Introduction
Colorectal cancer is the third most common cancer
globally and the second leading cause of cancer-related deaths
(1). According to the World Health
Organization (WHO) classification of tumors of the digestive system
(2), rectal cancer (RC) can be
histologically graded as well differentiated, moderately
differentiated, poorly differentiated and undifferentiated.
Different histopathological grades markedly influence treatment
strategies and prognosis (3). For
patients with early local tumor-stage RC and no lymph node or
distant metastasis (cT1N0M0), survival rates are relatively high
following endoscopic resection, local excision or segmental
resection, with chemotherapy not typically required postoperatively
(4). However, if post-endoscopic
resection pathology reveals high-risk recurrence factors, such as
moderate-to-poor differentiation, vascular invasion or positive
resection margins, clinical guidelines recommend additional
segmental resection and regional lymph node dissection (5). For rectal adenocarcinoma with moderate
differentiation and late-stage local tumor (T)-stage such as T4a
and T4b, local excision is prone to residual cancer, and
neoadjuvant therapy followed by radical total mesorectal excision
surgery is recommended (6). Poorly
differentiated or undifferentiated carcinomas indicate more
aggressive tumor biology, with a higher likelihood of early
metastasis and worse prognosis (7).
These cases often necessitate more aggressive treatment approaches
which may include additional surgical resection of the primary
lesion and regional lymphadenectomy. Postoperative adjuvant
radiotherapy or chemotherapy is often required to improve overall
therapeutic efficacy. Notable, the outcomes of salvage surgery for
patients with recurrence after local excision are limited. The
3-year overall survival rate is only ~31, and ~39% of patients
develop distant metastases (8).
However, Hahnloser et al (9)
reported that, when extended resection was performed within 30 days
of local excision, the 5-year survival rate was comparable with
that of patients who initially underwent radical resection.
Therefore, if the initial pathology following local excision
indicates poor differentiation, interval radical resection should
be considered. Nevertheless, this approach may compromise sphincter
preservation, leading certain patients to opt for adjuvant
chemoradiotherapy instead (10). In
summary, accurate preoperative prediction of the histological
differentiation of RC is of paramount importance, as it directly
influences surgical recommendations and patient decision-making
prior to the initial intervention.
Magnetic resonance imaging (MRI) is essential for
the non-invasive characterization of RC, providing superior soft
tissue resolution and offering more accurate insights into the
heterogeneity of the tumor compared with other imaging modalities
(11). However, pathological
differentiation is still primarily dependent on postoperative
pathological biopsy (12), and
there is insufficient evidence to demonstrate that conventional
imaging techniques can reliably differentiate pathological grading
of RC. Consequently, there is a pressing need for more intelligent
and sensitive diagnostic technologies to enable preoperative
assessment of RC differentiation. Radiomics, which extracts
numerous quantitative features from medical images, has shown
notable potential in evaluating RC characteristics (13). Existing studies have indicated that
radiomic features based on MRI of the primary tumor can predict the
pathological differentiation of other tumors. However, most of
these studies have focused primarily on the intratumoral features
(14,15), neglecting potentially valuable
information in the peritumoral region and requiring more clinical
interpretability which hinders the widespread application of
imaging models.
The peritumoral region includes endothelial cells,
fibroblasts, immune cells, other cell types and extracellular
components, forming the tumor microenvironment (16). The tumor microenvironment serves a
decisive role in tumor progression, treatment response and
metastasis. Therefore, radiomic features of the peritumoral
environment may provide crucial data for clinically assessing the
aggressive biological behavior of tumors. Whilst Liu et al
(17) reported that peritumoral
tissues in hepatocellular carcinoma (HCC) are associated with
pathological differentiation, systematic exploration of the
relationship between peritumoral tissues and pathological
differentiation in patients with RC is lacking, and the value of
peritumoral features in predicting RC pathological differentiation
remains controversial.
Therefore, the aim of the present study was to
construct a predictive model based on intratumoral and peritumoral
(within 5 mm of the tumor) radiomics features derived from
multiparametric MRI to achieve preoperative noninvasive
differentiation between well differentiated RC (adenomatous
structure ≥95%) and non-well differentiated RC. Furthermore, a
preoperative radiomics model that integrates the optimal features
from both intratumoral and 5-mm peritumoral regions was developed
and validated, with the goal of providing an imaging-based
reference for accurate preoperative assessment and clinical
decision-making in RC.
Materials and methods
Patient selection
The Medical Ethics Committee of the Affiliated
Hospital of North Sichuan Medical College (Nanchong, China)
approved the current retrospective study (approval no. 2024ER490-1)
and waived the requirement for informed consent from patients. The
data of 224 patients with RC who underwent surgery between January
2017 and February 2024 at the Affiliated Hospital of North Sichuan
Medical College were retrospectively analyzed. All patients
underwent preoperative MRI scanning, and the diagnosis of
adenocarcinoma was confirmed by pathological examination after
surgery.
The inclusion criteria were as follows: i) Primary
RC; ii) pathologically-confirmed adenocarcinoma; iii) postoperative
pathological findings with clear pathological differentiation; iv)
lesions which could be identified on MRI images and could be
successfully enlarged peritumorally; v) no radiotherapy prior to
surgery; and vi) radical, endoscopic resection or palliative
resection. The exclusion criteria were as follows: i) Lack of
description or formal report regarding the degree of tumor
differentiation (n=13); ii) incomplete MRI sequences or poor image
quality (n=31); iii) previous history of other tumors in
combination (n=6); and iv) pathological diagnosis of squamous,
carcinoid and neuroendocrine tumors (n=27). After appropriate
screening, a total of 224 patients were included in the present
study, who were randomized into training or validation groups in a
ratio of 7:3.
Pathological differentiation
analysis
Tumor specimens underwent hematoxylin and eosin (HE)
staining in the Department of Pathology at the Affiliated Hospital
of North Sichuan Medical College to determine the degree of
differentiation in RC. The samples were fixed with 4%
paraformaldehyde at 20–25°C for 24 to 48 h, followed by
dehydration, paraffin infiltration, embedding, and sectioning to a
thickness of 4–6 µm. Prior to HE staining, the sections are baked
at 60–65°C for 1 h, then subjected to deparaffinization using a
graded ethanol series and xylene, followed by rehydration. The
sections are subsequently stained with hematoxylin for 3–5 min and
counterstained with eosin for 1–3 min at room temperature. After
staining, the sections are dehydrated through a graded alcohol
series, cleared with xylene, and finally mounted with neutral gum.
The prepared samples can be observed for morphological analysis
under a light microscope. According to classification standards
(2), RC tumors were classified as
one of the following: i) Well differentiated, (grade 1), glandular
ductal structures account for >95% of the tumor; ii) moderately
differentiated (grade 2), glandular ductal structures account for
50–95% of the tumor; iii) poorly differentiated (grade 3),
glandular ductal structures account for 5–50% of the tumor; or iv)
undifferentiated (grade 4), not obvious undifferentiated features
and proportion of glandular ducts accounts for <5% of the tumor.
The latter category encompasses mucinous adenocarcinomas and
impression cell carcinomas. When RC tumors demonstrated different
differentiation results, the predominant differentiation determined
the final diagnosis. RC was classified according to the degree of
differentiation into well differentiated and non-well
differentiated.
MRI protocol
All patients with RC underwent MRI using a MAGNETOM
Skyra 3.0T (Siemens Healthineers) scanner prior to surgery.
Patients fasted for 4–6 h before the examination and underwent
bowel preparation 2 h before the scan. An abdominal phased-array
coil was used. Scanning sequences included T1_tse_tra,
t2_blade_tra_p2_320 and ep2d_diff_tra_b50-1000_TRACEW_DFC_MIX. The
specific parameters used were as follows: i) T1_tse_tra: Repetition
time (TR) of 613 msec, Echo time (TE) of 10 msec, inversion angle
of 160°, slice thickness of 3.5 mm, interslice distance of 3.85 mm,
matrix of 320×288, Field-of-view (FOV) of 190×190 mm and voxel size
of 0.6×0.6×3.5 mm; ii) T2_blade_tra_p2_320: TR of 5,010 msec, TE of
87 msec, inversion angle of 160°, slice thickness of 3.5 mm,
interslice distance of 3.85 mm, matrix of 320×320, FOV of 190×190
mm and voxel size of 0.6×0.6×3.5 mm; and iii)
ep2d_diff_tra_b50-1000_TRACEW_DFC_MIX: TR of 5,400 msec, TE of 75
msec, inversion angle of 90°, slice thickness of 3.5 mm, interslice
distance of 3.85 mm, matrix of 150×142, FOV of 301×301 mm, voxel
size of 2.0×2.1×3.5 mm and b-values of 50 and 1,000
s/mm2.
Data collection
The data collected included age, sex,
carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA
19-9), aspartate aminotransferase (AST), prealbumin,
albumin-to-globulin ratio, total bilirubin, direct bilirubin,
lactate, creatinine, leucine aminopeptidase, γ-glutamyl
transferase, adenosine deaminase, alkaline phosphatase, white blood
cells, red blood cells, serum albumin, hemoglobin, platelets,
neutrophils, lymphocytes, monocytes and the hemoglobin, albumin,
lymphocyte, and platelet score, all within 2 weeks before surgery.
Pathological data included differentiation grade, whilst imaging
data included the distance of the tumor from the anal verge, lesion
length, tumor-node-metastasis (TNM) stage, circumferential
resection margin (CRM) and extramural vascular invasion.
Imaging preprocessing, region of
interest (ROI) segmentation and peritumoral region dilation
Images were resampled at a voxel spacing of 1×1×1
mm3 before delineating the ROI and normalizing to
grayscale to compensate for voxel spatial differences and maintain
grayscale consistency, respectively. The entire ROI of T1-weighted
images (T1WI), T2-weighted images (T2WI) and diffusion-weighted
imaging (DWI) was outlined by a senior radiologist experienced in
diagnostic imaging of RC using open-source 3Dslicer software
(version 5.3.0; download.slicer.org/). Furthermore, and the entire
ROI of T1WI, T2WI and DWI was automatically analyzed using the
‘Margin’ module, which was used to automatically expand the 5-mm
region around the tumor to obtain a 3D segmented image.
Feature extraction
The image and the outlined ROI structure were
imported into 3D Slicer for feature extraction. The features
extracted by 3D Slicer were mainly performed using the open-source
plug-in (Pyradiomics v2.2.0; http://github.com/Radiomics/SlicerRadiomics). The
extracted features included mean, minimum, maximum, standard
deviation, skewness, kurtosis and others of the first-order
statistical features of the original image, as well as the surface
area, volume, surface area-to-volume ratio, sphericity, compactness
and 3D diameter of the morphological features. Moreover, the
following five types of texture features were extracted (18): i) Gray Level Cooccurence Matrix; ii)
Gray Level Run Length Matrix; iii) Gray Level Size Zone Matrix; iv)
Neighbouring Gray Tone Difference Matrix; and v) Gray Level
Dependence Matrix. Shape features were extracted from the original
images, while first-order and texture features were obtained from
both the original images and their filtered versions. The filtering
procedures included: Wavelet transformation (utilizing high-pass
(H) and low-pass (L) filters along the x, y, and z axes, generating
eight distinct decomposition combinations: HHH, HHL, HLH, LHH, LLL,
LLH, LHL, HLL) and Laplacian of Gaussian (LoG) filtering (σ=4.0,
5.0, and 6.0 mm). In this case, the wavelet transform image was
created using the default parameters of 3D Slicer. The wavelet
sequence used was Daubechies (Db4). A 3-layer decomposition was
performed. The fill mode was symmetric filling. Quantization of 64
bins was applied. The sampling mode was custom sampling, with
specific sampling densities of x-, y- and z-axis sampling intervals
of 4, 5 and 6 voxels, respectively. A total of 1,223 features were
extracted.
Imbalanced data processing
The dataset comprised 46 patients with well
differentiated tumors and 178 with non-well differentiated tumors.
However, the marked imbalance between the two groups could impair
classifier performance; therefore, to address this, the adaptive
synthetic (ADASYN) technique was employed to balance the dataset
(19). After resampling, the
proportion of well differentiated and non-well differentiated cases
reached 1:1, with the minority class increasing from 20.5% in the
original dataset to 50.8% after ADASYN processing. Specifically,
the number of patients with well differentiated and non-well
differentiated tumors was 184 and 178, respectively. Subsequently,
ReliefF (20) was used for
dimensionality reduction, selecting 20 notable distinguishing
features. The balanced dataset was randomly divided into a training
set (70%) and a testing set (30%) after five-fold
cross-validation.
Establishment and evaluation of the
predictive model
In the current study, three machine learning
algorithms, logistic regression, Light Gradient Boosting Machine
(LightGBM) Classifier and Gaussian Naive Bayes, were selected to
predict the pathological differentiation of patients with RC based
on imaging features in and around the 5-mm region of the tumor in
T1WI, T2WI and DWI. The model was constructed using k-fold
cross-validation on the training set as a resampling method (k=5)
and the hyperparameters were adjusted using grid search to build
the best model using the optimal parameters. The receiver operating
characteristic (ROC) curve was plotted with the true positive rate
as the vertical coordinate and the false positive rate as the
horizontal coordinate, and the predictive efficacy of the model was
evaluated by calculating the area under the receiver operating
curve (AUC) and validated in the test group. The accuracy,
sensitivity and specificity corresponding to the optimal thresholds
of each model were obtained by selecting the optimal thresholds
based on the Youden index. Confusion matrix metrics, such as AUC,
accuracy, sensitivity, specificity and F1 score, were used to
evaluate the prediction performance and stability of each model.
Following the identification of the optimal classification method
and imaging sequence for predicting the pathological
differentiation of RC through comparison with confusion matrix
metrics and standard deviations derived from forest plots, the
dataset was re-divided into a training set (70%), a validation set
(15%) and a test set (15%). The validation set was used to adjust
the model parameters and the test set was used to evaluate the
system performance. T1-LightGBM models of intratumor and intratumor
+ peritumor were constructed, and then ROC curves, learning curves,
calibration plots and decision curve analysis (DCA) were used to
further assess the clinical efficacy of the prediction models.
Calibration curves reflect how well the predicted probability
matches the actual outcome; DCA was used to assess the net benefit
of the model at different thresholds. Calibration was assessed
using the Hosmer-Lemeshow goodness-of-fit test, followed by
development of the Shapley Additive explanations (SHAP) overall
presentation model and single-sample interpretation.
Statistical analysis
Categorical data are presented as n (%), whilst
continuous variables are expressed as median (interquartile range).
Differences between well differentiated and non-well differentiated
groups were assessed using the Wilcoxon rank sum test, Fisher's
exact test or Pearson's χ2 test. A two-sided P<0.05
was considered to indicate a statistically significant difference.
Statistical analyses were performed using R 3.6.3 (https://www.r-project.org/) and Anaconda Python 3.7
(anaconda.com/).
Results
Patient characteristics
Postoperative pathological reports categorized the
224 patients into the well differentiated group (n=46) and the
non-well differentiated group (n=178). Baseline patient data are
presented in Table SI. CA 19-9,
AST and alanine aminotransferase were significantly associated with
pathological tissue differentiation (P<0.05), whilst no
significant differences were observed in other clinical indicators
or characteristics.
Feature analysis
A total of 1,224 features were extracted and
standardized. Table I presents the
application results of machine learning classification algorithms
on the balanced dataset generated using ADASYN. The T1-LightGBM
model demonstrated the best performance in the validation set.
 | Table I.Evaluation of the performance of
classification models on imbalance dataset using the adaptive
synthetic technique in the validation set. |
Table I.
Evaluation of the performance of
classification models on imbalance dataset using the adaptive
synthetic technique in the validation set.
| Model | AUC (SD) | Accuracy (SD) | Sensitivity
(SD) | Specificity
(SD) | PPV(SD) | NPV (SD) | F1 score (SD) | Cutoff (SD) |
|---|
| Logistic |
|
|
|
|
|
|
|
|
|
T1WI | 0.637 (0.056) | 0.542 (0.039) | 0.552 (0.134) | 0.512 (0.145) | 0.591 (0.037) | 0.474 (0.089) | 0.563 (0.084) | 0.445 (0.045) |
|
T2WI | 0.533 (0.104) | 0.521 (0.100) | 0.184 (0.121) | 0.799 (0.256) | 0.600 (0.261) | 0.535 (0.086) | 0.229 (0.106) | 0.544 (0.006) |
|
DWI | 0.615 (0.079) | 0.568 (0.046) | 0.291 (0.176) | 0.779 (0.170) | 0.558 (0.176) | 0.556 (0.060) | 0.356 (0.169) | 0.556 (0.072) |
| LightGBM |
|
|
|
|
|
|
|
|
|
T1WI | 0.756 (0.051) | 0.700 (0.074) | 0.707 (0.122) | 0.700 (0.204) | 0.730 (0.140) | 0.709 (0.053) | 0.700 (0.070) | 0.452 (0.291) |
|
T2WI | 0.601 (0.096) | 0.561 (0.098) | 0.536 (0.234) | 0.628 (0.163) | 0.578 (0.079) | 0.598 (0.162) | 0.521 (0.130) | 0.668 (0.260) |
|
DWI | 0.750 (0.073) | 0.658 (0.080) | 0.600 (0.258) | 0.704 (0.292) | 0.763 (0.160) | 0.646 (0.130) | 0.608 (0.142) | 0.380 (0.194) |
| GNB |
|
|
|
|
|
|
|
|
|
T1WI | 0.672 (0.107) | 0.621 (0.094) | 0.856 (0.056) | 0.421 (0.181) | 0.591 (0.130) | 0.741 (0.098) | 0.687 (0.074) | 0.001 (0.002) |
|
T2WI | 0.501 (0.108) | 0.471 (0.068) | 0.293 (0.131) | 0.668 (0.251) | 0.552 (0.255) | 0.493 (0.040) | 0.324 (0.099) | 0.107 (0.032) |
|
DWI | 0.645 (0.085) | 0.579 (0.076) | 0.540 (0.160) | 0.620 (0.150) | 0.587 (0.050) | 0.585 (0.150) | 0.546 (0.081) | 0.039 (0.027) |
Model development and evaluation
A total of three machine learning algorithms were
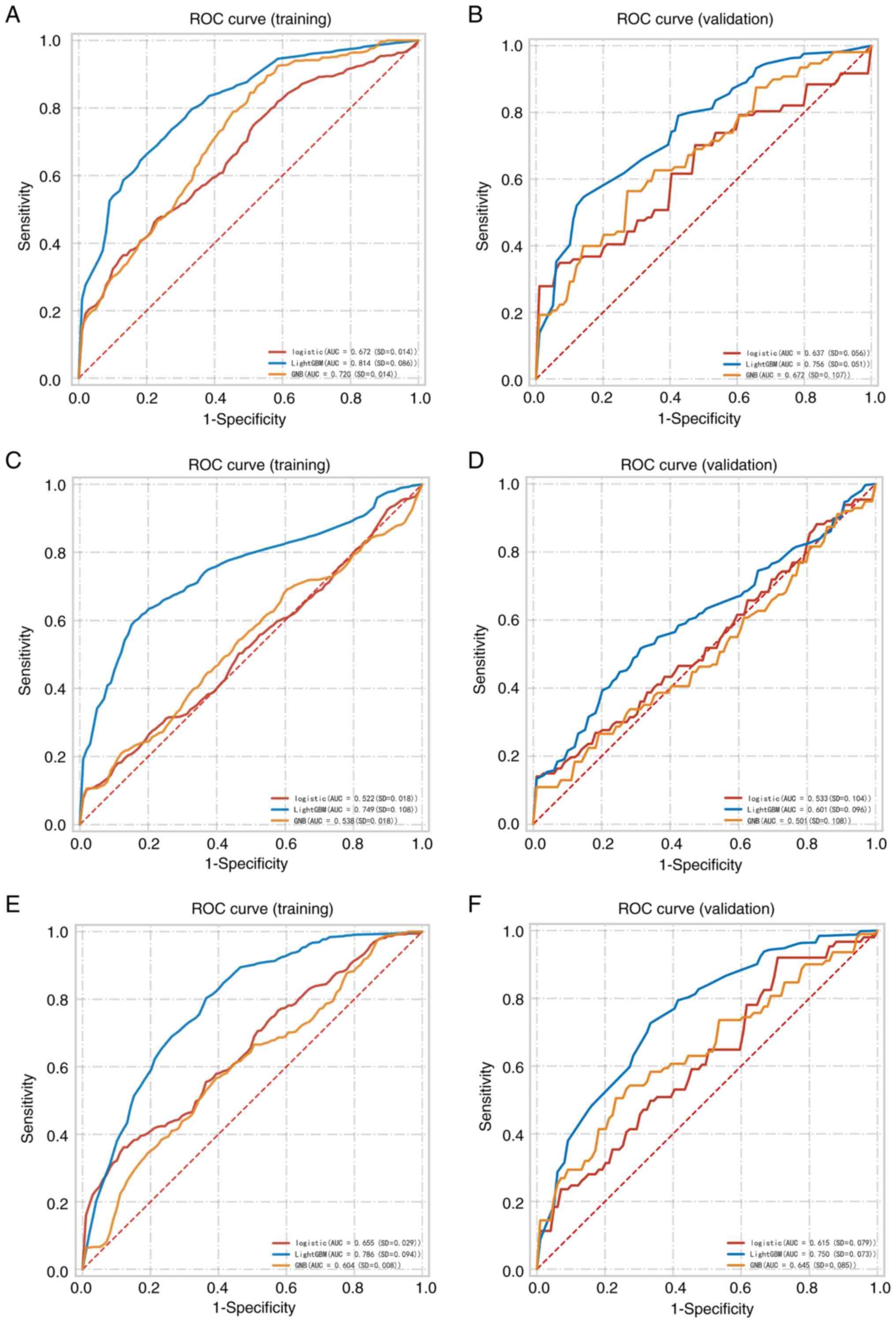
used to develop predictive models. Fig.
1 illustrates the ROC curves for the different models
constructed for the intratumoral + peritumoral features. The AUC
range for the models was 0.510-0.756 across all validation cohorts.
The T1-LightGBM model, which combined intratumoral and peritumoral
features, achieved AUC, accuracy, sensitivity, positive predictive
value (PPV), negative predictive value (NPV) and F1 score >0.7
in the validation set, outperforming the other eight models
(Table I). Moreover, the
T1-LightGBM model demonstrated the best predictive performance on
both training and validation sets (Fig.
1). The forest plot in Fig. S1
presents the ROC results for the nine models in predicting
pathological differentiation, indicating that the T1-LightGBM model
had the highest central value and the shortest error bars, further
confirming its superior stability.
The LightGBM model was chosen as the best model for
predicting pathological differentiation, and the intratumoral +
peritumoral and intratumoral prediction models of T1-LightGBM were
constructed using 5-fold cross-validation. The AUC, cutoff,
accuracy, sensitivity, specificity, PPV and NPV for the training,
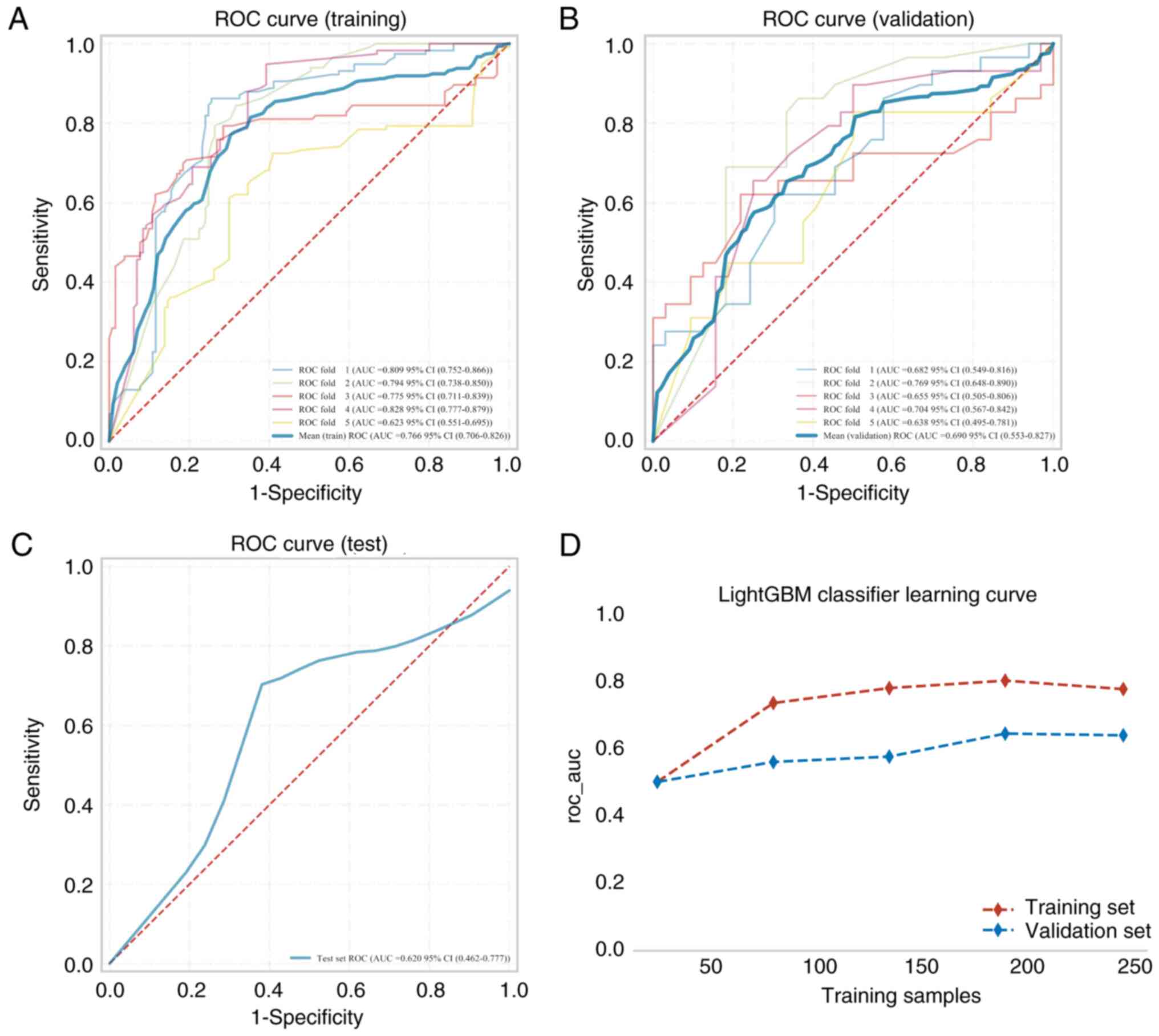
validation and test cohorts are presented in Table II, and the model ROC curves are
presented in Figs. 2 and 3. Assessment of the predictive performance
metrics indicated that the combined intratumoral + peritumoral
model outperformed the intratumoral-only model. Specifically, the
average AUC of the intratumoral + peritumoral model was 0.766 for
the training set, 0.690 for the validation set and 0.620 for the
test set (Fig. 2A-C; Table II), and the AUC of the training,
validation and test sets eventually stabilized at ~0.70. The AUC
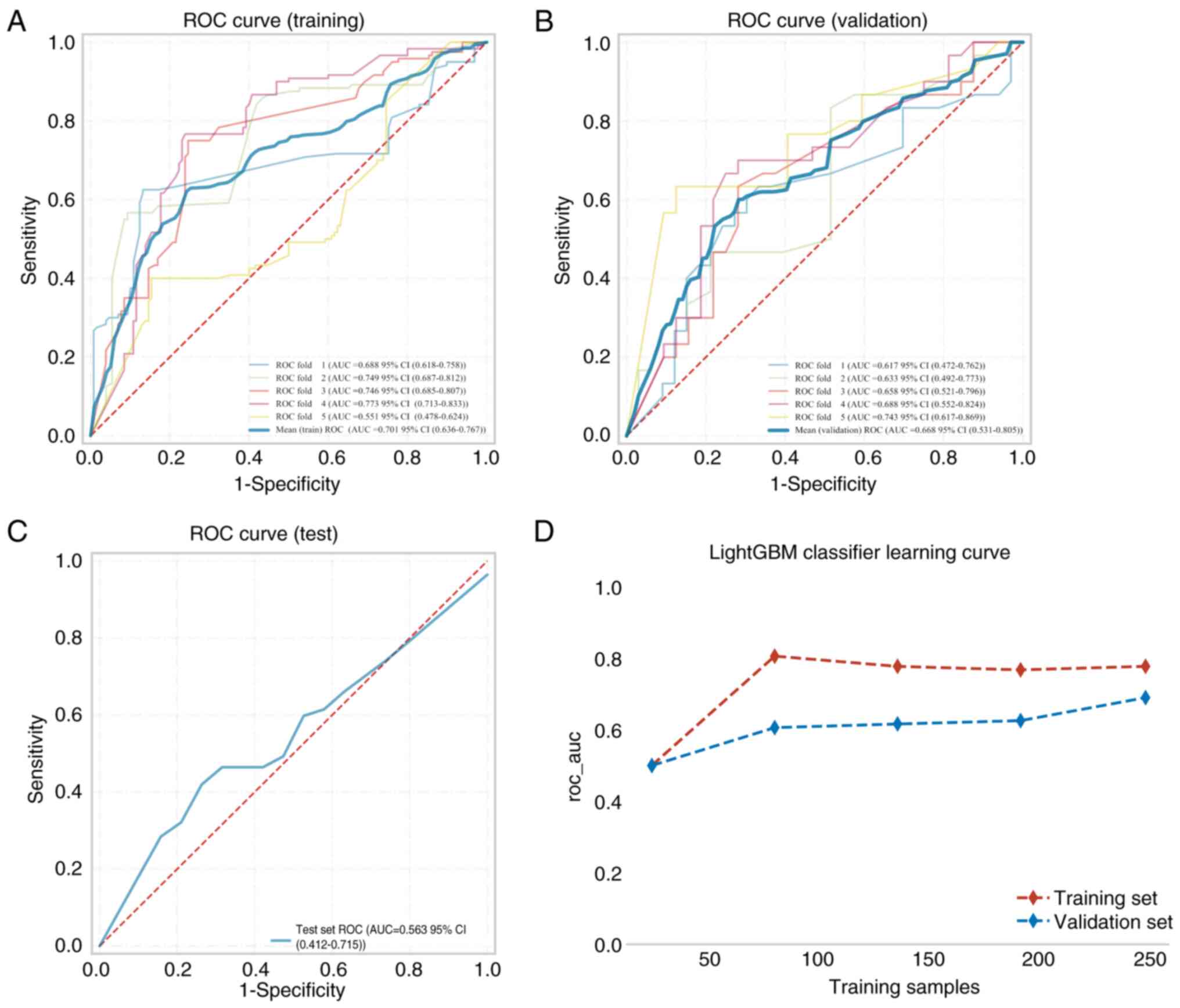
was 0.668 in the training set, 0.701 in the validation set and
0.563 in the test set for the intratumor model (Table II; Fig.
3A-C). Learning curves (Fig.
3D) were shown prior to assessing the accuracy of the model,
which indicated that the difference in error between the training
and the validation sets in the intratumoral + peritumoral 5-mm
model converged as the number of training samples increased. This
suggests that the model was not overfitted. Subsequently, the
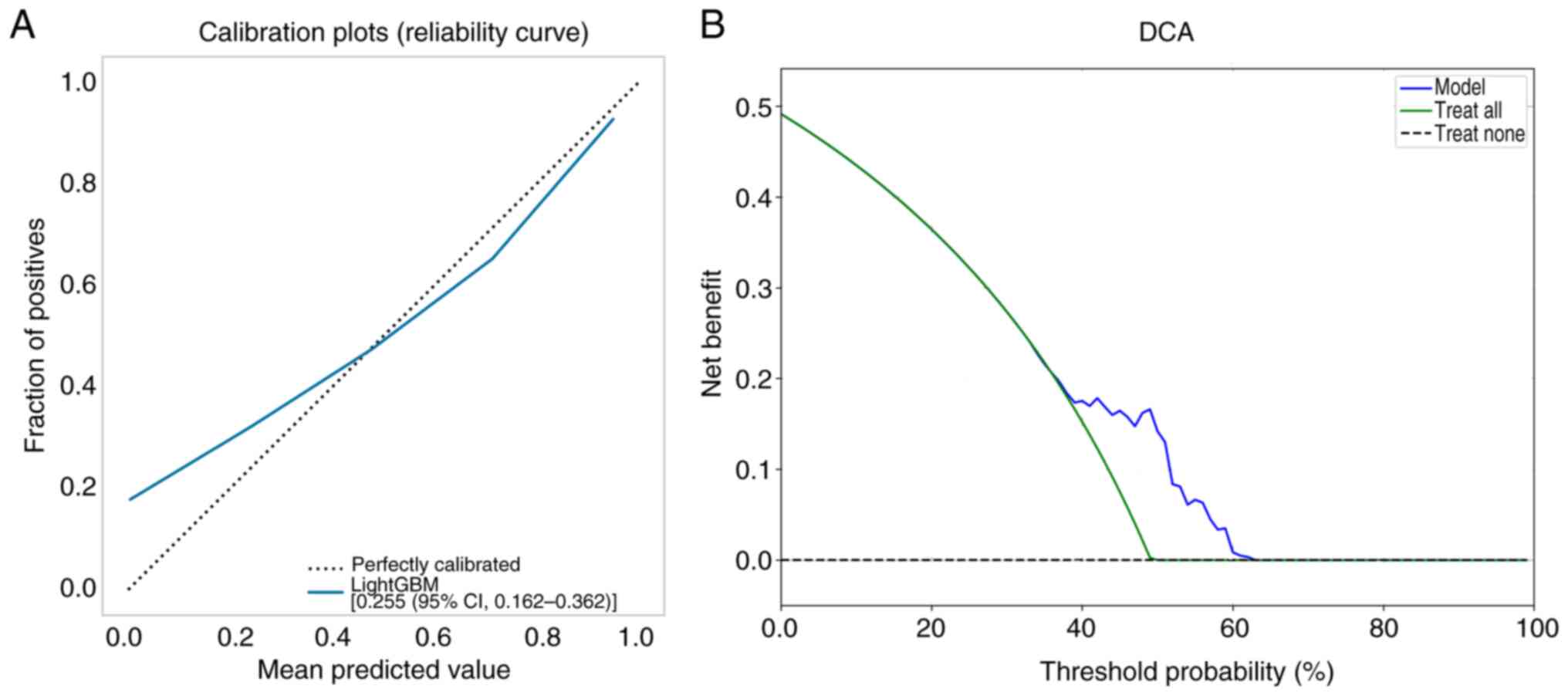
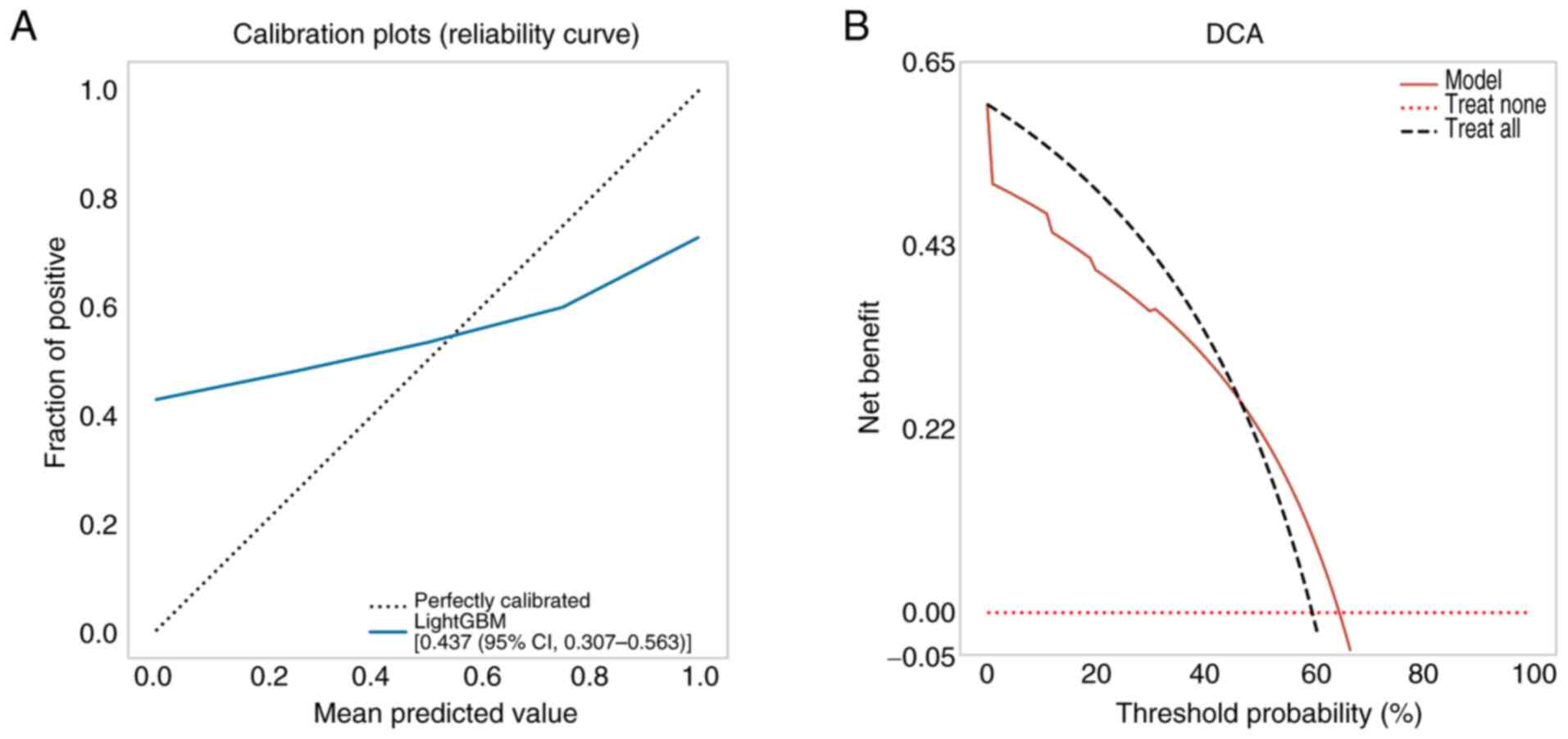
calibration curves of the validation set were used to assess the
accuracy of the model. The results revealed that the intratumoral +
peritumoral 5-mm T1-LightGBM model had notable agreement with the
predicted probability of observed pathological differentiation
(Fig. 4A). The Hosmer-Lemeshow test
demonstrated that the intratumoral + peritumoral T1-LightGBM model
was well-calibrated (P=0.064). Subsequently, a DCA of the model was
constructed in the study, indicating that the intratumoral +
peritumoral model had a more marked net benefit than all-or-none
pathological differentiation with a risk threshold of <64%
(Fig. 4B). By contrast, the
calibration curves and DCA of the intratumoral LightGBM model
(Fig. 5) performed slightly worse
than the intratumoral + peritumoral prediction model. In summary,
the LightGBM model may be used for classification modeling tasks in
this dataset.
 | Table II.Diagnostic performance of the Light
Gradient-Boosting Machine model constructed from intratumoral +
peritumoral and intratumoral features in predicting pathological
differentiation of rectal cancer. |
Table II.
Diagnostic performance of the Light
Gradient-Boosting Machine model constructed from intratumoral +
peritumoral and intratumoral features in predicting pathological
differentiation of rectal cancer.
| Model | AUC (95% CI) | Cutoff (95%
CI) | Accuracy (95%
CI) | Sensitivity (95%
CI) | Specificity (95%
CI) | PPV (95% CI) | NPV (95% CI) | F1 score (95%
CI) |
|---|
| Training set |
|
|
|
|
|
|
|
|
|
Intratumoral + | 0.690 | 0.383 | 0.661 | 0.731 | 0.599 | 0.626 | 0.725 | 0.668 |
|
peritumoral_ | (0.553-0.827) | (0.090-0.677) | (0.631-0.691) | (0.619-0.843) | (0.491-0.706) | (0.582-0.670) | (0.661-0.790) | (0.625-0.710) |
| 5
mm |
|
|
|
|
|
|
|
|
|
Intratumoral | 0.668 | 0.408 | 0.677 | 0.593 | 0.753 | 0.692 | 0.669 | 0.668 |
|
| (0.531-0.805) | (0.088-0.729) | (0.629-0.725) | (0.512-0.674) | (0.690-0.817) | (0.624-0.760) | (0.627-0.711) | (0.531-0.805) |
| Validation set |
|
|
|
|
|
|
|
|
|
Intratumoral + | 0.766 | 0.383 | 0.749 | 0.807 | 0.698 | 0.709 | 0.809 | 0.751 |
|
peritumoral_ | (0.706-0.826) | (0.090-0.677) | (0.701-0.798) | (0.719-0.895) | (0.616-0.780) | (0.655-0.763) | (0.733-0.886) | (0.702-0.800) |
| 5
mm |
|
|
|
|
|
|
|
|
|
Intratumoral | 0.701 | 0.408 | 0.728 | 0.622 | 0.827 | 0.771 | 0.711 | 0.701 |
|
| (0.636-0.767) | (0.088-0.729) | (0.681-0.776) | (0.490-0.753) | (0.768-0.886) | (0.719-0.823) | (0.650-0.772) | (0.636-0.767) |
| Test set |
|
|
|
|
|
|
|
|
|
Intratumoral + | 0.620 | 0.321 | 0.618 | 0.788 | 0.364 | 0.65 | 0.533 | 0.712 |
|
peritumoral_ | (0.462-0.777) |
|
|
|
|
|
|
|
| 5
mm |
|
|
|
|
|
|
|
|
|
Intratumoral | 0.563 | 0.349 | 0.571 | 0.357 | 0.786 | 0.625 | 0.55 | 0.455 |
|
| (0.412-0.715) |
|
|
|
|
|
|
|
Model interpretation
The present study used SHAP plots to elucidate the
performance of the intratumoral + peritumoral T1-LightGBM based
prediction model for pathological differentiation. Fig. S2A illustrates that the feature
wavelet_HHH_glszm_Size_Zone_Non_Uniformity_Normalized was the most
critical predictor for pathological differentiation. Furthermore,
Fig. S2B illustrates a specific
case of predicting pathological differentiation in patients with
RC, where the SHAP values of features are visualized as forces that
either augment or diminish the predictive assessment. The baseline
was 0.740, representing the average SHAP value across all
predictions. The patient falls into the ‘true negative’ group, with
SHAP values <0.740, resulting in a predicted probability of
17.0% for well differentiated RC. Notably, the feature
wavelet_HHH_glszm_Size_Zone_Non_Uniformity_Normalized has a
negative (blue) value, indicating its contribution to the
prediction of non-well differentiated RC in this patient.
Discussion
The present study developed and assessed multiple
MRI-based radiomics models, with a particular focus on the value of
peritumoral features for the preoperative, noninvasive prediction
of the pathological differentiation grade of RC. The results
demonstrated that the LightGBM model based on the T1-weighted
sequence and combining intratumoral features with those from a 5-mm
peritumoral shell, achieved the best performance. These findings
suggest that peritumoral radiomic features can provide clinically
meaningful additional information for assessing the differentiation
of RC.
MRI serves a critical role in pre- and
post-treatment evaluation of RC (21). T1WI effectively delineates
anatomical structures, whilst T2WI provides high-resolution soft
tissue contrast, highlighting differences in internal lesion
composition. DWI, particularly at high b-values (b ≥800
sec/mm2), is highly sensitive to Brownian motion of
water molecules within viable tissues and tumors, offering insights
into cellular activity. DWI also demonstrates superior sensitivity
in detecting small tumor lesions and pelvic lymph nodes (22). In the present study, radiomic
features were extracted from intratumoral and peritumoral regions
across these three sequences. The SHAP method was employed to
interpret model outputs, identifying the five most contributory
features for distinguishing pathological differentiation:
T1_wavelet_HHH_glszm_Size_Zone_Non_Uniformity_Normalized,
original_first_order_Median,
wavelet_HHH_glszm_Low_Gray_Level_Zone_Emphasis,
Original_shape_Maximum_2D_Diameter_Slice and
wavelet_HHH_glszm_High_Gray_Level_Zone_Emphasis. The top-ranked
feature, wavelet_HHH_glszm_Size_Zone_Non_Uniformity_Normalized,
quantifies gray-level intensity uniformity within tumors,
reflecting spatial aggregation of high-signal regions (23). This metric may be associated with
tumor angiogenesis or necrotic zone distribution (24), critical for evaluating structural
complexity and homogeneity (25).
Such heterogeneity aligns with the infiltrative growth patterns of
malignant RC, often manifesting as non-spherical morphology
(26). Collectively, these features
emphasize the importance of integrating morphological, statistical
and textural characteristics for comprehensive tumor
characterization.
Whilst aligning with established machine learning
methodologies, the novelty of the current study involves dataset
construction and feature selection. Prior research predominantly
focused on intratumoral regions, neglecting peritumoral
contributions. However, emerging evidence has highlighted the
prognostic value of peritumoral features in tumor biology, as
demonstrated in soft tissue sarcoma (27), head and neck squamous cell carcinoma
(28), gastric cancer (29), lung cancer (30,31)
and ovarian cancer (32). For
instance, Liu et al (17)
highlighted the predictive utility of peritumoral features in HCC
differentiation, whilst Barge et al (33) identified peritumoral texture
features in T1WI as key discriminators for tumor grading. Xu et
al (22) further reported that
models combining intratumoral and peritumoral (3 and 5 mm) features
outperformed single-region models in predicting lymphovascular
invasion in RC. Based on the aforementioned rationale, the current
study designated a 5-mm peritumoral region as the primary area for
analysis. This choice was informed, on the one hand, by clinical
consensus regarding the CRM in RC: A CRM of >1 mm is generally
considered a ‘safe margin’ (34),
although the optimal clearance remains controversial. A
meta-analysis reported that, whilst a margin of >1 mm improves
prognosis, a margin of >1 cm may yield superior oncological
outcomes (35). On the other hand,
prior peritumoral radiomics studies in lung cancer (36) and gliomas (37) suggested that regions closer to the
tumor harbor richer heterogeneity; as the boundary expands outward,
more normal tissue is included in the ROI, potentially diluting
meaningful structural differences. Therefore, a 5-mm peritumoral
band is likely to capture spatial heterogeneity whilst maximally
preserving imaging signals related to tumor biology. This provides
an effective basis for noninvasive, preoperative prediction of the
pathological grading of RC.
Previous research has explored the relationship
between functional MRI parameters and the pathological grading of
RC; however, the findings have been inconsistent. Kim et al
(38) reported that the Ktrans
value in the well differentiated group in their study was
significantly higher than in the moderately differentiated group
(0.127±0.032 vs. 0.084±0.036; P=0.036). By contrast, Shen et
al (39) reported that the
Ktrans value was significantly associated with the pathological
differentiation of RC (poor, 0.284±0.068 and moderately:
0.280±0.067 vs. well: 0.182±0.153, P=0.004), with the well
differentiated group showing lower Ktrans values than the
moderately differentiated group. However, the values between the
moderately and poorly differentiated groups were similar. n DWI,
apparent diffusion coefficient (ADC) values have also been used to
assess pathological grading. Curvo-Semedo et al (40) noted that poorly differentiated
tumors tended to exhibit lower ADC values compared with moderately
and highly differentiated tumors, although statistical significance
was not reached. Zhou et al (41) also reported no significant ADC
differences within the traditional WHO grading system. However,
when adopting the poorly differentiated clusters grading system
(42), ADC and intravoxel
incoherent motion (IVIM) parameters, such as perfusion fraction f
and pseudo-diffusion coefficient D*, demonstrated significant
differences between high- and low-grade tumors, with clear
correlations between perfusion metrics and tumor grade (r=0.842,
P<0.001; and r=0.356, P=0.011). This suggests that IVIM
parameters may be valuable for grading. Yuan et al (43) further extended the analysis to the
peritumoral region and reported that higher peritumoral-to-tumor
ADC ratios and ADCp mean values were associated with poor
differentiation, T3-4 stage and adverse features such as lymph node
metastasis (LNM), extranodal extension, tumor deposits and
lymphovascular invasion (LVI). They also reported that the
peritumoral-to-tumor ADC ratio outperformed tumor ADC in assessing
prognostic factors.
Clinically, treatment strategies for RC are
primarily based on TNM staging, which does not fully capture
inter-patient heterogeneity within the same stage. Pathological
differentiation serves as an important complementary indicator to
optimize risk stratification and therapeutic decision-making
(44,45). Numerous studies have reported that
poor differentiation is associated with higher local recurrence and
worse overall survival compared with well differentiation (46,47).
Therefore, it is classed as a high-risk feature in the European
Society for Medical Oncology and the National Comprehensive Cancer
Network (NCCN) guidelines (10).
The NCCN and the American Society of Colon and Rectal Surgeons
further specify in their local excision criteria for early RC that
appropriate candidates should have tumors of <3 cm,
well-to-moderately differentiated histology and no LVI (48). Additional studies support
incorporating tumor grade into risk assessment. Cho et al
(49) reported that in pT1 RC and
colon cancer, moderate/poor differentiation was significantly
associated with LNM (P=0.006 and P=0.002, respectively) and was an
independent predictor of LNM (odds ratio, 1.38-8.13; P=0.008).
Emile et al (50) developed
a risk-scoring model integrating tumor grade with nodal stage and
reported that, compared with well differentiated rectal
adenocarcinoma, poorly differentiated and undifferentiated tumors
had a markedly increased risk of LVI (more than threefold).
Considering the similar trends observed for moderately and poorly
differentiated tumors across functional imaging parameters, and the
frequent consideration in clinical guidelines and prior studies
that these categories share comparable aggressive biology, the
present study combined moderately and poorly differentiated RCs
into a ‘non-well differentiated’ group. This grouping strategy
helps clarify gradations of tumor aggressiveness, enhances the
ability of the model to identify clinically high-risk patients, and
provides more accurate guidance for treatment decisions. For
example, in mid-to-low RC initially staged as T3a/b by MRI, if the
model predicts moderate/poor differentiation, clinicians may
prioritize neoadjuvant chemoradiotherapy over immediate surgery, as
the true biological behavior may be more aggressive than suggested
by staging alone (51).
In addition to imaging features, studies by
Shibutani et al (52) and
Ryuk et al (53) reported
that serum CEA and CA 19-9 are independent prognostic factors for
RC. Elevated preoperative CEA levels were associated with tumor
size and T-stage, whilst high CA19-9 levels were reported to be
associated with tumor stage (54).
This is consistent with the findings of the present study, in which
elevated CA 19-9 levels demonstrated a significant association with
poorer differentiated RC (P=0.014), further supporting the
auxiliary value of serum biomarkers in assessing tumor biology.
Serum biomarkers reflect systemic tumor burden and metabolic status
(55), whereas radiomic features
quantify local tumor heterogeneity (24). Their combination could provide
multidimensional predictive information, enhancing accuracy and
clinical utility.
However, the current study has several limitations.
First, it adopted a single-center retrospective design which may
introduce selection bias, and the sample size was relatively small,
particularly for the independent test cohort. More multi-center
cohorts are needed for external validation to assess the robustness
and reproducibility of the predictive model and to strengthen the
conclusions of the present study. Second, radiomics features are
sensitive to scanning protocols and equipment parameters.
Consequently, standardized image acquisition and feature
harmonization should be promoted in the future to improve the
generalizability of the model. In addition, tumor region annotation
currently relies on manual delineation which may introduce
inter-observer variability. Developing automatic or semi-automatic
segmentation algorithms is an important direction for improvement.
Finally, the current study focused solely on radiomics features. In
future work, the plan is to explore integration with
clinicopathological indicators and molecular biomarkers, such as
genomic and transcriptomic features, to build multi-modal fusion
models, enabling more refined patient stratification and further
enhancing clinical translational value.
In conclusion, the present study highlights the
potential of peritumoral features as a non-invasive tool for
predicting RC differentiation. Notably, the integrated model
employing the LightGBM algorithm on T1-weighted sequences, which
combined both intratumoral and 5-mm peritumoral features,
demonstrated promising predictive potential for individualized RC
differentiation, thereby contributing to the development of
personalized treatment strategies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HL collected, analyzed and organized data, and
participated in drafting and revising the manuscript. HZ provided
supervision, guided the manuscript revision process and assisted in
data analysis. QZ performed data collection and operated
specialized software. JY contributed to the writing and revision of
the article. HL and JY conceived the study and designed the
methodology. HL and JY confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed according to the
guidelines of the Declaration of Helsinki and approved by the
Medical Ethics Committee of Affiliated Hospital of North Sichuan
Medical College (approval no. 2024ER490-1). The requirement for
informed consent was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board, : The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiang C, Liu Y, Xu C, Shen Y, Xu Q and Gu
L: Pathological features of lymph nodes around inferior mesenteric
artery in rectal cancer: A retrospective study. World J Surg Oncol.
19:1522021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Junginger T, Goenner U, Hitzler M, Trinh
TT, Heintz A, Wollschlaeger D and Blettner M: Long-term oncologic
outcome after transanal endoscopic microsurgery for rectal
carcinoma. Dis Colon Rectum. 59:8–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garcia-Aguilar J, Renfro LA, Chow OS, Shi
Q, Carrero XW, Lynn PB, Thomas CR Jr, Chan E, Cataldo PA, Marcet
JE, et al: Organ preservation for clinical T2N0 distal rectal
cancer using neoadjuvant chemoradiotherapy and local excision
(ACOSOG Z6041): Results of an open-label, single-arm,
multi-institutional, phase 2 trial. Lancet Oncol. 16:1537–1546.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Glynne-Jones R, Wyrwicz L, Tiret E, Brown
G, Rödel C, Cervantes A and Arnold D; ESMO Guidelines Committee, :
Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 29 (Suppl 4):iv2632018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ueno H, Hase K, Hashiguchi Y, Shimazaki H,
Yoshii S, Kudo SE, Tanaka M, Akagi Y, Suto T, Nagata S, et al:
Novel risk factors for lymph node metastasis in early invasive
colorectal cancer: A multi-institution pathology review. J
Gastroenterol. 49:1314–1323. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Doornebosch PG, Ferenschild FT, de Wilt
JH, Dawson I, Tetteroo GW and de Graaf EJ: Treatment of recurrence
after transanal endoscopic microsurgery (TEM) for T1 rectal cancer.
Dis Colon Rectum. 53:1234–1239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hahnloser D, Wolff BG, Larson DW, Ping J
and Nivatvongs S: Immediate radical resection after local excision
of rectal cancer: An oncologic compromise? Dis Colon Rectum.
48:429–437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fields AC, Lu P, Hu F, Hirji S, Irani J,
Bleday R, Melnitchouk N and Goldberg JE: Lymph node positivity in
T1/T2 rectal cancer: A word of caution in an era of increased
incidence and changing biology for rectal cancer. J Gastrointest
Surg. 25:1029–1035. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Horvat N, Carlos Tavares Rocha C, Clemente
Oliveira B, Petkovska I and Gollub MJ: MRI of rectal cancer: Tumor
staging, imaging techniques, and management. Radiographics.
39:367–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kleiner DE: Hepatocellular carcinoma:
Liver biopsy in the balance. Hepatology. 68:13–15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song M, Li S, Wang H, Hu K, Wang F, Teng
H, Wang Z, Liu J, Jia AY, Cai Y, et al: MRI radiomics independent
of clinical baseline characteristics and neoadjuvant treatment
modalities predicts response to neoadjuvant therapy in rectal
cancer. Br J Cancer. 127:249–257. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ameli S, Venkatesh BA, Shaghaghi M,
Ghadimi M, Hazhirkarzar B, Rezvani Habibabadi R, Aliyari Ghasabeh
M, Khoshpouri P, Pandey A, Pandey P, et al: Role of MRI-derived
radiomics features in determining degree of tumor differentiation
of hepatocellular carcinoma. Diagnostics (Basel). 12:23862022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu HF, Lu Y, Wang Q, Lu YJ and Xing W:
Machine learning-based CEMRI radiomics integrating LI-RADS features
achieves optimal evaluation of hepatocellular carcinoma
differentiation. J Hepatocell Carcinoma. 10:2103–2115. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirata E and Sahai E: Tumor
microenvironment and differential responses to therapy. Cold Spring
Harb Perspect Med. 7:a0267812017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu HF, Wang M, Wang Q, Lu Y, Lu YJ, Sheng
Y, Xing F, Zhang JL, Yu SN and Xing W: Multiparametric MRI-based
intratumoral and peritumoral radiomics for predicting the
pathological differentiation of hepatocellular carcinoma. Insights
Imaging. 15:972024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park H, Kim KA, Jung JH, Rhie J and Choi
SY: MRI features and texture analysis for the early prediction of
therapeutic response to neoadjuvant chemoradiotherapy and tumor
recurrence of locally advanced rectal cancer. Eur Radiol.
30:4201–4211. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim JH, Shin JK, Lee H, Lee DH, Kang JH,
Cho KH, Lee YG, Chon K, Baek SS and Park Y: Improving the
performance of machine learning models for early warning of harmful
algal blooms using an adaptive synthetic sampling method. Water
Res. 207:1178212021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khan TM, Xu S, Khan ZG and Uzair Chishti
M: Implementing multilabeling, ADASYN, and relieff techniques for
classification of breast cancer diagnostic through machine
learning: Efficient computer-aided diagnostic system. J Healthc
Eng. 2021:55776362021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bates DDB, Homsi ME, Chang KJ, Lalwani N,
Horvat N and Sheedy SP: MRI for rectal cancer: Staging, mrCRM,
EMVI, lymph node staging and post-treatment response. Clin
Colorectal Cancer. 21:10–18. 2022.PubMed/NCBI
|
|
22
|
Xu F, Hong J and Wu X: An integrative
clinical and intra- and peritumoral MRI radiomics nomogram for the
preoperative prediction of lymphovascular invasion in rectal
cancer. Acad Radiol. 32:3989–4001. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nayak P, Sinha S, Goda JS, Sahu A, Joshi
K, Choudhary OR, Mhatre R, Mummudi N and Agarwal JP: Computerized
tomography-based first order tumor texture features in non-small
cell lung carcinoma treated with concurrent chemoradiation: A
simplistic and potential surrogate imaging marker for survival. J
Cancer Res Ther. 19:366–375. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bernatowicz K, Amat R, Prior O, Frigola J,
Ligero M, Grussu F, Zatse C, Serna G, Nuciforo P, Toledo R, et al:
Radiomics signature for dynamic monitoring of tumor inflamed
microenvironment and immunotherapy response prediction. J
Immunother Cancer. 13:e0091402025. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Dai S, Wang Q, Chai X and Xian J:
Investigation of MRI-based radiomics model in differentiation
between sinonasal primary lymphomas and squamous cell carcinomas.
Jpn J Radiol. 39:755–762. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Yang Y, Wang T, Chen X, Tang M,
Deng J, Cai Z and Cui W: Intratumoral and peritumoral MRI-based
radiomics prediction of histopathological grade in soft tissue
sarcomas: A two-center study. Cancer Imaging. 23:1032023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin P, Xie W, Li Y, Zhang C, Wu H, Wan H,
Gao M, Liang F, Han P, Chen R, et al: Intratumoral and peritumoral
radiomics of MRIs predicts pathologic complete response to
neoadjuvant chemoimmunotherapy in patients with head and neck
squamous cell carcinoma. J Immunother Cancer. 12:e0096162024.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang Y, Wang H, Wu J, Chen C, Yuan Q,
Huang W, Li T, Xi S, Hu Y, Zhou Z, et al: Noninvasive imaging
evaluation of tumor immune microenvironment to predict outcomes in
gastric cancer. Ann Oncol. 31:760–768. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pérez-Morales J, Tunali I, Stringfield O,
Eschrich SA, Balagurunathan Y, Gillies RJ and Schabath MB:
Peritumoral and intratumoral radiomic features predict survival
outcomes among patients diagnosed in lung cancer screening. Sci
Rep. 10:105282020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Wang P, Xu J, Shi X, Yin T and Teng
F: Noninvasive radiomic biomarkers for predicting pseudoprogression
and hyperprogression in patients with non-small cell lung cancer
treated with immune checkpoint inhibition. Oncoimmunology.
13:23126282024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X, Wei M, Chen Y, Jia J, Zhang Y, Dai
Y, Qin C, Bai G and Chen S: Intratumoral and peritumoral MRI-based
radiomics for predicting extrapelvic peritoneal metastasis in
epithelial ovarian cancer. Insights Imaging. 15:2812024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barge P, Oevermann A, Maiolini A and
Durand A: Machine learning predicts histologic type and grade of
canine gliomas based on MRI texture analysis. Vet Radiol
Ultrasound. 64:724–732. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Scaife CL and Curley SA: Complication,
local recurrence, and survival rates after radiofrequency ablation
for hepatic malignancies. Surg Oncol Clin N Am. 12:243–255. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Margonis GA, Sergentanis TN,
Ntanasis-Stathopoulos I, Andreatos N, Tzanninis IG, Sasaki K,
Psaltopoulou T, Wang J, Buettner S, Papalois ΑE, et al: Impact of
surgical margin width on recurrence and overall survival following
R0 hepatic resection of colorectal metastases: A systematic review
and meta-analysis. Ann Surg. 267:1047–1055. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang X, Huang H, Du P, Wang L, Yin H and
Xu X: Intratumoral and peritumoral CT-based radiomics strategy
reveals distinct subtypes of non-small-cell lung cancer. J Cancer
Res Clin Oncol. 148:2247–2260. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng J, Liu J, Yue H, Bai H, Pan Y and
Wang J: Prediction of glioma grade using intratumoral and
peritumoral radiomic features from multiparametric MRI images.
IEEE/ACM Trans Comput Biol Bioinform. 19:1084–1095. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim HR, Kim SH and Nam KH: Association
between dynamic contrast-enhanced MRI parameters and prognostic
factors in patients with primary rectal cancer. Curr Oncol.
30:2543–2554. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen FU, Lu J, Chen L, Wang Z and Chen Y:
Diagnostic value of dynamic contrast-enhanced magnetic resonance
imaging in rectal cancer and its correlation with tumor
differentiation. Mol Clin Oncol. 4:500–506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Curvo-Semedo L, Lambregts DM, Maas M,
Beets GL, Caseiro-Alves F and Beets-Tan RG: Diffusion-weighted MRI
in rectal cancer: Apparent diffusion coefficient as a potential
noninvasive marker of tumor aggressiveness. J Magn Reson Imaging.
35:1365–1371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou B, Zhou Y, Tang Y, Bao Y, Zou L, Yao
Z and Feng X: Intravoxel incoherent motion MRI for rectal cancer:
Correlation of diffusion and perfusion characteristics with
clinical-pathologic factors. Acta Radiol. 64:898–906. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Barresi V, Reggiani Bonetti L, Branca G,
Di Gregorio C, Ponz de Leon M and Tuccari G: Colorectal carcinoma
grading by quantifying poorly differentiated cell clusters is more
reproducible and provides more robust prognostic information than
conventional grading. Virchows Arch. 461:621–628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yuan Y, Chen XL, Li ZL, Chen GW, Liu H,
Liu YS, Pang MH, Liu SY, Pu H and Li H: The application of apparent
diffusion coefficients derived from intratumoral and peritumoral
zones for assessing pathologic prognostic factors in rectal cancer.
Eur Radiol. 32:5106–5118. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lou S, Huang Y, Du F, Xue J, Mo G, Li H,
Yu Z, Li Y, Wang H, Huang Y, et al: Development and validation of a
deep learning-based pathomics signature for prognosis and
chemotherapy benefits in colorectal cancer: A retrospective
multicenter cohort study. Front Immunol. 16:16029092025. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang YQ, Chen XB, Cui YF, Yang F, Huang
SX, Li ZH, Ying YJ, Li SY, Li MH, Gao P, et al: Enhanced risk
stratification for stage II colorectal cancer using deep
learning-based CT classifier and pathological markers to optimize
adjuvant therapy decision. Ann Oncol. 36:1178–1189. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Oh SY, Park JY, Yang KM, Jeong SA, Kwon
YJ, Jung YT, Ma CH, Yun KW, Yoon KH, Kwak JY and Yu CS: Oncologic
outcomes of surgically treated colorectal cancer in octogenarians:
A comparative study using inverse probability of treatment
weighting (IPTW). BMC Gastroenterol. 25:2762025. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Y, Xu H, Chen G and Pan Y: Stratified
prognostic value of pathological response to preoperative treatment
in yp II/III rectal cancer. Front Oncol. 11:7951372021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tjandra JJ, Kilkenny JW, Buie WD, Hyman N,
Simmang C, Anthony T, Orsay C, Church J, Otchy D, Cohen J, et al:
Practice parameters for the management of rectal cancer (revised).
Dis Colon Rectum. 48:411–423. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cho SH, Park BS, Son GM, Kim HS, Kim SJ,
Park SB, Choi CW, Kim HW, Shin DH and Yun MS: Differences in
factors predicting lymph node metastasis between pT1 rectal cancer
and pT1 colon cancer: A retrospective study. Am Surg. 89:5829–5836.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Emile SH, Horesh N, Garoufalia Z, Gefen R,
Wignakumar A and Wexner SD: Development and validation of a
predictive score for preoperative detection of lymphovascular
invasion in rectal cancer. J Surg Oncol. 131:1081–1089. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Keller DS, Berho M, Perez RO, Wexner SD
and Chand M: The multidisciplinary management of rectal cancer. Nat
Rev Gastroenterol Hepatol. 17:414–429. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shibutani M, Maeda K, Nagahara H, Ohtani
H, Sakurai K, Toyokawa T, Kubo N, Tanaka H, Muguruma K, Ohira M and
Hirakawa K: Significance of CEA and CA19-9 combination as a
prognostic indicator and for recurrence monitoring in patients with
stage II colorectal cancer. Anticancer Res. 34:3753–3758.
2014.PubMed/NCBI
|
|
53
|
Ryuk JP, Choi GS, Park JS, Kim HJ, Park
SY, Yoon GS, Jun SH and Kwon YC: Predictive factors and the
prognosis of recurrence of colorectal cancer within 2 years after
curative resection. Ann Surg Treat Res. 86:143–151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang J, Wang X, Yu F, Chen J, Zhao S,
Zhang D, Yu Y, Liu X, Tang H and Peng Z: Combined detection of
preoperative serum CEA, CA19-9 and CA242 improve prognostic
prediction of surgically treated colorectal cancer patients. Int J
Clin Exp Pathol. 8:14853–14863. 2015.PubMed/NCBI
|
|
55
|
Farag CM, Antar R, Akosman S, Ng M and
Whalen MJ: What is hemoglobin, albumin, lymphocyte, platelet (HALP)
score? A comprehensive literature review of HALP's prognostic
ability in different cancer types. Oncotarget. 14:153–172. 2023.
View Article : Google Scholar : PubMed/NCBI
|