Introduction
Skin cutaneous melanoma (SKCM) is a highly
aggressive malignancy originating from melanocytes, with increasing
global incidence and poor prognosis despite recent advances in
immunotherapy and targeted therapy (1). In 2020, >320,000 new cases of SKCM
were diagnosed worldwide, with nearly 57,000 related deaths
(2). SKCM is primarily associated
with exposure to ultraviolet radiation, whether from natural
sunlight or indoor tanning practices (3). The management of SKCM involves various
therapeutic approaches, such as surgery, radiotherapy, targeted
therapy and immunotherapy, with the choice of treatment determined
according to the disease stage (4).
Although immune checkpoint inhibitors (ICIs) have revolutionized
treatment paradigms in advanced SKCM, notable inter-patient
heterogeneity leads to variable therapeutic responses, underscoring
the need for novel biomarkers to guide personalized therapy.
Programmed cell death (PCD) is a fundamental
biological process involved in tumor progression, immune evasion
and therapeutic resistance (5).
Various PCD modes, including apoptosis, pyroptosis, ferroptosis,
necroptosis, autophagy and the newly defined cuproptosis, have been
shown to influence cancer immunity and treatment outcomes (6). Increasing evidence indicates that
PCD-related genes (PRGs) serve multifaceted roles in modulating the
tumor microenvironment (TME) and shaping antitumor immunity
(7). For example, high expression
of ferroptosis regulators has been associated with immune cell
infiltration and response to ICIs in several cancer types (8,9). In
lung adenocarcinoma, a machine learning-derived PCD signature was
shown to associate with prognosis, immune infiltration levels and
immunotherapy sensitivity (10,11). A
PCD signature could also serve as a prognostic biomarker for
ovarian cancer (12), bladder
cancer (13) and hepatocellular
carcinoma (14). However, the
clinical relevance and prognostic value of PCD-related gene
expression in SKCM, particularly in the context of immunotherapy,
have not been fully elucidated.
With the advancement of high-throughput sequencing
and computational methods, machine learning has emerged as a
powerful tool for integrating multi-dimensional genomic data to
uncover clinically relevant biomarkers. Compared with traditional
statistical approaches, machine learning algorithms such as Lasso,
CoxBoost, survivalSVM and random survival forest offer greater
flexibility and accuracy in modeling complex gene-phenotype
relationships and building predictive models with high
generalizability (15,16).
In the present study, a novel PCD-related index
(PCDI) was developed for SKCM using integrative machine learning
strategies. The predictive efficacy of the PCDI was systematically
evaluated in terms of prognosis and response to immunotherapy,
providing a potential tool for precision oncology in patients with
SKCM.
Materials and methods
Data acquisition and
pre-processing
Transcriptomic and clinical data of patients with
SKCM were downloaded from The Cancer Genome Atlas (TCGA; TCGA-SKCM
dataset; n=425) database on November 10, 2023. Three independent
validation cohorts were retrieved from the Gene Expression Omnibus
(GEO, http://www.ncbi.nlm.nih.gov/geo/) database, namely,
GSE54467 (n=79) (17), GSE59455
(n=122) (18) and GSE65904 (n=210)
(19). Three immunotherapy datasets
[GSE91061 (20), GSE78220 (21) and IMvigor210 (22)] were used to assess the predictive
value of PCDI in treatment response.
A total of 15 types of PCD patterns (for example,
apoptosis, necroptosis, ferroptosis, pyroptosis and autophagy) were
included and PCD-related genes were curated from Molecular
Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp),
Kyoto Encyclopedia of Genes and Genomes (https://www.kegg.jp/), GeneCards (https://www.genecards.org/) and a recent study
(13) (Table SI). Differential expression
analysis (|log fold change| ≥1.5 and adjusted P<0.05) between
tumor and normal samples was conducted using the ‘limma’ R package
(version 3.64.3) (23) and
prognostic genes were screened using univariate Cox regression
analysis (P<0.05).
Construction of the PCDI
The present study employed an integrative machine
learning pipeline comprising 10 algorithms (Lasso, Ridge, CoxBoost,
random survival forest, survival-support vector machines, efficient
neural network, partial least squares regression for Cox, super
partial correlation, stepwise Cox and gradient boosting machine) to
develop and evaluate 77 model combinations. The present study
developed the PCDI in four steps following the methods of a
previous study (24): i) Univariate
Cox regression was used to investigate prognostic biomarkers
(TCGA); ii) a total of 77 algorithm combinations were then fitted
to the prediction model (TCGA); iii) all algorithm combinations
were carried out in GEO cohorts; and iv) the concordance (C)-index
was computed for each cohort. Detailed parameters of the 10 machine
learning algorithms are described in Data S1. The model with the highest
average C-index across TCGA and GEO cohorts was selected as the
optimal PCDI. Patients were stratified into high- and low-PCDI
(risk score) groups (cut-off value: 0.135) using the
‘surv_cutpoint’ function of the ‘survminer’ R package. Kaplan-Meier
survival analysis with log-rank test and time-dependent receiver
operating characteristic (ROC) curves were used to assess the
prognostic capability of PCDI. The clinical outcome of patients
with SKCM was analyzed through both univariate and multivariate Cox
regression models to identify potential risk factors associated
with the disease. Based on PCDI score and additional clinical
features (age, sex, TNM stage, T stage, N stage and M stage), the
‘nomogramEx’ program (https://cran.r-project.org/web/packages/nomogramEx/index.html)
was used to construct a predicting nomogram.
Evaluation of immunotherapeutic
value
Immune infiltration was evaluated using the
‘immunedeconv’ R package (version 1.0) (25), integrating CIBERSORT, TIMER and
other tools. Tumor microenvironment components (immune, stromal and
ESTIMATE scores) were assessed via the ‘estimate’ package (version
4.4.0) (26). To predict
immunotherapy response, the present study analyzed the tumor
mutation burden (TMB) (27), tumor
immune dysfunction and exclusion (TIDE) score (28), and immunophenoscore (29), as well as immune escape score
calculated with an immune escape gene set (CD47, ADAM10, HLA-G,
CD274, FASLG, CCL5, TGFB1, IL10, PTGER4) from a previous study
(30). TMB is an emerging biomarker
of sensitivity to ICIs and has been shown to be associated with
response to programmed cell death protein 1 (PD-1) and programmed
death-ligand 1 (PD-L1) blockade immunotherapy (26). TIDE is a computational tool used to
evaluate tumor immune escape mechanisms and predict the response to
ICI treatment. The immunophenoscore is a notable predictor of
response to anti-cytotoxic T-lymphocyte associated protein 4
(CTLA-4) and anti-PD-1 therapy. The immune escape score is an
indicator used to evaluate the ability of tumor cells to evade
immune system surveillance and killing. The predictive value of
PCDI was further validated in three independent immunotherapy
cohorts (GSE91061, GSE78220 and IMvigor210). Gene set enrichment
analysis (GSEA) and single sample GSEA (ssGSEA) were conducted to
explore pathway differences between high and low PCDI groups.
Subsequently, the ‘oncoPredict’ R package (version 1.2) (31) was employed to estimate the
IC50 values of therapeutic agents for SKCM cases,
utilizing data from the Genomics of Drug Sensitivity in Cancer
database (https://www.cancerrxgene.org/).
Human Protein Atlas (HPA)
database
The HPA database (http://www.proteinatlas.org) utilizes transcriptomic
and proteomic technologies to explore protein expression at the RNA
and protein levels in different human tissue types and organs. The
HPA database is a convenient way to explore the expression levels
of stratifin (SFN), which had the highest positive coefficient in
the formula for calculating PCDI score, in normal and tumor tissue.
Representative immunohistochemical images of SFN staining in normal
and SKCM tissues were obtained from the ‘Tissue’ and ‘Cancer’
sections of the HPA.
Cell culture
Melanoma cell lines (A375, A2058, MV3, Sk-mel-1 and
Sk-mel-28) and a normal skin-derived cell line (PIG1) were obtained
from The Cell Bank of Type Culture Collection of The Chinese
Academy of Sciences. These cells were cultured in RPMI-1640 medium
enriched with 10% FBS (Procell Life Science & Technology Co.,
Ltd.) and 1% antibiotic solution (Procell Life Science &
Technology Co., Ltd.). Cells were maintained at 37°C in an
atmosphere containing 5% CO2.
Cell transfection
To achieve SFN gene silencing, the melanoma A375 and
A2058 cell lines were separately transfected with si-SFN (cat. no.
siG000002810A-1-5; sequence 5′-GCAATTAGTATTGTTTGAAACAT-3′) or
si-negative control (cat. no. siN0000001-4-10; sequence
5′-GCAGGATGGGATAAGTCTAAA-3′) sequences provided by Guangzhou
RiboBio Co., Ltd. A total of 0.5 nmol siRNA was used for
transfection each time using Lipofectamine® 3000 (Thermo
Fisher Scientific, Inc.). A375 and A2058 cells were transfected
with siRNA at 37°C for 48 h before further experiments were
performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the aforementioned
cells (A375, A2058, MV3, Sk-mel-1, Sk-mel-28 and PIG-1) using
Triquick® Reagent (Beijing Solarbio Science &
Technology Co., Ltd.). According to the manufacturer's protocol,
complementary DNA (cDNA) for SFN was synthesized from the isolated
RNA employing the SweScript RT I FTTSt Strand cDNA Synthesis Kit
(cat. no. G3330-100; Wuhan Servicebio Technology Co., Ltd.). The
resulting cDNA was then used as a template for qPCR analysis
performed on a PCR system using 2X SYBR Green qPCR Master Mix (Low
ROX) (cat. no. G3321-05; Selleck China). The thermocycling
conditions were as follows: Initial denaturation at 95°C for 5 min;
followed by 35 cycles at 95°C for 30 sec, 55°C for 30 sec and 70°C
for 30 sec. Gene expression levels were quantified using the
2−ΔΔCq method and normalized to GAPDH expression
(32). Primer sequences used were
as follows: GAPDH forward (F), 5′-GTCTCCTCTGACTTCAACAGCG-3′ and
GAPDH reverse (R), 5′-ACCACCCTGTTGCTGTAGCCAA-3′; and SFN F,
5′-TCCACTACGAGATCGCCAACAG-3′ and SFN R,
5′-GTGTCAGGTTGTCTCGCAGCA-3′.
Cell Counting Kit-8 (CCK-8) assay
To evaluate cell viability, transduced A375 and
A2058 cells were seeded into 96-well plates and allowed to grow for
24, 48 or 72 h. After incubation, CCK-8 reagent (cat. no. G1613-1ML
Wuhan Servicebio Technology Co., Ltd.) was added to each well,
followed by a 4-h incubation at 37°C. Absorbance values were
measured at 450 nm using a microplate reader.
Statistical analysis
All statistical analyses were conducted using R
software (version 4.2.1; R Development Core Team). Differences
between continuous variables were assessed using either the
Wilcoxon rank-sum test or unpaired Student's t-test, depending on
data distribution. Pearson's correlation coefficients were
calculated to evaluate associations between two continuous
parameters. Kaplan-Meier survival curves were compared using the
two-sided log-rank test Univariate and multivariate Cox regression
analyses were performed to identify risk factors for the overall
survival of patients with SKCM. P<0.05 was considered to
indicate a statistically significant difference. All assays were
repeated three times.
Results
Identification of prognostic PRGs in
SKCM
To explore the prognostic value of PRGs in SKCM, the
present study first identified differentially expressed genes
(DEGs) between tumor and normal tissues from the TCGA-SKCM cohort.
A total of 1,638 genes were found to be significantly dysregulated
(|log2FC|>1.5; P<0.05) (Fig. S1A). By intersecting these DEGs with
a curated set of 1,254 PRGs, the present study obtained 117
differentially expressed PRGs (Fig.
S1B). Among them, univariate Cox regression identified 54 PRGs
that were significantly associated with overall survival (OS) in
patients with SKCM (Fig. S1C),
suggesting their potential utility for constructing a prognostic
model.
Integrative machine learning-based
PCDI
Using the 54 prognosis-associated PRGs, the present
study applied a machine learning integration strategy involving 77
algorithm combinations across TCGA and GEO datasets. Based on the
average C-index across training and validation cohorts, the Lasso
algorithm was identified as the optimal method, achieving the
highest predictive performance (Fig.
1A). The optimal λ parameter was selected when the partial
likelihood deviance reached its minimum in cross-validation and 13
PRGs were retained to construct the PCDI (Fig. 1B). The risk score for each patient
was calculated as: Risk score=(0.029 × SFNexp) + (−0.176
× IDSexp) + (−0.054 × SCFD1exp) + (−0.591 ×
TRIM34exp) + (−0.011 × CD74exp) + (−0.023 ×
FBXW7exp) + (−0.045 × CRIP1exp) + (−0.133 ×
IFNAR2exp) + (−0.051 × PIM2exp) + (−0.053 ×
CTSWexp) + (−0.008 × NUPR1exp) + (−0.029 ×
ICAM1exp) + (−0.024 × AIM2exp).
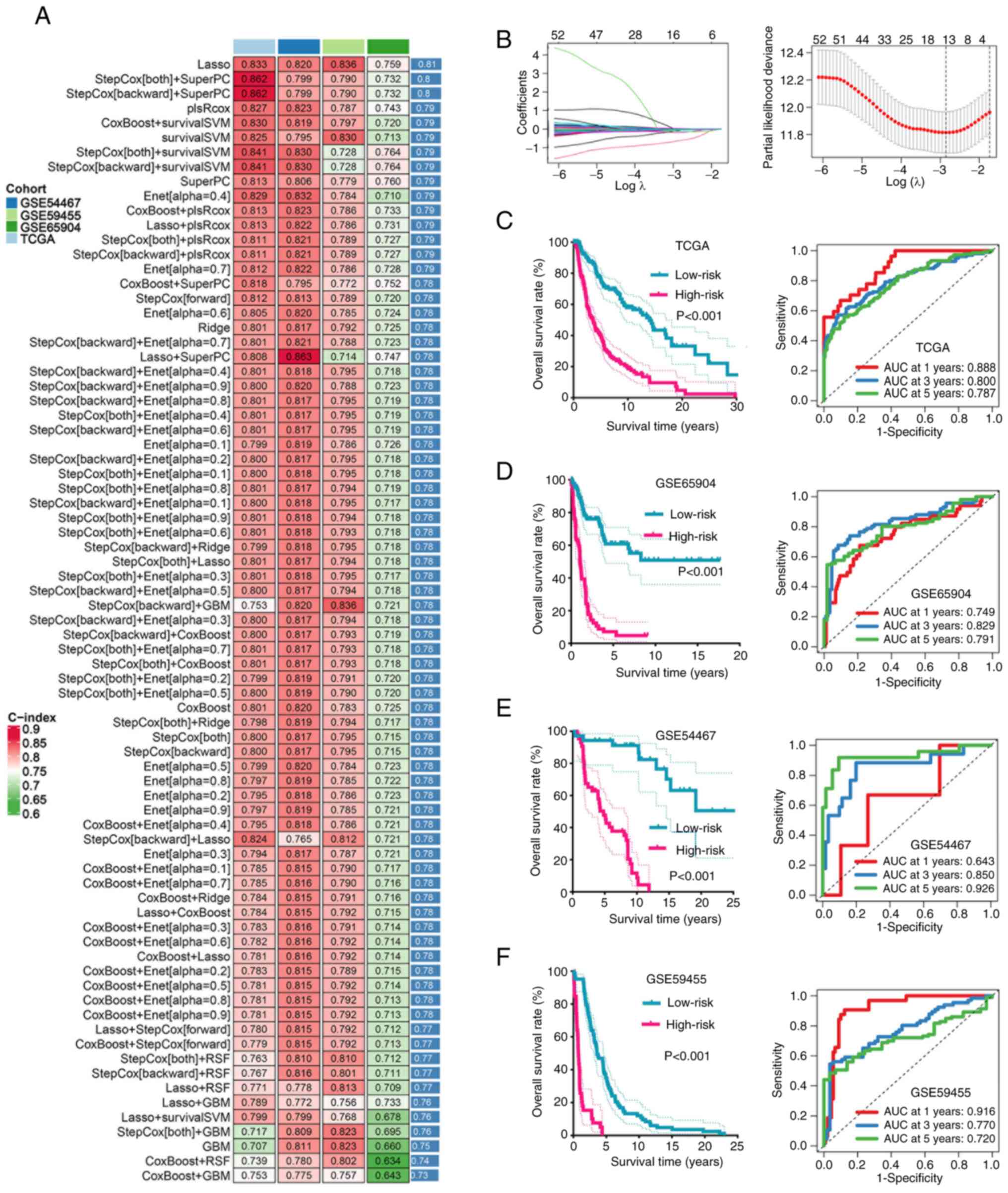 | Figure 1.Machine learning developed programmed
cell death-related signature. (A) The C-index of 77 types of
prognostic models of TCGA and GEO cohorts. (B) Determination of the
optimal λ was obtained when the partial likelihood deviance reached
the minimum value in the TCGA cohort and further generated Lasso
coefficients of the most useful prognostic genes. The survival
curve and their corresponding ROC curve in SKCM with different risk
score in (C) TCGA, (D) GSE65904, (E) GSE54467 and (F) GSE59455
dataset. C-index, concordance index; TCGA, The Cancer Genome Atlas;
GEO, Gene Expression Omnibus; GSE, gene expression data series;
SKCM, skin cutaneous melanoma; RSF, random survival forest;
survivalSVM, survival-support vector machines; Enet, efficient
neural network; plsRCox, partial least squares regression for Cox;
SuperPC, super partial correlation; GBM, gradient boosting machine;
AUC, area under the curve. |
Patients in the TCGA, GSE65904, GSE54467 and
GSE59455 cohorts were stratified into high- and low-risk groups
based on the optimal cut-off (0.1365). Kaplan-Meier survival
analyses demonstrated that patients in the high-risk group
consistently exhibited worse OS compared with that of the low-risk
group across all datasets (Fig.
1C-F). Time-dependent ROC analyses further confirmed the
predictive robustness of the model, with 1-, 3- and 5-year AUCs
>0.70 in the majority of cohorts. The AUC of the 1-, 3- and
5-year ROC curve was 0.888, 0.800 and 0.787 in the TCGA cohort
(Fig. 1C), 0.749, 0.829 and 0.791
in the GSE65904 cohort (Fig. 1D),
0.643, 0.850 and 0.926 in the GSE54467 cohort (Fig. 1E), and 0.916, 0.770 and 0.720 in the
GSE59455 cohort (Fig. 1F),
respectively.
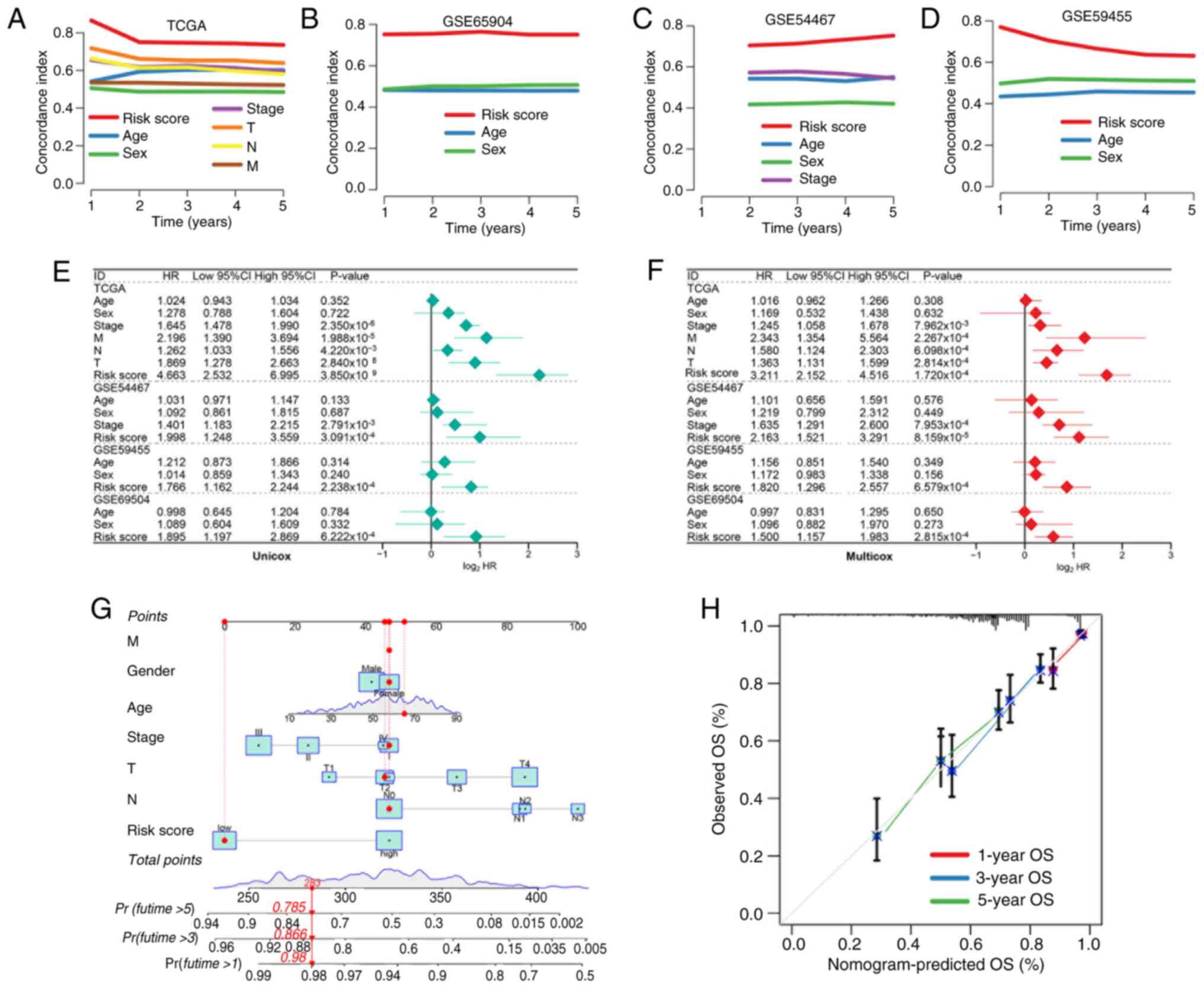
Evaluation of the performance of
PCDI
To further assess the clinical utility of the risk
signature, the present study compared its predictive accuracy
against standard clinical parameters. C-index analysis revealed
that the risk score consistently outperformed age, sex and TNM
stage in all cohorts (33)
(Fig. 2A-D). Univariate and
multivariate Cox regression analyses confirmed that the risk score
was an independent predictor of OS in SKCM (Fig. 2E and F). A prognostic nomogram
integrating the risk score and clinical features (for example, pT
and pN stage) was constructed (Fig.
2G) and calibration plots demonstrated notable agreement
between predicted and observed survival probabilities (Fig. 2H).
Immune landscape in high- and low-risk
groups
Due to the known association between PCD and immune
modulation, the present study investigated the immune infiltration
profiles of patients with SKCM with different risk scores. Multiple
deconvolution algorithms revealed significant negative correlations
between the risk score and immune cell abundance (Fig. 3A). Particularly, CD8+ T
cells, M1 macrophages and dendritic cells were significantly
negatively correlated with risk score in SKCM (Fig. 3B-D). ssGSEA analysis further
indicated lower immune cells and immune function scores, including
aDCs, B cells, CD8+ T cells, mast cells, NK cells, TILs,
T helper (Th) cells, Th2 cells, APC_co_stimulation, cytolytic
activity, inflammation promoting and T cell co-stimulation, human
leukocyte antigen (HLA), parainflammation and type II IFN response
in the high-risk group (Fig. 3E and
F). Patients with low-risk score also exhibited significantly
higher stromal, immune and ESTIMATE scores, which suggested a more
active tumor immune microenvironment (Fig. 3G).
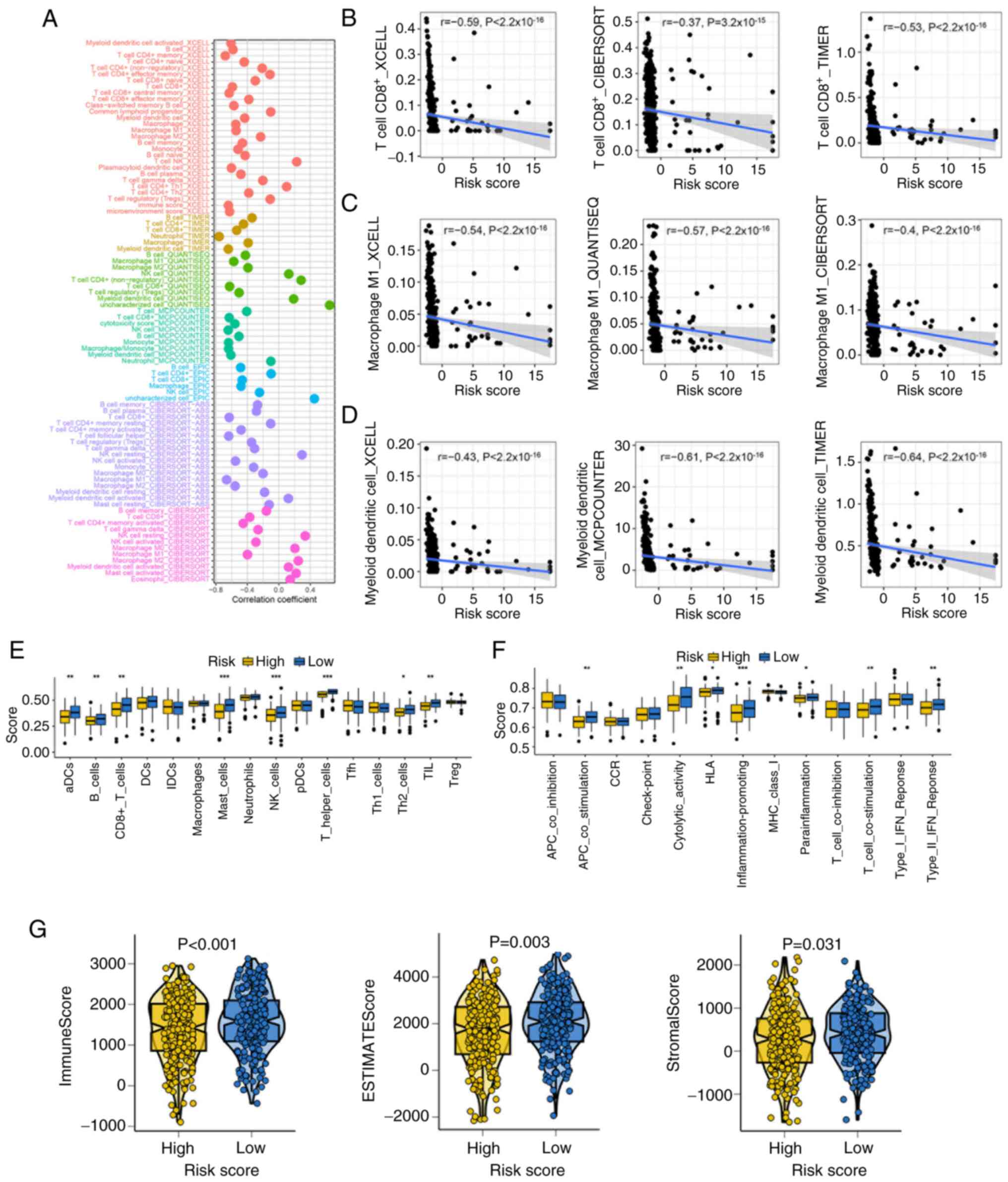 | Figure 3.Landscape of the immune infiltration
among various risk score groups. (A) Correlation between risk score
and immune cell abundance, determined using seven algorithms. The
correlation between the risk score and the number of (B)
CD8+ T cells, (C) M1 macrophages and (D) dendritic
cells. ssGSEA study assessing (E) immune cell and (F)
immune-related function levels across several risk score
categories. (G) The stromal, immune and ESTIMATE scores across
various risk score categories. *P<0.05, **P<0.01 and
***P<0.001. ssGSEA, single sample gene set enrichment analysis;
aDCs, antibody-drug conjugates; DCs, dendritic cells; NK, natural
killer; Tfh, T follicular helper cells; Th, T helper cells; TIL,
tumor infiltrating lymphocytes; Treg, regulatory T cells; APC,
antigen presenting cells; CCR, chemokine receptor; MHC, major
histocompatibility complex; iDCs, immature dendritic cells; pDCs,
plasmacytoid dendritic cells. |
Association between risk score and
immunotherapy response
To evaluate the predictive value of the risk
signature in immunotherapy response, the present study examined
multiple immune-related features. Patients with SKCM with low-risk
score had significantly higher expression of HLA-related genes and
immune checkpoint molecules compared with patients with SKCM and a
high-risk score (Fig. 4A and B).
Elevated expression levels of HLA-related genes predicted a higher
chance of immunotherapy benefits (34). Furthermore, they demonstrated
increased TMB score and PD-1/CTLA4 immunophenoscores, suggesting
enhanced tumor immunogenicity (Fig. 4C
and D). A marked response to immunotherapy is indicated by low
TIDE and immune escape scores (28,35).
In the present study, low-risk score patients with SKCM had lower
TIDE, immune escape and immune surveillance scores, indicating a
more favorable immunotherapy profile (Fig. 4E-G). These findings were further
validated in three independent melanoma immunotherapy cohorts. In
the IMvigor210, GSE78220 and GSE91061 datasets, the risk score of
non-responders in SKCM was notably greater compared with that of
responders, and low-risk patients exhibited higher response rates
and improved survival outcomes compared with high-risk patients
(Fig. 4H-P), confirming the ability
of the signature to stratify immunotherapy benefit.
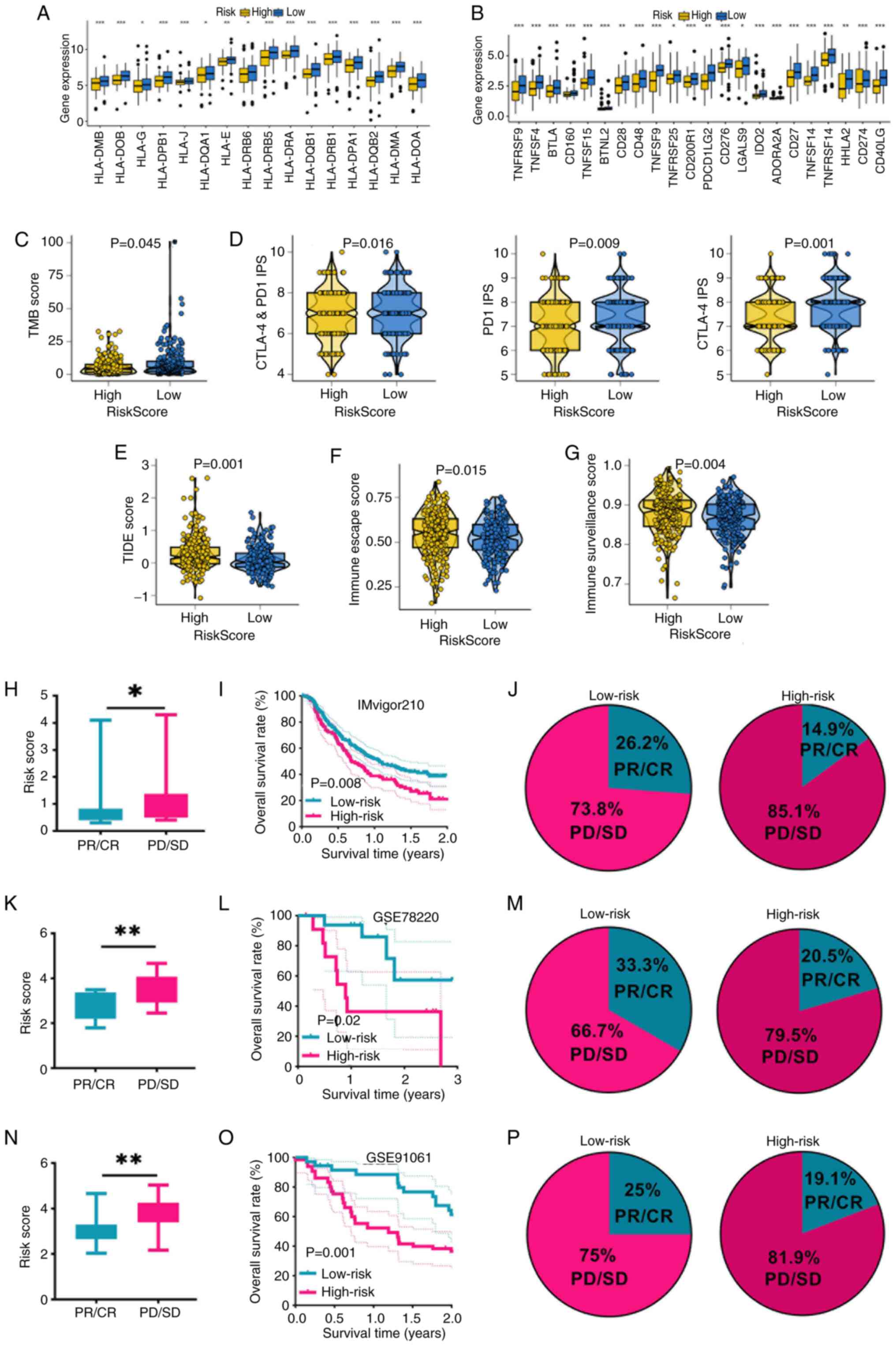 | Figure 4.Programmed cell death-related
signature acted as a biomarker for predicting the immunotherapy
benefits in SKCM. (A) Level of HLA-related genes, (B) immune
checkpoints, (C) TMB score, (D) PD1 and CTLA-4 immunophenoscore,
(E) TIDE score, (F) immunological escape score and (G) immune
surveillance score in patients with SKCM in different risk score
categories. (H) Risk score, (I) survival rates and (J)
immunotherapy response rate in patients with SKCM with different
risk score in the IMvigor210 cohort. (K) Risk score, (L) survival
rates and (M) immunotherapy response rate in patients with SKCM
with different risk scores in the GSE78220 cohort. (N) Risk score,
(O) survival rates and (P) immunotherapy response rate in patients
with SKCM with different risk scores in the GSE91061 cohort
*P<0.05, **P<0.01 and ***P<0.001. SKCM, skin cutaneous
melanoma; HLA, human leukocyte antigen; TMB, tumor mutation burden;
PD1, programmed cell death protein-1; CTLA4, cytotoxic T-lymphocyte
associated protein 4; TIDE, tumor immune dysfunction and exclusion;
GSE, gene expression data series; CR, complete response; PR,
partial response; SD, stable disease; PD, progressive disease. |
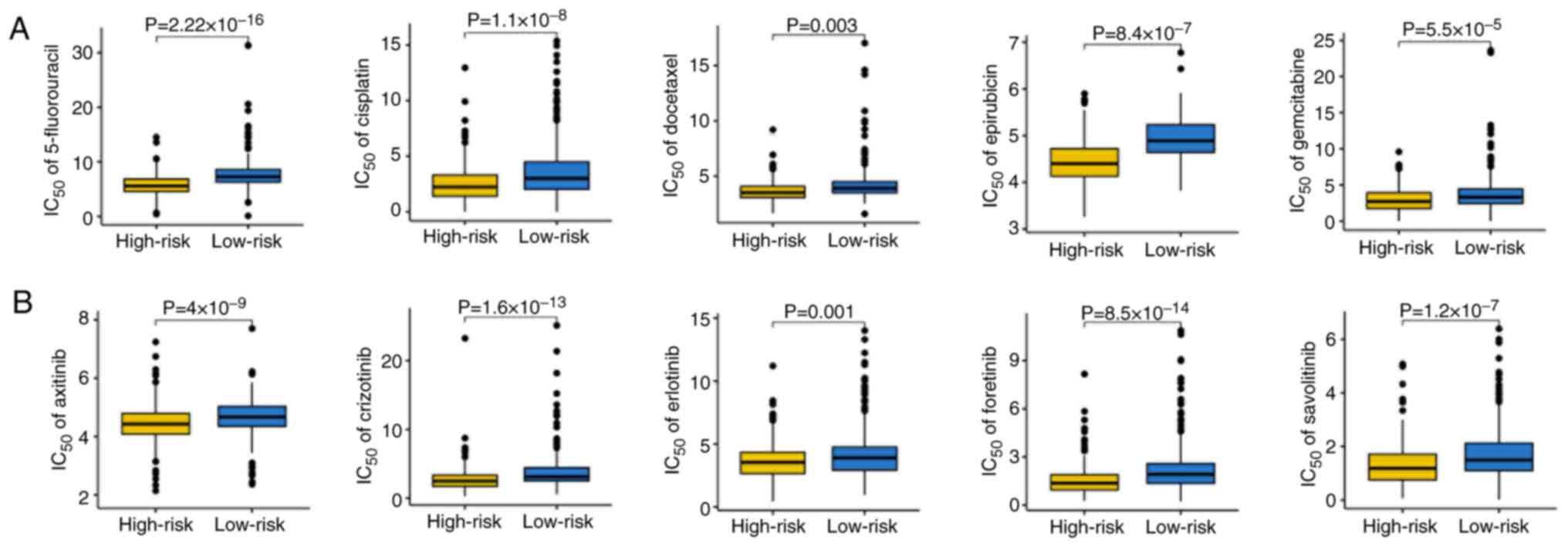
Predictive value for chemotherapy and
targeted therapy sensitivity
The present study next explored the association
between the risk score and drug sensitivity. Patients with low-risk
scores demonstrated significantly higher IC50 values for
commonly used chemotherapy agents, including 5-fluorouracil,
cisplatin, docetaxel, epirubicin and gemcitabine (Fig. 5A), and for several targeted
therapies such as axitinib, crizotinib, erlotinib, foretinib and
savolitinib (Fig. 5B). These
findings suggested that high-risk patients with SKCM may be more
sensitive to cytotoxic and targeted drugs, while low-risk patients
may derive greater benefit from immunotherapy.
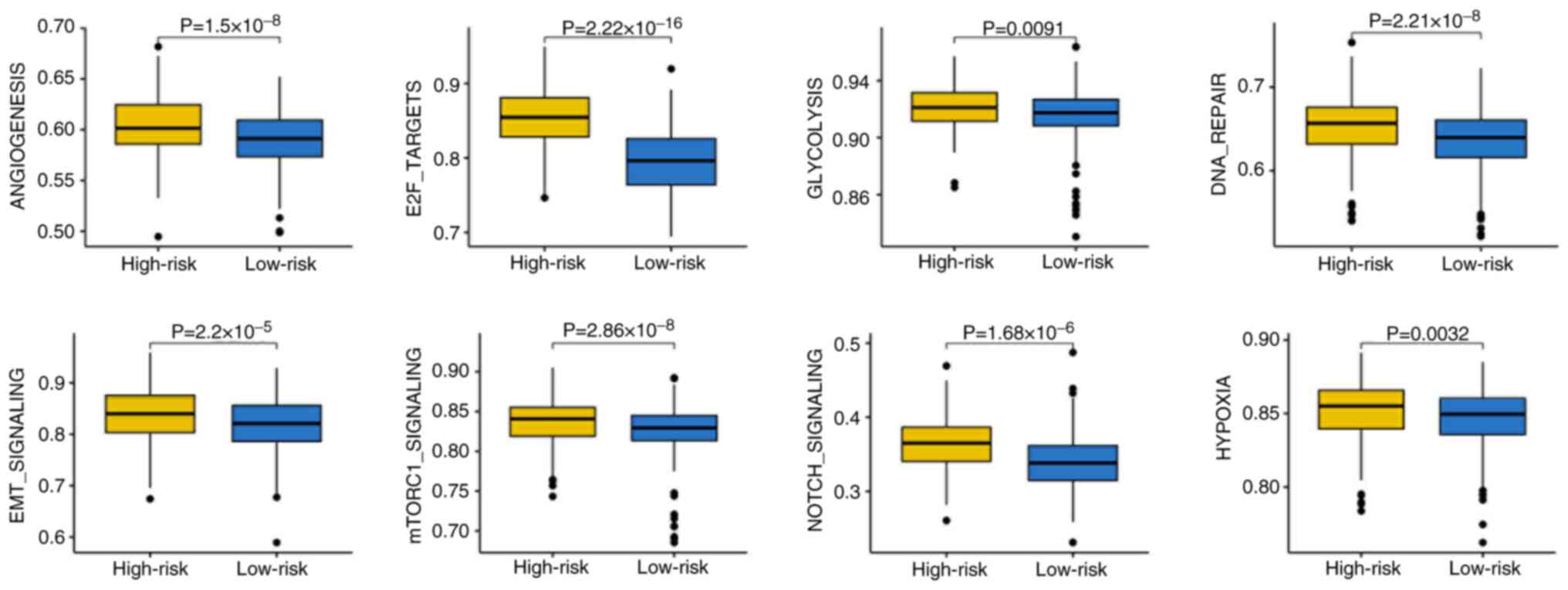
Enrichment of cancer hallmarks in
high-risk patients with SKCM
To elucidate the biological pathways associated with
different risk groups, the present study performed gene set
enrichment analysis. High-risk patients exhibited increased
enrichment scores for cancer-related hallmarks, including
‘angiogenesis’, ‘E2F targets’, ‘glycolysis’, ‘DNA repair’, ‘EMT
signaling’, ‘mTORC1 signaling’, ‘Notch signaling’ and ‘hypoxia’
(Fig. 6). These pathways are
well-known to contribute to tumor progression, immune evasion and
resistance to therapy, potentially explaining the poor prognosis
and lower immunotherapy benefit in high-risk patients.
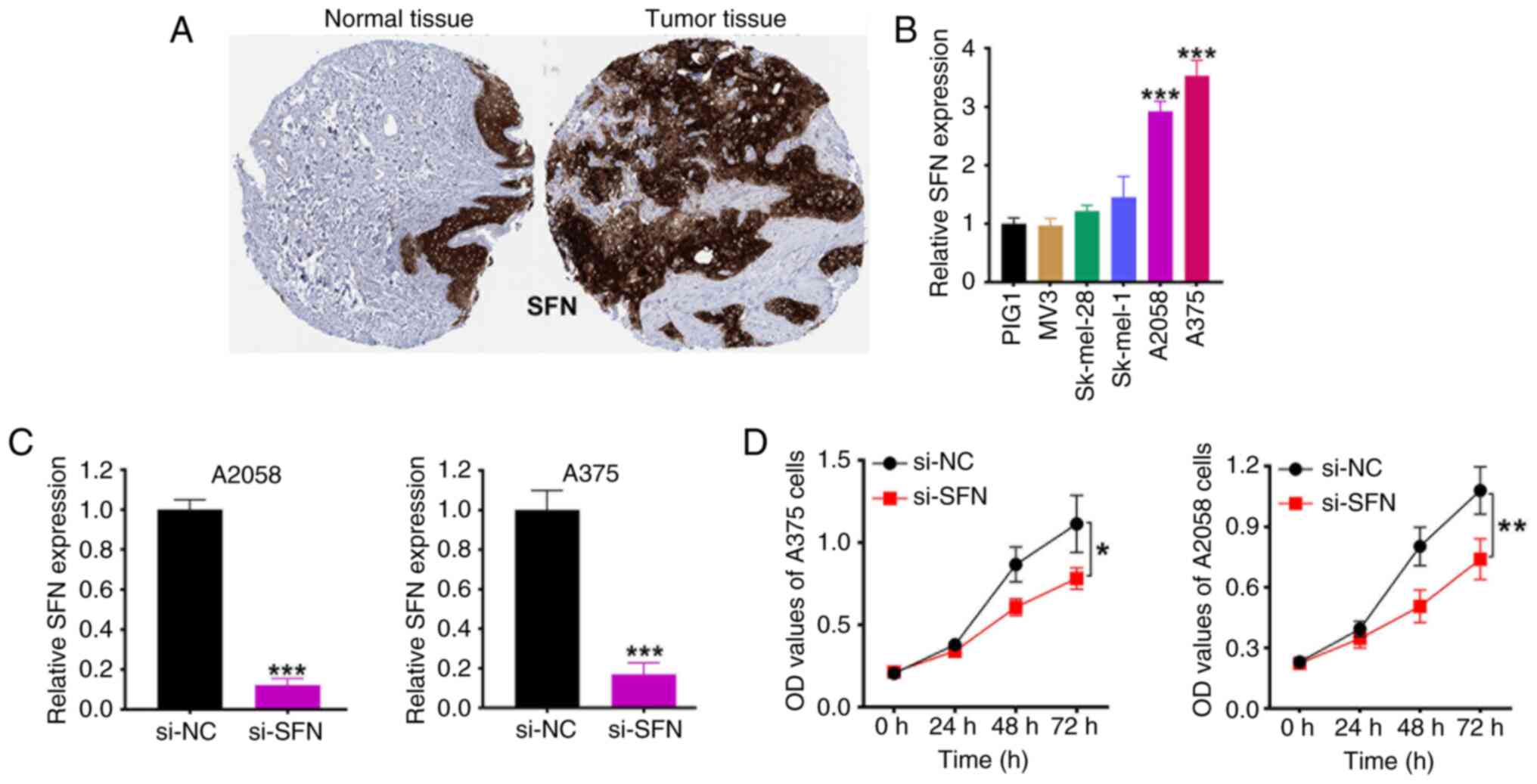
Validation of SFN as a functional
driver gene in SKCM
Among the 13 genes comprising the risk signature,
SFN had the highest positive coefficient and was consistently
upregulated in SKCM tissues. Immunohistochemistry confirmed
elevated SFN protein expression in SKCM compared with that in
normal skin (Fig. 7A). RT-qPCR also
demonstrated higher SFN expression in SKCM cell lines compared with
that in normal melanocytes of A375 and A2058 melanoma cells
(Fig. 7B). The SFN expression of
A375 and A2058 melanoma cells were then knocked down (Fig. 7C). Functionally, knockdown of SFN
significantly inhibited the proliferation of A375 and A2058
melanoma cells, as shown in the CCK-8 assays (Fig. 7D), supporting its role as an
oncogenic driver and validating its inclusion in the risk
model.
Discussion
In the present study, a robust PCDI was developed
and validated using integrated machine learning algorithms to
predict prognosis and immunotherapy response in patients with SKCM.
By integrating multi-cohort data and systematically evaluating 77
algorithm combinations, a 13-gene risk signature that stratifies
patients into distinct prognostic and immunological subtypes with
high accuracy and clinical relevance was identified.
The present study results demonstrated that the PCDI
effectively distinguishes patients with SKCM with poor outcomes
from those with favorable survival, as evidenced by consistent
results across TCGA and multiple independent GEO cohorts. The model
outperformed conventional clinical features such as age, sex and
TNM stage in prognostic prediction, which supports its potential
clinical utility (36,37). The integration of the risk score
into a nomogram further improved individual-level survival
prediction and calibration analyses confirmed its accuracy and
reliability (37).
Notably, the present study findings highlighted a
strong association between the PCDI and the tumor immune
microenvironment. High-risk patients exhibited reduced infiltration
of CD8+ T cells, M1 macrophages and antigen-presenting
cells, alongside lower stromal and immune scores, indicating a more
immunosuppressive tumor contexture (38,39).
Conversely, low-risk patients demonstrated a more immunoactive
landscape with higher expression of HLA genes, immune checkpoints
and immune cell infiltration, which suggests a favorable
environment for immunotherapeutic intervention (40).
The relevance of PCDI in predicting immunotherapy
response was further confirmed by its correlation with multiple
immune-related biomarkers. Low-risk patients exhibited higher TMB,
immunophenoscores for PD-1/CTLA-4 and lower TIDE and immune escape
scores compared with patients with a high-risk score, features
known to associate with enhanced response to immune checkpoint
blockade (21,41,42).
These findings were validated across three independent
immunotherapy-treated melanoma cohorts, where low-risk patients
demonstrated significantly improved response rates and OS. These
results suggested that PCDI could serve as a predictive biomarker
to guide immunotherapy decision-making in SKCM (43,44).
Furthermore, the drug sensitivity analysis in the
present study indicated that high-risk patients were more sensitive
to multiple chemotherapeutic and targeted agents, including
5-fluorouracil, cisplatin and several receptor tyrosine kinase
inhibitors (crizotinib, erlotinib and foretinib). This observation
implies a potential trade-off in treatment strategy: While low-risk
patients may benefit more from immunotherapy, high-risk patients
might demonstrate improved response to conventional or targeted
cytotoxic treatment (45). These
findings underscore the value of PCDI in guiding personalized
therapeutic strategies beyond immunotherapy.
Functional and pathway enrichment analyses revealed
that high-risk patients with SKCM were enriched in oncogenic
pathways such as glycolysis, E2F signaling, DNA repair, EMT and
hypoxia, all of which are known to contribute to tumor progression,
resistance to therapy and immune evasion (14–16).
These molecular characteristics likely explain the poor prognosis
and reduced immune responsiveness observed in the high-risk
group.
Among the 13 genes in the PCDI, SFN emerged as a
potential oncogenic driver in SKCM. The high expression levels of
SFN were validated at both the transcriptomic and protein levels
and functional assays confirmed its role in promoting melanoma cell
proliferation. These findings suggested that SFN may not only serve
as a biomarker but also represent a potential therapeutic target in
high-risk SKCM. A previous study reported that upregulation of SFN
was associated with the prognosis in ovarian cancer (46). Furthermore, SFN regulates cervical
cancer cell proliferation, apoptosis and metastasis progression
through Lin-11, Isl-1 and Mec-3 domain kinase 2/cofilin signaling
(47).
Despite the strengths of the present study,
including a large sample size, cross-cohort validation and
integration of multi-dimensional data, several limitations should
be acknowledged. First, the retrospective nature of the datasets
may introduce biases that could affect generalizability. Second,
although the immunotherapy response predictions were validated in
public cohorts, prospective clinical validation is warranted to
confirm the utility of PCDI in guiding treatment decisions. Lastly,
while SFN was functionally validated, the biological roles of other
model genes warrant further investigation.
Further understanding of the molecular mechanisms
through which PCDI-related genes contribute to melanoma progression
and immune modulation is essential. For instance, SFN, identified
as a key oncogenic driver in the present study model, is involved
in cell proliferation control. However, its downstream effectors in
the context of SKCM remain poorly characterized. Future studies
could explore whether SFN modulates tumor progression and immune
evasion pathways, such as PD-L1 expression or antigen presentation
machinery. While the present study PCDI demonstrated predictive
power across public immunotherapy cohorts, validation in
prospective, cohorts including treatment-naïve patients with SKCM
is imperative. Ideally, this would involve enrolling patients prior
to immunotherapy, obtaining baseline biopsies for transcriptomic
profiling, calculating PCDI scores and associating them with
clinical outcomes such as Response Evaluation Criteria in Solid
Tumors response, progression-free survival and OS.
In conclusion, the present study developed a novel
machine learning-based PCDI that robustly predicts prognosis and
immunotherapy response in SKCM. The index provides valuable
insights into tumor biology and immune phenotypes and holds
potential as a biomarker for personalized therapy in patients with
melanoma. Future studies integrating multi-omics and experimental
validation will further refine and extend the clinical utility of
the present study model.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XW prepared the original draft. XW and YZ performed
the investigation. YZ conducted the formal analysis and curated the
data. SY and HS acquired data, and reviewed and edited the
manuscript. WC and SS analyzed and interpreted the data, and were
involved in project administration. XW and YZ confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Long GV, Swetter SM, Menzies AM,
Gershenwald JE and Scolyer RA: Cutaneous melanoma. Lancet.
402:485–502. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Singh D, Laversanne M, Vignat J,
Vaccarella S, Meheus F, Cust AE, de Vries E, Whiteman DC and Bray
F: Global burden of cutaneous melanoma in 2020 and projections to
2040. JAMA Dermatol. 158:495–503. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopes F, Sleiman MG, Sebastian K, Bogucka
R, Jacobs EA and Adamson AS: UV exposure and the risk of cutaneous
melanoma in skin of color: A systematic review. JAMA Dermatol.
157:213–219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leonardi GC, Falzone L, Salemi R, Zanghì
A, Spandidos DA, Mccubrey JA, Candido S and Libra M: Cutaneous
melanoma: From pathogenesis to therapy (Review). Int J Oncol.
52:1071–1080. 2018.PubMed/NCBI
|
|
5
|
Newton K, Strasser A, Kayagaki N and Dixit
VM: Cell death. Cell. 187:235–256. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Hong M, Li Y, Chen D, Wu Y and Hu
Y: Programmed cell death tunes tumor immunity. Front Immunol.
13:8473452022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang T, Gu L, Kang X, Li J, Song Y, Wang
Y and Ma W: Programmed cell death disrupts inflammatory tumor
microenvironment (TME) and promotes glioblastoma evolution. Cell
Commun Signal. 22:3332024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Shou Y, Zhu R, Qiu Z, Zhang Q and
Xu J: Construction and validation of a ferroptosis-related
prognostic signature for melanoma based on single-cell RNA
sequencing. Front Cell Dev Biol. 10:8184572022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nedaeinia R, Dianat-Moghadam H,
Movahednasab M, Khosroabadi Z, Keshavarz M, Amoozgar Z and Salehi
R: Therapeutic and prognostic values of ferroptosis signature in
glioblastoma. Int Immunopharmacol. 155:1145972025. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Wang R, Hu D, Zhang C, Cao P and
Huang J: Machine learning reveals diverse cell death patterns in
lung adenocarcinoma prognosis and therapy. NPJ Precis Oncol.
8:492024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Cui Y, Zhou G, Zhang Z and Zhang
P: Leveraging mitochondrial-programmed cell death dynamics to
enhance prognostic accuracy and immunotherapy efficacy in lung
adenocarcinoma. J Immunother Cancer. 12:e0100082024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai X, Lin J, Liu L, Zheng J, Liu Q, Ji L
and Sun Y: A novel TCGA-validated programmed cell-death-related
signature of ovarian cancer. BMC Cancer. 24:5152024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y and Zhang Q: Leveraging programmed
cell death signature to predict clinical outcome and immunotherapy
benefits in postoperative bladder cancer. Sci Rep. 14:229762024.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gu X, Pan J, Li Y and Feng L: A programmed
cell death-related gene signature to predict prognosis and
therapeutic responses in liver hepatocellular carcinoma. Discov
Oncol. 15:712024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nayarisseri A, Khandelwal R, Tanwar P,
Madhavi M, Sharma D, Thakur G, Speck-Planche A and Singh SK:
Artificial intelligence, big data and machine learning approaches
in precision medicine & drug discovery. Curr Drug Targets.
22:631–655. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ngiam KY and Khor IW: Big data and machine
learning algorithms for health-care delivery. Lancet Oncol.
20:e262–e273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jayawardana K, Schramm SJ, Haydu L,
Thompson JF, Scolyer RA, Mann GJ, Müller S and Yang JY:
Determination of prognosis in metastatic melanoma through
integration of clinico-pathologic, mutation, mRNA, microRNA, and
protein information. Int J Cancer. 136:863–874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Budden T, Davey RJ, Vilain RE, Ashton KA,
Braye SG, Beveridge NJ and Bowden NA: Repair of UVB-induced DNA
damage is reduced in melanoma due to low XPC and global genome
repair. Oncotarget. 7:60940–60953. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cabrita R, Lauss M, Sanna A, Donia M,
Skaarup Larsen M, Mitra S, Johansson I, Phung B, Harbst K,
Vallon-Christersson J, et al: Tertiary lymphoid structures improve
immunotherapy and survival in melanoma. Nature. 577:561–565. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Riaz N, Havel JJ, Makarov V, Desrichard A,
Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH,
et al: Tumor and microenvironment evolution during immunotherapy
with nivolumab. Cell. 171:934–949.e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hugo W, Zaretsky JM, Sun L, Song C, Moreno
BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G,
et al: Genomic and transcriptomic features of response to Anti-PD-1
therapy in metastatic melanoma. Cell. 165:35–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rosenberg JE, Galsky MD, Powles T,
Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J,
Perez-Gracia JL, van der Heijden MS, et al: Atezolizumab
monotherapy for metastatic urothelial carcinoma: Final analysis
from the phase II IMvigor210 trial. ESMO Open. 9:1039722024.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H,
Wang L, Lu T, Zhang Y, Sun Z and Han X: Machine learning-based
integration develops an immune-derived lncRNA signature for
improving outcomes in colorectal cancer. Nat Commun. 13:8162022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sturm G, Finotello F and List M:
Immunedeconv: An R package for unified access to computational
methods for estimating immune cell fractions from bulk
RNA-sequencing data. Methods Mol Biol. 2120:223–232. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Palmeri M, Mehnert J, Silk AW, Jabbour SK,
Ganesan S, Popli P, Riedlinger G, Stephenson R, de Meritens AB,
Leiser A, et al: Real-world application of tumor mutational
burden-high (TMB-high) and microsatellite instability (MSI)
confirms their utility as immunotherapy biomarkers. ESMO Open.
7:1003362022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang
P and Liu XS: Large-scale public data reuse to model immunotherapy
response and resistance. Genome Med. 12:212020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Charoentong P, Finotello F, Angelova M,
Mayer C, Efremova M, Rieder D, Hackl H and Trajanoski Z: Pan-cancer
immunogenomic analyses reveal genotype-immunophenotype
relationships and predictors of response to checkpoint blockade.
Cell Rep. 18:248–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang
Z, Zhang Z, Xie J, Wang C, Chen D, et al: Single-cell landscape of
the ecosystem in early-relapse hepatocellular carcinoma. Cell.
184:404–421.e16. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Balch CM, Gershenwald JE, Soong SJ,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin A and Yan WH: HLA-G/ILTs targeted
solid cancer immunotherapy: Opportunities and challenges. Front
Immunol. 12:6986772021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin A, Zhang J and Luo P: Crosstalk
between the MSI status and tumor microenvironment in colorectal
cancer. Front Immunol. 11:20392020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
37
|
Gershenwald JE, Scolyer RA, Hess KR,
Sondak VK, Long GV, Ross MI, Lazar AJ, Faries MB, Kirkwood JM,
McArthur GA, et al: Melanoma staging: Evidence-based changes in the
American Joint Committee on Cancer eighth edition cancer staging
manual. CA Cancer J Clin. 67:472–492. 2017.PubMed/NCBI
|
|
38
|
Wang J, Yang F, Sun Q, Zeng Z, Liu M, Yu
W, Zhang P, Yu J, Yang L, Zhang X, et al: The prognostic landscape
of genes and infiltrating immune cells in cytokine induced killer
cell treated-lung squamous cell carcinoma and adenocarcinoma.
Cancer Biol Med. 18:1134–1147. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tien FM, Lu HH, Lin SY and Tsai HC:
Epigenetic remodeling of the immune landscape in cancer:
Therapeutic hurdles and opportunities. J Biomed Sci. 30:32023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun R, Limkin EJ, Vakalopoulou M, Champiat
S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S, Hollebecque
A, et al: A radiomics approach to assess tumour-infiltrating CD8
cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an
imaging biomarker, retrospective multicohort study. Lancet Oncol.
19:1180–1191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu D, Schilling B, Liu D, Sucker A,
Livingstone E, Jerby-Arnon L, Zimmer L, Gutzmer R, Satzger I,
Loquai C, et al: Integrative molecular and clinical modeling of
clinical outcomes to PD1 blockade in patients with metastatic
melanoma. Nat Med. 25:1916–1927. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gide TN, Quek C, Menzies AM, Tasker AT,
Shang P, Holst J, Madore J, Lim SY, Velickovic R, Wongchenko M, et
al: Distinct immune cell populations define response to anti-PD-1
monotherapy and anti-PD-1/Anti-CTLA-4 combined therapy. Cancer
Cell. 35:238–255.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang W, Soares J, Greninger P, Edelman EJ,
Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et
al: Genomics of drug sensitivity in cancer (GDSC): A resource for
therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.
41((Database Issue)): D955–D961. 2013.PubMed/NCBI
|
|
46
|
Hu Y, Zeng Q, Li C and Xie Y: Expression
profile and prognostic value of SFN in human ovarian cancer. Biosci
Rep. 39:BSR201901002019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du N, Li D, Zhao W and Liu Y: Stratifin
(SFN) regulates cervical cancer cell proliferation, apoptosis, and
cytoskeletal remodeling and metastasis progression through
LIMK2/cofilin signaling. Mol Biotechnol. 66:3369–3381. 2024.
View Article : Google Scholar : PubMed/NCBI
|