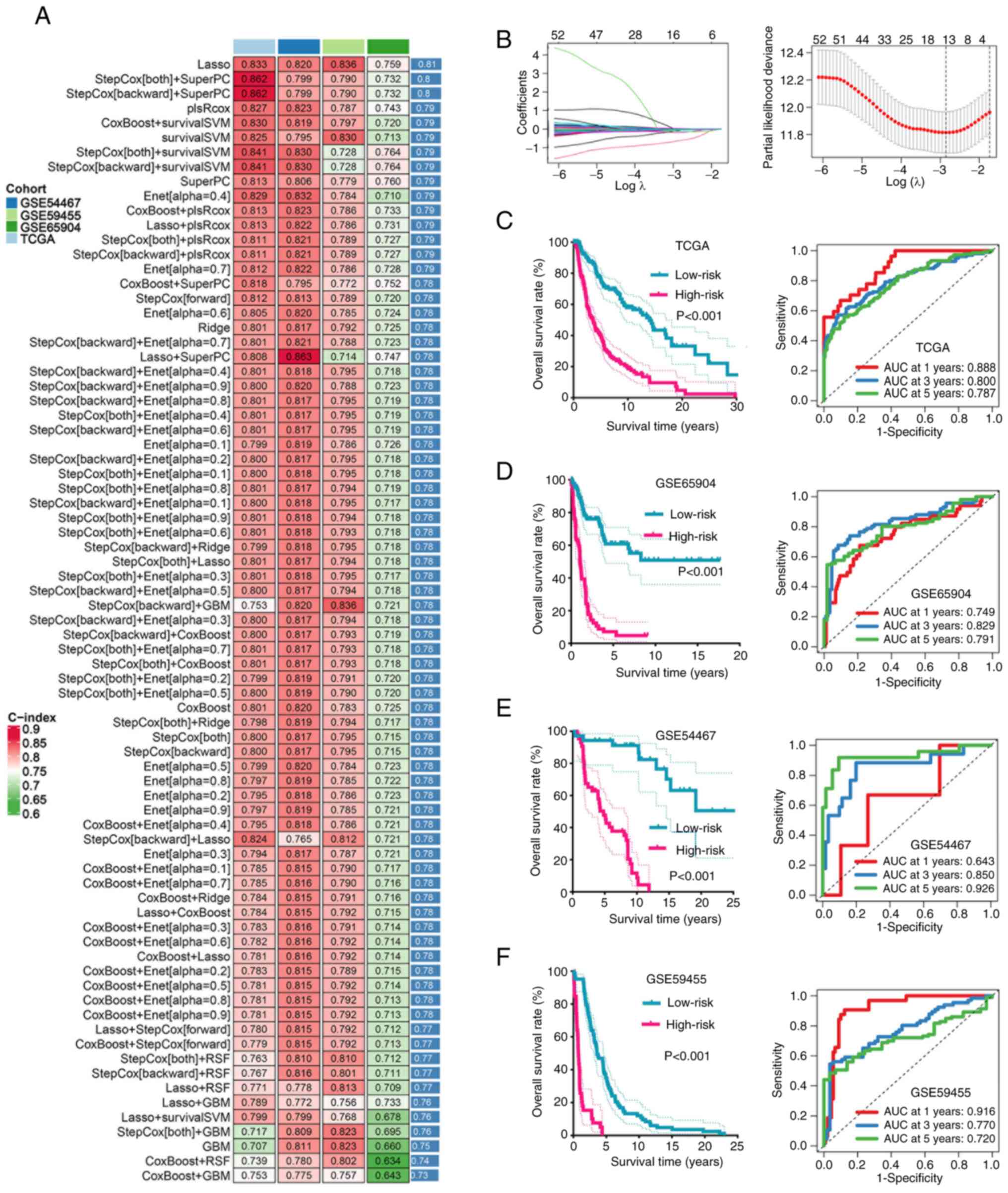
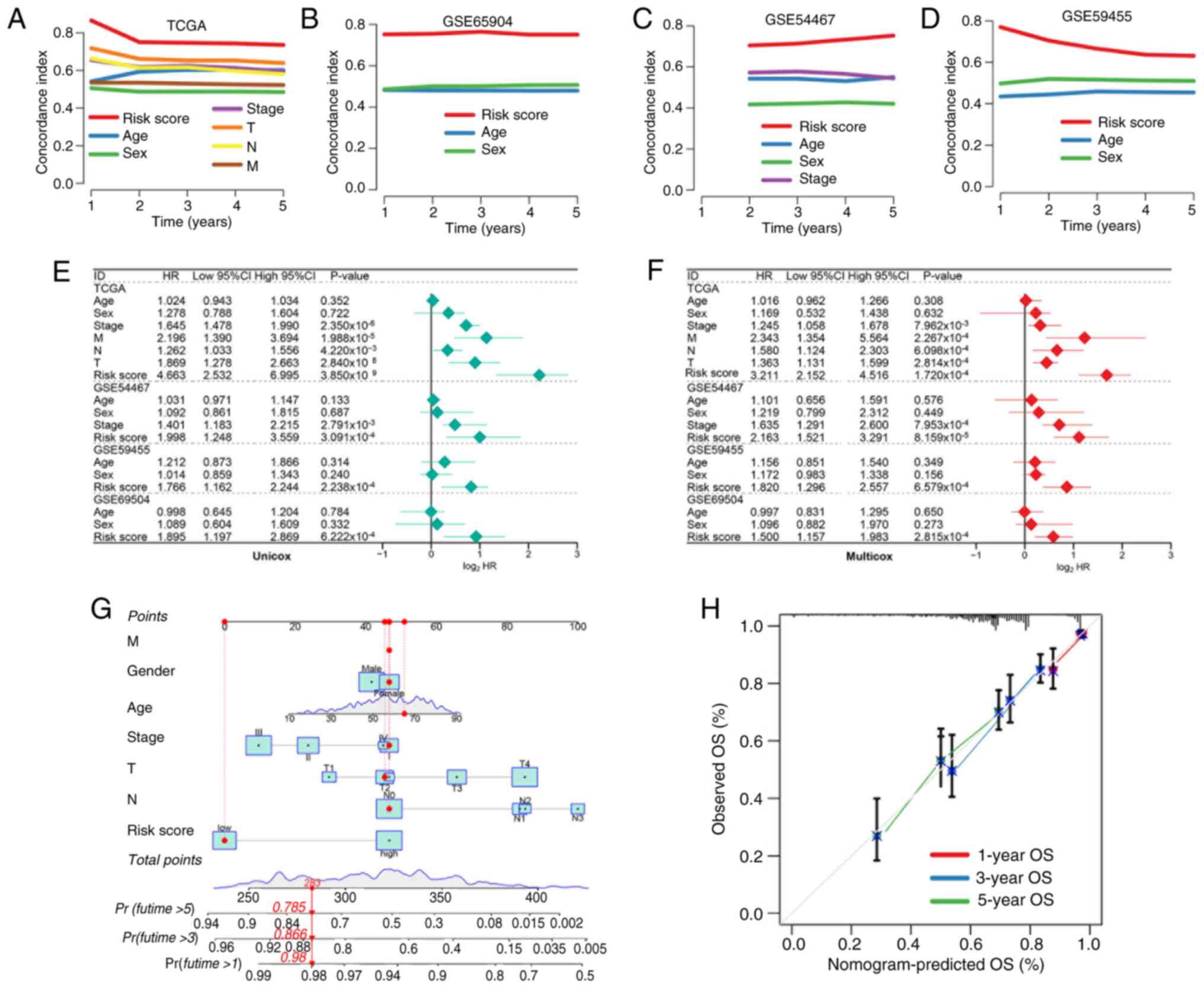
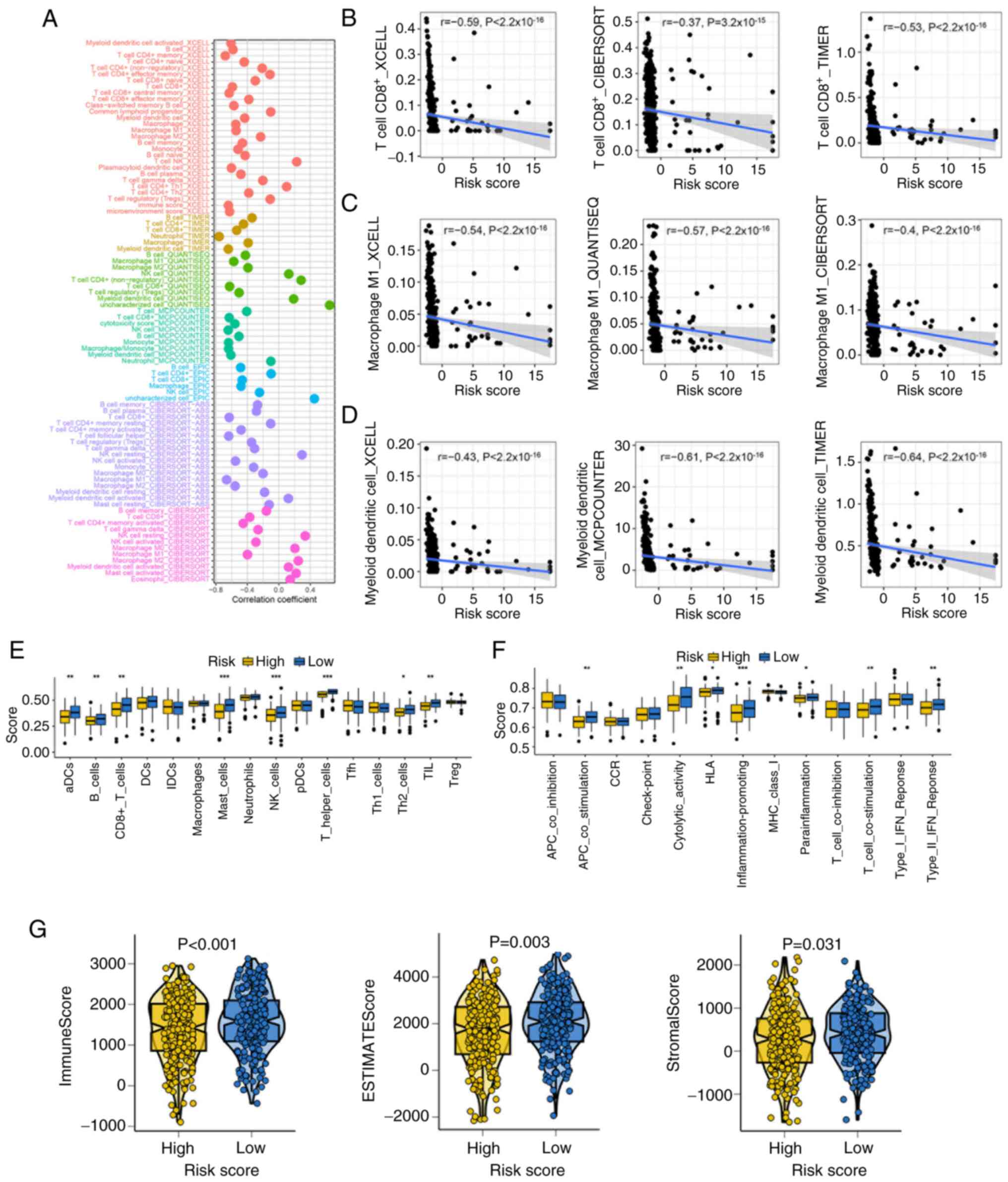
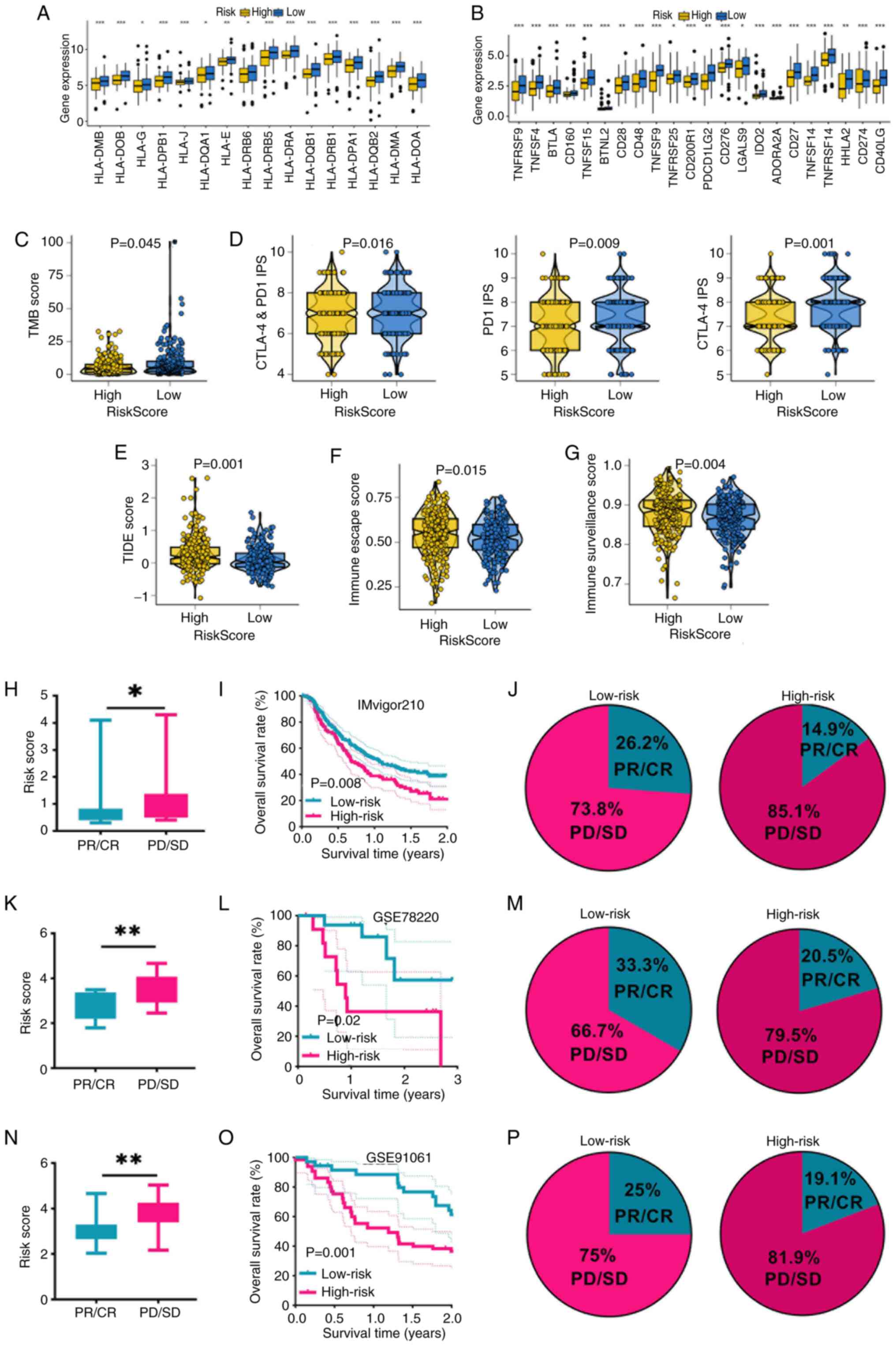
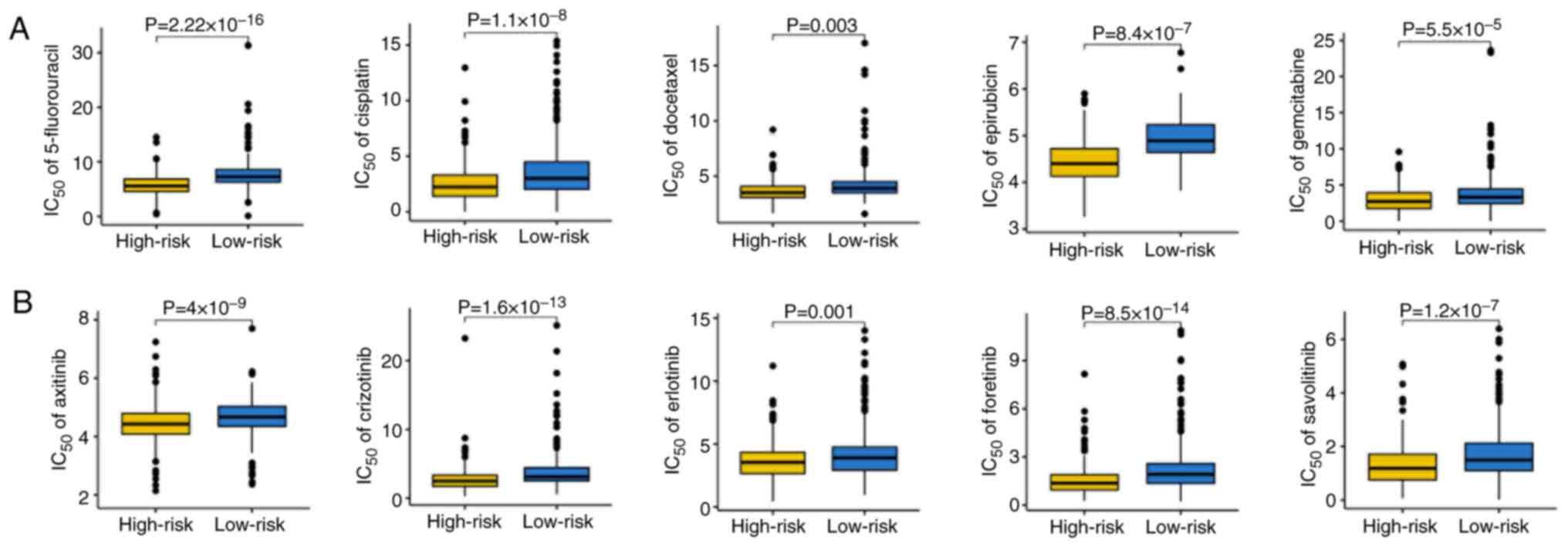
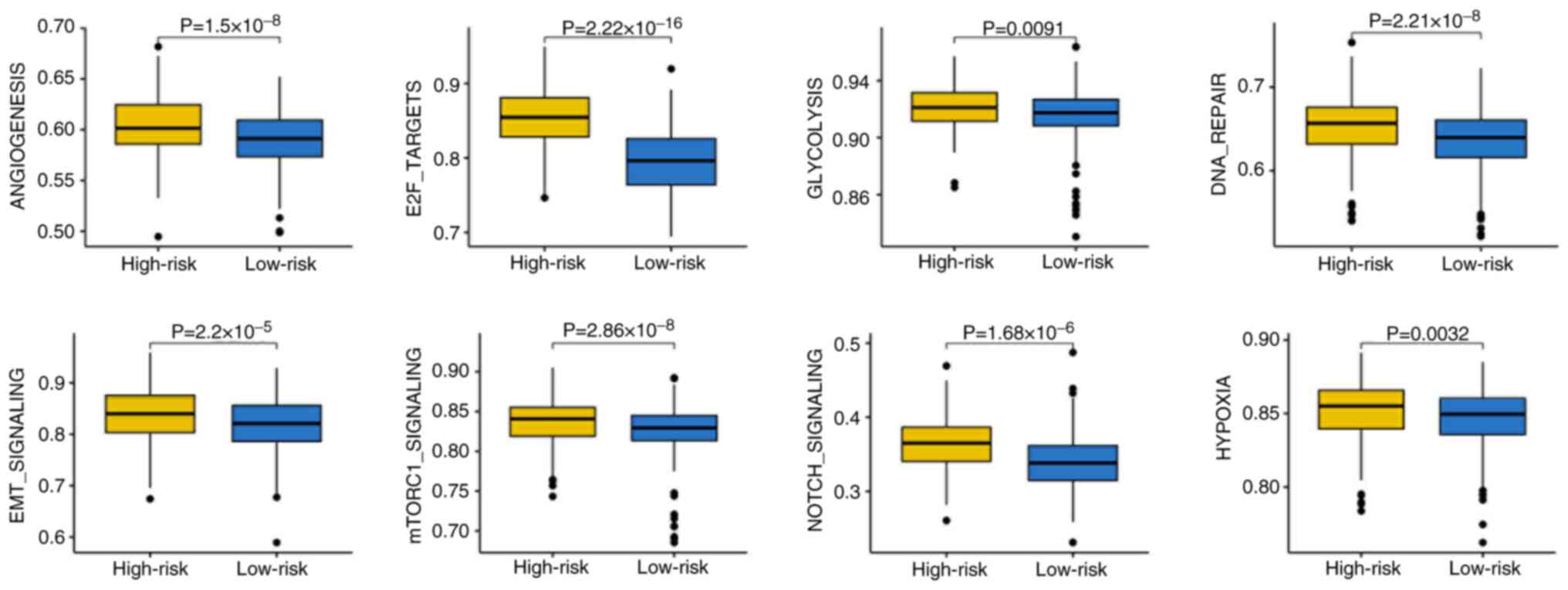
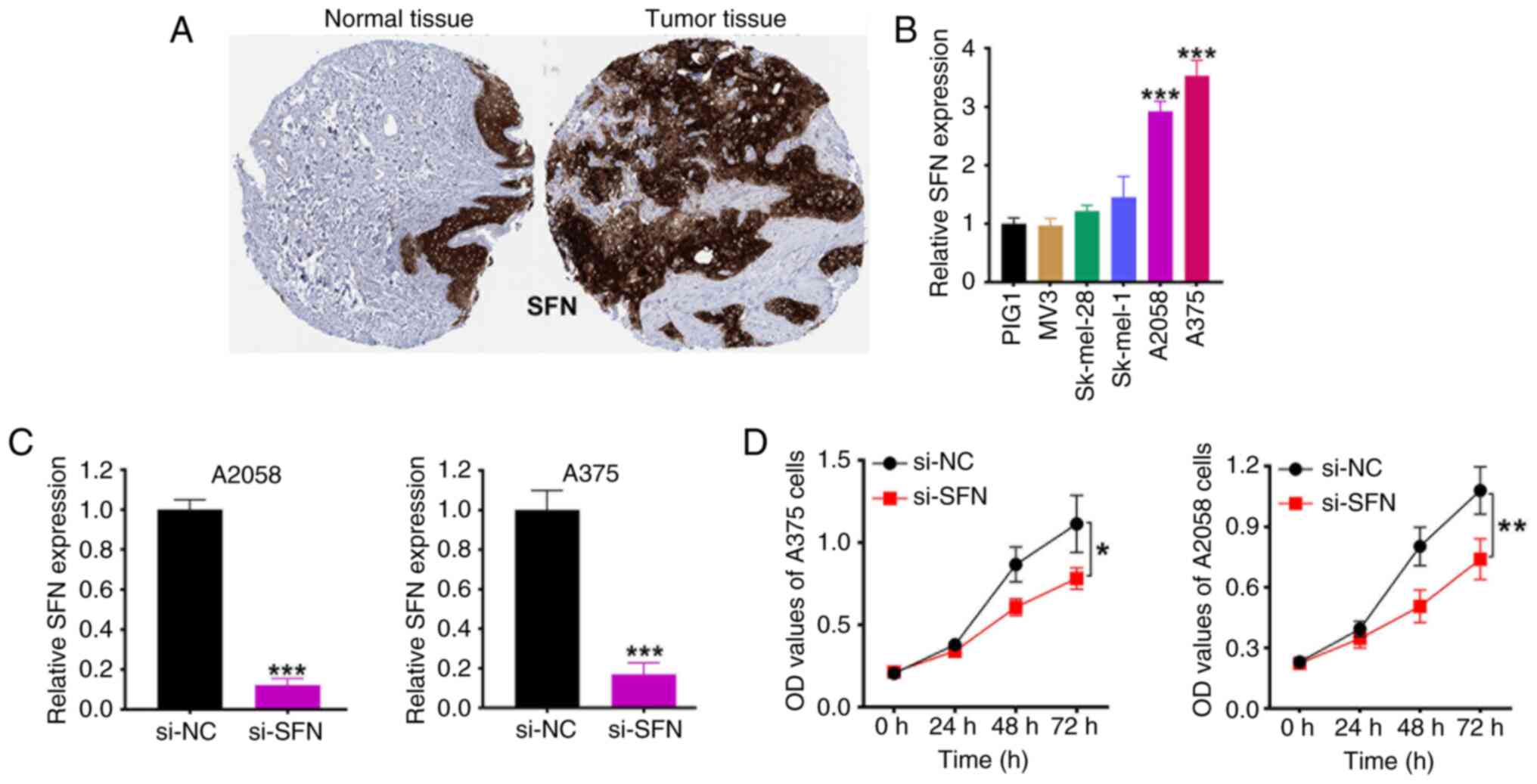
|
1
|
Long GV, Swetter SM, Menzies AM,
Gershenwald JE and Scolyer RA: Cutaneous melanoma. Lancet.
402:485–502. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Singh D, Laversanne M, Vignat J,
Vaccarella S, Meheus F, Cust AE, de Vries E, Whiteman DC and Bray
F: Global burden of cutaneous melanoma in 2020 and projections to
2040. JAMA Dermatol. 158:495–503. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopes F, Sleiman MG, Sebastian K, Bogucka
R, Jacobs EA and Adamson AS: UV exposure and the risk of cutaneous
melanoma in skin of color: A systematic review. JAMA Dermatol.
157:213–219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leonardi GC, Falzone L, Salemi R, Zanghì
A, Spandidos DA, Mccubrey JA, Candido S and Libra M: Cutaneous
melanoma: From pathogenesis to therapy (Review). Int J Oncol.
52:1071–1080. 2018.PubMed/NCBI
|
|
5
|
Newton K, Strasser A, Kayagaki N and Dixit
VM: Cell death. Cell. 187:235–256. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Hong M, Li Y, Chen D, Wu Y and Hu
Y: Programmed cell death tunes tumor immunity. Front Immunol.
13:8473452022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang T, Gu L, Kang X, Li J, Song Y, Wang
Y and Ma W: Programmed cell death disrupts inflammatory tumor
microenvironment (TME) and promotes glioblastoma evolution. Cell
Commun Signal. 22:3332024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Shou Y, Zhu R, Qiu Z, Zhang Q and
Xu J: Construction and validation of a ferroptosis-related
prognostic signature for melanoma based on single-cell RNA
sequencing. Front Cell Dev Biol. 10:8184572022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nedaeinia R, Dianat-Moghadam H,
Movahednasab M, Khosroabadi Z, Keshavarz M, Amoozgar Z and Salehi
R: Therapeutic and prognostic values of ferroptosis signature in
glioblastoma. Int Immunopharmacol. 155:1145972025. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Wang R, Hu D, Zhang C, Cao P and
Huang J: Machine learning reveals diverse cell death patterns in
lung adenocarcinoma prognosis and therapy. NPJ Precis Oncol.
8:492024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Cui Y, Zhou G, Zhang Z and Zhang
P: Leveraging mitochondrial-programmed cell death dynamics to
enhance prognostic accuracy and immunotherapy efficacy in lung
adenocarcinoma. J Immunother Cancer. 12:e0100082024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai X, Lin J, Liu L, Zheng J, Liu Q, Ji L
and Sun Y: A novel TCGA-validated programmed cell-death-related
signature of ovarian cancer. BMC Cancer. 24:5152024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y and Zhang Q: Leveraging programmed
cell death signature to predict clinical outcome and immunotherapy
benefits in postoperative bladder cancer. Sci Rep. 14:229762024.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gu X, Pan J, Li Y and Feng L: A programmed
cell death-related gene signature to predict prognosis and
therapeutic responses in liver hepatocellular carcinoma. Discov
Oncol. 15:712024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nayarisseri A, Khandelwal R, Tanwar P,
Madhavi M, Sharma D, Thakur G, Speck-Planche A and Singh SK:
Artificial intelligence, big data and machine learning approaches
in precision medicine & drug discovery. Curr Drug Targets.
22:631–655. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ngiam KY and Khor IW: Big data and machine
learning algorithms for health-care delivery. Lancet Oncol.
20:e262–e273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jayawardana K, Schramm SJ, Haydu L,
Thompson JF, Scolyer RA, Mann GJ, Müller S and Yang JY:
Determination of prognosis in metastatic melanoma through
integration of clinico-pathologic, mutation, mRNA, microRNA, and
protein information. Int J Cancer. 136:863–874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Budden T, Davey RJ, Vilain RE, Ashton KA,
Braye SG, Beveridge NJ and Bowden NA: Repair of UVB-induced DNA
damage is reduced in melanoma due to low XPC and global genome
repair. Oncotarget. 7:60940–60953. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cabrita R, Lauss M, Sanna A, Donia M,
Skaarup Larsen M, Mitra S, Johansson I, Phung B, Harbst K,
Vallon-Christersson J, et al: Tertiary lymphoid structures improve
immunotherapy and survival in melanoma. Nature. 577:561–565. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Riaz N, Havel JJ, Makarov V, Desrichard A,
Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH,
et al: Tumor and microenvironment evolution during immunotherapy
with nivolumab. Cell. 171:934–949.e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hugo W, Zaretsky JM, Sun L, Song C, Moreno
BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G,
et al: Genomic and transcriptomic features of response to Anti-PD-1
therapy in metastatic melanoma. Cell. 165:35–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rosenberg JE, Galsky MD, Powles T,
Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J,
Perez-Gracia JL, van der Heijden MS, et al: Atezolizumab
monotherapy for metastatic urothelial carcinoma: Final analysis
from the phase II IMvigor210 trial. ESMO Open. 9:1039722024.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H,
Wang L, Lu T, Zhang Y, Sun Z and Han X: Machine learning-based
integration develops an immune-derived lncRNA signature for
improving outcomes in colorectal cancer. Nat Commun. 13:8162022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sturm G, Finotello F and List M:
Immunedeconv: An R package for unified access to computational
methods for estimating immune cell fractions from bulk
RNA-sequencing data. Methods Mol Biol. 2120:223–232. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Palmeri M, Mehnert J, Silk AW, Jabbour SK,
Ganesan S, Popli P, Riedlinger G, Stephenson R, de Meritens AB,
Leiser A, et al: Real-world application of tumor mutational
burden-high (TMB-high) and microsatellite instability (MSI)
confirms their utility as immunotherapy biomarkers. ESMO Open.
7:1003362022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang
P and Liu XS: Large-scale public data reuse to model immunotherapy
response and resistance. Genome Med. 12:212020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Charoentong P, Finotello F, Angelova M,
Mayer C, Efremova M, Rieder D, Hackl H and Trajanoski Z: Pan-cancer
immunogenomic analyses reveal genotype-immunophenotype
relationships and predictors of response to checkpoint blockade.
Cell Rep. 18:248–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang
Z, Zhang Z, Xie J, Wang C, Chen D, et al: Single-cell landscape of
the ecosystem in early-relapse hepatocellular carcinoma. Cell.
184:404–421.e16. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Balch CM, Gershenwald JE, Soong SJ,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin A and Yan WH: HLA-G/ILTs targeted
solid cancer immunotherapy: Opportunities and challenges. Front
Immunol. 12:6986772021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin A, Zhang J and Luo P: Crosstalk
between the MSI status and tumor microenvironment in colorectal
cancer. Front Immunol. 11:20392020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
37
|
Gershenwald JE, Scolyer RA, Hess KR,
Sondak VK, Long GV, Ross MI, Lazar AJ, Faries MB, Kirkwood JM,
McArthur GA, et al: Melanoma staging: Evidence-based changes in the
American Joint Committee on Cancer eighth edition cancer staging
manual. CA Cancer J Clin. 67:472–492. 2017.PubMed/NCBI
|
|
38
|
Wang J, Yang F, Sun Q, Zeng Z, Liu M, Yu
W, Zhang P, Yu J, Yang L, Zhang X, et al: The prognostic landscape
of genes and infiltrating immune cells in cytokine induced killer
cell treated-lung squamous cell carcinoma and adenocarcinoma.
Cancer Biol Med. 18:1134–1147. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tien FM, Lu HH, Lin SY and Tsai HC:
Epigenetic remodeling of the immune landscape in cancer:
Therapeutic hurdles and opportunities. J Biomed Sci. 30:32023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun R, Limkin EJ, Vakalopoulou M, Champiat
S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S, Hollebecque
A, et al: A radiomics approach to assess tumour-infiltrating CD8
cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an
imaging biomarker, retrospective multicohort study. Lancet Oncol.
19:1180–1191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu D, Schilling B, Liu D, Sucker A,
Livingstone E, Jerby-Arnon L, Zimmer L, Gutzmer R, Satzger I,
Loquai C, et al: Integrative molecular and clinical modeling of
clinical outcomes to PD1 blockade in patients with metastatic
melanoma. Nat Med. 25:1916–1927. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gide TN, Quek C, Menzies AM, Tasker AT,
Shang P, Holst J, Madore J, Lim SY, Velickovic R, Wongchenko M, et
al: Distinct immune cell populations define response to anti-PD-1
monotherapy and anti-PD-1/Anti-CTLA-4 combined therapy. Cancer
Cell. 35:238–255.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang W, Soares J, Greninger P, Edelman EJ,
Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et
al: Genomics of drug sensitivity in cancer (GDSC): A resource for
therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.
41((Database Issue)): D955–D961. 2013.PubMed/NCBI
|
|
46
|
Hu Y, Zeng Q, Li C and Xie Y: Expression
profile and prognostic value of SFN in human ovarian cancer. Biosci
Rep. 39:BSR201901002019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Du N, Li D, Zhao W and Liu Y: Stratifin
(SFN) regulates cervical cancer cell proliferation, apoptosis, and
cytoskeletal remodeling and metastasis progression through
LIMK2/cofilin signaling. Mol Biotechnol. 66:3369–3381. 2024.
View Article : Google Scholar : PubMed/NCBI
|