Introduction
Cancer is among the most notable challenges to
global public health because of its high prevalence, molecular
heterogeneity and tendency towards therapy resistance. According to
the GLOBOCAN 2022 report, an estimated 19.96 million new cancer
cases and 9.74 million cancer deaths occurred worldwide in 2022
(1). The identification of the
following 14 cancer hallmarks in human tumors has markedly
influenced the understanding of malignant progression in recent
decades (2,3): Sustaining proliferative signaling,
evading growth suppressors, resisting cell death, enabling
replicative immortality, inducing or accessing vasculature,
activating invasion and metastasis, deregulating cellular
metabolism, avoiding immune destruction, genome instability and
mutation, tumor-promoting inflammation, senescent cells,
polymorphic microbiomes, nonmutational epigenetic reprogramming and
unlocking phenotypic plasticity. Despite major advances in targeted
therapies and immuno-oncology, clinical treatments continue to face
three major obstacles: i) Intrinsic and acquired therapy
resistance; ii) recurrence driven by the reactivation of dormant
cancer cells; and iii) metastasis resulting from tumor evolution.
These challenges highlight the urgent need for novel therapeutic
targets and precision interventions to improve clinical outcomes in
the future.
Ribosome-binding protein 1 (RRBP1), a key protein in
endoplasmic reticulum (ER)-ribosome binding, serves pleiotropic
roles in tumorigenesis and progression. Through its regulation of
organelle dynamics and protein synthesis, RRBP1 promotes malignant
phenotypes such as cell proliferation, invasion, metastasis and
chemoresistance. Notably, RRBP1 is upregulated in various cancer
types, such as breast cancer, colorectal cancer and bladder cancer,
and its expression levels are positively associated with advanced
clinical stages and poor prognosis (4–10).
Understanding the molecular mechanisms of RRBP1 provides insights
into dysregulated organelle communication and offers a potential
foundation for the development of therapies targeting subcellular
interactions. The present review systematically describes the roles
of RRBP1 in cancer, emphasizing its functional diversity. It first
outlines the structural features and functions of RRBP1, then
analyzes its expression and regulatory mechanisms across several
cancer types. Lastly, it evaluates the potential of RRBP1 as a
therapeutic target and proposes combined strategies targeting
organelle interaction networks to potentially enhance treatment
efficacy, with the goal of expanding future research directions in
precision oncology.
Function of RRBP1
RRBP1, also known as p180 because of its molecular
weight, is a ribosome-binding protein that localizes in the ER
membrane and is essential for the transport of newly synthesized
proteins (11). The gene structure
of RRBP1, featuring multiple splicing variants and its predicted
protein architecture, is detailed in Fig. 1. The present review used the Ensembl
Genome Browser (release 114; http://www.ensembl.org/) to display the genomic
structure of RRBP1, highlighting its splice variants, including the
MANE select transcript RRBP1-204 (Fig.
1A). Then the present review used AlphaFold (https://alphafold.ebi.ac.uk/) to predict the 3D
protein structure of RRBP1 (UniProt ID, Q9P2E9), emphasizing its
characteristic long α-helical segments and coiled-coil architecture
(Fig. 1B). RRBP1 consists of a
hydrophobic NH2-terminus and an acidic coiled
COOH-terminal structural domain. The former includes a
transmembrane structural domain and a highly conserved tandem
repeat sequence (ribosome-binding structural domain), whereas the
latter contains a lysine-rich region and a PDZ-binding structural
domain (12–14). These structures enable RRBP1 to
interact efficiently with ribosomes, ER and other cellular
components.
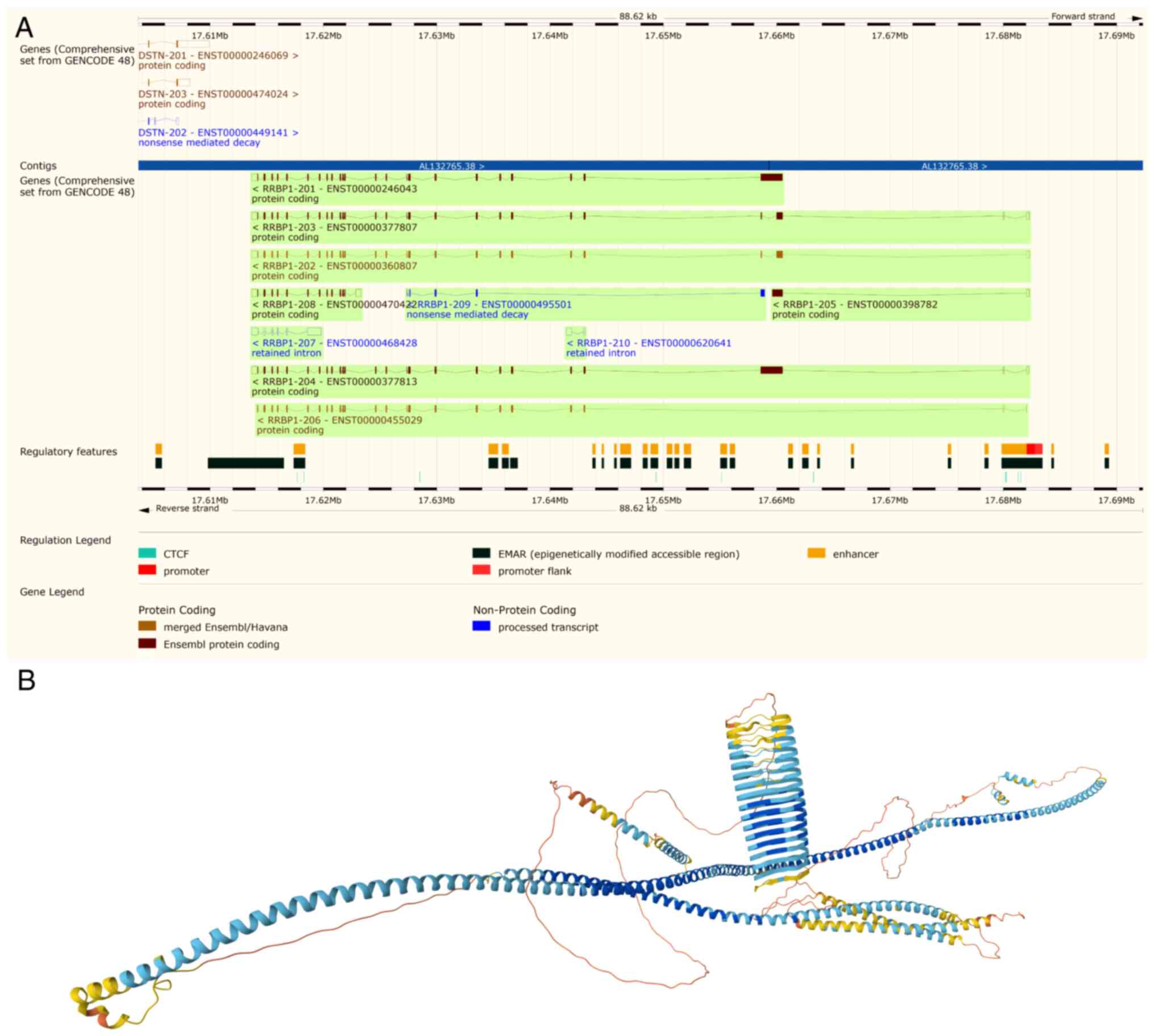 | Figure 1.Schematic of RRBP1 gene
structure and predicted 3D protein conformation. (A) RRBP1
gene from Ensembl, revealing various splicing variants, coding and
non-coding transcripts and regulatory features. The default
transcript (MANE Select) is RRBP1-204. Visualization adapted from
the Ensembl Genome Browser (RRBP1, release 114, EMBL-EBI,
http://www.ensembl.org). (B) Predicted protein
structure of RRBP1. The color gradient from blue to yellow
indicates the predicted local distance difference test confidence
score (blue, high confidence; yellow/orange, low confidence). The
structure reveals the characteristic long α-helical segments and
coiled-coil architecture of RRBP1. Predicted protein structure from
the AlphaFold Protein Structure Database (UniProt ID, Q9P2E9,
DeepMind and EMBL-EBI, http://alphafold.ebi.ac.uk). RRBP1, ribosome-binding
protein 1; EMBL-EBI, European Molecular Biology Laboratory-European
Bioinformatics Institute; MANE, matched annotation. |
Role of RRBP1 in the regulation of
protein synthesis and translation
Although RRBP1 was originally described as a
ribosomal receptor, this type III transmembrane protein on the ER
is also specifically involved in ER-dependent translational
regulatory mechanisms, such as the mRNA localization of placental
alkaline phosphatase, collagens Iα1 plus Iα2, IIIγ and
calreticulin, by directing their binding to ER membranes (15). Furthermore, RRBP1 promotes the
localization and anchoring of specific mRNAs through a
ribosome-independent mechanism (16). RRBP1 regulates the assembly of
multimer/protein complexes through its C-terminal structural
domain, thereby promoting translational activity (17). In collagen secretory cells, RRBP1
preferentially enhances the efficiency of secretory protein
biosynthesis by strengthening the association of ribosomes with the
ER membrane and it promotes the efficiency of multimer formation,
thus highlighting its central role in efficient translation in
specialized secretory cells (17).
Notably, the finding that RRBP1 deletion induces compensatory
upregulation of signal recognition particle (SRP) receptor subunits
(SRα/SRβ) potentially indicates a functional complementarity with
SRP-dependent pathways (15).
However, since the ER binding rate of Sec61β mRNA was not markedly
altered after knockdown of RRBP1, its ER localization may be
achieved through a pathway independent of RRBP1 and multiple
mechanisms may coordinate the targeting and synthesis of
tail-anchored proteins (18). For
example, in the case of Sec61β mRNA, localization can occur
independently of active translation yet is enhanced by translation,
its ORF sequence appears to harbor elements mediating ER anchoring
and some transcripts may engage translocon-bound ribosomes for
closer membrane proximity (18). In
addition, certain affected transmembrane proteins translocate from
the ER to the mitochondria, thereby revealing a potential role of
RRBP1 in cellular intercompartmental communication (15). Furthermore, yeast two-hybrid
analyses have indicated that the C-terminal region of RRBP1
interacts with the kinesin family member 5B (KIF5B) (19), which has markedly high expression in
a variety of cancer cell lines (13), such as MCF-7, HeLa and U2OS cells
(20). RRBP1, a receptor for KIF5B,
is involved in vesicle transport or mRNA localization of ER origin
and its role of combining ribosomes with kinesins suggests its
potential involvement in tumor transport or tumor cell invasion
processes.
Roles of RRBP1 in organelle
interactions
Mitochondria-ER contacts (MERCs), which serve as an
interface for communication between ER and mitochondria, are
involved in regulating mitochondrial energy metabolism homeostasis,
membrane phospholipid remodeling, programmed cell death signaling
and other core biological processes by mediating calcium-ion
dynamic homeostasis and the exchange of intermediates in lipid
metabolism (21). Abnormal function
of MERCs can interfere with the synergistic network of cellular
organelles and thus lead to tumorigenesis, neurodegenerative
diseases and cardiovascular system dysfunction (21); therefore, MERCs might have potential
value as targets for disease intervention. RRBP1 mediates MERC
formation through a specific transmembrane interaction between its
PDZ-binding domain and the PDZ domain of the mitochondrial protein
synaptojanin-2 binding protein (SYNJ2BP). This interaction notably
regulates mitochondrial DNA replication and spatial organization,
thereby modulating mitochondrial genomic stability and functional
dynamics (14,21,22).
Protein translation inhibitors such as puromycin disrupt the
RRBP1-SYNJ2BP interaction and specifically eliminate mitochondrial
contacts with ribosome-enriched rough ER (14). Knockdown of RRBP1 almost completely
eliminates ribosome-rich rough ER contact sites (riboMERCs),
leaving only certain smooth-type contact sites and leads to a
decrease in mitochondrial membrane potential; therefore, RRBP1
appears to be essential for mitochondrial function (21). Glycoprotein (Gp)78 ubiquitin ligase
activity regulates the size and tubular morphology of riboMERCs.
Gp78 upregulation induces the formation of extended riboMERCs in
COS-7 and HeLa cells in the presence of RRBP1 (21). This mechanism suggests that RRBP1
provides a structural basis and Gp78 finely regulates the
complexity of the contact sites through a ubiquitination-dependent
pathway. In addition, dynamin-related protein 1 deficiency disrupts
RRBP1-SYNJ2BP interactions and leads to defects in mitochondrial
DNA distribution (22).
Dynamin-related protein 1 deficiency indirectly affects ER lamellar
structure and RRBP1 function by interfering with mitochondrial
division dynamics (22), thus
suggesting a key role of ER-mitochondrial synergistic interactions
in mitochondrial genome regulation.
Furthermore, the ER-mitochondrial contact site also
serves a key role in mitochondrial autophagy. RRBP1 mediates
selective mitophagy induced by mitochondrial protein import stress
(MPIS) through dynamic subcellular relocalization (perinuclear
rough ER to peripheral ER). Under MPIS conditions, the
cytosolic-retained Nod-like receptor protein nucleotide-binding,
leucine-rich repeat X1 forms a complex with RRBP1 and drives
microtubule-associated protein 1 light chain 3 lipidation and
autophagosome formation. This process is independent of the
canonical phopsphatase and tensin homolog-induced putative kinase 1
(PINK1)-parkin RBR E3 ubiquitin protein ligase (PRKN) pathway and
preferentially uses ER-mitochondria contact sites as membrane
sources (23,24). RRBP1 exhibits dual functionality; it
assists in localized translational recovery during acute stress but
switches to a pro-autophagic mode under prolonged stress. The
interaction between RRBP1 and splicing factor, proline- and
glutamine-rich, suggests a potential role of RRBP1 in regulating
the subcellular localization of mitochondrially-associated mRNAs
during stress recovery (23).
Notably, PINK1 and PRKN are genetic risk factors for Parkinson's
disease and accumulating evidence suggests that mitochondrial
quality control defects are a key molecular basis for
neurodegeneration (24–27). Consistent with this possibility,
Sierksma et al (28) have
identified RRBP1 as a subthreshold Alzheimer's disease risk
gene. In parallel with its stress-adaptive function, RRBP1
modulates ER ribosome interactions in neurons. RRBP1 is enriched in
the ER tubules of neuronal axons and is virtually absent in
dendrites. Nerve growth factor neurotrophin-3 specifically enhances
ER-ribosome interactions through RRBP1, whereas brain-derived
neurotrophic factor/nerve growth factor has no such effect, thus
suggesting selective regulation of the signaling pathway (29). In addition, the identification of
RRBP1 in the neuromuscular junction has confirmed its specific
distribution in the synaptic region. RRBP1 may be involved in the
localization of synaptic proteins by binding the kinesin KIF5B,
which might serve as a novel candidate target for exploring the
mechanisms of neuromuscular diseases (30). Whether mediating MPIS-induced
mitochondrial autophagy or regulating axonal ER-ribosome dynamics,
RRBP1 functions as a sub-stable hub at the organelle-membrane
interface.
Roles of RRBP1 in lipid
metabolism
RRBP1 has been identified as a key gene in
the regulation of lipid levels in an integrated multi-omics study
(31). Observations that RRBP1
expression quantitative trait loci markedly co-localize with
genetic association signals for low-density lipoprotein, total
cholesterol and non-high-density lipoprotein suggest that
RRBP1 is an effector gene for lipid-related genetic
variation (31). RRBP1 is markedly
upregulated in adipose stem and progenitor cells in the
subcutaneous adipose tissue of mice with obesity induced by a
high-fat diet (32). In addition, a
cellular compartment in hepatocytes, denoted wrappER, has been
identified to functionally and structurally integrate all
hepatocyte systems and intracellular fatty acid elimination
pathways (33). When RRBP1
expression is inhibited in the liver, the uniform juxtaposition
between wrappER and mitochondria is disrupted and localized
abnormal bumps form (33). When the
complex comprising RRBP1 and SYNJ2BP is not functional, the
directional flow of fatty acids from the ER to the mitochondria is
impeded and lipids abnormally accumulate in liver cells; therefore,
the RRBP1-SYNJ2BP axis directly influences the balance of liver
lipid stability by modulating the physical properties of the
membrane-contact interface (33).
In parallel, RRBP1 silencing decreases very low-density lipoprotein
biosynthesis and triggers intrahepatic triglyceride and free fatty
acid accumulation, which in turn markedly increases lipid droplet
accumulation and the expression level of the lipid droplet marker
perilipin 2 (34). These findings
demonstrated that RRBP1 is important in regulating hepatic lipid
metabolic homeostasis by maintaining the structural integrity of
the contact between the ER and mitochondria.
Roles of RRBP1 in various types of
cancer
RRBP1 serves important regulatory roles in cancer
genesis and evolution. Due to its role in organelle interactions,
its molecular mechanisms and pathological associations are
receiving increasing attention (8,9,35–37).
RRBP1 influences tumor metabolic reprogramming and
microenvironmental adaptation by dynamically regulating substance
transport and signaling between subcellular compartments and
therefore might have translational value as a potential therapeutic
target. To comprehensively evaluate the contributions of genetic
alterations to RRBP1-driven oncogenesis, the present review
characterized the oncogenic status of RRBP1 across multiple cancer
types using the cBioPortal database for Cancer Genomics platform
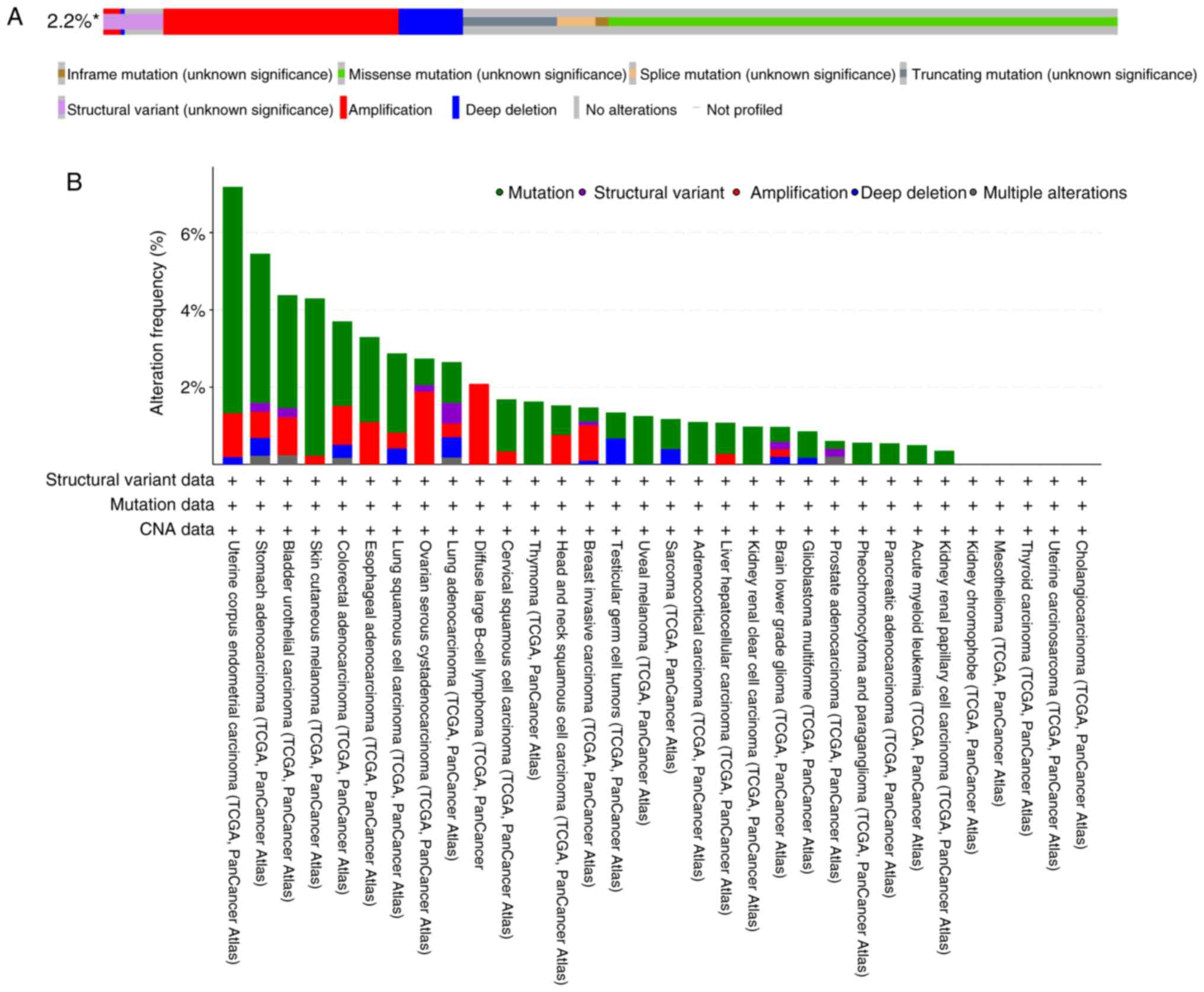
(https://www.cbioportal.org/) (Fig. 2). Based on The Cancer Genome Atlas
(TCGA) Pan-Cancer Atlas studies, the present review generated an
OncoPrint visualization map depicting the types and occurrence
frequencies of RRBP1 genetic alterations, while also
illustrating the lineage distribution characteristics of specific
RRBP1 mutations. Among 10,967 samples, 237 samples exhibited
RRBP1 alterations, accounting for ~2.2% of all cases
(Fig. 2A). These alterations were
primarily missense mutations. Among these, uterine corpus
endometrial carcinoma, stomach adenocarcinoma, bladder urothelial
carcinoma and skin cutaneous melanoma exhibited relatively high
alteration frequencies (>4%) (Fig.
2B). By contrast, because the RRBP1 gene in
cholangiocarcinoma, uterine carcinosarcoma, thyroid carcinoma,
mesothelioma and kidney chromophobe demonstrated no changes
(Fig. 2B), the oncogenic role of
RRBP1 in these cancer types may be limited.
The following section discusses research progress on
RRBP1 in tumors and explores the precision therapeutic potential of
targeting RRBP1 or its microenvironmental adaptors according to its
multiple regulatory mechanisms.
Lung cancer
A previous study reported that several mutated
genes, including RRBP1, have been identified to be enriched
primarily in nucleoplasmic localization and DNA repair-related
biological processes (38) and
therefore, may be involved in genetic susceptibility to lung
adenocarcinoma by influencing the DNA damage response mechanism. In
addition, RRBP1 is markedly upregulated in lung cancer tissues and
its upregulation is associated with early tumor stages (36). RRBP1 is also associated with the
unfolded protein response (UPR), a central mechanism through which
tumor cells respond to ER stress (ERS) (36). Cells rely on the ER to properly fold
and process secreted proteins and membrane proteins; however, when
protein folding is disrupted by various physiological or
pathological stimuli, such as deficient autophagy, energy
deprivation, inflammatory challenges and hypoxia, misfolded or
unfolded proteins accumulate, in the condition known as ERS
(39). To restore ER homeostasis,
cells activate an adaptive mechanism, the UPR, which is mediated by
three classic ER-resident sensors: Inositol-requiring enzyme 1α,
protein kinase R-like endoplasmic reticulum kinase (PERK) and
activating transcription factor 6 (ATF6) (40). The UPR decreases global protein
synthesis, enhances the production of molecular chaperones and
promotes the degradation of misfolded proteins, thereby alleviating
the burden on the ER (40).
Although the UPR is initially protective, a prolonged or unresolved
UPR may trigger apoptotic signals, thus serving dual roles in cell
survival and death. Tumor cells undergo persistent ERS because of
rapid proliferation and a harsh microenvironment and must rely on
the UPR to maintain their survival. Glucose-regulated protein 78
(GRP78), a regulator of the UPR, is markedly upregulated in lung
cancer. However, when GRP78 is upregulated, it dissociates from the
UPR signaling sensor PERK; therefore, PERK dimerizes and undergoes
autophosphorylation (36,41). Phosphorylated PERK further catalyzes
the phosphorylation of eukaryotic translation initiation factor
eukaryotic initiation factor 2α (eIF2α), thus leading to
translational repression, a core adaptive strategy enabling cells
to cope with mild ERS (Fig. 3)
(36,41). Tsai et al (36) have demonstrated that knockdown of
RRBP1 exacerbates ERS and markedly decreases cell viability and
tumor formation. This knockdown also markedly decreases ATF6 mRNA
levels; therefore, RRBP1 might be involved in the UPR by regulating
the mRNA stability of ATF6 (Fig. 3)
(36). By contrast, upregulation of
RRBP1 enhances the anti-apoptotic ability of tumor cells. RRBP1
protects cells against ERS-induced apoptosis by upregulating the
expression level of GRP78 and simultaneously decreasing the
accumulation of reactive oxygen species (ROS) (36). Therefore, RRBP1 regulates the UPR by
maintaining the mRNA stability of GRP78, which in turn helps lung
cancer cells adapt to ERS and chemotherapeutic stress and maintains
tumor survival and progression. Furthermore, in non-small cell lung
cancer, elevated expression of ubiquitin-specific processing
protease 35 (USP35) markedly upregulates RRBP1 protein levels
(41). USP35 stabilizes RRBP1
protein by directly binding RRBP1 and catalyzing its
deubiquitination modification, thereby inhibiting the
proteasome-mediated degradation pathway (Fig. 3) (41). The USP35/RRBP1 axis exerts a
protective effect through a dual mechanism; it enhances GRP78
expression and PERK phosphorylation levels (adaptive UPR signaling)
but also inhibits the CHOP-caspase3 apoptotic pathway, thus
markedly decreasing apoptosis (41).
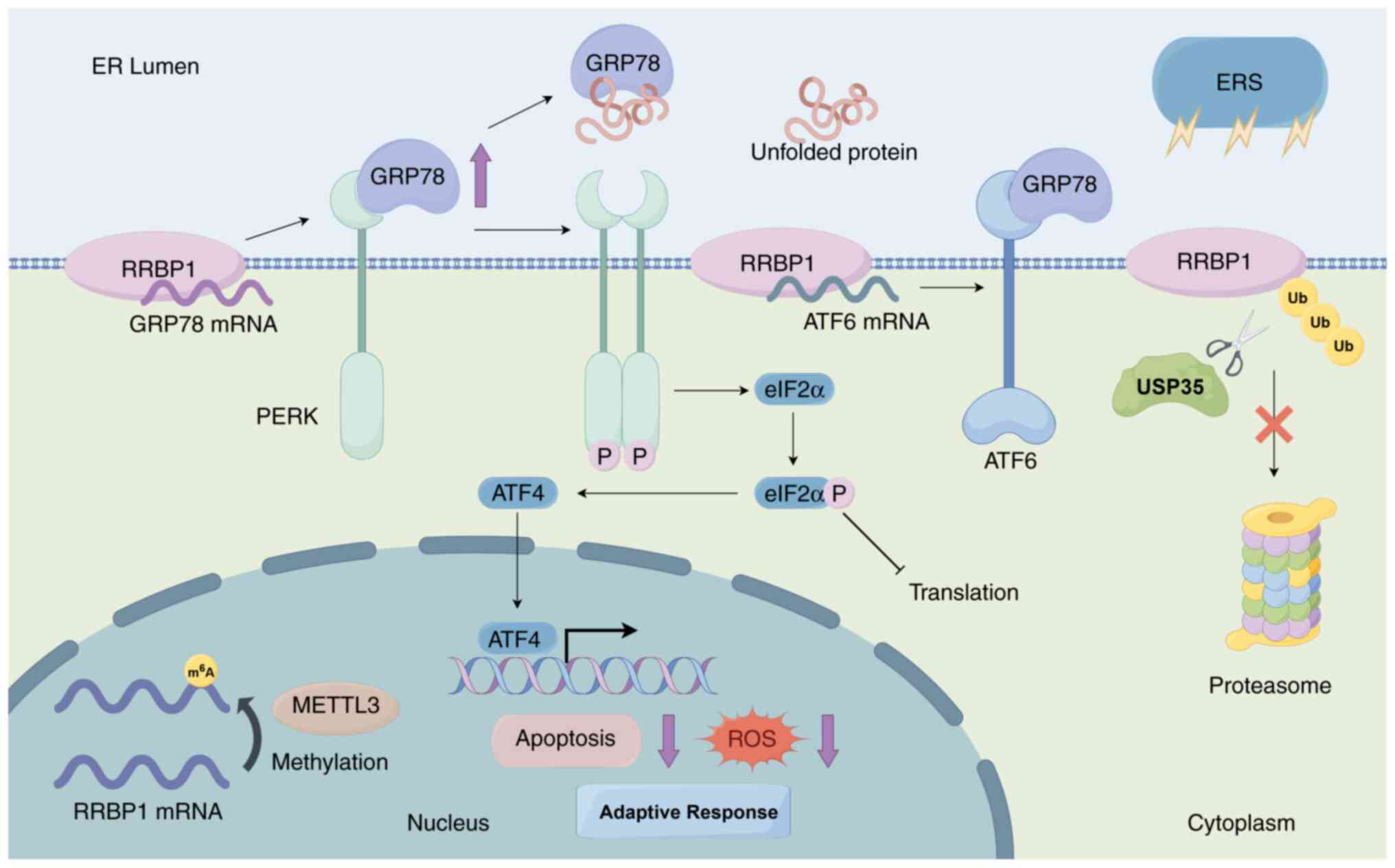 | Figure 3.RRBP1 adapts to ERS through the UPR
and enhancement of self-stabilization. RRBP1 upregulates GRP78
expression by stabilizing GRP78 mRNA and regulates ATF6 mRNA to
stabilize ATF6 for participation in the UPR. In addition, RRBP1
inhibits degradation pathways through METTL3-mediated m6A
methylation and USP35-mediated deubiquitination, thus enhancing
self-stability and promoting adaptive stress signaling. RRBP1,
ribosome-binding protein 1; Ub, ubiquitin; UPR, unfolded protein
response; ERS, endoplasmic reticulum stress; GRP78,
glucose-regulated protein 78; ROS, reactive oxygen species; METTL3,
methyltransferase-like 3; USP35, ubiquitin-specific processing
protease 35; ATF6, activating transcription factor 6; m6A,
N6-methyladenine; PERK, protein kinase R-like endoplasmic reticulum
kinase. |
Breast cancer
The mRNA and protein expression levels of RRBP1 in
breast cancer tissues are markedly higher compared with those in
normal tissues (4). RRBP1
expression is markedly associated with breast cancer histological
grade, human EGFR 2 (HER-2) status, p53 status and molecular
subtypes; furthermore, in patients with HER-2+ breast
cancer, increased RRBP1 expression is associated with worse overall
survival (OS) (4). The upregulation
of RRBP1 protein localizes primarily in the perinuclear region of
the cytoplasm (13). Normal breast
tissues demonstrate only weak to moderate cytoplasmic staining,
whereas the staining intensity is markedly enhanced in cancerous
tissues. RRBP1 expression is higher in malignant breast cancer
types compared with that in benign or proliferative lesions;
however, no gradient in RRBP1 expression has been observed in
benign or proliferative conditions (13). RRBP1 expression and lymph node
metastasis (LNM) are independent prognostic factors for OS in
patients with HER-2+ breast cancer, according to a
multivariate analysis (4). RRBP1
upregulation markedly influences survival in patients with
early-stage breast cancer, but this association has not been
observed in patients with advanced-stage types of cancer (4). RRBP1 promotes breast cancer cell
survival by participating in the UPR and this mechanism might
underlie its association with poor prognoses in patients with
HER-2+ breast cancer (4).
Liver cancer
In hepatocellular carcinoma (HCC) studies, RRBP1 has
been shown to serve a key role in high glucose-mediated tumor
malignant progression through the E2F transcription factor 1
(E2F1)/RRBP1 signaling pathway (42). He et al (42) have reported that high glucose
treatment markedly upregulates the mRNA and protein expression
levels of RRBP1 in HepG2 cells, whereas knockdown of RRBP1 markedly
inhibits cell proliferation, migration and invasive ability. RRBP1
has been suggested to be a core molecule mediating the pro-cancer
effect of high glucose. Furthermore, RRBP1 expression under
cellular stress conditions relies on an internal ribosome entry
site (IRES)-mediated translation mechanism (43). IRES elements are structured RNA
motifs within the 5′-untranslated regions of select mRNAs that
enable cap-independent initiation of translation (44). Under stress conditions, when
canonical cap-dependent protein synthesis is compromised, IRES
elements directly recruit the 40S ribosomal subunit, often with
assistance from IRES trans-acting factors, thereby sustaining the
translation of specific transcripts key for cell survival,
adaptation and reprogramming (44).
Notably, in cancer, dysregulated IRES-mediated translation and IRES
trans-acting factor activity have been implicated in tumorigenesis
and therapeutic resistance (45).
The 5′-untranslated region of RRBP1 mRNA has IRES activity, which
is markedly enhanced under chemotherapeutic drug treatment or serum
starvation (43). The La
autoantigen facilitates ribosome recruitment by binding the core
region of the IRES (−237 to −58 nt), whose two Gdf5 regulatory
region loop structures are essential for La protein binding
(43). In human HCC cells, RRBP1
protein levels are elevated under the aforementioned stress
conditions, but its mRNA transcripts do not markedly change, thus
suggesting that the enhancement of translational efficiency
originates from an IRES-dependent mechanism (43). Particularly, chemotherapeutic drugs
activate IRES-mediated translation by upregulating the expression
level of La protein, whereas serum starvation induces La protein
translocation from the nucleus to the cytoplasm, both of which work
together in activating IRES-mediated translation (43).
In summary, RRBP1 drives tumor malignant progression
in high glucose environments through the E2F1/RRBP1 signaling axis
and E2F1 directly binds RRBP1, and enhances its transcriptional
activity, thereby promoting HCC cell proliferation, migration and
invasion. Under chemotherapeutic or serum starvation conditions,
RRBP1 enhances the efficiency of protein synthesis through an
IRES-mediated translation mechanism, which in turn enhances the
survival of HCC cells in adverse conditions. This mechanism does
not depend on changes in mRNA transcript levels but instead relies
on translation regulation to achieve rapid stress adaptation.
Colorectal cancer
RRBP1 mRNA is higher in colorectal cancer (CRC)
tissues compared with normal tissues and RRBP1 protein is less
variable in normal tissues but more heterogeneous in cancerous
tissues (5). RRBP1 expression is
predictive of unfavorable survival outcomes; patients with high
RRBP1 expression have shorter disease-specific survival (5). The positive association between
chromosomal gains and RRBP1 mRNA levels (5) suggests the potential involvement of
RRBP1 in CRC progression as a driver gene. RRBP1 is also
associated with chemoresistance in CRC. The circular RNA (circRNA)
hsa_circ_0004085 is a key molecule mediating chemoresistance in
Fusobacterium nucleatum-infected colon cancer cells
(46). Furthermore, RRBP1 is a
direct binding protein of hsa_circ_0004085 (46). Hsa_circ_0004085 inhibits
chemotherapeutic drug-induced ERS-associated apoptosis by enhancing
the stability of GRP78 mRNA by binding RRBP1 and upregulating the
expression level of GRP78, thus promoting chemoresistance in tumor
cells (Fig. 4) (46). Furthermore, hsa_circ_0004085
activates the UPR by promoting the translocation of the sheared
form of ATF6 (ATF6p50) to the nucleus, thereby enhancing the
adaptive survival of tumor cells (46).
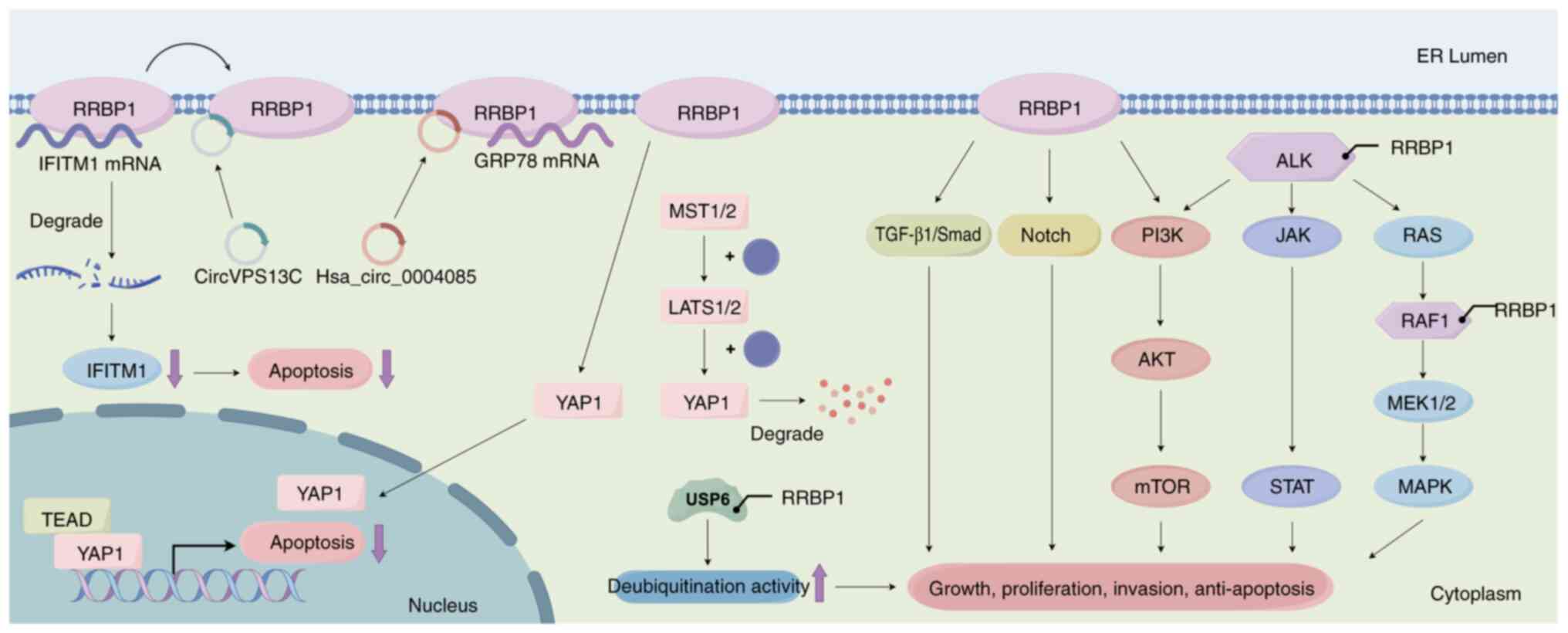 | Figure 4.RRBP1 promotes anti-apoptosis through
different signaling pathways, gene fusions and interactions with
circRNAs. RRBP1 activates various signaling pathways, thereby
promoting tumor cell proliferation and anti-apoptosis. It also
inactivates Hippo signaling and activates pro-survival and
anti-apoptotic genes. In addition, RRBP1 activates kinase
function through gene fusion (ALK, RAF1 and USP6),
promotes MAPK and other pathways and upregulates deubiquitination.
Furthermore, in the interaction between RRBP1 and circRNAs, binding
of RRBP1 to hsa_circ_0004085 stabilizes GRP78 and enhances cellular
adaptation to ERS. By contrast, circVPS13C inhibits the binding of
RRBP1 to IFITM1 mRNA by competitively binding RRBP1; subsequently,
degradation of IFITM1 mRNA leads to notable downregulation of its
protein expression level and further inhibits apoptosis. RRBP1,
ribosome-binding protein 1; YAP1, Yes-associated protein 1; ALK,
anaplastic lymphoma kinase; GRP78, glucose-regulated protein 78;
ERS, endoplasmic reticulum stress; IFITM1, interferon-induced
transmembrane protein 1; circRNA, circular RNA; hsa, Homo
sapiens; TEAD, transcription enhancement associated domain
family members; MST1/2, mammalian sterile 20-like kinase 1/2;
LATS1/2, large tumor suppressor 1/2. |
RRBP1 is involved in tumor progression, has
utility as a prognostic marker, is a driver gene in CRC and its
expression level is also closely associated with chromosomal
abnormalities. Furthermore, it regulates chemoresistance-related
pathways through interaction with hsa_circ_0004085. Therefore, CRC
treatments should integrate the role of RRBP1 at the molecular and
cellular levels to overcome the associated resistance
mechanisms.
Bladder cancer and upper tract
urothelial carcinoma (UTUC)
The mRNA and protein expression levels of RRBP1 in
bladder cancer tissues are markedly higher compared with that in
normal bladder tissues (6). RRBP1
is highly expressed in advanced-stage, LNM and basal squamous
subtype bladder cancer and is closely associated with poor
prognosis in patients (6–8). In addition, Luo et al (8) have identified that the RRBP1
gene region exhibits aberrant DNA methylation in UTUC, according to
methylation profiling microarray analysis, and its mRNA and protein
expression levels are upregulated in both tumor tissues and a UTUC
cell line. Experimental overexpression of RRBP1 has been found to
promote the proliferation of bladder cancer cells, whereas
downregulation of RRBP1 inhibits cell proliferation (6). This proliferative effect is associated
with the TGF-β1/SMAD signaling pathway, which is activated by RRBP1
upregulation (Fig. 4) (6). Wang et al (7) have reported that knockdown of RRBP1
results in the downregulation of C-C chemokine receptor type 7 and
subsequent inhibition of the migration and invasive ability of
bladder cancer cells. In addition, knockdown of RRBP1 inhibits the
proliferation, migration and invasive ability of UTUC cells
(8). The high expression level of
RRBP1 drives the malignant progression of tumors through a
multidimensional regulatory network. Molecular subtyping analysis
has revealed that the high expression level of RRBP1 is positively
associated with genes related to cell proliferation; therefore, it
might promote cell cycle progression through activation of the
PI3K/AKT and Notch signaling pathways (Fig. 4). Furthermore, RRBP1 upregulates
epithelial-mesenchymal transition (EMT) genes and basal cell
markers and suppresses luminal markers, thus contributing to the
acquisition of migratory and invasive properties of tumor cells and
a stem cell-like drug-resistant phenotype (8). The EMT is a reversible cellular
program in which epithelial cells lose their polarity and
intercellular adhesion through the downregulation of epithelial
markers such as E-cadherin and concurrently acquire mesenchymal
characteristics, including increased motility and the upregulation
of N-cadherin and vimentin (47).
This phenotypic shift facilitates tumor cell invasion through the
basement membrane and entry into the circulatory or lymphatic
systems, thus enabling metastatic dissemination. Notably, EMT is
closely associated with the acquisition of cancer stem cell-like
traits, including enhanced survival ability, resistance to
apoptosis and increased expression levels of drug efflux
transporters (47). These features
collectively contribute to resistance against chemotherapy and
immunotherapy. Furthermore, in a UTUC patient-derived organoid
model, high expression levels of RRBP1 have been associated with
chemoresistance to cisplatin, gemcitabine and epirubicin (8); therefore, it synergistically promotes
tumor cell survival by modulating ERS and yes-associated protein 1
(YAP1) signaling (Fig. 4) (8).
Overall, RRBP1 hypomethylation drives its
upregulation and subsequent participation in the malignant
progression of UTUC by promoting cell proliferation, the EMT
process and chemoresistance. Therefore, targeting RRBP1 or its
downstream effector pathways can reverse malignant tumor
phenotypes. In bladder cancer, RRBP1 serves a role in cell
proliferation through the TGF-β1/SMAD signaling pathway, whereas
cell migration and invasion involve the regulation of C-C chemokine
receptor type 7 and ERS, all of which may serve as potential
biomarkers for diagnosis and prognostic evaluation, thus providing
a precise therapeutic strategy for patients with high RRBP1
expression.
Prostate cancer
The mRNA and protein expression levels of RRBP1 are
higher in prostate cancer tissues compared with those in normal
cancer-adjacent tissues (9).
Patients with high (≥8) Gleason scores have higher RRBP1 expression
levels in tumor tissues compared with low Gleason scores (9). Furthermore, RRBP1 expression is
associated with the T stage of prostate cancer, LNM and
prostate-specific antigen level (9,48). The
median survival time is shorter in patients with prostate cancer
with higher compared with lower RRBP1 expression. Multifactorial
Cox regression analysis has indicated that RRBP1 is an independent
prognostic factor (9,48). Therefore, RRBP1 is closely
associated with tumor malignancy and progression and high RRBP1
expression can serve as a diagnostic marker and a predictor of poor
prognosis in prostate cancer. Methyltransferase-like 3 (METTL3), an
RNA methyltransferase that forms a core methyltransferase complex
with METTL14, is responsible for adding N6-methyladenine
(m6A) modifications, which is the most common internal
modification in eukaryotic RNAs (49,50).
Rather than functioning through a single pathway, m6A
modifications influence multiple RNA processes by recruiting reader
proteins such as YT521-B homology m6A RNA binding
protein F2, which recognize m6A-marked transcripts and
promote their degradation (51).
For example, m6A and its reader proteins have been shown
to regulate multiple steps of RNA metabolism, including alternative
pre-mRNA splicing (via the nuclear reader YTHDC1) (52), mRNA stability and degradation (via
YTHDF2) (51), and translation
efficiency through enhanced ribosome loading (via YTHDF1) (53). These findings supported that
METTL3-mediated m6A modification is a dynamic
epitranscriptomic mechanism shaping mRNA fate and cellular
function. Notably, abnormal METTL3 expression and the resulting
imbalance in m6A regulation have been associated with
pathological conditions, including tumorigenesis (54). RRBP1 is a key downstream
target gene regulated by METTL3 in an m6A-dependent
manner. METTL3 upregulates the expression level of RRBP1 by binding
and methylating RRBP1 mRNA and enhancing its stability (Fig. 3) (49). Furthermore,
m6A-sequencing has revealed that RRBP1 mRNA demonstrates
stronger m6A enrichment in tumor tissues compared with
non-tumor tissues, thus further supporting the molecular mechanism
of its regulation by METTL3 (49).
In summary, RRBP1 is a core effector molecule of the
METTL3/m6A signaling axis in prostate cancer and its
m6A-dependent stabilization promotes tumor
aggressiveness and is closely associated with poor patient
prognosis. Peptide inhibitors targeting this regulatory axis have
potential therapeutic value in the future.
Epithelioid inflammatory
myofibroblastic sarcoma (EIMS)
EIMS is a rare inflammatory myofibroblastic tumor
(IMT) subtype. IMT rarely recurs or metastasizes, whereas the
epithelioid variant of IMT, EIMS, which has distinctive
morphological features-for example, loosely arrayed round to
epithelioid tumor cells with vesicular nuclei, prominent nucleoli,
eosinophilic to amphophilic cytoplasm (55) and clinical aggressiveness, exhibits
poor prognosis (56,57). Chromosomal rearrangements are
frequently observed in IMT, including ALK rearrangement, a
genetic abnormality driving tumor cell proliferation in the form of
fusions of various chaperone genes such as TPM3-ALK, TPM4-ALK,
CLTC-ALK and EML4-ALK (56,58);
furthermore, the diversity of recombination patterns may be closely
associated with clinical heterogeneity (58). Notably, EIMS usually exhibits
nuclear membrane localization of Ran binding protein 2-ALK fusion
oncoproteins; in addition, novel RRBP1-ALK fusion proteins have
been identified in aggressive EIMS cases, showing cytoplasmic ALK
expression with perinuclear accentuation (Fig. 4) (56). The molecular structure and 3D
simulation structure of the RRBP1-ALK fusion protein are shown in
Fig. S1. The protein sequence of
RRBP1-ALK was obtained from the Ensembl database (https://www.ensembl.org/, release 114) and its domain
architecture was annotated using the Simple Modular Architecture
Research Tool (SMART; http://smart.embl.de/, accessed 12 August 2025). A
three-dimensional structural model was subsequently generated with
the AlphaFold server (https://alphafoldserver.com/, accessed 12 August
2025). Each ALK fusion partner retains a coiled-coil structural
domain in the ALK fusion protein, whereas RRBP1-ALK combines the
RRBP1 coiled-coil structural domain, the RRBP1 ER transmembrane
structural domain and the ribosome receptor structural domain
(56). The mechanism of the
formation of this fusion gene involves aberrant splicing of intron
20 of the RRBP1 gene; therefore, the RRBP1-ALK open
reading frame is composed of a 33-nucleotide sequence of
RRBP1 exon 20 fused to RRBP1 intron 20, which in turn
is fused to ALK exon 20 (56). This fusion pattern results in an ALK
protein with a cytoplasmic localization and an enhanced perinuclear
expression profile (56). EIMS
cases with RRBP1-ALK fusion exhibit a rapidly progressive
and aggressive disease course, with a median survival of only 2–10
months, which is markedly shorter compared with that of spindle
cell inflammatory myofibroblastoma with conventional ALK
fusion (56). Lee et al
(56) have suggested that
RRBP1-ALK is a novel recurrent oncogenic mechanism in
clinically aggressive EIMS. Although previous reports indicated
that the RRBP1-ALK fusion gene is seen only in EIMS,
subsequent reports have described the RRBP1-ALK fusion gene
in IMT (58,59). In a case report of recurrent IMT,
the 3′-end of exon 19 of the ALK gene was fused to the
5′-end of exon 10 of the RRBP1 gene (58). This case histologically revealed a
spindle cell type IMT, which differed from the morphological
features and clinical behavior of EIMS (58). Therefore, the RRBP1-ALK
fusion might have a heterogeneous pathogenic mechanism in different
IMT subtypes. A separate report presented a case with a
characteristic strong perinuclear ALK+ expression
pattern, in agreement with the expression profile of previously
reported cases of RRBP1-ALK fusion (56), and consisted of loosely organized
spindle cells on a mucus-like background (59). Furthermore, a recent study has
identified an RRBP1-USP6 fusion in low-grade myofibroblastic
sarcoma (Fig. 4) (60). This fusion arises from a T (17;20)
chromosomal translocation that juxtaposes exon 1 of the USP6
gene with exon 2 of the RRBP1 gene, thus marking the first
instance of RRBP1 being characterized as a fusion partner
for USP6 (60).
Structurally, this implies that the fusion transcript retains the
full coding sequence of USP6 together with its key
functional domains, including the ubiquitin-specific protease
catalytic core. RRBP1 functions as a regulatory module,
driving the abnormal expression of USP6. This finding has expanded
the genetic spectrum of USP6-associated neoplasms and is
potentially further links ER-ribosome pathway dysregulation and
malignant transformation in myofibroblastic tumors.
In conclusion, RRBP1 drives tumor malignancy
through fusion with ALK or USP6 genes and the
RRBP1-ALK fusion might enhance cancer cell survival and
invasiveness through ER localization, whereas the RRBP1-USP6
fusion aberrantly activates ubiquitination regulation, thus
suggesting an association between ER-ribosome pathway dysregulation
and tumor progression.
Pancreatic cancer
RRBP1 forms a fusion with the Raf1
gene (RRBP1-Raf1), which is markedly enriched in KRAS
wild-type tumors, according to the Arriba algorithm developed by
Uhrig et al (61). This
fusion retains the intact coding sequence of the Raf1 kinase
domain, whereas the transmembrane domain of RRBP1 anchors Raf1 to
the ER and drives its constitutive activation (61). Based on the structural features
described, it can be inferred that the breakpoint of the RRBP1-Raf1
fusion protein is located near the C-terminus of RRBP1, thereby
preserving its functional N-terminal domain and within the
N-terminal regulatory region of Raf1, maintaining the integrity of
its kinase domain. The structural preservation of the key domains
of both partners implies a breakpoint that occurs after the
functional segments of RRBP1 and before the key serine/threonine
kinase domain of Raf1, possibly resulting in a reading frame that
maintains the open reading frame and functional activity of the
fusion oncoprotein. Furthermore, functional studies demonstrated
that RRBP1-Raf1 expression in TP53-deficient MCF10A cells
and pancreatic ductal epithelial H6c7 cells markedly enhances
EGF-independent colony formation (61). After EGF withdrawal, the fusion
protein sustains MAPK pathway activation, as evidenced by markedly
elevated phosphorylation levels of MEK1/2 and ERK1/2 (Fig. 4) (61). In brief, the preservation of the
Raf1 kinase domain in this fusion might confer differential
sensitivity to specific Raf inhibitors. This finding expands the
molecular mechanisms underlying MAPK pathway hyperactivation in
pancreatic cancer and provides a novel therapeutic direction for
KRAS wild-type pancreatic cancer.
Oral squamous cell carcinoma
(OSCC)
RRBP1 is markedly upregulated in both early-stage
and late-stage cisplatin-resistant OSCC cell lines, as well as in
tumor tissues from chemotherapy-non-responsive patients vs. those
with chemotherapy-sensitive disease (62). Mechanistically, RRBP1 mediates
chemoresistance by regulating the Hippo signaling pathway effector
YAP1 (Fig. 4) (62). The Hippo signaling pathway, governed
by mammalian sterile 20-like kinase 1/2 and large tumor suppressor
(LATS) 1/2 kinases, is implicated in cancer chemoresistance. After
activation, LATS1/2 phosphorylates key residues of YAP1 and
transcriptional coactivator with PDZ-binding motif (TAZ) [YAP1
serine (Ser) 127 and Ser397], thus leading to their cytoplasmic
retention and subsequent degradation via the β-transducin
repeat-containing protein-dependent ubiquitin-proteasome system
(62). By contrast, Hippo pathway
inactivation results in dephosphorylated YAP1/TAZ evading
degradation, translocating to the nucleus, binding transcription
enhancement-associated domain family members (TEADs) and activating
pro-survival and anti-apoptotic genes (62). Notably, RRBP1 expression is strongly
associated with YAP1 target genes and RRBP1 knockdown markedly
decreases YAP1 and its downstream targets at both the mRNA and
protein levels (62). Furthermore,
RRBP1 depletion restores cisplatin sensitivity and enhances
apoptosis, as evidenced by increased expression levels of cleaved
PARP (62). Therefore, these
findings established RRBP1 as a central driver of cisplatin
resistance in OSCC, through a mechanism involving Hippo pathway
dysregulation, and highlight RRBP1 targeting as a therapeutic
strategy to potentially overcome cisplatin-induced chemoresistance
in advanced OSCC in the future.
Pituitary adenoma
In non-functioning pituitary adenomas (NFPAs), the
function of RRBP1 is uniquely modulated by the circRNA circVPS13C
and demonstrates a distinct regulatory mechanism (63). CircVPS13C competitively binds RRBP1
and disrupts its interaction with interferon-induced transmembrane
protein 1 (IFITM1) mRNA, thereby decreasing IFITM1 mRNA stability
and suppressing its expression (Fig.
4) (63). IFITM1, which is
known to regulate cell proliferation and migration, exerts
tumor-suppressive effects in pituitary tumors; overexpression of
IFITM1 markedly inhibits the proliferation of pituitary
tumor-derived folliculostellate cells and induces apoptosis
(63). Zhang et al (63) have revealed that RRBP1 silencing
markedly decreases IFITM1 mRNA and protein levels, whereas
overexpression of RRBP1 partially rescues the inhibitory effects of
circVPS13C on IFITM1. In contrast to its pro-tumorigenic roles in
other cancer types, such as lung cancer, bladder cancer and EIMS,
RRBP1 exhibits tissue-specific regulation in NFPAs by sustaining
the tumor-suppressive function of IFITM1 (63). This finding underscores the
functional diversity of RRBP1 in tumor microenvironments and
highlights the key regulatory role of the circVPS13C-RRBP1-IFITM1
axis in NFPA progression.
Other cancer types
In female reproductive system cancer types, RRBP1
exhibits marked upregulation with distinct clinicopathological
associations. For instance, in endometrial carcinoma, elevated
RRBP1 levels are strongly associated with advanced International
Federation of Gynecology and Obstetrics (FIGO) stages, poor
differentiation, deep myometrial invasion and LNM (37). Similarly, cervical squamous cell
carcinoma and epithelial ovarian cancer demonstrate markedly higher
RRBP1 expression in tumor tissues compared with normal tissues and
this elevated expression is associated with aggressive features
such as advanced FIGO staging, lymph node involvement and
unfavorable histotypes (35,64).
Notably, multivariate Cox analyses across these malignancies have
consistently identified RRBP1 as an independent prognostic factor
whose high expression predicts shortened OS and disease-free
survival (35,37,64).
In acute myeloid leukemia (AML), RRBP1 has been
reported as a potential leukemogenesis-associated molecule
regulated by exosomes derived from bone marrow microenvironmental
mesenchymal stromal cells (65).
Barrera-Ramirez et al (65)
have identified that mesenchymal stromal cell-derived exosomes in
patients with AML demonstrate diminished levels of microRNA
(miR)-339-3p, a direct RRBP1 suppressor, thereby derepressing RRBP1
transcription. This finding not only expands the oncogenic
repertoire of RRBP1 beyond solid tumors, but also suggests a novel
association between exosomal miRNA signaling and leukemic cell
regulation. To the best of our knowledge, no studies have directly
reported RRBP1 alterations or functional involvement in leukemias
other than AML or in lymphomas, to date. Thus, its role in these
hematological malignancies remains elusive and unreported,
warranting further investigation.
In esophageal cancer, RRBP1 is highly expressed at
both the mRNA and protein levels (10). Clinicopathological analysis has
demonstrated that high RRBP1 expression is markedly and positively
associated with the depth of tumor infiltration, LNM and advanced
TNM stage (10). Patients with high
RRBP1 expression exhibit a median survival of 43 months, in
contrast to the 56 months observed in low-expression cohorts,
thereby further validating its prognostic utility (10).
In bone metastasis, RRBP1 secreted by cancer cells
exerts a paradoxical role in modulating osteoblast differentiation
(66). Chen et al (66) have identified that RRBP1 is the only
highly abundant shared protein associated with osteoblast
differentiation in conditioned media of breast and prostate cancer
cells. Knockdown of RRBP1 in cancer cells enhances osteoblast
differentiation markers (ALP and bone γ-carboxyglutamate protein),
matrix mineralization and bone morphogenetic protein (BMP)
2/SMAD1/5/9 activation in pre-osteoblasts (66). These findings indicated that bone
metastatic cancer cells inhibit osteoblastic differentiation by
secreting RRBP1 and the mechanism may involve the regulation of ERS
levels and interference with the BMP/SMAD signaling pathway. These
findings provide a potential theoretical basis for targeting RRBP1
in the treatment of bone metastasis-associated bone lesions in the
future.
Beyond the aforementioned cancer types, several
tumor types have high RRBP1 alteration frequencies but lack
published evidence, to the best of our knowledge. Examples include
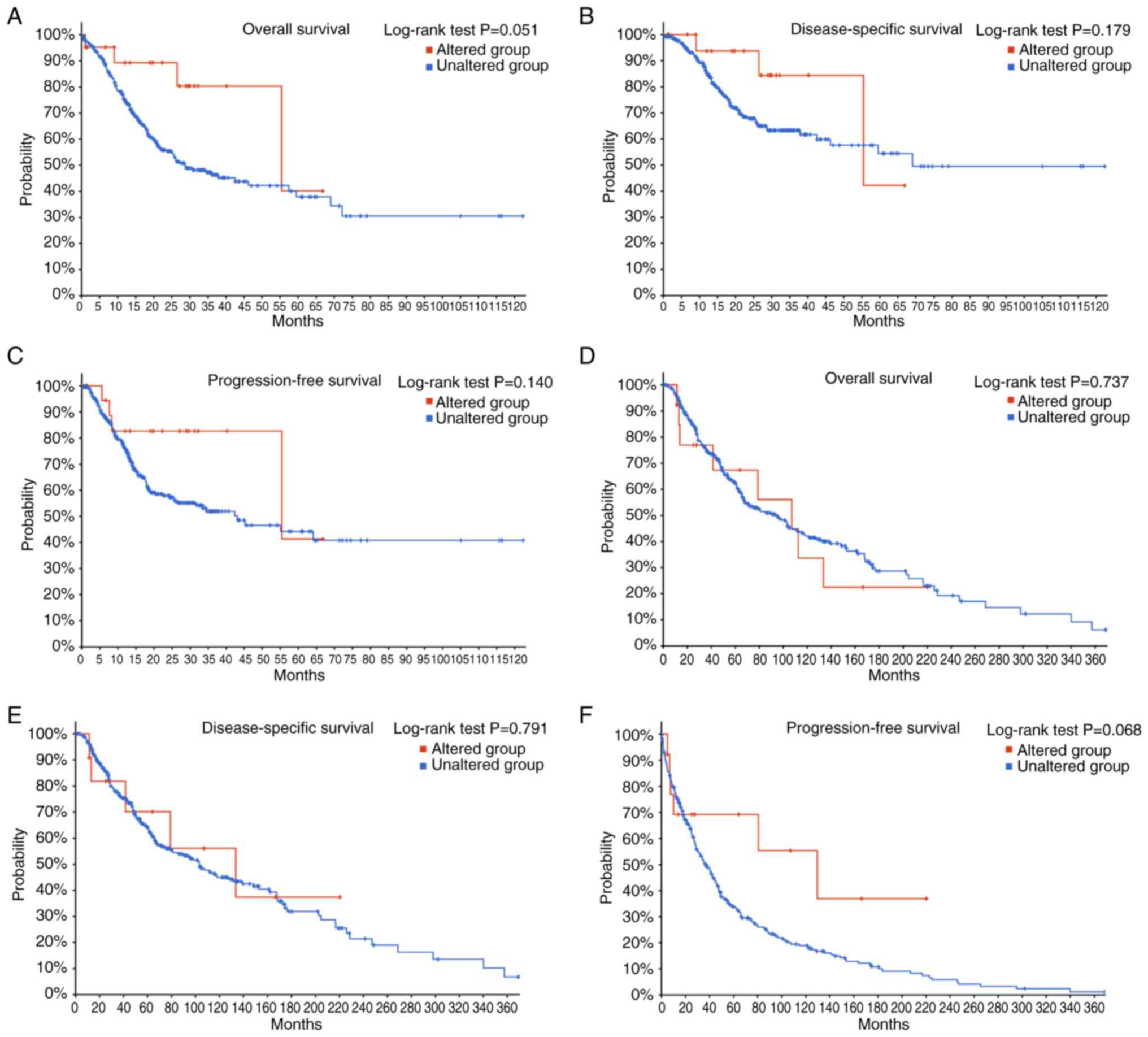
stomach adenocarcinoma and skin cutaneous melanoma (Fig. 2). The present review used the
cBioPortal database (TCGA; PanCancer Atlas; http://www.cbioportal.org/) to perform Kaplan-Meier
survival analyses for these cancer types and compared the survival
outcomes between patients with vs. without RRBP1 mutation
(Fig. 5). For stomach
adenocarcinoma, the RRBP1-altered group, as compared with
the unaltered group, demonstrated a trend toward improved OS,
disease-specific survival and progression-free survival (Fig. 5A-C). Similarly, in skin cutaneous
melanoma, patients with RRBP1 alterations also showed a
trend toward improved OS, disease-specific survival and
progression-free survival (Fig.
5D-F). Although these comparisons did not reach statistical
significance (log-rank P>0.05), the consistent direction of the
curves suggested potential biological relevance. One possible
explanation is that certain mutations in RRBP1 may impair
its function in protein transport and, therefore, slow tumor
progression or they may occur in molecular subtypes that are
intrinsically associated with a more favorable prognosis. However,
due to the limited number of mutated cases and potential
confounding clinical variables, these observations should be
interpreted with caution and require further exploration in studies
with larger sample sizes or functional studies.
Therapeutic targeting of RRBP1 in
cancer
The multifaceted roles of RRBP1 in tumorigenesis
and progression underscore its key position as a metastable hub at
organelle-membrane interfaces. RRBP1 serves complex and key roles
in multiple important molecular mechanisms in various types of
cancer (Fig. 6). Although its
upregulation is closely associated with tumor progression,
metastasis and chemotherapy resistance, its mechanistic basis
reveals environment-dependent regulatory mechanisms; therefore,
RRBP1 is a highly promising therapeutic target across cancer
types.
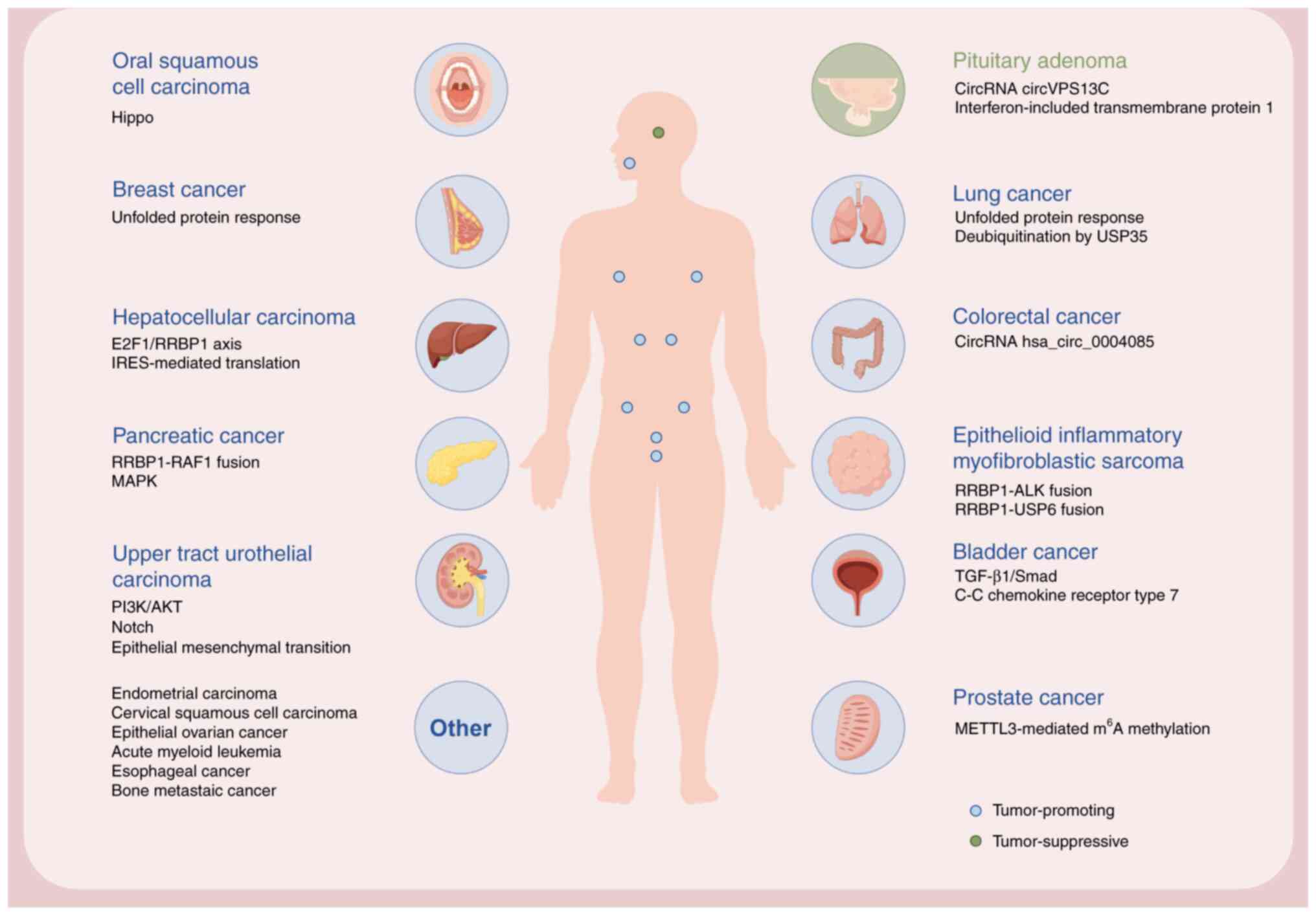 | Figure 6.RRBP1 expression in various human
organs and associated cancer types. Schematic diagram demonstrating
RRBP1-associated cancer types and representative molecular
mechanisms or keywords. Blue circles indicate tumor-promoting
roles, whereas green circles indicate tumor-suppressive roles.
‘Others’ include cancer types without specific organ illustration
(created in FigDraw). RRBP1, ribosome-binding protein 1, circRNA,
circular RNA; E2F1, E2F transcription factor 1; IRES, internal
ribosome entry site; USP35, ubiquitin-specific processing protease
35; m6A, N6-methyladenine; ALK, anaplastic lymphoma
kinase; METTL3, methyltransferase-like 3. |
Targeting RRBP1-mediated stress
adaptation
Stabilization of GRP78 inhibits ERS-associated
apoptosis and decreases ROS accumulation, thus markedly enhancing
cancer cell survival and chemoresistance (Fig. 3). In lung cancer, breast cancer and
CRCs, RRBP1 stabilizes GRP78 mRNA, thereby enhancing UPR-mediated
survival under ERS (4,36,46).
Small molecules targeting the GRP78-PERK-eIF2α axis, such as PERK
inhibitors (67), directly block
the signaling pathway but have the drawback of pancreatic toxicity.
Alternatively, by using a monoclonal antibody blocking RRBP1/GRP78
interaction, the regulation of UPR by RRBP1 can be decreased, thus
reversing chemoresistance.
Epigenetic modulation strategies
Enhancing self-stability through epigenetic
modification or inhibition of degradation pathways such as the
METTL3/m6A axis and USP35 deubiquitination promotes
adaptive stress signals (Fig. 3).
In lung cancer, the USP35 deubiquitination modification inhibits
the proteasome-mediated degradation pathway (41), whereas in prostate cancer, METTL3
stabilizes RRBP1 through m6A modification (49). Therefore, inhibition of the
METTL3/m6A axis, such as with small molecule peptides
(68), decreases the stability of
RRBP1 and blocks tumor progression. In addition, METTL3 inhibits
programmed cell death-ligand 1 (PD-L1) expression through
m6A modification and weakens the antitumor immune
response (69). Inhibition of
METTL3 remodels the tumor microenvironment and enhances the
efficacy of programmed cell death protein-1/PD-L1 blockade therapy.
Furthermore, small molecule inhibitors or gene editing systems
targeting USP35 can also decrease RRBP1 protein stability (70). However, functional synergy among USP
family members poses an important technical hurdle. Specific USP
isoforms, such as USP7, USP14 and USP22, can promote therapeutic
resistance by enhancing pro-survival protein stability, while other
members may participate in distinct regulatory processes (70). This functional heterogeneity not
only leads to a lack of specificity for targeted interventions but
may also trigger unintended off-target effects. Therefore, in-depth
analysis of the molecular basis of specific USP subtypes mediating
drug resistance phenotypes will provide a key theoretical basis for
the achievement of precision interventions in the future.
Pathway inhibition approaches
Activation of the TGF-β/SMAD, PI3K/AKT, Notch and
MAPK pathways, and inhibition of the Hippo pathway, drives tumor
proliferation and anti-apoptotic ability (Fig. 4) (6,8,61,62).
To address the multifaceted nature of the tumor, a layered
targeting strategy can be designed. First, small molecule
inhibitors or antibody drugs can be developed to specifically
target RRBP1 and eliminate its inhibition of the Hippo pathway and
synergistic activation effect of multiple pathways. Second, key
downstream nodes can be targeted, e.g., with YAP/TAZ-TEAD complex
blockers (71), dual PI3K/mTOR
inhibitors (72), Ras/Raf/MAPK
inhibitors (73) or neutralizing
antibodies to decrease ligand-receptor binding (74,75).
Third, epigenetic regulation can be integrated by co-delivery of
RRBP1 small interfering RNA with DNA methyltransferase inhibitors
via nanocarriers, thus reversing pro-oncogenic regulation. Notably,
because the RRBP1 pathways of activation and inhibition differ
among cancer types, precise guidance of treatment and localization
of its subcellular mode of action are necessary, in conjunction
with spatial transcriptomics technology to potentially achieve
precise spatiotemporal targeting in the future.
Fusion gene-targeted therapy
At the gene level, RRBP1 is abnormally
activated by gene fusions (ALK, Raf1 and USP6) and
subsequently regulates kinase function or deubiquitination;
therefore, RRBP1 is a potential therapeutic target (Fig. 4) (56,60,61).
For RRBP1-ALK/Raf1 fusion proteins arising in EIMS and pancreatic
cancer, selective kinase inhibitors can impair aberrant kinase
activity but must be optimized to overcome RRBP1-mediated drug
resistance (73,76). For RRBP1-USP6 fusions with abnormal
ubiquitination regulation, proteasome inhibitors or
deubiquitinating enzyme inhibitors can be explored (70). In addition, the rapid development of
gene editing technology in recent years is expected to overcome
existing therapeutic bottlenecks and provide more effective
treatment options through precise gene modification or modulation
strategies (77–81). Notably, one EIMS case report stated
that the RRBP1-ALK fusion+ case has notable
histological features, predominantly collagenous mesenchyme with
only localized areas of a mucus-like matrix (57). In contrast to the poor prognoses of
previous RRBP1-ALK fusion cases, which resulted in rapid
recurrence or death (56), this
case demonstrated a favorable prognosis with no recurrence or
metastasis during as many as 50 months of follow-up after
crizotinib-targeted therapy (57).
This prognostic difference may be closely associated with the
characteristics of the collagen fiber-dominated microenvironment in
the tumor stroma and the standardized use of ALK inhibitors after
surgery. Therefore, the development of therapeutic strategies
should be based on a combination of molecular features and tissue
microenvironmental characteristics. In addition, the type of
ALK fusion may be associated with the primary site of the
tumor. RRBP1-ALK fusions are observed predominantly in
abdominal EIMS, whereas rare fusion types such as echinoderm
microtubule-associated protein-like 4-ALK and
vinculin-ALK have been reported in central nervous system
cases (82). This molecular
heterogeneity highlights the need for molecular testing in the
diagnosis and classification of EIMS, particularly for the
differential diagnosis of cases involving rare sites. In summary,
gene fusions reveal molecular heterogeneity and clinical diversity
in different tumor types and the differences in pathogenic
mechanisms, histological features and therapeutic responses
highlight the key roles of precision medicine in diagnostic and
therapeutic decision-making. Future studies should further clarify
the interaction between the molecular characteristics of fusion
genes and the tumor microenvironment and establish improved
genotype-phenotype correlation models to potentially achieve truly
personalized treatment.
RNA-interaction disruption
RRBP1-RNA interactions have further revealed the
complex roles of RRBP1 in the regulation of tumor adaptation
(Fig. 4). In CRC, hsa_circ_0004085
binds RRBP1, stabilizes GRP78, enhances tumor cell adaptation to
ERS and promotes drug resistance, thereby inhibiting apoptosis
(46). For such RNA-interaction
networks, the development of targeted antisense oligonucleotides
(83), combined with specific
sequence design, chemical modification optimization, efficient
delivery systems and combined therapeutic strategies, can
potentially overcome the stability challenges posed by the cyclic
RNA structure, disrupt its binding to RRBP1 and restore apoptosis
susceptibility. However, in NFPA, overexpression of RRBP1 partially
reverses the inhibitory effect of circVPS13C on IFITM1, thereby
inducing apoptosis (63). This
duality challenges the traditional dichotomy of oncogenes and tumor
suppressor genes, highlights the high context dependence of RRBP1
function and emphasizes the need to design layered intervention
strategies based on microenvironmental characteristics.
The dual role of RRBP1 as a stress adaptor and
fusion partner presents both opportunities and challenges.
Targeting RRBP1-interacting pathways might enable precision therapy
by relying on the vulnerability of the upper and lower pathways.
However, both the tissue specificity of the RRBP1-ALK fusion and
the tumor suppressor function of IFITM1 by maintaining it in NFPA
caution against a one-size-fits-all solution (56–59,63).
Future research can prioritize functional stratification of the
RRBP1 network by using spatial multi-omics to determine how
organelle communication is rewired in specific tumor subtypes.
Despite the identification of RRBP1 as a promising
therapeutic candidate, to the best of our knowledge, no
interventional clinical trials directly targeting RRBP1 have been
registered in major trial repositories (ClinicalTrials.gov and World Health Organization
International Clinical Trials Registry Platform) to date. Current
therapeutic concepts remain largely preclinical and indirect,
focusing instead on upstream or downstream regulatory nodes such as
GRP78, METTL3, USP35 or oncogenic fusion partners (ALK, Raf1 and
USP6) (67–70,73,76).
These strategies underscore the translational potential of RRBP1
but also highlight that its development as a direct druggable
target is still at an early investigational stage.
RRBP1 cancer-independent activities
Beyond oncology, the multifaceted roles of RRBP1 in
diverse physiological and pathological settings highlight its key
function as a structural and metabolic coordinator. In the
musculoskeletal system, RRBP1 exhibits a regulatory role by driving
collagen biosynthesis and extracellular matrix (ECM) remodeling in
fibroblasts through miR-206-mediated post-transcriptional
regulation (84). Furthermore, its
dynamic expression during periodontal ligament stem cell
osteogenesis underscores its potential to promote osteogenesis
(85), possibly by promoting
ER-dependent ECM secretion. Paradoxically, the observation that
knockdown of RRBP1 in osteoblasts attenuates ALP and runt-related
transcription factor 2 expression in an osteoporosis model suggests
that bone formation is influenced by the environment (86). These findings have revealed that
RRBP1 is a multi-effect regulator spanning the environment of ECM
dynamics, osteogenic differentiation and bone formation. Therefore,
RRBP1 function can be considered an environmentally responsive
regulatory hub whose mode of action exhibits a dynamic balance
between physiological tissue remodeling and pathological bone
homeostatic imbalance.
Previous studies examining the relationship between
RRBP1 and hepatic lipid metabolism have focused on the functional
effects of RRBP1 expression regulation on ER-mitochondrial complex
formation and lipid metabolic output (21,32–34).
Although these functional gains or losses provide direct
mechanistic insights into the regulatory ability of RRBP1, they may
not fully reproduce the pathophysiological sequence observed in
vivo, whereby systemic metabolic alterations precede disruption
of lipid homeostasis. Notably, emerging investigations have begun
to bridge this knowledge gap by interrogating RRBP1 dynamics under
physiologically relevant metabolic states. For instance, a seminal
study has systematically characterized fasting-induced RRBP1
expression and subcellular localization patterns in both lean and
obese mouse models (87). Under
fasting, RRBP1 is specifically upregulated around the hepatic
portal vein and in the middle of the lobule and it drives specific
remodeling of rough surface ER-coated mitochondria, thereby
maintaining mitochondrial fatty acid oxidation function (87). By contrast, RRBP1 deficiency
inhibits fasting-induced ER-mitochondrial interactions and leads to
mitochondrial rounding, impaired β-oxidation and lipid accumulation
(87). These results have revealed
the molecular basis of RRBP1 in mediating metabolic adaptation
through dynamic regulation of ER-mitochondrial spatial
architecture. These findings have also clarified the role of RRBP1
as a key structural regulator that dynamically translates
physiological stimuli or pathological injury into structural and
functional remodeling of organelle networks. Future studies should
prioritize the elucidation of the upstream signaling pathways
regulating RRBP1 post-translational modifications and their
spatiotemporal coordination with other membrane contact site
regulators during metabolic adaptation.
Conclusions and perspectives
In cancer, RRBP1 drives malignancy by modulating
molecular signaling pathways and associated organelles, such as by
stabilizing the GRP78-PERK-eIF2α axis and inhibiting apoptosis,
activating pro-survival pathways (TGF-β/SMAD and PI3K/AKT) and
binding cyclic RNAs. In aggressive tumors, such as pancreatic
cancer and EIMS, this protein oncogenic role is further amplified
by fusion events (RRBP1-ALK and RRBP1-Raf1). Beyond
cancer, RRBP1 serves key central roles in several physiological and
pathological processes, such as the regulation of metabolic
homeostasis, regulation of neuronal function, homeostasis of the
cardiovascular system and remodeling of bone metabolism by
regulating the interactions between cellular organelles.
However, clinical translation faces challenges of
expression heterogeneity among cancer subtypes and the limitations
of current models, which are unable to reproduce systemic effects
due to a lack of cross-disease comparisons, since several previous
studies have relied on in vitro systemic or single-tissue
analyses (6,42,61,62).
To bridge these gaps, future research efforts should prioritize
multi-omics integration to decode the tissue-specific regulatory
networks and immune microenvironmental interactions of RRBP1.
Combining mechanistic understanding with advanced in vivo
models (humanized organoids or multi-tissue CRISPR screens) could
accelerate the development of organelle-targeted therapies and
pan-disease biomarkers, thereby potentially transforming precision
medicine from subcellular dynamics to clinical outcomes. Although
the cancer-promoting mechanism of RRBP1 has been gradually
revealed, its dynamic interaction network and targeted intervention
strategies can be further explored in depth, particularly to
overcome therapeutic resistance and microenvironmental remodeling.
Therefore, there is an urgent need for translational research in
this area.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82104603) and the Beijing Natural
Science Foundation (grant no. 7204322).
Availability of data and materials
Not applicable.
Authors' contributions
HH provided project administration, devised the
methodology, prepared Fig. 1,
Fig. 2, Fig. 3, Fig.
4, Fig. 5, Fig. 6 and Fig. S1 and drafted the manuscript. JO
conceived and designed the present review, acquired funding,
provided supervision, validated the data and reviewed the
manuscript. All authors read and approved the final manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanahan D: Hallmarks of Cancer: New
Dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang X, Sun S, Zhang X, Wu H, Tao W, Liu
T, Wei W, Geng J and Pang D: Expression of ribosome-binding protein
1 correlates with shorter survival in Her-2 positive breast cancer.
Cancer Sci. 106:740–746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan Y, Cao F, Guo A, Chang W, Chen X, Ma
W, Gao X, Guo S, Fu C and Zhu J: Endoplasmic reticulum
ribosome-binding protein 1, RRBP1, promotes progression of
colorectal cancer and predicts an unfavourable prognosis. Br J
Cancer. 113:763–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lv SW, Shi ZG, Wang XH, Zheng PY, Li HB,
Han QJ and Li ZJ: Ribosome binding Protein 1 correlates with
prognosis and cell proliferation in bladder cancer. Onco Targets
Ther. 13:6699–6707. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang M, Liu S, Zhou B, Wang J, Ping H and
Xing N: RRBP1 is highly expressed in bladder cancer and is
associated with migration and invasion. Oncol Lett.
20:2032020.PubMed/NCBI
|
|
8
|
Luo HL, Liu HY, Chang YL, Sung MT, Chen
PY, Su YL, Huang CC and Peng JM: Hypomethylated RRBP1 potentiates
tumor malignancy and chemoresistance in upper tract urothelial
carcinoma. Int J Mol Sci. 22:87612021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li T, Wang Q, Hong X, Li H, Yang K, Li J
and Lei B: RRBP1 is highly expressed in prostate cancer and
correlates with prognosis. Cancer Manag Res. 11:3021–3027. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Wang M, Zhang M, Li X, Zhu Z and
Wang H: Expression and significance of RRBP1 in esophageal
carcinoma. Cancer Manag Res. 10:1243–1249. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Savitz AJ and Meyer DI: 180-kD ribosome
receptor is essential for both ribosome binding and protein
translocation. J Cell Biol. 120:853–863. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wanker EE, Sun Y, Savitz AJ and Meyer DI:
Functional characterization of the 180-kD ribosome receptor in
vivo. J Cell Biol. 130:29–39. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Telikicherla D, Marimuthu A, Kashyap MK,
Ramachandra YL, Mohan S, Roa JC, Maharudraiah J and Pandey A:
Overexpression of ribosome binding protein 1 (RRBP1) in breast
cancer. Clin Proteomics. 9:72012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hung V, Lam SS, Udeshi ND, Svinkina T,
Guzman G, Mootha VK, Carr SA and Ting AY: Proteomic mapping of
cytosol-facing outer mitochondrial and ER membranes in living human
cells by proximity biotinylation. Elife. 6:e244632017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bhadra P, Schorr S, Lerner M, Nguyen D,
Dudek J, Förster F, Helms V, Lang S and Zimmermann R: Quantitative
proteomics and differential protein abundance analysis after
depletion of putative mRNA receptors in the ER membrane of human
cells identifies novel aspects of mRNA targeting to the ER.
Molecules. 26:35912021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui XA, Zhang H and Palazzo AF: p180
promotes the ribosome-independent localization of a subset of mRNA
to the endoplasmic reticulum. PLoS Biol. 10:e10013362012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ueno T, Kaneko K, Sata T, Hattori S and
Ogawa-Goto K: Regulation of polysome assembly on the endoplasmic
reticulum by a coiled-coil protein, p180. Nucleic Acids Res.
40:3006–3017. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui XA, Zhang H, Ilan L, Liu AX, Kharchuk
I and Palazzo AF: mRNA encoding Sec61β, a tail-anchored protein, is
localized on the endoplasmic reticulum. J Cell Sci. 128:3398–3410.
2015.PubMed/NCBI
|
|
19
|
Diefenbach RJ, Diefenbach E, Douglas MW
and Cunningham AL: The ribosome receptor, p180, interacts with
kinesin heavy chain, KIF5B. Biochem Biophys Res Commun.
319:987–992. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cardoso CM, Groth-Pedersen L, Høyer-Hansen
M, Kirkegaard T, Corcelle E, Andersen JS, Jäättelä M and Nylandsted
J: Depletion of kinesin 5B affects lysosomal distribution and
stability and induces Peri-nuclear accumulation of autophagosomes
in cancer cells. PLoS One. 4:e44242009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cardoen B, Vandevoorde KR, Gao G,
Ortiz-Silva M, Alan P, Liu W, Tiliakou E, Vogl AW, Hamarneh G and
Nabi IR: Membrane contact site detection (MCS-DETECT) reveals dual
control of rough mitochondria-ER contacts. J Cell Biol.
223:e2022061092024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ilamathi HS, Benhammouda S, Lounas A,
Al-Naemi K, Desrochers-Goyette J, Lines MA, Richard FJ, Vogel J and
Germain M: Contact sites between endoplasmic reticulum sheets and
mitochondria regulate mitochondrial DNA replication and
segregation. iScience. 26:1071802023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Killackey SA, Bi Y, Soares F, Hammi I,
Winsor NJ, Abdul-Sater AA, Philpott DJ, Arnoult D and Girardin SE:
Mitochondrial protein import stress regulates the LC3 lipidation
step of mitophagy through NLRX1 and RRBP1. Mol Cell.
82:2815–2831.e5. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Killackey SA, Bi Y, Philpott DJ, Arnoult D
and Girardin SE: Mitochondria-ER cooperation: NLRX1 detects
mitochondrial protein import stress and promotes mitophagy through
the ER protein RRBP1. Autophagy. 19:1601–1603. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ge P, Dawson VL and Dawson TM: PINK1 and
Parkin mitochondrial quality control: A source of regional
vulnerability in Parkinson's disease. Mol Neurodegener. 15:202020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu J, Liu W, Li R and Yang H: Mitophagy
in Parkinson's disease: From pathogenesis to treatment. Cells.
8:7122019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rüb C, Wilkening A and Voos W:
Mitochondrial quality control by the Pink1/Parkin system. Cell
Tissue Res. 367:111–123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sierksma A, Lu A, Mancuso R, Fattorelli N,
Thrupp N, Salta E, Zoco J, Blum D, Buée L, De Strooper B and Fiers
M: Novel Alzheimer risk genes determine the microglia response to
amyloid-β but not to TAU pathology. EMBO Mol Med. 12:e106062020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Koppers M, Özkan N, Nguyen HH, Jurriens D,
McCaughey J, Nguyen DTM, Li CH, Stucchi R, Altelaar M, MacGillavry
HD, et al: Axonal endoplasmic reticulum tubules control local
translation via P180/RRBP1-mediated ribosome interactions. Dev
Cell. 59:2053–2068.e9. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nazarian J, Bouri K and Hoffman EP:
Intracellular expression profiling by laser capture
microdissection: Three novel components of the neuromuscular
junction. Physiol Genomics. 21:70–80. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ramdas S, Judd J, Graham SE, Kanoni S,
Wang Y, Surakka I, Wenz B, Clarke SL, Chesi A, Wells A, et al: A
multi-layer functional genomic analysis to understand noncoding
genetic variation in lipids. Am J Hum Genet. 109:1366–1387. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tao T, Xu Y, Zhang CH, Zhang X, Chen J and
Liu J: Single-cell transcriptomic analysis and luteolin treatment
reveal three adipogenic genes, including Aspn, Htra1 and Efemp1.
Biochim Biophys Acta Mol Cell Biol Lipids. 1870:1595852025.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ilacqua N, Anastasia I, Aloshyn D,
Ghandehari-Alavijeh R, Peluso EA, Brearley-Sholto MC, Pellegrini
LV, Raimondi A, de Aguiar Vallim TQ and Pellegrini L: Expression of
Synj2bp in mouse liver regulates the extent of wrappER-mitochondria
contact to maintain hepatic lipid homeostasis. Biol Direct.
17:372022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Anastasia I, Ilacqua N, Raimondi A,
Lemieux P, Ghandehari-Alavijeh R, Faure G, Mekhedov SL, Williams
KJ, Caicci F, Valle G, et al: Mitochondria-rough-ER contacts in the
liver regulate systemic lipid homeostasis. Cell Rep. 34:1088732021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma J, Ren S, Ding J, Liu S, Zhu J, Ma R
and Meng F: Expression of RRBP1 in epithelial ovarian cancer and
its clinical significance. Biosci Rep. 39:BSR201906562019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP,
Jan YH, Lee CH, Hsiao YW, Yeh CT, Shen CN, et al: Endoplasmic
reticulum ribosome-binding protein 1 (RRBP1) overexpression is
frequently found in lung cancer patients and alleviates
intracellular stress-induced apoptosis through the enhancement of
GRP78. Oncogene. 32:4921–4931. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu S, Lin M, Ji H, Ding J, Zhu J, Ma R
and Meng F: RRBP1 overexpression is associated with progression and
prognosis in endometrial endometrioid adenocarcinoma. Diagn Pathol.
14:72019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fu F, Tao X, Jiang Z, Gao Z, Zhao Y, Li Y,
Hu H, Shen L, Sun Y and Zhang Y: Identification of germline
mutations in East-Asian young never-smokers with lung
adenocarcinoma by Whole-exome sequencing. Phenomics. 3:182–189.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang M and Kaufman RJ: Protein misfolding
in the endoplasmic reticulum as a conduit to human disease. Nature.
529:326–335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hetz C, Zhang K and Kaufman RJ:
Mechanisms, regulation and functions of the unfolded protein
response. Nat Rev Mol Cell Biol. 21:421–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang W, Wang M, Xiao Y, Wang Y, Ma L, Guo
L, Wu X, Lin X and Zhang P: USP35 mitigates endoplasmic reticulum
stress-induced apoptosis by stabilizing RRBP1 in non-small cell
lung cancer. Mol Oncol. 16:1572–1590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He Y, Huang S, Cheng T, Wang Y, Zhou SJ,
Zhang YM and Yu P: High glucose may promote the proliferation and
metastasis of hepatocellular carcinoma via E2F1/RRBP1 pathway. Life
Sci. 252:1176562020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gao W, Li Q, Zhu R and Jin J: La
autoantigen induces ribosome binding protein 1 (RRBP1) expression
through internal ribosome entry site (IRES)-Mediated translation
during cellular stress condition. Int J Mol Sci. 17:11742016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Godet AC, David F, Hantelys F, Tatin F,
Lacazette E, Garmy-Susini B and Prats AC: IRES Trans-acting
factors, key actors of the stress response. Int J Mol Sci.
20:9242019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Marques R, Lacerda R and Romão L: Internal
ribosome entry site (IRES)-Mediated translation and its potential
for Novel mRNA-based therapy development. Biomedicines.
10:18652022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hui B, Zhou C, Xu Y, Wang R, Dong Y, Zhou
Y, Ding J, Zhang X, Xu J and Gu Y: Exosomes secreted by
Fusobacterium nucleatum-infected colon cancer cells transmit
resistance to oxaliplatin and 5-FU by delivering hsa_circ_0004085.
J Nanobiotechnology. 22:622024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang Y, Hong W and Wei X: The molecular
mechanisms and therapeutic strategies of EMT in tumor progression
and metastasis. J Hematol Oncol. 15:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kuzuluk DG, Secinti IE, Erturk T, Hakverdi
S, Gorur S and Ozatlan D: Ribosome-binding protein-1 (RRBP1)
expression in prostate carcinomas and its relationship with
clinicopathological prognostic factors. Scott Med J. 69:83–87.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Feng Y, Li Z, Zhu J, Zou C, Tian Y, Xiong
J, He Q, Li W, Xu H, Zou C, et al: Stabilization of RRBP1 mRNA via
an m6A-dependent manner in prostate cancer constitutes a
therapeutic vulnerability amenable to small-peptide inhibition of
METTL3. Cell Mol Life Sci. 81:4142024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G:
Functions of N6-methyladenosine and its role in cancer. Mol Cancer.
18:1762019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yu L, Liu J, Lao IW, Luo Z and Wang J:
Epithelioid inflammatory myofibroblastic sarcoma: A
clinicopathological, immunohistochemical and molecular cytogenetic
analysis of five additional cases and review of the literature.
Diagn Pathol. 11:672016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lee JC, Li CF, Huang HY, Zhu MJ,
Mariño-Enríquez A, Lee CT, Ou WB, Hornick JL and Fletcher JA: ALK
oncoproteins in atypical inflammatory myofibroblastic tumours:
Novel RRBP1-ALK fusions in epithelioid inflammatory myofibroblastic
sarcoma. J Pathol. 241:316–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cheng N and Xue L: A case report of
RRBP1-ALK fusion gene-positive epithelioid inflammatory
myofibroblastic sarcoma with collagenous stroma and good prognosis.
Asian J Surg. 47:617–619. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu X, Li L, Zhang Y, Meng F, Xie H and
Duan R: A recurrent inflammatory myofibroblastic tumor patient with
two novel ALK fusions: A case report. Transl Cancer Res.
11:3379–3384. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vernemmen AIP, Samarska IV, Speel EM,
Riedl RG, Goudkade D, de Bruïne AP, Wouda S, van Marion AM,
Verlinden IV, van Lijnschoten I, et al: Abdominal inflammatory
myofibroblastic tumour: Clinicopathological and molecular analysis
of 20 cases, highlighting potential therapeutic targets.
Histopathology. 84:794–809. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Arcovito G, Crucitta S, Del Re M,
Caporalini C, Palomba A, Nozzoli F and Franchi A: Recurrent USP6
rearrangement in a subset of atypical myofibroblastic tumours of
the soft tissues: Low-grade myofibroblastic sarcoma or
atypical/malignant nodular fasciitis? Histopathology. 85:244–253.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Uhrig S, Ellermann J, Walther T, Burkhardt
P, Fröhlich M, Hutter B, Toprak UH, Neumann O, Stenzinger A, Scholl
C, et al: Accurate and efficient detection of gene fusions from RNA
sequencing data. Genome Res. 31:448–460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shriwas O, Arya R, Mohanty S, Mohapatra P,
Kumar S, Rath R, Kaushik SR, Pahwa F, Murmu KC, Majumdar SKD, et
al: RRBP1 rewires cisplatin resistance in oral squamous cell
carcinoma by regulating Hippo pathway. Br J Cancer. 124:2004–2016.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang W, Chen S, Du Q, Bian P, Chen Y, Liu
Z, Zheng J, Sai K, Mou Y, Chen Z, et al: CircVPS13C promotes
pituitary adenoma growth by decreasing the stability of IFITM1 mRNA
via interacting with RRBP1. Oncogene. 41:1550–1562. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhu J, Zhao R, Xu W, Ma J, Ning X, Ma R
and Meng F: Correlation between reticulum ribosome-binding protein
1 (RRBP1) overexpression and prognosis in cervical squamous cell
carcinoma. Biosci Trends. 14:279–284. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Barrera-Ramirez J, Lavoie JR, Maganti HB,
Stanford WL, Ito C, Sabloff M, Brand M, Rosu-Myles M, Le Y and
Allan DS: Micro-RNA profiling of exosomes from Marrow-derived
mesenchymal stromal cells in patients with acute myeloid leukemia:
Implications in leukemogenesis. Stem Cell Rev Rep. 13:817–825.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen R, Wang Y, Xu Y, He Y, Li Q, Xia C
and Zhang B: RRBP1 depletion of bone metastatic cancer cells
contributes to enhanced expression of the osteoblastic phenotype.
Front Oncol. 12:10051522022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hetz C, Axten JM and Patterson JB:
Pharmacological targeting of the unfolded protein response for
disease intervention. Nat Chem Biol. 15:764–775. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li Z, Feng Y, Han H, Jiang X, Chen W, Ma
X, Mei Y, Yuan D, Zhang D and Shi J: A stapled peptide inhibitor
targeting the binding Interface of N6-Adenosine-Methyltransferase
subunits METTL3 and METTL14 for cancer therapy. Angew Chem Int Ed
Engl. 63:e2024026112024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yu H, Liu J, Bu X, Ma Z, Yao Y, Li J,
Zhang T, Song W, Xiao X, Sun Y, et al: Targeting METTL3 reprograms
the tumor microenvironment to improve cancer immunotherapy. Cell
Chem Biol. 31:776–791.e7. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gao H, Xi Z, Dai J, Xue J, Guan X, Zhao L,
Chen Z and Xing F: Drug resistance mechanisms and treatment
strategies mediated by Ubiquitin-specific proteases (USPs) in
cancers: New directions and therapeutic options. Mol Cancer.
23:882024. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fu M, Hu Y, Lan T, Guan KL, Luo T and Luo
M: The Hippo signalling pathway and its implications in human
health and diseases. Signal Transduct Target Ther. 7:3762022.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
He Y, Sun MM, Zhang GG, Yang J, Chen KS,
Xu WW and Li B: Targeting PI3K/Akt signal transduction for cancer
therapy. Signal Transduct Target Ther. 6:4252021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bahar ME, Kim HJ and Kim DR: Targeting the
RAS/RAF/MAPK pathway for cancer therapy: From mechanism to clinical
studies. Signal Transduct Target Ther. 8:4552023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu S, Ren J and Ten Dijke P: Targeting
TGFβ signal transduction for cancer therapy. Signal Transduct
Target Ther. 6:82021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Shi Q, Xue C, Zeng Y, Yuan X, Chu Q, Jiang
S, Wang J, Zhang Y, Zhu D and Li L: Notch signaling pathway in
cancer: From mechanistic insights to targeted therapies. Signal
Transduct Target Ther. 9:1282024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shreenivas A, Janku F, Gouda MA, Chen HZ,
George B, Kato S and Kurzrock R: ALK fusions in the pan-cancer
setting: Another tumor-agnostic target? NPJ Precis Oncol.
7:1012023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhang H, Qin C, An C, Zheng X, Wen S, Chen
W, Liu X, Lv Z, Yang P, Xu W, et al: Application of the
CRISPR/Cas9-based gene editing technique in basic research,
diagnosis, and therapy of cancer. Mol Cancer. 20:1262021.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Frangoul H, Locatelli F, Sharma A, Bhatia
M, Mapara M, Molinari L, Wall D, Liem RI, Telfer P, Shah AJ, et al:
Exagamglogene autotemcel for severe sickle cell disease. N Engl J
Med. 390:1649–1662. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Macarrón Palacios A, Korus P, Wilkens BGC,
Heshmatpour N and Patnaik SR: Revolutionizing in vivo therapy with
CRISPR/Cas genome editing: Breakthroughs, opportunities and
challenges. Front Genome Ed. 6:13421932024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xu W, Zhang S, Qin H and Yao K: From bench
to bedside: Cutting-edge applications of base editing and prime
editing in precision medicine. J Transl Med. 22:11332024.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Healey N: Next-generation CRISPR-based
gene-editing therapies tested in clinical trials. Nat Med.
30:2380–2381. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kim EE, Park CK, Kang KM, Kwak Y, Park SH
and Won JK: Primary epithelioid inflammatory myofibroblastic
sarcoma of the brain with EML4::ALK fusion mimicking intra-axial
glioma: A case report and brief literature review. J Pathol Transl
Med. 58:141–145. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Çakan E, Lara OD, Szymanowska A, Bayraktar
E, Chavez-Reyes A, Lopez-Berestein G, Amero P and Rodriguez-Aguayo
C: Therapeutic antisense oligonucleotides in oncology: From bench
to bedside. Cancers (Basel). 16:29402024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Fry CS, Kirby TJ, Kosmac K, McCarthy JJ
and Peterson CA: Myogenic progenitor cells control extracellular
matrix production by fibroblasts during skeletal muscle
hypertrophy. Cell Stem Cell. 20:56–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zheng Y, Li X, Huang Y, Jia L and Li W:
Time series clustering of mRNA and lncRNA expression during
osteogenic differentiation of periodontal ligament stem cells.
PeerJ. 6:e52142018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang X, Wang Y, Zheng M, Wei Q, Zhang R,
Zhu K, Zhai Q and Xu Y: IMPC-based screening revealed that ROBO1
can regulate osteoporosis by inhibiting osteogenic differentiation.
Front Cell Dev Biol. 12:14502152024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Parlakgül G, Pang S, Artico LL, Min N,
Cagampan E, Villa R, Goncalves RLS, Lee GY, Xu CS, Hotamışlıgil GS
and Arruda AP: Spatial mapping of hepatic ER and mitochondria
architecture reveals zonated remodeling in fasting and obesity. Nat
Commun. 15:39822024. View Article : Google Scholar : PubMed/NCBI
|