influence tumorigenesis, and as the
primary source of ATP, mitochondria provide building blocks for
anabolism through cellular repair mechanisms, with their pivotal
role in regulated cell death signaling crucial for tumor
dissemination (93). For instance,
transmembrane guanylate cyclase natriuretic peptide receptor 1
facilitates GC lymph node metastasis by activating lipid droplet
lipolysis and enhancing mitochondrial oxidative phosphorylation
(OXPHOS) (94). Moreover, organic
cation transporter novel type 2 augments cancer stem-like traits in
HCC through increased fatty acid β-oxidation and OXPHOS activation
(95) (Fig. 3).
The OXPHOS metabolic pathway fulfills two critical
functions in driving tumor cell metastasis: i) Providing
bioenergetic demands in the form of ATP; and ii) Extracting carbon
from glucose for macromolecule synthesis, serving as a central hub
integrating catabolic and anabolic processes. Enzymes of the TCA
cycle in the mitochondrial matrix and transmembrane protein
complexes of the electron transport chain (ETC) constitute the core
machinery for this process. Within the mitochondrial electron
transport system, ETC complexes I–IV accept electrons from TCA
cycle-derived NADH and FADH2. Subsequent proton reflux
through Complex V (ATP synthase) into the mitochondrial matrix
generates ATP (96). Certain tumors
exhibit high dependency on OXPHOS-derived ATP, which can be
identified through specific genetic alterations (97). Metastasis-associated antigen 1 has
been identified as a novel regulator of ATP synthase via its
interaction with the ATP synthase F1 subunit α, promoting colon
cancer liver metastasis through mitochondrial bioenergetic
reprogramming and enhanced OXPHOS (98). Sorting nexin 17 facilitates HCC
proliferation and metastasis by interacting with STAT3, thereby
activating the STAT3/c-Myc signaling pathway to augment OXPHOS
(99).
CPT1A is a rate-limiting enzyme localized on the
mitochondrial outer membrane, and catalyzes the rate-limiting step
of FAO to generate ATP essential for tumor cell proliferation and
metastasis. CPT1A-dependent FAO represents a vital metabolic
pathway in malignancies. Elevated CPT1A expression in CRC (100) and PDAC (101) activates FAO and enhances
metastatic potential. Furthermore, ALKBH5 promotes CRC progression
by mediating CPT1A upregulation through m6A
demethylation, consequently facilitating M2 macrophage polarization
(102).
ROS exhibit a dual role in oncogenesis: At
low-to-moderate levels, mtROS function as crucial signaling
molecules that activate several protumor signaling pathways through
oxidative modification (103). For
instance, low-level mtROS stabilizes HIF-1α to enhance glycolysis
and facilitate metastasis (104),
whereas high-level mtROS conversely activates the SIRT3/PGC-1α axis
to promote OXPHOS (105).
Furthermore, fatty acid β-oxidation inherently generates ROS as
metabolic byproducts. Although FAO is recognized as a
tumor-promoting process, the metabolic crosstalk triggered by
FAO-derived ROS requires further mechanistic exploration. However,
when mtROS accumulation becomes excessive and surpasses the
cellular antioxidant defense capacity, it induces severe oxidative
stress, triggering cell death, necrosis and ferroptosis, resulting
in tumor-suppressive effects (106). For example, CST1 promotes gastric
cancer metastasis by recruiting OTUB1 to alleviate GPX4
ubiquitination, thereby enhancing GPX4 protein stability, reducing
ROS accumulation, and consequently inhibiting ferroptosis (107). Therefore, strategies aimed at
leveraging ROS to block tumor metastasis have emerged as a
promising therapeutic approach in oncology.
In recent years, metabolism in tumor metastasis has
attracted more attention. However, cancer cell metabolism is a
complex matter. The present review discusses, primarily based on
evidence from in vitro studies, several metabolic
alterations commonly found in cancer cells, including glycolysis,
FAO, amino acid metabolism and alterations in mitochondria. This
review systematically synthesizes evidence on how key genes target
metabolic enzymes or related signaling pathways to drive tumor
invasion and metastasis. It aims to establish the correlation
between tumor metastasis and cellular metabolism, and to identify
suitable targets at the intersection of metabolic pathways to
selectively disrupt metabolic networks and prevent tumor metastasis
(108,109). For instance, 2-deoxy-d-glucose can
inhibit glycolysis and induce cell death (110). However, monotherapy targeting
tumor progression still requires further exploration.
Digestive system neoplasms metastasis is one of the
leading causes of tumor-related mortality worldwide. The present
review identified common features in metabolic reprogramming across
gastrointestinal cancers. Beyond the classical Warburg effect,
metabolic alterations such as enhanced fatty acid β-oxidation,
elevated mitochondrial oxidative phosphorylation, and increased ROS
levels all contribute to varying degrees to tumor progression.
Furthermore, during the invasion or migration of digestive system
tumors, the expression levels of certain key metabolic enzymes are
upregulated; for instance, HK expression is increased in HCC
(75), colorectal cancer (76), PC (77) and GC (80). However, metabolic specificity also
exists among different gastrointestinal tumors, primarily stemming
from differences in tissue origin and the TME. For example,
enhanced bile acid metabolism promotes the progression of liver
cancer: Treatment with T-CDCA was reported to markedly increase the
proliferative capacity of HCC HepG2 cells, and downregulate the
expression of the tumor suppressor protein CEBPα in HCC (111). Additionally, metabolic
dysregulation mediated by gut microbiota serves a crucial role in
CRC, involving species such as Streptococcus bovis, Helicobacter
pylori and Escherichia coli (112).
Targeting a single metabolic pathway presents
notable challenges, whereas a combination of therapies
simultaneously targeting multiple tumor metabolites or metabolic
enzymes may enhance therapeutic efficacy. Studies have demonstrated
that mitochondria generate ATP through multiple pathways, serving
as the primary energy source for tumor metastasis. Precise
targeting of cancer-specific mitochondria can impair their capacity
for de-differentiation, proliferation and metastasis, thereby
contributing to improved treatment outcomes and overall prognosis
(113). As a result, the
individualized role of mitochondria in cancer should receive more
attention. The tumor metabolic microenvironment in vivo is
complex, and the present review summarizes only a part of the
complex events of cancer development. Therefore, researching the
function that mitochondrial metabolism serves in the TME can lead
to novel therapeutic approaches for cancer, including targeted
medication development.
Although preliminary progress has been made in
research on tumor metabolic reprogramming, notable knowledge gaps
and insufficient investigation remain regarding the role of
metabolic crosstalk in tumor metastasis, necessitating further
in-depth exploration: i) Most current studies rely on in
vitro cell models, which fail to fully recapitulate the
complexity and dynamic nature of the TME in vivo.
Consequently, conclusions derived from in vitro experiments
exhibit certain limitations; ii) tumor metabolism demonstrates high
heterogeneity, not only between primary and metastatic sites but
also within different regions of a single tumor. To date,
systematic studies on metabolic dynamics are still lacking; and
iii) although the present review summarizes several metabolic
enzymes and metabolites that regulate tumor cell metastasis, the
understanding of transcriptome-genome-metabolome crosstalk patterns
continues to evolve. Elucidating the mechanisms of metabolic
crosstalk in tumor cells will provide new opportunities for
effectively combating cancer.
Not applicable.
The present project was supported by the National Natural
Science Foundation of China (grant no. 32401139).
Not applicable.
JL conceived, organized and wrote the manuscript. WZ
revised the manuscript for important intellectual content. Data
authentication is not applicable. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
2
|
Cao M, Li H, Sun D and Chen W: Cancer
burden of major cancers in China: A need for sustainable actions.
Cancer Commun (Lond). 40:205–210. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clark AG and Vignjevic DM: Modes of cancer
cell invasion and the role of the microenvironment. Curr Opin Cell
Biol. 36:13–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu D, Shao F, Bian X, Meng Y, Liang T and
Lu Z: The evolving landscape of noncanonical functions of metabolic
enzymes in cancer and other pathologies. Cell Metab. 33:33–50.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan C, Li B and Simon MC: Moonlighting
functions of metabolic enzymes and metabolites in cancer. Mol Cell.
81:3760–3774. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lv L and Lei Q: Proteins moonlighting in
tumor metabolism and epigenetics. Front Med. 15:383–403. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martínez-Reyes I and Chandel NS: Cancer
metabolism: Looking forward. Nat Rev Cancer. 21:669–680. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML,
Chi HC, Tsai CY, Chung IH, Chen CY and Lin KH: Taurine up-regulated
gene 1 functions as a master regulator to coordinate glycolysis and
metastasis in hepatocellular carcinoma. Hepatology. 67:188–203.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He J, Li F, Zhou Y, Hou X, Liu S, Li X,
Zhang Y, Jing X and Yang L: LncRNA XLOC_006390 promotes pancreatic
carcinogenesis and glutamate metabolism by stabilizing c-Myc.
Cancer Lett. 469:419–428. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Robey RB and Hay N: Is Akt the ‘Warburg
kinase’?-Akt-energy metabolism interactions and oncogenesis. Semin
Cancer Biol. 19:25–31. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu F, Yan JJ, Gan Y, Chang Y, Wang HL, He
XX and Zhao Q: miR-885-5p negatively regulates warburg effect by
silencing hexokinase 2 in liver cancer. Mol Ther Nucleic Acids.
18:308–319. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin D, Hua L, Wang J, Liu Y and Li X: Long
Non-Coding RNA DUXAP8 facilitates cell viability, migration, and
glycolysis in non-small-cell lung cancer via regulating HK2 and
LDHA by Inhibition of miR-409-3p. Onco Targets Ther. 13:7111–7123.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo T, Peng S, Liu D and Li Y: Long
Non-coding RNA LOXL1-AS1 facilitates colorectal cancer progression
via regulating miR-1224-5p/miR-761/HK2 axis. Biochem Genet.
60:2416–2433. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu T, Li G, Wang C, Gong G, Wang L, Li C,
Chen Y and Wang X: MIR210HG regulates glycolysis, cell
proliferation, and metastasis of pancreatic cancer cells through
miR-125b-5p/HK2/PKM2 axis. RNA Biol. 18:2513–2530. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, She P, Wang C, Shi L, Zhang T,
Wang Y, Li H, Qian L and Li M: Hsa_circ_0001806 promotes glycolysis
and cell progression in hepatocellular carcinoma through
miR-125b/HK2. J Clin Lab Anal. 35:e239912021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding Z, Guo L, Deng Z and Li P: Circ-PRMT5
enhances the proliferation, migration and glycolysis of hepatoma
cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 19:269–279.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hong F, Deng Z, Tie R and Yang S:
Hsa_circ_0045932 regulates the progression of colorectal cancer by
regulating HK2 through sponging miR-873-5p. J Clin Lab Anal.
36:e246412022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng X, Shao J, Qian J and Liu S:
circRPS19 affects HK2-mediated aerobic glycolysis and cell
viability via the miR-125a-5p/USP7 pathway in gastric cancer. Int J
Oncol. 63:982023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S, Zhu K, Liu L, Gu J, Niu H and Guo J:
lncARSR sponges miR-34a-5p to promote colorectal cancer invasion
and metastasis via hexokinase-1-mediated glycolysis. Cancer Sci.
111:3938–3952. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang W, Xia Y, Hawke D, Li X, Liang J,
Xing D, Aldape K, Hunter T, Alfred Yung WK and Lu Z: PKM2
phosphorylates histone H3 and promotes gene transcription and
tumorigenesis. Cell. 150:685–696. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alquraishi M, Puckett DL, Alani DS,
Humidat AS, Frankel VD, Donohoe DR, Whelan J and Bettaieb A:
Pyruvate kinase M2: A simple molecule with complex functions. Free
Radic Biol Med. 143:176–192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Wang H, Wu B, Zhang C, Yu H, Li X,
Wang Q, Shi X, Fan C, Wang D, et al: Long Noncoding RNA LINC00551
suppresses glycolysis and tumor progression by regulating
c-Myc-mediated PKM2 expression in lung adenocarcinoma. Onco Targets
Ther. 13:11459–11470. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang Y, Zhang D, Zheng T, Yang G, Wang J,
Meng F, Liu Y, Zhang G, Zhang L, Han J, et al: lncRNA-SOX2OT
promotes hepatocellular carcinoma invasion and metastasis through
miR-122-5p-mediated activation of PKM2. Oncogenesis. 9:542020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren J, Li W, Pan G, Huang F, Yang J, Zhang
H, Zhou R and Xu N: miR-142-3p modulates cell invasion and
migration via PKM2-mediated aerobic glycolysis in colorectal
cancer. Anal Cell Pathol (Amst). 2021:99277202021.PubMed/NCBI
|
|
27
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Q, Pan X, Zhu D, Deng Z, Jiang R and
Wang X: Circular RNA MAT2B promotes glycolysis and malignancy of
hepatocellular carcinoma through the miR-338-3p/PKM2 axis under
hypoxic stress. Hepatology. 70:1298–1316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao Y, Yang F, Yang XA, Zhang L, Yu H,
Cheng X, Xu S, Pan J, Wang K and Li P: Mitochondrial metabolism is
inhibited by the HIF1α-MYC-PGC-1β axis in BRAF V600E thyroid
cancer. FEBS J. 286:1420–1436. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou M, He J, Li Y, Jiang L, Ran J, Wang
C, Ju C, Du D, Xu X, Wang X, et al: N6-methyladenosine
modification of REG1α facilitates colorectal cancer progression via
β-catenin/MYC/LDHA axis mediated glycolytic reprogramming. Cell
Death Dis. 14:5572023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen M, Cen K, Song Y, Zhang X, Liou YC,
Liu P, Huang J, Ruan J, He J, Ye W, et al:
NUSAP1-LDHA-Glycolysis-Lactate feedforward loop promotes Warburg
effect and metastasis in pancreatic ductal adenocarcinoma. Cancer
Lett. 567:2162852023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kotowski K, Rosik J, Machaj F, Supplitt S,
Wiczew D, Jabłońska K, Wiechec E, Ghavami S and Dzięgiel P: Role of
PFKFB3 and PFKFB4 in cancer: Genetic basis, impact on disease
development/progression, and potential as therapeutic targets.
Cancers (Basel). 13:9092021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lai S, Quan Z, Hao Y, Liu J, Wang Z, Dai
L, Dai H, He S and Tang B: Long Non-coding RNA LINC01572 promotes
hepatocellular carcinoma progression via sponging miR-195-5p to
enhance PFKFB4-mediated glycolysis and PI3K/AKT activation. Front
Cell Dev Biol. 9:7830882021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Holman GD: Chemical biology probes of
mammalian GLUT structure and function. Biochem J. 475:3511–3534.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wood IS and Trayhurn P: Glucose
transporters (GLUT and SGLT): Expanded families of sugar transport
proteins. Br J Nutr. 89:3–9. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu M, Yongzhi H, Chen S, Luo X, Lin Y,
Zhou Y, Jin H, Hou B, Deng Y, Tu L and Jian Z: The prognostic value
of GLUT1 in cancers: A systematic review and meta-analysis.
Oncotarget. 8:43356–43367. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Macheda ML, Rogers S and Best JD:
Molecular and cellular regulation of glucose transporter (GLUT)
proteins in cancer. J Cell Physiol. 202:654–662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shang R, Wang M, Dai B, Du J, Wang J, Liu
Z, Qu S, Yang X, Liu J, Xia C, et al: Long noncoding RNA SLC2A1-AS1
regulates aerobic glycolysis and progression in hepatocellular
carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol Oncol.
14:1381–1396. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu H, Zhang Q, Song Y, Hao Y, Cui Y,
Zhang X, Zhang X, Qin Y, Zhu G, Wang F, et al: Long non-coding RNA
SLC2A1-AS1 induced by GLI3 promotes aerobic glycolysis and
progression in esophageal squamous cell carcinoma by sponging
miR-378a-3p to enhance Glut1 expression. J Exp Clin Cancer Res.
40:2872021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cai K, Chen S, Zhu C, Li L, Yu C, He Z and
Sun C: FOXD1 facilitates pancreatic cancer cell proliferation,
invasion, and metastasis by regulating GLUT1-mediated aerobic
glycolysis. Cell Death Dis. 13:7652022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Song MY, Lee DY, Yun SM and Kim EH: GLUT3
promotes epithelial-mesenchymal transition via TGF-β/JNK/ATF2
signaling pathway in colorectal cancer cells. Biomedicines.
10:18372022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shi Y, Zhang Y, Ran F, Liu J, Lin J, Hao
X, Ding L and Ye Q: Let-7a-5p inhibits triple-negative breast tumor
growth and metastasis through GLUT12-mediated warburg effect.
Cancer Lett. 495:53–65. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Brahimi-Horn MC, Chiche J and Pouyssegur
J: Hypoxia and cancer. J Mol Med (Berl). 85:1301–1307. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Adams JM, Difazio LT, Rolandelli RH, Luján
JJ, Haskó G, Csóka B, Selmeczy Z and Németh ZH: HIF-1: A key
mediator in hypoxia. Acta Physiol Hung. 96:19–28. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nepal M, Choi HJ, Choi BY, Kim SL, Ryu JH,
Kim DH, Lee YH and Soh Y: Anti-angiogenic and anti-tumor activity
of Bavachinin by targeting hypoxia-inducible factor-1α. Eur J
Pharmacol. 691:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu L, Huan L, Guo T, Wu Y, Liu Y, Wang Q,
Huang S, Xu Y, Liang L and He X: LncRNA SNHG11 facilitates tumor
metastasis by interacting with and stabilizing HIF-1α. Oncogene.
39:7005–7018. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shi Z, Li G, Li Z, Liu J and Tang Y:
TMEM161B-AS1 suppresses proliferation, invasion and glycolysis by
targeting miR-23a-3p/HIF1AN signal axis in oesophageal squamous
cell carcinoma. J Cell Mol Med. 25:6535–6549. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao Q, Zhu Z, Xiao W, Zong G, Wang C,
Jiang W, Li K, Shen J, Guo X, Cui J, et al: Hypoxia-induced
circRNF13 promotes the progression and glycolysis of pancreatic
cancer. Exp Mol Med. 54:1940–1954. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xia X, Wang S, Ni B, Xing S, Cao H, Zhang
Z, Yu F, Zhao E and Zhao G: Hypoxic gastric cancer-derived exosomes
promote progression and metastasis via MiR-301a-3p/PHD3/HIF-1α
positive feedback loop. Oncogene. 39:6231–6244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jin L and Zhou Y: Crucial role of the
pentose phosphate pathway in malignant tumors. Oncol Lett.
17:4213–4221. 2019.PubMed/NCBI
|
|
51
|
Polat IH, Tarrado-Castellarnau M, Bharat
R, Perarnau J, Benito A, Cortés R, Sabatier P and Cascante M:
Oxidative pentose phosphate pathway enzyme 6-phosphogluconate
dehydrogenase plays a key role in breast cancer metabolism. Biology
(Basel). 10:852021.PubMed/NCBI
|
|
52
|
Lu M, Lu L, Dong Q, Yu G, Chen J, Qin L,
Wang L, Zhu W and Jia H: Elevated G6PD expression contributes to
migration and invasion of hepatocellular carcinoma cells by
inducing epithelial-mesenchymal transition. Acta Biochim Biophys
Sin (Shanghai). 50:370–380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen B, Cai T, Huang C, Zang X, Sun L, Guo
S, Wang Q, Chen Z, Zhao Y, Han Z, et al: G6PD-NF-κB-HGF signal in
gastric cancer-associated mesenchymal stem cells promotes the
proliferation and metastasis of gastric cancer cells by
upregulating the expression of HK2. Front Oncol. 11:6487062021.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen B, Hong Y, Gui R, Zheng H, Tian S,
Zhai X, Xie X, Chen Q, Qian Q, Ren X, et al: N6-methyladenosine
modification of circ_0003215 suppresses the pentose phosphate
pathway and malignancy of colorectal cancer through the
miR-663b/DLG4/G6PD axis. Cell Death Dis. 13:8042022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yuan M, Zhang X, Yue F, Zhang F, Jiang S,
Zhou X, Lv J, Zhang Z, Sun Y, Chen Z, et al: CircNOLC1 promotes
colorectal cancer liver metastasis by interacting with AZGP1 and
sponging miR-212-5p to regulate reprogramming of the oxidative
pentose phosphate pathway. Adv Sci (Weinh). 10:e22052292023.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Z, He Y, Li Y, Li J, Zhao H, Song G,
Miyagishi M, Wu S and Kasim V: NeuroD1 promotes tumor cell
proliferation and tumorigenesis by directly activating the pentose
phosphate pathway in colorectal carcinoma. Oncogene. 40:6736–6747.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tseng CW, Kuo WH, Chan SH, Chan HL, Chang
KJ and Wang LH: Transketolase regulates the metabolic switch to
control breast cancer cell metastasis via the α-ketoglutarate
signaling pathway. Cancer Res. 78:2799–2812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qin Z, Xiang C, Zhong F, Liu Y, Dong Q, Li
K, Shi W, Ding C, Qin L and He F: Transketolase (TKT) activity and
nuclear localization promote hepatocellular carcinoma in a
metabolic and a non-metabolic manner. J Exp Clin Cancer Res.
38:1542019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li M, Zhao X, Yong H, Xu J, Qu P, Qiao S,
Hou P, Li Z, Chu S, Zheng J and Bai J: Transketolase promotes
colorectal cancer metastasis through regulating AKT
phosphorylation. Cell Death Dis. 13:992022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang J, Li Z, Liu L, Wang Q, Li S, Chen
D, Hu Z, Yu T, Ding J, Li J, et al: Long noncoding RNA TSLNC8 is a
tumor suppressor that inactivates the interleukin-6/STAT3 signaling
pathway. Hepatology. 67:171–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zeng X, Guo H, Liu Z, Qin Z, Cong Y, Ren
N, Zhang Y and Zhang N: S100A11 activates the pentose phosphate
pathway to induce malignant biological behaviour of pancreatic
ductal adenocarcinoma. Cell Death Dis. 13:5682022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Anderson NM, Mucka P, Kern JG and Feng H:
The emerging role and targetability of the TCA cycle in cancer
metabolism. Protein Cell. 9:216–237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Woolbright BL, Rajendran G, Harris RA and
Taylor JA III: Metabolic flexibility in cancer: Targeting the
pyruvate dehydrogenase kinase: Pyruvate dehydrogenase axis. Mol
Cancer Ther. 18:1673–1681. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang G, Liu X, Xie J, Meng J and Ni X:
PDK-1 mediated Hippo-YAP-IRS2 signaling pathway and involved in the
apoptosis of non-small cell lung cancer cells. Biosci Rep.
39:BSR201820992019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sun J, Feng M, Zou H, Mao Y and Yu W:
Circ_0000284 facilitates the growth, metastasis and glycolysis of
intrahepatic cholangiocarcinoma through miR-152-3p-mediated PDK1
expression. Histol histopathol. 38:1129–1143. 2022.PubMed/NCBI
|
|
66
|
Wu K, Wang Z, Huang Y, Yao L, Kang N, Ge
W, Zhang R and He W: LncRNA PTPRG-AS1 facilitates glycolysis and
stemness properties of esophageal squamous cell carcinoma cells
through miR-599/PDK1 axis. J Gastroenterol Hepatol. 37:507–517.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He Z, Li Z, Zhang X, Yin K, Wang W, Xu Z,
Li B, Zhang L, Xu J, Sun G, et al: MiR-422a regulates cellular
metabolism and malignancy by targeting pyruvate dehydrogenase
kinase 2 in gastric cancer. Cell Death Dis. 9:5052018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Martin-Perez M, Urdiroz-Urricelqui U,
Bigas C and Benitah SA: The role of lipids in cancer progression
and metastasis. Cell Metab. 34:1675–1699. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bian X, Liu R, Meng Y, Xing D, Xu D and Lu
Z: Lipid metabolism and cancer. J Exp Med. 218:e202016062021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cheng C, Geng F, Cheng X and Guo D: Lipid
metabolism reprogramming and its potential targets in cancer.
Cancer Commun (Lond). 38:272018.PubMed/NCBI
|
|
71
|
Carbonetti G, Wilpshaar T, Kroonen J,
Studholme K, Converso C, d'Oelsnitz S and Kaczocha M: FABP5
coordinates lipid signaling that promotes prostate cancer
metastasis. Sci Rep. 9:189442019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yu CW, Liang X, Lipsky S, Karaaslan C,
Kozakewich H, Hotamisligil GS, Bischoff J and Cataltepe S: Dual
role of fatty acid-binding protein 5 on endothelial cell fate: A
potential link between lipid metabolism and angiogenic responses.
Angiogenesis. 19:95–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ohata T, Yokoo H, Kamiyama T, Fukai M,
Aiyama T, Hatanaka Y, Hatanaka K, Wakayama K, Orimo T, Kakisaka T,
et al: Fatty acid-binding protein 5 function in hepatocellular
carcinoma through induction of epithelial-mesenchymal transition.
Cancer Med. 6:1049–1061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang G, Bonkovsky HL, de Lemos A and
Burczynski FJ: Recent insights into the biological functions of
liver fatty acid binding protein 1. J Lipid Res. 56:2238–2247.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lin YX, Wu XB, Zheng CW, Zhang QL, Zhang
GQ, Chen K, Zhan Q and An FM: Mechanistic investigation on the
regulation of FABP1 by the IL-6/miR-603 signaling in the
pathogenesis of hepatocellular carcinoma. Biomed Res Int.
2021:85796582021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang H, Chen Y, Liu Y, Li Q, Luo J, Wang
L, Chen Y, Sang C, Zhang W, Ge X, et al: The lncRNA ZFAS1 regulates
lipogenesis in colorectal cancer by binding polyadenylate-binding
protein 2 to stabilize SREBP1 mRNA. Mol Ther Nucleic Acids.
27:363–374. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yu Y, Dong JT, He B, Zou YF, Li XS, Xi CH
and Yu Y: LncRNA SNHG16 induces the SREBP2 to promote lipogenesis
and enhance the progression of pancreatic cancer. Future Oncol.
15:3831–3844. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang H, Zhang Y, Guan X, Li X, Zhao Z, Gao
Y, Zhang X and Chen R: An integrated transcriptomics and proteomics
analysis implicates lncRNA MALAT1 in the regulation of lipid
metabolism. Mol Cell Proteomics. 20:1001412021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Corn KC, Windham MA and Rafat M: Lipids in
the tumor microenvironment: From cancer progression to treatment.
Prog Lipid Res. 80:1010552020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Che G, Wang W, Wang J, He C, Yin J, Chen
Z, He C, Wang X, Yang Y and Liu J: Sulfotransferase SULT2B1
facilitates colon cancer metastasis by promoting SCD1-mediated
lipid metabolism. Clin Transl Med. 14:e15872024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li H, Chen Z, Zhang Y, Yuan P, Liu J, Ding
L and Ye Q: MiR-4310 regulates hepatocellular carcinoma growth and
metastasis through lipid synthesis. Cancer Lett. 519:161–171. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu Y, Zhou Y, Gao H, Wang Y, Cheng Q, Jian
S, Ding Q, Gu W, Yao Y, Ma J, et al: LYAR promotes colorectal
cancer progression by upregulating FSCN1 expression and fatty acid
metabolism. Oxid Med Cell Longev. 2021:99797072021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Li X, Luo J, Mou K, Peng L, Zhou H, Lei Y,
Wang H, Zhao Z, Wang J, Wu J, et al: SDPR inhibits TGF-β induced
cancer metastasis through fatty acid oxidation regulation in
gastric cancer. Int J Biol Sci. 19:2999–3014. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhao P, Yuan F, Xu L, Jin Z, Liu Y, Su J,
Yuan L, Peng L, Wang C and Zhang G: HKDC1 reprograms lipid
metabolism to enhance gastric cancer metastasis and cisplatin
resistance via forming a ribonucleoprotein complex. Cancer Lett.
569:2163052023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Choi YK and Park KG: Targeting glutamine
metabolism for cancer treatment. Biomol Ther (Seoul). 26:19–28.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
De Vitto H, Perez-Valencia J and
Radosevich JA: Glutamine at focus: Versatile roles in cancer.
Tumour Biol. 37:1541–1558. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wu B, Chen Y, Chen Y, Xie X, Liang H, Peng
F and Che W: Circ_0001273 downregulation inhibits the growth,
migration and glutamine metabolism of esophageal cancer cells via
targeting the miR-622/SLC1A5 signaling axis. Thorac Cancer.
13:1795–1805. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Qian CJ, Tong YY, Wang YC, Teng XS and Yao
J: Circ_0001093 promotes glutamine metabolism and cancer
progression of esophageal squamous cell carcinoma by targeting
miR-579-3p/glutaminase axis. J Bioenerg Biomembr. 54:119–134. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Coloff JL, Murphy JP, Braun CR, Harris IS,
Shelton LM, Kami K, Gygi SP, Selfors LM and Brugge JS: Differential
glutamate metabolism in proliferating and quiescent mammary
epithelial cells. Cell metabolism. 23:867–880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhou X, Liu K, Cui J, Xiong J, Wu H, Peng
T and Guo Y: Circ-MBOAT2 knockdown represses tumor progression and
glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer.
J Exp Clin Cancer Res. 40:1242021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li W, Chen C, Zhao X, Ye H, Zhao Y, Fu Z,
Pan W, Zheng S, Wei L, Nong T, et al: HIF-2α regulates
non-canonical glutamine metabolism via activation of PI3K/mTORC2
pathway in human pancreatic ductal adenocarcinoma. J Cell Mol Med.
21:2896–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li Y, Li B, Xu Y, Qian L, Xu T, Meng G, Li
H, Wang Y, Zhang L, Jiang X, et al: GOT2 silencing promotes
reprogramming of glutamine metabolism and sensitizes hepatocellular
carcinoma to glutaminase inhibitors. Cancer Res. 82:3223–3235.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ghosh P, Vidal C, Dey S and Zhang L:
Mitochondria Targeting as an Effective Strategy for Cancer Therapy.
Int J Mol Sci. 21:33632020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Fu H, Zhang J, Chen H, Hou H, Chen H, Xie
R, Chen Y, Zhang J, Liu D, Yan L, et al: NPR1 promotes lipid
droplet lipolysis to enhance mitochondrial oxidative
phosphorylation and fuel gastric cancer metastasis. Adv Sci
(Weinh). 37:e032332025. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yang T, Liang N, Zhang J, Bai Y, Li Y,
Zhao Z, Chen L, Yang M, Huang Q, Hu P, et al: OCTN2 enhances
PGC-1α-mediated fatty acid oxidation and OXPHOS to support stemness
in hepatocellular carcinoma. Metabolism. 147:1556282023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Mookerjee SA, Gerencser AA, Nicholls DG
and Brand MD: Quantifying intracellular rates of glycolytic and
oxidative ATP production and consumption using extracellular flux
measurements. J Biol Chem. 293:12649–12652. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lissanu Deribe Y, Sun Y, Terranova C, Khan
F, Martinez-Ledesma J, Gay J, Gao G, Mullinax RA, Khor T, Feng N,
et al: Mutations in the SWI/SNF complex induce a targetable
dependence on oxidative phosphorylation in lung cancer. Nat Med.
24:1047–1057. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wang T, Sun F, Li C, Nan P, Song Y, Wan X,
Mo H, Wang J, Zhou Y, Guo Y, et al: MTA1, a novel ATP synthase
complex modulator, enhances colon cancer liver metastasis by
driving mitochondrial metabolism reprogramming. Adv Sci (Weinh).
10:e23007562023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liu Y, Tian W, Ge C, Zhang C, Huang Z,
Zhang C, Yang Y and Tian H: SNX17 mediates STAT3 activation to
promote hepatocellular carcinoma progression via a retromer
dependent mechanism. Int J Biol Sci. 21:2762–2779. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang YN, Zeng ZL, Lu J, Wang Y, Liu ZX, He
MM, Zhao Q, Wang ZX, Li T, Lu YX, et al: CPT1A-mediated fatty acid
oxidation promotes colorectal cancer cell metastasis by inhibiting
anoikis. Oncogene. 37:6025–6040. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Xu R, Liu Y, Ma L, Sun Y, Liu H, Yang Y,
Jin T and Yang D: NQO1/CPT1A promotes the progression of pancreatic
adenocarcinoma via fatty acid oxidation. Acta Biochim Biophys Sin
(Shanghai). 55:758–768. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Sun M, Yue Y, Wang X, Feng H, Qin Y, Chen
M, Wang Y and Yan S: ALKBH5-mediated upregulation of CPT1A promotes
macrophage fatty acid metabolism and M2 macrophage polarization,
facilitating malignant progression of colorectal cancer. Exp Cell
Res. 437:1139942024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Hayes JD, Dinkova-Kostova AT and Tew KD:
Oxidative stress in cancer. Cancer Cell. 38:167–197. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zeng Q, Lv C, Qi L, Wang Y, Hao S, Li G,
Sun H, Du L, Li J, Wang C, et al: Sodium selenite inhibits cervical
cancer progression via ROS-mediated suppression of glucose
metabolic reprogramming. Life Sci. 357:1231092024. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Castelli S, De Falco P, Ciccarone F,
Desideri E and Ciriolo MR: Lipid Catabolism and ROS in Cancer: A
Bidirectional Liaison. Cancers (Basel). 13:54842021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tiwari R, Mondal Y, Bharadwaj K, Mahajan
M, Mondal S and Sarkar A: Reactive Oxygen Species (ROS) and their
profound influence on regulating diverse aspects of cancer: A
concise review. Drug Dev Res. 86:e701072025. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li D, Wang Y, Dong C, Chen T, Dong A, Ren
J, Li W, Shu G, Yang J, Shen W, et al: CST1 inhibits ferroptosis
and promotes gastric cancer metastasis by regulating GPX4 protein
stability via OTUB1. Oncogene. 42:83–98. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Di Gregorio J, Petricca S, Iorio R,
Toniato E and Flati V: Mitochondrial and metabolic alterations in
cancer cells. Eur J Cell Biol. 101:1512252022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Vasan K, Werner M and Chandel NS:
Mitochondrial metabolism as a target for cancer therapy. Cell
Metab. 32:341–352. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Pajak B, Siwiak E, Sołtyka M, Priebe A,
Zieliński R, Fokt I, Ziemniak M, Jaśkiewicz A, Borowski R,
Domoradzki T and Priebe W: 2-Deoxy-d-Glucose and its analogs: From
diagnostic to therapeutic agents. Int J Mol Sci. 21:2342019.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Luo W, Guo S, Zhou Y, Zhu J, Zhao J, Wang
M, Sang L, Wang B and Chang B: Hepatocellular carcinoma: Novel
understandings and therapeutic strategies based on bile acids
(Review). Int J Oncol. 61:1172022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Gagnière J, Raisch J, Veziant J, Barnich
N, Bonnet R, Buc E, Bringer MA, Pezet D and Bonnet M: Gut
microbiota imbalance and colorectal cancer. World J Gastroenterol.
22:501–518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Behnam B and Taghizadeh-Hesary F:
Mitochondrial metabolism: A new dimension of personalized oncology.
Cancers (Basel). 15:40582023. View Article : Google Scholar : PubMed/NCBI
|