Introduction
Primary liver cancer (PLC) is the sixth most common
malignant tumor worldwide (1), and
primarily includes hepatocellular carcinoma (HCC) and intrahepatic
cholangiocarcinoma. In China, PLC had the second highest mortality
rate among all cancers in 2020 (2).
HCC is the most prevalent type of PLC, accounting for ~90% of PLC
cases (3). The most common causes
of HCC include viral hepatitis, metabolic disorders, alcohol
consumption and smoking (4).
Surgical resection is the standard treatment for
patients with early stage PLC. Recurrence occurs in ~50% of
patients who undergo surgical resection within 2 years and the
5-year survival rate ranges between 50 and 75% (5). Patients with HCC frequently lack
noticeable clinical symptoms, resulting in delayed medical
attention, and missed opportunities for surgical resection or liver
transplantation. Systematic therapies, including immunotherapy,
targeted therapy, local radiotherapy and chemotherapy, are often
used in patients with intermediate-to advanced-stage disease
(6,7). However, drug resistance is the primary
cause of treatment failure and can lead to tumor recurrence or
metastasis (8). Once recurrence and
metastasis occur, the prognosis of patients with HCC decreases.
Therefore, high recurrence and metastasis rates due to therapeutic
resistance remain major clinical issues.
Cancer stem cells (CSCs) are a rare subpopulation of
tumor cells capable of self-renewal and differentiation. CSCs are
resistant to chemotherapy and radiotherapy, and are associated with
tumorigenesis, recurrence and metastasis (9–11).
CSCs exhibit characteristics that are distinct from those of other
cancer cells and several surface markers have been identified in
previous studies, including CD44, CD133, epithelial cell adhesion
molecule (EpCAM) and hepatic leukemia factor (12–15).
MicroRNAs (miRNAs/miRs) are short endogenous
non-coding RNAs that regulate gene expression by binding to target
mRNAs (16). Previous studies have
reported that miRNAs serve key roles in tumor cell proliferation,
migration and invasion (17,18).
However, potential miRNAs associated with HCC-CSCs and their roles
in tumor therapy resistance, recurrence and metastasis remain to be
elucidated.
The present study aimed to identify potential miRNAs
associated with HCC-CSCs through a systematic review, data mining
and bioinformatics analysis. Subsequently, the expression of
candidate miRNAs and their biological functions were explored in
HCC-CSCs.
Materials and methods
Systematic review
The present study was reported as per the Preferred
Reporting Items for Systematic Reviews and Meta-Analyses protocol
(19) and registered in the
International Prospective Register of Systematic Reviews
(CRD42024508526).
Search strategy
All of the research articles used in the present
study were gathered from two publicly available literature
databases, PubMed (https://pubmed.ncbi.nlm.nih.gov/) and Web of Science
(https://www.webofscience.com/wos/),
with a search range from the date of establishment of the database
to January 15, 2024. A search strategy was collaboratively
developed and independently executed by two authors based on the
Population, Intervention, Comparison, Outcome and Study type
(PICOS) (20). The formulated PICOS
framework was as follows: i) The population was composed of
patients with HCC; ii) the intervention was the altered expression
levels of miRNAs in patients; iii) the comparison was between
groups, one with high miRNA expression and the other with low miRNA
expression; iv) the outcome was the difference in survival
prognosis between patients with HCC with low and high miRNA
expression; and v) the study method was comparative research. The
key words and search details are shown in Table SI.
Eligibility criteria
Research articles were included based on the
following criteria: i) Demonstrated significantly different miRNA
expression levels in patients with HCC (P<0.05); ii) compared
the prognoses of high and low miRNA expression groups; and iii)
described a clear follow-up time for patients. Studies were
excluded based on the following criteria: i) Did not use human
subjects; ii) did not perform a comparative analysis; iii) did not
include HCC; iv) did not have available data that could be
extracted; or v) were not original research projects.
Study selection and data
extraction
After excluding review articles and eliminating
duplicate items, the literature search results were imported into
Rayyan (https://www.rayyan.ai/), an online tool
for systematic literature reviews. Independent screening of titles
and abstracts was conducted using inclusion and exclusion criteria.
The miRNAs mentioned in the target articles were extracted and
arranged in tables that included the first author, year, country,
number of patients, follow-up period, type of evidence, miRNA and
expression.
Data mining analysis
Coremine Medical (http://www.coremine.com/medical/), an online
data-mining tool, was used to explore potential miRNAs associated
with CSCs, metastasis, recurrence and drug resistance in HCC. Key
words including ‘liver carcinoma’, ‘neoplasm recurrence’, ‘neoplasm
metastasis’, ‘drug resistance’ and ‘neoplastic stem cells’ were
chosen as the search strategy, and the associated genes were
downloaded and screened for miRNAs with a significant difference
(P<0.05), based on the analysis role of the web tool.
Bioinformatics analysis
The ONCO.IO website (https://onco.io/)
was used to explore miRNA-target gene interactions and their
biological processes in HCC. A regulatory network diagram was
created to illustrate these interactions. The Enrichr database
(https://maayanlab.cloud/Enrichr/) was
used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis. Box plots on the Encyclopedia of RNA
Interactomes (ENCORI) website (https://rnasysu.com/encori/) indicated the
differential expression levels of miRNAs between cancerous and
paracancerous tissue samples in liver HCC (LIHC). Kaplan-Meier
survival curves (https://www.kmplot.com) were used to assess the
effects of different miRNA expression levels on the survival of
patients with LIHC.
Cell culture and CSC culture
The Huh7 cell line was purchased from The Cell Bank
of Type Culture Collection of The Chinese Academy of Sciences and
confirmed by short tandem repeat profiling without discrepancies.
The cells were cultured in DMEM (cat. no. 11965118; Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS [cat. no.
E01010; Eva (Suzhou) Biopharmaceutical Technology Co., Ltd.] and 1%
penicillin-streptomycin (cat. no. C100C5; Suzhou Xinsaimei
Biotechnology Co., Ltd.). Cell culture was performed at 37°C in a
5% CO2 incubator. HCC-CSCs were developed from Huh7
cells using the protocol as described previously (21). The Huh7 cell line was cultured under
ultra-low attachment conditions with DMEM supplemented with 1%
penicillin-streptomycin, 0.5% N2 supplement (cat. no. 17502-048;
Gibco; Thermo Fisher Scientific, Inc.), 20 ng/ml basic fibroblast
growth factor (bFGF; cat. no. HY-P7004; MedChemExpress) and 20
ng/ml epidermal growth factor (EGF; cat. no. HY-P7109;
MedChemExpress) at 37°C in a 5% CO2 incubator for 5–7
days.
Tumor sphere assays and sphere
formation efficiency (SFE)
A total of 2,000 cells in the logarithmic growth
phase were selected, seeded into 24-well ultra-low adsorption
culture plates and cultured in DMEM supplemented with 1%
penicillin-streptomycin, 0.5% N2 supplement, 20 ng/ml bFGF, and 20
ng/ml EGF at 37°C in a 5% CO2 cell culture incubator for
6 days. Spherical cells were defined as those with diameters ≥100
µm. Imaging was performed using a Motic AE31E inverted microscope
(Motic Industrial Group Co., Ltd.). SFE was calculated on day 6
using the following formula: SFE=(number of spheres counted/number
of seeded cells) ×100.
miRNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
miRNAs were extracted and purified from Huh7 and
Huh7-CSCs using the miRcute miRNA Isolation Kit (cat. no. DP501;
Tiangen Biotech Co., Ltd.) according to the manufacturer's
protocol. The concentration and purity of the miRNAs were measured
using a NanoDrop 2000 spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc.). The RT-qPCR detection of miRNA was performed
using the poly(A) tailing-based method. RT was performed using the
miRNA 1st Strand cDNA Synthesis Kit (Tailing A; cat. no. MR201-01;
Vazyme Biotech Co., Ltd.). The reverse transcription for miRNA was
carried out at 37°C for 60 min, followed by an enzyme inactivation
step at 85°C for 5 sec. qPCR was performed using a Tap Pro
Universal SYBR™ qPCR Master Mix kit (cat. no. Q712-02;
Vazyme Biotech Co., Ltd.). The qPCR protocol consisted of an
initial denaturation step (95°C for 30 sec), followed by 40
amplification cycles (denaturation at 95°C for 30 sec and
annealing/extension at 60°C for 30 sec), and concluded with a melt
curve analysis step (95°C for 15 sec, 60°C for 60 sec and 95°C for
15 sec). U6 was used as the endogenous control and the
2−ΔΔCq method was used to analyze expression levels
(22). The primer sequences of the
detected genes were as follows: Hsa-miR-17-5p-q forward (F),
5′-GCCGCAAAGTGCTTACAGTGC-3′ and the reverse (R) primer for
miR-17-5p was the Universal reverse Q primer from the miRNA 1st
Strand cDNA Synthesis Kit; U6 F, 5′-CTCGCTTCGGCAGCACA-3′ and R,
5′-AACGCTTCACGAATTTGCGT-3′.
Statistical analysis
Statistical analyses were performed using the
Coremine, ONCO.IO, Enrichr, ENCORI and Kaplan-Meier plotter
databases. The ENCORI pan-cancer dataset (version 2024), comprising
10,546 samples from The Cancer Genome Atlas (TCGA), was analyzed.
The best-performing percentile was automatically selected as the
cutoff. The miRNA expression levels were quantified as reads per
million (RPM) and transformed using log2 (RPM + 0.01) to
normalize the data distribution. The RT-qPCR data are presented as
the mean ± Standard Error of the Mean and each experiment was
performed in triplicate unless otherwise noted. Differences in the
RT-qPCR results between the two groups were analyzed using unpaired
Student's t-test. Unless specified otherwise, P<0.05 was
considered to indicate a statistically significant difference.
Results
Study characteristics
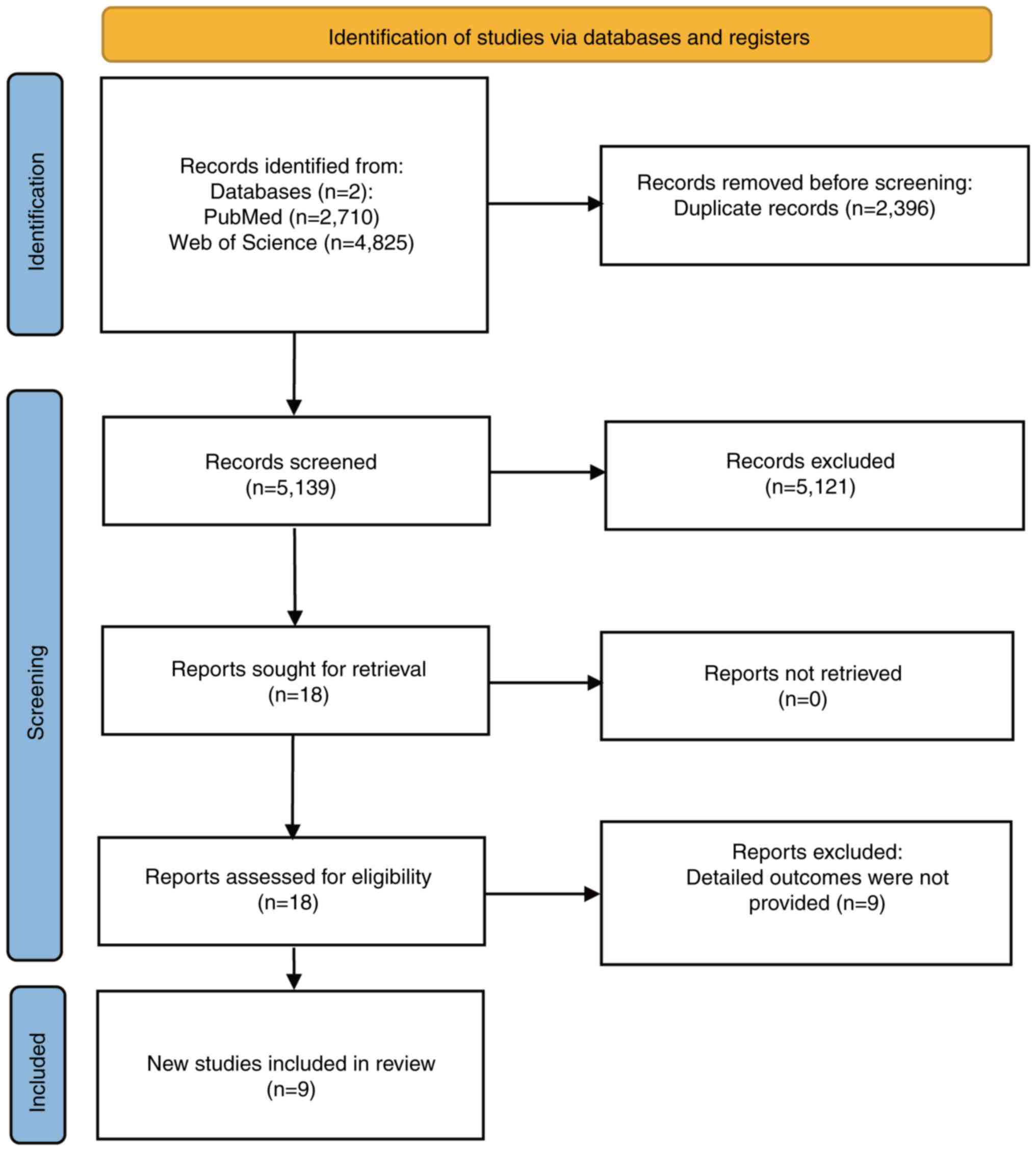
The initial literature search retrieved 7,535
research articles, with 2,710 from PubMed and 4,825 from Web of
Science. After further screening, 2,396 duplicates were removed,
and the titles and abstracts of the remaining 5,139 articles were
reviewed. A total of 5,121 articles failed to meet the inclusion
criteria, resulting in 18 articles selected for full reading and
review. Nine additional articles were excluded because they did not
meet the inclusion criteria and the remaining nine were included in
the study (23–31). The screening process is illustrated
in Fig. 1. These studies focused on
nine different miRNAs collected from the serum and tissues of
patients with HCC in China and South Korea. All nine miRNAs were
reported to have notable prognostic capability for patients with
HCC. The details of the nine studies are shown in Table I encompassing 1,318 patients with
HCC collectively.
 | Table I.Extracted data of the nine articles
selected for the systematic review. |
Table I.
Extracted data of the nine articles
selected for the systematic review.
| First author,
year | Country | No. of patients
(male, female, unknown) | Follow-up
period | Type of evidence
(95% CI) | miRNA | Expression | (Refs.) |
|---|
| Xie et al,
2018 | China | 149 (117, 30,
2) | 150 months | OS: HR, 0.072
(0.033–0.159); | miR-33a | Downregulated | (23) |
|
|
|
|
| P<0.001 PFS: HR,
0.194 |
| (HCC tissues) |
|
|
|
|
|
| (0.118–0.317);
P<0.001 |
|
|
|
| Zhuang et
al, 2015 | China | 182 (155, 27) | 656±393 days | OS: HR, 2.793
(1.550–5.033); | miR-128-2 | Upregulated | (24) |
|
|
|
|
| P=0.001 |
| (HCC serum) |
|
| Sun et al,
2015 | China | 60 (40, 20) | >24 months | DFS: HR, 2.681
(1.306–5.504); | miR-9 | Upregulated | (25) |
|
|
|
|
| P=0.007 |
| (HCC tissues) |
|
| Chen et al,
2012 | China | 66 (56, 10) | 100 months | OS: HR, 0.332
(0.139–0.793); | miR-203 | Downregulated | (26) |
|
|
|
|
| P=0.013 RFS: HR,
0.202 |
| (HCC tissues) |
|
|
|
|
|
| (0.064–0.638);
P=0.006 |
|
|
|
| Zhou et al,
2016 | China | 38 (30, 8) | 25 months | DFS: RR, 3.273
(1.107–9.679); | miR-375 | Downregulated | (27) |
|
|
|
|
| P=0.032 |
| (HCC tissues) |
|
| Luo et al,
2019 | China | 148 (100, 48) | 24 months | OS: HR, 2.226
(1.235–4.012); | miR-200c | Downregulated | (28) |
|
|
|
|
| P=0.008 RFS:
2.662 |
| (HCC tissues) |
|
|
|
|
|
| (1.618–4.38);
P<0.001 |
|
|
|
| Ha et al,
2019 | South | 289 (238, 51) | 151.4 months | IHRFS: HR, 1.89
(1.16–3.07); | miR-122 | Downregulated | (29) |
|
| Korea |
|
| P=0.010 DMFS: HR,
2.14 |
| (HCC tissues) |
|
|
|
|
|
| (1.05–4.36);
P=0.036 |
|
|
|
|
|
|
|
| RFS: HR, 2.17
(1.34–3.52); |
|
|
|
|
|
|
|
| P=0.002 |
|
|
|
| Zhu et al,
2012 | China | 266 (225, 41) | 81 months | OS: HR, 1.0
(0.7–1.4); P=0.901 | miR-29a-5p | Upregulated | (30) |
|
|
|
|
| TTR: HR, 0.5
(0.3–0.8); |
| (HCC tissues) |
|
|
|
|
|
| P=0.003 |
|
|
|
| Chen et al,
2012 | China | 120 (102, 18) | 46 months | OS: RR, 4.96
(1.78–13.82); | miR-17-5p | Upregulated | (31) |
|
|
|
|
| P=0.002 DFS: RR,
1.79 |
| (HCC tissues) |
|
|
|
|
|
| (1.14–2.98);
P=0.042 |
|
|
|
miR-17-5p: Potential miRNA associated
with CSCs, drug resistance, recurrence and metastasis in HCC
A total of 34 miRNAs, including miR-7-3HG, miR-100,
miR-25, miR-26B, miR-200A, miR-184, miR-190A, miR-21,miR-186,
miR-19A, miR-17, miR-196B, miR-22, Let-7d, miR-145, miR-183,
miR-34A,miR-17HG, miR-185, miR-107, Let-7c, miR-203A, miR-137,
miR-96, miR-191, miR-98, miR-4435-2HG, Let-7b, miR-31, miR-93,
miR-146A, miR-15B, miR-221, miR-192, associated with HCC, neoplasm
recurrence, neoplasm metastasis, drug resistance and CSCs were
identified in the Coremine database. The intersecting miRNA,
miR-17-5p, was selected by comparing the nine miRNAs from the
included studies with the 34 miRNAs identified in the Coremine
database (Fig. 2A). The ENCORI tool
demonstrated that miR-17-5p was expressed at significantly higher
levels in 370 cancerous tissues compared with that in 50 para-tumor
tissues (fold change, 2.08; Fig.
2B). Survival analysis of miR-17-5p, which included 163
patients who were followed-up for 48 months, was performed using
the Kaplan-Meier plotter online tool. The results of this analysis
revealed a significant association between miR-17-5p expression and
overall survival (months) in patients with HCC (Fig. 2C).
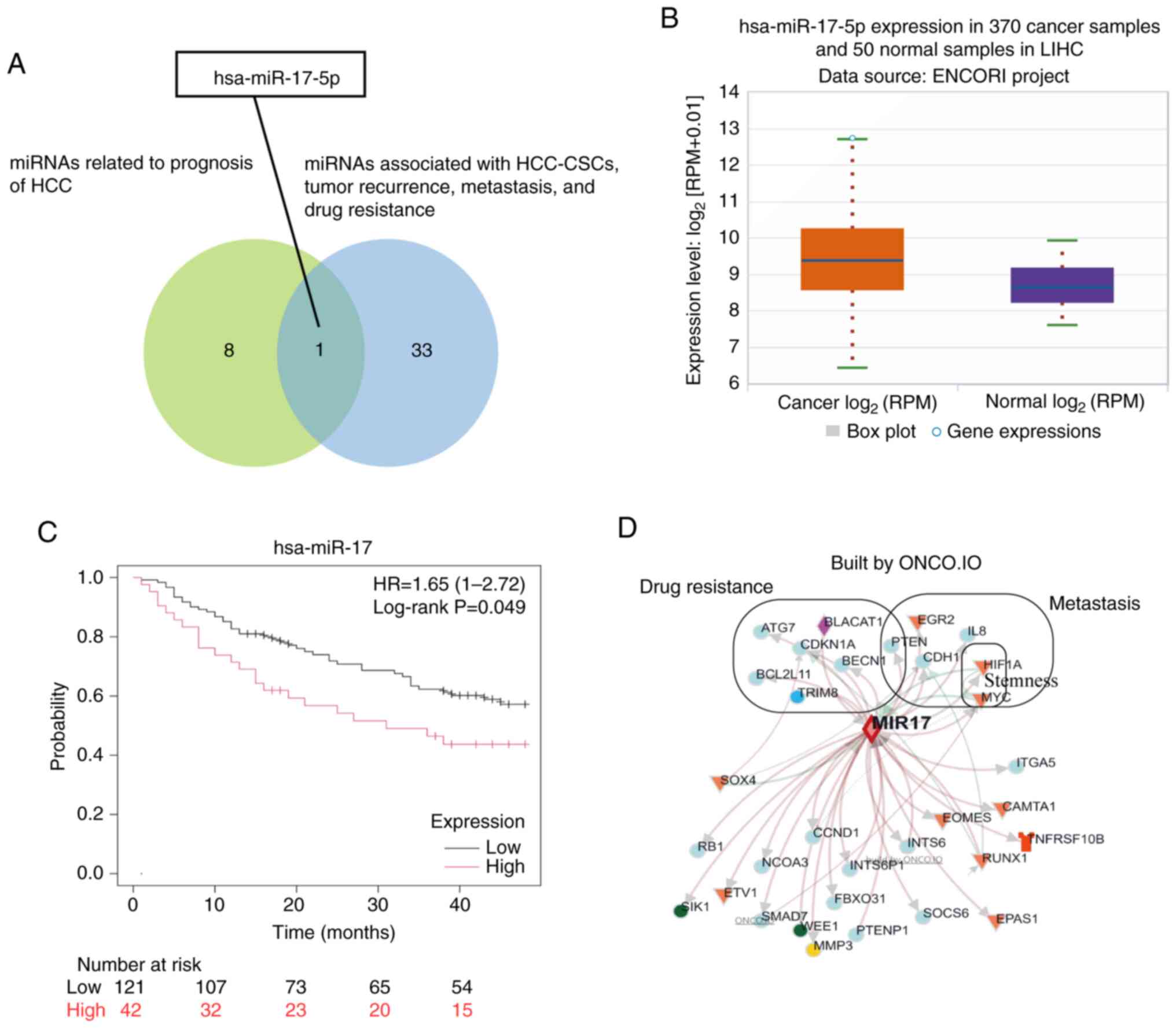 | Figure 2.miR-17-5p is a miRNA potentially
associated with CSCs, metastasis, recurrence and drug resistance in
HCC. (A) Venn diagram illustrating how the potential miRNA
(miR-17-5p) was selected by exploring the intersection between the
nine miRNAs associated with the prognosis of HCC, and the 34 miRNAs
associated with HCC-CSCs, metastasis, tumor recurrence and drug
resistance. (B) Box plot of miR-17-5p expression in LIHC. (C)
Survival analysis of miR-17-5p in LIHC. (D) Interaction network of
targeted genes of miR-17-5p, with the 12 genes involved in
metastasis, stemness and drug resistance specially marked.
miR/miRNA, microRNA; CSC, cancer stem cell; HCC, hepatocellular
carcinoma; LIHC, liver hepatocellular carcinoma; RPM, reads per
million. |
The interaction network of the target genes
regulated by miR-17-5p is shown in Fig.
2D. These 12 genes were directly identified through functional
analysis (pathogenic processes) using the ONCO.IO platform,
including two stemness-associated genes, seven drug
resistance-associated genes and six metastasis-associated genes.
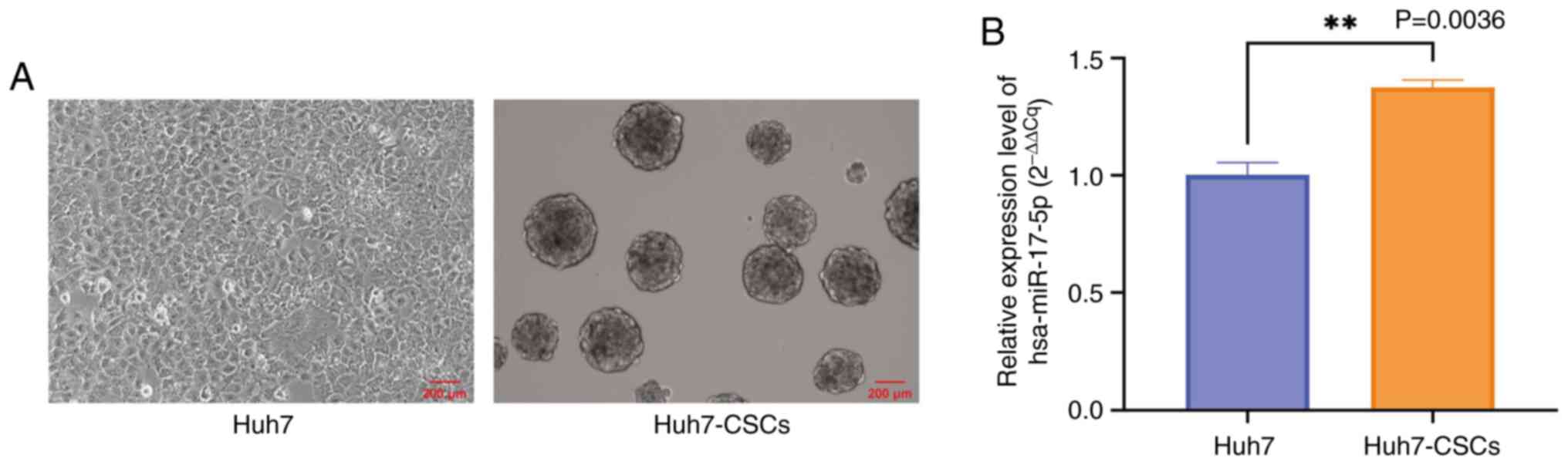
The combined list yielded 12 unique genes. Huh7-CSCs were
successfully developed from Huh7 cells. Spherical cells with a
diameter ≥100 µm were observed after 6 days of culturing in
ultra-low attachment conditions and the SFE of Huh7-CSCs was
calculated to be 4.7% (Fig. 3A).
Subsequently, the expression levels of miR-17-5p in Huh7-CSCs were
revealed to be significantly higher compared with those in Huh7
cells (Fig. 3B). The primary
biological processes regulated by miR-17-5p and its target genes,
including proliferation, invasion, migration, metastasis, stemness
and drug resistance, were explored using the ONCO.IO database
(Table II).
 | Table II.Role of microRNA-17-5p targeted genes
in biological processes of cancer. |
Table II.
Role of microRNA-17-5p targeted genes
in biological processes of cancer.
| Process | No. of genes | Targeted genes |
|---|
| Proliferation | 17 | RBL2, RB1, NCOA3,
SOCS6, UBE2C, ETV1, SIK1, CDKN1A, RND3, Myc, SMAD7, STAT3, Sox4,
CCND1, RELA, BLACAT1 and CDH1 |
| Invasion | 14 | PTEN, EGR2, ITGA5,
ITGB1, ETV1, IL8, PTENP1, CD274, Myc, Sox4, STAT3, HIF1A, BLACAT1
and RELA |
| Migration | 10 | RB1, PTEN, ETV1,
SIK1, PTENP1, Myc, INTS6, INTS6P1, Sox4 and RUNX1 |
| Metastasis | 6 | PTEN, EGR2, IL8,
HIF1A, Myc and CDH1 |
| Stemness | 2 | HIF1A and Myc |
| Drug
resistance | 7 | BLACAT1, ATG7,
CDKN1A, PTEN, BCL2L11, BECN1 and TRIM8 |
The Enrichr tool was used to explore the potential
pathways of the 12 miR-17-5p target genes involved in metastasis,
stemness and drug resistance. These 20 pathways are listed in
Table III after excluding those
associated with other diseases, including acute myeloid leukemia
and thyroid cancer. These 20 pathways are primarily involved in the
regulation of metabolism, cell fate, hepatitis viruses and the
immune checkpoint system. Certain pathways, including central
carbon metabolism and proteoglycans are closely associated with
cell stemness.
 | Table III.The 20 pathways of
microRNA-17-5p-targeted genes associated with metastasis, stemness
and drug resistance in HCC. |
Table III.
The 20 pathways of
microRNA-17-5p-targeted genes associated with metastasis, stemness
and drug resistance in HCC.
| Term | P-value | Genes |
|---|
| Pathways in
cancer |
2.75×10−7 | CDKN1A, BCL2L11,
CDH1, Myc, PTEN and HIF1A |
| Autophagy |
9.99×10−7 | BECN1, PTEN, HIF1A
and ATG7 |
| Central carbon
metabolism in cancer |
8.83×10−6 | Myc, PTEN and
HIF1A |
| miRNAs in
cancer |
2.54×10−5 | CDKN1A, BCL2L11,
Myc and PTEN |
| PI3K-AKT signaling
pathway |
4.27×10−5 | CDKN1A, BCL2L11,
Myc and PTEN |
| FOXO signaling
pathway |
5.79×10−5 | CDKN1A, BCL2L11 and
PTEN |
| Cellular
senescence |
9.73×10−5 | CDKN1A, Myc and
PTEN |
| Hepatitis B |
1.09×10−4 | EGR2, CDKN1A and
Myc |
| Hepatocellular
carcinoma |
1.21×10−4 | CDKN1A, Myc and
PTEN |
| Proteoglycans in
cancer |
2.18×10−4 | CDKN1A, Myc and
HIF1A |
| Mitophagy |
7.35×10−4 | BECN1 and
HIF1A |
| p53 signaling
pathway |
8.47×10−4 | CDKN1A and
PTEN |
| ErbB signaling
pathway |
1.15×10−3 | CDKN1A and Myc |
| PD-L1 expression
and PD-1 checkpoint pathway in cancer |
1.26×10−3 | PTEN and HIF1A |
| HIF-1 signaling
pathway |
1.87×10−3 | CDKN1A and
HIF1A |
| Cell cycle |
2.42×10−3 | CDKN1A and Myc |
| Apelin signaling
pathway |
2.94×10−3 | BECN1 and CDH1 |
| Apoptosis |
3.15×10−3 | BECN1 and
BCL2L11 |
| Hepatitis C |
3.84×10−3 | CDKN1A and Myc |
| JAK-STAT signaling
pathway |
4.08×10−3 | CDKN1A and Myc |
Discussion
Previous studies have identified ~2,000 miRNAs in
humans that regulate >60% of protein-coding genes and perform
post-transcriptional gene regulation by binding to target genes
(32,33). Systematic reviews have been used as
a high-evidence tool to detect potential miRNAs with diagnostic
and/or prognostic potential in the study and treatment of cancer.
For example, a meta-analysis of nine studies (involving 1,624 study
participants, of which 957 were patients with cervical cancer and
667 were healthy controls) revealed a marked upregulation of miR-21
expression in cervical cancer (34). In the present study, nine miRNAs
were found to be associated with the prognosis of patients with HCC
in a systematic review. In HCC tumor tissues, the expression levels
of miR-33a, miR-203, miR-375 and miR-200c were markedly
downregulated, whereas those of miR-9, miR-29a-5p and miR-17-5p
were markedly upregulated. Due to the post-transcriptional
regulation feature of miRNA, several miRNAs are abnormally
expressed during the development of HCC and can affect patient
prognosis. These miRNAs bind to multiple target genes and are
involved in numerous biological processes in HCC. For example,
miR-375, which is downregulated in HCC tissues, inhibits tumor
angiogenesis by targeting platelet-derived growth factor C.
Additionally, miR-375 has been shown to reduce sorafenib resistance
by regulating astrocyte elevated gene-1and sirtuin (SIRT)5
(35,36). By contrast, miR-9 is not only
upregulated in HCC tissues but also exhibits higher expression in
patients with HCC with early vascular invasion. Mechanistically,
miR-9 promotes HCC cell invasion and migration by targeting SIRT1,
FOXO1 and SWI/SNF-related, matrix-associated, actin-dependent
regulator of chromatin, subfamily D, member 2 (37). Data mining was combined with a
systematic review to further understand the potential impact of
these miRNAs on stemness, tumor metastasis, recurrence and drug
resistance in HCC. miR-17-5p was specifically identified as a
potential miRNA associated with these processes. A previous study
reported that miR-17-5p expression is strongly associated with the
number of tumor nodules, pathological grading of HCC and venous
infiltration (31). Other studies
have demonstrated that miR-17-5p induces hepatocarcinogenesis, and
promotes the proliferation and migration of HCC cells by targeting
Smad3, Runt-related transcription factor 3 and PTEN (38–40).
However, the role of miR-17-5p in HCC-CSCs remains to be
elucidated.
CSCs are gradually gaining popularity in research
because of their ability to drive tumor metastasis and recurrence
(41–43). However, the mechanisms underlying
stemness maintenance remain unclear. CSCs are a subpopulation of
cancer cells that remain dormant and may contribute to drug
resistance and the risk of recurrence (44,45).
By contrast, most HCC cells exhibit rapid proliferation and are
easily eliminated by DNA damage therapy. A previous study reported
that miR-17-5p expression is often higher in cancerous tissues
compared with that in paracancerous tissues (46). Notably, the expression levels of
miR-17-5p were significantly higher in HCC-CSCs compared with those
in HCC cells in the present study. This finding suggests that
miR-17-5p expression in HCC-CSCs may differ from that in the
majority of HCC cells and the high expression level of miR-17-5p
could be a unique biological feature that maintains the stemness of
HCC-CSCs.
To reveal the role of miR-17-5p in regulating
stemness, the target genes of miR-17-5p and their biological
processes were predicted using bioinformatics. Notably, miR-17-5p
may be involved in regulating stemness by targeting
hypoxia-inducible factor-1α (HIF1A) and Myc. Hypoxia is an
essential feature of the tumor microenvironment in the majority of
solid tumors, which contributes to tumor metabolism reprogramming
and leads to the failure of antitumor therapy (47). According to a previous study,
knocking down HIF-1α expression can inhibit the expression of
stemness-related genes (Oct3/4, Nanog, BMI-1 and Notch1), as well
as suppress self-renewal, migration and chemoresistance in liver
cancer cells under hypoxic conditions (48). Myc is one of the most common
oncogenes in human carcinogenesis, and acts as a bridge between
stem and tumor cells. The Myc family consists of c-Myc, n-Myc and
l-Myc, among which c-Myc serves a key role in HCC pathogenesis. A
previous study showed that miR-17-5p regulates the expression of
c-Myc to influence HCC development (49). The inactivation of Myc enables tumor
cells to de-differentiate into normal liver cell lineages. However,
upon its reactivation, these cells rapidly regain their malignant
characteristics. These tumor cells with stem cell properties are
CSCs. It has been demonstrated that in Myc-induced HCC, the
inactivation and reactivation of Myc can engage certain cells with
stem cell properties (50).
Myc and HIF1A, two stemness-maintaining genes, are
closely associated with central carbon metabolism (CCM) and
proteoglycans in tumors according to KEGG pathway analysis. CCM
primarily involves glycolysis and tricarboxylic acid cycle. CCM
reprogramming occurs in various tumors and serves as the primary
source of cellular energy (51).
The present study revealed that CCM is closely associated with
CSCs. Under aerobic conditions, CSCs exhibit dual metabolic modes,
utilizing glycolysis and oxidative phosphorylation (OXPHOS) as
energy sources that collectively drive tumor progression,
recurrence and drug resistance (52). Glycolysis is a key factor in
maintaining the stemness of HCC-CSCs. The upregulation of
glycolysis-associated genes in HCC-CSCs facilitates the expansion
of CSC populations, augments drug resistance and accelerates tumor
progression (53). For example,
hepatitis B virus X induces Bcl-2/adenovirus E1B 19 kDa-interacting
protein 3-like-dependent mitophagy, thereby upregulating glycolytic
metabolism. This metabolic shift modulates CSCs stemness through
elevated expression of cancer stemness-associated genes (such as
ATP-binding cassette subfamily G member 2, octamer-binding
transcription factor 4 and B-cell-specific Moloney murine leukemia
virus integration site 1), ultimately promoting tumor growth
(54). Furthermore, potassium
calcium-activated channel subfamily N member 4 potentiates
glycolysis in HCC-CSCs, upregulates stemness-associated
transcription factors, expands the HCC-CSCs population, and confers
resistance to radiotherapy and chemotherapy (55). OXPHOS serves as an alternative
energy source for CSCs. Emerging evidence has demonstrated that
HCC-CSCs exhibit more robust OXPHOS than HCC cell (56). Furthermore, the enhanced OXPHOS
levels in HCC-CSCs can promote stemness maintenance, thereby
increasing tumor metastasis. For example, organic cation/carnitine
transporter 2 activates peroxisome proliferator-activated
receptor-γ coactivator 1α signaling to potentiate OXPHOS, elevating
EpCAM/CD24 expression, augmenting CSC sphere-forming capacity, and
ultimately driving HCC cell proliferation, migration and invasion
(57).
Proteoglycans, macromolecules composed of core
proteins and glycosaminoglycan chains, have also been implicated in
CSCs (58). These molecules
contribute to the maintenance of the CSC phenotype and facilitate
tumor progression. Glypican-3, a heparan sulfate proteoglycan
subtype, modulates c-Myc activity to regulate the migratory,
invasive and CSC-forming capabilities of HCC cells within hypoxic
tumor microenvironments (59).
Hyaluronic acid (HA) belongs to the glycosaminoglycan family and
CD44 is an HA receptor and a surface marker of HCC-CSCs. TGF-β
affects the expression levels of pluripotent transcription factors
by regulating CD44 subtypes and targeting the MAPK signaling
pathway, mediates the formation ability of HCC-CSCs, promotes
epithelial-mesenchymal transition and increases the invasion and
migration of HCC cells (60). KEGG
pathway analysis indicated that miR-17-5p targeted genes associated
with stemness, metastasis and drug resistance mainly influenced
cell fate and metabolic reprogramming pathways to maintain CSC
survival in harsh microenvironments. Therefore, miR-17-5p may
represent a potential target that serves an essential role in
HCC-CSCs.
The present study had some limitations. First, the
nine studies selected in the systematic review only included
participants of Chinese and Korean ethnicities, which may have led
to bias and missed other potential miRNAs. However, inclusion of
only studies with Chinese and Korean ethnicities was due to the
current evidence base being limited and the reliability of the
present study findings was further validated using pan-ethnic
databases, such as TCGA. Second, a single HCC cell line (Huh7) was
used in the present study and only the expression levels of
miR-17-5p were explored in Huh7 and Huh7-CSCs, suggesting a
potential role for miR-17-5p in HCC-CSCs. In future studies, the
role of miR-17-5p will be explored in a larger population and
multiple cell lines. Direct investigations of miR-17-5p function in
drug resistance, metastasis and recurrence of HCC-CSCs will be
conducted, including miRNA mimic/inhibitor experiments, sphere
formation and drug resistance assays. In summary, miR-17-5p is a
miRNA potentially associated with CSCs, drug resistance, metastasis
and recurrence of HCC through cell fate and metabolic reprogramming
pathways. miR-17-5p exhibited higher expression in Huh7-CSCs
compared with that in Huh7 cells, and may target HIF1A and Myc to
maintain the stemness of HCC-CSCs. However, further experimental
studies are required to validate these hypotheses.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Southwest Medical
University Project (grant no. 2023ZYYJ08), the Sichuan Science and
Technology Program (grant no. 2022YFS0619), the National
Traditional Chinese Medicine Clinical Research Base Construction
Unit of the Affiliated Traditional Chinese Medicine Hospital of
Southwest Medical University (grant no. 2018-131), the Luzhou
Science and Technology Innovation Team (grant no. 2021-162-01) and
the Science and Technology Innovation Team of Affiliated
Traditional Chinese Medicine Hospital of Southwest Medical
University (grant no. 2022-CXTD-04).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YJ and XZ confirm the authenticity of all the raw
data. YJ, QP and XZ conducted the systematic review and network
analysis. YKC, XZ and JW designed the study. MP, PZD, DZ, XW, YKC
and JW supervised the experiments and discussion. YJ and XZ wrote
and edited the manuscript. YJ, XW and QP performed the cytological
experiments. XZ, DZ and MP performed the statistical analyses and
interpreted the data. DZ, XW and PZD were involved in reviewing and
editing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qi J, Li M, Wang L, Hu Y, Liu W, Long Z,
Zhou Z, Yin P and Zhou M: National and subnational trends in cancer
burden in China, 2005–20: An analysis of national mortality
surveillance data. Lancet Public Health. 8:e943–e955. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim E and Viatour P: Hepatocellular
carcinoma: Old friends and new tricks. Exp Mol Med. 52:1898–1907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anwanwan D, Singh SK, Singh S, Saikam V
and Singh R: Challenges in liver cancer and possible treatment
approaches. Biochim Biophys Acta Rev Cancer. 1873:1883142020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhong Y, Yang Y, He L, Zhou Y, Cheng N,
Chen G, Zhao B, Wang Y, Wang G and Liu X: Development of prognostic
evaluation model to predict the overall survival and early
recurrence of hepatocellular carcinoma. J Hepatocell Carcinoma.
8:301–312. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Makary MS, Khandpur U, Cloyd JM, Mumtaz K
and Dowell JD: Locoregional therapy approaches for hepatocellular
carcinoma: Recent advances and management strategies. Cancers
(Basel). 12:19142020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakano S, Eso Y, Okada H, Takai A,
Takahashi K and Seno H: Recent advances in immunotherapy for
hepatocellular carcinoma. Cancers (Basel). 12:7752020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Niu L, Liu L, Yang S, Ren J, Lai PBS and
Chen GG: New insights into sorafenib resistance in hepatocellular
carcinoma: Responsible mechanisms and promising strategies. Biochim
Biophys Acta Rev Cancer. 1868:564–570. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu J, Liao K and Zhou W: Exosomes regulate
the transformation of cancer cells in cancer stem cell homeostasis.
Stem Cells Int. 2018:48373702018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Wu G, Fu X, Xu S, Wang T, Zhang Q
and Yang Y: Aquaporin 3 maintains the stemness of CD133+
hepatocellular carcinoma cells by activating STAT3. Cell Death Dis.
10:4652019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bai X, Ni J, Beretov J, Graham P and Li Y:
Cancer stem cell in breast cancer therapeutic resistance. Cancer
Treat Rev. 69:152–163. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, Zhao S, Karnad A and Freeman JW:
The biology and role of CD44 in cancer progression: Therapeutic
implications. J Hematol Oncol. 11:642018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Glumac PM and LeBeau AM: The role of CD133
in cancer: A concise review. Clin Transl Med. 7:182018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiang DM, Sun W, Zhou T, Zhang C, Cheng Z,
Li SC, Jiang W, Wang R, Fu G, Cui X, et al: Oncofetal HLF
transactivates c-Jun to promote hepatocellular carcinoma
development and sorafenib resistance. Gut. 68:1858–1871. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akbari S, Kunter I, Azbazdar Y, Ozhan G,
Atabey N, Firtina Karagonlar Z and Erdal E: LGR5/R-Spo1/Wnt3a axis
promotes stemness and aggressive phenotype in hepatoblast-like
hepatocellular carcinoma cell lines. Cell Signal. 82:1099722021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu X, Wu Y, Zhou Z, Huang M, Deng W, Wang
Y, Zhou X, Chen L, Li Y, Zeng T, et al: Celecoxib inhibits the
epithelial-to-mesenchymal transition in bladder cancer via the
miRNA-145/TGFBR2/Smad3 axis. Int J Mol Med. 44:683–693.
2019.PubMed/NCBI
|
|
18
|
Lv T, Jiang L, Kong L and Yang J:
MicroRNA-29c-3p acts as a tumor suppressor gene and inhibits tumor
progression in hepatocellular carcinoma by targeting TRIM31. Oncol
Rep. 43:953–964. 2020.PubMed/NCBI
|
|
19
|
Moher D, Liberati A, Tetzlaff J and Altman
DG; PRISMA Group, : Preferred reporting items for systematic
reviews and meta-analyses: The PRISMA statement. PLoS Med.
6:e10000972009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Methley AM, Campbell S, Chew-Graham C,
McNally R and Cheraghi-Sohi S: PICO, PICOS and SPIDER: A comparison
study of specificity and sensitivity in three search tools for
qualitative systematic reviews. BMC Health Serv Res. 14:5792014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Torre-Healy LA, Berezovsky A and Lathia
JD: Isolation, characterization, and expansion of cancer stem
cells. Methods Mol Biol. 1553:133–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie RT, Cong XL, Zhong XM, Luo P, Yang HQ,
Lu GX, Luo P, Chang ZY, Sun R, Wu TM, et al: MicroRNA-33a
downregulation is associated with tumorigenesis and poor prognosis
in patients with hepatocellular carcinoma. Oncol Lett.
15:4571–4577. 2018.PubMed/NCBI
|
|
24
|
Zhuang L, Xu L, Wang P and Meng Z: Serum
miR-128-2 serves as a prognostic marker for patients with
hepatocellular carcinoma. PLoS One. 10:e01172742015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun J, Fang K, Shen H and Qian Y:
MicroRNA-9 is a ponderable index for the prognosis of human
hepatocellular carcinoma. Int J Clin Exp Med. 8:17748–17756.
2015.PubMed/NCBI
|
|
26
|
Chen HY, Han ZB, Fan JW, Xia J, Wu JY, Qiu
GQ, Tang HM and Peng ZH: miR-203 expression predicts outcome after
liver transplantation for hepatocellular carcinoma in cirrhotic
liver. Med Oncol. 29:1859–1865. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou N, Wu J, Wang X, Sun Z, Han Q and
Zhao L: Low-level expression of microRNA-375 predicts poor
prognosis in hepatocellular carcinoma. Tumour Biol. 37:2145–2152.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo C, Pu J, Liu F, Long X, Wang C, Wei H
and Tang Q: MicroRNA-200c expression is decreased in hepatocellular
carcinoma and associated with poor prognosis. Clin Res Hepatol
Gastroenterol. 43:715–721. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ha SY, Yu JI, Choi C, Kang SY, Joh JW,
Paik SW, Kim S, Kim M, Park HC, Park CK, et al: Prognostic
significance of miR-122 expression after curative resection in
patients with hepatocellular carcinoma. Sci Rep. 9:147382019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu HT, Dong QZ, Sheng YY, Wei JW, Wang G,
Zhou HJ, Ren N, Jia HL, Ye QH and Qin LX: MicroRNA-29a-5p is a
novel predictor for early recurrence of hepatitis B virus-related
hepatocellular carcinoma after surgical resection. PLoS One.
7:e523932012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen L, Jiang M, Yuan W and Tang H:
miR-17-5p as a novel prognostic marker for hepatocellular
carcinoma. J Invest Surg. 25:156–161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zenlander R, Salter H, Gilg S, Eggertsen G
and Stål P: MicroRNAs as plasma biomarkers of hepatocellular
carcinoma in patients with liver cirrhosis-A cross-sectional study.
Int J Mol Sci. 25:24142024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Friedman RC, Farh KKH, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gebrie A: Disease progression role as well
as the diagnostic and prognostic value of microRNA-21 in patients
with cervical cancer: A systematic review and meta-analysis. PLoS
One. 17:e02684802022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li D, Wang T, Sun FF, Feng JQ, Peng JJ, Li
H, Wang C, Wang D, Liu Y, Bai YD, et al: MicroRNA-375 represses
tumor angiogenesis and reverses resistance to sorafenib in
hepatocarcinoma. Cancer Gene Ther. 28:126–140. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang D and Yang J: MiR-375 attenuates
sorafenib resistance of hepatocellular carcinoma cells by
inhibiting cell autophagy. Acta Biochim Pol. 70:239–246.
2023.PubMed/NCBI
|
|
37
|
Chen Y, Xu H, Tang H, Li H, Zhang C, Jin S
and Bai D: miR-9-5p expression is associated with vascular invasion
and prognosis in hepatocellular carcinoma, and in vitro
verification. J Cancer Res Clin Oncol. 149:14657–14671. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu Z, Li X, Xu Y, Chen M, Chen W, Chen T,
Tang Q and He Z: microRNA-17 functions as an oncogene by
downregulating Smad3 expression in hepatocellular carcinoma. Cell
Death Dis. 10:7232019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Li F, Cheng J, Hou N, Pu Z, Zhang
H, Chen Y and Huang C: MicroRNA-17 family targets RUNX3 to increase
proliferation and migration of hepatocellular carcinoma. Crit Rev
Eukaryot Gene Expr. 33:71–84. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shan SW, Fang L, Shatseva T, Rutnam ZJ,
Yang X, Du W, Lu WY, Xuan JW, Deng Z and Yang BB: Mature miR-17-5p
and passenger miR-17-3p induce hepatocellular carcinoma by
targeting PTEN, GalNT7 and vimentin in different signal pathways. J
Cell Sci. 126:1517–1530. 2013.PubMed/NCBI
|
|
41
|
Liu YC, Yeh CT and Lin KH: Cancer stem
cell functions in hepatocellular carcinoma and comprehensive
therapeutic strategies. Cells. 9:13312020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim YJ, Yuk N, Shin HJ and Jung HJ: The
natural pigment violacein potentially suppresses the proliferation
and stemness of hepatocellular carcinoma cells in vitro. Int J Mol
Sci. 22:107312021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Prager BC, Xie Q, Bao S and Rich JN:
Cancer stem cells: The architects of the tumor ecosystem. Cell Stem
Cell. 24:41–53. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li L and Bhatia R: Molecular pathways:
Stem cell quiescence. Clin Cancer Res. 17:4936–4941. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song S, Ma D, Xu L, Wang Q, Liu L, Tong X
and Yan H: Low-intensity pulsed ultrasound-generated singlet oxygen
induces telomere damage leading to glioma stem cell awakening from
quiescence. iScience. 25:1035582021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zou R, Liu Y, Qiu S, Lu Y, Chen Y, Yu H,
Zhu H, Zhu W, Zhu L, Feng J and Han J: The identification of
N6-methyladenosine-related miRNAs predictive of hepatocellular
carcinoma prognosis and immunotherapy efficacy. Cancer Biomark.
38:551–566. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lequeux A, Noman MZ, Xiao M, Van Moer K,
Hasmim M, Benoit A, Bosseler M, Viry E, Arakelian T, Berchem G, et
al: Targeting HIF-1 alpha transcriptional activity drives cytotoxic
immune effector cells into melanoma and improves combination
immunotherapy. Oncogene. 40:4725–4735. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cui CP, Wong CCL, Kai AKL, Ho DWH, Lau
EYT, Tsui YM, Chan LK, Cheung TT, Chok KSH, Chan ACY, et al: SENP1
promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation
and SENP1/HIF-1α positive feedback loop. Gut. 66:2149–2159. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
EI Tayebi HM, Omar K, Hegy S, EI Maghrabi
M, EI Brolosy M, Hosny KA, Esmat G and Abdelaziz AI: Repression of
miR-17-5p with elevated expression of E2F-1 and c-MYC in
non-metastatic hepatocellular carcinoma and enhancement of cell
growth upon reversing this expression pattern. Biochem Biophys Res
Commun. 434:421–427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shachaf CM and Felsher DW: Tumor dormancy
and MYC inactivation: Pushing cancer to the brink of normalcy.
Cancer Res. 65:4471–4474. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ebrahimi KH, Gilbert-Jaramillo J, James WS
and McCullagh JSO: Interferon-stimulated gene products as
regulators of central carbon metabolism. FEBS J. 288:3715–3726.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Snyder V, Reed-Newman TC, Arnold L, Thomas
SM and Anant S: Cancer stem cell metabolism and potential
therapeutic targets. Front Oncol. 8:2032018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xia H, Huang Z, Xu Y, Yam JWP and Cui Y:
Reprogramming of central carbon metabolism in hepatocellular
carcinoma. Biomed Pharmacother. 153:1134852022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen YY, Wang WH, Che L, Lan Y, Zhang LY,
Zhan DL, Huang ZY, Lin ZN and Lin YC: BNIP3L-dependent mitophagy
promotes hbx-induced cancer stemness of hepatocellular carcinoma
cells via glycolysis metabolism reprogramming. Cancers (Basel).
12:6552020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fan J, Tian R, Yang X, Wang H, Shi Y, Fan
X, Zhang J, Chen Y, Zhang K, Chen Z and Li L: KCNN4 promotes the
stemness potentials of liver cancer stem cells by enhancing glucose
metabolism. Int J Mol Sci. 23:69582022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu G, Luo Q, Li H, Liu Q, Ju Y and Song
G: Increased oxidative phosphorylation is required for stemness
maintenance in liver cancer stem cells from hepatocellular
carcinoma cell line HCCLM3 cells. Int J Mol Sci. 21:52762020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang T, Liang N, Zhang J, Bai Y, Li Y,
Zhao Z, Chen L, Yang M, Huang Q, Hu P, et al: OCTN2 enhances
PGC-1α-mediated fatty acid oxidation and OXPHOS to support stemness
in hepatocellular carcinoma. Metabolism. 147:1556282023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang W, Han N, Xu Y, Zhao Y, Shi L, Filmus
J and Li F: Assembling custom side chains on proteoglycans to
interrogate their function in living cells. Nat Commun.
11:59152020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yao G and Yang Z: Glypican-3 knockdown
inhibits the cell growth, stemness, and glycolysis development of
hepatocellular carcinoma cells under hypoxic microenvironment
through lactylation. Arch Physiol Biochem. 130:546–554.
2024.PubMed/NCBI
|
|
60
|
Aguilar-Chaparro MA, Rivera-Pineda SA,
Hernández-Galdámez HV, Ríos-Castro E, Garibay-Cerdenares OL,
Piña-Vázquez C and Villa-Treviño S: Transforming growth factor-β
modulates cancer stem cell traits on CD44 subpopulations in
hepatocellular carcinoma. J Cell Biochem. 126:e700032025.
View Article : Google Scholar : PubMed/NCBI
|