Introduction
Breast cancer is the most common malignant tumor in
female patients (1); according to
data from the World Health Organization, in 2020, there were ~2.26
million new breast cancer cases worldwide, threatening their lives
by affecting both physical and psychological health (2,3). The
latest advancements in treatment have mainly focused on the use of
molecular biology and immunological approaches to develop highly
targeted therapies for different types of breast cancer. This has
been explored through inhibiting specific targets or molecules that
promote tumor progression (4). Due
to the presence of different subtypes and differences in treatment
modalities (5), it is common to
categorize breast cancers on the basis of expression patterns of
three receptors: Estrogen receptor, progesterone receptor and human
epidermal growth factor-2 (HER2) receptor, or as a triple-negative
breast cancer (TNBC). Most patients suffer from estrogen
receptor-positive breast cancers; the majority of patients have
estrogen receptor-positive breast cancer, accounting for >70% of
all cases (6), in vitro
models are primarily represented by MCF-7 cells. Endocrine therapy,
usually using tamoxifen (TAM), has been shown to markedly prolong
the survival of patients with breast cancer (7). TNBC, a subtype in which estrogen,
progesterone and HER2 receptors are not expressed, accounts for
12–17% of invasive breast cancers (8,9) and
83% of total mortalities (10).
Compared with other breast cancer subtypes, triple-negative breast
cancer (TNBC), represented by the MDA-MB-231 cell line in
vitro, has the highest recurrence and metastasis rates and is
exemplified by development at a young age and a poor prognosis
(11). Currently, there are no
effective treatments other than surgery, radiation and chemotherapy
therapy. Doxorubicin (DOX) is widely used to treat TNBC (12), as a chemotherapy drug, therefore in
the present study, both TAM and DOX were chosen to induce apoptosis
in breast cancer cells.
Multiple transcription factors have been reported to
regulate proliferation and apoptosis in breast cancer cells
(13,14). These transcription factors achieve
these effects by controlling protein translation and degradation.
Consequently, interactions between proteins and transcription
factors play a crucial role in the initiation and progression of
breast cancer (15,16). The transcription factor FoxO3a has
potent tumor suppressor effects in a variety of malignancies, such
as gastric, prostate and colorectal cancers (17–19).
RNA interference targeting FOXO3a was used to measure the impact of
simvastatin on FOXO3a-expressing cells (20). Therefore, the present study
performed FoxO3a knockdown. Previous studies have shown that low
expression of FoxO3a could inhibit the apoptosis of tumor cells
(21,22).
The application of proteomics allows for the
discovery of additional proteins, which can provide insight into
the mechanisms of disease and identify specific biomarkers that
induce apoptosis. In the present study, small interfering
(si)RNA-negative control (NC) and si-FoxO3a cel ls from two types
of breast cancer were treated with TAM and DOX to construct
estrogen-dependent and triple-negative si-FoxO3a breast cancer cell
lines. The aims were to validate the effect on drug-induced
apoptosis in breast cancer cells after interference with FoxO3a,
analyze the signaling pathways that serve a role in the regulation
of apoptosis after treatment via proteomics; and validate the
signaling pathway-associated differentially expressed proteins
(DEPs). The present study hypothesizes that targeting regulatory
proteins in protein digestion and absorption signaling pathway
could lead to timely detection of breast cancer metastasis and
improve treatment outcomes.
Materials and methods
Cell culture
Breast epithelial cells, MCF-10A, and breast cancer
cell lines MCF-7 and MDA-MB-231 were purchased from the Shanghai
Institute of Biological Sciences. The cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) and high-glucose
DMEM (both Gibco; Thermo Fisher Scientific, Inc.), supplemented
with 10% FBS (EvaCell) and 1% penicillin and streptomycin (Cytiva)
at 37°C with 5% CO2.
Cell transfection
A total of 2.5×105 cells/well (MCF-7 and
MDA-MB-231) were seeded into 6-well plates and incubated at 37°C
for 24 h. Liposomal transfection of siRNA was performed the
following day when the cell density reached 60% using the
transfection reagent Lipofectamine® 3000 (Thermo Fisher
Scientific, Inc.) for 24 h at 37°C. The siRNAs were designed by our
laboratory staff and synthesized by Guangzhou Ruibo Biotechnology
Co., Ltd. In addition, 1,750 ml basal medium, 5 µl siRNA, 5 µl
transfection reagent and 240 µl serum-free medium was added to each
well. At 8 h after transfection in serum-free medium, the medium
was replaced with complete medium and the cells were incubated for
a further 24 h. si-FoxO3a target sequence was
5′-AATGATGGGGGGCTGACTGAAA-3′, synthesized by RiboBio Biotechnology,
the initial siRNA transfection concentration is 50 nmol.
Reverse transcription quantitative PCR
(RT-qPCR)
Total cellular RNA was extracted with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) for reverse transcription (SureScript™ First-Strand cDNA;
Thermo Fisher Scientific, Inc.). The quality and concentration of
the extracted RNA was analyzed using spectrophotometry (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). Reverse
transcription conditions are as follows: 25°C 5 min, 42°C 15 min,
85°C 5 min. Blaze Taq SYBR® Green qPCR Mix 2.0 (Thermo
Fisher Scientific, Inc.) was used for RT-qPCR experiments.
Thermocycling conditions: 95°C pre-denaturation for 10 min, 95°C
denaturation for 15 sec, 60°C annealing for 1 min, 72°C extension
for 30 sec, repeated for 40 cycles, with three replicate wells per
sample, and the transcript levels were normalized to those of
GAPDH. Gene expression levels were analyzed using the
2−ΔΔCq method (23). The
following primers were used: GAPDH-forward (F):
5′-ACCCACTCCTCCACCTTTGAC-3′; GAPDH-reverse (R):
5′-TGTTGCTGTAGCCAAATTCGTT-3′; FoxO3a-F: 5′-CGGACAAACGGCTCACTCT-3′;
FoxO3a-R: 5′-GGACCCGCATGAATCGACTAT-3′;
Western blotting
Experiments were performed with MCF-7/MDA-MB-231
si-NC and si-FoxO3a cells treated with TAM/DOX for 24 h, which were
both washed twice with PBS and lysed on ice for 30 min with the
addition of lysis buffer(Beyotime Biotechnology). Centrifugation
was performed at 12,000 × g for 10 min at 4°C. The supernatant was
collected, and the protein concentration was determined using the
Enhanced BCA Protein Assay kit (Beyotime Institute of
Biotechnology). A total of 5 µg/lane protein per well, separated by
SDS-PAGE (5% concentrating and 5% separating gel), transferred to a
PVDF membrane and incubated overnight at 4°C with the following
primary antibodies: GAPDH (1:1,000; cat. no. P04406; Cell Signaling
Technology, Inc.), FoxO3a (1:1,000; cat. no. O43524; Cell Signaling
Technology, Inc.), trophoblast glycoprotein (TPBG; 1:1,000; cat.
no. Q13641; Cell Signaling Technology, Inc.), sodium-coupled
neutral amino acid symporter 2 (SLC38A2; 1:1,000; cat. no. DF4673;
Affinity Biosciences), asparagine synthetase (ASNS; 1:1,000; cat.
no. P08243; Cell Signaling Technology, Inc.), phosphorylated
(p)-FOXO3a (1:1,000; cat. no. O43524; Cell Signaling Technology,
Inc.) and zyxin (ZYX; 1:1,000; cat. no. Q15942; Cell Signaling
Technology, Inc.). The membrane was washed three times, incubated
with a goat anti-rabbit IgG antibody (1:1,000; cat. no. 210830803;
Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) for 1 h at room
temperature, and the results were detected with a chemiluminescent
ECL detection system(Thermo Fisher). The experiments all contained
three independent biological replicates, and three technical
replicates were used for each biological replicate to ensure data
reliability.
Water-soluble tetrazolium salt (WST-1)
assay
A total of 6×103 cells per well were
seeded in a 96-well plate, and three replicate wells were used for
each group. When the density reached 80–90%, the culture medium was
discarded, and 100 µl different concentrations of TAM/DOX were
added. Each well had 10 µl WST-1 reagent added 24 h after the drug
treatment, the reaction was carried out at 37°C for 15–30 min and
the optical density value was assessed at the wavelength of 460 nm.
The experiments all contained three independent biological
replicates, and three technical replicates were used for each
biological replicate to ensure data reliability.
Cell apoptosis
A total of 2.5×105 cells were seeded in a
6-well plate and digested with trypsin. Following centrifugation
for 10 min at 4°C, 12,000 × g the cells were resuspended in PBS,
centrifuged again and then, according to the protocol of the
FITC-coupled Annexin-V Apoptosis Detection Kit (BD Biosciences),
each sample was mixed with 490 µl 1X binding buffer and 5 µl each
of FITC and PI dyes. The cells were stained for 20 min at room
temperature, protected from light and assessed using flow cytometry
within 1 h. There were 10,000 cells in each group. The experiments
all contained three independent biological replicates, and three
technical replicates were used for each biological replicate to
ensure data reliability.
Hoechst staining
Hoechst 33342 is a dye that penetrates cell
membranes, causing living cells to fluoresce blue. When apoptosis
occurs with inconsistent nuclear fragmentation; the brightness of
the blue fluorescence is increased. A total of 10,000 si-NC and
si-FoxO3a cells were cultured for 6–8 h, the cells were treated
with the drugs for 24 h, the supernatant was discarded and the
cells were washed twice with precooled PBS. Hoechst 33342 staining
solution (Thermo Fisher Scientific, Inc.) was added, and the cells
were stained for 15–20 min at room temperature, before being washed
three times with precooled PBS at 4°C for 5 min each. The
fluorescent intensity of the cells was observed using fluorescence
microscopy. The experiments all contained three independent
biological replicates, and three technical replicates were used for
each biological replicate to ensure data reliability.
Wound healing assay
MCF-7 and MDA-MB-231 cells were seeded into 6-well
plates (3×105 and 5×105/well, respectively),
and the transfection and drug treatments were carried out as
aforementioned. After the cells grew to 90% confluence, a sterile
tip was used to scratch the cells. Cell debris was removed by
washing with PBS and the cells were cultured in serum-free medium
for 24 h. Images were acquired using a fluorescence microscope
(magnification, ×10) at 0 and 24 h after scratching. The relative
size of the wound area from 0 to 24 h was measured using Image J
software(National Institutes of Health; version no. 1.47) to
calculate the cell migration rate. The experiments all contained
three independent biological replicates, and three technical
replicates were used for each biological replicate to ensure data
reliability.
Proteomics processing
MCF-7 and MDA-MB-231 cells treated with TAM/DOX
drugs for 24 h were recorded as control (ctrl)-TAM and ctrl-DOX,
respectively. FoxO3a knockdown cells treated with drugs for 24 h
were recorded as si-TAM and si-DOX. After treatment, the cell
precipitate was collected, 20% trichloroacetic acid was added, the
mixture was vortexed well and the cells were left to precipitate
for 2 h at 4°C. The supernatant was removed by centrifugation at
4°C, 4,500 × g for 5 min, and the precipitate was washed with
precooled acetone 2–3 times. The precipitate was allowed to dry, a
final concentration of 200 mM triethylammonium bicarbonate was
added, the precipitate was broken by ultrasonication and it was
enzymatically digested overnight. Dithiothreitol was added to a
final concentration of 5 mM and reduced at 56°C for 30 min.
Iodine-acetyl carbamate was added to a final concentration of 11 mM
and incubated for 15 min at room temperature in the dark. The
confidence interval for the difference in the protein
quantification ratio between the si-FoxO3a and the si-NC group was
assessed as the multiplicity of differences, with a fold-change
>1.2 and P<0.05. The false discovery rate (FDR) for protein
identification and peptide-spectrum match identification in the
search database settings was 1%, revealing that different protein
networks existed in both cancer cell lines after interfering with
FoxO3a expression.
Liquid chromatography-mass
spectrometry analysis
Liquid chromatography-mass spectrometry analysis was
performed by Jingjie PTM Bio-Laboratory (Hangzhou) Co., Ltd. using
positive ionization mode. The primary mass spectrometry scan range
was 400–1,200 m/z with a resolution of 60,000. The fixed starting
point for the secondary mass spectrometry scan range was 110 m/z,
with a secondary scan resolution of 15,000. Mobile phase A was an
aqueous solution containing 0.1% formic acid and 2% acetonitrile;
mobile phase B was an aqueous solution containing 0.1% formic acid
and 90% acetonitrile. The liquid chromatography gradient settings
were as follows: 0–68 min, 6–23%B; 68–82 min: 23–32%B; 82–86 min:
32–80%B; 86–90 min: 80%B. Flow rate maintained constant at 500
nl/min.
Gene Ontology (GO), KEGG(Kyoto
Encyclopedia of Genes and Genomes) functional enrichment analysis,
and STRING protein interactions
GO (geneontology.org) and KEGG (kegg.jp) functional
enrichment analyses were performed based on secondary
categorization and Fisher's exact test was applied to calculate the
significance P-value with the aim of determining whether DEPs were
significantly enriched in certain functional types. The FDR value
in enrichment analysis is set to 1%. Directly copy or import the
identified differentially expressed proteins into the STRING
database (cn.string-db.org).
Statistical analyses
A Student's t-test (unpaired)was used to compare
differences between two groups, and a one-way ANOVA followed by
Tukey's post hoc test was used to compare differences between
multiple groups. Calculations were performed using SPSS Statistics
17.0 (IBM Corp.) software, with normality analysis being performed
using the Kolmogorov-Smirnov test. Three replications were
performed for all the experiments, and the data are expressed as
the mean ± standard deviation. P<0.05 was considered to indicate
a statistically significant difference.
Results
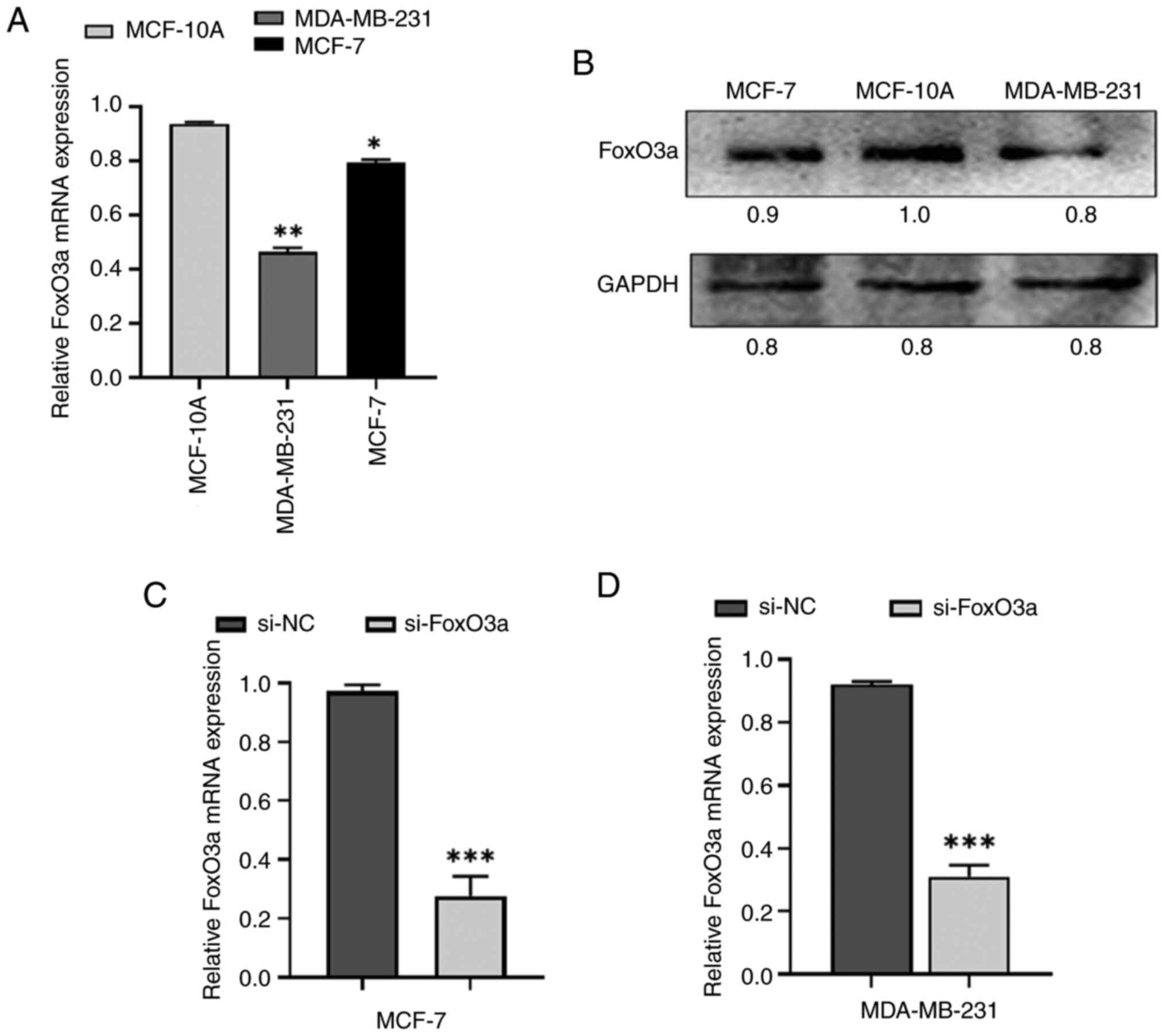
Assessment of baseline expression
levels of FoxO3a and successful knockdown of FoxO3a
FoxO3a expression was examined in MCF-7 and
MDA-MB-231 cells using RT-qPCR and western blotting, which
demonstrated that FoxO3a levels were detected in both cell types
(Fig. 1A and B). FoxO3a was
successfully knocked down by transfection of siRNA for 24 h.
RT-qPCR was used to assess the knockdown efficiency as shown in
Fig. 1C and D.
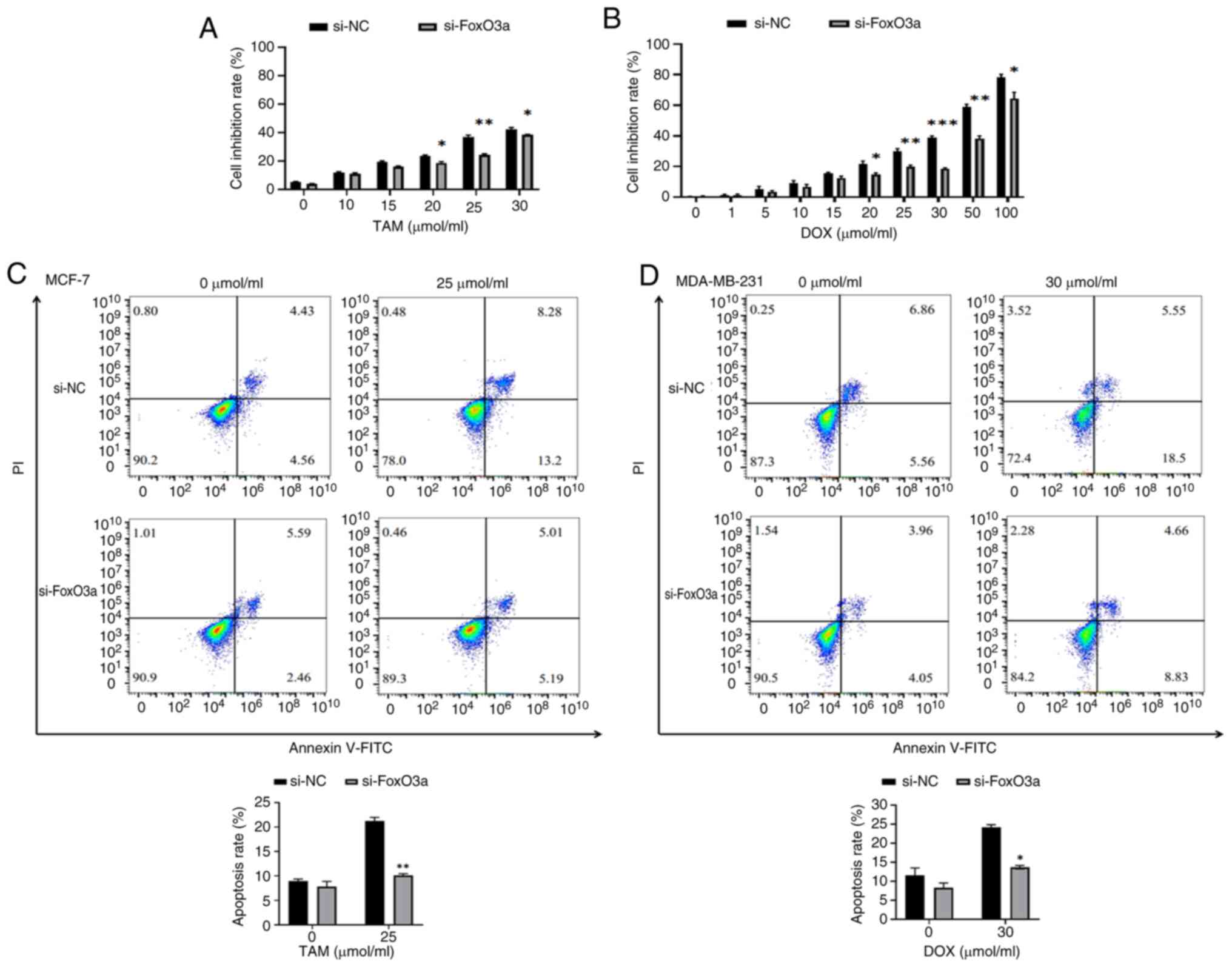
si-FoxO3a decreases the degree of
apoptosis
Annexin V/PI double staining demonstrates that
si-FoxO3a hinders the apoptosis of breast cancer cells by antitumor
drugs
The WST-1 assay was used to determine the optimal
concentrations of TAM and DOX. This method is easy to perform,
produces stable results and has low cytotoxicity. The results of
the WST-1 assay revealed that the apoptosis rate of si-FoxO3a MCF-7
and MDA-MB-231 cells by 25 µmol/ml and 30 µmol/ml TAM and DOX
respectively, was decreased compared with the si-NC cells (Fig. 2A and B). Flow cytometry was used to
analyze the proapoptotic effects of 0 and 25 µmol/ml TAM drug
treatments for 24 h on si-NC and si-FoxO3a MCF-7 cells, and 0 and
30 µmol/ml DOX drug treatments for 24 h on si-NC and si-FoxO3a
MDA-MB-231 cells. The results (Fig. 2C
and D) revealed that the apoptosis rates of the si-NC and
si-FoxO3a MCF-7 cells were 8.95±0.04% and 7.81±1.17%, respectively,
and the apoptotic rates of si-NC and si-FoxO3a MDA-MB-231 cells
were 11.56±2.22% and 8.31±0.20%, respectively, when cultured for 24
h without treatment. The differences were not found to be
statistically significant. These findings indicate that the
transfection of siRNA had no significant effect on the level of
apoptosis in either type of breast cancer cells. By contrast, when
treated with drugs for 24 h, the apoptosis rates of the si-NC and
si-FoxO3a treated MCF-7 cells were 21.22±0.88% and 10.10±0.44%,
respectively, and the apoptosis rates of si-NC and si-FoxO3a
MDA-MB-231 cells were 24.19±0.60% and 13.67±0.35%, respectively.
The level of apoptosis in si-FoxO3a cells was significantly reduced
after treatment with the drug, demonstrating that interfering with
FoxO3a decreases the sensitivity of cancer cells to the drug.
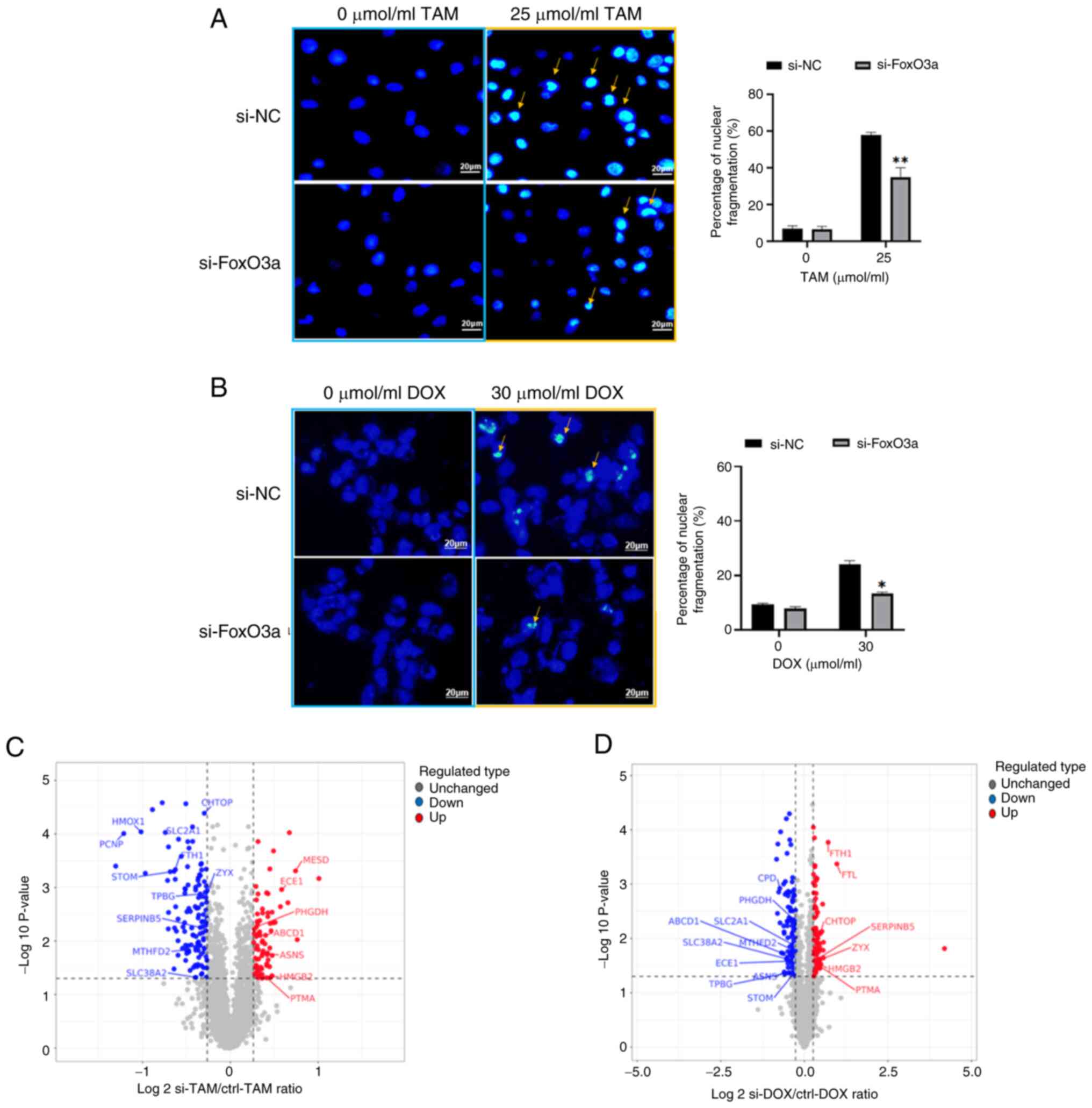
Hoechst staining demonstrates that
si-FoxO3a hinders the inhibitory effect of antitumor drugs on
breast cancer cells
si-NC cells and si-FoxO3a cells without drug
treatment presented with dark blue fluorescence staining of the
nuclei. After drug treatment for 24 h, the nuclear staining of the
two types of breast cancer cells was not homogeneous, as there was
some nuclear fragmentation, indicated by increased fluorescence
brightness. Furthermore, the levels of nuclear fragmentation in
si-FoxO3a cells were significantly reduced compared with si-NC
cells. This indicates reduced apoptosis of si-FoxO3a cells.
Quantification of the nuclear fragmentation levels is presented in
Fig. 3A and B.
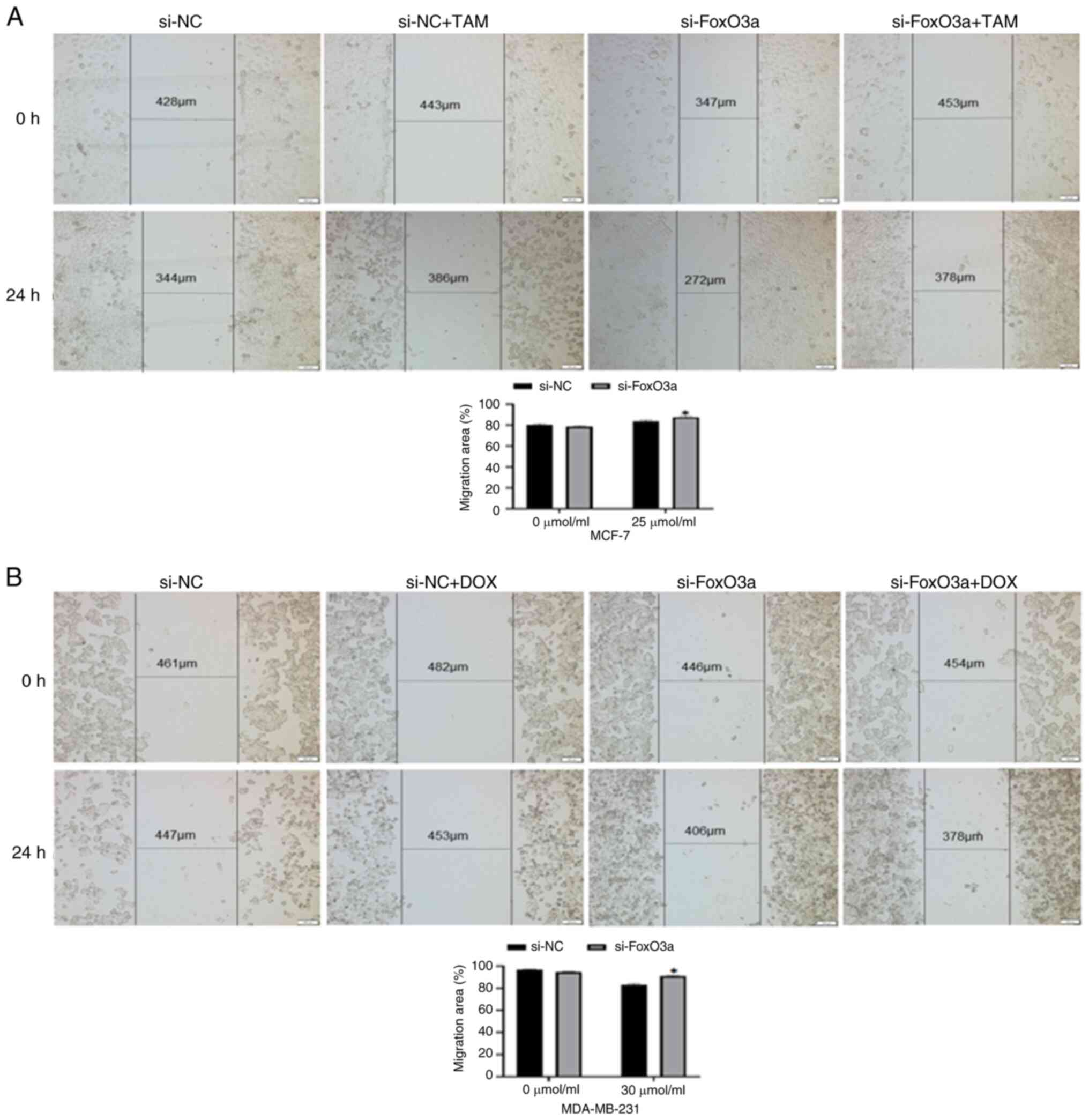
Wound healing assay demonstrates
enhanced migration of si-FoxO3a cells
Subsequently, the migration of MCF-7 and MDA-MB-231
si-NC and si-FoxO3a cells towards the scratched area was observed.
Without drug treatment, there was no significant difference in
migration rates between si-NC and si-FoxO3a cancer cells. Following
24 h treatment with TAM and DOX, respectively, the migration rate
of si-FoxO3a cells was significantly increased compared to the
si-NC group (Fig. 4A and B). This
indicates that FoxO3a knockdown reduces cancer cell death and
enhances their migratory capacity.
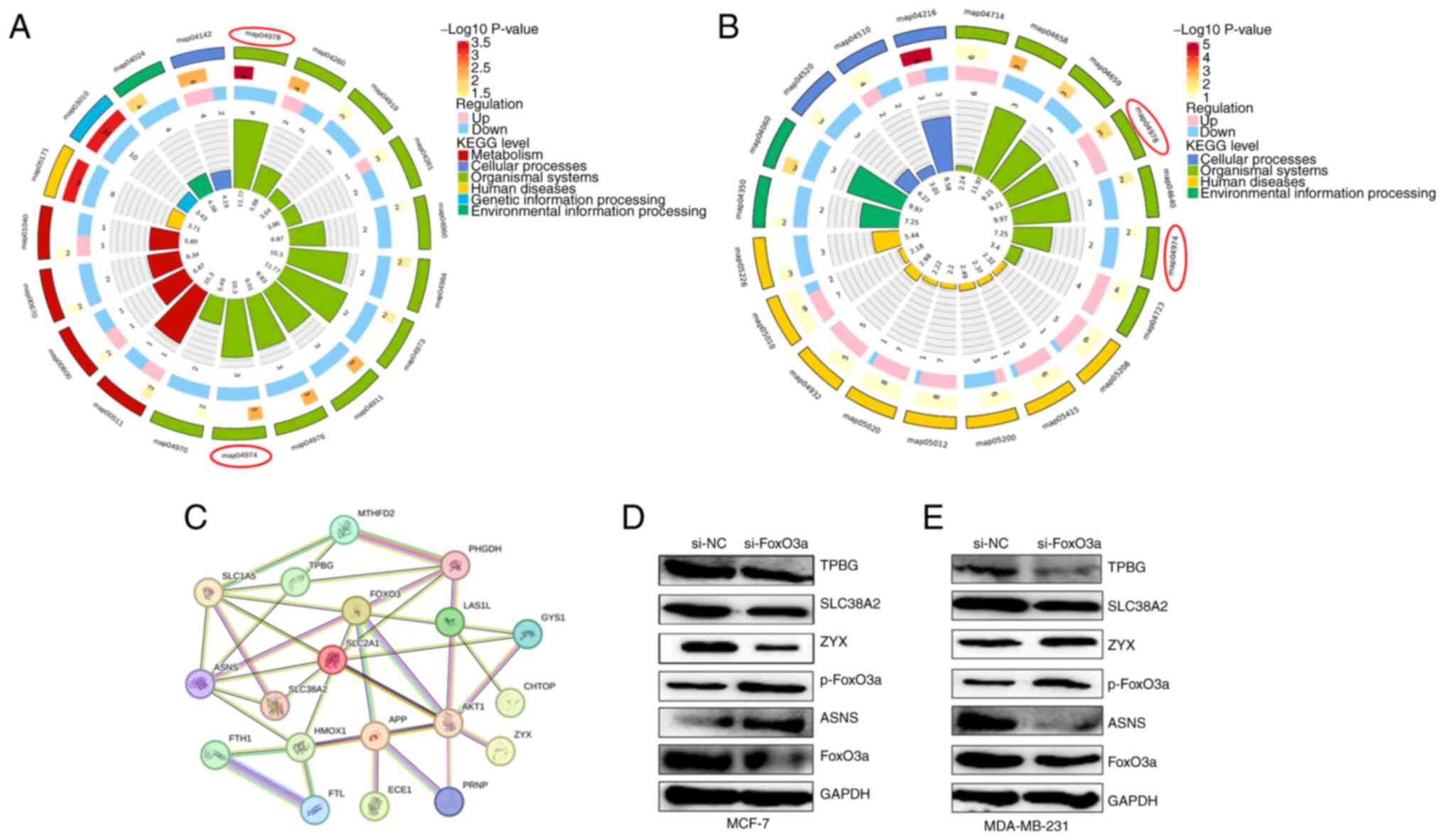
Proteomics analysis of DEPs in
si-FoxO3a
The study consisted of si-FoxO3a and si-NC groups
for label-free quantitative proteomics analysis, with three
biological replicates for each group. A screen for DEPs by the
ratio of quantitative values of DEPs in si-FoxO3a to si-NC cells
revealed 206 DEPs in MCF-7 and 207 DEPs in MDA-MB-231 cells, with
15 proteins in crossover and subcellular localization analysis. The
volcano plots (Fig. 3C and D) show
that TPBG, SLC38A2, ATP binding cassette subfamily D member 1,
endothelin converting enzyme 1, ASNS, ferritin heavy chain 1
(FTH1), ZYX, stomatin, solute carrier family 2 member 1 (SLC2A1),
serpin family B member 5, prothymosin α, phosphoglycerate
dehydrogenase, methylenetetrahydrofolate dehydrogenase
(NADP+ dependent) 2, high mobility group box 2 and
chromatin target of PRMT1 (CHTOP) were DEPs common to the two types
of cells after knockdown of FoxO3a. The presence of coregulatory
proteins could provide new perspectives on the treatment and
mitigation of drug resistance in both types of breast cancer.
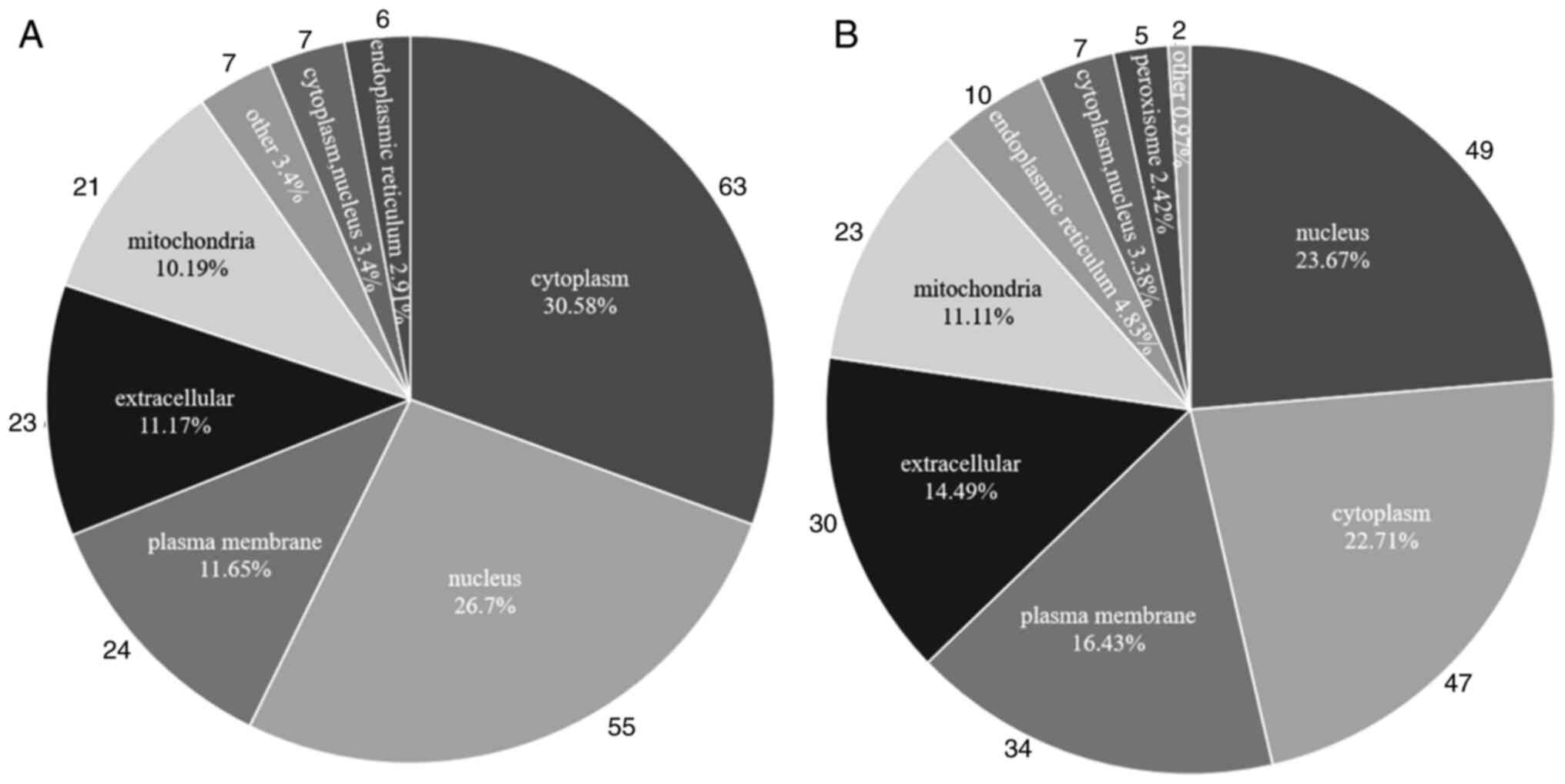
Subcellular localization analysis
Subcellular localization analysis was performed on
the identified DEPs. DEPs in MCF-7 cells were located mainly in the
nucleus (n=63), cytoplasm (n=55) and plasma membrane (n=24). DEPs
in MDA-MB-231 cells were located predominantly in the nucleus
(n=49), cytoplasm (n=47) and plasma membrane (n=34). Therefore, the
nucleus and cytoplasm are the regions where DEPs are predominantly
localized in these two types of breast cancer cells (Fig. 5A and B). This is a major region of
protein synthesis and energy transfer, therefore alterations of
proteins within this region can affect the structure and biological
function of other relevant proteins and thus regulate breast cancer
progression.
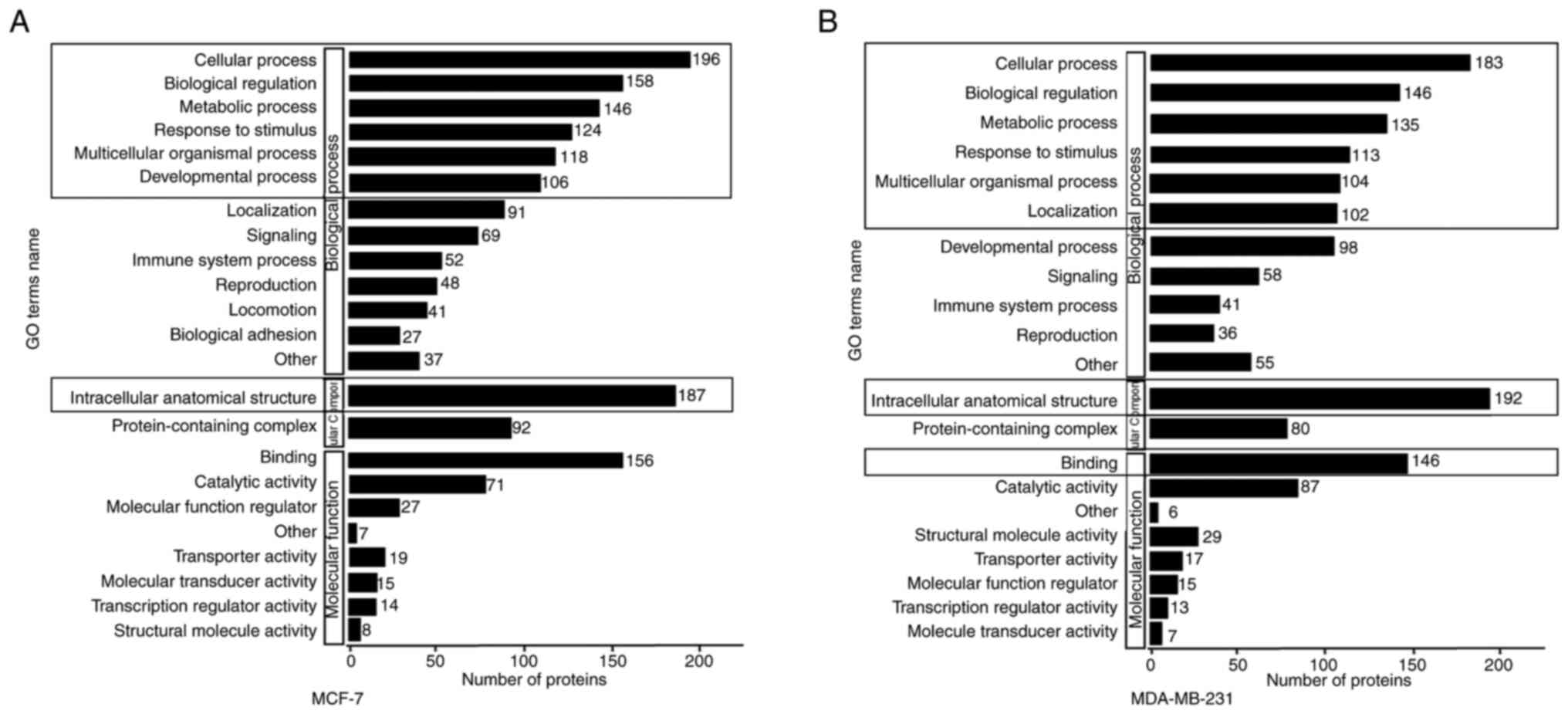
Gene Ontology (GO) secondary
classification map
The GO database (geneontology.org) is designed to
qualify and characterize gene and protein functions in three
categories: Biological process (BP), molecular function (MF) and
cellular component (CC). The y-axis represents the GO
classification, and the x-axis represents the number of proteins.
Fig. 6A and B shows the involvement
of >100 differentially expressed upregulated and downregulated
proteins in each of the three major classifications of the GO
database. The DEPs of the two breast cancer cell lines coregulate
BP processes such as ‘cellular process’, ‘biological regulation’,
‘metabolic process’, ‘response to stimulus’ and ‘multicellular
organismal processes’. These proteins are primarily localized to
the ‘intracellular anatomical structure’ and perform the MF of
‘binding’ (Fig. 6A and B).
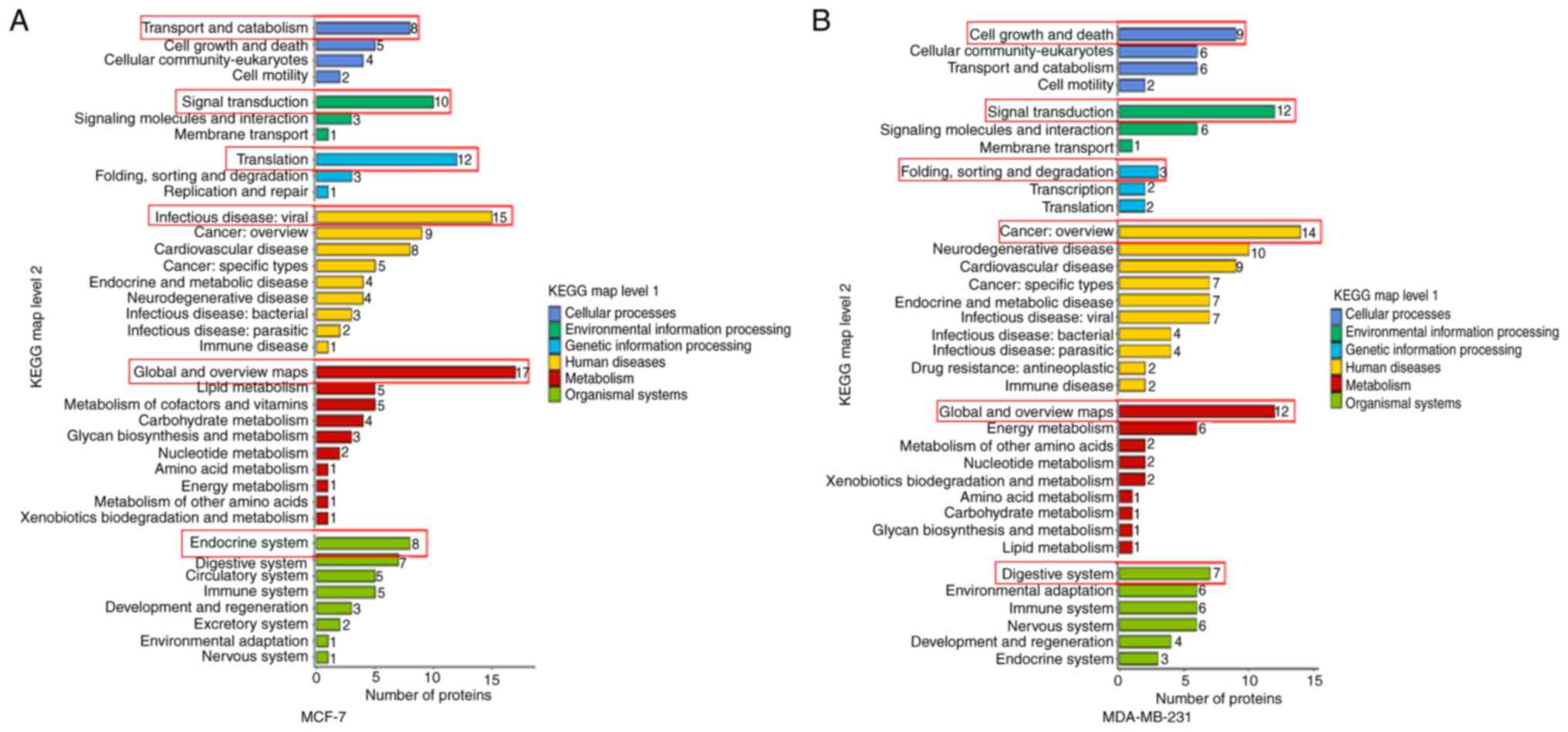
KEGG secondary classification map
As shown in Fig. 5,
MCF-7 and MDA-MB-231 cellular DEPs share common BPs and functions
across multiple KEGG secondary classifications. Most of these DEPs
were involved in the CP of ‘transport and catabolism’ and ‘cell
growth and death’; ‘signal transduction’ in EIP, suggesting a close
relationship with signaling pathways; ‘translation’ and ‘folding,
sorting and degradation’ in GIP, suggesting an association with
protein structure and function; ‘infectious disease: viral’ and
‘cancer: overview’ in HD; ‘global and overview maps’ in metabolism;
and the ‘endocrine system’ and ‘digestive system’ in OS (Fig. 7A and B). The description involved
the process of protein digestion and absorption.
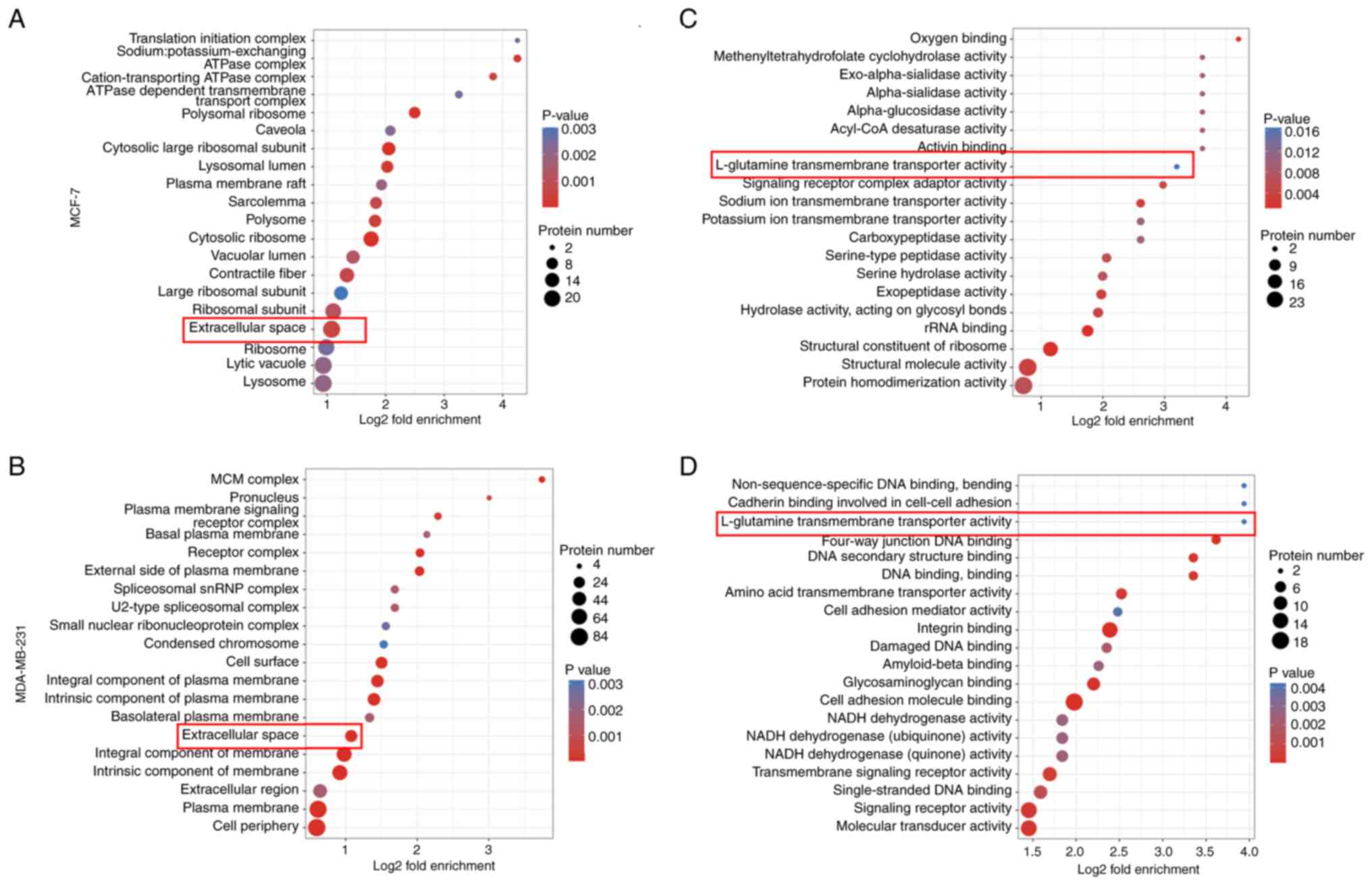
GO and KEGG pathway enrichment
analysis
To understand the functions of the DEPs, the GO
database was used to classify the enrichment of the screened DEPs.
The DEPs identified in MCF-7 cells were enriched mainly at the BP
level in ‘ribosome biogenesis’ and ‘protein localization to the
membrane’. The BP process of DEPs identified in MDA-MB-231 cells
was enriched mainly in the ‘cell surface receptor signaling
pathway’ and ‘regulation of the defense response’. Notably, the
common molecular functions of DEPs in both breast cancer cell lines
at the CC and MF levels, are mainly seen in the ‘extracellular
space’ and ‘L-glutamine transmembrane transporter activity’
(Fig. 8A-D). This suggests that
there are similar signaling pathway functions of the DEPs of two
si-FoxO3a breast cancer cell types and they are associated with
amino acid transport.
The results of KEGG enrichment analysis demonstrated
that DEPs in two si-FoxO3a breast cancer cell lines after treatment
with two drugs were enriched mainly in the ‘map04974 protein
digestion and absorption’ and ‘map04978 mineral absorption’
signaling pathways (Fig. 9A and B).
Considering the notable role that protein levels serve in the
development of breast cancer, and the combined subcellular
localization analysis and GO enrichment results, primarily
targeting the activity of ‘L-glutamine transmembrane transporter
activity’, demonstrated the role of transport proteins, the present
study focused on the preceding signaling pathway (map04974).
FoxO3a regulates protein digestion and
absorption signaling pathway-related proteins
Protein-protein interaction (PPI) network mapping
was performed for common DEPs and DEPs involved in apoptotic
processes, together with FoxO3a, in two types of breast cancer
cells. The findings indicated that the DEPs are closely related to
each other and are involved in the regulation of protein digestion
and absorption signaling pathways and apoptotic processes.
-Therefore, DEPs identified in MCF-7 and MDA-MB-231 cells involved
in the protein digestion and absorption signaling pathway and
apoptosis, were mapped together with FoxO3a. This was performed
using the STRING database to identify potential relationships
between the proteins (Fig. 9C).
Western blotting for verification of
the DEPs
The DEPs identified in the two types of breast
cancer cells were validated using western blotting, which revealed
that TPBG were downregulated and that p-FoxO3a levels were
increased in si-FoxO3a cells compared with si-NC cells. ASNS was
upregulated in MCF-7 cells and downregulated in MDA-MB-231 cells
treated with si-FoxO3a. In addition, ZYX was upregulated in MCF-7
cells, and downregulated in MDA-MB-231 cells. The validation
results were consistent with the proteomics results (Fig. 9D and E).
Discussion
Breast cancer, a malignant tumor with a high
incidence and mortality rate, is a serious threat to women's lives
and health (24). Drug resistance,
recurrence and metastasis affect its treatment and prognosis. Thus,
the search for reliable molecular markers can aid in the
implementation of targeted therapy strategies for breast cancer, to
overcome resistance to existing chemotherapeutic agents. The
estrogen receptor antagonist TAM can be used as a first-line
therapeutic agent as ~70% of breast cancers are estrogen
receptor-positive. DOX is one of the most active agents against
metastatic breast cancer cells, especially TNBC, and targeting
tumor cells by loading DOX-carrying liposomes onto the surface of
macrophages can enhance antitumor immune responses (25). Some side effects are associated with
prolonged drug use, and numerous efforts have been made by
researchers to mitigate these effects and improve their efficacy
(26). Existing studies have
demonstrated that the sirtuin 1/AKT pathway can overcome DOX
treatment resistance (27).
Exploring signaling pathways may be an effective approach to
improving drug efficacy. FoxO3a has gradually attracted the
attention of researchers as a transcription factor that has tumor
suppressor potential and is involved in multiple signaling
pathways. We have previously demonstrated its notable regulatory
role in a variety of malignant tumors (28), mainly involving regulating BPs such
as apoptosis, autophagy, DNA damage repair and angiogenesis to
influence tumor progression (29–31).
Our previous experiments revealed that FoxO3a can be altered during
drug-induced autophagy in MCF-7 cells (21). Therefore, the present study analyzed
the changes in signaling pathway proteins that occur when TAM and
DOX cause apoptosis in cancer cells. This was performed using
proteomic analysis of si-FoxO3a treated cells to identify the
metastatic process and provide a scientific basis for overcoming
therapeutic resistance.
FoxO3a expression was assessed in two breast cancer
cell lines before being successfully knocked down and evaluated
using RT-qPCR. Control and FoxO3a knockdown cell models of
estrogen-dependent MCF-7 cells and triple-negative MDA-MB-231
cells, were denoted as si-NC and si-FoxO3a, respectively. The
concentrations of 25 and 30 µmol/ml for TAM and DOX, respectively,
were determined using a WST-1 assay. The results demonstrated that
si-FoxO3a cells were more resistant to drug-induced apoptosis in
breast cancer cells compared with the si-NC cells, suggesting that
interference with FoxO3a can reduce cancer cell death and increase
their migration rate. The upper right quadrant of the flow
cytometry results may reflect a mixture of late apoptotic cells and
necrotic cells. Therefore, the present study further verified the
level of apoptosis using Hoechst staining, indicating the FoxO3a
transcription factor promotes drug-induced apoptosis in breast
cancer cells. This should be validated using FoxO3a overexpression
vectors in the future.
A study by Li et al (32) demonstrated that FoxO3a can be used
as a potential therapeutic target for breast cancer; therefore, the
development of FoxO3a-targeted drugs shows promise. Most of the
DEPs were localized in the nucleus and cytoplasm regions, which are
also the main regions for protein synthesis and energy transfer. GO
enrichment analysis revealed common molecular functions of DEPs in
both breast cancer cell lines at the CC and MF levels, mainly the
extracellular space and L-glutamine (Gln) transmembrane transporter
activity. These findings suggest that the DEPs are involved in the
same signaling pathways of both si-FoxO3a breast cancer cell lines
and are related to amino acid transport. This suggests modulating
associated signaling pathway proteins by knocking down FoxO3a may
be a key mechanism for drug-induced apoptosis in breast cancer
cells. KEGG enrichment analysis indicated that the two si-FoxO3a
breast cancer cell lines inhibited drug-induced apoptosis through
the protein digestion and absorption signaling pathway. DEPs were
selected to map the PPI network together with FoxO3a, and the
connections between the protein and FoxO3a were assessed. The
regulation of apoptosis in breast cancer cells following knock down
of FoxO3a expression may involve other or parallel pathways. As
indicated by GO and KEGG secondary classifications, differentially
expressed proteins in the two breast cancer cell lines jointly
regulate processes such as ‘biological regulation’, ‘metabolic
processes’, and ‘stimulus response’. Additionally, they are
involved in ‘translation’ and ‘folding, sorting, and degradation’
in GIP. These processes are largely unrelated to mineral
absorption, strongly suggesting an association with protein
digestion and absorption functions. Therefore, the validation of
proteins related to the protein digestion and absorption signaling
pathway was primarily focused on. DEPs TPBG, SLC38A2, ASNS, ZYX and
p-FoxO3a were selected for subsequent validation using western
blotting, which yielded results consistent with those of the
proteomic analysis. After FoxO3a is phosphorylated by kinases such
as AKT, it is translocated from the cell nucleus, leading to loss
of activity and reduced expression. At this point, p-FoxO3a levels
increase, and the AKT signaling pathway is generally activated.
After FoxO3a is phosphorylated by kinases such as AKT, it
translocated from the cell nucleus, leading to loss of activity and
reduced expression.
The end products of protein digestion are amino
acids, as seen in the cellular pathway diagram (Fig. 10) and the protein digestion and
absorption pathway is mainly involved in the regulation of multiple
amino acid transport proteins. The activation of cell signaling
pathways is closely related to protein hydrolysis and pro-apoptotic
processes, while the activation of protein digestion and absorption
signaling pathways is mainly related to the levels of various amino
acid transport proteins (33). TAM
and DOX can cause mitochondrial damage and thus induce apoptosis in
breast cancer cells. si-FoxO3a can impair this effect, as when Gln
is transported into the cell via SLC38A2, it inhibits the levels of
FoxO3a, research indicates that reduced FoxO3a expression leads to
excessive glutamine consumption by activated T cells, promoting the
tricarboxylic acid cycle and activating mTOR signaling to influence
transport processes (34).
Downregulating ASNS expression inhibits asparagine synthesis and
its downstream mTOR pathway to suppress TNBC progression, while
suppression of the mTOR signaling pathway affects FoxO3a levels
(35); therefore, si-FoxO3a also
affects ASNS expression by regulating the metabolism of various
amino acids. Furthermore, it may directly affect the levels of ZYX
and TPBG, and indirectly affects the levels of Wnt signaling, by
regulating mTOR (36). This
indicates a close relationship between FoxO3a and the protein
digestion and absorption signaling pathway. TPBG, SLC38A2, ASNS and
ZYX are associated with protein digestion and absorption signaling
pathways, hence these proteins were selected for validation.
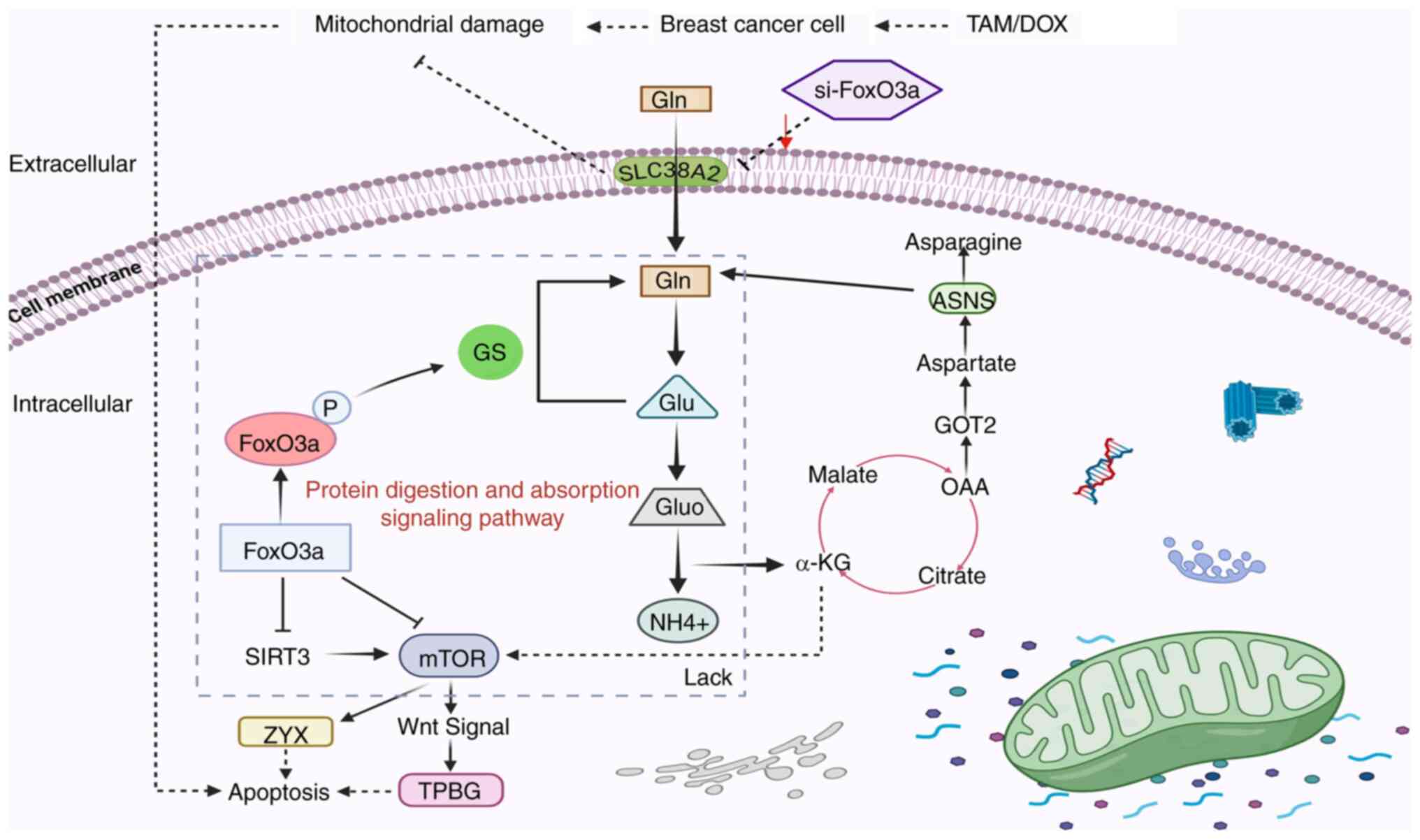 | Figure 10.FoxO3a regulates the protein
digestion and absorption signaling pathway. Antitumor drugs act on
breast cancer cells, inducing mitochondrial damage and triggering
the internal apoptotic program of cancer cells. si-FoxO3a impedes
the mitochondrial damage process and affects Gln transport into the
cell by decreasing the expression of the amino acid transport
protein SLC38A2. This leads to a decrease in the conversion of Gln
to glutamate, a decrease in the tricarboxylic acid cycle
intermediate product OAA and the catalytic function of ASNS cannot
be effectively performed. α-KG deficiency decreases mTOR
expression. FoxO3a suppresses mTOR expression, thereby inhibiting
ZYX expression and promoting cancer cell apoptosis. FoxO3 also
affects TPBG levels through regulation of the Wnt signaling pathway
to promote apoptosis in cancer cells. si, small interfering;
SLC38A2, solute carrier family 38 member 2; ASNS, asparagine
synthetase; α-KG, α-ketoglutarate; mTOR, mechanistic target of
rapamycin kinase; TPBG, trophoblastic glycoprotein; ZYX, zyxin;
Gluo, glutamate dehydrogenase; OAA, oxaloacetic acid; GS, glutamine
synthetase. |
Fig. 10 shows a
hypothetical model by which the aforementioned DEPs may participate
in the apoptosis process of breast cancer cells. The effect of
FoxO3a on SLC38A2 and TPBG requires further experimental
validation. TPBG is a transmembrane protein highly expressed mainly
in trophoblast and cancer cells (37). It promotes pancreatic cancer cell
metastasis through the Wnt signaling pathway, affecting the Wnt
downstream protein signaling cascade, causing increased cancer cell
migration and reducing apoptosis (38). It also induces lung cancer cell
invasion and epithelial-mesenchymal transition by inhibiting the
levels of microRNA-15b present (39). The ectopic expression of TPBG
markedly promotes the migration and invasive ability of two TNBC
lines (40). Studies have
demonstrated that TPBG is a therapeutic target for several
anticancer drugs currently in clinical development, mainly because
of its high expression in tumors and low expression in normal
tissues (41,42).
Tumor cells usually take up large quantities of
amino acids from the extracellular environment to sustain
proliferation and metastasis. Therefore, the upregulation of
certain amino acid transporter proteins to obtain large amounts of
amino acids acts as an important source of energy for tumor cells
(43). The SLC family of proteins
contains dozens of members that transport different amino acid
types (44). The membrane
transporter protein SLC38A2 is mainly responsible for the transport
of neutral amino acids, such as Gln (33), and is involved in a range of energy
metabolism processes. Therefore, the upregulation of SLC38A2 may
help maintain enlarged cell volume to sustain proliferation, thus
reducing cell apoptosis. Meanwhile, increased SLC38A2 expression
promotes glutamine-dependent cell proliferation and is associated
with low survival and resistance to TAM in patients with TNBC
(45,46).
Additional studies have demonstrated that the
development of drugs targeting SLC38A2 may be of value in some
hormone-resistant breast cancers (45,47),
demonstrating the importance of its role in future clinical
applications. ASNS uses glutamate to provide a nitrogen source to
enable aspartic acid to synthesize asparagine (48), and studies have demonstrated that
its high expression promotes breast cancer metastasis and
colorectal cancer progression (49,50).
The downregulation of ASNS inhibits the induction of cell cycle
arrest and suppresses cell proliferation (51). With this, its expression levels are
associated with that of apoptosis, thus harboring the potential to
serve as a prognostic marker for breast cancer (52). ASNS inhibition is essential for
targeting asparagine metabolism in cancer (53). Previous studies have demonstrated
that elevated asparagine levels catalyzed by ASNS are key for
breast cancer metastasis (53,54).
Inhibition of the Gln-associated pathway does not reduce the
viability of all cancer cell lines, even in Gln-dependent TNBC cell
lines (55). Therefore, the low
expression of ASNS in si-FoxO3a MDA-MB-231 cells may be due to the
substitution of other amino acids for Gln, which serve a role in
maintaining the growth of breast cancer cells, resulting in reduced
levels of ASNS. This is a hypothesis; therefore, further
verification is ultimately required.
ZYX is mainly responsible for the interaction
between cells and the extracellular matrix and is involved in the
organization and regeneration of the cytoskeleton. It is also
involved in in the regulation of adhesion-mediated extracellular
matrix signaling (56) to influence
the downstream pathways related to cell migration, invasion and
apoptosis. ZYX functions not only as an oncogenic protein, but also
as an antitumor protein in carcinogenesis (57). On one hand, ZYX promotes the
progression of hepatocellular carcinoma by activating the AKT/mTOR
signaling pathway (58). ZYX acts
as an inhibitor of the growth of colorectal cancer cells and is
associated with the levels of p53 (59). Therefore, its role in different
types of breast cancer warrants further investigation (60).
It is important to acknowledge the limitations of
the present study, as rescue experiments could not be performed due
to particular constraints, and only two cell lines were available
for experimentation. Additionally, due to a lack of in vivo
experiments to differentially validate the relationship between the
proteins, further exploration is needed to achieve this in the
future. Further limitations that should also be noted include the
need for verification of the association between FoxO3a and
proteins such as SLC38A2 and TPBG. This requires more exploration
in future studies. In addition, the proteomics analysis section of
the present study was performed by an external service provider.
The P-values reported were not corrected, and the lack of multiple
testing correction may have resulted in a higher false positive
rate.
Breast cancer development involves numerous proteins
and signaling molecules, and the results of the present study
indicate DEPs after knockdown of FoxO3a are involved in the amino
acid transport pathway and mainly serve a role in the protein
digestion and absorption signaling pathway (Fig. 10). In conclusion, knocking down
FoxO3a inhibits drug-induced apoptosis in breast cancer cells and
alters expression levels of proteins in the protein digestion and
absorption signaling pathways.
Acknowledgements
Not applicable.
Funding
Financial support was received for the research and/or
publication of the present article. Funding was received from the
Natural Science Project of Jilin Provincial Department of Science
and Technology (grant no. YDZJ202401085ZYTS), the Jilin Provincial
Department of Education Science and Technology Research Project
(grant no. JJKH20220460KJ), the Mekong College Student Scientific
Research and Innovation Fund Program (grant no. 2023JYMKZ001), and
the Jilin Province College Student Innovation and Entrepreneurship
Project (grant no. S202413706001).
Availability of data and materials
The data generated in the present study may be found
in ProteomeXchange via the iProX partner repository under accession
number PXD068299 or at the following URL: https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD068299.
Authors' contributions
LS and ZW wrote the manuscript, and designed and
performed the experiments. YL, JL, HT and XS performed the
experiments. CH, YZ, LC and SC analyzed the data. SC was involved
in the conception and design of the study. LS and ZW confirm the
authenticity of all the raw data. All authors reviewed the results,
and read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Katsura C, Ogunmwonyi I, Kankam HK and
Saha S: Breast cancer: Presentation, investigation and management.
Br J Hosp Med (Lond). 83:1–7. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Avti PK, Singh J, Dahiya D and Khanduja
KL: Dual functionality of pyrimidine and flavone in targeting
genomic variants of EGFR and ER receptors to influence the
differential survival rates in breast cancer patients. Integr Biol
(Camb). 15:zyad0142023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fortin J, Leblanc M, Elgbeili G, Cordova
MJ, Marin MF and Brunet A: The mental health impacts of receiving a
breast cancer diagnosis: A meta-analysis. Br J Cancer.
125:1582–1592. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ye F, Dewanjee S, Li Y, Jha NK, Chen ZS,
Kumar A, Vishakha Behl T, Jha SK and Tang H: Advancements in
clinical aspects of targeted therapy and immunotherapy in breast
cancer. Mol Cancer. 22:1052023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Castellote-Huguet P, Ruiz-Espana S,
Galan-Auge C, Santabarbara JM, Maceira AM and Moratal D: Breast
cancer diagnosis using texture and shape features in MRI. Annu Int
Conf IEEE Eng Med Biol Soc. 2023:1–4. 2023. View Article : Google Scholar
|
|
6
|
Nunnery SE and Mayer IA: Targeting the
PI3K/AKT/mTOR pathway in hormone-positive breast cancer. Drugs.
80:1685–1697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kotsopoulos J, Gronwald J, Huzarski T,
Aeilts A, Armel SR, Karlan B, Singer CF, Eisen A, Tung N, Olopade
O, et al: Tamoxifen and the risk of breast cancer in women with a
BRCA1 or BRCA2 mutation. Breast Cancer Res Treat. 201:257–264.
2023. View Article : Google Scholar
|
|
8
|
Masci D, Naro C, Puxeddu M, Urbani A,
Sette C, La Regina G and Silvestri R: Recent advances in drug
discovery for triple-negative breast cancer treatment. Molecules.
28:75132023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu C, Feng Q, Yang H, Wang G, Huang L, Bai
Q, Zhang C, Wang Y, Chen Y, Cheng Q, et al: A light-triggered
mesenchymal stem cell delivery system for photoacoustic imaging and
chemo-photothermal therapy of triple negative breast cancer. Adv
Sci (Weinh). 5:18003822018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hwang KT, Kim J, Jung J, Chang JH, Chai
YJ, Oh SW, Oh S, Kim YA, Park SB and Hwang KR: Impact of breast
cancer subtypes on prognosis of women with operable invasive breast
cancer: A population-based study using SEER database. Clin Cancer
Res. 25:1970–1979. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Al-Azhri J, Zhang Y, Bshara W, Zirpoli G,
McCann SE, Khoury T, Morrison CD, Edge SB, Ambrosone CB and Yao S:
Tumor expression of vitamin D receptor and breast cancer
histopathological characteristics and prognosis. Clin Cancer Res.
23:97–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Z, Zhou D, Yang J and Zhang D:
Doxorubicin promotes breast cancer cell migration and invasion via
DCAF13. FEBS Open Bio. 12:221–230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang D, Qiu T, Peng J, Li S, Tala Ren W,
Yang C, Wen Y, Chen CH, Sun J, et al: YB-1 is a positive regulator
of KLF5 transcription factor in basal-like breast cancer. Cell
Death Differ. 29:1283–1295. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katzenellenbogen BS, Guillen VS and
Katzenellenbogen JA: Targeting the oncogenic transcription factor
FOXM1 to improve outcomes in all subtypes of breast cancer. Breast
Cancer Res. 25:762023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Francois M, Donovan P and Fontaine F:
Modulating transcription factor activity: Interfering with
protein-protein interaction networks. Semin Cell Dev Biol.
99:12–19. 2020. View Article : Google Scholar
|
|
16
|
Liu TT, Yang H, Zhuo FF, Yang Z, Zhao MM,
Guo Q, Liu Y, Liu D, Zeng KW and Tu PF: Atypical E3 ligase ZFP91
promotes small-molecule-induced E2F2 transcription factor
degradation for cancer therapy. EBioMedicine. 86:1043532022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pang X, Zhou Z, Yu Z, Han L, Lin Z, Ao X,
Liu C, He Y, Ponnusamy M, Li P and Wang J: Foxo3a-dependent miR-633
regulates chemotherapeutic sensitivity in gastric cancer by
targeting Fas-associated death domain. RNA Biol. 16:233–248. 2019.
View Article : Google Scholar
|
|
18
|
Meng XY, Wang KJ, Ye SZ, Chen JF, Chen ZY,
Zhang ZY, Yin WQ, Jia XL, Li Y, Yu R and Ma Q: Sinularin stabilizes
FOXO3 protein to trigger prostate cancer cell intrinsic apoptosis.
Biochem Pharmacol. 220:1160112024. View Article : Google Scholar
|
|
19
|
Khoshinani HM, Afshar S, Pashaki AS,
Mahdavinezhad A, Nikzad S, Najafi R, Amini R, Gholami MH,
Khoshghadam A and Saidijam M: Involvement of miR-155/FOXO3a and
miR-222/PTEN in acquired radioresistance of colorectal cancer cell
line. Jpn J Radiol. 35:664–672. 2017. View Article : Google Scholar
|
|
20
|
Wolfe AR, Debeb BG, Lacerda L, Larson R,
Bambhroliya A, Huang X, Bertucci F, Finetti P, Birnbaum D, Van
Laere S, et al: Simvastatin prevents triple-negative breast cancer
metastasis in pre-clinical models through regulation of FOXO3a.
Breast Cancer Res Treat. 154:495–508. 2015. View Article : Google Scholar
|
|
21
|
Chen S, Li YQ, Yin XZ, Li SZ, Zhu YL, Fan
YY, Li WJ, Cui YL, Zhao J, Li X, et al: Recombinant adenoviruses
expressing apoptin suppress the growth of MCF-7 breast cancer cells
and affect cell autophagy. Oncol Rep. 41:2818–2832. 2019.PubMed/NCBI
|
|
22
|
Kang BG, Shende M, Inci G, Park SH, Jung
JS, Kim SB, Kim JH, Mo YW, Seo JH, Feng JH, et al: Combination of
metformin/efavirenz/fluoxetine exhibits profound anticancer
activity via a cancer cell-specific ROS amplification. Cancer Biol
Ther. 24:20–32. 2023.PubMed/NCBI
|
|
23
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
24
|
Khan MA, Sadaf Ahmad I, Aloliqi AA, Eisa
AA, Najm MZ, Habib M, Mustafa S, Massey S, Malik Z, et al: FOXO3
gene hypermethylation and its marked downregulation in breast
cancer cases: A study on female patients. Front Oncol.
12:10780512022. View Article : Google Scholar
|
|
25
|
Yang L, Zhang Y, Zhang Y, Xu Y, Li Y, Xie
Z, Wang H, Lin Y, Lin Q, Gong T, et al: Live macrophage-delivered
doxorubicin-loaded liposomes effectively treat triple-negative
breast cancer. ACS Nano. 16:9799–9809. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ansari L, Shiehzadeh F, Taherzadeh Z,
Nikoofal-Sahlabadi S, Momtazi-Borojeni AA, Sahebkar A and Eslami S:
The most prevalent side effects of pegylated liposomal doxorubicin
monotherapy in women with metastatic breast cancer: A systematic
review of clinical trials. Cancer Gene Ther. 24:189–193. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei Y, Guo Y, Zhou J, Dai K, Xu Q and Jin
X: Nicotinamide overcomes doxorubicin resistance of breast cancer
cells through deregulating SIRT1/Akt pathway. Anticancer Agents Med
Chem. 19:687–696. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Liu J, Bao D, Hu C, Zhao Y and Chen
S: Progress in the study of FOXO3a interacting with microRNA to
regulate tumourigenesis development. Front Oncol. 13:12939682023.
View Article : Google Scholar
|
|
29
|
Chen YF, Pandey S, Day CH, Chen YF, Jiang
AZ, Ho TJ, Chen RJ, Padma VV, Kuo WW and Huang CY: Synergistic
effect of HIF-1α and FoxO3a trigger cardiomyocyte apoptosis under
hyperglycemic ischemia condition. J Cell Physiol. 233:3660–3671.
2018. View Article : Google Scholar
|
|
30
|
Yadav RK, Chauhan AS, Zhuang L and Gan B:
FoxO transcription factors in cancer metabolism. Semin Cancer Biol.
50:65–76. 2018. View Article : Google Scholar
|
|
31
|
Liu H, Yin J, Wang H, Jiang G, Deng M,
Zhang G, Bu X, Cai S, Du J and He Z: FOXO3a modulates WNT/β-catenin
signaling and suppresses epithelial-to-mesenchymal transition in
prostate cancer cells. Cell Signal. 27:510–518. 2015. View Article : Google Scholar
|
|
32
|
Li H, Tang X, Sun Z, Qu Z and Zou X:
Integrating bioinformatics and experimental models to investigate
the mechanism of the chelidonine-induced mitotic catastrophe via
the AKT/FOXO3/FOXM1 axis in breast cancer cells. Biomol Biomed.
24:560–574. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gauthier-Coles G, Bröer A, McLeod MD,
George AJ, Hannan RD and Bröer S: Identification and
characterization of a novel SNAT2 (SLC38A2) inhibitor reveals
synergy with glucose transport inhibition in cancer cells. Front
Pharmacol. 13:9630662022. View Article : Google Scholar
|
|
34
|
Petry ÉR, Dresch DF, Carvalho C, Medeiros
PC, Rosa TG, de Oliveira CM, Martins LAM, Guma FCR, Marroni NP and
Wannmacher CMD: Oral glutamine supplementation relieves muscle loss
in immobilized rats, altering p38MAPK and FOXO3a signaling
pathways. Nutrition. 118:1122732024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan Q, Wang Q, Li J, Yin L, Liu S, Zu X
and Shen Y: CCT196969 inhibits TNBC by targeting the
HDAC5/RXRA/ASNS axis to down-regulate asparagine synthesis. J Exp
Clin Cancer Res. 44:2312025. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao X, Lai H, Li G, Qin Y, Chen R, Labrie
M, Stommel JM, Mills GB, Ma D, Gao Q and Fang Y: Rictor
orchestrates β-catenin/FOXO balance by maintaining redox
homeostasis during development of ovarian cancer. Oncogene.
44:1820–1832. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Park S, Yoo JE, Yeon GB, Kim JH, Lee JS,
Choi SK, Hwang YG, Park CW, Cho MS, Kim J, et al: Trophoblast
glycoprotein is a new candidate gene for Parkinson's disease. NPJ
Parkinsons Dis. 7:1102021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
He P, Jiang S, Ma M, Wang Y, Li R, Fang F,
Tian G and Zhang Z: Trophoblast glycoprotein promotes pancreatic
ductal adenocarcinoma cell metastasis through Wnt/planar cell
polarity signaling. Mol Med Rep. 12:503–509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su H, Yu S, Sun F, Lin D, Liu P and Zhao
L: LINC00342 induces metastasis of lung adenocarcinoma by targeting
miR-15b/TPBG. Acta Biochim Pol. 69:291–297. 2022.
|
|
40
|
Ye F, Liang Y, Wang Y, Yang RL, Luo D, Li
Y, Jin Y, Han D, Chen B, Zhao W, et al: Cancer-associated
fibroblasts facilitate breast cancer progression through exosomal
circTBPL1-mediated intercellular communication. Cell Death Dis.
14:4712023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu G, Leal M, Lin Q, Affolter T, Sapra P,
Bates B and Damelin M: Phenotype of TPBG gene replacement in the
mouse and impact on the pharmacokinetics of an antibody-drug
conjugate. Mol Pharm. 12:1730–1737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Stern PL, Brazzatti J, Sawan S and McGinn
OJ: Understanding and exploiting 5T4 oncofoetal glycoprotein
expression. Semin Cancer Biol. 29:13–20. 2014. View Article : Google Scholar
|
|
43
|
Kandasamy P, Zlobec I, Nydegger DT,
Pujol-Giménez J, Bhardwaj R, Shirasawa S, Tsunoda T and Hediger MA:
Oncogenic KRAS mutations enhance amino acid uptake by colorectal
cancer cells via the hippo signaling effector YAP1. Mol Oncol.
15:2782–2800. 2021. View Article : Google Scholar
|
|
44
|
Kandasamy P, Gyimesi G, Kanai Y and
Hediger MA: Amino acid transporters revisited: New views in health
and disease. Trends Biochem Sci. 43:752–789. 2018. View Article : Google Scholar
|
|
45
|
Morotti M, Zois CE, El-Ansari R, Craze ML,
Rakha EA, Fan SJ, Valli A, Haider S, Goberdhan DCI, Green AR and
Harris AL: Increased expression of glutamine transporter
SNAT2/SLC38A2 promotes glutamine dependence and oxidative stress
resistance, and is associated with worse prognosis in
triple-negative breast cancer. Br J Cancer. 124:494–505. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Morotti M, Bridges E, Valli A, Choudhry H,
Sheldon H, Wigfield S, Gray N, Zois CE, Grimm F, Jones D, et al:
Hypoxia-induced switch in SNAT2/SLC38A2 regulation generates
endocrine resistance in breast cancer. Proc Natl Acad Sci USA.
116:12452–12461. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Stretton C, Lipina C, Hyde R, Cwiklinski
E, Hoffmann TM, Taylor PM and Hundal HS: CDK7 is a component of the
integrated stress response regulating SNAT2 (SLC38A2)/System A
adaptation in response to cellular amino acid deprivation. Biochim
Biophys Acta Mol Cell Res. 1866:978–991. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shen Y, Li M, Xiong Y, Gui S, Bai J, Zhang
Y and Li C: Proteomics analysis identified ASNS as a novel
biomarker for predicting recurrence of skull base chordoma. Front
Oncol. 11:6984972021. View Article : Google Scholar
|
|
49
|
Du F, Chen J, Liu H, Cai Y, Cao T, Han W,
Yi X, Qian M, Tian D, Nie Y, et al: SOX12 promotes colorectal
cancer cell proliferation and metastasis by regulating asparagine
synthesis. Cell Death Dis. 10:2392019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Knott SRV, Wagenblast E, Khan S, Kim SY,
Soto M, Wagner M, Turgeon MO, Fish L, Erard N, Gable AL, et al:
Asparagine bioavailability governs metastasis in a model of breast
cancer. Nature. 554:378–381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang H, He X, Zheng Y, Feng W, Xia X, Yu X
and Lin Z: Down-regulation of asparagine synthetase induces cell
cycle arrest and inhibits cell proliferation of breast cancer. Chem
Biol Drug Des. 84:578–584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qin C, Yang X and Zhan Z: High expression
of asparagine synthetase is associated with poor prognosis of
breast cancer in Chinese population. Cancer Biother Radiopharm.
35:581–585. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nishikawa G, Kawada K, Hanada K, Maekawa
H, Itatani Y, Miyoshi H, Taketo MM and Obama K: Targeting
asparagine synthetase in tumorgenicity using patient-derived
tumor-initiating cells. Cells. 11:32732022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen W, Qin Y, Qiao L, Liu X, Gao C, Li
TR, Luo Y, Li D, Yan H, Han L, et al: FAM50A drives breast cancer
brain metastasis through interaction with C9ORF78 to enhance
L-asparagine production. Sci Adv. 11:eadt30752025. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Reis LMD, Adamoski D, Souza RO, Ascenção
CF, de Oliveira KR, Corrêa-da-Silva F, de Sá, Patroni FM, Dias MM,
Consonni SR, de Moraes-Vieira PMM, et al: Dual inhibition of
glutaminase and carnitine palmitoyltransferase decreases growth and
migration of glutaminase inhibition-resistant triple-negative
breast cancer cells. J Biol Chem. 294:9342–9357. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Partynska A, Gomulkiewicz A, Dziegiel P
and Podhorska-Okolow M: The role of zyxin in carcinogenesis.
Anticancer Res. 40:5981–5988. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kotb A, Hyndman ME and Patel TR: The role
of zyxin in regulation of malignancies. Heliyon. 4:e006952018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cai T, Bai J, Tan P, Huang Z, Liu C, Wu Z,
Cheng Y, Li T, Chen Y, Ruan J, et al: Zyxin promotes hepatocellular
carcinoma progression via the activation of AKT/mTOR signaling
pathway. Oncol Res. 31:805–817. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mohammadi H, Shakiba E, Rostampour R,
Bahremand K, Goodarzi MT, Bashiri H, Ghobadi KN and Asadi S: Down
expression of zyxin is associated with down expression of p53 in
colorectal cancer. Int J Mol Cell Med. 14:461–471. 2025.PubMed/NCBI
|
|
60
|
Ma B, Cheng H, Gao R, Mu C, Chen L, Wu S,
Chen Q and Zhu Y: Zyxin-Siah2-Lats2 axis mediates cooperation
between Hippo and TGF-β signalling pathways. Nat Commun.
7:111232016. View Article : Google Scholar : PubMed/NCBI
|